Abstract
Numerous studies have investigated the association of MIR499A rs3746444 polymorphism with breast cancer susceptibility, but the results have been inconsistent. In this work, we performed a meta-analysis to obtain a more reliable estimate of the association between the polymorphism and susceptibility to breast cancer. A comprehensive literature search was conducted on PubMed, Scopus, Web of Science (WoS), China National Knowledge Infrastructure (CNKI), VIP and Wanfang databases up to January 2020. A total of 14 studies involving 6,797 cases and 8,534 controls were included for analysis under five genetic models: homozygous (GG vs. AA), heterozygous (AG vs. AA), dominant (AG + GG vs. AA), recessive (GG vs. AA + AG) and allele (G vs. A). A statistically significant association was observed between the polymorphism and an increased breast cancer susceptibility under all genetic models (homozygous, OR = 1.33, 95% CI = 1.03–1.71, P = 0.03; heterozygous, OR = 1.08, 95% CI = 1.00–1.16, P = 0.04; dominant, OR = 1.15, 95% CI = 1.02–1.30; P = 0.03; recessive, OR = 1.35, 95% CI = 1.06–1.72, P = 0.01; allele, OR = 1.12, 95% CI = 1.00–1.26, P = 0.04). Subgroup analysis based on ethnicity suggested that significant association was present only among Asians, but not Caucasians. In conclusion, MIR499A rs3746444 polymorphism was significantly associated with breast cancer susceptibility among Asians, suggesting its potential use as a genetic risk marker in this population.
Subject terms: Genetic association study, Genetic markers, Cancer, Epidemiology, Genetics research
Introduction
Breast cancer is the most common type of cancer and the leading contributor to cancer-related deaths in women worldwide1. Early identification of individuals at risk of the cancer is the key to its prevention. Currently, genetic testing has emerged as a promising strategy for predicting breast cancer risk. The most regularly tested genes in breast cancer are BRCA1 and BRCA2, whose mutations have been unequivocally linked to a substantially elevated risk of the cancer. However, the overall prevalence of these high penetrance mutations is low, ranging from 0.2–0.3% in the general population to approximately 20% in breast cancer patients with a family history of the malignancy2. Thus, mutations in BRCA genes have a relatively low population attributable risk (PAR) and as such, the benefit of screening for these mutations at a population-wide scale is largely debatable3.
Over the past decades, the potential of genetic polymorphisms as markers for breast cancer risk assessment has become increasingly apparent4. In contrast to high penetrance mutations, genetic polymorphisms are very common in the general population, but are typically associated with a modest risk of cancer. However, it is believed that when a large number of such polymorphisms are combined, the magnitude of the risk conferred could be very significant5. In recent years, polymorphisms in microRNA genes have been widely investigated as the gene products play important roles in regulating the expression of many cancer-related genes6–8. Among the microRNA genes that have been frequently investigated is MIR499A, which encodes microRNA-499a. The functions and mechanism of miR-499a in breast cancer is not well understood, but it is known that miR-499a possesses both oncogenic and tumor suppressive functions in other cancer types. For example, in colorectal cancer, miR-499a could promote invasiveness and metastatic capabilities by targeting FOXO4 and PDCD4 tumor suppressor genes9. Similarly, in hepatocellular carcinoma (HCC), the microRNA was found to target MAPK6 to induce proliferation and migration of the cells10,11. However, in HCC, the microRNA was also observed to target the ETS1 oncogene to inhibit the cancer progression12. Additionally, in non-small cell lung cancer, miR-499a was found to exert its tumor suppressive function by targeting VAV3 oncogene13.
The mechanism of microRNA target recognition and selection is determined, at least in part, by its seed sequences, i.e. a conserved region essential for its binding to target mRNAs14. Thus, polymorphisms occurring in the seed region of a microRNA gene may contribute to its oncogenic or tumor suppressive functions and subsequently affect cancer risk or susceptibility. One such polymorphism is rs3746444, which results from an A-to-G substitution in the seed region of MIR499A. A number of studies have examined the association of the polymorphism with breast cancer susceptibility, but the results obtained have been inconsistent. For instance, while He et al.15 demonstrated that the variant allele of MIR499A rs3746444 polymorphism was significantly associated with an increased susceptibility to breast cancer, Morales et al.16 did not find any significant association. One of the major reasons for the inconsistency of study results in genetic association studies is the limited sample size and thus, low statistical power of the individual studies17,18. Therefore, in this work, we aimed to pool the study findings via a meta-analysis in order to derive a more precise estimate of the association between MIR499A rs3746444 polymorphism and breast cancer susceptibility.
Methodology
Literature search
A comprehensive literature search was performed on PubMed, Scopus, Web of Science (WoS), China National Knowledge Infrastructure (CNKI), VIP and Wanfang databases up to January 2020. No language restriction was imposed. The PubMed search terms used were as follows: [(breast neoplasms OR breast cancer) AND (MIR499A OR MIR-499 OR MIR-499A OR microRNA-499 OR microRNA-499A OR rs3746444) AND (polymorphism OR mutation OR variant OR variation)]. The search terms were modified appropriately for other databases.
Eligible studies were selected based on the following criteria: (i) those that investigated the association between MIR499A rs3746444 polymorphism and breast cancer susceptibility; (ii) case-control (including nested case-control) or cohort in design; and (iii) contained data on genotype and allele frequencies or sufficient data to derive this information. Non-human studies and duplicated reports were excluded. When overlapping data were found in multiple studies, only the most recent report or the one with the largest sample size was included.
Data extraction and quality assessment
Two investigators independently extracted the following information from eligible studies: First author, year of publication, country, number of cases and controls, genotype and allele frequencies, ethnicity of the subjects (Asian vs. Caucasian), and deviation of control genotype distribution from the Hardy-Weinberg equilibrium (HWE). If data on HWE was not reported, a goodness-of-fit test was used to determine whether the observed genotype frequencies conformed to the expected distribution. The quality of the eligible studies was assessed by using the Modified Newcastle-Ottawa Scale for Case-Control Studies of Genetic Association19. Studies with ≥5 stars were considered to be of high quality.
Statistical analysis
Quantitative synthesis of the data was performed by using Review Manager (version: 5.3.5), with the common allele/genotype used as the reference. The association between MIR499A rs3746444 polymorphism and breast cancer susceptibility was evaluated under five genetic models: homozygous (GG vs. AA), heterozygous (AG vs. AA), dominant (AG + GG vs. AA), recessive (GG vs. AA + AG) and allele (G vs. A) comparison models. Heterogeneity among the studies in each genetic model was assessed by using Cochran’s Q and I2 statistics. A P value of <0.1 and I2 value of >50% was interpreted as having significant heterogeneity. A fixed-effect method was used to calculate the pooled odds ratio (OR) and the corresponding 95% confidence interval (CI) for genetic models which did not show significant heterogeneity; otherwise a random-effect method was used. The significance of the genetic association was measured using Z test. A forest plot was constructed to graphically display the results. A subgroup analysis was performed based on ethnicity of the subjects (Asian vs. Caucasian) and methodology quality of the studies (high quality vs. low quality). Sensitivity analysis was performed by iteratively omitting one study at a time to determine the stability and robustness of the results. Publication bias was assessed by performing Begg’s test and Egger’s test using JASP (version 0.9.2.0), and by visually inspecting the funnel plot for asymmetry. For all analyses, statistically significance was assumed at P < 0.05, unless otherwise stated.
In silico analysis
Prediction of miR-499a target genes was performed using DIANA Tarbase version 7.0 (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index) and DIANA microT-CDS (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index) tools which have been integrated with miRBase (ver. 18) and Ensembl (ver. 69). Functional annotation of the predicted target genes as well as KEGG pathway enrichment analysis was then performed by using DIANA-miRPath version 3.0 (http://snf-515788.vm.okeanos.grnet.gr/)20–23. The DIANA-microT threshold used was 0.8 at a P value of 0.05. Fisher’s exact test (hypergeometric statistics) and false discovery rate (FDR) correction was used for the enrichment analysis. miRmut2Go (http://compbio.uthsc.edu/miR2GO/mir2goSNP.php) was used to compare the functional similarity between the wild type and variant alleles of the MIR499A rs3746444 polymorphism. Furthermore, RNAfold WebServer (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) was used to predict, based on minimum free energy (MFE) calculation, the secondary structures formed as a result of the A-to-G substitution. PolymiRTS Database 3.0 (http://compbio.uthsc.edu/miRSNP/) was used to predict whether the polymorphism affects its target recognition.
Results
Characteristics of the studies
The initial search strategy resulted in the identification of 1028 records (PubMed, N = 25; Scopus, N = 931; WoS, N = 59; CNKI, N = 7; VIP, N = 1; Wanfang, N = 5). After deduplication, 932 unique records were screened by title and abstract for relevance. A total of 15 records were identified as being potentially relevant; thus, full-text articles of 16 potentially relevant records were retrieved and checked for eligibility. The reference list of these records were also hand-searched to identify additional studies. Ultimately, 12 records (which comprised 13 studies) were included in the meta-analysis15,16,24–33. We also included preliminary data from our laboratory in the meta-analysis, making the total number of included studies 14. The search selection process is illustrated in Fig. 1.
Figure 1.
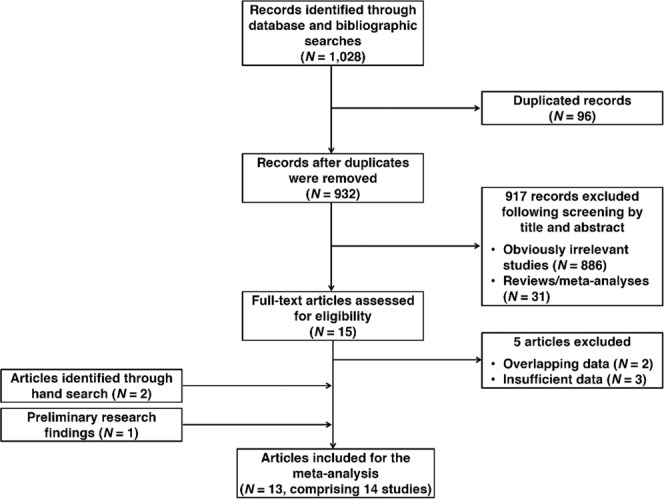
Flow diagram of study selection.
These 14 studies involved a total of 6,797 cases and 8,534 controls. Detailed characteristics of the included studies are shown in Table 1. Subjects in 10 of the studies belonged to Asian ethnicity, three other studies were conducted on Caucasians, while those in the remaining one study were Africans. Eight of the studies exhibited high methodological quality based on the Modified Newcastle-Ottawa Scale for Case-Control Studies of Genetic Association. The star ratings of the included studies are shown in Table 2.
Table 1.
Characteristics of the included studies.
| No. | Author | Year | Ethnicity | Population | No. of subjects (case/control) | HWE P value in controls |
|---|---|---|---|---|---|---|
| 1. | Hu et al.24 | 2009 | Asian | China | 1009/1093 | 0.057 |
| 2. | Catucci et al. (a)25 | 2010 | Caucasian | Italy | 756/1242 | 0.250 |
| 3. | Catucci et al. (b)25 | 2010 | Caucasian | Germany | 823/925 | 0.893 |
| 4. | Alshatwi et al.26 | 2012 | Asian | Saudi Arabia | 100/100 | 0.227 |
| 5. | Bansal et al.27 | 2014 | Asian | India | 121/164 | 0.002 |
| 6. | Omrani et al.28 | 2014 | Asian | Iran | 236/203 | <0.001 |
| 7. | Qi et al.29 | 2015 | Asian | China | 321/290 | 0.053 |
| 8. | He et al.15 | 2015 | Asian | China | 450/450 | 0.143 |
| 9. | Dai et al.30 | 2015 | Asian | China | 560/583 | 0.131 |
| 10. | Qian et al.32 | 2016 | African | Multiple | 1657/2028 | 0.288 |
| 11. | Afsharzadeh et al.33 | 2017 | Asian | Iran | 100/150 | 0.633 |
| 12. | Morales et al.16 | 2018 | Caucasian | Chile | 440/1048 | 0.836 |
| 13. | Doulah et al.31 | 2018 | Asian | Iran | 80/100 | 0.901 |
| 14. | Tan et al. | Unpublished | Asian | Malaysian | 144/158 | 0.165 |
Table 2.
Assessment of the quality of the included studies.
| Study | Selection | Comparability | Exposure | Total star | |||||
|---|---|---|---|---|---|---|---|---|---|
| Criteria 1 | Criteria 2 | Criteria 3 | Criteria 4 | Criteria 1 | Criteria 1 | Criteria 2 | Criteria 3 | ||
| Hu et al.24 | ★ | ★ | ★ | ★ | ★ | ★ | ★ | 7 | |
| Catucci et al. (a)25 | ★ | ★ | ★ | ★ | ★ | ★ | 6 | ||
| Catucci et al. (b)25 | ★ | ★ | ★ | ★ | ★ | ★ | 6 | ||
| Alshatwi et al.26 | ★ | ★ | ★ | ★ | 4 | ||||
| Bansal et al.27 | ★ | ★ | ★ | ★ | ★ | 5 | |||
| Omrani et al.28 | ★ | ★ | ★ | ★ | 4 | ||||
| Qi et al.29 | ★ | ★ | ★ | 3 | |||||
| He et al.15 | ★ | ★ | ★ | ★ | ★ | 5 | |||
| Dai et al.30 | ★ | ★ | ★ | ★ | ★ | ★ | 6 | ||
| Qian et al.32 | ★ | ★ | ★ | ★ | ★★ | ★ | 7 | ||
| Afsharzadeh et al.33 | ★ | ★ | ★ | 3 | |||||
| Morales et al.16 | ★ | ★ | ★ | ★ | ★★ | ★ | ★ | 8 | |
| Doulah et al.31 | ★ | ★ | ★ | 3 | |||||
| Tan et al. | ★ | ★ | ★ | ★ | 4 | ||||
Quantitative synthesis
The pooled association of MIR499A rs3746444 polymorphism with breast cancer susceptibility is summarized in Table 3. Overall, the polymorphism was found to be significantly associated with breast cancer susceptibility in all genetic models (homozygous model, OR = 1.33, 95% CI = 1.03–1.71, P = 0.03; heterozygous model, OR = 1.09, 95% CI = 1.01–1.17, P = 0.02; dominant model, OR = 1.15, 95% CI = 1.02–1.30, P = 0.03; recessive model, OR = 1.35, 95% CI = 1.06–1.72, P = 0.01; allele model, OR = 1.12, 95% CI = 1.00–1.26, P = 0.04) (Fig. 2). Subgroup analysis by ethnicity revealed that the statistically significant positive association was present only among Asians (P < 0.05 in all genetic models except allele model), but not among Caucasians (P > 0.05) (Table 3). In the allele model, a borderline lack of significance was observed (P = 0.08). In addition, when stratified by methodology quality of the studies, high quality studies exhibited a significant (or borderline lack of significant) positive association in all genetic models, whereas low quality studies showed a significant association only in recessive model (Table 3).
Table 3.
Summary of the association between MIR499A rs3746444 polymorphism and breast cancer susceptibility.
| Comparison model | No. of studies | No. of cases | No. of controls | Effect model | OR (95% CI) | P |
|---|---|---|---|---|---|---|
| Homozygous model | ||||||
| Overall | 14 | 4,704 | 6,085 | Random | 1.33 (1.03–1.71) | 0.03 |
| Asian | 10 | 2,147 | 2,420 | Random | 1.45 (1.01–2.07) | 0.04 |
| Caucasian | 3 | 1,363 | 2,219 | Fixed | 1.04 (0.79–1.37) | 0.79 |
| High quality | 8 | 4,097 | 5,367 | Fixed | 1.28 (1.09–1.51) | <0.01 |
| Low quality | 6 | 607 | 718 | Random | 1.23 (0.60–2.53) | 0.57 |
| Heterozygous model | ||||||
| Overall | 14 | 6,337 | 8,117 | Fixed | 1.08 (1.00–1.16) | 0.04 |
| Asian | 10 | 2,825 | 3,088 | Fixed | 1.18 (1.05–1.32) | <0.01 |
| Caucasian | 3 | 1,925 | 3,073 | Fixed | 1.04 (0.92–1.18) | 0.48 |
| High quality | 8 | 5,490 | 7,205 | Fixed | 1.07 (0.99–1.15) | 0.11 |
| Low quality | 6 | 847 | 912 | Random | 1.24 (0.86–1.79) | 0.26 |
| Dominant model | ||||||
| Overall | 14 | 6,797 | 8,534 | Random | 1.15 (1.02–1.30) | 0.03 |
| Asian | 10 | 3,121 | 3,291 | Random | 1.24 (1.02–1.50) | 0.03 |
| Caucasian | 3 | 2,019 | 3,215 | Fixed | 1.04 (0.93–1.17) | 0.47 |
| High quality | 8 | 5,816 | 7,533 | Fixed | 1.09 (1.02–1.18) | 0.02 |
| Low quality | 6 | 981 | 1,001 | Random | 1.27 (0.86–1.88) | 0.23 |
| Recessive model | ||||||
| Overall | 14 | 6,797 | 8,534 | Random | 1.35 (1.06–1.72) | 0.01 |
| Asian | 10 | 3,021 | 3,141 | Fixed | 1.60 (1.32–1.93) | <0.01 |
| Caucasian | 3 | 2,019 | 3,215 | Fixed | 1.02 (0.77–1.33) | 0.91 |
| High quality | 8 | 5,816 | 7,533 | Fixed | 1.16 (0.99–1.37) | 0.07 |
| Low quality | 6 | 981 | 1,001 | Random | 1.78 (1.03–3.08) | 0.04 |
| Allele model | ||||||
| Overall | 14 | 13,594 | 17,068 | Random | 1.12 (1.00–1.26) | 0.04 |
| Asian | 10 | 6,242 | 6,582 | Random | 1.17 (0.98–1.40) | 0.08 |
| Caucasian | 3 | 4,038 | 6,430 | Fixed | 1.03 (0.94–1.14) | 0.52 |
| High quality | 8 | 11,632 | 15,066 | Fixed | 1.11 (1.04–1.17) | <0.01 |
| Low quality | 6 | 1,962 | 2,002 | Random | 1.11 (0.78–1.58) | 0.56 |
Figure 2.
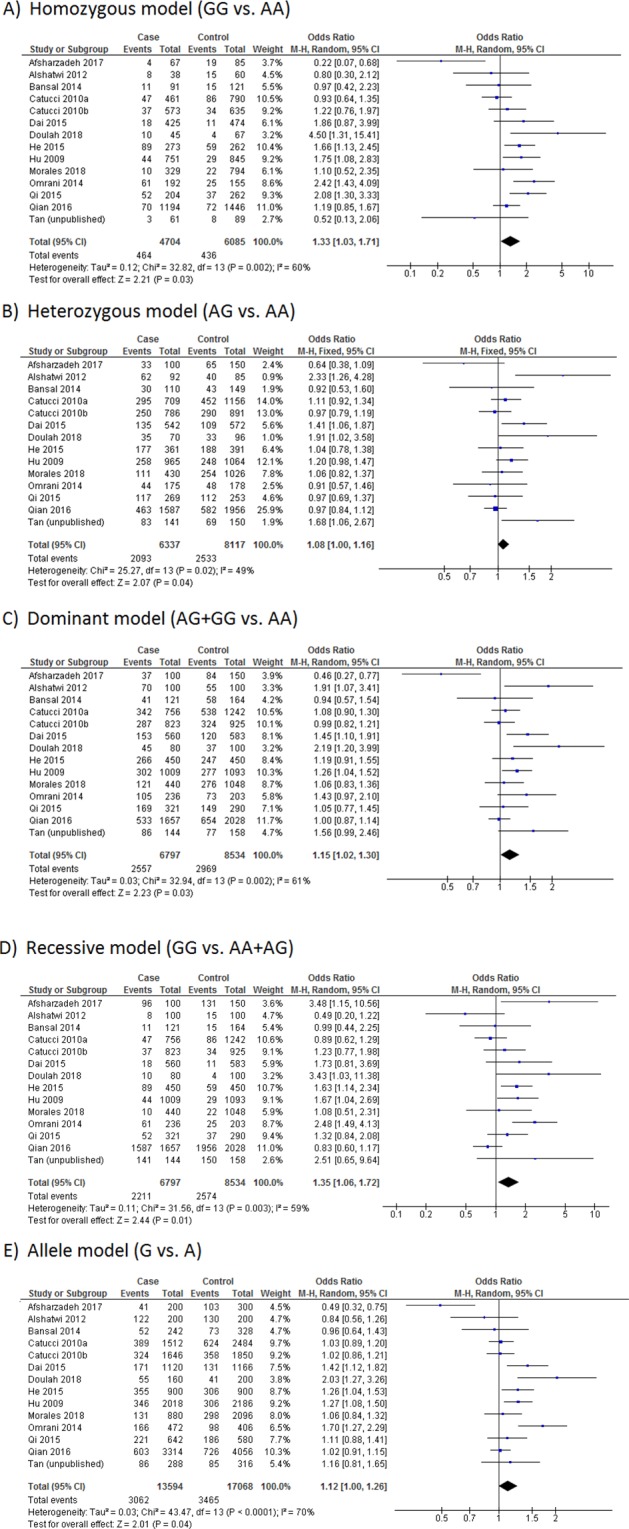
Forest plots of the association between MIR499A rs3746444 polymorphism and breast cancer susceptibility.
Sensitivity analysis
For heterozygous and recessive models, the results remained largely unchanged when any of the studies was removed from the analysis. However, in other models, the results were less stable. In particular, the statistical significance of homozygous, dominant and allele models diminished when data from Dai et al.30 was omitted. A similar observation was documented in homozygous and allele models when data from Hu et al.24 was removed. In addition, the homozygous model was also heavily influenced by Qi et al.29, while the allele model was heavily influenced by Doulah et al.31, He et al.15, Omrani et al.28 and Qi et al.29 Despite this, the direction and magnitude of the association were not affected by any of the studies, and the P values were still at the borderline of statistical significance.
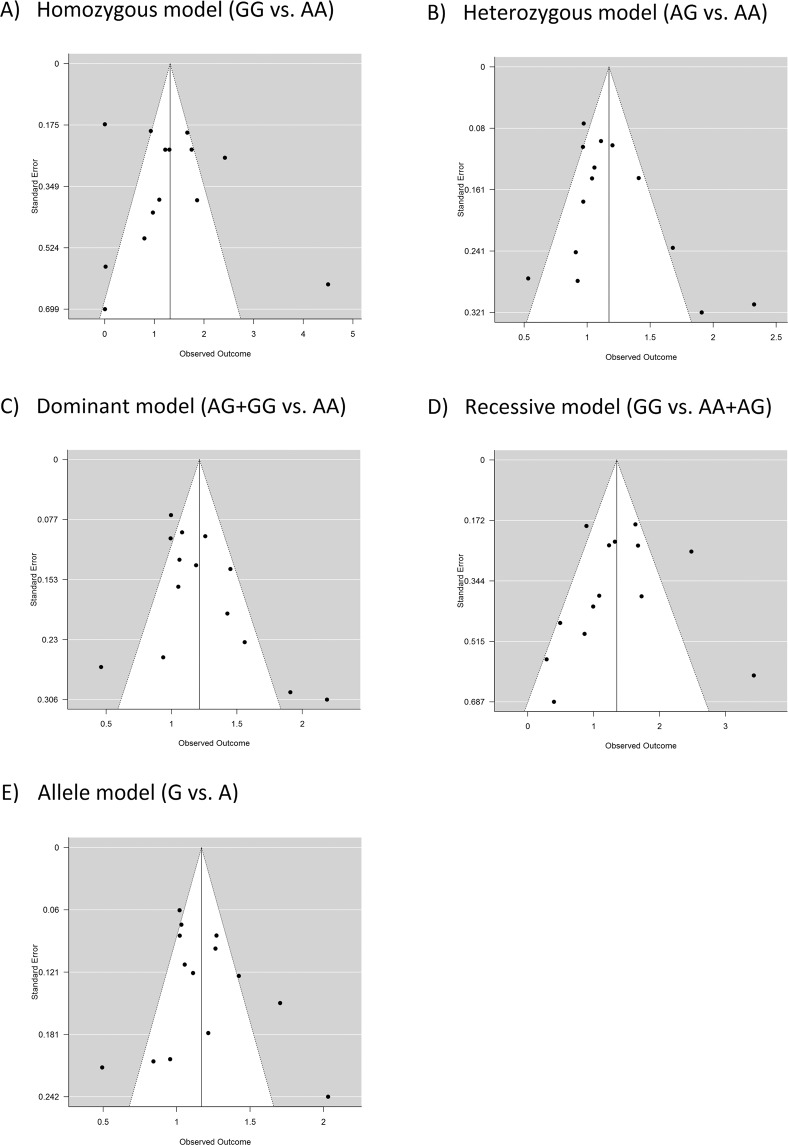
Publication bias
Funnel plots for publication bias are shown in Fig. 3. Visual inspection of the funnel plots did not reveal any evidence of publication bias. Formal tests using Begg’s and Egger’s methods also did not find any significant publication bias (homozygous model, Begg’s test P = 0.667, Egger’s test P = 0.787; heterozygous model, Begg’s test P = 0.388, Egger’s test P = 0.089; dominant model, Begg’s test P = 0.062, Egger’s test P = 0.055; recessive model, Begg’s test P = 0.279, Egger’s test P = 0.518; allele model, Begg’s test P = 0.518, Egger’s test P = 0.706).
Figure 3.
Funnel plots for assessing publication bias.
In silico analysis
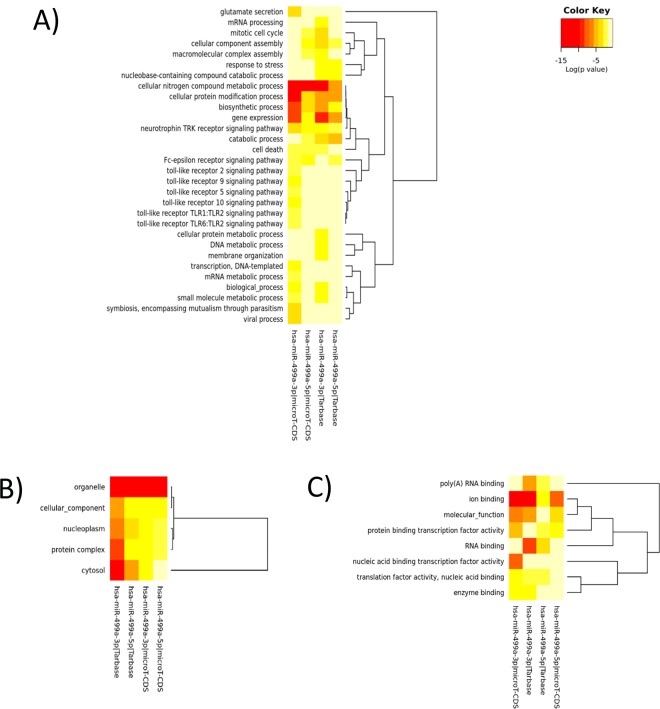
The DIANA Tarbase v7.0 and DIANA microT-CDS identified a total of 2,286 target genes for miR-499a. Of these, 1,169 were targeted by the 3p arm, 899 by the 5p arm, and 218 by both arms of miR-499a. Pathway enrichment analysis of these target genes revealed that transcriptional misregulation in cancer is the most common pathway involved (28 genes), followed by RNA transport (26 genes), ubiquitin mediated proteolysis (24 genes), cell cycle and hippo signaling pathway (22 genes) (Table 4). Functional annotation of target genes based on the biological processes, cellular components and molecular functions is shown in Fig. 4. It was noted that the most prominent biological process involved is the Toll-like receptor signaling pathway.
Table 4.
DIANA miRPath KEGG pathway enrichment analysis of the miR-499a target genes.
| KEGG pathway | KEGG pathway ID | p-value | Found genes | miRNAs |
|---|---|---|---|---|
| Transcriptional misregulation in cancer | hsa05202 | 2.15E-05 | 28 | 4 |
| Biotin metabolism | hsa00780 | 3.28E-05 | 1 | 1 |
| Thyroid hormone signaling pathway | hsa04919 | 0.000244899 | 19 | 4 |
| Cell cycle | hsa04110 | 0.000695201 | 22 | 4 |
| Sulfur relay system | hsa04122 | 0.003319243 | 2 | 2 |
| Hippo signaling pathway | hsa04390 | 0.005447739 | 22 | 4 |
| RNA transport | hsa03013 | 0.009415218 | 26 | 4 |
| Ubiquitin mediated proteolysis | hsa04120 | 0.021323824 | 24 | 4 |
| Hedgehog signaling pathway | hsa04340 | 0.027238434 | 11 | 3 |
| Prostate cancer | hsa05215 | 0.029004416 | 17 | 4 |
Figure 4.
Heat map showing (A) biological processes (B) cellular components and (C) molecular functions of predicted miR-499a target genes.
Analysis with PolymiRTS v3.0 showed that the A-to-G substitution in the rs3746444 polymorphism created new target sites for 763 genes and disrupted the target sites of 2,408 genes. Besides, miRmut2Go revealed that the A and G alleles had low functional similarity for biological processes (similarity score: 0.133) and molecular functions (similarity score: 0.221). No similarity score could be generated for cellular component as there was no significantly enriched GO term. In addition, analysis with RNAfold WebServer found that the rs3746444 polymorphism did not lead to significant effect on the folding (and thus, secondary structure formation) of the microRNA.
Discussion
MicroRNAs are an emerging class of post-transcriptional regulators which have been implicated in oncogenesis. Polymorphisms within microRNA genes may have an impact on an individual’s susceptibility to cancer. The MIR499A rs3746444 polymorphism, for example, has been shown to be significantly associated with risk of cancer of the respiratory, digestive, urinary and gynecological systems34. Over the past decade, a growing number of studies have examined the association between MIR499A rs3746444 polymorphism and breast cancer susceptibility, but the results were inconsistent and often contradictory. In this work, we addressed this irregularity by pooling data from 14 previous studies which comprised 15,331 subjects (6,797 cases and 8,534 controls) via a meta-analysis. We found that the polymorphism was significantly associated with an increased breast cancer susceptibility under all five genetic models investigated.
One of the possible explanations for this observation is that the two alleles of the rs3746444 polymorphism resulted in different levels of miR-499a. In fact, it has been demonstrated previously that the variant GG genotype of the polymorphism had the lowest delta CT value (which implied a high expression level), followed by AG and AA genotypes, although it was not clear whether the difference was statistically significant26. It has also been reported in an in vitro study that miR-499a originating from the two alleles of rs3746444 polymorphism reduced the expression of tumor suppressor genes to different extents35. Considering these previous findings, we postulate that the variant allele of the polymorphism gave rise to a higher level of miR-499a and caused a significant decrease in the expression of tumor suppressor genes, thereby leading to an increase in breast cancer susceptibility. Besides, our in silico analysis showed that the A-to-G substitution in the rs3746444 polymorphism disrupted the target sites and created new target sites for a large number of genes. Genes targeted by the different alleles of the polymorphism are involved in different biological processes and have different molecular functions. We postulate that the A-to-G substitution resulted in a higher affinity of miR-499a for tumor suppressor genes, which could explain the increased breast cancer susceptibility associated with the variant allele. Nonetheless, the above postulations require further investigations as miR-499a is known to regulate not only tumor suppressor genes, but also oncogenes9,10,12,13.
In the present meta-analysis, a random-effect method was used in several genetic models as significant heterogeneity was observed among the studies. We addressed the heterogeneity by performing subgroup analysis according to the ethnicity of the subjects (Asians vs. Caucasians) as well as the methodological quality of the included studies. In most genetic models, subgroup analysis by ethnicity reduced the statistical heterogeneity. Interestingly, we also observed that significant association was only present in Asians, but not in Caucasians, under most genetic models. This discrepancy could be attributed to the heterogeneity in linkage disequilibrium among subjects of different ethnicities, which is a common concern in genetic association studies involving multiple populations36,37. Moreover, genetic association is known to be affected by gene-gene, gene-environment and gene-nutrient interactions, which might explain the disagreement in study findings between the two subgroups38,39. It is also worthy of mention that among the 14 studies included in the meta-analysis, only three were conducted among Caucasians. Hence, it remains a possibility that the lack of significance among Caucasians was a false negative observation due to a relatively weak statistical power40. Only one of the included studies was conducted among Africans; thus, subgroup analysis was not performed on Africans.
Besides, we noted that statistical heterogeneity was eliminated when only studies of high methodological quality were included in the analysis. A high quality study was defined as one which reported and fulfilled at least five of the following criteria: (i) cases were confirmed as having the malignancy, (ii) no evidence of selection bias among cases, (iii) population-based controls and genotypic distribution that conformed to HWE, (iv) appropriate control selection, (v) cases and controls were of the same ethnicity, (vi) no evidence of population stratification, (vii) genotypes validated using an independent method and investigators were blinded to the subject status, (viii) used the same genotyping method for all subjects, and (ix) genotyping call rate >99%19. It is unknown which of the criteria contributed to the heterogeneity. However, all low quality studies did not fulfill and/or report criteria (v), (vi), (viii) and (ix) – the former two were related to ethnic descent, which again highlighted the influence of ethnicity on the genetic association.
In this work, two formal tests were employed to assess publication bias, namely Begg’s and Egger’s tests. Both tests are commonly used in meta-analysis of genetic association studies41. However, there are inherent limitations associated with each test. Begg’s test does not take into account between-study heterogeneity and is more accurate when the number of included studies is large42. On the other hand, Egger’s test tend to give false-positive results and is more suitable for studies with continuous outcomes43,44. We included the two tests in our analysis to complement the limitations of each other. Both tests indicated that there was no significant publication bias in all genetic models.
There are several limitations of this meta-analysis. First, we reported only crude estimates of genetic association and did not measure gene-gene or gene-environment interactions because not all included studies contained this information. Second, the number of studies included was relatively small, especially for studies in non-Asian populations. Therefore, the association of MIR499A rs3746444 polymorphism with breast cancer susceptibility among non-Asian populations remained unclear. Nevertheless, a major strength of the present work is that it included a larger number of studies and subjects compared to previous reports on this topic45,46. Thus, the present meta-analysis provided an updated and integrated estimate of the association between the polymorphism and breast cancer risk. In addition, we performed in silico analysis to predict the functional impact of the polymorphism, which may help to clarify the mechanisms by which MIR499A rs3746444 influences breast cancer susceptibility.
In conclusion, we provided quantitative evidence that MIR499A rs3746444 polymorphism was associated with an elevated breast cancer susceptibility among Asians, but not among Caucasians. Additional studies is required to better clarify the clinical impact of this genetic association. Future work which takes into account gene-gene, gene-environment and gene-nutrient interactions is warranted for a more precise evidence and to further elucidate the underlying mechanism of breast cancer susceptibility.
Acknowledgements
This work was supported by the Geran Galakan Penyelidik Muda (Young Researcher Incentive Grant) of Universiti Kebangsaan Malaysia (No. GGPM-2018–045) and the Fundamental Research Grant Scheme of the Ministry of Education Malaysia (No. FRGS/1/2019/SKK08/UKM/02/9).
Author contributions
S.C.T. conceived and designed the study, screened and selected the eligible studies for meta-analysis, collected and extracted the data, appraised the study quality, performed statistical analysis, and wrote the manuscript. P.Y.L. independently screened and selected the studies, extracted the data for analysis, and provided statistical expertise. J.F. performed literature search in Chinese databases, retrieved Chinese publications, and translated eligible Chinese studies into English. M.F.M.M. performed in silico analysis. E.A.M.H. provided critical revision of the draft manuscript. R.J. provided significant input and feedback on the draft manuscript. All authors have read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Nelson, H. D. et al. Risk assessment, genetic counseling, and genetic testing for BRCA-related cancer: systematic review to update the U.S. Preventive Services Task Force Recommendation. (Agency for Healthcare Research and Quality (US), 2013). [PubMed]
- 3.Lippi G, Mattiuzzi C, Montagnana M. BRCA population screening for predicting breast cancer: for or against? Ann. Transl. Med. 2017;5:275–275. doi: 10.21037/atm.2017.06.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wendt C, Margolin S. Identifying breast cancer susceptibility genes - a review of the genetic background in familial breast cancer. Acta Oncol. 2019;58:135–146. doi: 10.1080/0284186X.2018.1529428. [DOI] [PubMed] [Google Scholar]
- 5.Cuzick J, Brentnall A, Dowsett M. SNPs for breast cancer risk assessment. Oncotarget. 2017;8:99211–99212. doi: 10.18632/oncotarget.22278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anfossi S, Fu X, Nagvekar R, Calin GA. MicroRNAs, Regulatory Messengers Inside and Outside Cancer Cells. Adv. Exp. Med. Biol. 2018;1056:87–108. doi: 10.1007/978-3-319-74470-4_6. [DOI] [PubMed] [Google Scholar]
- 7.Teoh SL, Das S. The Role of MicroRNAs in Diagnosis, Prognosis, Metastasis and Resistant Cases in Breast Cancer. Curr. Pharm. Des. 2017;23:1845–1859. doi: 10.2174/1381612822666161027120043. [DOI] [PubMed] [Google Scholar]
- 8.Das, S. et al. Micro-RNA and the Features of Metabolic Syndrome: A Narrative Review. MRMC20, (2020). [DOI] [PubMed]
- 9.Liu X, et al. MicroRNA-499-5p promotes cellular invasion and tumor metastasis in colorectal cancer by targeting FOXO4 and PDCD4. Carcinogenesis. 2011;32:1798–1805. doi: 10.1093/carcin/bgr213. [DOI] [PubMed] [Google Scholar]
- 10.Xiang Z, Wang S, Xiang Y. Up-regulated microRNA499a by hepatitis B virus induced hepatocellular carcinogenesis via targeting MAPK6. PLoS ONE. 2014;9:e111410. doi: 10.1371/journal.pone.0111410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sulaiman SA, Abu N, Ab-Mutalib N-S, Low TY, Jamal R. Signatures of gene expression, DNA methylation and microRNAs of hepatocellular carcinoma with vascular invasion. Future Oncol. 2019;15:2603–2617. doi: 10.2217/fon-2018-0909. [DOI] [PubMed] [Google Scholar]
- 12.Wei W, et al. MicroRNA-1 and microRNA-499 downregulate the expression of the ets1 proto-oncogene in HepG2 cells. Oncology Reports. 2012;28:701–706. doi: 10.3892/or.2012.1850. [DOI] [PubMed] [Google Scholar]
- 13.Li, M. et al. Overexpression of miR-499-5p inhibits non-small cell lung cancer proliferation and metastasis by targeting VAV3. Sci Rep6, (2016). [DOI] [PMC free article] [PubMed] [Retracted]
- 14.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He B, et al. Associations of polymorphisms in microRNAs with female breast cancer risk in Chinese population. Tumour Biol. 2015;36:4575–4582. doi: 10.1007/s13277-015-3102-2. [DOI] [PubMed] [Google Scholar]
- 16.Morales S, et al. Genetic Variants in pre-miR-146a, pre-miR-499, pre-miR-125a, pre-miR-605, and pri-miR-182 Are Associated with Breast Cancer Susceptibility in a South American Population. Genes. 2018;9:427. doi: 10.3390/genes9090427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan SC. Low penetrance genetic polymorphisms as potential biomarkers for colorectal cancer predisposition. J Gene Med. 2018;20:e3010. doi: 10.1002/jgm.3010. [DOI] [PubMed] [Google Scholar]
- 18.Tan SC, Ankathil R. Genetic susceptibility to cervical cancer: role of common polymorphisms in apoptosis-related genes. Tumour Biol. 2015;36:6633–6644. doi: 10.1007/s13277-015-3868-2. [DOI] [PubMed] [Google Scholar]
- 19.Yang P, Liu H, Lin J, Yang H. The Association of rs4753426 Polymorphism in the Melatonin Receptor 1B (MTNR1B) Gene and Susceptibility to Adolescent Idiopathic Scoliosis: A Systematic Review and Meta-analysis. Pain Physician. 2015;18:419–431. [PubMed] [Google Scholar]
- 20.Vlachos IS, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Z, et al. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum. Mutat. 2009;30:79–84. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- 25.Catucci I, et al. Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Hum. Mutat. 2010;31:E1052–1057. doi: 10.1002/humu.21141. [DOI] [PubMed] [Google Scholar]
- 26.Alshatwi Ali A., Shafi Gowhar, Hasan Tarique N., Syed Naveed Ahmed, Al-Hazzani Amal A., Alsaif Mohammed A., Alsaif Abdulaziz A. Differential Expression Profile and Genetic Variants of MicroRNAs Sequences in Breast Cancer Patients. PLoS ONE. 2012;7(2):e30049. doi: 10.1371/journal.pone.0030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bansal C, et al. Common genetic variants in pre-microRNAs and risk of breast cancer in the North Indian population. Ecancermedicalscience. 2014;8:473. doi: 10.3332/ecancer.2014.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omrani M, et al. hsa-mir-499 rs3746444 gene polymorphism is associated with susceptibility to breast cancer in an Iranian population. Biomark Med. 2014;8:259–267. doi: 10.2217/bmm.13.118. [DOI] [PubMed] [Google Scholar]
- 29.Qi P, et al. Associations of miRNA polymorphisms and expression levels with breast cancer risk in the Chinese population. Genet. Mol. Res. 2015;14:6289–6296. doi: 10.4238/2015.June.11.2. [DOI] [PubMed] [Google Scholar]
- 30.Dai Zhi-Ming, Kang Hua-Feng, Zhang Wang-Gang, Li Hong-Bao, Zhang Shu-Qun, Ma Xiao-Bin, Lin Shuai, Wang Meng, Feng Yan-Jing, Liu Kang, Liu Xing-Han, Xu Peng, Dai Zhi-Jun. The Associations of Single Nucleotide Polymorphisms in miR196a2, miR-499, and miR-608 With Breast Cancer Susceptibility. Medicine. 2016;95(7):e2826. doi: 10.1097/MD.0000000000002826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doulah A, Salehzadeh A, Mojarrad M. Association of single nucleotide polymorphisms in miR- 499 and miR-196a with susceptibility to breast cancer. Trop. J. Pharm Res. 2018;17:319. doi: 10.4314/tjpr.v17i2.17. [DOI] [Google Scholar]
- 32.Qian F, et al. Genetic variants in microRNA and microRNA biogenesis pathway genes and breast cancer risk among women of African ancestry. Hum. Genet. 2016;135:1145–1159. doi: 10.1007/s00439-016-1707-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Afsharzadeh SM, Mohaddes Ardebili SM, Seyedi SM, Karimian Fathi N, Mojarrad M. Association between rs11614913, rs3746444, rs2910164 and occurrence of breast cancer in Iranian population. Meta Gene. 2017;11:20–25. doi: 10.1016/j.mgene.2016.11.004. [DOI] [Google Scholar]
- 34.Yang, X., Li, X. & Zhou, B. A Meta-Analysis of miR-499 rs3746444 Polymorphism for Cancer Risk of Different Systems: Evidence From 65 Case-Control Studies. Front. Physiol. 9, (2018). [DOI] [PMC free article] [PubMed]
- 35.Torruella-Loran I, et al. MicroRNA Genetic Variation: From Population Analysis to Functional Implications of Three Allele Variants Associated with Cancer. Hum. Mutat. 2016;37:1060–1073. doi: 10.1002/humu.23045. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y-J, Papasian CJ, Liu J-F, Hamilton J, Deng H-W. Is Replication the Gold Standard for Validating Genome-Wide Association Findings? PLOS ONE. 2008;3:e4037. doi: 10.1371/journal.pone.0004037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura T. East meets West: ethnic differences in prostate cancer epidemiology between East Asians and Caucasians. Chin J Cancer. 2012;31:421–429. doi: 10.5732/cjc.011.10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nadeem A, et al. Gene-gene, gene-environment, gene-nutrient interactions and single nucleotide polymorphisms of inflammatory cytokines. World J Diabetes. 2015;6:642–647. doi: 10.4239/wjd.v6.i4.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdullah N, et al. Predicting type 2 diabetes using genetic and environmental risk factors in a multi-ethnic Malaysian cohort. Public Health. 2017;149:31–38. doi: 10.1016/j.puhe.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 40.Vadillo MA, Konstantinidis E, Shanks DR. Underpowered samples, false negatives, and unconscious learning. Psychon Bull Rev. 2016;23:87–102. doi: 10.3758/s13423-015-0892-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee YH. Meta-Analysis of Genetic Association Studies. Ann Lab Med. 2015;35:283. doi: 10.3343/alm.2015.35.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi X, et al. Effect Comparison between Egger’s Test and Begg’s Test in Publication Bias Diagnosis in Meta-Analyses: Evidence from a Pilot Survey. International Journal of Research Studies in Biosciences. 2017;5:14–20. [Google Scholar]
- 43.Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295:676–680. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- 44.Cochrane Handbook for Systematic Reviews of Interventions (Version 5.1.0). (2011).
- 45.Mu K, et al. Meta-analysis of the association between three microRNA polymorphisms and breast cancer susceptibility. Oncotarget. 2017;8:68809–68824. doi: 10.18632/oncotarget.18516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, et al. Association between three functional microRNA polymorphisms (miR-499 rs3746444, miR-196a rs11614913 and miR-146a rs2910164) and breast cancer risk: a meta-analysis. Oncotarget. 2017;8:393–407. doi: 10.18632/oncotarget.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]