Abstract
Background & Aims
The epithelia of the intestine and colon turn over rapidly and are maintained by adult stem cells at the base of crypts. Although the small intestine and colon have distinct, well-characterized physiological functions, it remains unclear if there are fundamental regional differences in stem cell behavior or region-dependent degenerative changes during aging. Mesenchyme-free organoids provide useful tools for investigating intestinal stem cell biology in vitro and have started to be used for investigating age-related changes in stem cell function. However, it is unknown whether organoids maintain hallmarks of age in the absence of an aging niche. We tested whether stem cell–enriched organoids preserved the DNA methylation-based aging profiles associated with the tissues and crypts from which they were derived.
Methods
To address this, we used standard human methylation arrays and the human epigenetic clock as a biomarker of age to analyze in vitro–derived, 3-dimensional, stem cell–enriched intestinal organoids.
Results
We found that human stem cell–enriched organoids maintained segmental differences in methylation patterns and that age, as measured by the epigenetic clock, also was maintained in vitro. Surprisingly, we found that stem cell–enriched organoids derived from the small intestine showed striking epigenetic age reduction relative to organoids derived from colon.
Conclusions
Our data validate the use of organoids as a model for studying human intestinal aging and introduce methods that can be used when modeling aging or age-onset diseases in vitro.
Keywords: Intestinal Stem Cells, Organoids, Epigenetic Clock, Aging, DNA Methylation
Abbreviations used in this paper: DNAm, age based on DNA methylation; FDR, false discovery rate; ISC, intestinal stem cell; PBS, phosphate-buffered saline; PCA, principal component analysis; SI, small intestine; TPCL, Translational Pathology Core Laboratory
Graphical abstract
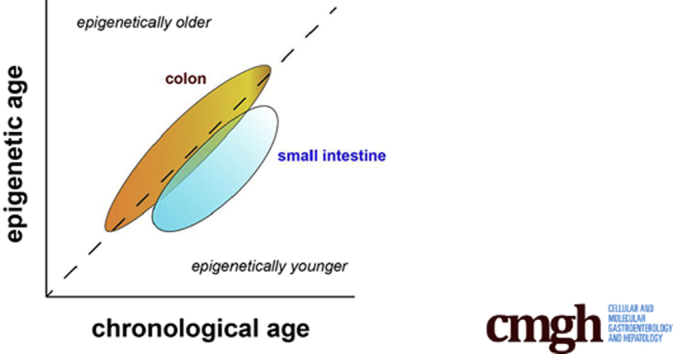
Summary.
We found that human intestinal organoids maintain the age of the patient from whom they are derived, as measured by the epigenetic clock. Unexpectedly, we found that crypts and spheroids derived from small intestine showed striking epigenetic age reduction, relative to the colon.
Maintenance of the gut epithelium throughout life relies on tissue-specific stem cells that reside at the base of invaginating crypts in both the small intestine (SI) and colon. These intestinal stem cells (ISCs), marked by the Wnt target gene Lgr5, rapidly divide to maintain the stem cell population or differentiate to generate specialized absorptive and secretory epithelial cells.1
Although the SI and colon have distinct, well-characterized digestive functions, it remains unclear to what extent there are regional differences in stem cell behavior, especially during aging. The majority of data on stem cells in the mammalian digestive tract are derived from studying the mouse SI; however, a major caveat to using mice to investigate intestinal physiology relevant to human beings is species-specific differences in regional biology.2 It is imperative that we use reliable in vitro models to study human gut stem cell behavior because human in vivo analyses generally are impractical. Three-dimensional organoids, self-organizing cultures of ISCs, and their progeny have transformed our ability to study human intestinal physiology and stem cell function.3, 4, 5, 6 Recent evidence has suggested that organoids derived from human colon and terminal ileum maintain region-specific DNA methylation patterns and regional gene expression.7 In addition, mouse colon organoids aged in vitro show global methylation changes similar to those seen in the aging mouse colon.8 However, although there is potential for organoids to be used as models for aging,9 the degree to which organoids faithfully recapitulate aging has yet to be determined.
Within the human gastrointestinal tract, aging is associated with altered motility, inflammation, barrier dysfunction, and an increased risk of colon cancer.10, 11, 12 It is unclear, however, to what extent changes in stem cell function underlie age-related functional changes, especially in human beings. In the mouse SI, aging is associated with decreased crypt repair after irradiation, decreasing ISC numbers, slower contribution of Lgr5+ cells to the intestinal epithelium as determined by in vivo fate-mapping experiments, and decreased organoid-forming capacity in vitro.13, 14, 15 In addition, decreased Wnt signaling in aged animals impaired the ability of mouse ISCs to maintain homeostasis and contributed to decreased organoid-forming capacity in vitro.16,17 By contrast, dietary restriction, a lifespan extending intervention, enhanced stem cell numbers and improved regenerative capacity, whereas short-term fasting enhances organoid formation in aged mice.14,18,19 These data suggest that ISC behavior likely is impacted by the aging process, but the molecular changes that occur with age in human beings and how these vary from individual to individual are unknown.
To address the impact of aging on human ISCs, we wanted to confirm whether mesenchyme-free, 3-dimensional organoid cultures maintain hallmarks of age in the absence of an aging intestinal microenvironment. There are many hallmarks of aging, including cellular senescence, telomere shortening, DNA damage, and gene expression and epigenetic changes.20 At the epigenetic level, genome-wide DNA methylation changes are pervasive during aging, with increased variability in methylation observed over time—a process known as epigeneticdrift.21, 22, 23, 24 Although many of the changes that occur with age are stochastic and vary from individual to individual, the methylation status of a distinct subset of CpG sites in the genome is tightly correlated with age, such that these sites can be used as an epigenetic clock to accurately predict the age of human tissues.25,26 In addition, epigenetic clocks can predict lifespan.27 Although these epigenetic clocks show tight correlations between chronological age and predicted age based on DNA methylation across populations, overprediction and underprediction of age can be biologically and clinically meaningful. For example, patients with conditions associated with early onset of age-related clinical features, such as Down syndrome, Werner syndrome, or human immunodeficiency virus, showed a premature age advancement according to the epigenetic clock.28, 29, 30, 31 Conversely, lifespan-extending perturbations such as dietary restriction caused a decrease in the ticking rate of recently developed epigenetic clocks for mice, indicating that the clock may reflect a biologically relevant state of aging.32,33
Here, we use the pan-tissue epigenetic clock by Horvath26 as a biomarker of aging to determine if human intestinal organoids derived from SI and colon are a viable model for studying stem cell behavior during aging, given the absence of an aging niche. We focused on the multitissue estimator because it applies to developmental samples and allows one to compare aging rates from different tissues and cell types.26
We found that intestinal organoids from human SI and colon have global methylation patterns and epigenetic ages similar to the primary crypt cells from which they were derived. Unexpectedly, however, we also observed a marked decrease in the epigenetic aging rate in human SI, when compared with colon, in both crypt epithelial cells and stem cell–enriched organoids (hereafter referred to as spheroids). Strikingly, the change in the rate of aging in the SI appears in midlife. Taken together, using a well-established molecular hallmark of age, our data show significant differences in the human SI and colon, findings that are preserved in organoids in vitro.
Results
Regional Differences in Methylation Patterns Are Maintained In Vitro
To begin a comparison of the aging of the proximal and distal regions of the human intestine, we assayed DNA methylation patterns of the SI (duodenum and jejunum segments) and colon. SI specimens were obtained from individuals aged 0–85 years, and colonic specimens were collected from patients aged 0–80 years (Table 1). Genomic DNA was isolated from the SI (n = 14) and colonic (n = 12) mucosal segments, consisting of epithelium, lamina propria, and muscularis mucosae, in addition to the stem cell–containing crypts of the SI (n = 18) and colon (n = 18) samples (Figure 1A). All samples used were pathologically normal. Subsequently, genome-wide methylation was assayed using commercially available methylation arrays (see Materials and Methods section).
Table 1.
Summary of Human Small Intestine and Colon Specimens for DNA Methylation Analysis
| ID number | Tissue type | Cell type analyzed | Reason for surgery | Sex | Age/age range, y |
|---|---|---|---|---|---|
| 1 | Fetal small intestine | Mucosa | Fetal abortion | M | 0 |
| 2 | Duodenum | Mucosa | Unknown | F | 50–55 |
| 3 | Duodenum | Mucosa | Unknown | F | 75–80 |
| 4 | Duodenum | Mucosa | Unknown | F | 65–70 |
| 5 | Jejunum | Mucosa | Developmental abnormality | F | 0–5 |
| 6 | Duodenum | Mucosa | Unknown | F | 60–65 |
| 7 | Duodenum | Mucosa | Unknown | M | 70–75 |
| 8 | Duodenum | Mucosa | Inflammatory bowel disease | F | 17 |
| 9 | Duodenum | Mucosa | Pancreatic cancer | F | 35–40 |
| 10 | Jejunum | Mucosa | Organ donation | F | 68 |
| 11 | Duodenum | Mucosa | Pancreatic mass | F | 16–20 |
| 12 | Jejunum | Mucosa | Organ donation | M | 17 |
| 13 | Jejunum | Mucosa | Organ donation | M | 35 |
| 14 | Duodenum | Mucosa | Pancreatic cancer | F | 80–85 |
| 2 | Duodenum | Crypt | Unknown | F | 50–55 |
| 3 | Duodenum | Crypt | Unknown | F | 75–80 |
| 4 | Duodenum | Crypt | Unknown | F | 65–70 |
| 5 | Jejunum | Crypt | Developmental abnormality | F | 0–5 |
| 6 | Duodenum | Crypt | Unknown | F | 60–65 |
| 9 | Duodenum | Crypt | Pancreatic cancer | F | 35–40 |
| 10 | Jejunum | Crypt | Organ donation | F | 68 |
| 11 | Duodenum | Crypt | Pancreatic mass | F | 16–20 |
| 12 | Jejunum | Crypt | Organ donation | M | 17 |
| 13 | Jejunum | Crypt | Organ donation | M | 35 |
| 14 | Duodenum | Crypt | Pancreatic cancer | F | 80–85 |
| 15 | Duodenum | Crypt | Organ donation | M | 71 |
| 16 | Duodenum | Crypt | Unknown | M | 55–60 |
| 17 | Duodenum | Crypt | Unknown | M | 75–80 |
| 18 | Duodenum | Crypt | Unknown | F | 65–70 |
| 19 | Duodenum | Crypt | Unknown | M | 50–55 |
| 20 | Duodenum | Crypt | Biliary stricture, pancreatic dysplasia | M | 55–60 |
| 21 | Jejunum | Crypt | Organ donation | M | 47 |
| 10 | Colon | Mucosa | Organ donation | F | 68 |
| 12 | Colon | Mucosa | Organ donation | M | 17 |
| 13 | Colon | Mucosa | Organ donation | M | 35 |
| 22 | Colon | Mucosa | Unknown | F | 70–75 |
| 23 | Colon | Mucosa | Unknown | M | 30–35 |
| 24 | Colon | Mucosa | Colorectal cancer | F | 75–80 |
| 25 | Colon | Mucosa | Diverticulitis | F | 65–70 |
| 26 | Colon | Mucosa | Colorectal cancer | F | 65–70 |
| 27 | Colon | Mucosa | Colorectal cancer | F | 70–75 |
| 28 | Colon | Mucosa | Colorectal cancer | M | 70–75 |
| 29 | Colon | Mucosa | Diverticulitis | F | 65–70 |
| 30 | Colon | Mucosa | Unknown | F | 75–80 |
| 10 | Colon | Crypt | Organ donation | F | 68 |
| 12 | Colon | Crypt | Organ donation | M | 17 |
| 13 | Colon | Crypt | Organ donation | M | 35 |
| 21 | Colon | Crypt | Organ donation | M | 47 |
| 22 | Colon | Crypt | Unknown | F | 70–75 |
| 24 | Colon | Crypt | Colorectal cancer | F | 75–80 |
| 26 | Colon | Crypt | Colorectal cancer | F | 65–70 |
| 27 | Colon | Crypt | Colorectal cancer | F | 70–75 |
| 28 | Colon | Crypt | Colorectal cancer | M | 70–75 |
| 29 | Colon | Crypt | Diverticulitis | F | 65–70 |
| 31 | Colon | Crypt | Unknown | M | 55–60 |
| 32 | Colon | Crypt | Diverticulitis | F | 65–70 |
| 33 | Colon | Crypt | Unknown | F | 75–80 |
| 34 | Colon | Crypt | Developmental abnormality | F | 0.5 |
| 35 | Colon | Crypt | Colorectal cancer | M | 61–65 |
| 36 | Colon | Crypt | Colorectal cancer | M | 56–60 |
| 37 | Colon | Crypt | Colorectal cancer | M | 51–55 |
| 38 | Colon | Crypt | Colorectal cancer | M | 61–65 |
F, female; M, male.
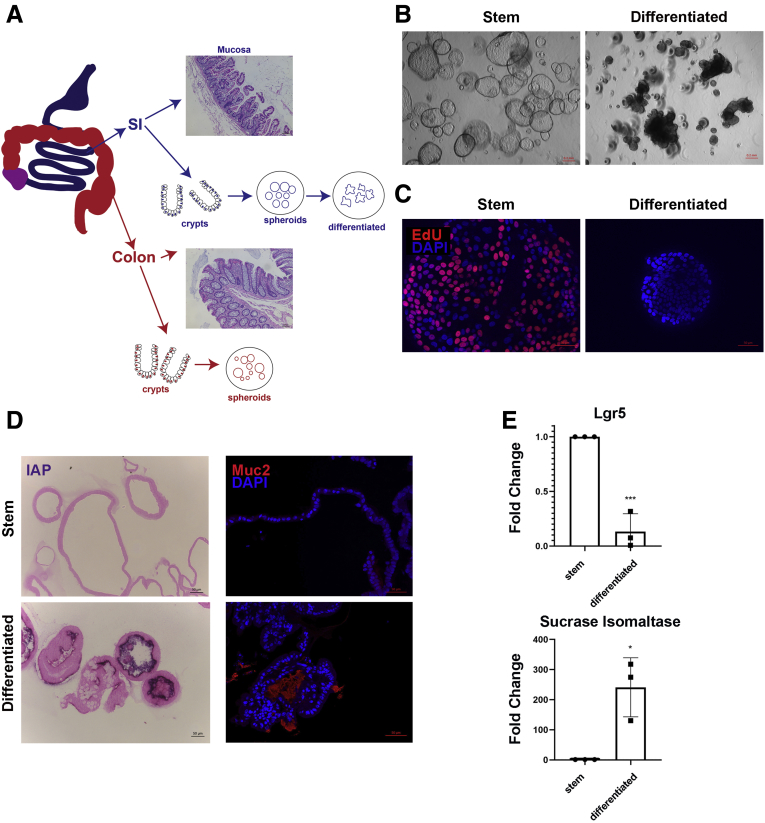
Figure 1.
Analysis of stem cell and differentiation markers in human intestinal spheroids. (A) Schematic of samples used. Representative H&E-stained sections of mucosae shown. Scale bar: 100 μm. Mucosa, isolated crypts, and in vitro organoids were used for methylation arrays for both human SI and colon. (B) Brightfield images of undifferentiated duodenal spheroids (left) and differentiated spheroids (right). Scale bars: 200 μm. (C) Sixteen-hour 5-ethynyl-20-deoxyuridine incorporation in whole-mount undifferentiated spheroids and spheroids after 4 days of differentiation. Note loss of proliferation upon differentiation. Scale bar: 50 um. (D) Nitro-blue tetrazolium and 5-bromo-4-chloro-3′-indolyphosphate NBT/BCIP intestinal alkaline phosphatase (IAP) stain (purple) identifying enterocytes (left) and immunofluorescence staining for mucin-2 identifying Goblet cells (right) on 5-μm sections of duodenal differentiated spheroids. Scale bars: 50 μm. (E) Quantitative polymerase chain reaction for stem cell marker Lgr5 and enterocyte marker sucrase isomaltase in undifferentiated and differentiated spheroids derived from n = 3 duodenal specimens. Results are means ± SD. Samples were run in triplicate. Expression was normalized to RPL13A transcript levels. Statistics: Student t test: *P < .05, ***P < .001. DAPI, 4′,6-diamidino-2-phenylindole.
To determine whether regional differences in epigenetic patterns are maintained in vitro for more proximal tissues, we conducted differential methylation analysis of mucosa, crypts, and spheroids from both the SI (duodenum and jejunum) and colon.4,7,34 Spheroids are enriched for cells harboring stem cell characteristics—they divide rapidly and are capable of multilineage differentiation.4 Upon reduction of Wnt3A and withdrawal of other growth factors, SI spheroids change morphology to dense folded organoids, stop dividing, down-regulate Lgr5, and begin to express markers of mature absorptive and secretory lineages (Figure 1B–E).
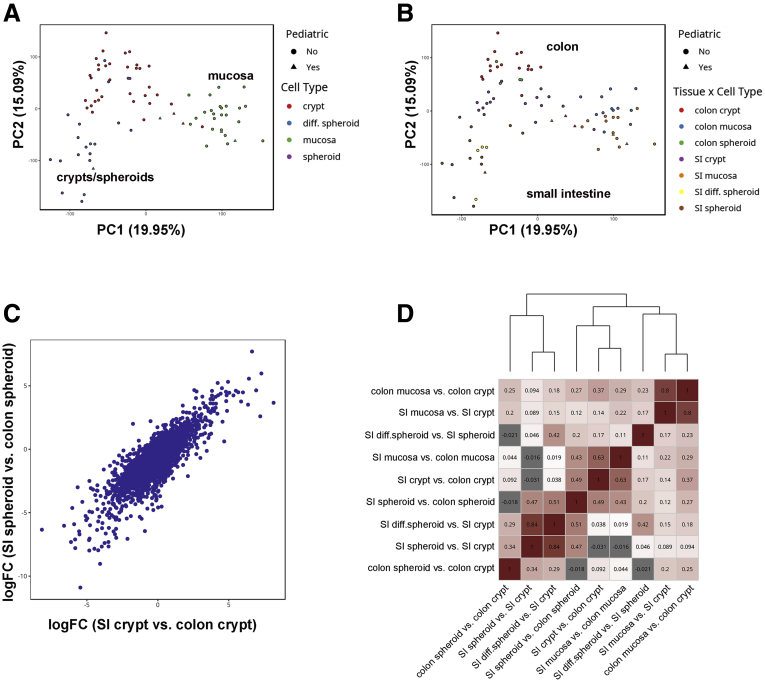
Principal component analysis (PCA) of promoter methylation data from these samples showed clustering by cell type, with separation of whole mucosa from crypts and spheroids (Figure 2A). PCA analysis also showed separation by tissue type, with SI clustering away from the colon (Figure 2B). SI and colon spheroids grouped closer to isolated crypts from the same region, although there were no distinct clusters (Figure 2B). Pediatric and adult samples showed similar behavior in PCA plots.
Figure 2.
Differential methylation analysis of human intestinal and colonic mucosa, crypts, and organoids. (A and B) PCA of methylation at promoters. (A) PCA showing clustering by cell type (mucosa, crypts, spheroids). (B) PCA for cell and tissue type, showing clustering by tissue (small intestine vs colon) and cell type (mucosa, crypts, spheroids). (C) Log-fold-change (logFC) plot showing correlation of regional differences in methylation levels at CpG sites within promoters between crypts (x-axis) and spheroids (y-axis). The biweight midcorrelation coefficient is 0.49. (D) Pairwise correlations between logFC values for the 9 comparisons used in differential methylation of promoters. Correlations were computed using biweight midcorrelation, a robust alternative to Pearson correlation. Average-linked hierarchical clustering was used to create a clustering dendrogram from Euclidean distances between each vector of pairwise correlations. diff, differentiated; PC, Principal Component.
We also determined whether the differences in methylation levels between colon and SI spheroids were correlated with the differences observed between colon and SI crypts. These analyses showed a strong positive correlation between the log fold change values of these 2 comparisons, providing additional evidence that the differences between the colon and SI are maintained in vitro (Figure 2C). Moreover, we correlated the log fold change values of all of the pairwise comparisons with each other and observed 3 distinct clusters (Figure 2D). One cluster, as expected, highlighted that tissue differences (SI vs colon) were correlated across mucosa, crypts, and spheroids. A second cluster emphasized methylation differences between mucosa vs crypts, and a third cluster highlighted differences between in vitro spheroids and primary crypts from both organs. In other words, comparing spheroids with isolated crypts from the same tissue showed a signature, an overall change in methylation patterns, that results from deriving in vitro cultures. Importantly, the changes in methylation between crypts and spheroids were conserved between the SI and colon, suggesting a possible, common in vitro methylation signature (Figure 2D). Taken together, these data show that regional differences in methylation patterns are largely preserved in culture. These data comparing duodenum and jejunum with colon are consistent with recent reports comparing primary tissues and organoids from the terminal ileum and colon.7
DNA Methylation Age Is Reduced in Crypts From the Small Intestine
DNA methylation patterns change dynamically with age.21,23 Having confirmed that spheroids derived and maintained in vitro recapitulate regional differences in methylation patterns, we wanted to determine whether SI and colon cultures derived in vitro accurately reflect the age of the tissue from which they were derived. Several algorithms, referred to as epigenetic clocks, use DNA methylation changes to predict chronological age for multiple species. By using methylation array data sets from thousands of samples from >50 tissue and cell types, the Horvath26 epigenetic clock identified 353 CpG sites that accurately estimate age based on methylation levels across all reported human cell and tissue types, except sperm. As such, the Horvath26 clock can be used as a biomarker of aging. Therefore, we used the methylation status of 353 CpGs that comprise the Horvath26 clock to calculate the predicted age based on DNA methylation (DNAm age) of human SI (n = 14) and colon (n = 12) mucosa, as well as isolated SI (n = 18) and colon (n = 18) crypts (Table 1).
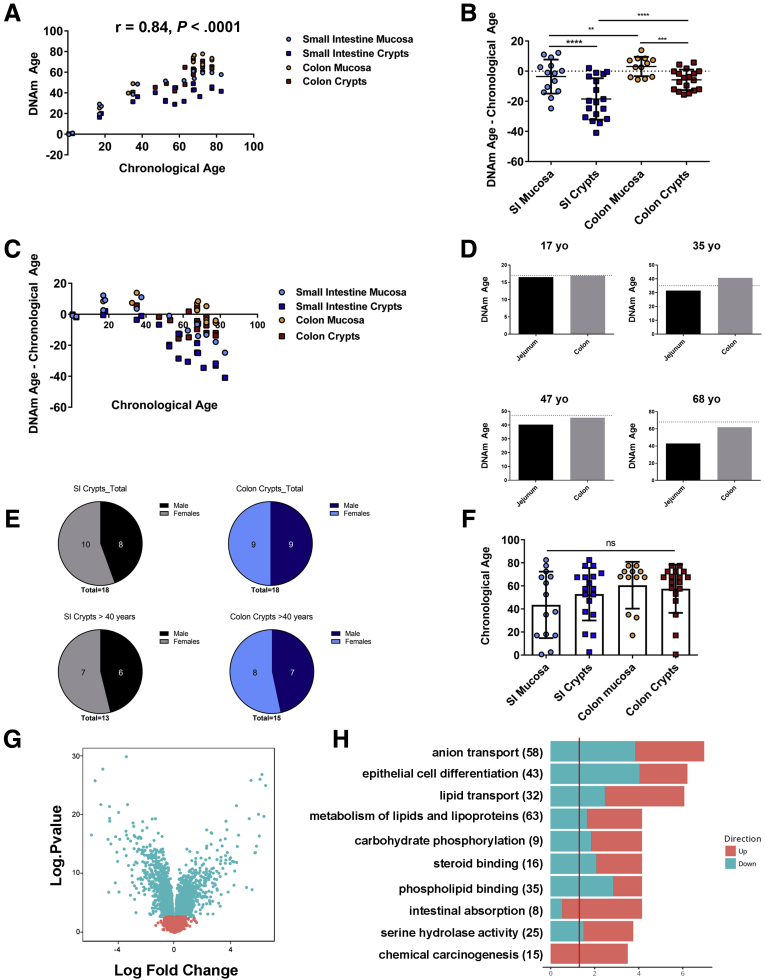
Across all samples, there was a linear relationship between chronological age and predicted DNAm age, as expected, with a Pearson correlation coefficient of r = 0.84 (P < .0001) (Figure 3A). We next examined the linear relationship between predicted and chronological age for each tissue and cell type separately. In SI, the correlation coefficient was higher for mucosa (r = 0.94; P < .0001) than for crypts (r = 0.86; P < .0001). In colon, the correlation coefficient was similar for mucosa and crypts (r = 0.96; P < .0001; and r = 0.95; P < .0001, respectively).
Figure 3.
Epigenetic clock analysis of human small intestine and colon shows a decreased ticking rate in crypt cells from the small intestine. (A) Scatter plot of DNAm age based on the epigenetic clock (y-axis), vs chronological age (x-axis) for mucosa and crypts isolated from human small intestine and colon. Pearson correlation coefficient = 0.84; P < .0001. (B) Plot of DNAm age deceleration (the absolute difference between DNAm age and chronological age) for mucosa and crypts isolated from SI and colon. Data are presented as means ± SD. Statistics were derived from a linear mixed-effects model: **P < .01, ***P < .001, ****P < .0001. (C) Relationship between chronological age (x-axis) and the absolute difference between DNAm age and chronological age (y-axis). Note the age-dependent increase in DNAm deceleration for SI crypts. (D) DNAm-based age prediction (y-axis) for paired jejunum and transverse colon crypts from 4 individuals. Dotted line indicates an individual’s chronological age. Data points also were included in the cumulative analysis in panel C. (E) Venn diagrams show distributions of males and females among crypt specimens used for epigenetic aging analysis. Proportions of each gender in all samples (top); proportions of each sex in the samples from those older than age 40 (bottom). (F) Plot of chronological age for samples used for epigenetic age analysis, by category. Data are represented as means ± SD. (G) Differential gene expression between SI and colon crypts. The x-axis shows the log fold change and the y-axis shows the -log10P value of differential expression. Blue dots show genes that were significantly different (FDR < 0.01), red dots show genes that were not significantly different. A total of 3028 differentially expressed genes were identified. (H) Gene ontology and pathway analysis of the 3028 genes expressed differentially between human SI and colon crypts. The x-axis shows the -log10P value of the overlap between up-regulated and down-regulated differentially expressed genes and each gene set on the y-axis. The vertical line is the significance cut-off value at P < .05. yo, year old.
We then compared the DNAm ages of SI and colon mucosa with that of crypts isolated from each organ by calculating the absolute difference between DNAm age and chronological age, a difference known as DNA age acceleration or deceleration (Figure 3B). Remarkably, crypts isolated from the SI showed a significant age deceleration compared with surrounding mucosa (mean error, -18.5 vs -3.4 y, respectively; P < 1.6 × 10-7). In addition, SI crypts had a significant age deceleration when compared with that seen for colon crypts (mean error, -18.5 vs -5.8 y, respectively; P < 6.6 × 10-6). There was also a decreased epigenetic age in colon crypts compared with whole colonic mucosa (mean error, -5.8 vs +3.1 y, respectively; P < 1.34 × 10-4); however, the magnitude of this difference was smaller than that detected for SI. We also observed that the SI mucosa appeared to age more slowly than the colon mucosa (mean error, -3.4 vs +3.1, respectively; P < 5.3 × 10-3), but the degree of age deceleration was smaller in SI mucosa, and the colon mucosa actually showed mild age acceleration. Taken together, these data indicate that the ticking rate of the epigenetic clock for epithelial cells within SI crypts is considerably slower than for either of the other cells contained within the SI mucosa or cells within colonic crypts.
Small Intestine DNAm Aging Rate Slows in Midlife
Although an apparent age deceleration was observed in SI crypts, the ticking rate of the epigenetic clock in SI is in sync with the chronological clock until middle age (approximately 40 years), when it begins to slow. Specifically, we found that the degree of DNAm age deceleration in SI samples, crypts in particular, increased with greater chronological age (Figure 3C). There is a negative correlation between age prediction error and chronological age for SI crypts (r = -0.90; P < 1 × 10-4). A similar trend was seen in SI mucosa, although to a lesser extent than in crypts (r = -0.78; P = .001). Although there was a negative correlation between DNAm age prediction error and chronological age for both colon crypts and mucosa (r = -0.47; P = .05; and r = -0.69; P = .01, respectively), these were much less significant than the trends noted in the SI. Taken together, these data suggest that the ticking rate of the epigenetic clock is largely in sync with the chronological clock through older age for colon, in contrast to the age deceleration observed in the SI. Importantly, the age deceleration observed in the SI is not owing to an inability of the clock to predict age for the SI.
DNA Methylation Age Deceleration for Small Intestine Is Observed in Paired Intestinal Samples
To address the confounding factor of different underlying medical histories in SI vs colon surgical samples and to account for interindividual variation, we analyzed 4 paired jejunum and transverse colon segments from organ donors immediately post-mortem for DNAm age analysis. The predicted DNAm age of jejunum crypts was younger than the DNAm age of paired colon crypts for all 3 older adult donors (ages, 35, 47, and 68 years), consistent with our findings that crypts isolated from the SI showed decreased epigenetic aging rates when compared with colon crypts (Figure 3D). By contrast, the 17-year-old donor showed congruence between DNAm and chronological age for both jejunum and colon.
Age and Sex Do Not Likely Contribute to Tissue Differences in DNAm Age
DNAm aging profiles are influenced by sex, and the epigenetic aging rate for women tends to be lower than men, at least for blood cells.25,35 Given that we uncovered a slowing in epigenetic aging for SI midlife (∼40 years), it was important to consider how males and females were distributed between our younger (<40 year) and older (>40 year) populations. Overall, for SI and colon crypt samples, our samples included almost equal proportions of male and female samples. Notably, this distribution did not change dramatically after age 40 (Figure 3E), indicating that sex is likely not a major contributing factor to the slowed ticking rate of the epigenetic clock at middle age for SI crypts. We also noted that there was no significant difference in the age distribution for mucosa and crypts from SI and colon (Figure 3F).
To further characterize which variables were significant predictors of DNA methylation, we fitted a linear mixed-effects model to predict DNAm age as a function of chronological age, sex, batch, tissue, and cell type. We found that chronological age was a strong predictor of methylation age (P < 3.14 × 10-16), as expected, whereas sex (P < .33) and batch (P < .04) were not significant, using a strict cut-off value of P < .01 for statistical significance. The combined influence of tissue and cell type (ie, SI crypts, SI mucosa, and so forth) were also a highly significant predictor of DNA methylation age (P < 3.1 × 10-10).
By using our gene expression data, we then asked whether changes in gene expression could provide insight into the observed differences in chronological aging rates between the SI and colon, as determined by the epigenetic clock. We identified 3028 differentially expressed genes between SI (n = 16) and colon (n = 6) crypts (false discovery rate [FDR] < 0.01) (Figure 3G). Gene ontology analysis showed the most striking differences in gene expression between SI and colon crypts were related to anion transport, epithelial differentiation, lipid transport and metabolism, and intestinal absorption (Figure 3H), likely reflecting functional differences between the 2 tissues. However, this analysis did not show enrichment for any previously characterized aging-associated genes.
DNA Methylation Age Deceleration of Human Small Intestine Is Maintained in Cultured Spheroids
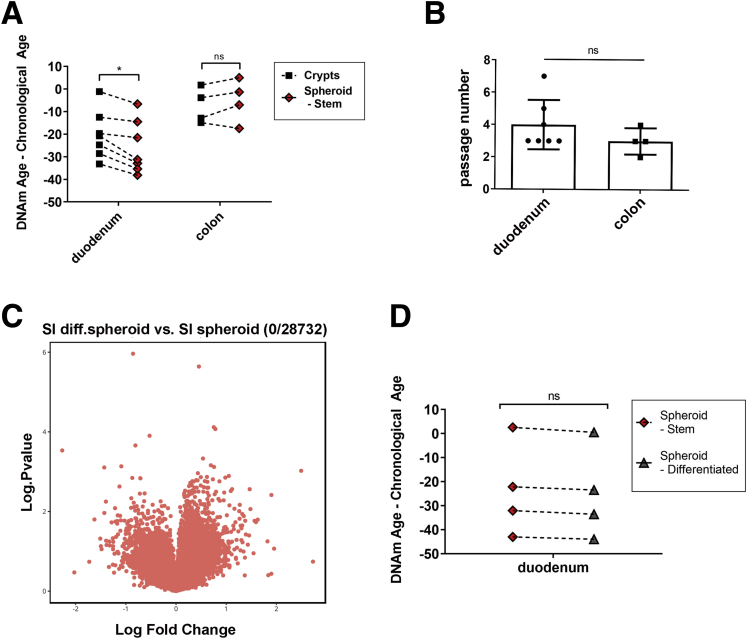
There is potential for organoids to be used as models for aging9; however, whether in vitro culture conditions alter or reset aging profiles for SI or colon, thereby minimizing detection of age-associated functional changes, is not clear. To determine if human ISCs cultured in vitro maintain epigenetic aging profiles similar to cells found in primary uncultured epithelial crypts, we compared the DNAm age of duodenal and colonic crypts with their corresponding spheroid cultures, analyzed at early passages (Figure 4A and B). Consistent with our previous data, duodenal spheroids showed DNAm age deceleration similar to the crypts from which they were derived, although there was a slightly younger DNAm age in SI spheroids compared with crypts. In colon, we found that spheroids also have a DNAm age similar to the crypts from which they were derived, however, in contrast to SI, the DNAm ages of colon spheroids and crypts were close to the chronological age of the donor. Importantly, these data indicate that age, as measured by DNA methylation patterns, is largely maintained in culture. In addition, these results confirm regional differences in epigenetic aging and suggest potential differences in ISC aging, as reflected by DNAm, in closely related tissues.
Figure 4.
DNA methylation age deceleration of human small intestine is maintained in cultured spheroids and during differentiation. (A) The difference between DNAm age and chronological age for paired crypts and spheroids derived from duodenum and colon. Dotted lines indicate paired samples isolated from the same individual. Statistics were derived from the nonparametric paired Wilcoxon signed-rank test. (B) Passage number of SI vs colon spheroids used for DNAm aging analysis. Data are represented as means ± SD. (C) Volcano plot for differential methylation analysis identifies zero differentially methylated promoters (FDR < 0.01) between duodenal stem cell–enriched spheroids and differentiated cultures. X-axis indicates log fold change, the y-axis indicates the log P value. (D) The difference between DNAm age and chronological age for paired duodenal stem cell–enriched and differentiated spheroids derived from the same sample (n = 4). Dotted line indicates paired samples. Statistics were derived from the nonparametric paired Wilcoxon signed-rank test. *P < .05.
DNA Methylation Age Does Not Change During Differentiation In Vitro
To investigate whether the DNAm age, as reported by the epigenetic clock, is altered during stem cell differentiation in culture, we subjected duodenal spheroids to differentiation conditions for 4 days (Figure 1B–E). We first analyzed the global methylation pattern of differentiated cells and compared them with the undifferentiated spheroids from which they were derived. Differentiated cultures clustered closely with stem cell–enriched spheroids from the same tissue (Figure 2B). Similar to Kraiczy et al,7 we found no differentially methylated regions (FDR <0.01) between SI-derived stem cell–enriched spheroids (n = 12) and differentiated (n = 4) organoids (Figure 4C), although our small sample size and stringent statistical cut-off limited our ability to detect subtle changes in methylation. Interestingly, when stem cell–enriched spheroids from duodenum were allowed to differentiate for 4 days in vitro, the observed decreased epigenetic age was maintained (Figure 4D), indicating that youthful progenitor cells may be responsible for maintaining the lower epigenetic age of SI epithelium as a whole. In addition, these data underscore the observation that proliferation rates do not impact the epigenetic clock significantly.26
Discussion
It is well known that lifestyle and environmental factors can influence aging, and there is increasing interest in understanding the relationship between interindividual variability in aging rates and susceptibility to age-associated pathologies, including cancer and neurodegenerative disease. In this study, we sought to determine whether human SI and colon organoids can be used as reliable tools to investigate stem cell–intrinsic aging in vitro. DNA methylation-based age predictions now provide us with a tool to quantify aging rates across individuals and between tissue types.25,26,36 The Horvath26 multitissue epigenetic clock shows tight linear correlations between DNAm age and chronological age across tissues. However, some tissues, such as cerebellum, show a decreased DNAm aging rate, while other organs, such as breast, show an increased DNAm aging rate, indicating that not all tissues within the same individual age similarly.26,37,38
Although previous studies have indicated that regional DNA methylation patterns and gene expression are maintained in intestinal organoids,7,39,40 it was not obvious that the methylation status of sites used for age estimation would be unchanged in vitro. Although there was a slight epigenetic age reduction in SI spheroids compared with SI crypts, our data suggest that in vitro stem-like spheroids largely show DNAm aging rates similar to the crypts from which they were derived. In addition, we observed that DNAm age is preserved during differentiation because there were no significant differences between highly proliferative stem cell–containing spheroids and differentiating organoids containing nonproliferating cells, further supporting the notion that epigenetic clocks do not simply reflect differences in cell division rates.26 Therefore, our data establish that intestinal organoids are valid models for studying stem cell–intrinsic aging, even in the absence of a mesenchymal niche. Importantly, our data indicate that epigenetic clocks can be a useful tool for assessing the degree to which intestinal diseases, particularly those with a strong inflammatory component, correlate with acceleration of aging hallmarks.41
Our data also suggest fundamental differences in SI vs colonic DNAm aging profiles, at least within epithelial cells of the crypts, which is surprising given the similarities shared between the SI and colon. Specifically, we found that the correlation between DNAm and chronological age is lower in SI crypts compared with SI mucosa, while DNAm and chronological age are highly correlated for both crypts and mucosa in colon. Interestingly, the difference between DNAm age in SI crypts and mucosa is enhanced after midlife. The ticking rate of the epigenetic clock for epithelial cells within SI crypts is considerably slower than for cells within colonic crypts, even when SI and colon were taken from the same person. The magnitude of the DNAm age reduction we observed for the SI is considerably larger than that reported previously for other tissues, providing a striking example of an organ for which DNAm age diverges from chronological age in an age-dependent fashion.26,37,38 Given that the effect of cell composition on DNAm age prediction has been shown to be minimal,26,42 we do not expect variation in the composition of epithelial cells with age to explain the observed differences in DNAm age between SI and colon crypts. Unfortunately, differential gene expression analysis could not explain the differences in DNAm aging between SI and colon, although our analysis was limited by small sample size. That being said, there are limited data in mouse and rat models to suggest more robust expression of metabolism-associated genes, immune response, and inflammatory genes in colon compared with SI during aging, which suggest that gene expression changes with age may differ by region, although further investigation in human samples is necessary.43,44
Much of what we know about ISC behavior comes from studies in the SI, particularly in mice, and little is known about how ISCs in the human colon compare with those from the SI, especially during aging. Recent findings have highlighted age-related changes in the mammalian intestine and have begun to address the role stem cells may play in functional decline.14, 15, 16, 17 Specifically, ISCs from mouse, and potentially human beings, show a decrease in regenerative capacity as a result of decreased Wnt signaling.16,17 Our data suggest that human SI and colon crypt cells differ with respect to epigenetic age; however, whether this is physiologically relevant, as determined by functional changes with age, remains to be tested. Although a comparison of age-related changes in ISCs from the SI and colon using organoids is theoretically feasible, colonic spheroids are consistently less proliferative and less robust than SI-derived cultures when grown under identical conditions (S.L. and D.L.J., unpublished data). Therefore, interpretation of experiments comparing growth and differentiation of SI and colon spheroids from young and old patients may be limited by intrinsic regional differences in stem cell function45 and preclude a direct comparison of the effects of aging on SI and colon stem cell behavior. However, based on our current results, we predict that age-related functional changes in human colon stem cells will be more dramatic than any detected for SI stem cells. Our findings lay the foundation for characterizing such functional differences in human SI and colon stem cells, which may eventually shed light on the differences in the incidence of age-associated diseases between these 2 organs, including adenocarcinoma.
Materials and Methods
Experimental Procedures
Human bowel tissue
De-identified and discarded surgical specimens were retrieved from the University of California Los Angeles Translational Pathology Core Laboratory (TPCL). After clinical surgical pathology evaluation, normal adjacent tissue was obtained. Experimentation using TPCL-derived human tissues was approved by the University of California Los Angeles Institutional Review Board, which waived the informed consent requirement for specimens acquired from the TPCL (IRB 11-002504). The age of the human specimen was provided as a 5-year age range, according to TPCL practices. Procurement of paired jejunum and colon from anonymous post-mortem donors was facilitated by the OneLegacy Foundation (Los Angeles, CA). Samples are summarized in Table 1.
Intestinal mucosa and crypt isolation
Tissues were washed in phosphate-buffered saline (PBS), and mucosa was isolated by removal of outer muscle layers using surgical scissors. Mucosae were cut into 1-cm pieces and washed with PBS until the supernatant was clear. Small intestine and colonic crypts were isolated using modified existing methods.4,6 Specimens were incubated at 4°C in 8 mmol/L EDTA and 1 mmol/L dithiothreitol for 30 minutes to 1 hour, with gentle agitation. Supernatant was removed and replaced with fresh PBS. Specimens were vortexed to release crypts. Crypts were collected into 15-mL tubes and the process was repeated to collect 4–6 fractions. Fractions were centrifuged at 100 × g to pellet crypts. SI crypts subsequently were filtered with 100-um mesh to remove villus domains and epithelial debris. Isolated crypts were resuspended in basic medium containing Advanced Dulbecco's Modified Eagle Medium/F12, 1× Glutamax, 10 mmol/L HEPES buffer, and 1× penicillin/streptomycin (all ThermoFisher Scientific, Waltham, MA) for spheroid cultures, or resuspended in PBS for DNA lysis.
Genomic DNA Extraction
Duodenal and colonic mucosal specimens were chopped into 5-mm pieces and homogenized; crypts were isolated as described earlier. Homogenized mucosa and isolated crypts were lysed and genomic DNA was isolated using standard phenol–chloroform extraction with ethanol precipitation. DNA was stored at -20°C before downstream processing.
Bisulfite Conversion and Methylation Arrays
Bisulfite conversion of 500 ng DNA per sample was conducted using the EZ-96 DNA Methylation Gold Kit (Zymo Research, Irvine, CA). Samples subsequently were processed for Illumina (San Diego, CA) HumanMethylation450 BeadChips and Infinium MethylationEPIC arrays according to the manufacturers’ instruction, and specimens were randomized across arrays. Methylation status was reported as the β value (range, 0–1, with 0 = unmethylated and 1 = fully methylated). For differential methylation analysis and principle component analysis the M value was used.46
Differential Methylation Analysis
Array preprocessing was run with the RnBeads package.47 Probes outside CpG contexts, on sex chromosomes, or in single-nucleotide polymorphisms or with missing values were removed. The greedycut algorithm in RnBeads was used to remove probes with low detection score P values, and probes with a SD less than 0.005 also were removed. Only probes shared between the HumanMethylation450K and HumanMethylationEPIC chips were retained. Background correction was performed with the out-of-band method implemented by ENMix (National Institute of Environmental Health Sciences, Research Triangle Park, NC),48 and arrays were normalized using β mixture quantile dilation (BMIQ).49 β values were averaged across promoters (1.5 kb before transcription start site to 0.5 kb after the transcription start site), gene bodies, or CpG islands and converted to M values. M values were corrected for batch effect using the parametric empiric Bayes method from ComBat.50 Differential methylation was assessed using linear modeling with the limma package,51 with age and sex included as covariates, and a cut-off FDR of <0.01 was used to determine significance.
DNA Methylation Age Analysis Using the Epigenetic Clock
After standard quality control, all samples were submitted to the epigenetic clock software,26 and the predicted DNA methylation age was compared with the chronological age for each sample. For each tissue sample for which the precise age of the donor was not given, the midpoint of the 5-year age range was assigned as the chronological age. Age correlation was calculated using the Pearson correlation coefficient between chronological age and predicted DNAm age. DNAm age acceleration/deceleration was calculated as the absolute difference between chronological age and DNAm age and was analyzed using a linear mixed-effects model with a random effect for patient ID to account for multiple samples coming from the same individuals. Paired analysis was conducted using the paired Wilcoxon rank-sum test.
RNA Extraction and Gene Expression Microarray Analysis
Samples were homogenized and lysed in TRIzol (Invitrogen, Carlsbad, CA) and RNA was isolated using the RNA Clean and Concentrator kit (Zymo Research, Irvine, CA). RNA integrity was analyzed on the Agilent 2200 TapeStation (Santa Clara, CA), and RNA integrity number greater than 7.0 was used as a quality control threshold for RNA quality. Gene expression was quantified on Illumina HT12 v4 microarrays and were preprocessed using the lumi pipeline (Robert H. Lurie Comprehensive Cancer Center, Chicago, IL).52 Expression values were normalized using the variance-stabilized transformation,53 and robust spline normalization was used for interarray normalization. Probes with a detection score P value greater than .01 were dropped, as were probes that were unannotated. Duplicated probes for the same transcript were dropped using the collapseRows function54 from the WGCNA package (Department of Human Genetics, UCLA, Los Angeles, CA). Differential expression was performed using limma,51 with age and sex included as covariates. Gene ontology enrichment was assessed using Enrichr (Department of Pharmacology and Systems Therapeutics, Icahn School of Medicine at Mount Sinai, New York, NY).55
In Vitro Organoid Cultures
To generate cystic 3-dimensional spheroids, isolated crypts were seeded at 150 crypts/25 uL of Growth Factor Reduced Matrigel (356231; BD Biosciences, San Jose, CA) in a 48-well plate. Matrigel was allowed to solidify for 15 minutes at 37°C. Cells were overlaid with 250 uL of slightly modified stem cell media,4,6 consisting of 50% Wnt3A conditioned medium (see later), 50% Advanced Dulbecco's Modified Eagle Medium/F12, 1× penicillin/streptomycin, 1× Glutamax, 10 mmol/L HEPES, 1 mmol/L N-acetyl-L-cysteine (A9165; Sigma, St. Louis, MO), 1× N2 supplement (17502; Life Technologies, Carlsbad, CA), 1× B27 minus VitA (12587; Life Technologies), 50 ng/mL epidermal growth factor (316-09; Peprotech), 1 ug/mL human R-spondin (4645-RS; R&D, Minneapolis, MN), 100 ng/mL murine noggin (250-38; Peprotech, Rocky Hill, NJ), 10 mmol/L nicotinamide (N0636; Sigma), 10 nmol/L gastrin (G9145; Sigma), 10 umol/L SB202190 (S7067; Sigma), and 100 nmol/L prostaglandin E2 (P5640; Sigma). Medium was changed every 2–3 days. Spheroids were passaged every 5–7 days by dissociation for 3 minutes in TrypLE Express (Life Technologies) at 37°C followed by reseeding in Matrigel. For 2 days after splitting, 10 umol/L Rock inhibitor Y27632 (Sigma) was added to the medium. To trigger differentiation of cells within SI spheroids, nicotinamide, SB202190, and PGE2 were withdrawn; Wnt3A conditioned medium was reduced to 10%; and 10 umol/L DAPT γ secretase inhibitor (D5942; Sigma) was added for 4 days of differentiation.4 Brightfield images of organoids were acquired using the Zeiss AxioVert A1 TL/RL microscope with the AxioCam MRm camera. Images were analyzed using ZEN software (Zeiss, Oberkochen, Germany).
Wnt3A Conditioned Medium
L Wnt-3A cells (CRL-2647; American Type Culture Collection, Manassas, VA) were cultured until 80%–90% confluence in Dulbecco’s modified Eagle medium + 10% fetal bovine serum + 1× penicillin/streptomycin. Cells then were split to approximately 10% density and cultured for 4 days in t75 flasks. Day 4 supernatant was sterile-filtered. Fresh medium was replaced for 3 more days. Day 7 supernatant was sterile-filtered, and combined with day 4 medium.4 Aliquoted stocks were stored at -20°C.
5-Ethynyl-2’-Deoxyuridine Labeling
Spheroids were split to chamber slides (62407-355; ThermoFisher Scientific) in stem cell medium containing Y27632 (Y0503; Sigma). Ethynyl-2’-deoxyuridine labeling was performed using the Click-iT ethynyl-2’-deoxyuridine Alexa 594 Imaging Kit (C10339; Invitrogen). For proliferation analysis in stem cell vs differentiation conditions: 2 days after splitting, half of the wells were changed to differentiation medium, as described earlier, and half were maintained in stem cell medium. Cells were treated with 50 umol/L ethynyl-2’-deoxyuridine for the last 16 hours before fixation.34 Organoids were fixed in 4% paraformaldehyde and the Click-iT reaction was conducted according to the manufacturer’s protocols. Samples were mounted in Vectashield with 4′,6-diamidino-2-phenylindole (H-1200; Vector Laboratories, Burlingame, CA) to mark nuclei.
Histologic Analysis and Immunofluorescence Staining
Organoids were fixed in 5% neutral buffered formalin for 30 minutes, embedded in HistoGel specimen processing gel (HG-4000-012; ThermoFisher Scientific) before embedding in paraffin, and sectioned at 5 um. Intestinal alkaline phosphatase was detected using nitroblue tetrazolium chloride 5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) according to published methods.56 For immunostaining on paraffin sections, slides were dewaxed in xylene, rehydrated, and heat-mediated antigen retrieval was conducted with citrate buffer (pH 6). For Goblet cells, sections were immunostained with mucin-2 primary antibody (sc-15334, 1:50 dilution; Santa Cruz Biotechnology, Dallas, TX). Secondary staining was conducted with Alexa 568 goat anti-rabbit IgG (A-11011, 1:1000 dilution; ThermoFisher Scientific). Samples were mounted in Vectashield with 4′,6-diamidino-2-phenylindole (H-1200; Vector Laboratories) to mark nuclei. Images were acquired using the Zeiss Axio Observer.Z1 microscope with Apotome, with an AxioCam 503 mono camera (Zeiss). Images were analyzed using ZEN software.
Reverse Transcription and Quantitative Polymerase Chain Reaction
To isolate total RNA, samples were lysed in TRIzol reagent (15596026; Invitrogen), followed by chloroform extraction and isopropanol precipitation. RNA was DNase-treated and reverse-transcribed to complementary DNA using the iScript Reverse Transcription Supermix (15596026; Bio-Rad, Hercules, CA). Quantitative reverse-transcription polymerase chain reaction was conducted using SSO Advanced Universal SYBR Green Supermix (Bio-Rad) and gene-specific primers (see the oligonucleotide primers used were as follows:). Data were obtained on the CFX96 Real Time System (Bio-Rad) using manufacturer recommended device settings. Samples were analyzed in triplicate. The human ribosomal gene RPL13A was used as reference for quantification of gene expression. Fold-change of messenger RNA expression between undifferentiated spheroids and differentiated organoids was calculated using the delta delta Ct method. The oligonucleotide primers used were as follows: RPL13A, 5’-GCCCTACGACAAGAAAAAGCG-3’ (forward)/5’-TACTTCCAGCCAACC TCGTGA-3’ (reverse); Lgr5, 5’-ACCAGACTATGCCTTTGGAAAC-3’ (forward)/5’- TTCCCAGGGAGTGGATTCTAT-3’ (reverse); and sucrase isomaltase, 5’- TTTTGGCATCCAGATTCGAC-3’ (forward)/5’-ATCCAGGCAGCCAAGAATC-3’ (reverse).
Data Availability
Genome-wide data has been deposited in Gene Expression Omnibus (GEO) with the following accession numbers: GSE141254 (DNA methylation) and GSE141255 (expression).
All authors had access to the study data and reviewed and approved the final manuscript.
Acknowledgments
The authors would like to thank Jiafang Wang for technical assistance. The authors would like to thank the University of California Los Angeles Neuroscience Genomics Core for assistance with Illumina methylation assays, and the University of California Los Angeles Translational Pathology Core Laboratory for facilitating procurement of human specimens and histology support. The authors also would like to acknowledge OneLegacy, the organ donors, and their families for giving the gift of knowledge by their generous donations, and Dr Ali Zarrinpar and Coney Lee for coordinating tissue retrieval from OneLegacy.
Footnotes
Author contributions Leanne Jones and Sophia Lewis designed the study; Sophia Lewis procured and isolated all human bowel specimens and conducted all in vitro organoid experimentation; Daniel Nachun designed and performed differential methylation analysis; Sophia Lewis and Daniel Nachun analyzed epigenetic clock data with help from Steve Horvath; Martin Martin provided technical support and resources; and Leanne Jones and Sophia Lewis wrote the manuscript with assistance from Giovanni Coppola and Daniel Nachun, followed by additional comments from Steve Horvath and Martin Martin.
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by a University of California Los Angeles Broad Stem Cell Research Center Predoctoral Training Grant (S.L.), a University of California Los Angeles Broad Stem Cell Research Center Innovation Award (D.L.J.); the Rose Hills Foundation (S.L. and D.L.J.); the CURE: Digestive Diseases Research Center at University of California Los Angeles Pilot and Feasibility Study Award P30 DK 41301 (D.L.J.); and the University of California Los Angeles Clinical and Translational Science Institute/Broad Stem Cell Research Center/David Geffen School of Medicine Regenerative Medicine Theme Award (D.L.J.).
References
- 1.Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 2.Tetteh P.W., Kretzschmar K., Begthel H., van den Born M., Korving J., Morsink F., Farin H., van Es J.H., Offerhaus G.J.A., Clevers H. Generation of an inducible colon-specific Cre enzyme mouse line for colon cancer research. Proc Natl Acad Sci U S A. 2016;113:11859–11864. doi: 10.1073/pnas.1614057113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato T., Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 4.Jung P., Sato T., Merlos-Suárez A., Barriga F.M., Iglesias M., Rossell D., Auer H., Gallardo M., Blasco M.A., Sancho E., Clevers H., Batlle E. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 5.Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 6.Sato T., Stange D.E., Ferrante M., Vries R., Van Es J.H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 7.Kraiczy J., Nayak K.M., Howell K.J., Ross A., Forbester J., Salvestrini C., Mustata R., Perkins S., Andersson-Rolf A., Leenen E., Liebert A., Vallier L., Rosenstiel P.C., Stegle O., Dougan G., Heuschkel R., Koo B.-K., Zilbauer M. DNA methylation defines regional identity of human intestinal epithelial organoids and undergoes dynamic changes during development. Gut. 2019;68:49–61. doi: 10.1136/gutjnl-2017-314817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao Y., Kang B., Petkovich D.A., Bhandari Y.R., In J., Stein-O'Brien G., Kong X., Xie W., Zachos N., Maegawa S., Vaidya H., Brown S., Chiu Yen R.-W., Shao X., Thakor J., Lu Z., Cai Y., Zhang Y., Mallona I., Peinado M.A., Zahnow C.A., Ahuja N., Fertig E., Issa J.-P., Baylin S.B., Easwaran H. Aging-like spontaneous epigenetic silencing facilitates Wnt activation, stemness, and BrafV600E-induced tumorigenesis. Cancer Cell. 2019;35:315–328.e316. doi: 10.1016/j.ccell.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J.L., Todhunter M.E., LaBarge M.A., Gartner Z.J. Opportunities for organoids as new models of aging. J Cell Biol. 2018;217:39–50. doi: 10.1083/jcb.201709054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Man A.L., Bertelli E., Rentini S., Regoli M., Briars G., Marini M., Watson A.J.M., Nicoletti C. Age-associated modifications of intestinal permeability and innate immunity in human small intestine. Clin Sci. 2015;129:515–527. doi: 10.1042/CS20150046. [DOI] [PubMed] [Google Scholar]
- 11.Ren W.-Y., Wu K.-F., Li X., Luo M., Liu H.-C., Zhang S.-C., Hu Y. Age-related changes in small intestinal mucosa epithelium architecture and epithelial tight junction in rat models. Aging Clin Exp Res. 2014;26:183–191. doi: 10.1007/s40520-013-0148-0. [DOI] [PubMed] [Google Scholar]
- 12.Bhutto A., Morley J.E. The clinical significance of gastrointestinal changes with aging. Curr Opin Clin Nutr Metab Care. 2008;11:651–660. doi: 10.1097/MCO.0b013e32830b5d37. [DOI] [PubMed] [Google Scholar]
- 13.Martin K., Potten C.S., Roberts S.A., Kirkwood T.B. Altered stem cell regeneration in irradiated intestinal crypts of senescent mice. J Cell Sci. 1998;111:2297–2303. doi: 10.1242/jcs.111.16.2297. [DOI] [PubMed] [Google Scholar]
- 14.Mihaylova M.M., Cheng C.-W., Cao A.Q., Tripathi S., Mana M.D., Bauer-Rowe K.E., Abu-Remaileh M., Clavain L., Erdemir A., Lewis C.A., Freinkman E., Dickey A.S., La Spada A.R., Huang Y., Bell G.W., Deshpande V., Carmeliet P., Katajisto P., Sabatini D.M., Yilmaz Ö.H. Fasting activates fatty acid oxidation to enhance intestinal stem cell function during homeostasis and aging. Cell Stem Cell. 2018;22:769–778.e4. doi: 10.1016/j.stem.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Igarashi M., Miura M., Williams E., Jaksch F., Kadowaki T., Yamauchi T., Guarente L. NAD+ supplementation rejuvenates aged gut adult stem cells. Aging Cell. 2019;18 doi: 10.1111/acel.12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nalapareddy K., Nattamai K.J., Kumar R.S., Karns R., Wikenheiser-Brokamp K.A., Sampson L.L., Mahe M.M., Sundaram N., Yacyshyn M.-B., Yacyshyn B., Helmrath M.A., Zheng Y., Geiger H. Canonical Wnt signaling ameliorates aging of intestinal stem cells. Cell Rep. 2017;18:2608–2621. doi: 10.1016/j.celrep.2017.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pentinmikko N., Iqbal S., Mana M., Andersson S., Cognetta A.B., Suciu R.M., Roper J., Luopajärvi K., Markelin E., Gopalakrishnan S., Smolander O.-P., Naranjo S., Saarinen T., Juuti A., Pietiläinen K., Auvinen P., Ristimäki A., Gupta N., Tammela T., Jacks T., Sabatini D.M., Cravatt B.F., Yilmaz Ö.H., Katajisto P. Notum produced by Paneth cells attenuates regeneration of aged intestinal epithelium. Nature. 2019;571:398–402. doi: 10.1038/s41586-019-1383-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Igarashi M., Guarente L. mTORC1 and SIRT1 cooperate to foster expansion of gut adult stem cells during calorie restriction. Cell. 2016;166:436–450. doi: 10.1016/j.cell.2016.05.044. [DOI] [PubMed] [Google Scholar]
- 19.Yilmaz Ö.H., Katajisto P., Lamming D.W., Gültekin Y., Bauer-Rowe K.E., Sengupta S., Birsoy K., Dursun A., Yilmaz V.O., Selig M., Nielsen G.P., Mino-Kenudson M., Zukerberg L.R., Bhan A.K., Deshpande V., Sabatini D.M. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraga M.F., Ballestar E., Paz M.F., Ropero S., Setien F., Ballestar M.L., Heine-Suñer D., Cigudosa J.C., Urioste M., Benitez J., Boix-Chornet M., Sanchez-Aguilera A., Ling C., Carlsson E., Poulsen P., Vaag A., Stephan Z., Spector T.D., Wu Y.-Z., Plass C., Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones M.J., Goodman S.J., Kobor M.S. DNA methylation and healthy human aging. Aging Cell. 2015;14:924–932. doi: 10.1111/acel.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heyn H., Li N., Ferreira H.J., Moran S., Pisano D.G., Gomez A., Diez J., Sanchez-Mut J.V., Setien F., Carmona F.J., Puca A.A., Sayols S., Pujana M.A., Serra-Musach J., Iglesias-Platas I., Formiga F., Fernandez A.F., Fraga M.F., Heath S.C., Valencia A., Gut I.G., Wang J., Esteller M. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci U S A. 2012;109:10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maegawa S., Hinkal G., Kim H.S., Shen L., Zhang L., Zhang J., Zhang N., Liang S., Donehower L.A., Issa J.-P.J. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannum G., Guinney J., Zhao L., Zhang L., Hughes G., Sadda S., Klotzle B., Bibikova M., Fan J.-B., Gao Y., Deconde R., Chen M., Rajapakse I., Friend S., Ideker T., Zhang K. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen B.H., Marioni R.E., Colicino E., Peters M.J., Ward-Caviness C.K., Tsai P.-C., Roetker N.S., Just A.C., Demerath E.W., Guan W., Bressler J., Fornage M., Studenski S., Vandiver A.R., Moore A.Z., Tanaka T., Kiel D.P., Liang L., Vokonas P., Schwartz J., Lunetta K.L., Murabito J.M., Bandinelli S., Hernandez D.G., Melzer D., Nalls M., Pilling L.C., Price T.R., Singleton A.B., Gieger C., Holle R., Kretschmer A., Kronenberg F., Kunze S., Linseisen J., Meisinger C., Rathmann W., Waldenberger M., Visscher P.M., Shah S., Wray N.R., McRae A.F., Franco O.H., Hofman A., Uitterlinden A.G., Absher D., Assimes T., Levine M.E., Lu A.T., Tsao P.S., Hou L., Manson J.E., Carty C.L., LaCroix A.Z., Reiner A.P., Spector T.D., Feinberg A.P., Levy D., Baccarelli A., van Meurs J., Bell J.T., Peters A., Deary I.J., Pankow J.S., Ferrucci L., Horvath S. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 2016;8:1844–1865. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gross A.M., Jaeger P.A., Kreisberg J.F., Licon K., Jepsen K.L., Khosroheidari M., Morsey B.M., Swindells S., Shen H., Ng C.T., Flagg K., Chen D., Zhang K., Fox H.S., Ideker T. Methylome-wide analysis of chronic HIV infection reveals five-year increase in biological age and epigenetic targeting of HLA. Mol Cell. 2016;62:157–168. doi: 10.1016/j.molcel.2016.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horvath S., Garagnani P., Bacalini M.G., Pirazzini C., Salvioli S., Gentilini D., Di Blasio A.M., Giuliani C., Tung S., Vinters H.V., Franceschi C. Accelerated epigenetic aging in Down syndrome. Aging Cell. 2015;14:491–495. doi: 10.1111/acel.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maierhofer A., Flunkert J., Oshima J., Martin G.M., Haaf T., Horvath S. Accelerated epigenetic aging in Werner syndrome. Aging (Albany NY) 2017;9:1143–1152. doi: 10.18632/aging.101217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horvath S., Levine A.J. HIV-1 infection accelerates age according to the epigenetic clock. J Infect Dis. 2015;212:1563–1573. doi: 10.1093/infdis/jiv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang T., Tsui B., Kreisberg J.F., Robertson N.A., Gross A.M., Yu M.K., Carter H., Brown-Borg H.M., Adams P.D., Ideker T. Epigenetic aging signatures in mice livers are slowed by dwarfism, calorie restriction and rapamycin treatment. Genome Biol. 2017;18:57. doi: 10.1186/s13059-017-1186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petkovich D.A., Podolskiy D.I., Lobanov A.V., Lee S.-G., Miller R.A., Gladyshev V.N. Using DNA methylation profiling to evaluate biological age and longevity interventions. Cell Metab. 2017;25:954–960.e956. doi: 10.1016/j.cmet.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foulke-Abel J., In J., Yin J., Zachos N.C., Kovbasnjuk O., Estes M.K., de Jonge H., Donowitz M. Human enteroids as a model of upper small intestinal ion transport physiology and pathophysiology. Gastroenterology. 2016;150:638–649.e638. doi: 10.1053/j.gastro.2015.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horvath S., Gurven M., Levine M.E., Trumble B.C., Kaplan H., Allayee H., Ritz B.R., Chen B., Lu A.T., Rickabaugh T.M., Jamieson B.D., Sun D., Li S., Chen W., Quintana-Murci L., Fagny M., Kobor M.S., Tsao P.S., Reiner A.P., Edlefsen K.L., Absher D., Assimes T.L. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17:171. doi: 10.1186/s13059-016-1030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horvath S., Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19:371–384. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- 37.Horvath S., Mah V., Lu A.T., Woo J.S., Choi O.-W., Jasinska A.J., Riancho J.A., Tung S., Coles N.S., Braun J., Vinters H.V., Coles L.S. The cerebellum ages slowly according to the epigenetic clock. Aging (Albany NY) 2015;7:294–306. doi: 10.18632/aging.100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sehl M.E., Henry J.E., Storniolo A.M., Ganz P.A., Horvath S. DNA methylation age is elevated in breast tissue of healthy women. Breast Cancer Res Treat. 2017;164:209–219. doi: 10.1007/s10549-017-4218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Middendorp S., Schneeberger K., Wiegerinck C.L., Mokry M., Akkerman R.D.L., van Wijngaarden S., Clevers H., Nieuwenhuis E.E.S. Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells. 2014;32:1083–1091. doi: 10.1002/stem.1655. [DOI] [PubMed] [Google Scholar]
- 40.Kraiczy J., Ross A.D.B., Forbester J.L., Dougan G., Vallier L., Zilbauer M. Genome-wide epigenetic and transcriptomic characterization of human-induced pluripotent stem cell-derived intestinal epithelial organoids. Cell Mol Gastroenterol Hepatol. 2019;7:285–288. doi: 10.1016/j.jcmgh.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franceschi C., Garagnani P., Parini P., Giuliani C., Santoro A. Inflammaging: a new immune–metabolic viewpoint forage-related diseases. Nat Rev Endocrinol. 2018;14:576–590. doi: 10.1038/s41574-018-0059-4. [DOI] [PubMed] [Google Scholar]
- 42.Lu A.T., Xue L., Salfati E.L., Chen B.H., Ferrucci L., Levy D., Joehanes R., Murabito J.M., Kiel D.P., Tsai P.-C., Yet I., Bell J.T., Mangino M., Tanaka T., McRae A.F., Marioni R.E., Visscher P.M., Wray N.R., Deary I.J., Levine M.E., Quach A., Assimes T., Tsao P.S., Absher D., Stewart J.D., Li Y., Reiner A.P., Hou L., Baccarelli A.A., Whitsel E.A., Aviv A., Cardona A., Day F.R., Wareham N.J., Perry J.R.B., Ong K.K., Raj K., Lunetta K.L., Horvath S. GWAS of epigenetic aging rates in blood reveals a critical role for TERT. Nat Commun. 2018;9:387. doi: 10.1038/s41467-017-02697-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Englander E.W. Gene expression changes reveal patterns of aging in the rat digestive tract. Ageing Res Rev. 2005;4:564–578. doi: 10.1016/j.arr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 44.Steegenga W.T., De wit N.J., Boekschoten M.V., Ijssennagger N., Lute C., Keshtkar S., Bromhaar M.M., Kampman E., de Groot L.C., Muller M. Structural, functional and molecular analysis of the effects of aging in the small intestine and colon of C57BL/6J mice. BMC Med Genomics. 2012;5:38. doi: 10.1186/1755-8794-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cramer J.M., Thompson T., Geskin A., LaFramboise W., Lagasse E. Distinct human stem cell populations in small and large intestine. PLoS One. 2015;10 doi: 10.1371/journal.pone.0118792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du P., Zhang X., Huang C.-C., Jafari N., Kibbe W.A., Hou L., Lin S.M. Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Assenov Y., Müller F., Lutsik P., Walter J., Lengauer T., Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods. 2014;11:1138–1140. doi: 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Z., Niu L., Li L., Taylor J.A. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res. 2016;44:e20. doi: 10.1093/nar/gkv907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teschendorff A.E., Marabita F., Lechner M., Bartlett T., Tegner J., Gomez-Cabrero D., Beck S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 51.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Du P., Kibbe W.A., Lin S.M. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 53.Lin S.M., Du P., Huber W., Kibbe W.A. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36:e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller J.A., Cai C., Langfelder P., Geschwind D.H., Kurian S.M., Salomon D.R., Horvath S. Strategies for aggregating gene expression data: the collapseRows R function. BMC Bioinformatics. 2011;12:322. doi: 10.1186/1471-2105-12-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma'ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.San Roman A.K., Aronson B.E., Krasinski S.D., Shivdasani R.A., Verzi M.P. Transcription factors GATA4 and HNF4A control distinct aspects of intestinal homeostasis in conjunction with transcription factor CDX2. J Biol Chem. 2015;290:1850–1860. doi: 10.1074/jbc.M114.620211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genome-wide data has been deposited in Gene Expression Omnibus (GEO) with the following accession numbers: GSE141254 (DNA methylation) and GSE141255 (expression).
All authors had access to the study data and reviewed and approved the final manuscript.