Abstract
Gene fusions that contribute to oncogenicity can be explored for identifying cancer biomarkers and potential drug targets. To investigate the nature and distribution of fusion transcripts in cancer, we examined the transcriptome data of about 9,000 primary tumors from 33 different cancers in TCGA (The Cancer Genome Atlas) along with cell line data from CCLE (Cancer Cell Line Encyclopedia) using ChimeRScope, a novel fusion detection algorithm. We identified several fusions with sense (canonical, 39%) or antisense (non-canonical, 61%) transcripts recurrent across cancers. The majority of the recurrent non-canonical fusions found in our study are novel, unexplored, and exhibited highly variable profiles across cancers, with breast cancer and glioblastoma having the highest and lowest rates, respectively. Overall, 4,344 recurrent fusions were identified from TCGA in this study, of which 70% were novel. Additional analysis of 802 tumor-derived cell line transcriptome data across 20 cancers revealed significant variability in recurrent fusion profiles between primary tumors and corresponding cell lines. A subset of canonical and non-canonical fusions was validated by examining the structural variation evidence in whole-genome sequencing (WGS) data or by Sanger sequencing of fusion junctions. Several recurrent fusion genes identified in our study show promise for drug repurposing in basket trials and present opportunities for mechanistic studies.
Keywords: pan-cancer analysis, antisense fusion, TCGA, CCLE, fusion transcripts, primary tumor-cell line comparison, cancer, ChimeRScope, therapeutic targets, recurrent fusions
Introduction
Genomic instability, a hallmark of cancer, leads to the accumulation of dynamic changes in the genome manifested as structural variants (SVs) that include quantitative (copy number variations [CNVs]), positional (translocations), or orientational (inversions) rearrangements. Fusion genes, frequently observed in cancer, are the consequences of such structural rearrangements of the genome1 resulting in the concatenation of two different genes or gene fragments. Fusion transcripts that originate from fusion genes are likely unique to a cancer type, which can be exploited to understand the underlying mechanisms of malignancy and can serve as effective diagnostic or prognostic markers.2,3 These chimeras could contribute to oncogenicity by altering the expression of tumor suppressor or proto-oncogenes, or by modifying the original function of a protein resulting in an abnormal chimeric protein that stimulates tumorigenesis. Examples of established cancer-specific biomarkers include the nucleophosmin-anaplastic lymphoma tyrosine kinase (NPM-ALK) fusion transcript for the identification of NPM-ALK-positive anaplastic large cell lymphoma (ALCL), and BCR-ABL1 fusion in chronic myeloid leukemia.4,5 Chimeric genes have also been used for molecular subtyping of cancers, leading to the development of precisely targeted interventions, including heterogeneous cancers such as prostate cancer.6 Also, cancer-specific fusions can serve as ideal therapeutic targets leading to effective treatment strategies. Dasatinib, imatinib, and ponatinib are drugs that target the fusion gene BCR-ABL1 in chronic myeloid leukemia.7,8
Although fusion transcripts in cancer resulting from chromosomal aberrations were identified more than six decades ago,9,10 recent advances in next-generation sequencing (NGS) technologies have helped to advance this field immensely. Although fusions were initially found to be more prevalent in liquid cancers, later they were discovered to be prominent in solid tumors.11,12 Furthermore, fusion transcripts were initially assumed to be exclusive to cancer cells, but several reports identified a large number of fusion transcripts in non-cancerous cells,13,14 suggesting that they are prevalent in normal cells as well. Another less explored area is the concept of the transcriptionally induced fusions resulting from a spin-off of transcriptional deregulation, including read-through transcription of neighboring genes or cis-splicing or trans-splicing events.15, 16, 17, 18, 19 This notion expands the space for a large number of fusion transcripts that were earlier unexplored due to the lack of supporting evidence at the structural variation level in the genome. Finally, evidence of natural antisense transcript (NAT) expression for at least 38% of the annotated transcripts in the cancer genome further adds to the pool of unexplored fusions containing antisense transcripts.20
The advances in NGS technologies have fueled the prospects of discovering therapeutically actionable fusion transcripts in cancer cells, leading to the development of a plethora of algorithms to detect fusions from both genomic and transcriptomic sequences.21, 22, 23 Most of the popular fusion detection algorithms rely on the alignment of NGS reads to the reference transcriptome,24,25 but they do not compensate for the extensive perturbations in the transcriptomes resulting from structural variations prevalent in the cancer genomes. This alignment technique potentially misses several true fusion events due to poor alignment of reads from the perturbed cancer transcriptome to the reference sequences. Of the several algorithms available for identifying fusion transcripts from RNA sequencing (RNA-seq), a novel alignment-free algorithm, ChimeRScope, developed recently by our group, performed superiorly compared to other popular tools with higher specificity and sensitivity.16 ChimeRScope employs short k-mer-based unique transcript fingerprints to serve as a reference to match with k-mers from cancer transcriptome reads and identify fusion transcripts in cancer cells that harbor frequent chromosomal abnormalities and mutations.
Using ChimeRScope, we explored the fusion landscape of 8,883 primary tumor transcriptomes and 802 cancer cell lines, using TCGA (The Cancer Genome Atlas, https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) and CCLE (Cancer Cell Line Encyclopedia, https://www.broadinstitute.org/ccle) datasets across 33 cancers. TCGA harbors the most extensive cancer omics data with more than 11,000 patient samples spanning across 33 cancer types and provides a platform for integrative molecular analysis of cancer. Although the fusion landscape of several major cancer types from TCGA was analyzed before,26, 27, 28, 29, 30 the fusion detection methods employed were primarily alignment-based with limited sensitivity and the overlap among these reports was sparse.25,31,32 Importantly, most of these studies were limited only to fusions with evidence at the genomic level or to canonical fusions (both partners in the sense orientation). In contrast, we used ChimeRScope in this study to explore the entire gamut of fusions, including the canonical and non-canonical (one or both transcripts in antisense orientation) types as well as the potentially transcription-induced fusions (with lacking evidence of fusion at the genome level). We also screened for fusions that are recurrent in the common cancer cell lines (CCLE) as a means to corroborate fusions that are co-occurring in corresponding primary tumors. Fusions in CCLE that are also detected in TCGA patients serve as a valuable resource to carry out mechanistic studies aimed at exploring the functional significance of these events. Furthermore, the fusions identified in cell lines could be rapidly tested to understand the mechanistic bases that generate different species of fusions in cancer. The novelties of our work include the use of an alignment-free fusion prediction tool,23 comprehensive analysis of different types of fusion transcripts that include canonical and non-canonical fusions, and detection of potentially transcriptome-induced fusions. In addition to fusion detection from 33 cancer types, we also experimentally validated some of the predicted fusions by Sanger sequencing of fusion transcripts identified from CCLE cell lines. To our knowledge, the catalog of fusions and the analysis results presented in this report are the most comprehensive and insightful to date. Data from this study present a vast pool of common unreported fusion events across several cancer types and non-canonical transcripts whose relevance and clinical significance are yet to be explored.
Results
Fusion Gene Distribution in Tumor and Normal Samples
We comprehensively screened 8,883 primary tumor and 730 normal adjacent-tissue samples spanning 33 cancer types from TCGA in this study (Table 1). BRCA (breast invasive carcinoma) followed by LGG (brain lower grade glioma), LUAD (lung adenocarcinoma), LUSC (lung squamous cell carcinoma), and PRAD (prostrate adenocarcinoma) are the most dominant types analyzed, with 500 or more tumor samples in each cancer type. CHOL (cholangiocarcinoma), DLBC (lymphoid neoplasm diffuse large B cell lymphoma), and UCS (uterine carcinosarcoma) are among those with the smallest sample sizes, with approximately 50 tumors. The sample sizes of matched adjacent-normal and tumor tissues are not consistent across cancer types in TCGA. BRCA and kidney renal clear cell carcinomas (KIRCs) are among the highest number of matched adjacent-normal samples, but some cancers have few or none. To compensate for the lack of normal samples for some cancer types, fusions identified in any normal sample were filtered out from fusions identified in any tumor sample. Another rationale to remove all fusions identified from adjacent normal tissues was the possibility of library construction or alignment artifacts that could lead to false positives.33 A few well-known cancer-associated fusions, such as TMPRSS2-ERG and BCR-ABL1, were also identified in some normal samples. TMPRSS2-ERG fusion was identified in three matching normal and tumor samples in prostate adenocarcinoma; hence, these cases could have resulted from sample contamination with the tumor. Similarly, BCR-ABL1 fusion, the prognostic marker for chronic myeloid leukemia, was identified in one of the normal samples in KIRC. To include the well-characterized cancer-associated fusions found in the normal samples, we retained all of the cancer fusions cataloged in the COSMIC (https://cancer.sanger.ac.uk/cosmic/fusion) database. In addition to these inconsistencies, we also identified 202 fusions from normal samples that were identified as tumor-associated fusions by a recent report28 (Figure S1). Since we identified these fusions in the control samples, they were also eliminated from further analyses.
Table 1.
List of TCGA and CCLE samples analyzed in this study
| TCGA-Acronym | Cancer | Tumor Samples Processed (%) | Normal-Adjacent Samples Processed (%) | Cell Lines Processed (%) |
|---|---|---|---|---|
| TCGA-ACC | adrenocortical carcinoma | 79 (0.9) | 0 | 0 |
| TCGA-BLCA | bladder urothelial carcinoma | 408 (4.6) | 19 (2.6) | 26 (3.2) |
| TCGA-BRCA | breast | 1,094 (12.3) | 113 (15.5) | 52 (6.5) |
| TCGA-CESC | cervical squamous cell carcinoma and endocervical adenocarcinoma | 304 (3.4) | 3 (0.4) | 22 (2.7) |
| TCGA-CHOL | cholangiocarcinoma | 36 (0.4) | 9 (1.2) | 0 |
| TCGA-COAD | colon adenocarcinoma | 244 (2.7) | 41 (5.6) | 58 (7.2) |
| TCGA-DLBC | lymphoid neoplasm diffuse large B cell lymphoma | 48 (0.5) | 0 | 54 (6.7) |
| TCGA-ESCA | esophageal carcinoma | 161 (1.8) | 11 (1.5) | 25 (3.1) |
| TCGA-GBM | glioblastoma | 117 (1.32) | 5 (0.7) | 0 |
| TCGA-HNSC | head and neck squamous cell carcinoma | 458 (5.2) | 44 (6.0) | 32 (4) |
| TCGA-KICH | kidney chromophobe | 65 (0.7) | 24 (3.3) | 0 |
| TCGA-KIRC | kidney renal clear cell carcinoma | 474 (5.3) | 72 (9.9) | 21 (2.6) |
| TCGA-KIRP | kidney renal papillary cell carcinoma | 288 (3.2) | 32 (4.4) | 0 |
| TCGA-LAML | acute myeloid leukemia | 131 (1.5) | 0 | 0 |
| TCGA-LGG | brain lower grade glioma | 508 (5.7) | 0 | 63 (7.9) |
| TCGA-LIHC | liver hepatocellular carcinoma | 371 (4.2) | 50 (6.8) | 30 (3.7) |
| TCGA-LUAD | lung adenocarcinoma | 506 (5.7) | 59 (8.1) | 0 |
| TCGA-LUSC | lung squamous cell carcinoma | 501 (5.6) | 49 (6.7) | 184 (22.9) |
| TCGA-MESO | mesothelioma | 86 (1) | 0 | 1 (0.1) |
| TCGA-OV | ovarian | 305 (3.4) | 0 | 45 (5.6) |
| TCGA-PAAD | pancreatic adenocarcinoma | 182 (2) | 4 (0.5) | 41 (5.1) |
| TCGA-PCPG | pheochromocytoma and paraganglioma | 178 (2) | 3 (0.4) | 0 |
| TCGA-PRAD | prostate adenocarcinoma | 497 (5.6) | 52 (7.1) | 7 (0.9) |
| TCGA-READ | rectum adenocarcinoma | 94 (1.1) | 10 (1.4) | 0 |
| TCGA-SARC | sarcoma | 259 (2.9) | 2 (0.3) | 38 (4.7) |
| TCGA-SKCM | skin cutaneous melanoma | 103 (1.2) | 1 (0.1) | 51 (6.4) |
| TCGA-STAD | stomach adenocarcinoma | 375 (4.2) | 32 (4.4) | 37 (4.6) |
| TCGA-TGCT | testicular germ cell tumors | 150 (1.7) | 0 | 0 |
| TCGA-THCA | thyroid carcinoma | 426 (4.8) | 58 (7.9) | 12 (1.5) |
| TCGA-THYM | thymoma | 119 (1.3) | 2 (0.3) | 0 |
| TCGA-UCEC | uterine corpus endometrial carcinoma | 180 (2.0) | 35 (4.8) | 3 (0.4) |
| TCGA-UCS | uterine carcinosarcoma | 56 (0.6) | 0 | 0 |
| TCGA-UVM | uveal melanoma | 80 (0.9) | 0 | 0 |
Percent contribution of samples analyzed in each dataset is included in the parenthesis.
To limit our analyses to high-confidence fusions, only the recurring fusions (those occurring more than once within or across any cancer type) were analyzed in this study. After removing fusions from normal adjacent samples and fusions that did not pass our stringent filters, 4,344 recurrent fusions were identified. They include canonical, non-canonical, or read-through types. Of all of the predicted fusions across 33 cancer types, breast carcinoma accounted for the largest share (57.4%), followed by LUSC (9.8%), HNSC (head and neck squamous cell carcinoma) (4.1%), and BLCA (bladder urothelial carcinoma) (4%) (Table S1). We also noted significant differences in the percentage of samples containing at least one recurrent fusion across cancer types. For example, LUSC, UCS, and SARC (sarcoma) contained the highest number of samples with at least one recurrent fusion (>90%), while UVM (uveal melanoma), OV (ovarian serous cystadenocarcinoma), and THCA (thyroid carcinoma) had the lowest number of samples with at least one recurrent fusion (<40%).
Analysis of Mixed Species of Fusion Transcripts
We identified four distinct species of fusion transcripts among those predicted by ChimeRScope. These include fusions with (1) both partner transcripts in the sense orientation; (2) the 5′ partner in sense and the 3′ partner in antisense orientation; (3) the 5′ partner in antisense and the 3′ partner in sense orientation; and (4) both partner transcripts in antisense orientation. The first class of fusions is referred to as “canonical” because the intended functions of both genes are likely to be preserved. However, the last three species of fusions have at least one partner transcript in antisense orientation, which could lead to a variety of functional alterations. These groups are together referred to as ‘non-canonical’ fusions. The percentage of canonical and non-canonical fusions varied vastly across the cancer types studied both in TCGA and CCLE data (Figure 1A; Table S1).
Figure 1.
Representation of Different Types of Fusions Identified in TCGA and CCLE
33 cancers were analyzed in TCGA and 20 in CCLE. Each cancer in TCGA is color-coded based on the organ system. (A) Pie charts illustrate fusion percentage in the cancers of organ systems analyzed in this study. The percentage of recurrent canonical (red) and non-canonical (blue) fusions in each cancer is represented by pie charts and is calculated without considering the read-through fusions identified in each cancer type. (B) The table represents the total number of samples analyzed and the percentage of samples with canonical or non-canonical fusions in each cancer. Note that the percentages are overlapping, as a sample may contain both canonical and non-canonical recurrent fusions.
Overall, canonical fusions were predicted at a much higher rate than non-canonical fusions in most cancers with two striking extremities: glioblastoma showed the highest number of canonical fusions (100%) while breast cancer had the highest number of non-canonical fusions (79%) in TCGA samples. UVM and THYM (thymoma) also displayed a very high fraction (>90%) of canonical fusions. In general, a very high percentage of CCLE samples contained recurrent fusions across all cancers compared to those of TCGA (Figure 1B). Results from CCLE samples mostly corroborate the results from TCGA with a few exceptions. MESO (mesothelioma) and PAAD (pancreatic adenocarcinoma) had more non-canonical fusions in CCLE than did corresponding primary tumors in TCGA. Alternatively, OV and BRCA had more canonical fusions in CCLE samples than in TCGA. Most of the CCLE samples showed an even distribution of canonical and non-canonical fusions. In contrast, TCGA samples contained predominantly canonical fusions with a relatively small proportion (less than 10%) of non-canonical ones.
Frequency of Recurrent Fusions in TCGA Samples
Among the canonical fusions identified by ChimeRScope across 33 cancer types in TCGA, 1,665 fusions were recurrent (n ≥ 2) either within or across cancers, and we focused our further analyses on these recurrent fusions. The top 40 recurrent fusion gene pairs and their frequencies across cancers are presented in Figure 2, and a full list of the recurrent fusions is provided in Table S2. Frequent fusions partnering with a kinase (mitogen-activated protein kinase-activated protein kinase 5 [MAPKAPK5]), a receptor (cholinergic receptor muscarinic 3 [CHRM3]), a transporter (solute carrier organic anion transporter family member 1A2 [SLCO1A2]), or an ATP-binding cassette subfamily C member 9 (ABCC9) were detected in more than 13 cancer types. Among the fusions represented in Figure 2, those containing G protein-coupled receptor (MAS-related G protein-coupled receptor member X3 [MRGPRX3]), SLCO1A2, and CHRM3 were recurrent, with more than 50 occurrences (indicated by red and green boxes) within a cancer type (Table S2).
Figure 2.
Heatmap Representing the Top 40 Recurrent Canonical Fusions Predicted in TCGA Tumors
Normalized fusion frequencies across cancers are represented as a heatmap.
ChimeRScope predictions revealed several new associations for known cancer fusions (Table S2) with unknown cancer types. For example, our analysis shows that PTPRK-RSPO3 (protein tyrosine phosphatase receptor type K and R-spondin 3) gene fusion known to be associated with colon cancer34,35 was also identified in READ (rectum adenocarcinoma) and STAD (stomach adenocarcinoma) along with the COAD (colon adenocarcinoma), which suggests that these gene rearrangements might be a common genetic event in intestinal cancers. FGFR3-TACC3 (fibroblast growth factor receptor 3 and transforming acidic coiled-coil containing protein 3) fusion associated with multiple cancers33, 36, 37 was also found to be recurrent in BLCA, ESCA (esophageal carcinoma), CESC (cervical squamous cell carcinoma and endocervical adenocarcinoma), HNSC, LIHC (liver hepatocellular carcinoma), and LUSC. The occurrence of FGFR3-TACC3 fusion in CESC, ESCA, and LIHC has not been reported previously, signifying that this common fusion is prevalent in more tissue types than previously discovered. Other FGFR fusions partnering with BicC family RNA binding protein 1 (BICC1), shootin 1 (SHTN1), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and arginyltransferase 1 (ATE1) have also been detected in multiple cancers from our analysis. Several of these FGFR fusions have been identified as activating fusions that have been linked to activation of downstream genes.38 These fusions could be investigated as an attractive actionable target for chemotherapy. Following the same trend, ETV6-NTRK3 (ETS variant 6 and neurotrophic tyrosine kinase, receptor, type 3) fusions were also identified in multiple cancers including breast, colon, skin, and thyroid cancers. This ETV6 fusion with NTRK3 has also been identified as oncogenic by the activation of the Ras-MAPK and phosphatidylinositol 3-kinase (PI3K)-AKT pathways.39 In contrast, most of the ESR1 (estrogen receptor 1) fusions except for ESR1-CCDC170 (coiled-coil domain containing 170) were found to be exclusively present in breast cancer. ESR1-CCDC170 was also identified in uterine cancers. This fusion was reported to be enriched in luminal-B tumors,40 which increases the aggressiveness of estrogen receptor (ER)+ breast cancer cell phenotype by enhancing cell migration, invasion, and reducing endocrine sensitivity.40 Since these fusions were also identified in uterine cancers, a similar hormone-dependent phenotype linked to this fusion may exist in uterine carcinoma. The ESR1 locus was reported to have several amplifications in TCGA datasets.41 Several other fusion partners of ESR1 were also identified in breast cancer exclusively, including TMEM12, collagen (COL1A1, COL3A1), actin (ACTB, ACTG1), and IGFBP5 (insulin-like growth factor [IGF] binding protein 5).
The genes participating in recurrent canonical fusions were enriched by various oncogenic signaling pathways. Ingenuity Pathway Analysis (IPA) revealed the enrichment of these fusion genes in several signaling pathways, including EIF2 (eukaryotic translation initiation factor 2), TGFB1 (transforming growth factor [TGF] beta 1), VEGF (vascular endothelial growth factor), HER2 (human epidermal growth factor receptor 2), and estrogen receptor signaling, suggesting that most of these fusions have the potential for oncogenicity regardless of the tissue type (Table S3).
Understanding the landscape of genes forming recurrent fusions either within or across cancers can provide significant insight into the mechanisms of fusion gene formation. Frequencies of genes participating in canonical and non-canonical fusions in TCGA indicate that some genes are frequently fused with the same partner. For example, CHRM3 was one of the most frequent fusion gene partners in 15 cancers in both canonical and non-canonical fusions (Figure S2A).
Frequency of Recurrent Non-canonical Fusions in TCGA
Out of the 2,605 recurrent non-canonical fusions in TCGA, several fusions were identified in 10 or more cancers (Table S4). Interestingly, note that although most of the cancers in TCGA had recurrent non-canonical fusions, these fusions were detected from a small percentage of samples. IL34 was one among the top recurrent gene among non-canonical fusions identified in eight cancers (Figure S2B). IL34 expression has been associated with the progression of tumor growth, metastasis, and poor prognosis in several cancers.42, 43, 44 IPA of transcripts participating in recurrent non-canonical fusions revealed the enrichment of several important cancer-related pathways, including eIF2 and eIF4 (eukaryotic initiation factors), GP6 (glycoprotein VI), mTOR (mammalian target of rapamycin), ILK (integrin-linked kinase), and HIF1a (hypoxia-inducible factor 1) signaling pathways (Table S5). Some genes in these pathways are important targets for chemotherapy45 and could be explored for identifying actionable targets.
Distribution of Fusion Breakpoints in the Exons of Partner Genes
To understand the breakpoint preference among canonical and non-canonical fusions, we classified fusions based on the distance of breakpoint (at the fusion junction) from the participating exon boundaries as E-E (both breakpoints close to the exon boundaries), E-M or M-E (only one of the breakpoints near the boundary), and M-M (both breakpoints away from the exon boundaries) (refer to Materials and Methods). If the breakpoint in exons occurs randomly, the proportion of each class should be one-third, but if the breakage event happens preferentially near exon boundaries, there should be a higher proportion of E-E fusions. Our results showed that canonical fusions in most cancers in TCGA belonged to the E-E category except for UVM and BRCA, which exhibited the highest numbers of fusions in the E-M category (Figure S3A). UVM (25%), BRCA (23%), OV (18%), and kidney cancers (KIRC, 14% and KIRP [kidney renal papillary cell carcinoma], 16%) had some of the highest frequency of fusions falling into the M-M region. Since the majority of breakpoints in canonical fusions fall into the E-E regions, these fusion events are likely influenced by the intragenic splicing mechanism as suggested in an earlier report.14 Since fusions involving genes in opposite transcriptional orientation (antisense) would have different intron-exon structures, we predicted that most of the fusion junctions would not be in the standard exon-intron junctions (E-E regions). As expected, fusion breakpoints in the non-canonical fusions were frequent in the E-M and M-M region when compared to those of the canonical fusions (Figure S3B).
Kinase Partners with Fusion Transcripts from Tumor Samples
Several reports suggest the heavy involvement of kinases in fusion transcripts is an indicator of oncogenic driver events in some cancers.28,33 To investigate the type and frequency of kinases predicted by ChimeRScope, we compared our fusion transcripts against known human kinases. The percentages of kinases identified among canonical and non-canonical fusions in TCGA are listed in Table S6. Approximately 7% of the recurrent canonical fusions identified from tumor samples contained a kinase gene. Thyroid carcinoma had a very high percentage of recurrent canonical kinase fusions when compared to the rest of the cancers (>60%) (Figure S4). An earlier report33 also identified a high percentage of recurrent kinase fusions in thyroid cancer, indicating that kinase fusions could be explored as biomarkers or therapeutic targets in this cancer. Although the number of recurrent canonical fusions detected in GBM (glioblastoma multiforme) and CHOL was rather low, more than 15% of the fusions identified in these cancers contained a kinase. Several kinase fusions were recurrent across multiple cancers, with some fusions identified in five or more cancer types (Table S7). Several kinases including SMG1 (nonsense-mediated mRNA decay associated PI3K related kinase), ERBB2 (erb-b2 receptor tyrosine kinase 2), RET (rearranged during transfection), and FGFR2 (fibroblast growth factor 2) were promiscuous with more than three partners across cancer types (Figure S5). MAPKAPK5 specifically fused with ACAD10 (acyl-CoA [coenzyme A] dehydrogenase family member 1) in 22 occurrences across 16 cancer types. EML4 (echinoderm microtubule-associated protein-like 4) fusions with ALK (ALK receptor tyrosine kinase) that were previously associated with non-small-cell lung cancer (NSCLC) were identified in LUAD along with KIRP and THCA at low frequency. A different ALK fusion, STRN (Striatin)-ALK was also expressed in KIRP and THCA at low frequencies. Pathway analysis for these recurrent kinase fusions revealed several highly enriched pathways related to cancer, including nuclear factor κB (NF-κB) signaling, FGF signaling, regulation of the epithelial-mesenchymal transition pathway, PTEN signaling, FAK signaling, gap junction signaling, HER-2 signaling in breast cancer, IGF-1 signaling, and ERK (extracellular signal-regulated kinase)/MAPK signaling (Table S8), signifying that these fusion genes could affect oncogenesis.
Compared to recurrent canonical fusions, significantly few kinases were identified among non-canonical fusions in TCGA (Table S9). A fusion containing PRKCA (protein kinase C alpha) was the most frequent, identified in five of the cancers screened. Half of the cancers did not have recurrent non-canonical fusions that involved a kinase gene, indicating that this is a rare phenomenon in most cancers. Understanding the mechanism of non-canonical fusion formation would help address this bias in kinase frequency among these fusion types.
Fusions Containing an Oncogene or Tumor Suppressor Gene
Since alteration of oncogene or tumor suppressor functions can be key to the initiation and progression of cancer, we also screened for these fusions in our dataset. Of the recurrent canonical fusions identified in TCGA samples, 211 fusions contained at least one oncogene or tumor suppressor gene (TSG) (Table S10). Fusions containing PRKAR1A (protein kinase cyclic AMP [cAMP]-dependent type I regulatory subunit, oncogene/TSG), FGFR3 (oncogene), MKL1 (oncogene/TSG), NCOR2 (nuclear receptor corepressor 2, TSG), CCDC6 (coiled-coil domain containing 6, TSG), and SUZ12 (oncogene/TSG) were identified in more than four cancer types. CCDC6-RET fusions in thyroid and CBFB-MYH11 (core-binding factor, beta subunit and myosin heavy chain 11) fusions in LAML (acute myeloid leukemia) were recurrent. CCDC6-RET fusion was identified in lung adenocarcinoma and colon cancer, consistent with earlier reports.33 We also identified other fusion partners for RET: ERC1 (ELKS/RAB6-interacting/CAST family member 1)-RET (BRCA, THCA) and NCOA4 (nuclear receptor coactivator 4)-RET (THCA). RET fusions seem to be recurrent in thyroid cancer, providing strong justification for targeted treatment approaches in this cancer. SUZ12 participated in recurrent fusions across 11 cancer types. PRKAR1A fusions were recurrent across and within several cancer types. PRKAR1A fused with multiple partners; out of these, COBW domain-containing proteins (CBWD3, CBWD5, CBWD6) were identified in several cancers, including lung and bladder cancer.
The percentage of TSG/oncogene fusions among the recurrent non-canonical fusions predicted was similar to those in the canonical fusions, although the majority of these fusions were identified from BRCA (Table S11). Fusions containing VHL (von Hippel-Lindau tumor suppressor), AXIN2 (axin 2, tumor suppressor), and KDM5A (lysine-specific demethylase 5A, oncogene) were identified in three or more cancers. Several fusions containing B2M (β-2 microglobulin), an important tumor suppressor in immune response, were detected in breast cancer, warranting the need to investigate the functional consequence of these fusions. Another interesting observation of breast cancer was the occurrence of ErbB2 fusions. BRCA samples that are HER2 positive or the HER2-E PAM50 subtype had statistically higher ErbB2 fusions (Fisher’s exact test p = 0.004 and 0.002, respectively). HER2A-amplified BRCA also had a higher percentage of ErbB2 fusions (Fisher’s exact test p = 0.0001), which were reflected in higher ErbB2 expression (Fisher’s exact test p = 1.2E−05). Overexpression of ErbB2, a receptor tyrosine kinase with intrinsic tyrosine kinase activity, is associated with breast cancer metastasis and lower survival rates.46 These observations indicate the need to investigate ErbB2 fusions in breast cancer and their association with gene expression and patient survival.
Recurrent Fusions in Oncogenic Pathways
To investigate oncogenic pathways that are affected by fusions in each TCGA cancer type, we cataloged recurrent fusions identified across 10 important oncogenic pathways (selection based on a previous report47). RTKRAS, NOTCH, PI3K, and HIPPO signaling pathways were found to be the most affected oncogenic pathways by recurrent canonical fusions (Figure 3A; Table S12). Thyroid cancer had more than 50% of the recurrent canonical fusions in the RTK (receptor tyrosine kinase)-RAS pathway, with a high frequency of CCDC-RET fusion. The frequency of non-canonical fusions in these selected oncogenic pathways was relatively less compared to canonical fusions, except for BRCA (Figure 3B). Interestingly, fewer than 4% of the genes in these oncogenic pathways participated in canonical recurrent fusions, whereas participation in recurrent non-canonical fusions was even less (2%).
Figure 3.
Frequency of Recurrent Canonical and Non-canonical Fusions in TCGA among Nine Oncogenic Pathways
Gene frequencies were normalized against sample size, recurrent fusion frequency in each cancer, and genes in each pathway. (A) Canonical fusions. (B) Non-canonical fusions.
Distribution of Read-Through Fusions across Cancers
Along with fusion transcripts resulting from structural rearrangements or trans-splicing events, we also analyzed fusions that resulted from read-through events. We identified 351 such events across 33 cancers, with sparse representation from most cancer types, out of which 74 were recurrent (Table S13). OV, ESCA, and LAML mainly contributed to the pool of read-through fusions (>40%) while some cancers have none (Table S1). Several common fusions were identified across multiple cancers. For example, ZCCHC8-RSRC2 fusion was detected in five cancers (BLCA, BRCA, CESC, HNSC, and PAAD) at very low frequency (Table S13). This read-through fusion has been recently reported in acute lymphoblastic leukemia48 and other cancers27 but it has not been detected before in pancreatic and cervical cancer.
Comparison of Recurrent Fusions across Cancer Types in TCGA Samples
We compared the landscape of recurrent fusions across all cancers in TCGA to understand the similarities and differences among them. The similarities among cancers are determined based on their recurrent fusion profile in multiple cancers. The fusion profiles of various cancers were vastly diverse, with high variability in both abundance and frequency of individual fusions. BRCA, LUSC, HNSC, and BLCA had some of the highest frequency of fusions (>600 in each), while MESO, KICH (kidney chromophobe), GBM, and UVM had the lowest number of fusions (<30 in each) (Table S1). Cancers also exhibited variability in the abundance of non-canonical fusions. Cancers that generally had higher canonical fusions also had higher non-canonical fusions, with a few exceptions.
Large variations in the chromosomal location of genes participating in canonical and non-canonical fusions were identified from our analysis. For example, canonical fusions were frequent in the genes located on chromosomes 1, 4, 6, 7, 16, and 17 in more than half of the cancers in TCGA (Figure S6A). LUAD and LUSC had a large fraction of genes on chromosome 21 that participated in canonical fusions. Since several other chromosomes with high fusion occurrence were also shared between these cancers, it would be beneficial to investigate these regions further to identify hotspots for gene fusion. Genes on chromosomes 1, 3, 17, 12, and 19 were frequently involved in non-canonical fusions in more than half of the cancers in TCGA (Figure S6B).
We constructed an all-to-all cancer similarity matrix using recurrent canonical fusions to compare and contrast the cancers based on their fusion transcript profiles. The highest similarity was observed between LUSC and THYM (similarity score = 0.84) (Figure S7A). LUSC and THYM shared 39 recurrent fusions, including RAB3IP (interactor of the Ras-like GTPase Rab3A) and CHRM3 as recurrent 5′ fusion partners with other genes in both cancers. RAB3IP and CHRM3 are both proto-oncogenes associated with colon and liver metastasis.49, 50, 51 We also identified RAB3IP fusions in gastrointestinal (GI) cancers, including STAD, COAD, CHOL, and LIHC. LUSC displayed the highest degree of similarity with other cancer types (average similarity across all cancers = 0.36), suggesting that LUSC shares a fusion landscape with several other cancer types. It is important to investigate such fusion signatures across tumors to understand common pathways contributing to cancer development. GBM exhibited the least degree of similarity across cancers (average similarity = 0.27), which is most likely due to the low frequency of unique fusions detected in these samples.
Hierarchical clustering of recurrent canonical fusions to classify similar TCGA tumors identified several distinct clusters (Figure S7B). Breast and female reproductive cancers (OV and UCS) and GI tract cancers (ESCA, STAD, LIHC, PAAD, and READ) clustered as two separate groups, indicating similarities in the fusion profiles among these cancers. Several cancers originating from similar cells or organ systems also clustered together. For example, uterine (UCEC [uterine corpus endometrial carcinoma] and UCS), kidney (KIRC and KIRP), lung (LUAD and LUSC), and melanoma of the skin and eye (SKCM [skin cutaneous melanoma] and UVM) exhibited similar fusion profiles. In addition, cancers with squamous histology (LUSC, HNSC, CESC, and BLCA) clustered together, indicating similarities in the fusion profile.
Since several GI cancers in TCGA clustered together, showing a similarity in recurrent canonical fusion profile, we further analyzed the recurrent fusions identified in the GI tract cancers. Fusions involving CHRM3 were expressed in several GI tract cancers studied. CHMR3 is reported to play an essential role in regulating cell proliferation and migration of several cancers, including colon and gastric cancers.52,53 In addition to GI cancers, ovarian, cervical, and Merkel cell cancer have shown evidence of activated muscarinic receptor expression leading to cell proliferation, survival, migration, and angiogenesis.54,55 Pathway analysis of common fusion transcripts across cancers of the GI tract using IPA has identified the TR (thyroid hormone receptor)/RXR (retinoid X receptor) activation pathway as enriched (Benjamini-Hochberg [BH] multiple testing correction p = 0.003). RXR is a master regulator and plays a central role in nuclear signaling, and a truncated RXR has been associated with oncogenicity.56
Clustering of non-canonical recurrent fusions identified across cancers also revealed a similar pattern, with some notable differences (Figures S8A and S8B). In this case, endocrine cancers (PCPG [pheochromocytoma and paraganglioma] and ACC [adrenocortical carcinoma]) were clustered together, but uterine, kidney, lung, and melanomas were found to be distributed in different clusters. Cancers with squamous histology clustered together for both recurrent canonical and non-canonical fusions, indicating that they exhibited similar profiles for both the fusion types.
Recurrent Druggable Fusions
To evaluate the therapeutic implications of the recurrent fusions identified, we screened for recurrent gene fusions that were druggable. A fusion was defined as druggable if the evidence supporting the role of fusion was reported in DEPO,57 oncoKB,58 or Cancer Genome Interpreter databases.59 In total, 25 unique druggable fusions were identified across the canonical fusions, of which FGFR2 and RET can be potential targets in three fusions each (Table S14). A total of 14 genes participating in canonical fusions identified by ChimeRScope were targeted by 36 drugs in 21 different cancers. As shown in Figure 4, FGFR3 is the most common drug target among the six tumors. Both THCA and LUAD have the highest number of drugs (US Food and Drug Administration [FDA] approved or in various stages of clinical trials), targeting five genes in each tumor. UCS has the lowest number of targeted drug-gene interactions, with only one targeted gene, ESR1. The most frequent drug among the dataset is AZD4547, which targets FGFR genes (FGFR2 and FGFR3). ESR1 was a common drug target among both BRCA and UCS. Among the recurrent non-canonical fusions, ErBB2 and ESR1 fusions were druggable and were only identified in breast cancer samples.
Figure 4.
List of TCGA Cancers and Genes Participating in Recurrent Canonical Fusions Targeted by Drugs
Drugs include both those approved by the FDA and those in various stages of clinical trials. 14 genes participating in canonical fusions were targeted by 36 drugs in 21 different cancers as shown in the figure.
We also identified several fusions that could be repurposed to target the same genes but in different cancers. For example, loratinib is an FDA-approved drug for metastatic NSCLC that targets the ALK gene. We identified ALK gene fusions in other cancers, including KIRP, LUAD, and THCA (Table S14). These findings open up opportunities for repurposing drugs across pan-cancer targets in basket trials.
Fusions Identified from Cancer Cell Lines
To compare against the predicted recurrent fusions from TCGA primary tumor data, we also analyzed transcriptome-sequencing data from 802 common laboratory cell lines (from CCLE) that originated from 20 different tissues (Table 1; Table S15). The sample sizes for CCLE cell lines varied, with lung squamous cell carcinoma having the largest size and mesothelioma having the smallest size. Colon, pancreatic, esophageal, and lymphoma cell lines had higher than average fusion frequency per cell line. Thyroid, skin, and bladder cancer cell lines had the least number of fusions. We identified 2,524 unique recurrent fusions from these cell lines with approximately equal distribution of canonical and non-canonical fusions (Figure 1; Tables S16 and S17).
The percentage of non-canonical fusions in CCLE also varied across cancers, similar to the trend observed in primary tumor data from TCGA. Though MESO had only one cell line analyzed, 96% of the total fusions detected turned out to be non-canonical in that cell line. MESO, PAAD, and PRAD cell lines showed a higher percentage of non-canonical fusions than canonical fusions (Figure 1; Table S15). SARC, LUSC, OV, and THCA cell lines had less than 10% of non-canonical fusions, with SARC showing almost negligible fusions (0.4%).
A large number of recurrent canonical fusions (91 fusions) were identified in more than 35% of the cancer cell lines analyzed across cancers, indicating that fusions present in cell lines are more pervasive across cancers than the ones identified in primary tumors. A few of the recurrent canonical fusions identified in cell lines were also found to be ubiquitous in all of the cancer types we analyzed (Table S16). Two of these recurrent canonical fusions were also identified previously in non-cancerous cells or tissues, which probably points toward a non-oncogenic role for these common fusions.14
Canonical fusions in CCLE were mostly formed from the genes located on chromosomes 5, 6, 10, 14, 20, and X in 60%–70% of the cancers analyzed (Figure S9A). Some chromosomes (chromosomes 5, 10, 15, and 20) were frequent hotspots for both canonical and non-canonical fusions in CCLE (Figure S9B). OV and BRCA cell lines had chromosomes 4, 10, 13, 20, and 22 frequently participating in non-canonical fusions. We also identified a high frequency of chromosome Y genes in non-canonical fusions in lung squamous cell carcinoma.
In general, non-canonical fusions in CCLE also followed the same trend as canonical fusions, with some fusions occurring in more than 60% of the cell lines analyzed (Table S17). None of the non-canonical recurrent fusions in CCLE was reported earlier in non-cancerous cells or tissues.14 Hierarchical clustering analysis of recurrent canonical or non-canonical fusions in CCLE revealed different patterns, except for some cancers of squamous origin (ESCA and HNSC), which clustered together in both groups (Figures S10A, S10B, S11A, and S11B).
Comparison of Fusions Identified in TCGA and CCLE
We sought to compare the fusions shared between primary tumor (TCGA) and cancer cell line (CCLE) samples (1) to corroborate the predictions in two orthogonal systems, and (2) to experimentally validate their existence using cell line cultures. The pool of recurrent fusions identified from CCLE samples was vastly different from those identified in TCGA tumors, with minimal overlap (Figures S12A and S12B). Of the common fusions identified between the two groups, canonical fusions were found to be more common (9%) than non-canonical fusions (2%) (Table S18). Hierarchical clustering analysis of recurrent fusions identified in CCLE and TCGA also revealed separate clusters for primary tumor and cell lines for both canonical and non-canonical fusions, indicating that fusions in cell lines are vastly different from the primary tumors (Figures 5A–5D).
Figure 5.
Similarity Matrix and Hierarchical Clustering of Recurrent Fusions Identified from TCGA and CCLE
(A) Similarity matrix of recurrent canonical fusions predicted from TCGA and CCLE. Cancers were positioned according to their similarity in the fusion landscape. (B) Unsupervised hierarchical clustering of canonical recurrent fusions in TCGA and CCLE. (C) Similarity matrix of recurrent non-canonical fusions predicted from TCGA and CCLE. Cancers were positioned according to their similarity in the fusion landscape. (D) Unsupervised hierarchical clustering of non-canonical recurrent fusions in TCGA and CCLE.
We also compared fusions identified in specific cell lines to corresponding primary tumor data in TCGA to identify tumor-specific fusions. Relatively few fusions were common between cell line primary tumors, and the majority of these common fusions were non-recurrent canonical fusions (Figure S13). Breast cancer had the largest number of common fusions, followed by LUSC, COAD, and HNSC. In contrast to other cancers, breast cancer also had a high frequency of recurrent non-canonical fusions that were common between CCLE and TCGA samples.
Among the total fusions identified in this study, most of them are novel fusions (90%) with only a 10% overlap with those reported in TCGA (Figure S14). The majority of these novel fusions were non-recurrent and non-canonical fusions. Most of the reported fusions in TCGA that are also common between cell lines and TCGA primary tumors were non-recurrent canonical fusions. This suggests that the canonical fusions identified in our study are reliable, warranting further investigation of their functional significance in cancer biology.
Verification of the Predicted Fusions Using the Whole-Genome Sequencing (WGS) Data
As fusion transcripts mostly originate due to perturbations in the genome structure, we sought to verify the structural variations around the fusion genes, where corresponding WGS data are available for TCGA samples. Using the BreakDancer tool, WGS samples from skin, bladder, breast, glioblastoma, kidney, and thyroid cancer were analyzed (n = 88). Fusions were considered validated when BreakDancer reported the presence of structural variation for at least one of the participating fusion transcripts. Of the total fusions identified in these samples, 12% of canonical fusions and 18.6% of the non-canonical fusions were validated by BreakDancer (Table S19). However, most cancers had more than half of the fusions validated except for breast cancer and glioblastoma. Large variations in the percentage of validated fusions were observed, with breast cancer being an outlier with very low validation percentage. Among the samples screened for structural variations using BreakDancer, inter-chromosomal translocations (CTXs) and insertions (INSs) were relatively rare compared to other structural variations within a cancer (Table S20). Breast cancer displayed the highest percentage of both intra-chromosomal translocations (ITXs) and inversions (INVs), accounting for the majority of non-canonical fusions identified in these samples.
Since structural variation tools have limitations in identifying complex structural variations often associated with fusions, we also validated our predicted fusions by detecting discordant readpairs mapped to the chromosome locus in the WGS data. More than 90% of the fusions identified in the selected TCGA samples were validated using this method (Table S21). BreakDancer validated less than 20% of the fusions in the same samples, possibly due to the low sensitivity in detecting complex structural variations.60
Experimental Validation of Fusion Predictions by Sanger Sequencing
We further validated a selected set of our predicted recurrent fusions using several CCLE cell lines by PCR amplification and Sanger sequencing (Figure 6). Primers were designed to amplify fusion junctions for sequencing. Eight canonical and seven non-canonical fusions identified in the RNA-seq data from nine cell lines (A549, SKBR3, BT-549, UACC-893, BT-474, SKES1, A673, ZR-75, and HDMB) were selected for validation. BT-549, SKES1, A549, and BT-474 had more than one fusion selected for validation (Table S22). Thirteen of the 15 total fusion transcripts tested contained chimeric transcripts in the correct orientation as predicted by ChimeRScope and confirmed by Sanger sequencing. We were also able to validate the existence of non-canonical fusions (at least one partner in antisense orientation: GBA-MTX1, DNHD1-RRP8, PRDM4-PWP1, RPL39L-ST6GAL1, BOP1-MROH1, SPDYE3-UPK3B) in CCLE cell lines, indicating that ChimeRScope can accurately predict fusions containing antisense genes along with canonical fusions (Figure S15).
Figure 6.
ChimeRScope Predictions along with PCR and Sanger Sequencing Results for Selected Fusions
(A) Agarose gel electrophoresis of amplicons containing fusion junctions for CLTC-VMP1 and PRKAR1A-PLCB1. (B) Graphical output from ChimeRScope Examiner illustrating the fusion events for CLTC-VMP1 and PRKAR1A-PLCB1. Name of the fusion partners, their original orientations, transcripts that might be involved in the fusion event, along with the highlighted region specifying the region near the fusion junction from each fusion partner are shown. (C and D) Sanger sequencing results confirming the fusions predicted by ChimeRScope for VMP1-CLTC and PLCB1-PRKAR1A fusions respectively.
Breast Cancer Fusion Profiles: Do They Affect Gene Expression, or Reflect Clinical Status? A Case Study
The frequency of recurrent fusions identified within canonical and non-canonical fusions varied across cancers, with breast cancer exhibiting the highest number of recurrent fusions of both kinds (Figure 1; Table S1). Within these recurrent fusions, non-canonical fusions were more prevalent than canonical fusions in this cancer (Table S1). BRCA samples in TCGA exhibited vast differences in the number of fusions identified per sample. To investigate the clinical and molecular characteristics of breast cancer samples exhibiting varied fusion frequencies, we categorized these samples into high or low fusion groups based on their fusion frequency. We did not detect any major differences in survival between these two groups (Kaplan-Meier estimates, p = 0.08, data not shown). We also compared their gene expression profiles to identify the functional significance of their fusion status. Gene expression analysis using edgeR identified 170 genes that were differentially regulated between breast cancer samples with high or low fusion status (false discovery rate [FDR] corrected p ≤ 0.05, absolute log2 fold change ≥ 1). Gene set enrichment analysis of these differentially regulated genes revealed several pathways related to hormone regulation, along with the regulation of RNA splicing through spliceosome (Table S23).
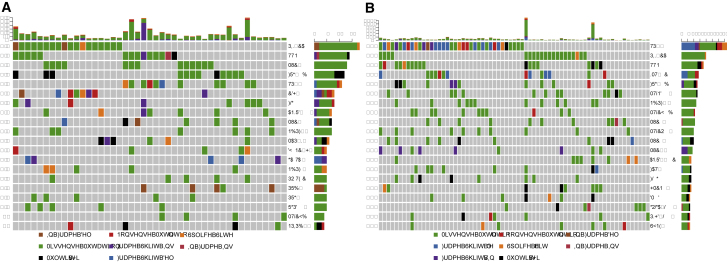
Mutual exclusivity of mutations and fusions was recently identified as an important driver of genes in cancer.28 To investigate whether these selected groups with high or low fusion frequency in breast cancer were driven by either fusions or mutations, the mutation profile of these samples was investigated. TP53 mutations were found to be significantly lower in the samples with high fusion category (23%) when compared to low fusion samples (51%, adjusted p = 0.01) (Figure 7). The high fusion category had very similar overall mutation frequency compared to low fusion samples.
Figure 7.
Oncoplots of the Mutated Genes in Breast Cancer Samples Containing High or Low Fusion Frequency
The mutation frequency for each gene is shown on the y axis of each plot. The vertical barplot on the top of each plot represents the total number of gene mutations in each sample. The horizontal bars represent the frequency and type of mutations detected. (A) High fusion group. (B) Low fusion group.
Discussion
This study analyzed 8,883 primary tumor samples spanning 33 cancer types from the TCGA database along with 802 cancer cell lines from CCLE using ChimeRScope and identified several novel fusion transcripts. We limited our analysis only to fusion transcripts that were recurrent (n ≥ 2) either within or across cancers and generated a catalog of frequent high-confidence fusions across pan-cancer primary tumor and cell line samples. Overall, 4,344 fusions were identified from TCGA in this study, of which 30.6% were also reported in the TCGA fusion database (https://www.tumorfusions.org/). The high overlap of our data with the reported fusions in TCGA is reassuring since the majority of other fusion detection methods have limited overlap of predicted fusions (2% among all methods and a maximum of 26% across any two methods25,31,61). ChimeRScope also detected fusions in several of the normal samples, which is consistent with a recent report that fusions are also common in non-tumor cells.14
An interesting observation from this study is the identification of a large number of non-canonical fusions that contained antisense transcripts. Only 6 of the 2,605 non-canonical recurrent fusions were also reported in the TCGA fusion database. We also compared our unique fusion list with other fusion databases62, 63, 64, 65 and found a very similar pattern with a higher number of reported canonical fusions than non-canonical fusions (Figure S14). This observation suggests that most of the fusions identified in our pan-cancer study are novel. These fusions need to be investigated further to determine their functions and impacts on cancer initiation, progression, or treatment resistance.
Compared to all other cancers in TCGA, breast cancer exhibited a very large number of non-canonical fusions in our analysis (78%). This observation is consistent with earlier reports that suggest breast cancer harbors a large number of structural variations compared to other cancers, specifically intrachromosomal translocations and inversions being the most common types.32,66 We also detected a high incidence of inversions and intrachromosomal variations in the BRCA-WGS data, which supports a large number of non-canonical fusions identified in breast cancer compared to other cancers (Table S20). A recent report on the transcriptional costs of structural variation in breast cancer identified the presence of genes in opposite transcriptional orientation resulting in stable antisense transcription.67 The authors were able to identify fusion pairs with genes that had transcriptional orientation either toward or away from each other. We also identified similar fusion patterns with 3′-3′ and 5′-5′ oriented transcripts from our analysis. The presence of fusions containing antisense genes varied across cancer types, indicating that the mechanisms that control the formation of such fusions are possibly unique to each cancer. We also found that several of these fusions recur within and across cancers, suggesting that they are not random transcriptional artifacts. These fusions could either result from frequent structural variations in the genome or due to antisense gene expression. The pervasive expression of antisense transcripts has been reported in many cancers, which accounts for about 38% of the annotated transcripts.20,68 These NATs (non-coding natural antisense transcripts [ncNATs]) regulate transcription and have been identified in several diseases, including cancer.69, 70, 71 Since ncNAT expression is higher in cancerous tissues than in normal adjacent tissues,69 their involvement in oncogenicity through fusions cannot be ruled out.
We were able to verify the genomic bases supporting the majority of the recurrent fusions in selected samples that had WGS data by identifying the structural variation associated with these fusions, which suggests that these fusions resulted from an underlying genomic event.72 We validated most of the fusions identified by ChimeRScope through discordant read mapping (2,991 fusions validated of 3,000 fusions from 88 TCGA samples) and Sanger sequencing (13 of 15 fusions validated from cell line data from CCLE) of fusion junctions. These data reemphasize the high accuracy of ChimeRScope in predicting fusions from RNA-seq reads. Alternatively, the structural validation tool BreakDancer validated less than 20% of these fusions, possibly due to inherent limitations in its sensitivity or the challenges associated with resolving complex events.60 Nevertheless, BreakDancer was selected as a structural variation detection tool for validation because this tool was able to detect more translocations, deletions, and inversions compared to other popular tools, such as SVDetect, DELLY, and Meerkat.
Fusion verification using discordant reads from WGS files identified very few recurrent fusions with no underlying genomic support, which is consistent with the reported literature. Often, fusion detection studies in different cancer types are limited to only those fusions having genomic structural variation support.27 This is largely due to the well-established notion that fusions are typically associated with chromosomal rearrangements,73,74 and the possibility of sequencing artifacts that might interfere with fusion detection algorithms.75 Although the majority of the fusions arise through a genomic structure-altering event, several studies have indicated that transcriptional fusions arise without an underlying genomic event,19,76 meaning that chimeric transcripts contribute to about half of the total genes in humans.13 A recent investigation of transcriptomic and WGS data of 27 cancer types from TCGA identified that 18% of the transcript fusions did not contain any structural variation at the genomic level.77 Similarly, several novel transcripts including read-through transcripts identified in BRCA did not have any structural variation support.67 Since our study was limited to only recurrent fusions, their existence is valid because a majority of them had supporting genomic evidence. We speculate that non-recurrent fusions that are often random in nature might arise without a basis at the genomic structure level. The exact mechanisms by which such chimeric transcripts arise without an underlying genomic event are poorly understood and warrant further investigation.
We also identified several transcriptional read-through fusions that are recurrent in various cancers. These fusions also exhibited different patterns across cancer types. OV, LAML, STAD, and ESCA had the highest percentage of read-through fusions in TCGA. We did not identify any fusions belonging to this category in prostate cancer despite several reports of read-through fusions by others.78, 79, 80, 81 This is probably because most of these studies analyzed non-TCGA samples, and the one study that did analyze TCGA samples used only a limited dataset of 44 prostate cancer and adjacent control samples.79 In our study, we also detected several read-through fusions in prostate samples, but none has passed our stringent filtering criteria.
Several kinase fusions recurring across cancers were identified in our analysis. Kinases, in addition to being important in oncogenesis, are also attractive drug targets.82 Our data reconfirm the previous observation that thyroid cancer is one of the cancers that have the highest percentage of recurrent kinase fusions.33 Consistent with other reports, CCDC6-RET fusion was found to be the most recurrent in thyroid cancer33,83 and was also identified in colon cancer and lung adenocarcinoma at a very low frequency. We found the SMG1 transcript to be highly promiscuous with several fusion partners across cancers. SMG1 is involved in nonsense-mediated mRNA decay (NMD) as part of the mRNA surveillance complex that degrades RNA with a premature stop codon and regulates the cellular RNA abundance.84,85 Understanding the possible involvement of SMG1 fusions in the accumulation of non-canonical fusions containing antisense transcripts requires further investigation.
The fusion profile of breast cancer samples exhibited stark differences with other cancers. Breast cancer alone has contributed more than 50% of the total fusions identified in our analysis. It is also interesting that most of the recurrent fusions (78%) were recurrent only within breast cancer and are not found in other TCGA cancer samples (Table S2). We also identified BRCA samples displaying huge variation in the fusion frequencies, suggesting that some of these BRCA samples had higher genomic aberrations, or had defective RNA splicing machinery that resulted in the extraordinary number of fusion transcripts. In addition, TP53 mutations were almost doubled in patients with low fusions, indicating that mutations on major oncogenes might be mutually exclusive with fusions. A recent study also found that fusions and mutations in driver genes are mutually exclusive across cancer types in TGCA.28 Differentially expressed pathways between the two groups with high or low fusion frequencies identified genes associated with the negative regulation of mRNA splicing in the high fusion cohort. This deregulation could be vital for fusion transcript generation since splicing defects can result in trans-splicing events that give rise to novel fusion transcripts.86, 87, 88
Analysis of fusions in 802 tumor-derived cell lines from CCLE using ChimeRScope revealed a high frequency of recurrent fusions across several cell lines irrespective of the tissue of origin. Several reports, including a recent study on recurrent fusions across multiple cancer types, also identified fusions that were frequent across cell lines, but the mechanism behind this phenomenon is unexplored.89, 90, 91 We also noted huge variability in recurrent fusion profiles among primary tumors and cancer cell lines. We were able to identify only 150 (including both canonical and non-canonical) recurrent fusions that were common between TCGA and CCLE. Although limited in numbers, these common fusions could be exploited to understand their role in cancer biology and their potential use as druggable targets. Cancer cell lines have been widely used as in vitro tumor models, and several studies have attempted to find cell lines that have the closest genetic identity to primary tumors.92, 93, 94, 95 Most of these studies have compared gene expression profiles, copy number variations, and mutation profiles but have not investigated the differences in fusion profiles among the primary tumor and tumor-derived cell lines. When compared to primary tumors, BRCA had the highest frequency of overlapping fusions. These common fusions in BRCA were recurring non-canonical fusions, indicating that non-canonical fusions were also common in cell lines. We also identified several ubiquitous fusions in all of the cell lines analyzed. It would be interesting to inspect the functional consequence of these fusions and study the probable mechanism of fusion formation using cell line models.
Conclusions
Our analysis using ChimeRScope identified several recurrent fusion transcripts that are common across cancers, reemphasizing the need for identifying therapy focused on actionable targets. Basket trials in cancer, targeting common therapeutic targets independent of tissue of origin, is an emerging strategy that is driven by the genomic profile of the patients. Our data also identified several novel recurrent non-canonical fusions containing antisense transcripts that warrant further investigation. We also identified significant differences in the fusions harbored by common tumor-derived cell lines compared to primary tumors, indicating that cell lines might not represent the complete fusion repertoire in patients. Fusions identified in cell lines that are common with the primary tumors could be exploited for functionally validating clinically actionable fusions.
Materials and Methods
Level 1 paired-end RNA sequencing data (aligned BAM files) from TCGA and CCLE were downloaded from the Genomic Data Commons (GDC) data portal. Aligned BAM files from 8,883 TCGA samples across 33 tumor types along with 802 commonly used cancer cell lines (from CCLE) were analyzed for this study (Table 1). We also downloaded aligned BAM files from adjacent non-tumor (referred to as normal) tissues from the same patients to serve as control groups. These BAM files were generated by TCGA using a two-pass alignment method with STAR 2.4.2a.96 Unaligned reads to the reference genome GRCh38 were extracted using SAMtools 1.3 and bedtools 2.24.97,98 These unmapped reads in fastq format were used as input to predict fusion transcripts using ChimeRScope.16 A flowchart summarizing the methods, in brief, is illustrated in Figure S16. The ChimeRScope method contains four functional modules that include Builder, Scanner, Sweeper, and Examiner. First, we used the ChimeRScope Builder to create a gene fingerprint (GF) library for GRCh38 transcriptome with k-mers, where k = 17. ChimeRScope Scanner and Sweeper modules were executed using this GF library as the reference for the k-mers from the unmapped reads using default parameters. Fusions identified in the tumor samples were compared against the fusions from any adjacent normal tissue samples, and common fusions were removed to retain only tumor-specific fusions. However, fusions reported in the COSMIC database (https://cancer.sanger.ac.uk) and TICdb database (https://genetica.unav.edu/TICdb/) were used as a positive fusion list to retain clinically relevant fusions despite their presence in the adjacent normal tissue. ChimeRScope Sweeper output was further filtered to remove fusions with fusion event supporting reads (FESRs) <5 to minimize false positives. Furthermore, the adaptor contaminations, homologous genes, and sequences with NCBI-BLAST alignment scores <200 were all filtered out as suggested by ChimeRScope.16 Since we also wanted to evaluate the profile of read-through fusions (resulting from the fusion of two adjacent genes in the same coding orientation) that could also be oncogenic, we analyzed them separately. Lastly, the ChimeRScope Examiner module was run with default parameters, and more stringent filters (as described below) were applied to the ChimeRScope Examiner output to further remove potential false positives.
Distribution of reads across the fusion junction: Distribution of reads across the fusion junction: Only fusions containing at least five reads (FESRs) covering the fusion junction and the flanking regions are retained.16 In-house Python scripts were used to filter out fusions supported by FESRs that do not exhibit the minimum distribution at the participating exon of both genes in a given fusion pair.
Distance from the exon boundaries: the participating exons were divided into three regions: region I, region II, and region III (see Figure S17A). Regions I and III represent the first and last quartiles of an exon, respectively, and hence are closer to exon terminals (designated as E), while the middle half corresponds to region II that is farther from exon terminals (designated as M). Based on this, a fusion was categorized as E-E when both exon breakpoints are in the regions I or III (i.e., near the terminals), E-M when only one of the breakpoints is near the exon terminal, and M-M when both breakpoints are away from the exon terminals. Figure S17B explains the categories and their relationship with the exon regions.
Fusions mapping to the known non-coding RNAs: the fusion sequence reported by ChimeRScope is approximately 100 bases upstream and also downstream of the fusion junction. The fusion sequence was aligned against the NCBI’s non-redundant nucleotide sequence (nr) transcriptome reference to remove any possible non-coding RNA matches. After applying all of these filters, we limited our analysis to only the high confidence recurrent fusions (n ≥ 2) within or across cancers. These fusions were further analyzed to identify potential protein-coding in-frame fusions. Transcripts participating in fusions were compared against the cancer gene census from the COSMIC database (https://cancer.sanger.ac.uk/ downloaded in October 2017) and known human kinases to identify kinases, oncogenes, or tumor suppressors, to focus on genes relevant to cancer. We analyzed the fusion profiles of genes likely to be cancer drivers or therapeutic targets in 10 canonical pathways: cell cycle, Hippo, Myc, Notch, Nrf2, PI3K/Akt, RTK-RAS, TGF-β signaling, p53, and β-catenin/Wnt.47 Recurrent fusions identified in TCGA were further categorized as druggable when they were identified as therapeutic targets from DEPO57 (http://depo-dinglab.ddns.net), oncoKB58 (https://oncokb.org), or the Cancer Genome Interpreter database59 (http://www.cancergenomeinterpreter.org) to understand their functional relevance to chemotherapy. IPA (https://www.ingenuity.com) was used to identify pathways enriched by transcripts that are associated with fusions.
Similarity of Fusions across Cancers
A probability-based similarity measure was derived between all pairwise combinations of cancers based on the predicted proportion of fusions that are shared between cancers. Let A and B represent the set of fusions in cancer A and B, respectively. The union of these two sets represents all unique fusions in both sets, denoted as A ∪ B. The size of the union (sample space) is determined as
The assumption here is that the probability distribution is uniform across the sample space. The shared number of fusions between the two cancers is the intersection of A and B, denoted as A ∩ B. The probability of each cancer is the fraction of fusions in that particular set compared to all fusions in the union. The “specific mutual information” theory measure gives a relative amount of information shared between two random variables when these variables take on specific values.99 The similarity measure for two cancers A and B, called Sim(A, B), was adopted from the information theory and is derived as
The similarity score, here, is the information shared between two cancers, based on the shared fusions. The value of Sim(A, B) will always be between 0 and 1, where 0 implies no similarity whereas 1 represents 100% similarity between the two cancers. Hierarchical clustering with complete linkage was used to cluster cancers based on the distance matrix using the R package factoextra (https://cran.r-project.org/package=factoextra).100
Canonical and Non-canonical Fusions
ChimeRScope was able to predict many fusion transcripts that are fully or partly antisense (mapped to the direction opposite to the reference transcript orientation) with high confidence. Such fusions were identified by mapping sequences at the fusion junction to the reference transcript sequences using the bl2seq module of BLAST. Fusions with both partner transcripts mapped to their reference transcripts in the same orientation were categorized as canonical, whereas when one or both of the transcripts were mapped in an antisense orientation, such fusions were categorized as non-canonical. These non-canonical fusions were also manually curated by inspecting the reads mapped to these transcripts in corresponding BAM files.
Verification of Fusion Transcript Predictions Using Whole-Genome Data
The TCGA WGS data on 88 samples from breast, glioblastoma, kidney, bladder, and skin cancers were downloaded. BreakDancer (version 1.1)101 was run on the WGS data, and somatic rearrangements were identified. A fusion predicted by ChimeRScope was validated when the evidence of somatic rearrangements in the genome was within a window size of 1 Mb from the fusion breakpoint. Fusions in the same WGS samples were also validated using SAMtools97 by identifying discordant reads mapping to partner genes as reported previously.28 Briefly, discordant reads were extracted from the WGS reads using SAMtools, and the genomic locations of these reads were compared to the predicted fusion breakpoints from ChimeRScope. A Python script was used to match the position of the discordant reads in the WGS BAM file with a fusion breakpoint, and a match is considered when the position of the discordant read is within 1 MB of a fusion breakpoint. The python script used for WGS validation is hosted on GitHub (https://github.com/unmc-abrar/Python-Match-SV).
Validation of Predicted Fusions by Sanger Sequencing
Fusions identified by ChimeRScope in selected cell line data from CCLE were also validated using PCR amplification and Sanger sequencing. Fusions were selected for validation based on their recurrent nature and or functional significance. A total of eight canonical and seven non-canonical fusions identified in the RNA-seq data from eight CCLE cell lines and one in-house dataset were selected for validation (Table S22). These selected cell lines were cultured under standard conditions, and total RNA was extracted as per standard protocol using a RNeasy mini kit (QIAGEN). RNA extracted from these cell lines was converted into cDNA using a first-strand cDNA synthesis kit (Thermo Fisher Scientific) as per the manufacturer’s protocol, followed by PCR using the oligonucleotide primers designed using Vector NTI Advance (version 11.5.4) or Primer3Plus102 (Table S22). PCR was performed for each primer set using either the standard Taq PCR kit (New England Biolabs) or the One Taq PCR kit with GC buffer (New England Biolabs) depending on the GC content of the target sequences. All PCR products were analyzed on 2% agarose gels. The PCR products of the expected amplicon size were extracted from the agarose gel using the Zymoclean gel DNA recovery kit (Zymo Research). All amplicons extracted from the agarose gel were Sanger sequenced using Applied Biosystems (ABI) 3730 DNA analyzer as per the manufacturer’s protocol.
Case Study: Breast Cancer
Breast cancer was selected for a detailed study, as ChimeRScope has predicted by far the highest number of fusions in this cancer in our analysis. Breast cancer samples analyzed in this study were divided into four quartiles based on the number of predicted fusions sorted in ascending order, and samples belonging to the first and fourth quartiles were identified as low and high fusion groups, respectively. These two groups were further analyzed for differences in their clinical characteristics, gene expression, and structural variation profiles. Differences in clinical features were analyzed using Fisher’s exact test. The R package edgeR was used to identify differentially expressed genes between the two groups.103 A gene was considered to be differentially expressed when the FDR corrected p value was below or equal to 0.05 and the absolute log2 fold change was above 1. Gene set enrichment analysis (GSEA) was used to identify enriched upregulated and downregulated pathways among the high and low fusion groups.104 An enrichment score for the differentially expressed gene set was calculated, which is a weighted Kolmogorov-Smirnov-like statistic, and a positive (negative) normalized enrichment score (NES) was computed.104 The nominal p value of the NES was calculated based on 1,000 permutations and a GO term with a NES score ≥1.5, and a nominal p value ≤0.05 was considered significant.105 Somatic mutations for the breast cancer TCGA cohort were extracted from the MAF file available for download from the TCGA GDC website. A Bioconductor package, maftools, was used to analyze and estimate the mutation load among high or low fusion samples in breast cancer.106
Author Contributions
N.N.V. and C.G. designed the study. N.N.V., A.A., J.K.B., and N.K.M. performed data analysis. Y.L. designed ChimeRScope and helped with its implementation. S.R., M.J.K., S.M., K.K.B., V.B., and S.S.J. provided cell lines and helped with validation. N.N.V. and C.G. wrote the manuscript with input from all authors.
Acknowledgments
The authors are grateful to Sanjit Pandey for providing systems administrative support to Linux and Windows servers, and to the Bioinformatics and Systems Biology core at the University of Nebraska Medical Center (UNMC). The authors also acknowledge the Holland Computing Center of the University of Nebraska-Lincoln for computational resources, which receives support from the Nebraska Research Initiative. This work was supported by the National Institutes of Health, United States (grants 5P20GM103427, 1P30GM127200, 2P01AG029531, 5P30CA036727, and 5P30GM110768) and by the National Science Federation, United States EPSCoR Award (grant OIA-1557417).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.01.023.
Contributor Information
Neetha Nanoth Vellichirammal, Email: n.nanothvellichiram@unmc.edu.
Chittibabu Guda, Email: babu.guda@unmc.edu.
Supplemental Information
References
- 1.Inaki K., Hillmer A.M., Ukil L., Yao F., Woo X.Y., Vardy L.A., Zawack K.F., Lee C.W., Ariyaratne P.N., Chan Y.S. Transcriptional consequences of genomic structural aberrations in breast cancer. Genome Res. 2011;21:676–687. doi: 10.1101/gr.113225.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitelman F., Johansson B., Mertens F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 3.Mertens F., Johansson B., Fioretos T., Mitelman F. The emerging complexity of gene fusions in cancer. Nat. Rev. Cancer. 2015;15:371–381. doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- 4.Dreazen O., Klisak I., Jones G., Ho W.G., Sparkes R.S., Gale R.P. Multiple molecular abnormalities in Ph1 chromosome positive acute lymphoblastic leukaemia. Br. J. Haematol. 1987;67:319–324. doi: 10.1111/j.1365-2141.1987.tb02353.x. [DOI] [PubMed] [Google Scholar]
- 5.Morris S.W., Kirstein M.N., Valentine M.B., Dittmer K.G., Shapiro D.N., Saltman D.L., Look A.T. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baccarani M., Saglio G., Goldman J., Hochhaus A., Simonsson B., Appelbaum F., Apperley J., Cervantes F., Cortes J., Deininger M., European LeukemiaNet Evolving concepts in the management of chronic myeloid leukemia: recommendations from an expert panel on behalf of the European LeukemiaNet. Blood. 2006;108:1809–1820. doi: 10.1182/blood-2006-02-005686. [DOI] [PubMed] [Google Scholar]
- 8.Soverini S., Mancini M., Bavaro L., Cavo M., Martinelli G. Chronic myeloid leukemia: the paradigm of targeting oncogenic tyrosine kinase signaling and counteracting resistance for successful cancer therapy. Mol. Cancer. 2018;17:49. doi: 10.1186/s12943-018-0780-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowley J.D. Letter: a new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 10.Breg W.R., Miller D.A., Allderdice P.W., Miller O.J. Identification of translocation chromosomes by quinacrine fluorescence. Am. J. Dis. Child. 1972;123:561–564. doi: 10.1001/archpedi.1972.02110120085007. [DOI] [PubMed] [Google Scholar]
- 11.Mitelman F., Johansson B., Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat. Genet. 2004;36:331–334. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- 12.Edwards P.A., Howarth K.D. Are breast cancers driven by fusion genes? Breast Cancer Res. 2012;14:303. doi: 10.1186/bcr3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X., Zhao L., Jiang H., Wang W. Short homologous sequences are strongly associated with the generation of chimeric RNAs in eukaryotes. J. Mol. Evol. 2009;68:56–65. doi: 10.1007/s00239-008-9187-0. [DOI] [PubMed] [Google Scholar]
- 14.Babiceanu M., Qin F., Xie Z., Jia Y., Lopez K., Janus N., Facemire L., Kumar S., Pang Y., Qi Y. Recurrent chimeric fusion RNAs in non-cancer tissues and cells. Nucleic Acids Res. 2016;44:2859–2872. doi: 10.1093/nar/gkw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varley K.E., Gertz J., Roberts B.S., Davis N.S., Bowling K.M., Kirby M.K., Nesmith A.S., Oliver P.G., Grizzle W.E., Forero A. Recurrent read-through fusion transcripts in breast cancer. Breast Cancer Res. Treat. 2014;146:287–297. doi: 10.1007/s10549-014-3019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chwalenia K., Facemire L., Li H. Chimeric RNAs in cancer and normal physiology. Wiley Interdiscip. Rev. RNA. 2017;8:e1427. doi: 10.1002/wrna.1427. [DOI] [PubMed] [Google Scholar]
- 17.Greger L., Su J., Rung J., Ferreira P.G., Lappalainen T., Dermitzakis E.T., Brazma A., Geuvadis Consortium Tandem RNA chimeras contribute to transcriptome diversity in human population and are associated with intronic genetic variants. PLoS ONE. 2014;9:e104567. doi: 10.1371/journal.pone.0104567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parra G., Reymond A., Dabbouseh N., Dermitzakis E.T., Castelo R., Thomson T.M., Antonarakis S.E., Guigó R. Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res. 2006;16:37–44. doi: 10.1101/gr.4145906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia Y., Xie Z., Li H. Intergenically spliced chimeric RNAs in cancer. Trends Cancer. 2016;2:475–484. doi: 10.1016/j.trecan.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balbin O.A., Malik R., Dhanasekaran S.M., Prensner J.R., Cao X., Wu Y.M., Robinson D., Wang R., Chen G., Beer D.G. The landscape of antisense gene expression in human cancers. Genome Res. 2015;25:1068–1079. doi: 10.1101/gr.180596.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q., Xia J., Jia P., Pao W., Zhao Z. Application of next generation sequencing to human gene fusion detection: computational tools, features and perspectives. Brief. Bioinform. 2013;14:506–519. doi: 10.1093/bib/bbs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beccuti M., Carrara M., Cordero F., Donatelli S., Calogero R.A. The structure of state of art gene fusion-finder algorithms. OA Bioinformatics. 2013;1:2. [Google Scholar]
- 23.Latysheva N.S., Babu M.M. Discovering and understanding oncogenic gene fusions through data intensive computational approaches. Nucleic Acids Res. 2016;44:4487–4503. doi: 10.1093/nar/gkw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrara M., Beccuti M., Cavallo F., Donatelli S., Lazzarato F., Cordero F., Calogero R.A. State of art fusion-finder algorithms are suitable to detect transcription-induced chimeras in normal tissues? BMC Bioinformatics. 2013;14(Suppl 7):S2. doi: 10.1186/1471-2105-14-S7-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S., Vo A.D., Qin F., Li H. Comparative assessment of methods for the fusion transcripts detection from RNA-seq data. Sci. Rep. 2016;6:21597. doi: 10.1038/srep21597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu X., Wang Q., Tang M., Barthel F., Amin S., Yoshihara K., Lang F.M., Martinez-Ledesma E., Lee S.H., Zheng S., Verhaak R.G.W. TumorFusions: an integrative resource for cancer-associated transcript fusions. Nucleic Acids Res. 2018;46(D1):D1144–D1149. doi: 10.1093/nar/gkx1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshihara K., Wang Q., Torres-Garcia W., Zheng S., Vegesna R., Kim H., Verhaak R.G. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34:4845–4854. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Q., Liang W.W., Foltz S.M., Mutharasu G., Jayasinghe R.G., Cao S., Liao W.W., Reynolds S.M., Wyczalkowski M.A., Yao L. Driver fusions and their implications in the development and treatment of human cancers. Cell Rep. 2018;23:227–238.e3. doi: 10.1016/j.celrep.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim P., Zhou X. FusionGDB: fusion gene annotation DataBase. Nucleic Acids Res. 2019;47(D1):D994–D1004. doi: 10.1093/nar/gky1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee M., Lee K., Yu N., Jang I., Choi I., Kim P., Jang Y.E., Kim B., Kim S., Lee B. ChimerDB 3.0: an enhanced database for fusion genes from cancer transcriptome and literature data mining. Nucleic Acids Res. 2017;45(D1):D784–D789. doi: 10.1093/nar/gkw1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abate F., Acquaviva A., Paciello G., Foti C., Ficarra E., Ferrarini A., Delledonne M., Iacobucci I., Soverini S., Martinelli G., Macii E. Bellerophontes: an RNA-seq data analysis framework for chimeric transcripts discovery based on accurate fusion model. Bioinformatics. 2012;28:2114–2121. doi: 10.1093/bioinformatics/bts334. [DOI] [PubMed] [Google Scholar]
- 32.Yang L., Luquette L.J., Gehlenborg N., Xi R., Haseley P.S., Hsieh C.H., Zhang C., Ren X., Protopopov A., Chin L. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell. 2013;153:919–929. doi: 10.1016/j.cell.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stransky N., Cerami E., Schalm S., Kim J.L., Lengauer C. The landscape of kinase fusions in cancer. Nat. Commun. 2014;5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seshagiri S., Stawiski E.W., Durinck S., Modrusan Z., Storm E.E., Conboy C.B., Chaudhuri S., Guan Y., Janakiraman V., Jaiswal B.S. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han T., Schatoff E.M., Murphy C., Zafra M.P., Wilkinson J.E., Elemento O., Dow L.E. R-Spondin chromosome rearrangements drive Wnt-dependent tumour initiation and maintenance in the intestine. Nat. Commun. 2017;8:15945. doi: 10.1038/ncomms15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams S.V., Hurst C.D., Knowles M.A. Oncogenic FGFR3 gene fusions in bladder cancer. Hum. Mol. Genet. 2013;22:795–803. doi: 10.1093/hmg/dds486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh D., Chan J.M., Zoppoli P., Niola F., Sullivan R., Castano A., Liu E.M., Reichel J., Porrati P., Pellegatta S. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y.M., Su F., Kalyana-Sundaram S., Khazanov N., Ateeq B., Cao X., Lonigro R.J., Vats P., Wang R., Lin S.F. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tognon C., Knezevich S.R., Huntsman D., Roskelley C.D., Melnyk N., Mathers J.A., Becker L., Carneiro F., MacPherson N., Horsman D. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 40.Veeraraghavan J., Tan Y., Cao X.X., Kim J.A., Wang X., Chamness G.C., Maiti S.N., Cooper L.J., Edwards D.P., Contreras A. Recurrent ESR1-CCDC170 rearrangements in an aggressive subset of oestrogen receptor-positive breast cancers. Nat. Commun. 2014;5:4577. doi: 10.1038/ncomms5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holst F., Hoivik E.A., Gibson W.J., Taylor-Weiner A., Schumacher S.E., Asmann Y.W., Grossmann P., Trovik J., Necela B.M., Thompson E.A. Recurrent hormone-binding domain truncated ESR1 amplifications in primary endometrial cancers suggest their implication in hormone independent growth. Sci. Rep. 2016;6:25521. doi: 10.1038/srep25521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baghdadi M., Endo H., Takano A., Ishikawa K., Kameda Y., Wada H., Miyagi Y., Yokose T., Ito H., Nakayama H. High co-expression of IL-34 and M-CSF correlates with tumor progression and poor survival in lung cancers. Sci. Rep. 2018;8:418. doi: 10.1038/s41598-017-18796-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franzè E., Dinallo V., Rizzo A., Di Giovangiulio M., Bevivino G., Stolfi C., Caprioli F., Colantoni A., Ortenzi A., Grazia A.D. Interleukin-34 sustains pro-tumorigenic signals in colon cancer tissue. Oncotarget. 2017;9:3432–3445. doi: 10.18632/oncotarget.23289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ségaliny A.I., Mohamadi A., Dizier B., Lokajczyk A., Brion R., Lanel R., Amiaud J., Charrier C., Boisson-Vidal C., Heymann D. Interleukin-34 promotes tumor progression and metastatic process in osteosarcoma through induction of angiogenesis and macrophage recruitment. Int. J. Cancer. 2015;137:73–85. doi: 10.1002/ijc.29376. [DOI] [PubMed] [Google Scholar]
- 45.Semenza G.L. HIF-1 and mechanisms of hypoxia sensing. Curr. Opin. Cell Biol. 2001;13:167–171. doi: 10.1016/s0955-0674(00)00194-0. [DOI] [PubMed] [Google Scholar]
- 46.Tan M., Yao J., Yu D. Overexpression of the c-erbB-2 gene enhanced intrinsic metastasis potential in human breast cancer cells without increasing their transformation abilities. Cancer Res. 1997;57:1199–1205. [PubMed] [Google Scholar]
- 47.Sanchez-Vega F., Mina M., Armenia J., Chatila W.K., Luna A., La K.C., Dimitriadoy S., Liu D.L., Kantheti H.S., Saghafinia S. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018;173:321–337.e10.. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y., Easton J., Shao Y., Maciaszek J., Wang Z., Wilkinson M.R., McCastlain K., Edmonson M., Pounds S.B., Shi L. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat. Genet. 2017;49:1211–1218. doi: 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Belo A., Cheng K., Chahdi A., Shant J., Xie G., Khurana S., Raufman J.P. Muscarinic receptor agonists stimulate human colon cancer cell migration and invasion. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;300:G749–G760. doi: 10.1152/ajpgi.00306.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Bruijn D.R., dos Santos N.R., Kater-Baats E., Thijssen J., van den Berk L., Stap J., Balemans M., Schepens M., Merkx G., van Kessel A.G. The cancer-related protein SSX2 interacts with the human homologue of a Ras-like GTPase interactor, RAB3IP, and a novel nuclear protein, SSX2IP. Genes Chromosomes Cancer. 2002;34:285–298. doi: 10.1002/gcc.10073. [DOI] [PubMed] [Google Scholar]
- 51.Hur K., Cejas P., Feliu J., Moreno-Rubio J., Burgos E., Boland C.R., Goel A. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut. 2014;63:635–646. doi: 10.1136/gutjnl-2012-304219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie G., Cheng K., Shant J., Raufman J.P. Acetylcholine-induced activation of M3 muscarinic receptors stimulates robust matrix metalloproteinase gene expression in human colon cancer cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296:G755–G763. doi: 10.1152/ajpgi.90519.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kodaira M., Kajimura M., Takeuchi K., Lin S., Hanai H., Kaneko E. Functional muscarinic m3 receptor expressed in gastric cancer cells stimulates tyrosine phosphorylation and MAP kinase. J. Gastroenterol. 1999;34:163–171. doi: 10.1007/s005350050238. [DOI] [PubMed] [Google Scholar]
- 54.Shah N., Khurana S., Cheng K., Raufman J.P. Muscarinic receptors and ligands in cancer. Am. J. Physiol. Cell Physiol. 2009;296:C221–C232. doi: 10.1152/ajpcell.00514.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu H., Xia H., Tang Q., Xu H., Wei G., Chen Y., Dai X., Gong Q., Bi F. Acetylcholine acts through M3 muscarinic receptor to activate the EGFR signaling and promotes gastric cancer cell proliferation. Sci. Rep. 2017;7:40802. doi: 10.1038/srep40802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou H., Liu W., Su Y., Wei Z., Liu J., Kolluri S.K., Wu H., Cao Y., Chen J., Wu Y. NSAID sulindac and its analog bind RXRα and inhibit RXRα-dependent AKT signaling. Cancer Cell. 2010;17:560–573. doi: 10.1016/j.ccr.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun S.Q., Mashl R.J., Sengupta S., Scott A.D., Wang W., Batra P., Wang L.B., Wyczalkowski M.A., Ding L. Database of evidence for precision oncology portal. Bioinformatics. 2018;34:4315–4317. doi: 10.1093/bioinformatics/bty531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chakravarty D., Gao J., Phillips S.M., Kundra R., Zhang H., Wang J., Rudolph J.E., Yaeger R., Soumerai T., Nissan M.H. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol. 2017;2017:1–6. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamborero D., Rubio-Perez C., Deu-Pons J., Schroeder M.P., Vivancos A., Rovira A., Tusquets I., Albanell J., Rodon J., Tabernero J. Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med. 2018;10:25. doi: 10.1186/s13073-018-0531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alaei-Mahabadi B., Bhadury J., Karlsson J.W., Nilsson J.A., Larsson E. Global analysis of somatic structural genomic alterations and their impact on gene expression in diverse human cancers. Proc. Natl. Acad. Sci. USA. 2016;113:13768–13773. doi: 10.1073/pnas.1606220113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y., Wu N., Liu J., Wu Z., Dong D. FusionCancer: a database of cancer fusion genes derived from RNA-seq data. Diagn. Pathol. 2015;10:131. doi: 10.1186/s13000-015-0310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mitelman F., Johansson B., Mertens F.E. 2018. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer.https://mitelmandatabase.isb-cgc.org [Google Scholar]
- 63.Gorohovski A., Tagore S., Palande V., Malka A., Raviv-Shay D., Frenkel-Morgenstern M. ChiTaRS-3.1-the enhanced chimeric transcripts and RNA-seq database matched with protein-protein interactions. Nucleic Acids Res. 2017;45(D1):D790–D795. doi: 10.1093/nar/gkw1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Novo F.J., de Mendíbil I.O., Vizmanos J.L. TICdb: a collection of gene-mapped translocation breakpoints in cancer. BMC Genomics. 2007;8:33. doi: 10.1186/1471-2164-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Forbes S.A., Beare D., Boutselakis H., Bamford S., Bindal N., Tate J., Cole C.G., Ward S., Dawson E., Ponting L. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stephens P.J., McBride D.J., Lin M.L., Varela I., Pleasance E.D., Simpson J.T., Stebbings L.A., Leroy C., Edkins S., Mudie L.J. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shlien A., Raine K., Fuligni F., Arnold R., Nik-Zainal S., Dronov S., Mamanova L., Rosic A., Ju Y.S., Cooke S.L., ICGC Breast Cancer Working Group, Oslo Breast Cancer Research Consortium Direct transcriptional consequences of somatic mutation in breast cancer. Cell Rep. 2016;16:2032–2046. doi: 10.1016/j.celrep.2016.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lai J., Lehman M.L., Dinger M.E., Hendy S.C., Mercer T.R., Seim I., Lawrence M.G., Mattick J.S., Clements J.A., Nelson C.C. A variant of the KLK4 gene is expressed as a cis sense-antisense chimeric transcript in prostate cancer cells. RNA. 2010;16:1156–1166. doi: 10.1261/rna.2019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wenric S., ElGuendi S., Caberg J.H., Bezzaou W., Fasquelle C., Charloteaux B., Karim L., Hennuy B., Frères P., Collignon J. Transcriptome-wide analysis of natural antisense transcripts shows their potential role in breast cancer. Sci. Rep. 2017;7:17452. doi: 10.1038/s41598-017-17811-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luo J.H., Ren B., Keryanov S., Tseng G.C., Rao U.N., Monga S.P., Strom S., Demetris A.J., Nalesnik M., Yu Y.P. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology. 2006;44:1012–1024. doi: 10.1002/hep.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Han Y., Liu Y., Gui Y., Cai Z. Long intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder. J. Surg. Oncol. 2013;107:555–559. doi: 10.1002/jso.23264. [DOI] [PubMed] [Google Scholar]
- 72.Johansson B., Mertens F., Schyman T., Björk J., Mandahl N., Mitelman F. Most gene fusions in cancer are stochastic events. Genes Chromosomes Cancer. 2019;58:607–611. doi: 10.1002/gcc.22745. [DOI] [PubMed] [Google Scholar]
- 73.Tomlins S.A., Rhodes D.R., Perner S., Dhanasekaran S.M., Mehra R., Sun X.W., Varambally S., Cao X., Tchinda J., Kuefer R. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 74.Soda M., Choi Y.L., Enomoto M., Takada S., Yamashita Y., Ishikawa S., Fujiwara S., Watanabe H., Kurashina K., Hatanaka H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 75.Houseley J., Tollervey D. Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS ONE. 2010;5:e12271. doi: 10.1371/journal.pone.0012271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frenkel-Morgenstern M., Lacroix V., Ezkurdia I., Levin Y., Gabashvili A., Prilusky J., Del Pozo A., Tress M., Johnson R., Guigo R., Valencia A. Chimeras taking shape: potential functions of proteins encoded by chimeric RNA transcripts. Genome Res. 2012;22:1231–1242. doi: 10.1101/gr.130062.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fonseca, N.A., He, Y., Greger, L., PCAWG-3, Brazma, A., and Zhang, Z. Comprehensive genome and transcriptome analysis reveals genetic basis for gene fusions in cancer. bioRxiv, doi.org/10.1101/148684.
- 78.Rickman D.S., Pflueger D., Moss B., VanDoren V.E., Chen C.X., de la Taille A., Kuefer R., Tewari A.K., Setlur S.R., Demichelis F., Rubin M.A. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res. 2009;69:2734–2738. doi: 10.1158/0008-5472.CAN-08-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao S., Løvf M., Carm K.T., Bakken A.C., Hoff A.M., Skotheim R.I. Novel transcription-induced fusion RNAs in prostate cancer. Oncotarget. 2017;8:49133–49143. doi: 10.18632/oncotarget.17099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nacu S., Yuan W., Kan Z., Bhatt D., Rivers C.S., Stinson J., Peters B.A., Modrusan Z., Jung K., Seshagiri S., Wu T.D. Deep RNA sequencing analysis of readthrough gene fusions in human prostate adenocarcinoma and reference samples. BMC Med. Genomics. 2011;4:11. doi: 10.1186/1755-8794-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang J., White N.M., Schmidt H.K., Fulton R.S., Tomlinson C., Warren W.C., Wilson R.K., Maher C.A. INTEGRATE: gene fusion discovery using whole genome and transcriptome data. Genome Res. 2016;26:108–118. doi: 10.1101/gr.186114.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arslan M.A., Kutuk O., Basaga H. Protein kinases as drug targets in cancer. Curr. Cancer Drug Targets. 2006;6:623–634. doi: 10.2174/156800906778742479. [DOI] [PubMed] [Google Scholar]
- 83.Celestino R., Sigstad E., Løvf M., Thomassen G.O., Grøholt K.K., Jørgensen L.H., Berner A., Castro P., Lothe R.A., Bjøro T. Survey of 548 oncogenic fusion transcripts in thyroid tumors supports the importance of the already established thyroid fusions genes. Genes Chromosomes Cancer. 2012;51:1154–1164. doi: 10.1002/gcc.22003. [DOI] [PubMed] [Google Scholar]
- 84.Kervestin S., Jacobson A. NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hug N., Longman D., Cáceres J.F. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016;44:1483–1495. doi: 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lei Q., Li C., Zuo Z., Huang C., Cheng H., Zhou R. Evolutionary insights into RNA trans-splicing in vertebrates. Genome Biol. Evol. 2016;8:562–577. doi: 10.1093/gbe/evw025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guerra E., Trerotola M., Dell’ Arciprete R., Bonasera V., Palombo B., El-Sewedy T., Ciccimarra T., Crescenzi C., Lorenzini F., Rossi C. A bicistronic CYCLIN D1-TROP2 mRNA chimera demonstrates a novel oncogenic mechanism in human cancer. Cancer Res. 2008;68:8113–8121. doi: 10.1158/0008-5472.CAN-07-6135. [DOI] [PubMed] [Google Scholar]
- 88.Yuan C., Liu Y., Yang M., Liao D.J. New methods as alternative or corrective measures for the pitfalls and artifacts of reverse transcription and polymerase chain reactions (RT-PCR) in cloning chimeric or antisense-accompanied RNA. RNA Biol. 2013;10:958–967. doi: 10.4161/rna.24570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu Y.P., Liu P., Nelson J., Hamilton R.L., Bhargava R., Michalopoulos G., Chen Q., Zhang J., Ma D., Pennathur A. Identification of recurrent fusion genes across multiple cancer types. Sci. Rep. 2019;9:1074. doi: 10.1038/s41598-019-38550-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nome T., Thomassen G.O., Bruun J., Ahlquist T., Bakken A.C., Hoff A.M., Rognum T., Nesbakken A., Lorenz S., Sun J. Common fusion transcripts identified in colorectal cancer cell lines by high-throughput RNA sequencing. Transl. Oncol. 2013;6:546–553. doi: 10.1593/tlo.13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sakarya O., Breu H., Radovich M., Chen Y., Wang Y.N., Barbacioru C., Utiramerur S., Whitley P.P., Brockman J.P., Vatta P. RNA-seq mapping and detection of gene fusions with a suffix array algorithm. PLoS Comput. Biol. 2012;8:e1002464. doi: 10.1371/journal.pcbi.1002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Domcke S., Sinha R., Levine D.A., Sander C., Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ross D.T., Perou C.M. A comparison of gene expression signatures from breast tumors and breast tissue derived cell lines. Dis. Markers. 2001;17:99–109. doi: 10.1155/2001/850531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nickerson M.L., Witte N., Im K.M., Turan S., Owens C., Misner K., Tsang S.X., Cai Z., Wu S., Dean M. Molecular analysis of urothelial cancer cell lines for modeling tumor biology and drug response. Oncogene. 2017;36:35–46. doi: 10.1038/onc.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li H., Wawrose J.S., Gooding W.E., Garraway L.A., Lui V.W., Peyser N.D., Grandis J.R. Genomic analysis of head and neck squamous cell carcinoma cell lines and human tumors: a rational approach to preclinical model selection. Mol. Cancer Res. 2014;12:571–582. doi: 10.1158/1541-7786.MCR-13-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smadja F., McKeown K.R., Hatzivassiloglou V. Translating collocations for bilingual lexicons: a statistical approach. Comput. Linguist. 1996;22:1–38. [Google Scholar]
- 100.Kassambara A. 2015. factoextra: extract and visualize the results of multivariate data analyses. R package version 1.0.3.https://www.google.com/?gws_rd=ssl [Google Scholar]
- 101.Chen K., Wallis J.W., McLellan M.D., Larson D.E., Kalicki J.M., Pohl C.S., McGrath S.D., Wendl M.C., Zhang Q., Locke D.P. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chow L.M., Endersby R., Zhu X., Rankin S., Qu C., Zhang J., Broniscer A., Ellison D.W., Baker S.J. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell. 2011;19:305–316. doi: 10.1016/j.ccr.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mayakonda A., Koeffler H.P. Maftools: efficient analysis, visualization and summarization of MAF files from large-scale cohort based cancer studies. bioRxiv. 2016 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.