Abstract
MicroRNAs are ~20 nt long small noncoding RNAs that are processed from stem-looped precursors and function mainly as posttranscriptional regulators of protein coding genes through binding to 3′-untranslated regions of messenger RNAs to inhibit the translation or cause RNA degradation. It is predicted microRNAs could regulate up to half of all human genes and are proved to play important roles in human diseases including cancer. They bind to target mRNAs based on complementary binding which is dominated by the so-called “seed” region which are the 5′ 2–8 bases of the microRNA. Due to the small size in nature, even a single nucleotide variation in the precursor region especially those located in the seed regions could show big influence. Here, I summarized and reviewed the current knowledge of these single nucleotide alterations in microRNAs in human cancer including (i) common SNPs in the precursor region, (ii) isomiRs, (iii) somatic mutations of microRNAs. Briefly, this is an underexploited field and clearly, warrants further studies to reveal their biological and clinical significances. I believe they will be key to advancing personalized medicine.
Keywords: microRNAs, Single nucleotide alterations, SNPs, Isomirs, Somatic mutations
1. Introduction
Since the discovery of the first microRNA in 1993, this type of small non-coding RNAs has attracted the attention of scientists in nearly all fields of researches and has proved themselves as a superstar [1,2]. They have been reported to participate in various biological development processes and cell activities and are implicated in human diseases with diagnostic and therapeutic potential [3,4]. According to the microRNA registry miRbase (mirbase.org), the number of annotated human microRNAs has been expanded from 56 in miRBase release 1.0 in December 2002 to 2656 in the current release and is expected to be again growing in the future. They are scattered on all chromosomes except the Y chromosome in humans and locates in intergenic, intronic or exonic regions as either single microRNA or miR clusters [5]. They are briefly small 20–24 nucleotide noncoding RNAs derived from primary microRNA transcripts and processed by a two-step cleavage involving enzymes like Drosha and Dicer. The mature microRNAs are believed to function mainly as, although not exclusively, negative regulators of target genes through RNA decay (which is major) or translation inhibition [6]. Canonically, they bind to 3′-UTR of target genes through a complementary sequence of the 2–8 nucleotides in their 5′ end (so-called seed region) [7]. With the development of many microRNA target prediction algorithms, it is estimated that nearly half or even more of all human genes are directly regulated by microRNAs [8]. As such interesting and important players in cellular processes, microRNAs have been implicated in nearly all types of human cancers [[9], [10], [11]]. Considering the relatively small size of microRNAs and especially the small size of seed regions which are critical for microRNA target recognition, even a single nucleotide variation in the pre-miRNA regions particularly those in the mature and seed regions may greatly influence either the maturation, the binding affinity/stability or even target selectivity of microRNAs. Here, I summarized the current knowledge of microRNA biogenesis, targets recognition through canonical and non-canonical binding, and microRNAs in human cancers and gave a focused review on common SNPs in pre-microRNAs, isomiRs and somatic mutations of microRNAs. I found this is still by far a not fully explored field and warrants further investigations. Considering the great potential of both microRNAs and genome variations in precision medicine [[12], [13], [14], [15], [16], [17]], I believe future studies in this field are of great importance and significance.
2. MicroRNA biogenesis and target recognition
MicroRNAs are small noncoding RNAs that are 19–24 nt long. They are transcribed mainly through the RNA polymerase II and following the transcription the canonical biogenesis of microRNAs is a two-step process (cropping and dicing) which includes one cleavage event in the nucleus by RNase III enzyme Drosha and the other in the cytoplasm by another RNase III enzyme Dicer [18]. Drosha process the pri-microRNA into a ~60–100 nt stem-loop structured microRNA precursor (pre-microRNA). Then pre-microRNAs are exported into the cytoplasm with the help of exportins and then Dicer will cleave the pre-microRNA into a double stranded RNAs. Except this Drosha-dependent and Dicer-dependent canonical pathway, other non-canonical pathways of microRNA biogenesis have also been reported [19,20]. For example, some microRNAs are Drosha independent or Dicer independent. Good example of Drosha-independent microRNAs are mirtrons which are microRNAs derived from the introns of mRNAs and produced directly through RNA splicing. m7G (7-methylguanosine)-capped pre-microRNAs are another example which does not need Drosha cleavage but could directly be exported to the cytoplasm for Dicer processing. Also, it has been found that biogenesis of miR-451 is one rare event that requires Drosha but not Dicer and was cleaved by Ago2 instead in the cytoplasm [20].
After mature microRNAs are formed, they are incorporated into the RISC complex. Most microRNAs target the 3′-UTR regions of mRNAs through complementary Watson-Crick pairing of their seed region which is the 5′ 2–8 base of the mature microRNA. They either inhibits the translation or cause mRNA decapping and degradation. Many microRNA target prediction algorithms based on this theory have been developed and widely accepted including TargetScan, Pictar, RNA22, PITA etc [8]. However, it was found that all these algorithms showed a high false positive prediction rates and it was reported that up to 50% of the predicted targets are false positives [21]. This indicates that microRNAs may function through other direct or indirect mechanisms. Interestingly, it was found that some microRNAs could bind to the 5′-UTR, the CDS or the promoter regions to even upregulate transcription or enhance translation [22]. miR10a that binds to the 5′UTR, miR373 binding to promoter and miR34a binding to both 5′-UTR and 3′-UTR are all good examples for these nonclassical binding [[23], [24], [25]]. What is more, some argue that microRNAs may target transcription factors and exert their function through a two-layer miR-TF-mRNA axis [[26], [27], [28], [29], [30]]. All in all, microRNAs in human cells play important roles and they regulate gene expression through several different types of regulatory mechanisms.
3. Common single nucleotide polymorphisms (SNPs) in pre-microRNAs
Single nucleotide polymorphisms frequently called SNPs, are the most common type of genetic variations in human. It was estimated that there are more than 100 million SNPs in human genome and about 7.5 million of them are common SNPs (SNPs with minor allele frequency (MAF) no less than 5%) [31]. With limitations of samples size and statistical power, past case-control studies have also revealed the biological significance and functionality of common SNPs but not rare SNPs. To begin with, I first obtained all SNP data located in the pre-microRNAs from Ensemble and ranked them with MAF and number of related publications. In brief, there are in total 322 SNPs located in the microRNA precursor regions including 37 in the seed regions, 65 in the mature regions and the rest 220 in the precursor regions (Supplementary Table 1). Surprisingly, only 16 of these common SNPs were well studied with more than 10 related publication up to date (Table 1), with only 1 located in the seed regions and 3 in the mature regions and 12 in precursors.
Table 1.
List of well-studied SNPs located in the pre-microRNA regions.
| Gene name | Variant name | Minor allele frequency | SNP positions | # of related publications |
|---|---|---|---|---|
| MIR196A2 | rs11614913 | 0.332668 | in mature | 425 |
| MIR499A | rs3746444 | 0.183506 | in seed | 289 |
| MIR499B | rs3746444 | 0.183506 | in mature | 289 |
| MIR149 | rs2292832 | 0.386581 | in precursor | 157 |
| MIR27A | rs895819 | 0.363818 | in precursor | 136 |
| MIR608 | rs4919510 | 0.363818 | in mature | 82 |
| MIR423 | rs6505162 | 0.497804 | in precursor | 77 |
| MIR1908 | rs174561 | 0.279553 | in precursor | 31 |
| MIR492 | rs2289030 | 0.114417 | in precursor | 25 |
| MIR605 | rs2043556 | 0.259585 | in precursor | 24 |
| MIR149 | rs71428439 | 0.14397 | in precursor | 15 |
| MIR449B | rs10061133 | 0.122204 | in mature | 13 |
| MIR604 | rs2368392 | 0.323083 | in precursor | 12 |
| MIR1307 | rs7911488 | 0.303714 | in precursor | 12 |
| MIR618 | rs2682818 | 0.242412 | in precursor | 12 |
| MIR202 | rs12355840 | 0.31889 | in precursor | 11 |
3.1. miR196A2 rs11614913
rs11614913 located in the mature miR-196a-3p is the most widely studied microRNA SNP so far. It was first discovered as a genetic variant impacting patients’ survival in non-small cell lung cancer [32] and breast cancer [33]. Since then, it has been implicated in various human cancers including breast, lung, prostate, colorectal, gastric, head and neck, hepatocellular carcinomas, etc [[34], [35], [36], [37], [38], [39], [40], [41]]. Scientists found this SNP influenced the pre-microRNA processing and maturation. Also, this position underwent somatic mutation in breast cancer in 14% of all patients and correlates with higher expression of miR196a [42]. However, conflicting results from large case-control studies have been observed for this SNP and it seems that this SNP has population and cancer type specific effect. For example, C allele is correlated with high risk of hepatocellular carcinoma [43] while T allele is correlated with high risk of Glioma, prostate cancer and colorectal carcinoma [[44], [45], [46]]. In a meta-analysis of 46 studies involving 20673 cases and 25143 controls, it was discovered that C allele showed significant association with cancer risk in Asians but not in Caucasians [38]. In another study, it was shown that rs11614913 T allele frequency is significantly low in African population compared to Non-African population [47].
3.2. miR499 rs3746444
The miR499 locus encodes two microRNAs miR499a (transcribing from the forward strand) and miR499b (transcribing from the reverse strand). rs3746444 is located in the seed region of miR499a-3p while in the mature region of miR499b-5p. It was first discovered together with miR196a2 SNP rs11614913 from a case-control study as risk variants for breast cancer involving 1009 cases and 1093 controls [33]. It also shows a population and caner type specific effects. In Chen 2014, T allele predicts risk for esophageal cancer while C allele for all other cancers [48]. Mi et al. found that C allele is associated with increased cancer risk in Asian but not in Caucasians [46].
3.3. miR149 SNPs rs2292832 and rs71428439
These two SNPs are the only common SNPs (MAF = 0.39 for rs2292832 and 0.14 for rs71428439) located in a single microRNA locus which are both well studied up to date. They are only 2 base pair away from each other and locates in the 3’ end of the pre-miR149. Rs71428439 G allele showed reduced level of maturation of miR149 compared with the A allele [49] and predicts high risk for lung cancer and hepatocellular carcinomas [50,51]. rs2292832 was found to be related to risk of multiple cancers including colorectal cancer, liver cancer and breast cancer [[52], [53], [54], [55]] and in a recent study [56], it was shown that this SNP may have a population specific effect in gastric cancer patients. While T allele carriers in Caucasians displayed decreased risk of gastric cancer, they have increased risk among Asians.
3.4. miR423 rs6505162
rs6505162 is located in the precursor region of miR423 and was discovered to influence the maturation and expression of miR423 [57,58]. It ranks #1 in minor allele frequency (49.78%, see Table 1) of all the 322 SNPs located in the pre-microRNA regions, which indicates its high prevalence in human diseases related to miR423. Up to date, miR423 SNP rs6505162 has been implicated in multiple cancers including esophageal, breast, ovarian and colorectal cancer [[59], [60], [61]]. Interestingly, there is a significant interaction between rs6505162 and age in esophageal cancers and the protective effect was more than twice among younger patients than older ones [59].
Except for the above summarized four well studied miR SNPs and several others, biological and clinical significance for the majority of these genetic variants located in microRNA precursor regions remain unclear. Most interesting SNPs among these include multiple common SNPs located in a single microRNA locus (e.g. SNPs for miR6891, miR5189, miR4719, miR7150, miR6839, miR548AJ2 and miR5700) and SNPs showing high minor allele frequencies (e.g. rs2663345, rs71974827). Whether they influenced the maturation, expression of microRNAs or targets selection need to be revealed. From past studies, we could imagine these SNPs may also show population, gender, age and cancer type specific effects and researches on these could lead to significant progression in precision medicine.
4. Isomirs
microRNAs were typically annotated as a single defined sequence before Landgraf et al. [62] have found that the same microRNA could have several variants which differ in length and sequence. The development of deep sequencing has enabled the identification of isomirs, which are microRNA variants that are heterogeneous in length or sequence. Importantly and interestingly, isomirs are not rare and was estimated to contribute to half of the miRnome in human cells and for each microRNA the contribution of isomirs to the expression levels varies from 0% to nearly 100% [63,64]. Some isomirs may affect stability of microRNAs and also target selection [65]. However, up to date, the functionality and biological and clinical significance of majority of these isomirs remain unclear and is yet to be revealed in the future.
Isomirs could be classified to 4 main classes: 5′ isomiRs, 3′ isomiRs, polymorphic isomiRs and mixed type isomiRs and they were generated through multiple Drosha or Dicer cleavage, 5′ or 3′ trimming, nucleotide addition, removal or RNA editing [66]. It was found that [67] some isomiRs expression are more abundant and show superior performance as biomarkers than their canonical forms in cancer. Scientists then found that isomer expression could be used to classify breast cancer subtypes and some isomirs could be used as biomarkers in breast cancer and even showed improved performances compared with the traditional gene expression profiling [68]. Recently, scientists used 50 5′ isomir as a panel and showed the expression of these isomirs could effectively classify tumors from 32 different tumor types with high sensitivity [69]. Individual isomir have been validated to be involved in cancer progression and also have diagnostic values in cancer. For example, 5’ isomir of miR140-3p suppress breast cancer proliferation and migration [70], while miR196a isomirs are significantly different between cancer and normal tissues and is diagnostic in laryngeal cancer patients [71]. Isomirs also showed gender or race specific expression patterns [72]. One study from triple negative breast cancer revealed that a total of 21 isomiRs are differentially expressed between white patients and black patients including miR-183-5p isomiRs and they then verified each isomir has distinct effect in cancer cell through microarray analyses of the transcriptome [73]. Later, they performed a more through analyses of isomirs and corresponding mRNA targets in triple negative breast cancer and discovered that genes from key metastasis linked pathways associated with isomirs in a race dependent manner [74]. In another study which analyzed isomirs in prostate cancer TCGA cohort that contains 562 samples from 472 patients, scientists detected a total of more than 3000 isomiRs, of which 524 are differential between tumor and normal and a large portion (1519, ~50%) were specifically abundant in white patients [75]. Aberrant expression of isomirs could be potential cancer biomarkers. In one study, it was reported that some isomirs could be predictive of prostate cancer metastasis [76] and in another, isomirs expression in the urine extracellular vesicles could be used as a non-invasive method to detect prostate cancer [77].
5. Somatic mutations in microRNAs
Somatic mutations also lead to single nucleotide variations in microRNAs. At present, there is one database SomamiR which included data from more than 600 samples in 50 different type of cancers and mapped ~2500 somatic mutations in microRNAs including ~1800 located in pre-microRNA regions, 644 in mature regions and 181 in seed regions [78,79]. Up to date, there is still limited understanding of these mutations which could be due to overall low mutation rate and there are still no case control studies which contain enough number of samples to reach statistical significance for these mutations. One recently published paper in lung cancer [80] analyzing the TCGA cohort which contain ~500 lung adenocarcinomas and ~500 lung squamous cell carcinomas identified more than 1000 mutation affecting ~500 microRNAs. They identified 10 significantly over mutated hotspot microRNA genes including miR379. Future studies which include more samples or focus on other cancer types will lead to more comprehensive understanding of these somatic mutations in microRNAs.
6. Concluding remarks
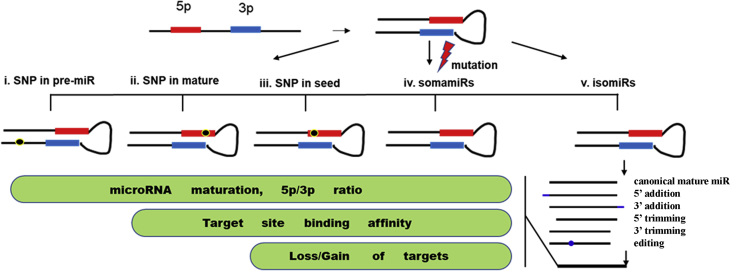
In this review, I summarized the current knowledge of single nucleotide variations in human microRNAs including the single nucleotide polymorphisms with minor allele frequencies higher than 5%, the isomiRs, and somatic mutations in microRNA genes. A diagrammatic summary was shown in Fig. 1. In general, this field is not fully explored with too much emphasis on only several star SNPs. Majority of the common SNPs in microRNA regions are not studied, while it is only at the starting phase for the studies on isomiRs and microRNA somatic mutations. Considering the important role of microRNAs in gene regulation and their significance in human diseases, further studies are in need. This review also provided some hint for future studies in this field. For example, researches in the future should take into consideration whether there is age, gender, race/ethnic, tissue/cancer type or other relative factor(s) specific role for a given variation. Knowledge obtained from these studies will surely improve the understanding of microRNAs in human diseases and aid in further development of precision medicine.
Fig. 1.
Diagrammatic summary of single nucleotide variations (SNVs) in microRNAs. SNVs can be classified into three categories: single nucleotide polymorphisms (SNPs), somatic mutations (somamiRs) and isomiRs. SNPs in pre-miRs, mature miRs or within the seed sequences may (1) influence the biogenesis/maturation of the mature miRs and cause change in 5p/3p ratios, (2) strengthen or reduce the binding affinity for the mature miRs to bind to their targets or (3) change miRs' targetome through loss or gain of binding sites. SomamiRs, which are somatically mutated microRNAs, could be considered as a combination of all possible SNPs. IsomiRs are bona fide microRNA variants or isoforms that result from either addition, depletion/trimming or microRNA editing and usually show race, gender, population, and disease subtype dependencies. All of these SNVs modulate the intracellular microRNA-mRNA interaction network.
Declaration of competing interest
There is no competing interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2020.02.003.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Maqbool R., Ul Hussain M. MicroRNAs and human diseases: diagnostic and therapeutic potential. Cell Tissue Res. 2014;358(1):1–15. doi: 10.1007/s00441-013-1787-3. [DOI] [PubMed] [Google Scholar]
- 4.Nicolas F.E., Lopez-Martinez A.F. MicroRNAs in human diseases. Recent Pat. DNA Gene Sequences. 2010;4(3):142–154. doi: 10.2174/187221510794751659. [DOI] [PubMed] [Google Scholar]
- 5.Negrini M., Ferracin M., Sabbioni S., Croce C.M. MicroRNAs in human cancer: from research to therapy. J. Cell Sci. 2007;120(Pt 11):1833–1840. doi: 10.1242/jcs.03450. [DOI] [PubMed] [Google Scholar]
- 6.Cai Y., Yu X., Hu S., Yu J. A brief review on the mechanisms of miRNA regulation. Dev. Reprod. Biol. 2009;7(4):147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pillai R.S. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11(12):1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riffo-Campos A.L., Riquelme I., Brebi-Mieville P. Tools for sequence-based miRNA target prediction: what to choose? Int. J. Mol. Sci. 2016;17(12) doi: 10.3390/ijms17121987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikitina E.G., Urazova L.N., Stegny V.N. MicroRNAs and human cancer. Exp. Oncol. 2012;34(1):2–8. [PubMed] [Google Scholar]
- 10.Farazi T.A., Hoell J.I., Morozov P., Tuschl T. MicroRNAs in human cancer. Adv. Exp. Med. Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruan K., Fang X., Ouyang G. MicroRNAs: novel regulators in the hallmarks of human cancer. Canc. Lett. 2009;285(2):116–126. doi: 10.1016/j.canlet.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 12.Genome variation in precision medicine. Nat. Genet. 2016;48(7):701. doi: 10.1038/ng.3614. [DOI] [PubMed] [Google Scholar]
- 13.Detassis S., Grasso M., Del Vescovo V., Denti M.A. microRNAs make the call in cancer personalized medicine. Front. cell Dev. Biol. 2017;5:86. doi: 10.3389/fcell.2017.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saumet A., Mathelier A., Lecellier C.H. The potential of microRNAs in personalized medicine against cancers. BioMed Res. Int. 2014;2014:642916. doi: 10.1155/2014/642916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabbri M. MicroRNAs and cancer: towards a personalized medicine. Curr. Mol. Med. 2013;13(5):751–756. doi: 10.2174/1566524011313050006. [DOI] [PubMed] [Google Scholar]
- 16.Hudson T.J. Genome variation and personalized cancer medicine. J. Intern. Med. 2013;274(5):440–450. doi: 10.1111/joim.12097. [DOI] [PubMed] [Google Scholar]
- 17.Agrawal S., Khan F. Human genetic variation and personalized medicine. Indian J. Physiol. Pharmacol. 2007;51(1):7–28. [PubMed] [Google Scholar]
- 18.Kim V.N. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6(5):376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 19.Miyoshi K., Miyoshi T., Siomi H. Many ways to generate microRNA-like small RNAs: non-canonical pathways for microRNA production. Mol. Genet. Genom. : MGG. 2010;284(2):95–103. doi: 10.1007/s00438-010-0556-1. [DOI] [PubMed] [Google Scholar]
- 20.Yang J.S., Lai E.C. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol. Cell. 2011;43(6):892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinzon N., Li B., Martinez L., Sergeeva A., Presumey J., Apparailly F., Seitz H. microRNA target prediction programs predict many false positives. Genome Res. 2017;27(2):234–245. doi: 10.1101/gr.205146.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ni W.J., Leng X.M. Dynamic miRNA-mRNA paradigms: new faces of miRNAs. Biochem. Biophys. Rep. 2015;4:337–341. doi: 10.1016/j.bbrep.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orom U.A., Nielsen F.C., Lund A.H. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30(4):460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Place R.F., Li L.C., Pookot D., Noonan E.J., Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. U.S.A. 2008;105(5):1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee I., Ajay S.S., Yook J.I., Kim H.S., Hong S.H., Kim N.H., Dhanasekaran S.M., Chinnaiyan A.M., Athey B.D. New class of microRNA targets containing simultaneous 5'-UTR and 3'-UTR interaction sites. Genome Res. 2009;19(7):1175–1183. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin C.C., Chen Y.J., Chen C.Y., Oyang Y.J., Juan H.F., Huang H.C. Crosstalk between transcription factors and microRNAs in human protein interaction network. BMC Syst. Biol. 2012;6:18. doi: 10.1186/1752-0509-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen K., Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 2007;8(2):93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 28.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319(5871):1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 29.Martinez N.J., Walhout A.J. The interplay between transcription factors and microRNAs in genome-scale regulatory networks. Bioessays : News and Reviews in Molecular, Cellular and Developmental Biology. 2009;31(4):435–445. doi: 10.1002/bies.200800212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nazarov P.V., Reinsbach S.E., Muller A., Nicot N., Philippidou D., Vallar L., Kreis S. Interplay of microRNAs, transcription factors and target genes: linking dynamic expression changes to function. Nucleic Acids Res. 2013;41(5):2817–2831. doi: 10.1093/nar/gks1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hao K., Schadt E.E., Storey J.D. Calibrating the performance of SNP arrays for whole-genome association studies. PLoS Genet. 2008;4(6) doi: 10.1371/journal.pgen.1000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Z., Chen J., Tian T., Zhou X., Gu H., Xu L., Zeng Y., Miao R., Jin G., Ma H. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J. Clin. Invest. 2008;118(7):2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Z., Liang J., Wang Z., Tian T., Zhou X., Chen J., Miao R., Wang Y., Wang X., Shen H. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum. Mutat. 2009;30(1):79–84. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- 34.Yan W., Gao X., Zhang S. Association of miR-196a2 rs11614913 and miR-499 rs3746444 polymorphisms with cancer risk: a meta-analysis. Oncotarget. 2017;8(69):114344–114359. doi: 10.18632/oncotarget.22547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sibin M.K., Harshitha S.M., Narasingarao K.V., Dhananjaya I.B., Dhaval P.S., Chetan G.K. Effect of rs11614913 polymorphism on mature miR196a2 expression and its target gene HOXC8 expression in human glioma. J. Mol. Neurosci. : MN. 2017;61(2):144–151. doi: 10.1007/s12031-016-0855-z. [DOI] [PubMed] [Google Scholar]
- 36.Li M., Li R.J., Bai H., Xiao P., Liu G.J., Guo Y.W., Mei J.Z. Association between the pre-miR-196a2 rs11614913 polymorphism and gastric cancer susceptibility in a Chinese population. Genet. Mol. Res. : GMR. 2016;15(2) doi: 10.4238/gmr.15027516. [DOI] [PubMed] [Google Scholar]
- 37.Song Z.S., Wu Y., Zhao H.G., Liu C.X., Cai H.Y., Guo B.Z., Xie Y.A., Shi H.R. Association between the rs11614913 variant of miRNA-196a-2 and the risk of epithelial ovarian cancer. Oncol. Lett. 2016;11(1):194–200. doi: 10.3892/ol.2015.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang Z., Li Y., He X., Jiu T., Wei J., Tian F., Gu C. Quantitative assessment of the association between miR-196a2 rs11614913 polymorphism and cancer risk: evidence based on 45,816 subjects. Tumour Biol. : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(7):6271–6282. doi: 10.1007/s13277-014-1822-3. [DOI] [PubMed] [Google Scholar]
- 39.Wan D., Gu W., Xu G., Shen C., Ding D., Shen S., Wang S., Gong X., He S., Zhi Q. Effects of common polymorphisms rs2910164 in miR-146a and rs11614913 in miR-196a2 on susceptibility to colorectal cancer: a systematic review meta-analysis. Clin. Transl. Oncol.: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2014;16:792–800. doi: 10.1007/s12094-013-1150-x. 9. [DOI] [PubMed] [Google Scholar]
- 40.Yuan Z., Zeng X., Yang D., Wang W., Liu Z. Effects of common polymorphism rs11614913 in Hsa-miR-196a2 on lung cancer risk. PloS One. 2013;8(4) doi: 10.1371/journal.pone.0061047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J., Wang Q., Liu H., Shao N., Tan B., Zhang G., Wang K., Jia Y., Ma W., Wang N. The association of miR-146a rs2910164 and miR-196a2 rs11614913 polymorphisms with cancer risk: a meta-analysis of 32 studies. Mutagenesis. 2012;27(6):779–788. doi: 10.1093/mutage/ges052. [DOI] [PubMed] [Google Scholar]
- 42.Zhao H., Xu J., Zhao D., Geng M., Ge H., Fu L., Zhu Z. Somatic mutation of the SNP rs11614913 and its association with increased MIR 196A2 expression in breast cancer. DNA Cell Biol. 2016;35(2):81–87. doi: 10.1089/dna.2014.2785. [DOI] [PubMed] [Google Scholar]
- 43.Akkiz H., Bayram S., Bekar A., Akgollu E., Ulger Y. A functional polymorphism in pre-microRNA-196a-2 contributes to the susceptibility of hepatocellular carcinoma in a Turkish population: a case-control study. J. Viral Hepat. 2011;18(7):e399–407. doi: 10.1111/j.1365-2893.2010.01414.x. [DOI] [PubMed] [Google Scholar]
- 44.Dou T., Wu Q., Chen X., Ribas J., Ni X., Tang C., Huang F., Zhou L., Lu D. A polymorphism of microRNA196a genome region was associated with decreased risk of glioma in Chinese population. J. Canc. Res. Clin. Oncol. 2010;136(12):1853–1859. doi: 10.1007/s00432-010-0844-5. [DOI] [PubMed] [Google Scholar]
- 45.Haerian M.S., Haerian B.S., Molanaei S., Kosari F., Sabeti S., Bidari-Zerehpoosh F., Abdolali E. MIR196A2 rs11614913 contributes to susceptibility to colorectal cancer in Iranian population: a multi-center case-control study and meta-analysis. Gene. 2018;669:82–90. doi: 10.1016/j.gene.2018.05.082. [DOI] [PubMed] [Google Scholar]
- 46.Mi Y., Ren K., Zou J., Bai Y., Zhang L., Zuo L., Okada A., Yasui T. The association between three genetic variants in MicroRNAs (Rs11614913, Rs2910164, Rs3746444) and prostate cancer risk. Cell. Physiol. Biochem. : Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018;48(1):149–157. doi: 10.1159/000491671. [DOI] [PubMed] [Google Scholar]
- 47.Rawlings-Goss R.A., Campbell M.C., Tishkoff S.A. Global population-specific variation in miRNA associated with cancer risk and clinical biomarkers. BMC Med. Genom. 2014;7:53. doi: 10.1186/1755-8794-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C., Yang S., Chaugai S., Wang Y., Wang D.W. Meta-analysis of Hsa-mir-499 polymorphism (rs3746444) for cancer risk: evidence from 31 case-control studies. BMC Med. Genet. 2014;15:126. doi: 10.1186/s12881-014-0126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding S.L., Wang J.X., Jiao J.Q., Tu X., Wang Q., Liu F., Li Q., Gao J., Zhou Q.Y., Gu D.F. A pre-microRNA-149 (miR-149) genetic variation affects miR-149 maturation and its ability to regulate the Puma protein in apoptosis. J. Biol. Chem. 2013;288(37):26865–26877. doi: 10.1074/jbc.M112.440453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu J., Lv S., An J., Lu C. Pre-miR-149 rs71428439 polymorphism is associated with increased cancer risk and AKT1/cyclinD1 signaling in hepatocellular carcinoma. Int. J. Clin. Exp. Med. 2015;8(8):13628–13633. [PMC free article] [PubMed] [Google Scholar]
- 51.Fang C., Li X.P., Chen Y.X., Wu N.Y., Yin J.Y., Zhang W., Zhou H.H., Liu Z.Q. Functional miRNA variants affect lung cancer susceptibility and platinum-based chemotherapy response. J. Thorac. Dis. 2018;10(6):3329–3340. doi: 10.21037/jtd.2018.05.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan X.M., Xiao X., Qin H.J., Zhang Z., Li Z.H., Gao L.B., Jia J. MicroRNA variants and colorectal cancer risk: a meta-analysis. Genet. Mol. Res. : GMR. 2016;15(3) doi: 10.4238/gmr.15038478. [DOI] [PubMed] [Google Scholar]
- 53.Wang X.H., Wang F.R., Tang Y.F., Zou H.Z., Zhao Y.Q. Association of miR-149C>T and miR-499A>G polymorphisms with the risk of hepatocellular carcinoma in the Chinese population. Genet. Mol. Res. : GMR. 2014;13(3):5048–5054. doi: 10.4238/2014.July.4.20. [DOI] [PubMed] [Google Scholar]
- 54.Wei W.J., Lu Z.W., Li D.S., Wang Y., Zhu Y.X., Wang Z.Y., Wu Y., Wang Y.L., Ji Q.H. Association of the miR-149 Rs2292832 polymorphism with papillary thyroid cancer risk and clinicopathologic characteristics in a Chinese population. Int. J. Mol. Sci. 2014;15(11):20968–20981. doi: 10.3390/ijms151120968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng Y., Duan F., Song C., Zhao X., Dai L., Cui S. Systematic evaluation of cancer risk associated with rs2292832 in miR149 and rs895819 in miR27a: a comprehensive and updated metaanalysis. Oncotarget. 2016;7(16):22368–22384. doi: 10.18632/oncotarget.8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L., Liu Q., Wang F. Association between miR-149 gene rs2292832 polymorphism and risk of gastric cancer. Arch. Med. Res. 2018;49(4):270–277. doi: 10.1016/j.arcmed.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 57.Zhao H., Gao A., Zhang Z., Tian R., Luo A., Li M., Zhao D., Fu L., Dong J.T., Zhu Z. Genetic analysis and preliminary function study of miR-423 in breast cancer. Tumour Biol. : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36(6):4763–4771. doi: 10.1007/s13277-015-3126-7. [DOI] [PubMed] [Google Scholar]
- 58.Su X., Hu Y., Li Y., Cao J.L., Wang X.Q., Ma X., Xia H.F. The polymorphism of rs6505162 in the MIR423 coding region and recurrent pregnancy loss. Reproduction. 2015;150(1):65–76. doi: 10.1530/REP-15-0007. [DOI] [PubMed] [Google Scholar]
- 59.Ye Y., Wang K.K., Gu J., Yang H., Lin J., Ajani J.A., Wu X. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Canc. Prev. Res. (Phila) 2008;1(6):460–469. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith R.A., Jedlinski D.J., Gabrovska P.N., Weinstein S.R., Haupt L., Griffiths L.R. A genetic variant located in miR-423 is associated with reduced breast cancer risk. CANCER GENOMICS PROTEOMICS. 2012;9(3):115–118. [PubMed] [Google Scholar]
- 61.Jia W., Zeng L., Luo S., Bai F., Zhong R., Wu L., Huang G.L., Pu X. Association of microRNA-423 rs6505162 C>A polymorphism with susceptibility and metastasis of colorectal carcinoma. Medicine. 2018;97(6) doi: 10.1097/MD.0000000000009846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haseeb A., Makki M.S., Khan N.M., Ahmad I., Haqqi T.M. Deep sequencing and analyses of miRNAs, isomiRs and miRNA induced silencing complex (miRISC)-associated miRNome in primary human chondrocytes. Sci. Rep. 2017;7(1):15178. doi: 10.1038/s41598-017-15388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karali M., Persico M., Mutarelli M., Carissimo A., Pizzo M., Singh Marwah V., Ambrosio C., Pinelli M., Carrella D., Ferrari S. High-resolution analysis of the human retina miRNome reveals isomiR variations and novel microRNAs. Nucleic Acids Res. 2016;44(4):1525–1540. doi: 10.1093/nar/gkw039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neilsen C.T., Goodall G.J., Bracken C.P. IsomiRs--the overlooked repertoire in the dynamic microRNAome. Trends Genet. : TIG (Trends Genet.) 2012;28(11):544–549. doi: 10.1016/j.tig.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 66.Muller H., Marzi M.J., Nicassio F., IsomiRage From functional classification to differential expression of miRNA isoforms. Front. Bioeng. Biotechnol. 2014;2:38. doi: 10.3389/fbioe.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu C.W., Evans J.M., Huang S., Mahoney D.W., Dukek B.A., Taylor W.R., Yab T.C., Smyrk T.C., Jen J., Kisiel J.B. A Comprehensive Approach to Sequence-oriented IsomiR annotation (CASMIR): demonstration with IsomiR profiling in colorectal neoplasia. BMC Genom. 2018;19(1):401. doi: 10.1186/s12864-018-4794-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lan C., Peng H., McGowan E.M., Hutvagner G., Li J. An isomiR expression panel based novel breast cancer classification approach using improved mutual information. BMC Med. Genom. 2018;11(Suppl 6):118. doi: 10.1186/s12920-018-0434-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang S., Zheng Z., Chen P., Wu M. Tumor classification and biomarker discovery based on the 5'isomiR expression level. BMC Canc. 2019;19(1):127. doi: 10.1186/s12885-019-5340-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salem O., Erdem N., Jung J., Munstermann E., Worner A., Wilhelm H., Wiemann S., Korner C. The highly expressed 5'isomiR of hsa-miR-140-3p contributes to the tumor-suppressive effects of miR-140 by reducing breast cancer proliferation and migration. BMC Genom. 2016;17:566. doi: 10.1186/s12864-016-2869-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saito K., Inagaki K., Kamimoto T., Ito Y., Sugita T., Nakajo S., Hirasawa A., Iwamaru A., Ishikura T., Hanaoka H. MicroRNA-196a is a putative diagnostic biomarker and therapeutic target for laryngeal cancer. PloS One. 2013;8(8) doi: 10.1371/journal.pone.0071480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo L., Liang T., Yu J., Zou Q. A comprehensive analysis of miRNA/isomiR expression with gender difference. PloS One. 2016;11(5) doi: 10.1371/journal.pone.0154955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Telonis A.G., Loher P., Jing Y., Londin E., Rigoutsos I. Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucleic Acids Res. 2015;43(19):9158–9175. doi: 10.1093/nar/gkv922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Telonis A.G., Rigoutsos I. Race disparities in the contribution of miRNA isoforms and tRNA-derived fragments to triple-negative breast cancer. Canc. Res. 2018;78(5):1140–1154. doi: 10.1158/0008-5472.CAN-17-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Magee R.G., Telonis A.G., Loher P., Londin E., Rigoutsos I. Profiles of miRNA isoforms and tRNA fragments in prostate cancer. Sci. Rep. 2018;8(1):5314. doi: 10.1038/s41598-018-22488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watahiki A., Wang Y., Morris J., Dennis K., O'Dwyer H.M., Gleave M., Gout P.W. MicroRNAs associated with metastatic prostate cancer. PloS One. 2011;6(9) doi: 10.1371/journal.pone.0024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koppers-Lalic D., Hackenberg M., de Menezes R., Misovic B., Wachalska M., Geldof A., Zini N., de Reijke T., Wurdinger T., Vis A. Noninvasive prostate cancer detection by measuring miRNA variants (isomiRs) in urine extracellular vesicles. Oncotarget. 2016;7(16):22566–22578. doi: 10.18632/oncotarget.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bhattacharya A., Ziebarth J.D., Cui Y. SomamiR: a database for somatic mutations impacting microRNA function in cancer. Nucleic Acids Res. 2013;41(Database issue):D977–D982. doi: 10.1093/nar/gks1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bhattacharya A., Cui Y. SomamiR 2.0: a database of cancer somatic mutations altering microRNA-ceRNA interactions. Nucleic Acids Res. 2016;44(D1):D1005–D1010. doi: 10.1093/nar/gkv1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Galka-Marciniak P., Urbanek-Trzeciak M.O., Nawrocka P.M., Dutkiewicz A., Giefing M., Lewandowska M.A., Kozlowski P. Somatic mutations in miRNA genes in lung cancer-potential functional consequences of non-coding sequence variants. Cancers. 2019;11(6) doi: 10.3390/cancers11060793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.