Abstract
Objective
It is well established that the liver-specific miR-122, a bona fide tumor suppressor, plays a critical role in lipid homeostasis. However, its role, if any, in amino acid metabolism has not been explored. Since glutamine (Gln) is a critical energy and anaplerotic source for mammalian cells, we assessed Gln metabolism in control wild type (WT) mice and miR-122 knockout (KO) mice by stable isotope resolved metabolomics (SIRM) studies.
Methods
Six-to eight-week-old WT and KO mice and 12- to 15-month-old liver tumor-bearing mice were injected with [U–13C5,15N2]-L-Gln, and polar metabolites from the liver tissues were analyzed by nuclear magnetic resonance (NMR) imaging and ion chromatography-mass spectrometry (IC-MS). Gln-metabolism was also assessed in a Gln-dependent hepatocellular carcinoma (HCC) cell line (EC4). Expressions of glutaminases (Gls and Gls2) were analyzed in mouse livers and human primary HCC samples.
Results
The results showed that loss of miR-122 promoted glutaminolysis but suppressed gluconeogenesis in mouse livers as evident from the buildup of 13C- and/or 15N-Glu and decrease in glucose-6-phosphate (G6P) levels, respectively, in KO livers. Enhanced glutaminolysis is consistent with the upregulation of expressions of Gls (kidney-type glutaminase) and Slc1a5, a neutral amino acid transporter in KO livers. Both Gls and Slc1a5 were confirmed as direct miR-122 targets by the respective 3′-UTR-driven luciferase assays. Importantly, expressions of Gls and Slc1a5 as well as glutaminase activity were suppressed in a Gln-dependent HCC (EC4) cell line transfected with miR-122 mimic that resulted in decreased 13C-Gln, 13C-á-ketoglutarate, 13C-isocitrate, and 13C-citrate levels. In contrast, 13C-phosphoenolpyruvate and 13C-G6P levels were elevated in cells expressing ectopic miR-122, suggesting enhanced gluconeogenesis. Finally, The Cancer Genome Atlas—Liver Hepatocellular Carcinoma (TCGA-LIHC) database analysis showed that expression of GLS is negatively correlated with miR-122 in primary human HCCs, and the upregulation of GLS RNA is associated with higher tumor grade. More importantly, patients with higher expressions of GLS or SLC1A5 in tumors exhibited poor survival compared with those expressing lower levels of these proteins.
Conclusions
Collectively, these results show that miR-122 modulates Gln metabolism both in vitro and in vivo, implicating the therapeutic potential of miR-122 in HCCs that exhibit relatively high GLS levels.
Keywords: Glutaminase, Glutaminolysis, Gluconeogenesis, Krebs cycle, miRNA-122, miR-122, SIRM, Slc1a5
Abbreviations: HCC, hepatocellular carcinoma; Gln, glutamine; Glu, glutamate; Glc, glucose; KO, germ-line knockout; LKO, liver-specific knockout; HSQC, heteronuclear single quantum coherence spectroscopy; 3′UTR, 3′-untranslated region; SIRM, stable isotope resolved metabolomics; TCGA-LIHC, The Cancer Genome Atlas—Liver and Hepatocellular Cancer; WT, wild type.
Highlights
-
•
miR-122, the most abundant liver specific microRNA and a potent tumor suppressor, regulates glutamine metabolism.
-
•
SIRM analysis showed enhanced glutaminolysis and impeded gluconeogenesis in the livers of miR-122 KO mice.
-
•
Gls, a key enzyme involved in glutaminolysis and a miR-122 target is upregulated in miR-122 KO livers.
-
•
Ectopic miR-122 expression suppressed glutaminolysis but enhanced gluconeogenesis in a glutamine dependent HCC cell line.
-
•
Expression of MIR-122 negatively correlated with that of GLS in human primary HCCs.
1. Introduction
miR-122 is an abundant conserved liver-specific multi-functional microRNA [1,2]. This multi-functional microRNA regulates hepatitis C virus replication [3,4], circadian rhythm [5], liver polyploidy [6], and detoxification of xenobiotics such as acetaminophen [7]. Circulating miR-122 has been established as a sensitive and specific biomarker for liver injury in mice and humans [8,9]. Our studies with liver-specific (LKO) and germ-line knockout (KO) mice have shown that miR-122 is essential for liver metabolic homeostasis and maintenance of hepatocyte differentiation state, and its loss of function causes development of nonalcoholic steatohepatitis [10]. miR-122 expression is downregulated in hepatocellular carcinoma (HCC) cell lines, and its downregulation in primary HCCs correlates with poor prognosis and metastasis [11]. The development of spontaneous liver tumors in miR-122 LKO and KO mice confirmed its role as a tumor suppressor [10,12]. To understand the underlying molecular mechanism of action of this critical microRNA, we have identified its targets in mouse and human liver transcriptomes using Argonaute-HITS-CLIP analysis [13]. We found that most of the miR-122 targets are located in the 3′-UTRs and coding regions.
It is now well established that mammalian cells use glutamine (Gln) as an alternative energy source of glucose for adenosine triphosphate (ATP) production and as an anaplerotic source for biomass generation [14]. Gln and its metabolites support several crucial cellular pathways including the tricarboxylic acid (TCA) cycle, redox balance, mTOR activation, and biosynthesis of nucleotides, amino acids, fatty acids, and aminosugars [15,16]. As such, Gln, the most abundant circulating amino acid in human plasma, supports inexorable growth of cancer cells [14]. Gln deprivation induces apoptosis in rapidly proliferating Gln-dependent cells such as intestinal mucosal cells and many cancer cells [17].
Major factors for Gln utilization include SLC1A5, a Gln transporter for uptaking Gln into the cells, and the mitochondrial glutaminase (GLS), the gatekeeper enzyme of glutaminolysis. Upregulation of SLC1A5 expression has been observed in many neoplasms and is associated with poor patient survival [18,19]. Mitochondrial GLS catabolizes Gln to glutamate and ammonia. Glutamate is then converted to α-ketoglutarate by glutamate dehydrogenase (GDH) or transaminases, followed by oxidation via the TCA cycle [16]. Two forms of glutaminase encoded by different genes are identified, which are broadly expressed kidney-type GLS and the liver-type glutaminase, GLS2, which is specifically expressed in the liver, pancreas, and brain [20]. GLS2 has been reported to suppress or promote tumor, depending on the tumor type [21]. In contrast, GLS expression is frequently upregulated in many cancer types [14,22]. Many cancer cells switch from GLS2 to GLS with advanced pathological states [23]. Currently, CB-839, a GLS inhibitor, is undergoing clinical trials for multiple cancer types [24].
The role of miR-122 in cholesterol and triglyceride metabolism is well-documented [25]. However, the role of this potent tumor suppressor in the liver metabolism of amino acids such as Gln is unknown. In this study we report that miR-122 modulates Gln metabolism in the liver and tumors by regulating the expression of Gls and Slc1a5.
2. Materials and methods
2.1. Animal models
miR122 fl/fl (control), miR-122 fl/fl; Alb-Cre (liver-specific knockout or LKO), and miR-122−/− (KO) mice were previously generated in our laboratory [10]. All animals were housed in a temperature-controlled room under a 12-hour light/dark cycle and under pathogen-free conditions. All animal studies were reviewed and approved by the Ohio State University Institutional Laboratory Animal Care and Use Committee.
2.2. Human tissue samples (HCC and matching liver tissue)
De-identified tissue specimens (HCC and benign adjacent liver) were procured from the Human Co-operative Tissue Network and stored at −80 °C until use. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of The Ohio State University institutional Review Board (Study ID-2004C0081).
2.3. Cell line and transfection
Gln-dependent mouse HCC cell line (EC4) cells were obtained from Dr. Dean Felsher and cultured in Dulbecco Modified Eagle Medium (DMEM) (Corning) supplemented with 10% fetal bovine serum (FBS, Sigma) and 1% penicillin-streptomycin (Corning). For SIRM analysis, these cells were cultured in glutamine (Gln)-free DMEM (Thermo Fisher Scientific, catalogue # A1443001), 10% dialyzed FBS (cat# 26-400-044, GIBCO), 1% penicillin-streptomycin, and 3 mM [U–13C,15N]-Gln (Cambridge Isotope Laboratories). For ectopic miR-122 expression, these cells were transfected with 50 nM of miR-122 mimic (Dharmacon Catalogue# C-300591-05) or negative control mimic (Dharmacon, Catalogue# CN-001000-01-50) using RNAimax (Invitrogen) following the manufacturer's protocol. Hepa1-6 cell line obtained from Dr. Gretchen Darlington was grown in the same media containing regular Gln.
2.4. Stable isotopes resolve metabolomics studies in mice and EC4 cell line
miR-122 LKO or KO mice and age-matched control mice (6–8 weeks old) were injected with [U–13C5,15N2]-Gln [7 mg in 0.1 mL of phosphate-buffered saline (PBS)] through the tail vein three times as described in [17] and euthanized 15 min after the last injection. KO mice bearing tumors (∼12 months old) were injected with the Gln tracer as just described, and microscopic tumors and benign livers from the same mouse were snap-frozen and pulverized; the metabolites were extracted and subjected to NMR analysis.
To delineate the role of miR-122 in Gln metabolism, Gln-dependent mouse hepatoma (EC4) cells were transfected with 50 nM of miR-122 mimic (Dharmacon Catalogue# C-300591-05) or control miR mimic (Dharmacon, Catalogue# CN-001000-01-50). After 24 h, cells were incubated with Gln-free DMEM supplemented with Gln, 10% dialyzed FBS, 1% penicillin-streptomycin, and 3 mM [U–13C5,15N2]-Gln. The tracer media were collected at different times and flash frozen for subsequent de-proteinization using 80% acetone and analysis by NMR [26]. Cells were harvested after 72 h, washed with cold PBS, quenched with acetonitrile:H2O (2:1.5), then extracted as described following.
Polar metabolites were extracted from pulverized mouse liver and tumor tissues as well as EC4 cells by the solvent partitioning method with a final CH3CN:H2O:CHCl3 (2:1.5:1, v/v) ratio, as described previously [27]. The polar extracts were lyophilized before reconstitution in D2O for NMR analysis or in H2O for ion chromatography-Fourier transform mass spectrometry (IC-FTMS) analysis.
Polar extracts for ion chromatography-mass spectrometry (IC-MS) were reconstituted in 20 μL ultrapure deionized water (EMD Millipore), of which 10 μL was injected for IC-MS as previously described [28]. All analyses were performed on a Dionex ICS-5000+ ion chromatography interfaced to a Thermo Fusion Orbitrap Tribrid mass spectrometer (Thermo Fisher Scientific). IC-FTMS was performed using an IonPac AS11-HC-4 μm RFIC&HPIC (2 × 250 mm) column and an IonPac AG11-HC-4 μm guard column (2 × 50 mm) [28]. The column flow rate was kept at 0.38 mL/min with column temperature at 35 °C and 0.06 mL/min methanol added post column as a makeup solvent to aid vaporization in the heated electrospray ionization (HESI) unit. The HESI vaporizer temperature was 400 °C with the sheath gas set at 35 (arbitrary units) and the auxiliary nitrogen flow set at 4 (arbitrary units). The column was initially equilibrated for 8 min with 1 mM KOH, followed by 1 mM KOH for 2 min after 10 μL of sample was injected. The KOH gradient program used to elute samples included ramping up to 40 mM KOH from 2 to 25 min, and to 100 mM from 25 to 39.1 min, at 100 mM to 50 min, and ramping down to 1 mM KOH at 50.1 min and at 1 mM KOH to 52.5 min. KOH suppression was achieved with a Dionex AERS 500 2-mm suppressor with an external AXP pump supplying regenerant at a flow rate of 0.75 mL/min and injected into the Orbitrap mass spectrometer via HESI. Mass spectra were recorded at a resolution of 450,000 (achieving a resolution of ∼360,000 at 400 m/z) from 80 to 700 m/z mass scan range, with detection in the negative ion mode voltage using the following settings: HESI = 2800 V; ion transfer tube temperature = 300 °C; automatic gain control (AGC) = 2 × 105; maximal injection time = 100 millisec. Peak areas were integrated and exported to Excel via the Thermo TraceFinder (version 3.3) software package. Peak areas were corrected for natural abundance as previously described [29].
Fractional enrichment (fraction) was calculated as the fraction of the natural abundance corrected signal of each isotopologue normalized to the sum of all isotopologues for each metabolite and averaged across all replicates. The metabolites were quantified using either natural abundance corrected peak intensities (Ion intensity/mg protein/104) or the total amount of metabolite (μmol/g protein); the latter was achieved by reference to an external standard mixture.
Polar extracts for NMR were dissolved in D2O containing 27.5 nmol DSS, which served both as an internal chemical shift reference and as a concentration standard. NMR spectra were recorded in 1.7 mm NMR tubes at 14.1 T/288 K with a 3-mm inverse triple resonance (HCN) cold probe on an Agilent DD2 spectrometer using automation. 1D 1H PRESAT NMR spectra were recorded with an acquisition time of 2 sec and a relaxation delay of 3 sec during which the residual HOD resonance was suppressed using a weak transmitter pulse. 1D 1H-{13C} HSQC NMR spectra were acquired with an acquisition time of 0.2 sec and a relaxation delay of 1.8 s, using adiabatic 13C decoupling during the acquisition time; 4096 transients were co-added. Free induction decays were processed using MNOVA. All NMR spectra were zero filled and apodized using a Gaussian window function, for PRESAT spectra with 0.5 Hz and for HSQC spectra with 4 Hz line broadening, baseline corrected using a simple polynomial, phased, and referenced to internal DSS (0 ppm). The peak intensities were calibrated by the DSS peak and then normalized by total mass of protein. 13C-enriched compounds were identified by comparison to in-house databases.
1D proton spectra were recorded using PRESAT with an acquisition time of 2 sec and a relaxation delay of 4 s, during which the residual water signal was saturated. Spectra were apodized using a 1 Hz line broadening exponential, baseline corrected, phased, and referenced to internal DSS. Peaks were deconvoluted using MNOVA or integrated by manual line fitting. Isotopomer distributions were determined as the fraction 13C according to Equation (1) [30]:
Spectra were paired by treatment (e.g., WT vs. KO or miR-122 mimic vs. negative control mimic) and overlaid for comparison.
2.5. RNA isolation and real-time quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Total RNA was isolated using TRIzol (Life technologies, #15596018) followed by DNase I treatment (NEB, M0303). For gene expression assay, DNase I-treated total RNA was reverse transcribed into cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems), and qRT-PCR was performed using SYBR Green chemistry. The expression was normalized to that of 18S rRNA. All real-time reactions, including controls without cDNA, were run in triplicate in a thermocycler. Relative expression was calculated using the comparative CT method.
Primer sequences for qRT-PCR assay
| Hsa-GLS-RTF | 5′-TGACTTGTGAATCAGCCAGTGTG |
| Hsa-GLS-RTR | 5′-TGTTGCCCATCTTATCCAGAGG |
| Mmu-Gls-RTF | 5′-GGATGAAGCACTACACTTTGGACAC |
| Mmu-Gls-RTR | 5′-CGAGATTCTTGTGGACAGTCTGG |
| Mmu-Slc1a5-RTF | 5′-CCATCACCTCCATCAACGACTC |
| Mmu-Slc1a5-RTR | 5′-AAGGCAGCAGACACCAGATTGG |
| mPck2-RTF | 5′-CAGGAGCCACACCTGTTATT |
| mPck2-RTR | 5′-GATCAGGAGGCCAAGGTTATC |
| mFbp2-RTF | 5′-ACTGCCATCAAAGCCATCTC |
| mFbp2-RTR | 5′-CACCTCATCTCCTGTCACATTC |
| 18S rRNA-RTF | 5′-GTAACCCGTTGAACCCCATT |
| 18S rRNA-RTR | 5′-CCATCCAATCGGTAGTAGCG |
2.6. Western blot analysis
Cell or tissue lysates prepared in lysis buffer (Cell Signaling Technology) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 10% or 4%–12% acrylamide gel from BioRad), transferred onto nitrocellulose membranes, and probed with primary antibodies specific for GLS (abcam # ab93434), vinculin (Proteintech # 66305-1-Ig), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Proteintech, #60004-I-Ig). Proteins of interest were detected by IRDye secondary antibodies (LICOR) and visualized by Odyssey CLx Imaging System (LICOR), and signals were quantified by Image Studio Software (Ver. 5.2).
2.7. Generation of luciferase reporter plasmids and luciferase reporter assay
3′-UTRs of Gls and Slc1a5 harboring the miR-122 binding site were amplified from mouse liver DNA with specific primers and cloned into downstream of Renilla luciferase CDS in psi-CHECK2 (Promega) dual luciferase reporter vector (Promega) at the Xho I and Not I sites in the multiple cloning site (MCS). Deletion or point mutant of individual miR-122 binding sites was generated by PCR using specific primers as described earlier [13]. psi-CHECK2 plasmids (50 ng DNA) harboring miR-122 target 3′UTRs [wild type (WT) or mutants] or empty vector were then co-transfected with miR-122 or scrambled (NC) RNA mimics (25 nM) into EC4 (for Gls) or Hepa1-6 (for Slc1a5) cells. Relative luciferase [Renilla to Firefly (internal control)] activity was measured after 48 h per the manufacturer's protocol (Promega).
Primer sequences for cloning into psi-CHECK2 vector
| mmu-Gls-3′-UTR-F: 5′ CCGCTCGAGTTAGCTTTAGTTTACACATA (Forward primer with XhoI site) |
| mmu-Gls-3′-UTR-R: 5′ ATAAGAATGCGGCCGCTCTTTTTCTGAGCAACTAAC (Reverse oligo with Not I site) |
| mmu-Gls-Del-F: 5′ATCTCGAGATGCTTCTGTTTATTTTTAAA (Forward primer for deletion of miR122 binding site in Gls 3′-UTR) |
| mmu-Gls-Del-R: 5′ ATGCGGCCGCCCTTTTCTTAATATCAGAGTT (Reverse primer for deletion of miR122 binding site) |
| mmu-Slc1a5-F: 5′ AGC TTT GTT TAA ACT GGA GAA ATG GAC TGG ATG A (Forward primer with PMEI site) |
| mmu-Slc1a5-R: 5′ ATA AGA ATG CGG CCG CTT TGA CAC CAT TTT ATT GCT (Reverse primer with Not I RE site) |
| mmu-Slc1a5-Del1: 5′ AGC TTT GTT TAA ACG GAA TCT AGG GGT CGG TGT C (Forward primer for deletion of site 1) |
| mmu-Slc1a5-Mut2-F: 5′ ATG ACA GAA ATG TGA GGC ATG AGT CTC G (Forward primer for mutation of site 2) |
| mmu-Slc1a5-Mut2-R: 5′ AGC TTT GTT TAA ACT GGA GAA ATG GAC TGG ATG A (Reverse primer for mutation of site 2) |
| mmu-Slc1a5-Del3: 5′ ATA AGA ATG CGG CCG CAC ATT TAT TCT TGG CTT TCC (Reverse primer for deletion of site 3) |
2.8. Preparation of mitochondrial extract and glutaminase activity assay
Glutaminase activity was analyzed following a published protocol [31]. EC4 cells were seeded in six-well plates in triplicate and after 24 h were transfected with miR-122 mimic and negative control mimic (50 nM). The following day, cells were washed twice with ice-cold PBS and harvested using a cell scraper. Then, cells were collected by centrifugation at 700×g for 5 min at 4 °C. The pellet was resuspended in the homogenization buffer containing 200 mM sucrose, 10 mM Tris/MOPS (pH 7.4), and 1 mM EGTA/Tris and homogenized in a dounce homogenizer. The homogenate was passed through a 3-mL syringe using an 18-gauge 1½-inch needle and expelled back into the conical tube on ice with a 27-gauge ½-inch needle. These steps were repeated five times. The cellular extract was centrifuged for 5 min at 600×g at 4 °C. The supernatant was centrifuged at 10,000×g, 4 °C for 5 min. The mitochondrial pellet was resuspended in the homogenization buffer and placed on ice, and the protein concentration was determined using the BCA method (Pierce).
Glutaminase activity was measured by a two-step method. First, Gln was hydrolyzed to glutamate and ammonia by glutaminase. Next, glutamate dehydrogenase was used to catalyze oxidative deamination of glutamate to form α-ketoglutarate and reduced nicotinamide adenine dinucleotide (NADH). To completely oxidize the glutamate, α-ketoglutarate is removed by adding hydrazine as it is formed. Typically, extracts containing glutaminase (25–30 ug protein) were added to the initial assay mix (20 mM Gln, 0.15 M KH2PO4, 0.2 mM ethylenediamine tetraacetic acid (EDTA), and 50 mM Tris–acetate, pH 8.6), and the mixture was incubated at 37 °C for 1 h. The reaction was stopped by adding 10 μL of 3N HCl to inactivate the glutaminase followed by the addition of the reaction mix containing 6.3 U/mL of purified bovine liver glutamate dehydrogenase, 0.08 M Tris–acetate (pH 9.4), 0.2 M hydrazine, 0.25 mM ADP, and 2 mM NAD+. The samples were mixed and incubated at room temperature for 45 min, and the absorbance was recorded at 340 nm against water blank. Relative activity in miR-122 mimic transfected cells relative to NC mimic transfected cells were determined.
| GLS activity = μmol/L/min = (O.D. × 1000 × Total assay volume in μL) ÷ (Time in min × Molar extinction coefficient in mM × volume of sample used in assay) |
2.9. Analysis of TCGA-LIHC data
TCGA-LIHC data was downloaded using Xena (https://xena.ucsc.edu/). The association between gene expression and patient overall survival (OS) was tested with Cox proportional hazard regression models. The Mann–Whitney U test was used to test the difference of expression levels of miR-122 target genes in tumors and adjacent normal tissues. Spearman correlation analysis was used to test the correlation between miR-122 and its target genes (Gls, Slc1a5). The association between gene expression and patient OS was tested using the Cox proportional hazard regression model using gene expression as continuous variables. To display the relationship between gene expression and OS, the gene expression was dichotomized to “high” and “low” expression based on their median expression, and then the log-rank test was used to generate the Kaplan–Meier survival curves; the p-values on the plots were from the Cox proportional hazard tests to keep all the information from the gene expression.
2.10. Statistical analysis
qRT-PCR and transfection analysis were performed in triplicate. The data were presented as means ± standard deviations (SDs). The majority of the experiments were repeated at least twice with multiple replicates. Statistical significance was calculated using a Student t test if there were at least three replicates, which were corrected for false discovery rate (FDR) to obtain Q values when appropriate. A P or Q value of ≤0.05 was considered significant.
2.11. Software
BioVinci - Version: 1.1.5, r20181005 - Data visualization, analytics.
2.12. Data availability
The datasets generated during and/or analyzed during the current study (TCGA-LIHC data) are available in the Xena Website (https://xena.ucsc.edu/).
3. Results
3.1. NMR analysis identifies enhanced glutaminolysis in miR-122 KO livers
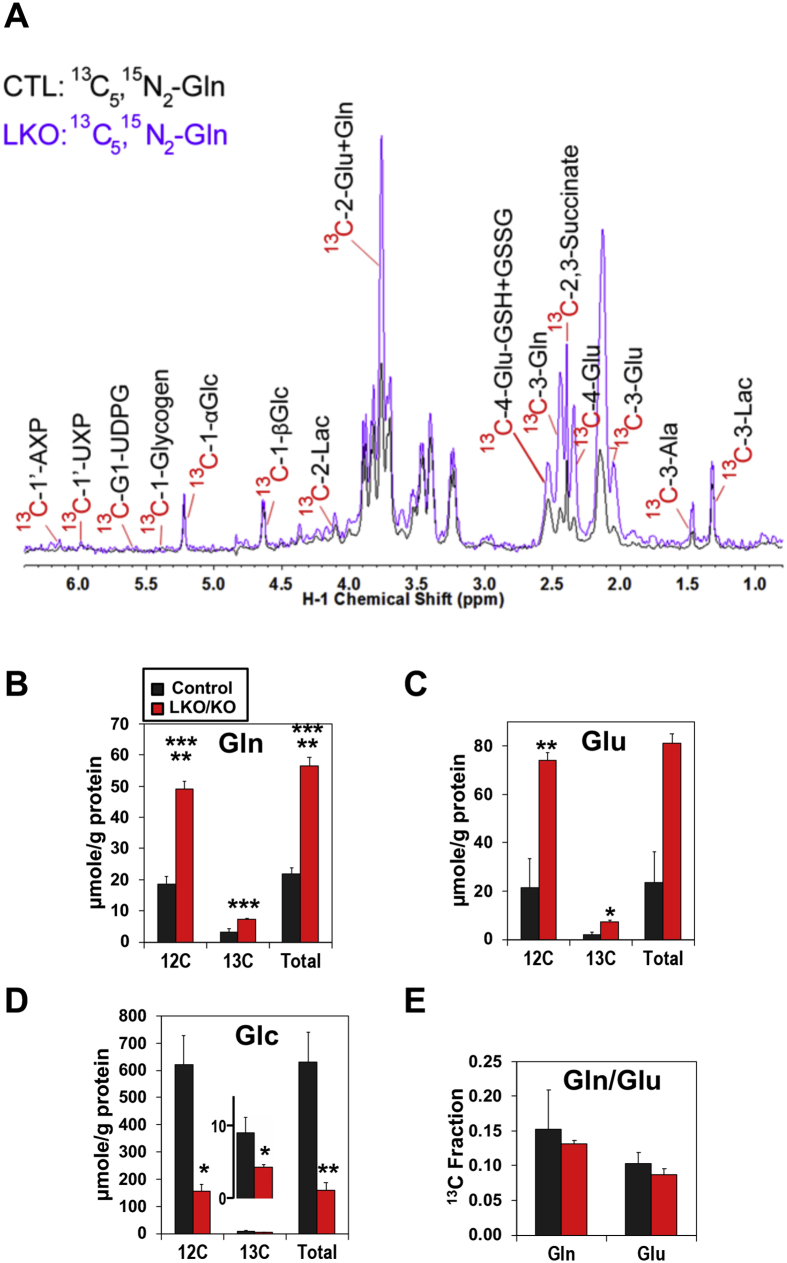
To elucidate the role of miR122 in hepatic Gln metabolism, 6- to 8-week-old LKO (Mir122fl/fl, Alb-Cre) or KO (Mir122−/−) mice and age-matched control (Mir122fl/fl) mice were injected with 13C5,15N2-Gln, and polar metabolites extracted from the livers were subjected to NMR analysis (see methods for details). To avoid confusion, we abbreviated liver-specific and germline miR-122-depleted mice as KO mice throughout the text, as identical results were obtained with both. Analysis by both 1D 1H and 1D 1H{13C} HSQC NMR enabled comparison of 13C-labeled metabolites derived from [13C5,15N2]-Gln via different metabolic pathways in the liver extracts of WT and KO mice (representative data shown in Figure 1). These included 13C-lactate, -alanine (Gls + malic enzyme (ME)-mediated Krebs cycle); 13C-succinate, -glutamate, (Gls + Krebs cycle), 13C-GSH/GSSG (Gls + Krebs cycle + glutathione synthesis), 13C-glucose (Gls + Krebs cycle + gluconeogenesis), 13C-glycogen (Gls + Krebs cycle + gluconeogenesis + glycogen synthesis), 13C-glycine (Gls + Krebs cycle + gluconeogenesis + one-carbon metabolism), and 13C-ribose residue of adenine (AXP), guanine (GXP), and uracil (UXP) nucleotides as well as UDP-glucose (UDPG) (Gls + Krebs cycle + gluconeogenesis + pentose phosphate pathway) (see Figure 5 for scheme).
Figure 1.
Figure 5.
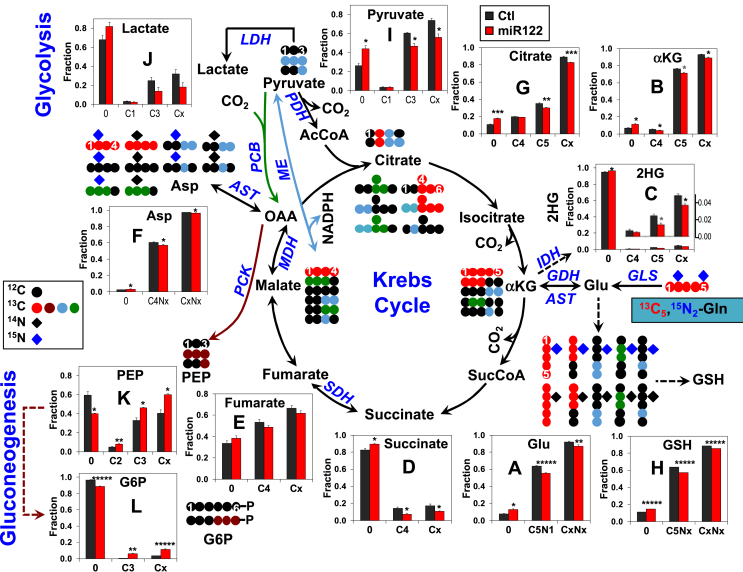
Fractional enrichment analysis by IC-MS showed blockade of glutaminolysis and enhanced gluconeogenesis by ectopic expression of miR-122 in a Gln-dependent HCC (EC4) cell line. Growing EC4 Cells were transfected with miR-122 mimic (miR122) or NC mimic (Ctl) (50 nM) for 24 h. Medium was then replaced with Gln-, glucose-, and pyruvate-free DMEM containing 13C5,15N2-Gln and dialyzed FBS and cells were cultured for another 24 h and harvested. The polar metabolite was extracted and subjected to IC-MS analysis. Fractional enrichment (fraction) was calculated as the fraction of the natural abundance corrected signal of each isotopologue divided by the summed signals of all isotopologues for each metabolite and averaged across all replicates. Fractional enrichment was reduced for 13C and/or 15N labeled glutamate (A), α-ketoglutarate (B), 2-hydroxyglutarate (2HG), (C), succinate (D), fumarate (E), aspartate (F), pyruvate (I), lactate (J), citrate (G), and glutathione (GSH), (H) and increased for 13C labeled phosphoenolpyruvate (PEP, K) and glucose-6-phosphate (G6P, L) in miR-122 mimic transfected cells versus negative control mimic transfected cells. Cx and CxNx refer to total fractions of labeled products. * (gray), *, **, ***, and ***** denote Q values of <0.1, 0.05, 0.01, 0.005, and 0.0005, respectively, with n = 3.
We noted that the levels of both 13C-glutamate (Glu) and -Gln were higher in the livers of KO mice compared with the control mice (Figure 1B, C, D) with no significant changes in the fractional enrichment (Figure 1E). These results suggest enhanced glutaminolysis in miR-22-depleted livers. We also noted that existing unlabeled (12C)-Gln and-Glu levels were much higher in KO than control livers (Figure 1B, C). Moreover, decreased levels of 13C- and total glucose (Glc, Figure 1D) were noted in KO mice compared with the control mice, which correlated with decreased blood glucose in miR-122-depleted mice (data not shown). These data indicate enhanced hepatic glutaminolysis and inefficient conversion of Gln to glucose in KO livers. Compared with the adjacent benign livers, analysis of Gln metabolism in liver tumors developed spontaneously in KO mice after 12 months showed that 13C-Gln to -Glu conversion was further enhanced in some tumors (data from one tumor-bearing mouse shown in Figure S1), suggesting enhancement of glutaminolysis in HCCs.
3.2. Ion chromatography mass spectrometry (IC-MS) analysis shows enhanced conversion of Gln to Krebs cycle metabolites in the liver of miR-122 KO mice
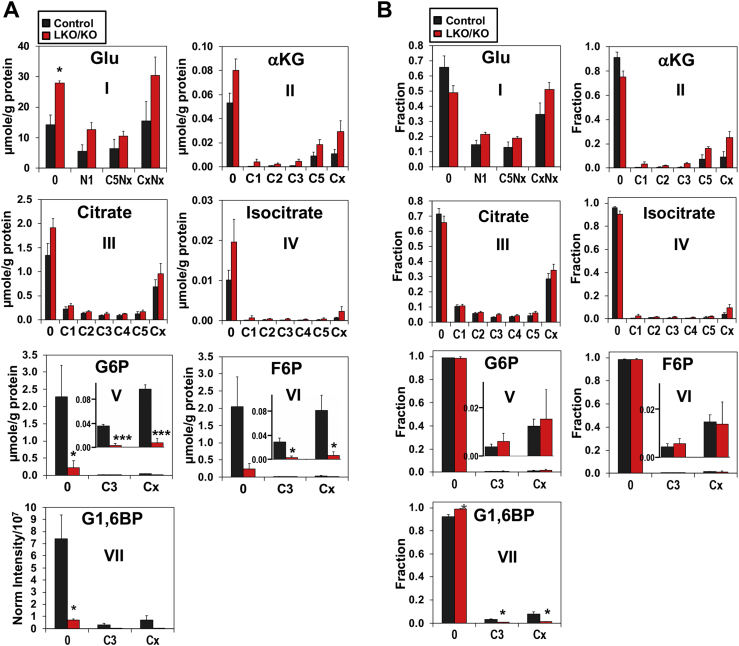
We next quantified polar metabolites extracted from liver tissues by IC-MS, which showed the level (Figure 2A) and fractional distribution (Figure 2B) of 13C- and/or 15N-isotopologues of Glu, á-ketoglutarate, isocitrate, and citrate in both KO and control livers. The 13C-labeling patterns of these metabolites are consistent with [13C5,15N2]-Gln metabolism via glutaminolysis and the Krebs cycle. For example, [13C5, Nx]-Glu (C5Nx, A-I) and [13C5]-á-ketoglutarate (C5, A-II) represented the first and second products of glutaminolysis, whereas 13C4-citrate (C4, A-III) was the subsequent product of the Krebs cycle (see Figure 5 for scheme). We also noted a significant presence of [13C1]-citrate (C1, A-III), -isocitrate (C1, A-IV), and -á-ketoglutarate (C1, A-II), which are presumably derived from the ME-mediated Krebs cycle reactions (see Figure 5 for scheme). Moreover, [15N1]-Glu was a product of transamination via aspartate aminotransferase (AST) and/or oxidation via glutamate dehydrogenase (GDH). The abundance of this Glu isotopologue (A-I) in KO livers indicates high activity of one or both enzymes. When comparing KO to control livers, elevated levels (Figure 2A) and fractional enrichment (Figure 2B) of many of the labeled products and the summed total (Cx, CxNx) of Glu, á-ketoglutarate, citrate, and isocitrate were evident, which is consistent with enhanced glutaminolysis plus the Krebs cycle in miR-122-depleted livers, based on Equation (2) (cf. Figure 1 legend).
Figure 2.
Ion chromatography-mass spectrometry (IC-MS) analysis of glutamate and other Krebs cycle metabolites derived from labeled Gln showed enhanced glutaminolysis but inhibition of gluconeogenesis in the livers of miR-122 KO mice. The polar extracts from the livers were analyzed by IC-MS. Elevated levels (A) and fractional distribution (B) of selected and total labeled isotopologues (Cx or CxNx; x ≥ 1) of (I) Glu, (II) α-ketoglutarate, (III) citrate, and (IV) isocitrate were evident in miR-122 KO mouse livers compared with control mouse livers. Also noted were the decreased levels with little changes in the fractional enrichment of 13C3- (C3) and total labeled isotopologues (Cx) of glucose-6-phosphate (G6P) (V) and frutose-6-phosphate (F6P) (VI) in miR122 LKO/KO mouse livers compared with control mouse livers. * (gray), *, and *** denote Q values of ≤0.1, 0.05, and 0.005, respectively, with n = 3–4.
In contrast, we found much reduced levels (Figure 2A–V,VI) of 13C3- and total 13C labeled isotopologues of G6P and F6P in KO liver with little changes in fractional enrichment (Figure 2B-V-VI), which points to inefficient gluconeogenesis fueled by both labeled Gln and unlabeled sources according to Equation (2). It should be noted that although many of the changes in 13C- and/or 15N- labeling of individual metabolites did not reach statistical significance, the same trend was observed for all metabolites in the same pathway pointing to the altered pathway.
3.3. miR-122 regulates expressions of Gls and Slc1a5
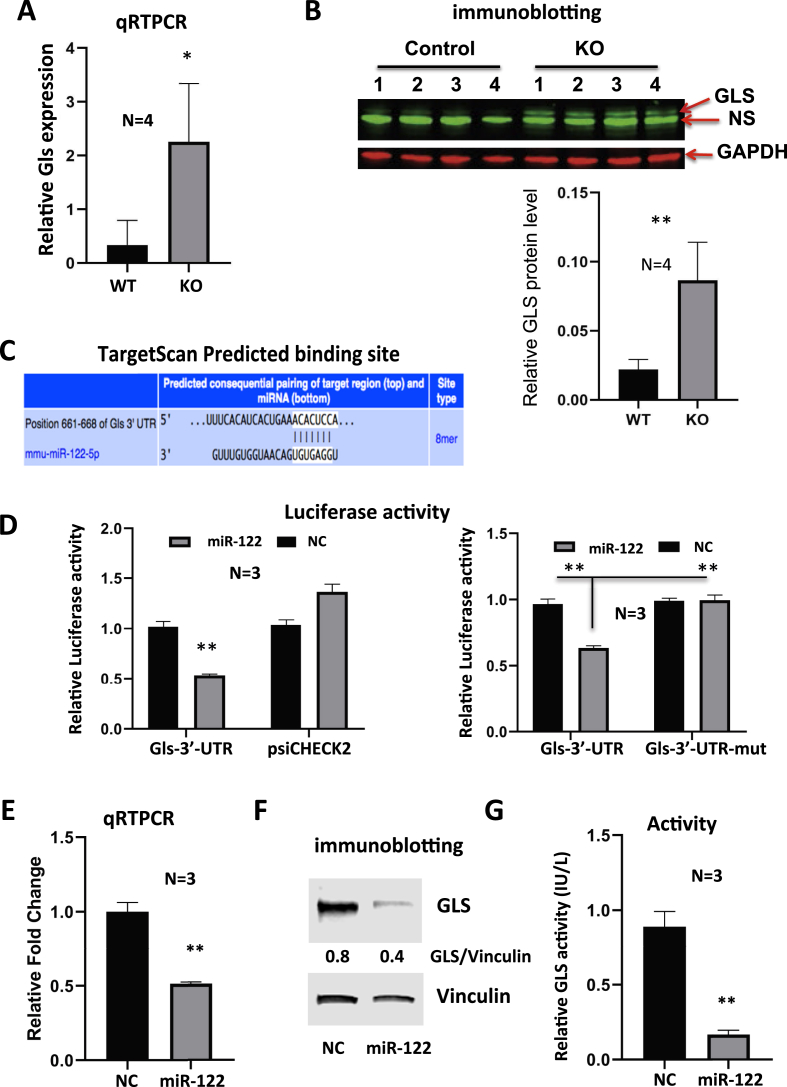
To understand why glutaminolysis is enhanced in miR-122 KO mice, we next determined if the expression of glutaminase is regulated by miR-122. Indeed, qRT-PCR and immunoblot analyses showed both RNA and protein levels of Gls, the kidney-type glutaminase was significantly elevated in the livers of miR-122 KO mice compared with the WT mice (Figure 3A, B), suggesting its role in activating Gln metabolism in miR-122-depleted livers. Next, we sought to determine whether Gls is a direct miR-122 target. Our recent AGO-CLIP analysis [13] has identified an 8 mer miR-122 seed match in the Gls 3′-UTR, which is also predicted by TargetScan (Figure 3C, upper panel). We, therefore, cloned the 3′-UTR region of Gls spanning or depleted of this site into psi-CHECK2 vector downstream of renilla luciferase coding sequence (CDS) and transfected Hepa cells with each of these vectors along with miR-122 or negative control (NC) mimic. Relative luciferase activity (renilla to firefly) measured 48 h post-transfection was significantly inhibited in cells expressing psi-CHECK2 harboring Gls-3′-UTR-WT miR-122 site co-transfected with miR-122 mimic compared with those co-transfected with NC mimic control (Figure 3D). This inhibition was reversed in cells co-transfected with psi-CHECK2 harboring Gls-3′UTR lacking miR122 cognate site (Figure 3D). As expected, miR-122 NC mimic had no effect on luciferase activity in psi-CHECK2 transfected cells. These data validated Gls as a miR-122 target. We also found significant suppression of Gls RNA and protein levels (Figure 3E, F) as well as glutaminase activity in EC4 cells expressing ectopic miR-122 (Figure 3G). These results confirmed suppression of Gls activity by direct targeting of miR-122.
Figure 3.
miR-122 suppresses Gls expression by direct targeting.A, B. qRT-PCR analysis of Gls mRNA (A) and immunoblot analysis of GLS protein (B) in miR122 KO and control mice. NS denotes nonspecific band. C. TargetScan predicted miR-122 seed match in mouse Gls 3′-UTR. D. Luciferase assays in EC4 cells co-transfected with psi-CHECK2 vector harboring the wild type (WT) or mutant Gls-3′UTR deleted of miR-122 cognate site and miR-122 mimic or NC mimic (negative control). E, F. qRT-PCR (E) and immunoblot (F) analyses of Gls after ectopic expression of miR-122 mimic or NC mimic in EC4 cells. G. Glutaminase activity assay in the mitochondrial extract of EC4 cells transfected with miR-122 mimic or NC mimic (See methods for details). * and **denote P values of ≤0.05 and 0.01, respectively (n = 3–4).
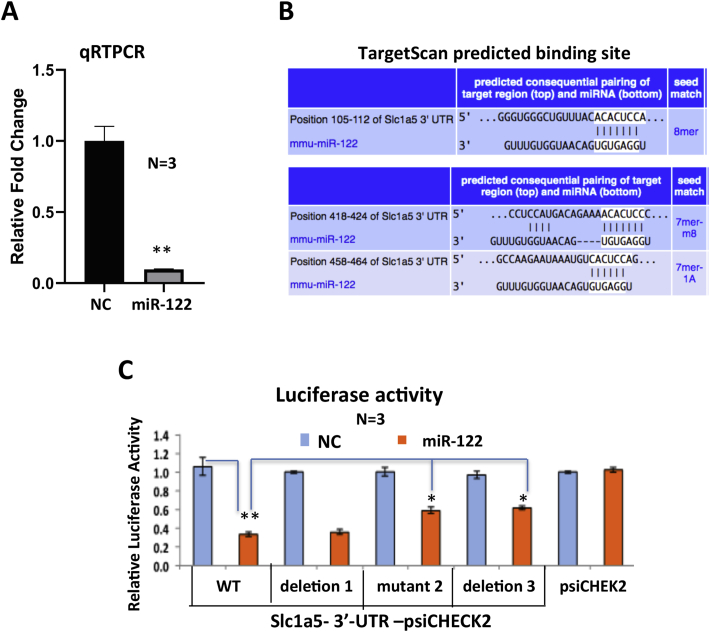
In addition, microarray data (GSE20610) [10] showed significant increase in expression of the Gln transporter, Slc1a5, in miR-122 KO livers compared with the WTs. Indeed, endogenous Slc1a5 expression in EC4 cells was suppressed by ectopic miR-122 expression (Figure 4A). As TargetScan predicted three miR-122 binding sites on 3′-UTR of Slc1a5 (Figure 4B), we next queried whether Slc1a5 is a direct miR-122 target by luciferase assay in Hepa cells transfected with psi-CHECK2 vector harboring WT or mutants of each of the three predicted miR-122 binding sites. Relative luciferase activity was inhibited in cells co-transfected with psi-CHECK2-Slc1a5-3′UTR-WT and miR-122 mimic compared with those co-transfected with the NC mimic (Figure 4C), suggesting that Slc1a5 is a miR-122 target. Among three cognate sites, mutation of site 2 or deletion of site 3 could partially rescue luciferase activity from miR-122 inhibition, whereas deletion of site 1 had no effect (Figure 4C). These results indicate that sites 2 and 3 contribute to miR-122-mediated inhibition of Slc1a5 expression. Collectively, these results indicate that miR-122 inhibits two key factors involved in Gln metabolism in hepatic cells, namely, Gls and Slc1a5.
Figure 4.
miR-122 represses Slc1a5 expression by direct targeting.A. qRT-PCR analysis of Slc1a5 mRNA expression after miR-122 mimic or NC mimic expression in HCC cells. B. miR-122 seed matches in Slc1a5 predicted by TargetScan. C. Validation of two of three sites in Slc1a5-3′-UTR as miR122 targets by luciferase assay driven by wild type (WT) and mutant Slc1a5 3′-UTR co-transfected with miR-122 mimic or NC mimic. * and **denote P values of ≤0.05 and 0.01, respectively (n = 3–4).
3.4. Ectopic miR-122 expression blocks glutaminolysis and enhances gluconeogenesis in a Gln-dependent HCC cell line
Stable isotope resolved metabolomics (SIRM) data showed that Gln catabolism was enhanced in miR-122-depleted livers in vivo (Figure 1, Figure 2). To verify if miR-122 indeed regulates Gln metabolism, Gln-dependent mouse EC4 cells were transfected with miR-122 mimic or NC mimic followed by incubation in media containing [13C5,15N2]-Gln and IC-MS analysis of the extracted polar metabolites (see methods for details). We detected 13C and/or 15N products (Figure 5) that are consistent with glutaminolytic, Krebs cycle, gluconeogenesis, and AST/GDH activities, as observed in mouse livers in vivo. These include [13C5, N1]-Glu and [13C5]-á-ketoglutarate (glutaminolysis), [13C4]-citrate, -succinate, -fumarate, [13C4,15Nx]-Asp (Krebs cycle), [13C3]-PEP, -G6P (gluconeogenesis), [13C5]-Glu, [15N1]-Glu, and [13Cx,15Nx]-Asp (AST/GDH). In addition, the presence of [13C3]- pyruvate and -lactate along with their [13C1]-isotopologues confirmed the malic enzyme (ME) activity. Moreover, incorporation of the glutaminolytic products (e.g., [13C5,N1]-Glu) into glutathione (GSH) was evident.
Fractional enrichment analysis of these products demonstrated that ectopic miR-122 expression in EC4 cells blocked glutaminolysis and subsequent Gln metabolism in the Krebs cycle. These included reduced enrichment in [13C5,N1]-Glu (A), [13C5]-α-ketoglutarate (B), 2-hydroxyglutarate (2HG) (C), an alternative product of isocitrate dehyderogenase (IDH), and [13C4]-succinate/fumarate (D, E) (Figure 5) in the miR-122 mimic transfecting cells compared with the negative control. Reduced fractional enrichment of [13C5,Nx]-GSH (Figure 5H) in miR-122 mimic transfected cells as a result of attenuated glutaminolysis was also evident. We further noted that miR-122 ectopic expression increased enrichment in [13C3]-phosphoenolpyruvate (PEP) (Figure 5K) and glucose-6-phosphate (Figure 5L), suggesting enhanced gluconeogenesis. However, reduced enrichment in the glycolytic products [13C3]-pyruvate (Figure 5I) and -lactate (Figure 5J) was evident, which points to inefficient glucose utilization. Alternatively, changes in labeled pyruvate and lactate could result from attenuated ME activity by ectopic miR-122 expression. Reduced efficiency of glucose utilization can create a metabolic stress to the cells [32].
The fractional enrichment data correlated with the data on absolute levels of the aforementioned metabolites. For example, we observed a nonstatistically significant decrease in [13C5,N1]-Glu (A), [13C5]-α-ketoglutarate (B) [13C4]-isocitrate (C), and -citrate levels (D) (Figure 6) and a significant increase in [13C3]-G6P and -PEP levels (Figure 6E,F) in EC4 cells transfected with miR-122 mimic. The total levels and fractional enrichment of 13C (Cx) or 13C,15N (CxNx) labeled products for these metabolites also showed a similar trend as individual labeled isotopologues (Figure 5, Figure 6). Together, these data demonstrated that miR-122 blocked glutaminolysis while enhanced gluconeogenesis in Gln-dependent HCC cells.
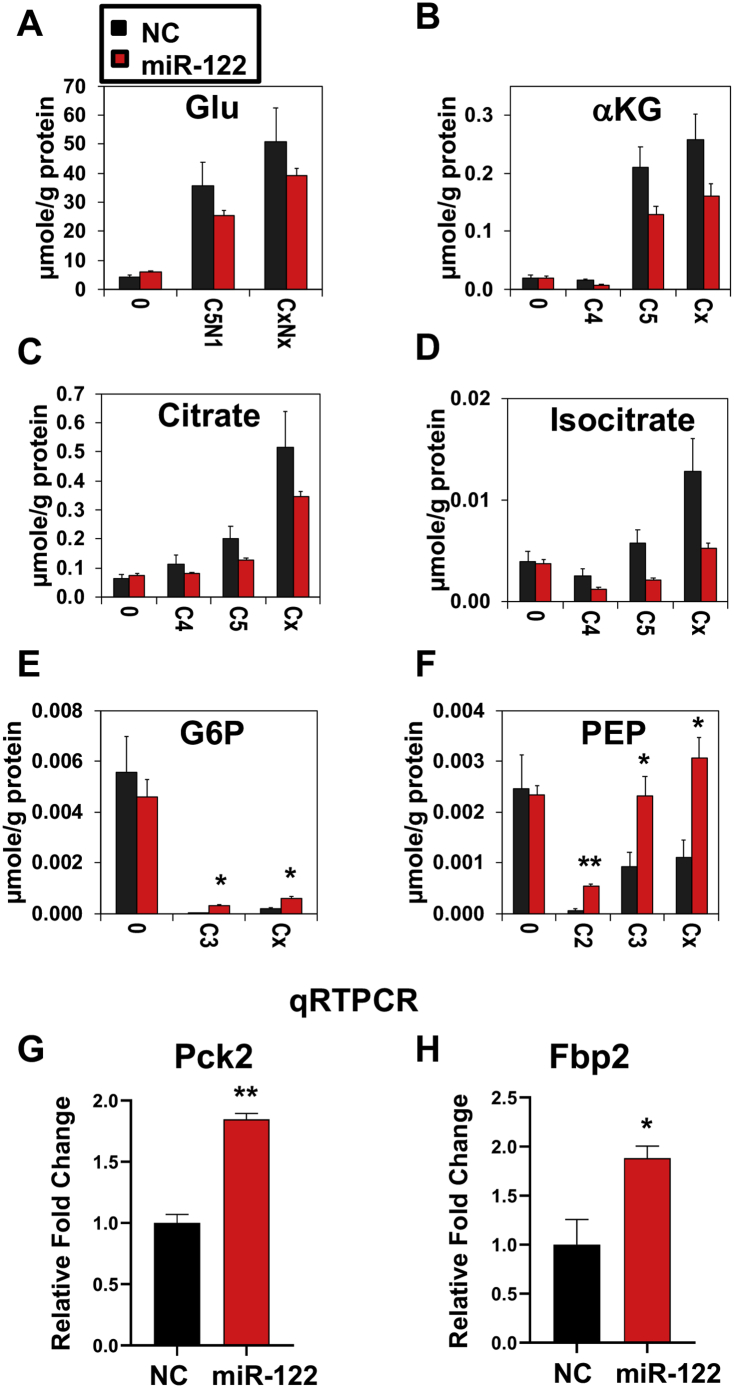
Figure 6.
Quantification of total amounts by IC-MS is consistent with blockade of glutaminolysis and enhanced gluconeogenesis by ectopic miR-122 mimic expression in Gln-dependent EC4 cells. Total amounts of metabolites were quantified via IC-MS analysis as described in Methods. A-F. Depletion of selected 13C- and/or 15N- labeled metabolites (μmol/g protein) including glutamate (A), α-ketoglutarate (B), citrate (C), and isocitrate (D), as well as accumulation gluconeogenic products including G6P (E) and PEP (F) in EC4 cells transfected with miR122 mimic (miR122) or NC mimic (Ctl). Cx and CxNx refer to total levels of labeled products. * and ** denote Q values of 0.05, 0.01, respectively with n = 3. qRT-PCR analysis showed elevated gene expression of key gluconeogenic enzymes such as Pck2 (G) and Fbp2 (H) after transfecting EC4 cells with miR-122 mimic (miR-122) or NC mimic (NC). * and ** denote P values of ≤0.05 and 0.01, respectively (n = 3).
We further examined the expression of genes that code irreversible and vital steps of gluconeogenesis, phosphoenolpyruvate carboxykinase (PCK), and fructose-1,6-bisphosphatase (FBP). PCK catalyzes the de-carboxylation and phosphorylation of oxaloacetate (OAA) to PEP, whereas FBP transforms fructose-1,6-biphosphate to fructose-6-phosphate. qRT-PCR analysis showed significant upregulation of Pck2 (G) and Fbp2 (H) (Figure 6) in EC4 cells transfected with miR-122 mimic, which might be involved in enhanced gluconeogenesis after ectopic miR-122 expression in EC4 cells.
3.5. Upregulation of GLS or SLC1A5 expression in primary human HCCs inversely correlates with miR-122 expression
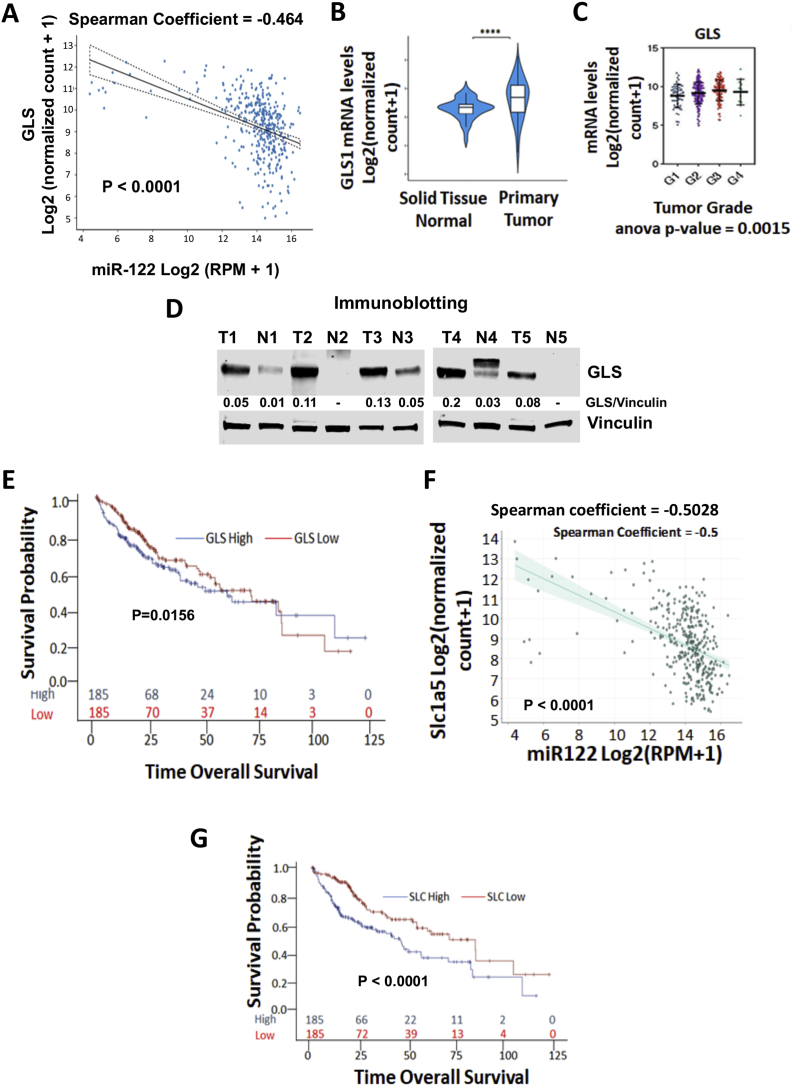
Next, we analyzed TCGA-LIHC database containing expression data for 50 normal and 362 primary HCC specimens to determine if there is any correlation between GLS and miR-122 levels in primary human HCCs. The Spearman coefficient of −0.4603 (P < 0.0001) indicates an inverse relationship between miR-122 and GLS expressions (Figure 7A). TCGA-LIHC database analysis also showed significantly higher GLS expressions in tumors compared with the benign livers in patients with HCC (Figure 7B). Furthermore, GLS levels increased in tumors with higher grades (Figure 7C). Analysis of a few primary HCC samples by immunoblotting confirmed elevated GLS protein levels in tumors compared with adjacent benign liver tissues (Figure 7D). The association between gene expression and patient OS tested using Cox proportional hazard regression model showed that the OS of patients with HCC with higher GLS levels in tumors exhibited significantly decreased survival (P = 0.0156) relative to those with lower GLS levels (Figure 7E). As observed in miR-122 KO mouse liver (Figure S2A), GLS2 expression is significantly suppressed in human HCCs compared with benign liver tissues (P < 0.0001) (Figure S2B). Furthermore, GLS2 expression directly correlated with miR-122 levels in tumors (P = 0.0012) (Figure S2C). However, GLS2 suppression in tumors did not correlate with patient prognosis (data not shown).
Figure 7.
GLS expression and its correlation with miR-122 expression in primary human HCCs and poor survival in HCC patients. A. TCGA-LIHC database analysis of GLS and miR-122 expressions in 50 normal liver and 362 primary HCC specimens. B.GLS expression in tumors and normal liver tissues. C.GLS expression levels in HCC with different grades. D. Immunoblot analysis of GLS in human primary HCC (T) and matching benign liver (N) tissues from 5 patients. E. Association of GLS expression with HCC patients' survival in TCGA-LIHC data analyzed using the Cox proportional hazard regression model but displayed with Kaplan–Meier survival curves (p-value is from the test with Cox proportional regression model). F. Correlation between expression of miR-122 and SLC1A5. G. Association of SLC1A5 expression with HCC patients' survival using TCGA-LIHC data (p-value is from the test with Cox proportional regression model). Detailed statistical analysis is provided in Methods.
TCGA-LIHC data analysis did not show a significant difference in SLC1A5 expression between tumor and benign liver tissues (data not shown). However, an inverse correlation was observed between miR-122 and SLC1A5 expressions (Spearman coefficient = −0.5028, P < 0.0001) (Figure 7F). Cox proportional hazard regression models showed that upregulation of SLC1A5 RNA level correlated with poorer patient survival (P < 0.001) (Figure 7G). Collectively, these data suggest upregulation of expression of two miR-122 targets involved in Gln metabolism, (GLS) and transport (SLC1A5), is associated with poor HCC patient survival.
4. Discussion
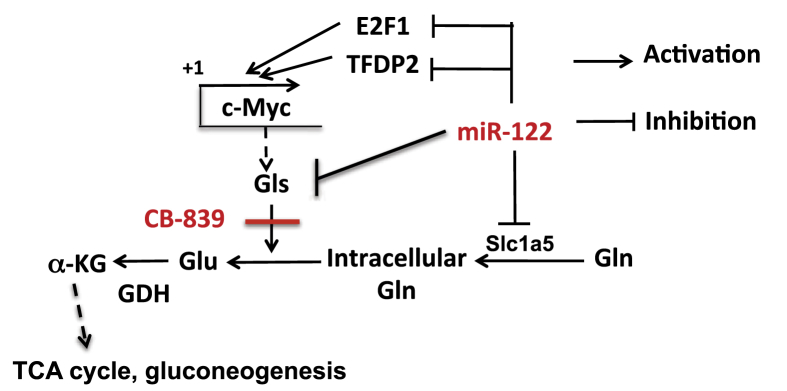
miR-122 is the most abundant liver-specific tumor suppressor microRNA in mammals and plays an essential role in lipid homeostasis. However, its role in amino acid and glucose metabolism is poorly understood. In the present study, we presented multiple experimental evidences demonstrating its critical role in glutaminolysis in mouse livers, and in a Gln-dependent HCC (EC4) cell line. Consequently, Krebs cycle activity was attenuated due to compromised anaplerotic input from Gln, which can, in turn, block cancer cells’ inexorable growth capacity [33]. The importance of activated glutaminolysis in supporting HCC growth is evident from further enhancement of Gln utilization in some spontaneous liver tumors developed in miR-122 KO mice (Figure S1). We confirmed Gls, the gatekeeper enzyme for glutaminolysis, and Slc1a5, a Gln transporter that facilitates the entry of Gln into the cells [18], as miR-122 targets. Thus, the blockade of glutaminolysis in EC4 cells by ectopic miR-122 expression is, at least in part, due to downregulation of the expression Gls and a Slc1a5. Inhibition of Gln metabolism after ectopic expression of miR-122 in EC4 cells that do not express miR-122 complemented the in vivo Gln SIRM data observed in miR-122 KO livers. We believe that upregulation of the kidney-type glutaminase Gls plays a key role in enhanced glutaminolysis in miR-122 KO mice, as microarray data [GSE20610] showed that the expression of the liver-type isoform Gls2 is downregulated in the mutant livers [10]. An oncogenic role of GLS and tumor suppressor function of GLS2 have been reported [22]. Upregulation of kidney-type glutaminase Gls and downregulation of liver-type Gls2 observed in miR-122 KO mice suggest that miR-122 KO hepatocytes have acquired the characteristics of dedifferentiated hepatocytes that are predisposed to hepatocellular cancer. Several miRNAs such as miR-200c [34], miR-513c [35], and miR-23a/b [36] have been reported to regulate Gls level in different cancers other than HCCs. However, the expressions of none of these miRNAs are significantly deregulated in miR-122 KO livers (data not shown). Nevertheless, a higher abundance of miR-122 could be the mechanism of relatively reduced level of Gls, the kidney isoform. It has been demonstrated that c-Myc promotes Gln metabolism in cells by upregulating expression of both Gls and Slc1a5 [19]. We have previously shown that c-Myc is transcriptionally activated in miR-122 KO hepatocytes due to de-repression of miR-122 targets, E2f1 and Tfdp2 [37]. Thus, apart from direct targeting of Gls and Slc1a5, upregulation of c-Myc in KO livers or in a subset of primary HCCs could be an additional mechanism for activating these genes and promoting glutaminolysis. A model delineating probable mechanisms of regulation of Gln metabolism by miR-122 is shown in Figure 8. It is possible miR-122 could inhibit growth of tumor cells that expressing high GLS but are resistant to CB-839, a potent inhibitor of GLS activity [24].
Figure 8.
A model depicting regulation of Gln metabolism by miR-122. miR-122 regulates Gln metabolism by directly targeting Gls and Slc1a5, and indirectly by regulating c-Myc expression through targeting E2f1 and Tfdp2 that activate c-Myc gene expression [37].
In addition, our SIRM analysis uncovered regulation of gluconeogenesis by miR-122 as demonstrated by the activation of the gluconeogenic pathway by ectopic miR-122 expression in Gln-dependent HCC (EC4) cells and its inhibition in miR-122-depleted livers. Upregulation of Pck2 and Fbp2, two key genes that catalyze irreversible steps in EC4 cells expressing miR-122, suggests their potential involvement in miR-122-mediated activation of gluconeogenesis. Blood glucose homeostasis is maintained via regulation of metabolic flux through the hepatic gluconeogenic pathway. Reduced gluconeogenesis in miR-122 KO mice could be a reason for lower blood glucose level in the mutant mice (unpublished data). The liver is the main organ for gluconeogenesis, which is negatively regulated by insulin and positively by glucagon [38]. It will be important to elucidate the mechanism underlying systemic glucose homeostasis by miR-122 in future studies. Apart from miR-122, several other miRNAs are reported to regulate gluconeogenesis in different tissues [39]. However, RNA-seq data showed that none of these miRNAs are significantly altered in miR-122 KO livers. Interestingly, our previous study showed that the upregulation of miR-23a in a dietary NASH-induced HCC mouse model and in primary human HCCs impeded gluconeogenesis by suppressing expression of its targets glucose-6-phosphate, catalytic subunit (G6PC), a key gluconeogenic enzyme, and the transcription factor PGC1α [40]. Thus, downregulation of miR-122 and upregulation of miR-23a may act in concert to block gluconeogenesis in hepatocellular cancer.
It has been demonstrated that many tumors, especially those overexpressing c-MYC, are addicted to Gln as an energy source [15,41]. Furthermore, our findings from a mouse liver and HCC cell line study implicate that miR-122 mediated the regulation of glutaminolysis in human HCCs, as we found an inverse correlation of GLS and SLC1A5 expressions with that of miR-122 in primary human HCCs. CB-839, a specific inhibitor of Gls, has demonstrated efficacy in blocking tumor development in preclinical models [24] and is undergoing clinical trials alone or in combination with chemo-, radiation-, immuno- or targeted therapy in different solid tumors such as renal, breast, lung, and colon cancers [42]. We have shown that miR-122 delivery using recombinant AAV8 virus inhibited c-MYC-induced liver tumor growth [10] where Gls is highly upregulated [17]. In the future, it would be of interest to test the efficacy of CB-839 in combination with miR-122 mimic in preclinical HCC models that exhibit high GLS expression (Figure 8).
5. Conclusions
Our study demonstrated that miR-122, the most abundant liver-specific miRNA and a potent tumor suppressor, regulates Gln metabolism. SIRM analysis showed that glutaminolysis is enhanced whereas gluconeogenesis is suppressed in miR-122 KO mouse liver. Increased expression of Gls, the kidney-type glutaminase, correlated with the enhancement of Gln metabolism in miR-122 KO mice livers. We established Gls and Slc1a5 as direct miR-122 targets. Ectopic expression of miR122 resulted in the suppression of glutaminolysis and enhancement of gluconeogenesis in a Gln-dependent HCC cell line. Expressions of GLS and SLC1A5 are inversely correlated with MIR-122 in human primary HCCs.
Funding
This work was supported, in part, by R01CA193244, R01CA086078, DK088076 grants and a Pelotonia Idea grant (to KG); and 1U24DK097215-01A1, 5R01ES22191, and P01CA163223-01A1 (to TWMF).
Author contribution
D.S.G., T.F. and K.G. wrote the manuscript. K.G. conceived the project. D.S.G., T.C., T.F. and K.G., designed experiments. D.S.G., T.C., P.H. and M.A. performed experiments. D.S.G., T.C., K.-y.T., M.A., V.K.C., X.Z. and T.F. analyzed the data. K.-y.T., X.Z., V.K.C., T.F. and K.G. edited the manuscript. All authors reviewed the manuscript.
Acknowledgements
We thank Dr. Dean Felsher for providing EC4 cells, Dr. Juan M Barahas for generating Slc1a5-3′-UTR-psi-CHECK2 constructs and luciferase assays, Dr. Shu-hao Hsu and Dr. Bo Wang with injection of 13C-glutamine through tail vein, Dr. Huban Kutay for mouse colony maintenance and Pipasha Biswas for technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.01.003.
Contributor Information
Dipanwita Sengupta, Email: dipannwitasg@gmail.com.
Teresa Cassel, Email: terricassel@gmail.com.
Kun-yu Teng, Email: tky382002@gmail.com.
Mona Aljuhani, Email: mona.aljuhani@osumc.edu.
Vivek K. Chowdhary, Email: vivek.chowdhary@umassmed.edu.
Peng Hu, Email: peng.hu@osumc.edu.
Xiaoli Zhang, Email: xiaoli.zhang@osumc.edu.
Teresa W.-M. Fan, Email: twmfan@gmail.com.
Kalpana Ghoshal, Email: Kalpana.ghoshal@osumc.edu.
Conflicts of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Thakral S., Ghoshal K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Current Gene Therapy. 2015;15(2):142–150. doi: 10.2174/1566523214666141224095610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandiera S., Pfeffer S., Baumert T.F., Zeisel M.B. miR-122--a key factor and therapeutic target in liver disease. Journal of Hepatology. 2015;62(2):448–457. doi: 10.1016/j.jhep.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Jopling C.L. Regulation of hepatitis C virus by microRNA-122. Biochemical Society Transactions. 2008;36(Pt 6):1220–1223. doi: 10.1042/BST0361220. [DOI] [PubMed] [Google Scholar]
- 4.Luna J.M., Scheel T.K., Danino T., Shaw K.S., Mele A., Fak J.J. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 2015;160(6):1099–1110. doi: 10.1016/j.cell.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gatfield D., Le Martelot G., Vejnar C.E., Gerlach D., Schaad O., Fleury-Olela F. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes & Development. 2009;23(11):1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu S.H., Delgado E.R., Otero P.A., Teng K.Y., Kutay H., Meehan K.M. MicroRNA-122 regulates polyploidization in the murine liver. Hepatology. 2016 doi: 10.1002/hep.28573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowdhary V., Teng K.Y., Thakral S., Zhang B., Lin C.H., Wani N. miRNA-122 protects mice and human hepatocytes from acetaminophen toxicity by regulating cytochrome P450 family 1 subfamily A member 2 and family 2 subfamily E member 1 expression. American Journal Of Pathology. 2017;187(12):2758–2774. doi: 10.1016/j.ajpath.2017.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K., Zhang S., Marzolf B., Troisch P., Brightman A., Hu Z. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(11):4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antoine D.J., Dear J.W., Lewis P.S., Platt V., Coyle J., Masson M. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology. 2013;58(2):777–787. doi: 10.1002/hep.26294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu S.H., Wang B., Kota J., Yu J., Costinean S., Kutay H. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. Journal of Clinical Investigation. 2012;122(8):2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai S., Nasser M.W., Wang B., Hsu S.H., Datta J., Kutay H. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. Journal of Biological Chemistry. 2009;284(46):32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai W.C., Hsu S.D., Hsu C.S., Lai T.C., Chen S.J., Shen R. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. Journal of Clinical Investigation. 2012;122(8):2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luna J.M., Barajas J.M., Teng K.Y., Sun H.L., Moore M.J., Rice C.M. Argonaute CLIP defines a deregulated miR-122-bound transcriptome that correlates with patient survival in human liver cancer. Molecules and Cells. 2017;67(3):400–410 e7. doi: 10.1016/j.molcel.2017.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hensley C.T., Wasti A.T., DeBerardinis R.J. Glutamine and cancer: cell biology, physiology, and clinical opportunities. Journal of Clinical Investigation. 2013;123(9):3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Still E.R., Yuneva M.O. Hopefully devoted to Q: targeting glutamine addiction in cancer. British Journal of Cancer. 2017;116(11):1375–1381. doi: 10.1038/bjc.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altman B.J., Stine Z.E., Dang C.V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nature Reviews Cancer. 2016;16(10):619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuneva M.O., Fan T.W., Allen T.D., Higashi R.M., Ferraris D.V., Tsukamoto T. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metabolism. 2012;15(2):157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y., Zhao T., Li Z., Wang L., Yuan S., Sun L. The role of ASCT2 in cancer: a review. European Journal of Pharmacology. 2018;837:81–87. doi: 10.1016/j.ejphar.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Dang C.V., Le A., Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clinical Cancer Research. 2009;15(21):6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katt W.P., Lukey M.J., Cerione R.A. A tale of two glutaminases: homologous enzymes with distinct roles in tumorigenesis. Future Medicinal Chemistry. 2017;9(2):223–243. doi: 10.4155/fmc-2016-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J., Zhang C., Lin M., Zhu W., Liang Y., Hong X. Glutaminase 2 negatively regulates the PI3K/AKT signaling and shows tumor suppression activity in human hepatocellular carcinoma. Oncotarget. 2014;5(9):2635–2647. doi: 10.18632/oncotarget.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mates J.M., Campos-Sandoval J.A., Marquez J. Glutaminase isoenzymes in the metabolic therapy of cancer. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2018;1870(2):158–164. doi: 10.1016/j.bbcan.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Yu D., Shi X., Meng G., Chen J., Yan C., Jiang Y. Kidney-type glutaminase (GLS1) is a biomarker for pathologic diagnosis and prognosis of hepatocellular carcinoma. Oncotarget. 2015;6(10):7619–7631. doi: 10.18632/oncotarget.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang Y., Stine Z.E., Xia J., Lu Y., O'Connor R.S., Altman B.J. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. Journal of Clinical Investigation. 2015;125(6):2293–2306. doi: 10.1172/JCI75836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore K.J., Rayner K.J., Suarez Y., Fernandez-Hernando C. The role of microRNAs in cholesterol efflux and hepatic lipid metabolism. Annual Review of Nutrition. 2011;31:49–63. doi: 10.1146/annurev-nutr-081810-160756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lane A.N., Fan T.W. NMR-based stable isotope resolved metabolomics in systems biochemistry. Archives of Biochemistry and Biophysics. 2017;628:123–131. doi: 10.1016/j.abb.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun R.C., Fan T.W., Deng P., Higashi R.M., Lane A.N., Le A.T. Noninvasive liquid diet delivery of stable isotopes into mouse models for deep metabolic network tracing. Nature Communications. 2017;8(1):1646. doi: 10.1038/s41467-017-01518-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan T.W., Warmoes M.O., Sun Q., Song H., Turchan-Cholewo J., Martin J.T. Distinctly perturbed metabolic networks underlie differential tumor tissue damages induced by immune modulator beta-glucan in a two-case ex vivo non-small-cell lung cancer study. Cold Spring Harb Mol Case Stud. 2016;2(4):a000893. doi: 10.1101/mcs.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moseley H.N., Sperling L.J., Rienstra C.M. Automated protein resonance assignments of magic angle spinning solid-state NMR spectra of beta1 immunoglobulin binding domain of protein G (GB1) Journal of Biomolecular NMR. 2010;48(3):123–128. doi: 10.1007/s10858-010-9448-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane A.N., Fan T.W., Higashi R.M. Isotopomer-based metabolomic analysis by NMR and mass spectrometry. Methods in Cell Biology. 2008;84:541–588. doi: 10.1016/S0091-679X(07)84018-0. [DOI] [PubMed] [Google Scholar]
- 31.Kenny J., Bao Y., Hamm B., Taylor L., Toth A., Wagers B. Bacterial expression, purification, and characterization of rat kidney-type mitochondrial glutaminase. Protein Expression and Purification. 2003;31(1):140–148. doi: 10.1016/s1046-5928(03)00161-x. [DOI] [PubMed] [Google Scholar]
- 32.Khan M.W., Chakrabarti P. Gluconeogenesis combats cancer: opening new doors in cancer biology. Cell Death & Disease. 2015;6 doi: 10.1038/cddis.2015.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson N.M., Mucka P., Kern J.G., Feng H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell. 2018;9(2):216–237. doi: 10.1007/s13238-017-0451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu F., Li Y., Liu G. MicroRNA-200c exacerbates the ischemia/reperfusion injury of heart through targeting the glutaminase (GLS)-mediated glutamine metabolism. European Review for Medical and Pharmacological Sciences. 2017;21(14):3282–3289. [PubMed] [Google Scholar]
- 35.Xia H.L., Lv Y., Xu C.W., Fu M.C., Zhang T., Yan X.M. MiR-513c suppresses neuroblastoma cell migration, invasion, and proliferation through direct targeting glutaminase (GLS) Cancer Biomarkers. 2017;20(4):589–596. doi: 10.3233/CBM-170577. [DOI] [PubMed] [Google Scholar]
- 36.Gao P., Tchernyshyov I., Chang T.C., Lee Y.S., Kita K., Ochi T. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang B., Hsu S.H., Wang X., Kutay H., Bid H.K., Yu J. Reciprocal regulation of microRNA-122 and c-Myc in hepatocellular cancer: role of E2F1 and transcription factor dimerization partner 2. Hepatology. 2014;59(2):555–566. doi: 10.1002/hep.26712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hatting M., Tavares C.D.J., Sharabi K., Rines A.K., Puigserver P. Insulin regulation of gluconeogenesis. Annals of the New York Academy of Sciences. 2018;1411(1):21–35. doi: 10.1111/nyas.13435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reyes R.K., Motiwala T., Jacob S.T. Regulation of glucose metabolism in hepatocarcinogenesis by microRNAs. Gene Expression. 2014;16(2):85–92. doi: 10.3727/105221614X13919976902093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang B., Hsu S.H., Frankel W., Ghoshal K., Jacob S.T. Stat3-mediated activation of microRNA-23a suppresses gluconeogenesis in hepatocellular carcinoma by down-regulating glucose-6-phosphatase and peroxisome proliferator-activated receptor gamma, coactivator 1 alpha. Hepatology. 2012;56(1):186–197. doi: 10.1002/hep.25632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wise D.R., DeBerardinis R.J., Mancuso A., Sayed N., Zhang X.Y., Pfeiffer H.K. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proceedings of the National Academy of Sciences of the U S A. 2008;105(48):18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song M., Kim S.H., Im C.Y., Hwang H.J. Recent development of small molecule glutaminase inhibitors. Current Topics in Medicinal Chemistry. 2018;18(6):432–443. doi: 10.2174/1568026618666180525100830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study (TCGA-LIHC data) are available in the Xena Website (https://xena.ucsc.edu/).