Abstract
Background: In search of novel biomarkers of response to bevacizumab in metastatic colorectal cancer (mCRC), we analyzed the expression and prognostic role of several proteins related to angiogenesis. Methods: A retrospective, multicenter study on 80 surgical samples from mCRC patients treated in first line with bevacizumab plus chemotherapy was accomplished. The following proteins were analyzed by immunohistochemistry: hERG1 potassium channel, β1-integrin, pAKT, NFkB, HIF-1α, HIF-2α, p53, VEGF-A, GLUT-1, and CA-IX. Data were analyzed in conjunction with the clinicopathological characteristics of the patients, KRAS status, response to bevacizumab, and follow-up. Results: (1) All the proteins were expressed in the samples, with statistically significant associations between HIF-1α and gender, HIF-2α and left colon, hERG1 and VEGF-A, β1-integrin and HIF-2α, GLUT-1 and both HIF-1α and HIF-2α, and CA-IX and VEGF-A. (2) At the univariate analysis, positivity for hERG1, VEGF-A, and the active form of HIF-2α (aHIF-2α), and the G3 histological grade showed a positive impact on progression-free survival (PFS). (3) hERG1 and aHIF-2α maintained their positive impact on PFS at the multivariate analysis. (4) hERG1 behaved as a protective factor for PFS independently on KRAS status. Conclusions: hERG1 and aHIF-2α might help to identify patients who would benefit from bevacizumab treatment.
Introduction
The management of metastatic colorectal cancer (mCRC) has deeply changed in the last 20 years thanks to the use of either anti-EGFR antibodies or antiangiogenic therapies, mainly anti–VEGF-A antibodies. According to NCCN guidelines (Version 2.2017), the main determinants for the therapeutic choices are the localization of the tumor (right colon, left colon, or rectum), in addition to its molecular features (MSI, CIMP, BRAF mutations, RAS mutational status, SCNA expression) [1]. For example, RAS wild-type left-sided tumors can be treated with anti-EGFR antibodies plus chemotherapy, whereas right-sided tumors, either RAS wild type or mutated, are preferentially treated with chemotherapy, with the eventual addition of anti–VEGF-A antibodies, similarly to all the RAS mutated tumors [1]. To accomplish proper antiangiogenic therapies, different agents with peculiar mechanisms of action have been developed in the last years and tested in several clinical trials [2,3].
The first antiangiogenic factor to be developed was a humanized anti–vascular endothelial growth factor (VEGF)-A monoclonal antibody, bevacizumab (BV) [4,5]. Thanks to numerous clinical trials, the combination of BV with a fluoropyrimidine-based chemotherapy emerged to be an efficient option in both first and second lines [[6], [7], [8], [9], [10], [11], [12]]. Moreover, several randomized studies showed that a prolonged inhibition of angiogenesis beyond the clinical evidence of disease progression could improve mCRC patients' survival. Hence, it is becoming mandatory to identify appropriate biomarkers associated with a positive response to BV, to orientate treatment choice also beyond first line, with the aim to optimize the cost/benefit ratio of antiangiogenic strategies for each mCRC patient [13]. However, in spite of several attempts, no clinically validated appropriate biomarkers were found, and the choice of the second-line treatment is mainly based on physicians' preferences. In fact, although the plasma levels of VEGF-A were first addressed as a putative indicator of response to BV in mCRC, several retrospective analyses failed to confirm these findings [14]. Two single-nucleotide polymorphisms of the VEGF-A gene [1154G.A (rs1570360) and 1405C.G (rs2010963)] were found to be associated with an increase of both overall (OS) and progression-free (PFS) survival [15]. However, their effectiveness to identify patients more suitable to respond to anti–VEGF-A agents still needs to be confirmed in larger trials. The same lack of confirmation in larger clinical settings also occurred for the expression of VEGF-A unrelated biomarkers which were found to be associated with survival outcomes: apolipoprotein E, vitamin D-binding protein, and angiotensinogen [16]. Finally, two different studies indicated that a high serum lactic dehydrogenase pretreatment level behaves as a predictor of efficacy of first-line BV-based therapy [17,18]. However, an Italian Group for the Study of Digestive Tract Cancers (GISCAD) phase II prospective trial failed to confirm the predictive values of pretreatment serum lactic dehydrogenase [19]. Overall, clinically validated biomarkers capable to select patients that would likely respond to BV plus chemotherapy, and could be also suitable to continue BV-based therapies, are still needed [20].
Among the various angiogenesis-related cancer biomarkers, we studied the potassium channel encoded by the human ether-a-go-go-related gene 1 (KCNH2 or hERG1), a protein widely expressed in several types of human cancers, including CRC [[21], [22], [23]]. In particular, hERG1 is the starting hub of a proangiogenic signaling pathway activated by cell adhesion to the extracellular matrix that regulates the expression of both HIF-1α and Hif-2α through the phosphorylation of Akt and the ensuing activation of NFkB [24]. Such pathway leads to VEGF-A secretion and hence sustains angiogenesis and metastatic spread in CRC preclinical models [24]. The relevance of this pathway also emerged by the finding that the combined treatment with hERG1 blockers and BV inhibits both local tumor growth and metastatic spread in preclinical models of CRC [24,25]. Furthermore, hERG1 expression associates with that of the glucose transporter GLUT-1 and the membrane carbonic anhydrase IX (CA-IX) in primary samples from TNM stages I-III CRC patients and can predict disease relapse [26,27].
Based on the above preclinical data, we analyzed the expression of several proangiogenic proteins, directly or indirectly linked to hERG1 channels and their downstream signaling pathway, in surgical samples from mCRC patients and tested their prognostic impact for response to BV-based therapies.
Materials and Methods
Patients
A multicenter retrospective study was conducted on 80 mCRC patients treated in first line with BV plus chemotherapy between June 2010 and June 2017 in three Italian institutions. The selection of patients was performed by medical oncologists of the hospitals involved in the study (Azienda Ospedaliero-Universitaria Careggi, Florence; Campus Bio-Medico University, Rome; Spedali Civili Hospital, Brescia). Afterwards, paraffin-embedded samples were retrieved from the Pathological Anatomy Sections of the above-mentioned institutions: i) 35 samples were collected from archives of the Section of Pathological Anatomy, Department of Experimental and Clinical Medicine, University of Florence, Florence; ii) 36 samples from the Section of Pathological Anatomy, Department of Pathology, Campus Bio-Medico University of Rome; and iii) 9 samples from the Pathology Section of the Spedali Civili Hospital, Brescia. All the patients enrolled in the study were classified as cTNM stage IV; paraffin-embedded samples were classified as pTNM stage IV (i.e., with synchronous metastases defined as developed within 3-12 months from surgery) in 76 samples, while 4 patients were operated for a TNM stage III disease (and therefore developed metachronous metastases after 12 months).
Best response and progression were defined according to RECIST criteria. KRAS mutation analysis was performed by either MALDI-TOF (Sequenom) or pyrosequencing.
The demographic and clinicopathological characteristics and survival as well as KRAS molecular status of the patients enrolled in the study are reported in Supplementary Table S1.
Immunohistochemistry (IHC)
Eighty formalin-fixed, paraffin-embedded mCRC samples belonging to TNM stage IV (76 samples) and III (4 samples) that further progressed to TNM stage IV were analyzed for the expression of the following proteins: hERG1, β1-integrin, pAKT, NFkB, HIF-1α, HIF-2α, p53, VEGF-A, GLUT-1, and CA-IX. IHC was carried out on 7-μm sections on positively charged slides. After dewaxing and rehydrating the sections, endogenous peroxidases were blocked with a 1% H2O2 solution in phosphate-buffered saline (PBS). Subsequently, antigen retrieval was performed with different procedures, depending on the antibody used: 1) by treatment with proteinase K (5 μg/ml) in PBS at 37°C for 5 minutes (for hERG1, VEGF-A, GLUT-1, and CA-IX staining) or 2) by heating the samples in a microwave oven at 600 W in citrate buffer pH 6.0 for 15 (for pAKT, NFkB, HIF-1α, HIF-2α, and p53 staining) or 20 minutes (for β1-integrin staining). The following antibodies were used: anti-hERG1 monoclonal antibody (MCK Therapeutics; 0,005 μg/μl), anti-BETA1 integrin [monoclonal antibody (4B7R) to Integrin beta 1, Abcam, 1:35], anti-pAKT [monoclonal antibody p-Akt1/2/3 (B-5), Santa Cruz Biotechnology, 1:100], anti-NFkB [polyclonal antibody anti-NFkB p65 (A): SC-109, Santa Cruz Biotechnology, 1:100], anti-VEGF-A [polyclonal antibody anti-VEGF-A (A-20), Santa Cruz Biotechnology, 1:100], anti-Glut-1 (polyclonal rabbit anti-human GLUT1 H-43, Santa Cruz Biotechnology, 1:100), anti-HIF-1α [monoclonal antibody HIF-1 alpha Antibody (HA111), Novus Biologicals, 1:100], anti-HIF-2 alpha (polyclonal antibody HIF-2 ALPHA/EPAS; Novus Biologicals, 1:100), anti–CA-IX monoclonal antibody (monoclonal murine antibody M75, described in [28]), and anti-p53 monoclonal antibody (Dako Cytomation, 1:50). Incubation with the primary antibodies was carried out overnight at 4°C, except for the anti-p53 antibody, which was incubated for 30 minutes at room temperature, and for anti–β1-integrin antibody, which was incubated for 2 hours at room temperature. Immunostaining was performed with a commercially available kit (PicTure max kit; Invitrogen) according to the manufacturer's instructions.
Scoring System Assessment
Immunohistochemistry slides were scored by two independent operators (J.I. and E.L.). A specific scoring system was applied for each protein. Previously published scoring systems [26] were applied to hERG1, p53, VEGF-A, GLUT-1, and CA-IX. Samples were considered positive when the percentage of stained cells was higher than the assigned threshold: for hERG1 and β1-integrin, a cutoff of 50% was applied; for p53, VEGF-A, CA-IX, and pAKT, a cutoff of 10% was used; and for GLUT-1, a cutoff of 1% was set. The localization of staining was evaluated for NFkB, HIF-1α, and HIF-2α, and only samples where at least 1% of tumor nuclei was labeled were considered positive. Normal epithelium, stroma, and areas of necrosis were not estimated. The applied scoring system is shown in Supplementary Table S2.
Statistical Analysis
Throughout the manuscript, continuous variables were expressed as mean ± SD, and categorical ones were expressed as absolute and relative frequencies. The correlations between continuous and categorical variables were analyzed with Pearson's correlation coefficient, while for categorical variables, the Pearson χ2 test or the Fisher's exact test was used when appropriate. P value of χ2 test was applied when all frequencies were >5; P value of Fisher's exact test was used when at least one frequency was ≤5. In order to define the differences in either progression of disease rate or survival rate between the levels of markers and levels of combined ones, the univariate Cox's model was adopted, including log-rank test and Kaplan-Meier (KM) curves. Hazard ratios and 95% Wald's confidence intervals were computed. A multivariate Cox's model (backward selection; covariates were retained in the model if the P value was < .05) was computed to define the possible markers, or combination of markers, associated with either the progression of the disease or the death of the patient. A two-sided P value < .05 was considered to be statistically significant.
Results
Clinical Characteristics
The mean age of the patients was 65.8 ± 1.2 years (range 34-79), with the majority of patients belonging to the age group ≥65.8, in agreement with the characteristics of the disease [29]. Of the 80 patients, 30 (37.5%) were females and 50 (62.5%) were males. Thirty-four tumors (43%) were located in the right colon (right colon + transverse), 29 (36%) in the left colon (left colon + sigmoid colon), and 13 (16%) in the rectum. Sixty tumors (75%) were classified as G2, 4 (5%) as G1, and 9 (11%) as G3. Twenty-six (33%) had wild-type KRAS, and 47 (59%) had KRAS mutations. Forty-five out of 80 (56%) patients had disease progression, and 50/80 (62%) died during the time of follow-up. In accordance to RECIST criteria, 9 (11%) patients had a complete response, 18 (23%) presented a partial response, 31 (39%) had stable disease, and 16 (20%) underwent progression. The distribution of the demographic and clinicopathological characteristics and survival of the cohort are shown in Supplementary Table S1.
Immunohistological Findings
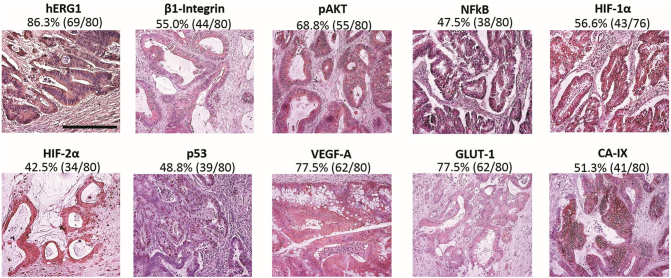
The expression of the following proteins was determined by IHC on paraffin-embedded samples: hERG1, β1-Integrin, pAKT, NFkB, HIF-1α, HIF-2α, p53, VEGF-A, GLUT-1, and CA-IX. Representative pictures of each protein, along with the percentage of patients positive to the biomarker (according to the scoring system described in Materials and Methods and summarized in Supplementary Table S2), are shown in Figure 1. hERG1 staining was detected mainly on the plasma membrane, with a weak staining also at the cytoplasmic level, due to the presence of immature forms of hERG1 in the cytoplasm as previously described [30]. β1-Integrin was detected in the cytoplasm of tumor cells because the antibody recognizes a β1-integrin specific intracellular epitope, although the main localization of the protein is on the plasma membrane. pAKT and VEGF-A were expressed in the cytoplasm of neoplastic cells, as expected. NFkB, HIF-1α, and HIF-2α staining could be detected either in the cytoplasm or in the nuclei of tumor cells, but only samples displaying a nuclear staining were considered positive since the nuclear localization mirrors an “active” protein (aHIF-1α and aHIF-2α, respectively). p53 labeled tumor nuclei. GLUT-1 stained plasma membrane and cytoplasm and CA-IX were expressed only on the plasma membrane of neoplastic cells. All the samples were scored as described in Materials and Methods and summarized in Supplementary Table S2.
Figure 1.
Representative images of samples expressing the markers of interest. IHC experiments and scoring assessment were performed as described in Materials and Methods and Supplementary Table S2. Bar: 100 μm. The marker under investigation, percentages, and absolute values of samples with a positive expression or with expression of the active form of the marker are reported on the top of each images.
Relationships Between Biological Markers and Clinical Characteristics
A hERG1 positive signal was observed in 86.3% (69/80) of patients. β1-Integrin was detected in 55% (44/80) of samples, pAKT in 69% (55/80), NFkB in 48% (38/80), aHIF-1α in 56.6% (43/76), aHIF-2α in 42% (34/80), p53 in 49% (39/80), VEGF-A in 78% (62/80), GLUT-1 in 78% (62/80), and CA-IX in 51% (41/80) (Figure 1). Patients expressing hERG1, β1-integrin, and VEGF-A were 77% (34/80) also positive for either aHIF-1α or aHIF-2α, or both. Thirty of 80 samples were positive for hERG1 although negative for β1-integrin, and 73% of them were positive for VEGF-A. Overall, the pathway linking hERG1 to VEGF-A is apparently operative in mCRC primary samples (Supplementary Figure S1). Indeed, only four patients were negative for both hERG1 and VEGF-A.
Statistically significant associations emerged between aHIF-1α and gender (P = .020; aHIF-1α and male gender) and between aHIF-2α and tumor localization (P = .004; HIF-2α active form and left colon) (Table 1).
Table 1.
Statistically Significant Associations Between Biological Markers and Clinical Characteristics
| Parameter/Marker | Marker | P Value | ||
|---|---|---|---|---|
| Gender | iHIF-1α | aHIF-1α |
.020* |
|
| Female | 17 (22%) | 11 (15%) | ||
| Male | 16 (21%) | 32 (42%) | ||
| Localization | iHIF-2α | aHIF-2α | .004* | |
| Right colon | 25 (54%) | 12 (36%) | ||
| Left colon | 10 (22%) | 19 (58%) | ||
| Rectum | 11 (24%) | 2 (6%) | ||
| VEGF-A | hERG1 − | hERG1 + |
.049^ |
|
| Negative | 5 (6%) | 13 (16%) | ||
| Positive | 6 (8%) | 56 (70%) | ||
| CA-IX − | CA-IX + |
.024^ |
||
| Negative | 13 (16%) | 5 (6%) | ||
| Positive | 26 (32%) | 36 (45%) | ||
| HIF-2α | β1-integrin − | β1-Integrin + |
.016* |
|
| Inactive | 26 (33%) | 20 (25%) | ||
| Active | 10 (13%) | 24 (30%) | ||
| GLUT-1 | iHIF-1α | aHIF-1α |
.045* |
|
| Negative | 11 (15%) | 6 (12%) | ||
| Positive | 22 (25%) | 37 (49%) | ||
| iHIF-2α | aHIF-2α |
.003^ |
||
| Negative | 16 (20%) | 2 (3%) | ||
| Positive | 30 (38%) | 32 (40%) | ||
Absolute values and percentages (in parentheses) are indicated. P value of χ2 test (indicated with *) is reported when all frequencies are >5; P value of Fisher's exact test (indicated with ^) is reported when at least one frequency is ≤5. “a”: samples with active protein; “I”: samples with inactive protein.
The following statistically significant associations between molecular markers emerged: hERG1 and VEGF-A (P = .049; positive hERG1 and positive VEGF-A), β1-integrin and HIF-2α (P = .016; positive β1-integrin and aHIF-2α), GLUT-1 and HIF-1α (P = .045; positive GLUT-1 and aHIF-1α), GLUT-1 and HIF-2α (P = .003; positive GLUT-1 and aHIF-2α), and CA IX and VEGF-A (P = .024, positive CA-IX and positive VEGF-A) (Table 1).
Impact on Survival
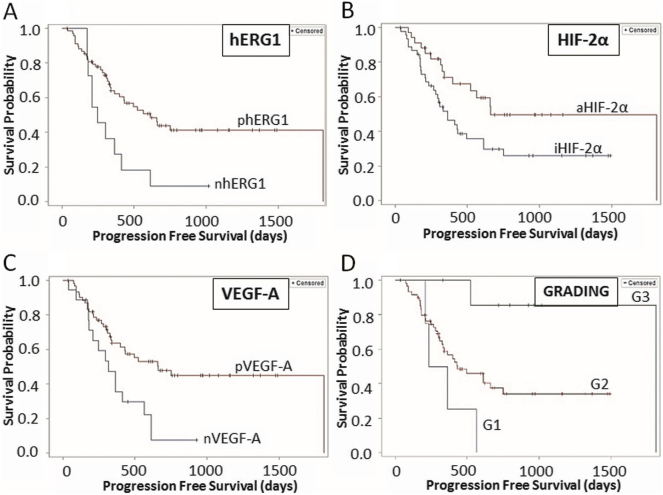
The cohort under study had a median PFS of 336 ± 43 days and a median OS of 804 ± 39 days. The univariate analysis showed that hERG1, aHIF-2α, and VEGF-A had a significant impact on PFS, with hazard ratio (HR) values <1, indicating their role as protective factors (Table 2). The KM curves relative to the three biomarkers (Figure 2) clearly show that patients positive for any of the three markers have a longer PFS compared to negative patients.
Table 2.
Univariate PFS Analysis
| HR (95% CI) | P (LR Test) | |
|---|---|---|
| hERG1 | ||
| Negative | 1 (Ref) | .015 |
| Positive | 0.41 (0.20-0.84) | |
| HIF-2α | ||
| Inactive | 1 (Ref) | .027 |
| Active | 0.49 (0.26-0.92) | |
| GLUT-1 | ||
| Negative | 1 (Ref) | .402 |
| Positive | 0.75 (0.39-1.46) | |
| KRAS status | ||
| Wild type | 1 (Ref) | .601 |
| Mutated | 0.84 (0.44-1.60) | |
| NFkB | ||
| Inactive | 1 (Ref) | .168 |
| Active | 0.65 (0.35-1.20) | |
| β1-Integrin | ||
| Negative | 1 (Ref) | .427 |
| Positive | 0.79 (0.43-1.42) | |
| HIF-1α | ||
| Inactive | 1 (Ref) | .683 |
| Active | 0.88 (0.48-1.62) | |
| pAKT | ||
| Negative | 1 (Ref) | .422 |
| Positive | 0.77 (0.41-1.45) | |
| VEGF-A | ||
| Negative | 1 (Ref) | .008 |
| Positive | 0.42 (0.22-0.80) | |
| p53 | ||
| Negative | 1 (Ref) | .688 |
| Positive | 1.13 (0.62-2.04) | |
| CA-IX | ||
| Negative | 1 (Ref) | .417 |
| Positive | 0.78 (0.43-1.42) | |
| Gender | ||
| Female | 1 (Ref) | .322 |
| Male | 1.37 (0.73-2.56) | |
| Grading | ||
| G3 | 1 (Ref) | .013 |
| G1 | 16.44 (1.81-149.10) | |
| G2 | 7.65 (1.04-56.07) | |
| Localization | ||
| Rectum | 1 (Ref) | 1.000 |
| Right colon | 0.78 (0.33-1.84) | |
| Left colon | 1.06 (0.32-3.54) |
Univariate analysis was performed using Cox's model, including log-rank test and KM curves. HRs and 95% Wald's confidence intervals were computed as described in Materials and Methods section. P values of log-rank test, HRs, and 95% Wald's confidence intervals are reported. Median PFS: 336 ± 43 days. Significant P values are in bold and underlined.
Figure 2.
KM curves of PFS. (A) hERG1 (negative: 10, positive: 35); (B) HIF-2α (inactive: 30, active: 15); (C) VEGF-A (negative: 15, positive: 30); (D) grading (G1: 4, G2: 34, G3: 2). Only statistically significant curves are reported. “n”: negative samples; “p”: positive samples; “a”: samples with active protein; “I”: samples with inactive protein.
Among the clinicopathological features (see Supplementary Table S1), only the tumor grading had a significant impact on PFS, so that patients with a less differentiated (G3) primary tumor had a better response to BV (Table 2; see also the KM curve in Figure 2).
The multivariate analysis, using hERG1, aHIF-2α, VEGF-A ,and all the clinicopathological features, showed that hERG1 positivity and the presence of an active form of HIF-2α significantly impacted on PFS. In particular, both hERG1 positivity (P = .012; HR: 0.362; 95% Wald confidence limits: 0.163-0.803) and aHIF-2α (P = .027; HR: 0.400; 95% Wald confidence limits: 0.178-0.902) correlated with a longer PFS, hence behaving as biomarkers of a positive response to treatment with BV (Table S3). To confirm the impact of the two biomarkers (hERG1 and aHIF-2α), we performed another multivariate PFS analysis with all the proangiogenic markers and the KRAS status. From this analysis, only hERG1 positivity showed a statistically significant impact. Also, in this case, hERG1 behaved as a protective factor for treatment with BV (P = .002; HR: 0.302; 95% Wald confidence limits: 0.139-0.655) (Supplementary Table S4).
None of the tested markers and clinicopathological characteristics had an impact on OS (median OS 804 ± 39 days), and no statistically significant association was found between best response (defined according to RECIST Criteria) and the expression of any of the proteins tested and clinicopathological features.
Patients were hence stratified into four groups on the basis of the positivity or negativity for any biological marker including KRAS mutational status, and a risk analysis was performed. Statistically significant differences in PFS emerged for the combinations hERG1-pAKT, hERG1-NFkB, hERG1–aHIF-1α, hERG1–aHIF-2α, hERG1–VEGF-A, and hERG1-KRAS (Supplementary Table S3). In particular, 1) in the combinations hERG1-pAKT, hERG1-NFkB and hERG1–HIF-1α, patients expressing hERG1 had a longer PFS than hERG1-negative subjects, regardless of the other marker (Supplementary Figure 2S); 2) in the combinations hERG1–aHIF-2α and hERG1–VEGF-A, patients positive for either biomarker(s) had a longer PFS compared to patients negative for both proteins; 3) in the combination KRAS-hERG1, patients positive for hERG1, either with a mutated or wild-type KRAS, had a longer PFS compared to patients that do not express hERG1 (Figure 3, A, B, and C).
Figure 3.
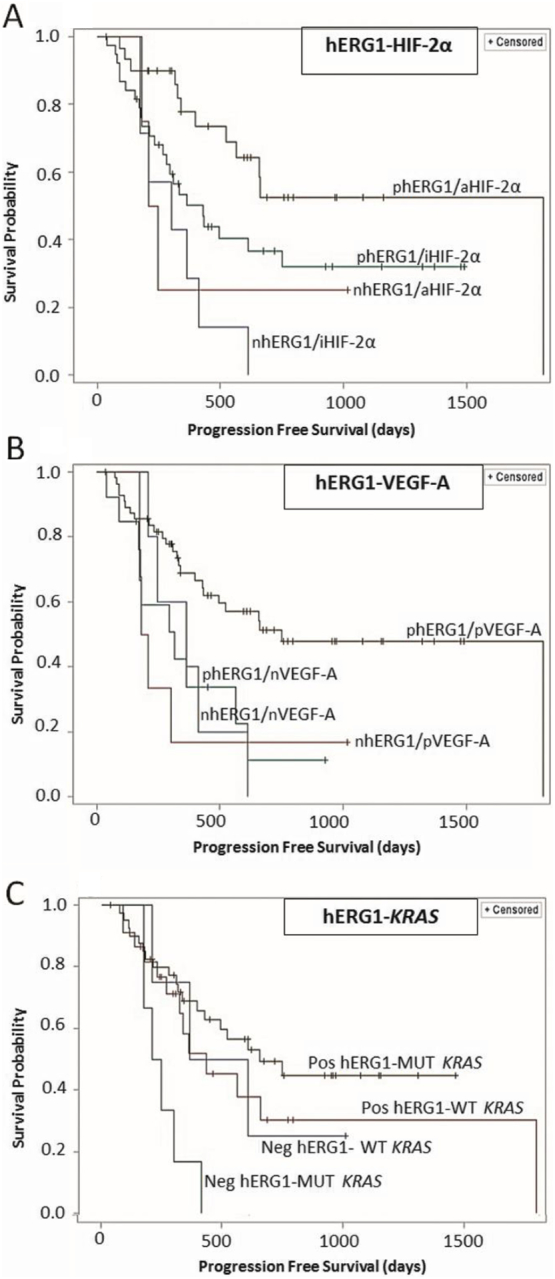
KM curves of PFS obtained from the risk analysis. (A) Combination of hERG1 and HIF-2α. Combinations are defined as follows: 00: hERG1 negative–HIF-2α inactive; 01: hERG1 negative–HIF-2α active; 10: hERG1 positive–HIF-2α inactive; 11: both hERG1 and HIF-2α active. hERG1 and HIF-2α (00: 7, 11: 30). (B) Combination of hERG1–VEGF-A. 00: hERG1 negative–VEGF-A negative; 01: hERG1 negative–VEGF-A positive; 10: hERG1 positive–VEGFA negative; 11: both hERG1 and VEGF-A positive. hERG1 and VEGF-A (00: 5, 11: 56). (C) Combination of hERG1 and KRAS status. For KRAS status, 01 indicates wild-type KRAS and hERG1 positivity; 10 identifies mutated KRAS samples negative for hERG1 expression; samples harboring KRAS mutations and positive for hERG1 are indicated with 11. Number of patients: 00 (3), 01 (12), 10 (7), and 11 (19). Only statistically significant curves are reported. “n”: negative samples; “p”: positive samples; “a”: samples with active protein; “I”: samples with inactive protein.
Discussion
The identification of biomarkers correlated with a positive response to BV plus chemotherapy in mCRC patients is still a big challenge. Appropriate biomarker(s) would help to select those patients eligible for BV treatment in first line and its prolongation in second line. We got strong preclinical data showing the existence, in CRC cells, of a proangiogenic signaling pathway centered on the expression and activity of hERG1 potassium channels [26]. Based on these data, we performed a multicenter retrospective study in which several angiogenesis-related proteins linked to such pathway were analyzed in samples belonging to mCRC patients treated in first line with BV plus chemotherapy. The main results of our study were as follows: 1) all the members of the signaling pathway downstream to hERG1 channels are expressed in CRC primary samples, showing different associations among them; 2) both hERG1 and an active form of HIF-2α are prognostic biomarkers of positive response to BV plus chemotherapy, as emerging from univariate and multivariate PFS analyses; and 3) the impact of hERG1 on the positive response to BV plus chemotherapy occurs irrespective of the KRAS mutational status of the tumor sample.
The different biomarkers belonging to the signaling pathway previously identified to drive proangiogenic and prometastatic behaviors in CRC cell lines turned out to be expressed with statistically significant associations (Table 1 and Figure S1) in mCRC human primary samples. This finding indicates that such pathway is operative also in vivo. Some specific associations merit attention. For instance, the associations between VEGF-A and both hERG1 and CA-IX not only confirm what was previously detected in stage I-III CRC [24] but further stress the relevance of the functional network between hERG1, angiogenesis, and pH-regulating mechanisms. Such network is emerging to drive tumor malignancy in several cancer types including CRC [[24], [25], [26], [27],[31], [32]]. A strong correlation between β1-integrin and HIF-2α and between GLUT-1 and both HIF-1α and HIF-2α also emerged. The former association agrees with the finding that hypoxia and HIF (s) induce β1-integrin expression and activation [33]. The association between GLUT-1 and HIF-1α is expected since the transcription of the GLUT-1 gene is known to be under the control of HIF-1α [34]. It is surprising that we did not find an association between GLUT-1 and hERG1 as previously found in TNM stage I-III CRC patients, where the two proteins are inversely associated and a low GLUT-1 with high hERG1 expression identifies a patient subgroup with a worse prognosis [24]. We proposed a model in which GLUT-1 expression is high until hypoxia is present within the tumor mass, to decline after reestablishment of normoxia, thanks to hERG1 overexpression and the ensuing hERG1-triggered angiogenesis [24]. On the contrary, in the cohort analyzed in the present paper (i.e., only TNM stage IV, metastatic samples, treated in first line with BV), GLUT-1 expression is higher than in TNM stage I-III patients (77.5% vs. 34.8%), as expected [35]. Furthermore, the association between hERG1 and GLUT-1 is lost, suggesting that metastatic lesions gather an independent angiogenic state, in which the balance and interplay of the two markers are no longer present, and both reach high expression levels. aHIF-2α association with tumor localization (much HIF-2α active forms in left-sided tumors) is conceivably related to the different biomolecular features of right- and left-sided lesions [36]. For example, HIF-2α is known to regulate the expression of the EGFR gene, which shows higher levels in the tumors of the left colon [37]. Furthermore, our data showing the relevance of HIF-2α give further support to the notion that, in colorectal carcinomas, HIF-2α plays a key role in the angiogenesis, greater than HIF-1α [[37], [38], [39]].Overall, the expression and correlation data we obtained support, in the human clinical setting, the conclusion we derived from model cells in vitro, i.e., the relevance of hERG1 in driving angiogenesis and the metastatic process in CRC, and its potential therapeutic translatability.
Furthermore, such data give a mechanistic explanation to the main results of the present paper, i.e., the positive impact on response to BV of some components of the above signaling pathway, in particular hERG1 and the active form of HIF-2α. The impact of the different proangiogenic biomarkers on patients' outcome was analyzed evaluating different clinical parameters of response (PFS, OS, and best response), but a statistically relevant association emerged only when evaluating the PFS. This is not a surprise since several studies reported PFS as an appropriate surrogate of OS and thus a good indicator for response to therapy [40].
At the univariate analysis, three proteins, hERG1, HIF-2α, and VEGF-A, showed a significant impact, with patients positive for any of the three markers having a longer PFS. Other biomarkers, such as CA-IX or GLUT-1, shown to have a negative or positive impact, respectively, on OS in TNM stages I-III cases [24] lose their relevance on the response to antiangiogenesis treatment. Also, tumor localization had no impact on PFS besides its correlation with aHIF-2α and its relevance in therapeutic choice. While VEGF-A had no impact in TNM stages I-III, its expression positively impacted on PFS of stage IV patients treated with BV. This fact disagrees with what was reported by Hedge et al. [14], who showed that baseline levels of VEGF-A have a negative impact (HR >1) on survival of mCRC patients treated with BV. Such discrepancy may suggest different roles of the intracellular and extracellular (serum) VEGF-A in driving the response to BV treatment.
Different from VEGF-A, both hERG1 and aHIF-2α maintained their significant, positive impact on PFS at the multivariate analysis, hence configuring as independent prognostic factors of positive response to BV. The impact of hERG1 and aHIF-2α emerged also when analyzing the combined expression of the different biomarkers. Notably, mCRC patients whose lesions were positive for both HIF-2α and hERG1 had a lower risk of progression during BV plus chemotherapy treatment. From these results, it is tempting to speculate that hERG1 is upstream to HIF-2α and of the whole proangiogenic signaling pathway, thus confirming what was previously shown in CRC cells. In the latter model, in fact, hERG1 activity positively regulates HIF-2α expression and in turn VEGF-A secretion [26]. This implies that hERG1-positive patients with more aHIF-2α and greater VEGF-A secretion would benefit from the block of VEGF-A through treatment with BV, as shown by the survival analysis reported.
The relevance of hERG1 and its downstream pathway also emerges from the finding that hERG1-positive patients with KRAS mutations have a longer PFS than patients harboring the same mutation but negative for hERG1. This result might have a clinical relevance because the evaluation of KRAS mutational status is the only approved predictive biomarker to define the treatment options for mCRC patients [41].
Conclusions
Overall, the results provided in this paper suggest the use of hERG1 and aHIF-2α immunoreactivity to select patients suitable to be treated with BV, in addition to chemotherapy, both in first and second line, irrespective of the KRAS status or localization of the tumor. In other words, the simultaneous presence of both proteins behaves as a prognostic biomarker and, once validated in an appropriate prospective clinical study, could be proposed as a predictive factor for response to therapy with antiangiogenic agents such as BV.
Funding
This work was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC, grant no. 1662) to A. A. to perform the experiments reported. The sponsor had no involvement in study design; in the collection, analysis, and interpretation of data; in the writing of the manuscript; and in the decision to submit the manuscript for publication.
Additional Information
Ethics Approval and Consent to Participate
The study protocol was approved by the Ethical Committee of AOUC (BIO.14.033). All the patients were enrolled after informed written consent. The study was performed in accordance with the Declaration of Helsinki.
Availability of Data and Material
The datasets created and analyzed during the current study are available from the corresponding author on reasonable request.
Declaration of Interest
None.
Authors' Contribution
J. I. performed the IHC experiments, evaluated the slides, analyzed the data, and wrote the manuscript; E. L. performed the IHC experiments, evaluated the slides, analyzed the data, and wrote the manuscript; L. T. performed statistical analyses; G. Petroni performed the IHC experiments; L. A. treated and followed up the patients and retrieved clinical data; L. M. performed the pathological evaluation of the samples and retrieved the paraffin-embedded samples; G. Perrone performed the pathological evaluation of the samples and retrieved the paraffin-embedded samples; D. C. operated the patients and identified the patients to be enrolled; M. F. retrieved histopathological data; M. M. A. retrieved clinical data; M. C. retrieved histopathological data; A. G. retrieved clinical data and identified the patients to be enrolled; V. V. performed the pathological evaluation of the samples and retrieved the paraffin-embedded samples; L. B. supervised the statistical analyses; R. C. operated the patients; F. D. C. supervised the study and revised the manuscript; A. A. conceived the study, analyzed the data, wrote the manuscript, and supervised the study.
All the authors read and approved the final version of the manuscript.
Footnotes
Declaration of Interest: None
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2020.01.001.
Appendix A. Supplementary data
Supplementary information
References
- 1.Wells A, Messersmith MD. Systemic management of colorectal cancer. J Natl Compr Canc Netw. 2017;15:699–702. doi: 10.6004/jnccn.2017.0077. [DOI] [PubMed] [Google Scholar]
- 2.Sobrero A, Bruzzi P. Incremental advance or seismic shift? The need to raise the bar of efficacy for drug approval. J Clin Oncol. 2009;27(35):5868–5873. doi: 10.1200/JCO.2009.22.4162. [DOI] [PubMed] [Google Scholar]
- 3.Wagner AD, Arnold D, Grothey AA, Haerting J. Unverzagt S. Anti-angiogenic therapies for metastatic colorectal cancer. Cochrane Database Syst Rev. 2009;3 doi: 10.1002/14651858.CD005392.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrara N, Hillan KJ, Gerber HP, Novotny W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat Rev Drug Discov. 2004;3(5):391–400. doi: 10.1038/nrd1381. [DOI] [PubMed] [Google Scholar]
- 5.Mulcahy MF, Benson AB., 3rd Bevacizumab in the treatment of colorectal cancer. Expert Opin Biol Ther. 2005;5(7):997–1005. doi: 10.1517/14712598.5.7.997. [DOI] [PubMed] [Google Scholar]
- 6.Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J. HeimW, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 7.Giantonio BJ, Catalano PJ, Meropol NJ, O'Dwyer PJ, Mitchell EP, Alberts SR. Bevacizumab in combination with oxaliplatin, fluorouracil, and leucovorin (FOLFOX4) for previously treated metastatic colorectal cancer: results from the Eastern Cooperative Oncology Group Study E3200. J Clin Oncol. 2007;25(12):1539–1544. doi: 10.1200/JCO.2006.09.6305. [DOI] [PubMed] [Google Scholar]
- 8.Saltz LB, Clarke S, Dı ´az-Rubio E, Scheithauer W, Figer A, Wong R, et al. Bevacizumab in combination with oxaliplatin-based chemotherapy as first-line therapy in metastatic colorectal cancer: a randomized phase III study. J Clin Oncol 2008; 26: 2013–2019Erratum in J Clin Oncol (2009) 27:653. [DOI] [PubMed]
- 9.Loupakis F, Bria E, Vaccaro V, Cuppone F, Milella M, Carlini P. Magnitude of benefit of the addition of bevacizumab to first-line chemotherapy for metastatic colorectal cancer: meta-analysis of randomized clinical trials. J Exp Clin Cancer Res. 2010;29(1):58. doi: 10.1186/1756-9966-29-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennouna J, Sastre J, Arnold D, Österlund P, Greil R, Van Cutsem E. Continuation of bevacizumab after first progression in metastatic colorectal cancer (ML18147): a randomised phase 3 trial. Lancet Oncol. 2013;14(1):29–37. doi: 10.1016/S1470-2045(12)70477-1. [DOI] [PubMed] [Google Scholar]
- 11.Loupakis F, Cremolini C, Masi G, Lonardi S, Zagonel V, Salvatore L. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N Engl J Med. 2014;371(17):1609–1618. doi: 10.1056/NEJMoa1403108. [DOI] [PubMed] [Google Scholar]
- 12.Masi G, Salvatore L, Boni L, Loupakis F, Cremolini C, Fornaro L. BEBYP Study Investigators. Continuation or reintroduction of bevacizumab beyond progression to first-line therapy in metastatic colorectal cancer: final results of the randomized BEBYP trial. Ann Oncol. 2015;26(4):724–730. doi: 10.1093/annonc/mdv012. [DOI] [PubMed] [Google Scholar]
- 13.Mousa L, Salem ME, Mikhail S. Biomarkers of angiogenesis in colorectal cancer. Biomark Cancer. 2015;7(Suppl 1):13–19. doi: 10.4137/BIC.S25250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegde PS, Jubb AM, Chen D, Li NF, Meng YG, Bernaards C. Predictive impact of circulating vascular endothelial growth factor in four Phase III trials evaluating bevacizumab. Clin Cancer Res. 2013;19:929–937. doi: 10.1158/1078-0432.CCR-12-2535. [DOI] [PubMed] [Google Scholar]
- 15.Formica V, Palmirotta R, Del Monte G, Savonarola A, Ludovici G, De Marchis ML. Predictive value of VEGF gene polymorphisms for metastatic colorectal cancer patients receiving first-line treatment including fluorouracil, irinotecan. and bevacizumab Int J Colorectal Dis. 2011;26:143–151. doi: 10.1007/s00384-010-1108-1. [DOI] [PubMed] [Google Scholar]
- 16.Martin P, Mullen MP, Scaife C, Tosetto M, Nolan B, Wynne K. Predicting response to vascular endothelial growth factor inhibitor and chemotherapy in metastatic colorectal cancer. BMC Cancer. 2014;14:887. doi: 10.1186/1471-2407-14-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scartozzi M, Giampieri R, Maccaroni E, Del Prete M, Faloppi L, Bianconi M. Pretreatment lactate dehydrogenase levels as predictor of efficacy of first-line bevacizumab-based therapy in metastatic colorectal cancer patients. Br J Cancer. 2012;106(5):799–804. doi: 10.1038/bjc.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marmorino F, Salvatore L, Barbara C, Allegrini G, Antonuzzo L, Masi G. Serum LDH predicts benefit from bevacizumab beyond progression in metastatic colorectal cancer. Br J Cancer. 2017;116:318–323. doi: 10.1038/bjc.2016.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giampieri R, Puzzoni M, Daniele B, Ferrari D, Lonardi S, Zaniboni A. First-line FOLFIRI and bevacizumab in patients with advanced colorectal cancer prospectively stratified according to serum LDH: final results of the GISCAD (Italian Group for the Study of Digestive Tract Cancers) CENTRAL (ColorEctalavastiNTRiAlLdh) trial. Br J Cancer. 2017;117:1099–1104. doi: 10.1038/bjc.2017.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duda DG, Munn L. Jain R. Can we identify predictive biomarkers for anti-angiogenic therapy of cancer using mathematical modeling. J Natl Cancer Inst. 2013;762–765(36):105. doi: 10.1093/jnci/djt114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arcangeli A, Crociani O, Lastraioli E, Masi A, Pillozzi S, Becchetti A. Targeting ion channels in cancer: a novel frontier in antineoplastic therapy. Curr Med Chem. 2009;16(1):66–93. doi: 10.2174/092986709787002835. [DOI] [PubMed] [Google Scholar]
- 22.Jehel J, Schweizer PA, Katus HA, Thomas D. Novel roles for hERG K(+) channels in cell proliferation and apoptosis. Cell Death Dis. 2011;2:e193. doi: 10.1038/cddis.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lastraioli E, Lottini T, Bencini L, Bernini M. Arcangeli A. hERG1 potassium channels: novel biomarkers in human solid cancers. Biomed Res Int. 2015;2015:896432. doi: 10.1155/2015/896432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crociani O, Zanieri F, Pillozzi S, Lastraioli E, Stefanini M, Fiore A. hERG1 channels modulate integrin signaling to trigger angiogenesis and tumor progression in colorectal cancer. Sci Rep. 2013;3:3308. doi: 10.1038/srep03308. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Crociani O, Lastraioli E, Boni L, Pillozzi S, Romoli MR, D'Amico M. hERG1 channels regulate VEGF-A secretion in human gastric cancer: clinicopathological correlations and therapeutical implications. Clin Cancer Res. 2014;20(6):1502–1512. doi: 10.1158/1078-0432.CCR-13-2633. [DOI] [PubMed] [Google Scholar]
- 26.Lastraioli E, Bencini L, Bianchini E, Romoli MR, Crociani O, Giommoni E. hERG1 channels and Glut-1 as independent prognostic indicators of worse outcome in stage I and II colorectal cancer: a pilot study. Transl Oncol. 2012;5:105–112. doi: 10.1593/tlo.11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muratori L, Petroni G, Antonuzzo L, Boni L, Iorio J, Lastraioli E. hERG1 positivity and Glut-1 negativity identifies high-risk TNM stage I and II colorectal cancer patients. regardless of adjuvant chemotherapy. Onco Targets Ther. 2016;9:6325–6332. doi: 10.2147/OTT.S114090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pastoreková S, Parkkila S, Parkkila AK, Opavský R, Zelník V, Saarnio J. Carbonic anhydrase IX, MN/CA IX: analysis of stomach complementary. DNA sequence and expression in human and rat alimentary tracts. Gastroenterology. 1997;112(2):398–408. doi: 10.1053/gast.1997.v112.pm9024293. [DOI] [PubMed] [Google Scholar]
- 29.Steele SR, Park GE, Johnson EK, Martin MJ, Stojadinovic A, Maykel JA, Causey MW. The impact of age on colorectal cancer incidence, treatment, and outcomes in an equal-access health care system. Dis Colon Rectum. 2014 Mar;57(3):303–310. doi: 10.1097/DCR.0b013e3182a586e7. [DOI] [PubMed] [Google Scholar]
- 30.Guasti L, Crociani O, Redaelli E, Pillozzi S, Polvani S, Masselli M. Identification of a posttranslational mechanism for the regulation of hERG1 K+ channel expression and hERG1 current density in tumor cells. Mol Cell Biol. 2008;28:5043–5060. doi: 10.1128/MCB.00304-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lastraioli E, Guasti L, Crociani O, Polvani S, Hofmann G, Witchel H. herg1 gene and HERG1 protein are overexpressed in colorectal cancers and regulate cell invasion of tumor cells. Cancer Res. 2004;64(2):606–611. doi: 10.1158/0008-5472.can-03-2360. [DOI] [PubMed] [Google Scholar]
- 32.Iorio J, Petroni G, D'Amico M, Lastraioli E, Arcangeli A. The cross talk between KCa3.1 potassium channel and NHE1 Na+/H+ exchanger regulates adhesion-dependent pH fluctuations in CRC cells. 5th Annual Meeting of the International Society of Cancer. Metabolism. 2018:113–114. [Google Scholar]
- 33.Keely S, Glover LE, MacManus CF, Campbell EL, Scully MM, Furuta GT. Selective induction of integrin beta1 by hypoxia-inducible factor: implications for wound healing. FASEB J. 2009;23(5):1338–1346. doi: 10.1096/fj.08-125344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Semenza G. Signal transduction to hypoxia-inducible factor 1. Biochem Pharmacol. 2002;64(5–6):993–998. doi: 10.1016/s0006-2952(02)01168-1. [DOI] [PubMed] [Google Scholar]
- 35.Furudoi A, Tanaka S, Haruma K, Yoshihara M, Sumii K, Kajiyama G. Clinical significance of human erythrocyte glucose transporter 1 expression at the deepest invasive site of advanced colorectal carcinoma. Oncology. 2001;60:162–169. doi: 10.1159/000055314. [DOI] [PubMed] [Google Scholar]
- 36.Lee MS, Menter DG, Kopetz S. Right versus left colon cancer biology: integrating the consensus molecular subtypes. J Natl Compr Canc Netw. 2017;15:411–419. doi: 10.6004/jnccn.2017.0038. [DOI] [PubMed] [Google Scholar]
- 37.Zhao J, Du F, Shen G, Zheng F, Xu B. The role of hypoxia-inducible factor-2 in digestive system cancers. Cell Death Dis. 2015 Jan;6(1) doi: 10.1038/cddis.2014.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshimura H, Dhar DK, Kohno H, Kubota H, Fujii T, Ueda S. Prognostic impact of hypoxia-inducible factors 1alpha and 2alpha in colorectal cancer patients: correlation with tumor angiogenesis and cyclooxygenase-2 expression. Clin Cancer Res. 2004;10(24):8554–8560. doi: 10.1158/1078-0432.CCR-0946-03. [DOI] [PubMed] [Google Scholar]
- 39.Imamura T, Kikuchi H, Herraiz MT, Park DY, Mizukami Y, Mino-Kenduson M. HIF-1α and HIF-2α have divergent roles in colon cancer. Int J Cancer. 2009;124:763–771. doi: 10.1002/ijc.24032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang PA, Bentzen SM, Chen EX, Siu LL. Surrogate end points for median overall survival in metastatic colorectal cancer: literature-based analysis from 39 randomized controlled trials of first-line chemotherapy. J Clin Oncol. 2007 Oct 10;25(29):4562–4568. doi: 10.1200/JCO.2006.08.1935. [DOI] [PubMed] [Google Scholar]
- 41.Holch J, Stintzing S, Heinemann V. Treatment of metastatic colorectal cancer: standard of care and future perspectives. Visc Med. 2016;32(3):178–183. doi: 10.1159/000446052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information
Data Availability Statement
The datasets created and analyzed during the current study are available from the corresponding author on reasonable request.