Abstract
Background
Dysregulation of polycomb chromobox (CBX) proteins that mediate epigenetic gene silencing contributes to the progression of human cancers. Yet their roles in clear cell renal cell carcinoma (ccRCC) remain to be explored.
Methods
The expression of CBX4 and its clinical significance were determined by qRT-PCR, western blot, immunohistochemistry and statistical analyses. The biological function of CBX4 in ccRCC tumor growth and metastasis and the underlying mechanism were investigated using in vitro and in vivo models.
Findings
CBX4 exerts oncogenic activities in ccRCC via interaction with HDAC1 to transcriptionally suppress tumor suppressor KLF6. CBX4 expression is increased in ccRCC and correlated with poor prognosis in two independent cohorts containing 840 patients. High CBX4 expression is significantly associated with Fuhrman grade and tumor lymph node invasion. CBX4 overexpression promotes tumor growth and metastasis, whereas CBX4 knockdown results in the opposite phenotypes. Mechanistically, CBX4 downregulates KLF6 via repressing the transcriptional activity of its promoter. Further studies show that CBX4 physically binds to HDAC1 to maintain its localization on the KLF6 promoter. Ectopic expression of KLF6 or disruption of CBX4-HDAC1 interaction attenuates CBX4-mediated cell growth and migration. Furthermore, CBX4 depletion markedly enhances the histone deacetylase inhibitor (HDACi)-induced cell apoptosis and suppression of tumor growth.
Interpretation
Our data suggest CBX4 as an oncogene with prognostic potential in ccRCC. The newly identified CBX4/HDAC1/KLF6 axis may represent a potential therapeutic target for the clinical intervention of ccRCC.
Keywords: CBX4, KLF6, HDAC1, Combinative therapy, Clear cell renal cell carcinoma
Research in context.
Evidence before this study
CBX proteins are responsible for the epigenetic silencing of genes involved in tumor progression. CBX4 has been demonstrated to function as both tumor suppressor and oncogene in human cancers. But its clinical significance and biological function in clear cell renal cell carcinoma (ccRCC) remain unclear.
Added value of this study
Using clinical samples, cell culture and mice models, we showed that CBX4 was upregulated in ccRCC to exert oncogenic activities to promote tumor growth and metastasis. Patients with high expression of CBX4 were likely to survived shorter. We also demonstrated that CBX4 was able to interact with HDAC1 to suppress the transcriptional activity of tumor suppressor KLF6 promoter. Combinative treatment of CBX4 knockdown and HDAC inhibitor is of great lethal effect on ccRCC cell survival.
Implications of all the available evidence
CBX4 is a promising prognostic factor in ccRCC. Targeting CBX4/HDAC1/KLF6 axis may provide clinical benefit for the treatment of patients with ccRCC.
Alt-text: Unlabelled box
1. Introduction
Renal cell carcinoma (RCC) is the second leading cause of death in human urological malignancies, accounting for 2-3% of human cancers [1]. The global burden of RCC is approximately 403,262 new cases and 175,098 deaths per year, with an annual rate of increase of approximately 0.7% [2]. Clear cell renal cell carcinoma (ccRCC), the most prevalent subtype of RCC, represents about 75% of RCC [3]. Today, surgical resection followed by observation represents the clinical protocol for non-metastatic ccRCC. Up to 60% of non-metastatic patients will experience recurrence and metastatic disease within 5 years. The 5-year survival rate of metastatic ccRCC dramatically drops down to 10% due to the chemoresistance [4]. Although remarkable progress has been made recently in the clinical application of immunotherapies, the characterization of genetic alterations and a better understanding of the detailed mechanisms behind the ccRCC progression are urgently needed.
Polycomb group (PcG) proteins are major transcriptional repressors that epigenetically modify chromatin to participate in regulations of cell differentiation, senescence, and survival [5]. Polycomb chromobox (CBX) proteins belong to PcG family and include five members (CBX2, 4, 6, 7, 8). CBX proteins canonically function through the polycomb repressive complex 1 (PRC1) to exert anti-tumor or pro-tumor activities, depending on the cellular context [6]. CBX4, regulated by p63, protects epithelial stem cells from senescence through PRC1-dependent repression of the Ink4a locus [7]. CBX4 was identified as a SUMO E3 ligase. Several substrates, such as DNA methyltransferase 3a (Dnmt3a) [8], PR/SET domain 16 (Prdm16) [9] and BMI1 [10], have been identified as CBX4 downstream. Overexpression of CBX4 has also been found in breast cancer [11] and osteosarcoma [12]. Functionally, CBX4 promotes hepatocellular carcinoma via sumoylating HIF-1α to upregulate vascular endothelial growth factor (VEGF) [13]. CBX4 suppresses colorectal cancer metastasis by recruiting histone deacetylase 3 (HDAC3) to the Runx2 promoter to inhibit its expression [14]. However, the role of CBX4 in ccRCC remains unclear.
In this study, we showed that CBX4 played an oncogenic role in ccRCC. CBX4 expression was increased and correlated with unfavorable prognosis. CBX4 promoted cell proliferation and migration by interacting with HDAC1 to transcriptionally repress the expression of tumor suppressor Kruppel-like factor 6 (KLF6). Inhibition of CBX4 was able to increase cell apoptosis induced by the treatment of HDAC inhibitor Trichostatin A (TSA). These findings suggest CBX4 as a promising prognostic factor and a potential therapeutic target in ccRCC.
2. Materials and methods
2.1. Patients
To determine the expression of CBX4 in ccRCC, 26 pairs of fresh specimens were obtained from The First Affiliated Hospital of Sun Yat-sen University and Sun Yat-sen University Cancer Center (SYSUCC). Furthermore, 306 pairs of paraffin-embedded tissues of ccRCC patients diagnosed between Jan 2010 to Dec 2012 in SYSUCC were collected. All samples were anonymous. None of the patients had received radiotherapy or chemotherapy before surgery. This study was approved by Institute Research Ethics Committee of The First Affiliated Hospital of Sun Yat-sen University and SYSUCC. The upregulation of CBX4 in ccRCC was confirmed in The Cancer Genome Atlas (TCGA) dataset (http://www.cbioportal.org) and the Oncomine dataset (https://www.oncomine.org).
2.2. Immunohistochemistry (IHC) and scoring
Tissue microarray (TMA) was constructed, using 306 ccRCC tissues and the adjacent nontumorous kidney tissues. The TMA sections were dewaxed by xylene and graded alcohols. Tissues were hydrated and washed in PBS buffer. Incubation of 3% hydrogen peroxide in methanol for 20 min was used to inhibit the endogenous peroxidase. The tissues were then blocked by avidin–biotin blockage using a biotin-blocking kit (DAKO, Darmstadt, Germany). After incubated with CBX4/KLF6 antibody overnight in a moist chamber at 4 °C, slides were then washed in PBS buffer, incubated with goat anti-rabbit antibody for 40 min, and finally developed with DAB and counterstained with hematoxylin. Expression of CBX4 and KLF6 was evaluated by H-score method as following: percentage score (“0” (0%), “1” (1%–25%), “2” (26%–50%), “3” (51%–75%), or “4” (76%–100%) was multiplied by the staining intensity score(“0” (negative staining), “1” (weak staining), “2” (moderate staining), or “3” (strong staining)).
2.3. Chromatin immunoprecipitation assay (ChIP)
ChIP was performed according to the manufactory instruction of ChIP kit (Millipore, 17-10,085 & 17-10,086). Briefly, ACNH cells transfected with CBX4 overexpression vector or siRNAs were cultured in 100 mm plates to grow to 70% - 80% confluence. Cells were fixed by fixative solution (1/10th the volume of growth medium volume), and then treated with stop solution (1/20th the volume of growth medium volume). Cells were centrifugated and resuspended in ChIP buffer to collect nuclear pellet. The cell lysate was then sonicated and incubated with 5 mg of CBX4 antibody at 4 °C overnight. After incubating with protein A/G agarose beads for 4 h at 4 °C, the bound DNA-protein complexes were eluted and washed to reverse the cross-links. DNAs were extracted and purified for PCR detection. The primers for the KLF6 promoters are shown in Supplementary Table 1.
2.4. Luciferase reporter assay
ACHN cells were co-transfected with CBX4 overexpression vector or siRNAs, and 500 ng of psiCHECK-2-KLF6-3′-UTR wild-type or mutant reporters. Cells were collected 36 h after transfection and analyzed with the Dual-Luciferase Reporter Assay System (Promega, CA, USA).
2.5. Animal model
For tumor growth model, 1 × 107 ACHN and RCC4 cells with CBX4 overexpression or 786-O cells with CBX4 depletion were subcutaneously injected into the right flanks of four-weeks-aged male BALB/c-nude mice (six mice per group). The tumor sizes were measured with calipers and calculated with the formula: Volume (mm3) = [width2 (mm2) × length (mm)]/2. After 25 days, all tumors were dissected and weighed. For metastasis model, 5 × 105 cells with CBX4 overexpression or 786-O cells with CBX4 depletion were injected via the tail vein. After 5 weeks, the lungs of mice were dissected and fixed in 4% paraformaldehyde for 24 h to make paraffin-embedded tissue block. The lung tissues were stained with hematoxylin and eosin (HE). Tumors metastasizing to the lung was counted and quantified. All animal studies were approved by the Medical Experimental Animal Care Commission of The First Affiliated Hospital of Sun Yat-sen University.
2.6. Statistical analysis
Data are mean ± SEM from three independent experiments. The significance between groups was calculated by the Student's t-test. Survival analyses were conducted by Kaplan–Meier analyses (log-rank test). If the P-value was less than 0.05, the differences were considered statistically significant.
3. Results
3.1. CBX4 is upregulated and correlated with poor outcomes in ccRCC
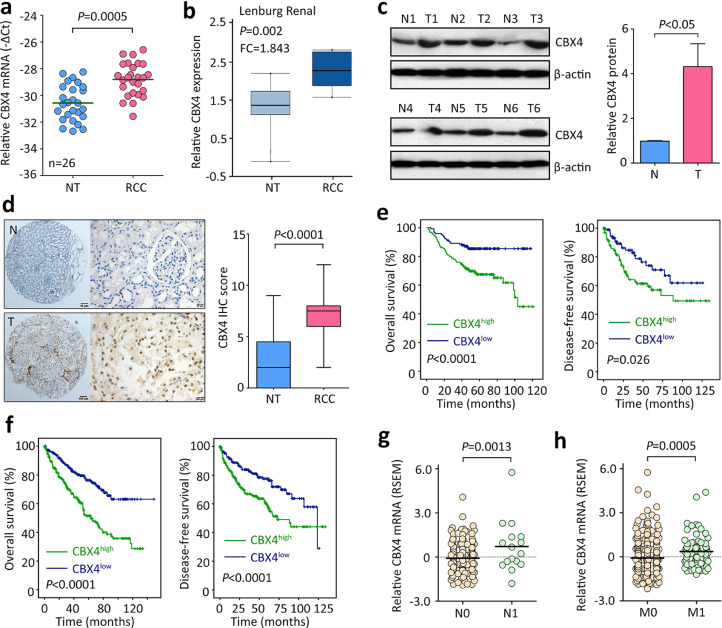
The expression of CBX4 in ccRCC samples was determined by qRT-PCR, western blot and IHC. Twenty-six pairs of fresh tissues were collected. The mRNA expression level of CBX4 in ccRCC tissues was significantly higher than that in nontumorous tissues (Fig. 1A). This was validated by Lenburg Renal study from Oncomine dataset (Fig. 1B). In consistent, CBX4 protein expression was noticeably increased by 4.1 folds in ccRCC tissues, compared to adjacent kidney tissues (Fig. 1C). The upregulation of CBX4 was further confirmed by IHC in a large cohort containing 306 paraffin-embedded samples. Results showed that CBX4 was mainly expressed in the nucleus in cancer cells. Its expression in ccRCC was much higher (Fig. 1D). Based on the median IHC score (3.5), patients were divided into two groups: CBX4 high and CBX4 low. High expression of CBX4 was closely associated with higher Fuhrman grade, tumor sarcomatoid differentiation and lymph node invasion in SYSUCC cohort (Supplementary Table 2). In TCGA cohort, ccRCC patients with higher pathological grade or advanced clinical stage had more CBX4 mRNA expression (Supplementary Fig. 1).
Fig. 1.
CBX4 expression is upregulated and correlated with poor survivals in RCC. a. The mRNA expression of CBX4 in 26 pairs of RCC and corresponding adjacent nontumorous tissues was examined by qRT-PCR. b. The increase of CBX4 mRNA in RCC tissues was confirmed in Lenburg Renal study from oncomine dataset. c. The protein level of CBX4 was determined in 26 paired of RCC (T) and nontumorous (N) tissues. Representative images of western blot (left panel) and the statistics (right panel) were presented. d. A cohort of 306 patients was recruited. Paraffin-embedded tissues were used for IHC assays to evaluate the expression of CBX4 protein in clinical samples. Representative images of IHC (left panel) and the comparison of CBX4 IHC score (right panel) were shown. e. Kaplan-Meier survival analyses were conducted to indicate the prognostic value of CBX4 in RCC. A total of 306 RCC patients were divided into two groups according to the median of IHC score. f. The prognostic implication of CBX4 in RCC was validated in TCGA cohort containing 534 patients. g. In TCGA cohort, the expression of CBX4 mRNA was compared between patients with (N1) or without (N0) lymph node metastasis. h. CBX4 mRNA expression was higher in RCC patients with distant tumor metastasis (M1) than those without (M0).
The prognostic value of CBX4 in ccRCC was next determined. Compared with those with low CBX4, patients with high CBX4 expression were likely to survive shorter and experience tumor progression in SYSUCC cohort (Fig. 1E). The median months of overall and disease-free survivals in high CBX4 group were 83.9 and 81.5, whereas they were respectively 104.5 and 94.1 in low CBX4 groups. Multivariate analyses using Cox regression model revealed that CBX4, as well as Fuhrman grade, Lymph node invasion and T stage, was an independent factor for overall survival in ccRCC (Supplementary Table 3). Prognostic analyses of TCGA data validated that CBX4 expression was reversely associated with clinical outcomes (Fig. 1F). Patients with high CBX4 expression were frequently with tumor lymph node and distant metastasis (Fig. 1F&H). These data suggest CBX4 serve as a promising factor in ccRCC.
3.2. CBX4 promotes cell proliferation in ccRCC
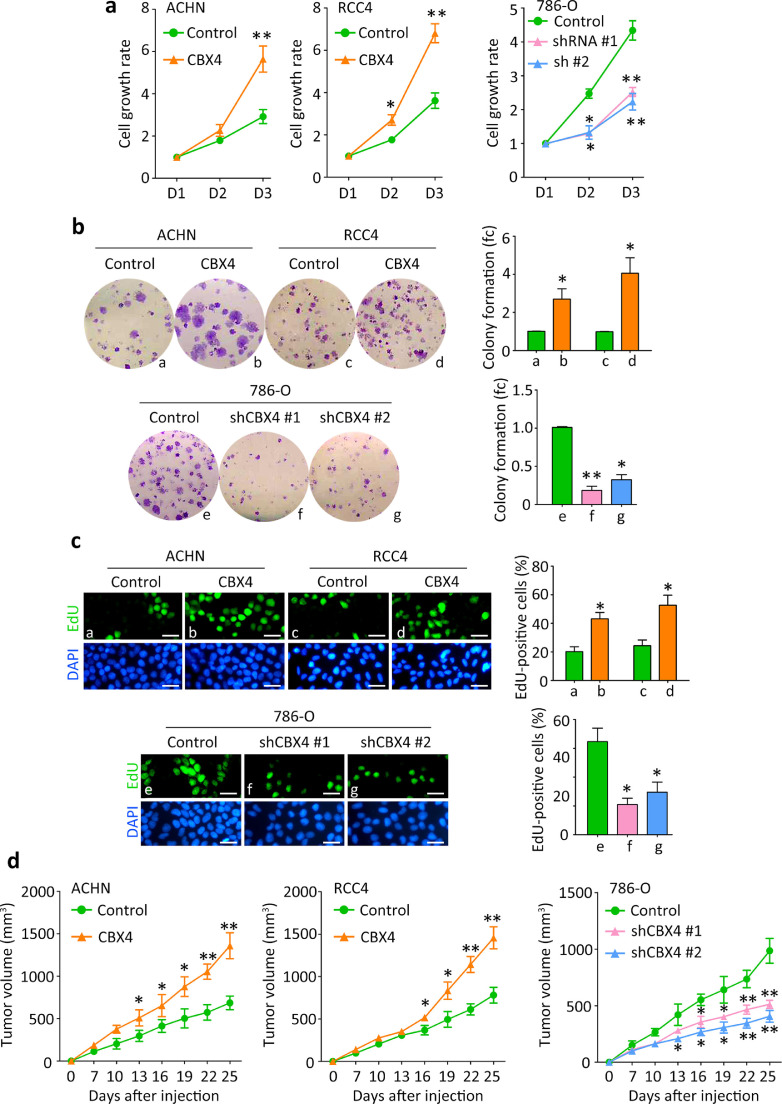
Gain-of-function and loss-of-function assays were performed to reveal the role of CBX4 in the progression of ccRCC. According to the basal line of CBX4 expression in ccRCC cell lines (data not shown), stable cell lines were established using CBX4 overexpression vector in ACHN and RCC4 cells, and CBX4 shRNAs in 786-O cells. The mRNA and protein expression of CBX4 in stable cell lines were examined by qRT-PCR and western blot (Supplementary Figure 2). MTT assays showed that ectopic expression of CBX4 increased, whereas knockdown of CBX4 decreased the cell viabilities of ccRCC cells (Fig. 2A). CBX4-mediated cell proliferation was examined by colony formation and EdU assays. Overexpression of CBX4 markedly enhanced the ability of clonogenicity of ACHN and RCC4 cells. Silence of CBX4 in 786-O cells remarkably reduced the colonies (Fig. 2B). EdU-positive cells were more frequently depicted in CBX4-expressing cells but rarely seen in CBX4-depleted cells (Fig. 2C).
Fig. 2.
CBX4 promotes cell proliferation and tumor growth in RCC. a. Cells transfected with CBX4 overexpression vector or siRNAs were cultured into 96-well plates for 3 days. The cell viabilities were determined by MTT assays. b. The stable cells were cultured in 6-well plates for 10 days. Colonies were stained by 0.05% crystal violet and counted (left panel). The related fold change (fc) of colony was shown (right panel). fc, fold change. c. The effect of CBX4 on cell proliferation in RCC was further confirmed by EdU staining. Scale bar = 50 μm. d.In vivo models were used to determine the role of CBX4 in tumor growth. Stable cells were injected into the flank of the mice for 25 days. The volumes of tumors were measured every 3 days and indicated by growth curves. All *P<0.05, **P<0.01 (Student's t-test).
In vivo subcutaneous model was used to determine the effect of CBX4 on tumor growth. Tumors were successfully formed in 6/6 and 4/6 of mice injected with CBX4-expressing and control ACHN and RCC4 cells, respectively. 786-O-bearing tumors were found in 3/6 and 5/6 of mice in CBX4 silence and control groups. Overexpression of CBX4 significantly promoted, whereas knockdown of CBX4 attenuated tumor growth (Fig. 2D). When the mice weights in all groups showed no difference, the tumors were much heavier in the CBX4-overexpression groups but much lighter in the CBX4-depletion groups, compared to the corresponding control groups (Supplementary figure 3).
3.3. CBX4 facilitates cell migration in ccRCC
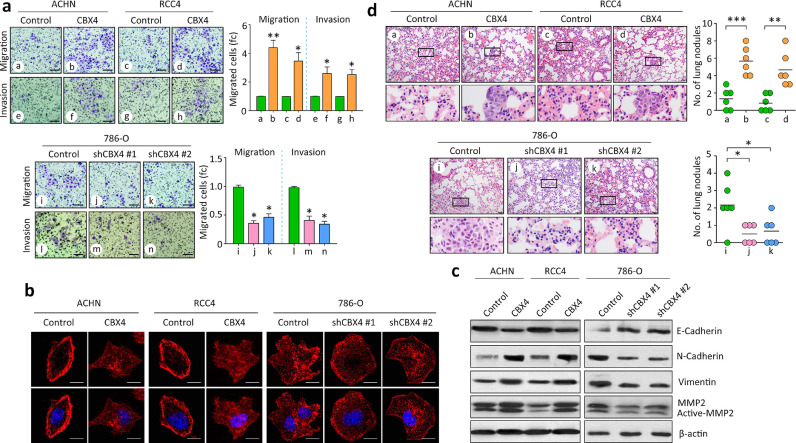
Since clinical data revealed that CBX4 expression was correlated with metastatic features in ccRCC, the effect of CBX4 on cell migration was next determined. As shown by the Transwell assays, CBX4 enhanced the cell migration and invasion in ACHN and RCC4 cells. CBX4 shRNAs weakened the movement of 786-O cells (Fig. 3A). Cell shapes were indicated by phallotoxin staining to visualize cytoskeleton F-actin. The formation of cell pseudopodium was increased by CBX4 overexpression but impaired by CBX4 silencing (Fig. 3B). Western blot showed that the expression of mesenchymal markers Vimentin, N-cadherin and MMP2 was induced by CBX4, but reduced by CBX4 shRNAs in ccRCC cells. In contrast, the expression of epithelial markers E-cadherin was inhibited by CBX4 overexpression (Fig. 3C). This indicates CBX4 may trigger the epithelial-to-mesenchymal transition (EMT) process. In vivo models were applied to confirm the pro-metastatic effect of CBX4 in ccRCC. Injection of ACHN and RCC4 cells with CBX4 overexpression formed more nodules in the lung, compared with the control cells. shRNAs against CBX4 attenuated the lung metastasis in mice injected with 786-O cells (Fig. 3D). Collectively, these data suggest CBX4 an oncogene in ccRCC.
Fig. 3.
CBX4 facilitates tumor metastasis in RCC. a. Stable cells with CBX4 overexpression or knockdown were used for migration and invasion assays. Cells were stained by 0.05% crystal violet and counted under a microscope (left panel). The related fold changes (fc) of migrated cells or invaded cells were indicated (right panel). *P<0.05, **P<0.01 (Student's t-test). Scale bar = 50 μm. b. Cells cultured in 12-well plates were stained by TRITC-labeled phalloidin to show F-actin (red) to indicate the cell pseudopodium (denoted by arrows). Scale bar = 10 μm. The number of cell pseudopodium was counted and compared. *P<0.05 (Student's t-test). c. Proteins derived from stable cells were subjected to western blot to examine the expression of EMT markers, including E-Cadherin, N—Cadherin, Vimentin and MMP2. d. Cells were injected in to null mice via tail vein. Lung nodules were counted under a microscope and showed by HE staining. Representative images (left panel) and the statistical analyses were shown. *P<0.05, **P<0.01 (Student's t-test).
3.4. CBX4 transcriptionally suppresses KLF6 to exert oncogenic activities
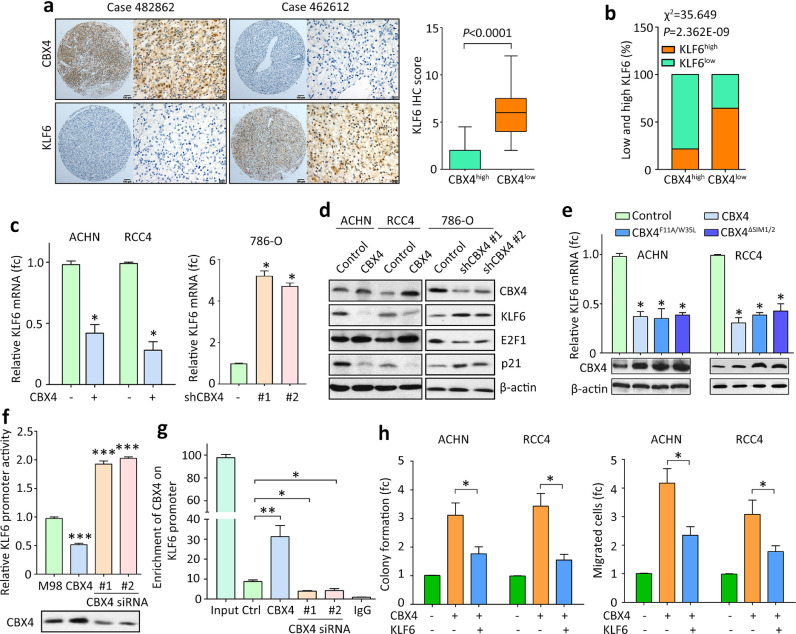
The underlying mechanism through which CBX4 exerted oncogenic activities was next determined. According to the published data (GSE6344), CBX4 expression was closed related to Kruppel like factors, such as KLF2, KLF3, KLF6 and KLF8 in ccRCC tissues (Supplementary figure 4A). In another study, treatment of CBX4 shRNAs resulted in increased expression of KLF6 (Supplementary figure 4B). Since KLF6 had been reported as a tumor suppressor in ccRCC [15], it was chosen as the downstream effector of CBX4 in this study. In 26 pairs of clinical fresh specimens, CBX4 mRNA expression was reversely associated with KLF6 mRNA (Supplementary figure 5). In SYSUCC cohort, patients with high CBX4 expression frequently expressed less KLF6 (Fig. 4A). Determined by IHC, low CBX4 expression was significantly associated with high KLF6 expression (Fig. 4B). In TCGA cohort, the genomic alterations of CBX4 and KLF6 was mutually exclusive (Supplementary Fig. 6A). Samples with overexpression of CBX4 likely have a significant downregulation of KFL6 (Supplementary Fig. 6B). Patients with high expression of KLF6 survived longer than those with low KLF6, but patients with both high CBX4 expression and low KLF6 expression had the worse overall and disease-free survivals (Supplementary Fig. 7). The effect of CBX2, CBX6, CBX7 and CBX8 on the expression of KLF6 was next determined. TCGA data showed that the expression of CBX6, CBX7 and CBX8 was significantly associated with KLF6 (Supplementary Fig. 8A-D). However, the mRNA expression of KLF6 in 786-O cells treated with siRNAs for CBX2, CBX6, CBX7 or CBX8 remained unchanged (Supplementary Fig. 8E&F). These data implied a potential link between CBX4 and KLF6 in ccRCC.
Fig. 4.
CBX4 exhibits oncogenic functions via transcriptional suppression of KLF6 in RCC. a. The correlation of CBX4 and KLF6 expression in RCC samples was determined. Representative images of CBX4 and KLF6 IHC in RCC were presented (left panel). The IHC score of KLF6 was indicated in cases with high or low expression of CBX4. b. Statistical analyses showed that RCC patients with low expression of CBX4 was frequently accompanied with high KLF6 level in SYSUCC cohort. c. Cells were transfected with CBX4 overexpression vectors or siRNAs for 36 h. The KLF6 mRNA levels were determined by qRT-PCR. fc, fold change. *P<0.05 (Student's t-test). d. The expression of KLF6, as well as its downstream targets E2F1 and p21, was examined in cells with CBX4 overexpression or silence. e. Cells were transfected with overexpression vector of CBX4, CBX4F11A/W35L or CBX4ΔSIM1/2 for 36 h. The KLF6 mRNA expression was examined by qRT-PCR. fc, fold change. *P<0.05 (Student's t-test). f. Dual luciferase report assays were used to evaluate the impact of CBX4 on the transcriptional activity of KLF6 promoter in ACHN cells. ***P<0.001 (Student's t-test). g. ChIP assays were performed to detect the direct binding of CBX4 on KLF6 promoter in ACHN cells. The enrichment of CBX4 on KLF6 promoter was determined by qRT-PCR. *P<0.05, **P<0.01. h. KLF6 was overexpressed in CBX4-expressing RCC cells. Rescue experiments were performed to examine the role of KLF6 in CBX4-mediated cell growth and migration. fc, fold change. *P<0.05 (Student's t-test).
The regulation of KLF6 by CBX4 was next examined in ccRCC cell lines. Ectopic expression of CBX4 suppressed, whereas depletion of CBX4 induced the expression of KLF6 mRNA (Fig. 4C). Consistently, the protein level of KLF6, as well as its well-known downstream E2F1 and p21, was altered by the change of CBX4 expression (Fig. 4D). Current literatures suggested the chromodomain and SUMO E3 ligase activity were important for CBX proteins to exert transcriptional activities. According to a previous study [14], deletion mutant of SUMO interacting motifs (CBX4ΔSIM1/2) and chromodomain mutant (CBX4F11A/W35L) containing both CBX4F11A and CBX4W35L mutations were constructed. After transfection of CBX4F11A/W35L or CBX4ΔSIM1/2 in ACHN and RCC4 cells for 36 h, KLF6 mRNA was determined by qRT-PCR. Results showed that both mutants were capable of suppressing KLF6 expression (Fig. 4E), indicating that CBX4 modulates the expression of KLF6 via the manner independent of its chromodomain and SUMO E3 ligase activity. Dual luciferase reporter and ChIP assays were performed to determine whether CBX4 was capable of transcriptionally modulating KLF6. Results showed that increased CBX4 significantly inhibited, while decreased CBX4 stimulated the transcriptional activity of the KLF6 promoter (Fig. 4F). The enrichment of CBX4 on the KLF6 promoter was induced by CBX4 overexpression but reduced by CBX4 siRNAs (Fig. 4G). The profound impact of CBX4 on KLF6 expression prompted us to test whether the alteration of KLF6 would rescue CBX4-promoted malignant phenotypes. KLF6 expression was restored to the basal line in cells with CBX4 overexpression or depletion. Upon the overexpression of KLF6 in CBX4-expressing cells, the downregulation of tumor suppressor p21 and epithelial marker E-cadherin, as well as the upregulation of mesenchymal marker Vimentin, was partially rescued (Supplementary figure 9), indicating that KLF6 inhibited the EMT process induced by CBX4. Strikingly, CBX4-promoted colony formation and cell migration were attenuated by the re-expression of KLF6 (Fig. 4H). The inhibition of cell proliferation and migration by CBX4 shRNA was abolished by KLF6 shRNA, while the promotion of CBX4-induced cell growth was attenuated by E2F1 knockdown and p21 overexpression (Supplementary Fig. 10). Collectively, these data indicated that CBX4 exhibited oncogenic activities in ccRCC via KLF6 suppression.
3.5. CBX4-mediated KLF6 suppression requires HDAC1
Previous studies showed that CBX4 cooperated with histone deacetylases (HDACs) to transcriptionally repress target genes [14, 16]. Whether HDAC was required for the KLF6 suppression by CBX4 was next determined. Dual luciferase reporter assays demonstrated that siRNAs for HDAC1, HDAC2 and HDAC9 significantly affected the CBX4-mediated inhibitory of KLF6 promoter activity (Fig. 5A and Supplementary Fig. 11). Strikingly, knockdown of HDAC1 almost fully restored the inhibitory effect of CBX4 on KLF6 promoter (Fig. 5A). In TCGA cohort, the genomic alterations of HDAC1 and KLF6 was mutually exclusive (Supplementary figure 6A). HDAC1 mRNA expression was positively but not significantly correlated with CBX4 mRNA (Supplementary figure 6B&C). High expression of HDAC1 was correlated with poor overall and disease-free survivals (Supplementary figure 12A). The prognosis of ccRCC patients could be distinguished by the combination of CBX4 and HDAC1 mRNA expression (Supplementary Fig. 12B).
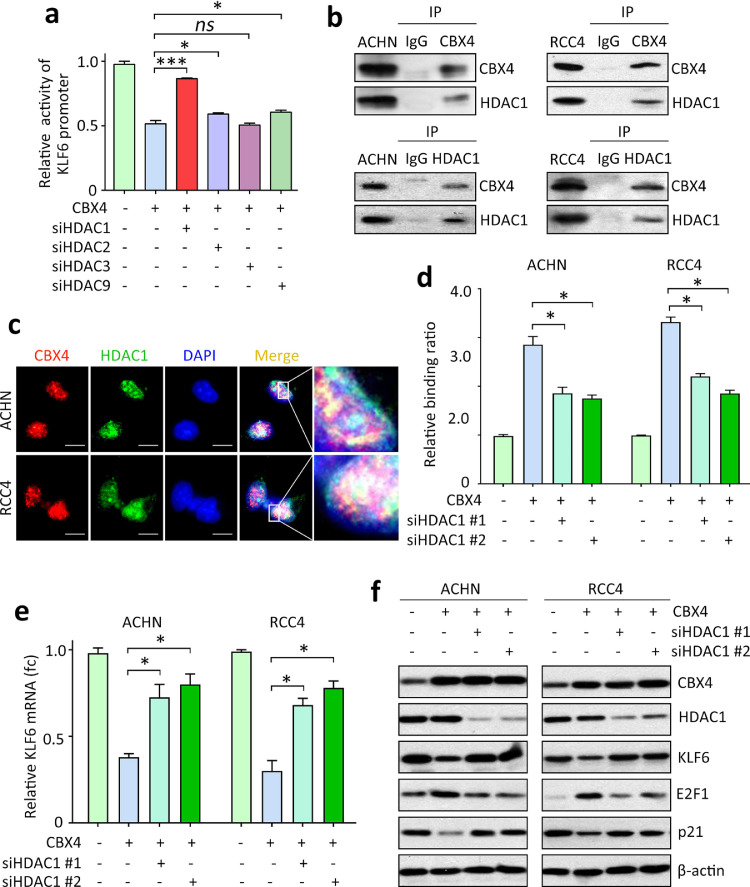
Fig. 5.
HDAC1 is required for CBX4-mediated KLF6 suppression in RCC. a. siRNAs for HDAC1, HDAC2, HDAC3 and HDAC9 were introduced into ACHN cells with CBX4 overexpression. The effects of siRNAs on the activity of KLF6 promoter were examined by luciferase reporter assays. *P<0.05, ***P<0.001 (Student's t-test). ns, non-significant. b. The interaction between CBX4 and HDAC1 was determined by co-IP experiments in ACHN and RCC4 cells. c. Immunofluorescence was used to indicate the colocalization of CBX4 and HDAC1 in the nuclear of RCC cells. Scale bar = 10 μm. d. Cells with CBX4 overexpression were transfected with two HDAC1 siRNAs (siHDAC1 #1 and #2). The binding ratio of CBX4 on KLF6 promoter was determined by ChIP assays. *P<0.05 (Student's t-test). e. The mRNA expression of KLF6 in ACHN and RCC4 cells were examined by qRT-PCR, following the transfection of HDAC1 siRNA for 36 h. *P<0.05 (Student's t-test). f. The expressions of CBX4, HDAC1 and KLF6, as well as E2F1 and p21, were examined by western blot.
The interaction between CBX4 and HDAC1 was examined. Endogenous CBX4 and HDAC1 were detectable in the precipitate mediated by the antibody of CBX4 or HDAC1 in both ACHN and RCC4 cells (Fig. 5B). The nuclear colocalization of CBX4 and HDAC1 in the nucleus of ccRCC cells was observed by confocal immunofluorescence (Fig. 5C). Further studies showed that the interaction of CBX4 and HDAC1 did not interrupt the binding of CBX4 to PRC1 proteins (Supplementary figure 13). CBX4 knockdown markedly weakened the binding of HDAC1 to KLF6 promoter (Supplementary figure 14). When HDAC1 was knocked down in ACHN and RCC4 cells, the binding of CBX4 onto KLF6 promoter was markedly decreased (Fig. 5D). Consequently, the mRNA expression of KLF6 in CBX4-expressing cells with HDAC1 depletion was upregulated (Fig. 5E). The protein expression of KLF6, as well as its downstream effector E2F1 and p21, was rescued to the basal line, compared with the control expression (Fig. 5F). Our data further showed that interruption of CBX4 and HDAC1 binding using HDAC1 siRNAs significantly increased the H3K27 acetylation but not methylation (Supplementary figure 15). These findings suggested that CBX4 interacted with HDAC1 to modulate the expression of KLF6 in ccRCC.
3.6. CBX4 depletion enhances HDACi-induced cell death
Since CBX4 exerts functions partly via HDACs, the effect of HDAC inhibitor (HDACi) on the CBX4-mediated phenotypes was next determined. Trichostatin A (TSA) and SAHA, specific HDACis that had been demonstrated to induce cell apoptosis in ccRCC cells [17], was used in our study. According to the previous report [17], 2 ng/mL TSA and 1.25 μM SAHA were used in our in vitro experiments. Combinative treatment of CBX4 siRNA and HDACi resulted in a dramatically decrease of colony formation in 786-O cells (Fig. 6A). Annexin/V assays showed a significant increase of cell apoptosis, compared with CBX4 siRNA or HDACi alone (Fig. 6B). The depolarization of mitochondrial outer membrane was next examined by detecting the reduction of mitochondrial membrane potential (MMP). Results showed that the combined treatment remarkably lowered the mitochondrial transmembrane potential (Fig. 6C), followed by an obvious release of cytochrome c from mitochondria to cytoplasm (Fig. 6D). Furthermore, the cleavages of apoptotic markers PARP and Caspase-3 were upregulated (Fig. 6D). The therapeutic effect of combinative treatment was partially attenuated by the knockdown of KLF6 in 786-O cells (Supplementary figure 16). Cells with CBX4 depletion were subcutaneously injected in nude mice to determine the in vivo effect of the combination treatment. Seven days after injection, TSA (1 mg/kg mouse body weight) was given to the mice once daily for five consecutive days per week for total 15 days. Compared with the control or signal treatment groups, tumor growth in “shCBX4 and TSA” group was significantly attenuated (Fig. 6E). The treatment did not appear to have a noticeable effect on body weight in mice (Fig. 6F). On Day 25, mice were sacrificed and the tumor weights were measured. As expected, the combination of TSA and CBX4 shRNA significantly reduced the weights of xenograft tumor, compared with other groups (Fig. 6F). These data indicated that the CBX4 depletion-mediated anti-proliferative effect was enhanced by the treatment of HDACi in ccRCC.
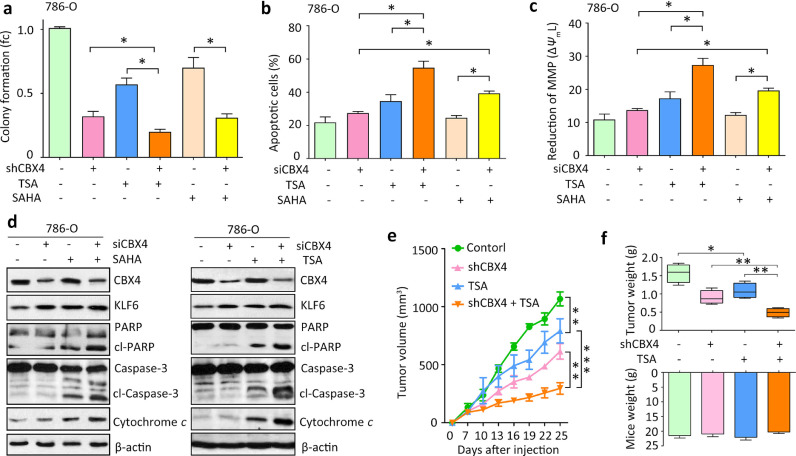
Fig. 6.
CBX4 deficiency enhances the lethal effect of HDACi in RCC. a. Stable786-O cells were treated with HDACi (2 ng/mL TSA and 1.25 μM SAHA) or both. Colony formation was used to evaluate the effects of the three treatments on the cell growth. fc, fold change. b. Cell apoptosis was assessed in cells treated with CBX4 siRNA, HDACi or both via flow cytometry using Annexin V/PI staining. c. Reduction of mitochondrial membrane potential (MMP) was induced in cells treated as in A. The MMP collapse (ΔΨmL) was measured by flow cytometry after staining the cells with DioC6 and quantitative analysis of ΔΨmL was shown. d. Apoptotic markers including PARP and its cleaved form (cl-PARP), Caspase 3 and its cleaved form (cl-Caspase 3) and cytosolic cytochrome c, were examined by western blot in 786-O cells. e. Nude mice were inoculated with 1 × 107 of 786-O cells with or without CBX4 overexpression. After the formed tumor was palpable, mice were randomly divided into four groups as indicated in the curve. TSA (1 mg/kg mouse body weight) was given to the “TSA” and “shCBX4 + TSA” groups, once daily for five consecutive days per week for 15 days. The tumor volumes were calculated every three days. Data are mean ± SEM, **P<0.01, ***P<0.001 (One way ANOVA). f. On day 25, mice were sacrificed, and the tumor weights and mice body weights were measured and indicated. All data represented mean ± SEM of three independent experiments. All *P<0.05, **P<0.01, ***P<0.01 (Student's t-test).
4. Discussion
As the most common malignancy of the adult kidney, ccRCC represents one of the lethal threats to human's life [2]. Unfortunately, the molecular mechanism of the development and progression of ccRCC remains unclear to date, despite of lots of efforts in identification of aberrant signaling pathways in this deadly disease [3]. Dysregulation of PcG proteins have been implicated in ccRCC [18, 19]. Here we showed that CBX4 expression was upregulated in clinical samples and correlated with poor outcomes. In vitro and in vivo data demonstrated that CBX4 played an oncogenic role in ccRCC via interacting with HDAC1 to suppress the transcription of tumor suppressor KLF6.
One of the main goals of current cancer studies is to search for potential biomarkers for the prognostic prediction of patients with malignancies. The prognostic significances of CBX members have been well described in human cancers. Overexpression of CBX2 in leukemia [20], high grade serous ovarian carcinoma [21] and prostate cancer [22] was associated with poor survivals. High CBX6 expression was linked to worse overall survival in hepatocellular carcinoma [23], but better recurrence-free survival in breast carcinoma [24]. Reduced expression of CBX7 in breast cancer [25] and colon carcinoma [26] was found to be a negative prognostic factor. CBX8 upregulation in hepatocellular carcinoma [27] and breast cancer [28] predicted unfavorable prognosis of patients. For CBX4, paradoxical conclusions were reported. On one hand, CBX4 expression was increased in hepatocellular carcinoma [13], breast cancer [11] and osteosarcoma [12], and correlated with poor clinical outcomes. On the other hand, CBX4 expression was decreased in colorectal carcinoma, and connected with better survivals [14]. In this study, CBX4 was upregulated at both mRNA and protein levels in ccRCC. Patients with high expression of CBX4 were accompanied with worse overall and disease-free survivals. This may be due to that elevated CBX4 expression was associated with higher Fuhrman grade, tumor sarcomatoid differentiation and lymph node invasion responsible for the disease progression. Taken together, these data suggest that CBX proteins be differently expressed in human cancers and be promising biomarkers for the prediction of postsurgical outcomes.
Current literatures suggest CBX proteins may function as both oncogene and tumor suppressor, dependent on the cell contents. The investigation of the biological function of CBX4 in ccRCC demonstrated that CBX4 exerted oncogenic activities to promote tumor growth and metastasis via transcriptionally downregulating KLF6. Two alternative mechanisms have been demonstrated for CBX4 to exhibit biological functions: transcriptional repression and post-transcriptional sumoylation. Current literatures indicate that CBX4 functions as both a transcriptional repressor and a SUMO E3 ligase through its N-terminal chromodomain and two SUMO-interacting motifs (SIMs), respectively. SIMs-dependent, rather than polycomb-dependent, function of CBX4 contributes to its effect on CBX2 polySUMOylation [20], increased Prdm16 protein expression [9] and enhanced hypoxia-induced VEGF expression [13]. On the other hand, the N-terminal chromodomain, but not the SIMs, of CBX4 was required for its transcriptional repression of link4a locus [29], nonepidermal lineage genes [7] and transcriptional factor Runx2 [14] to protect cells from senescence, differentiation and death. In our study, CBX4 interacts with HDAC1 to inhibit the transcriptional activity of KLF6 independent of its SIMs and PRC1-binding domain. These data are consistent with a previous study that showed CBX4 inhibited Runx2 transcription in both SIMs and PRC1-independent fashion [14]. Interestingly, CBX7, lack of SIMs, interacted with HDAC2 to suppress CCNE1 gene expression [30]. Collectively, our data suggest that the effect of gene repress of CBX proteins in human cancers does not depend on catalytic activities.
KLF6, a member of Kruppel-like factor (KLF) family that is a group of zinc finger transcription factors, is a putative tumor suppressor in human cancers. In ccRCC, KLF6 exerts antitumor effects in several studies [31, 32]. However, a latest study showed that KLF6 was responsible for the upregulation of lipid metabolism genes to support ccRCC progression [33]. According to the TCGA data, KLF6 expression in ccRCC tissues were significantly higher than that in nontumorous tissues. However, as shown in our supplementary figure 6 and 7, KLF6 mRNA expression was reversely correlated with oncogenic CBX4. High expression of KLF6 mRNA was associated with better overall and disease-free survivals of ccRCC patients in TCGA dataset, which may finely support the tumor suppressor role of KLF6 in ccRCC. Knockdown of KLF6 in ccRCC cells, using CRISPR-Cas9 system, resulted in enhanced cell proliferation and migration (Supplementary figure 17). Furthermore, in rescue experiments using two KLF6 shRNAs, our data showed KLF6 knockdown markedly attenuated the CBX4 shRNA-induced suppression of cell proliferation and migration. Similar to our results, KLF6 mRNA was also increased in TCGA glioblastoma tissues. However, KLF6 was identified as a tumor suppressor to inhibit glioblastoma by several research groups [34, 35]. Collectively, KLF6 may exert both anti-tumorigenic and pro-tumorigenic activities in ccRCC cells. It should be noted that KLF6 splice variant 1 (KLF6-SV1) exhibits oncogenic activities in many kinds of human cancers [36, 37]. Previous studies implied that KLF6-SV1’s activity antagonizes the wild-type KLF6’s (KLF6) tumor-suppressive effects. It should be of great interest to investigate whether KLF6-SV1 plays roles in the development and progression of ccRCC, and also in CBX4-mediated malignant phenotypes in our future studies.
In our study, treatment of TSA (a specific inhibitor of HDAC proteins) enhanced the attenuation of tumor growth mediated by CBX4 depletion. According to the current literatures, KLF6 transcriptional activation is associated with increased histone H3 acetylation [38]. A single lysine-to-arginine point mutation (K209R) reduces acetylation of KLF6 and abrogates its capacity to modulate gene expression [39]. Interestingly, TSA treatment is able to increase the acetylation of histone H3 to increase the nuclear localization of KLF6 and its transcriptional activity [40]. These data indicate that HDACi treatment may enhance the histone H3 acetylation at the promoter region of KLF6 to promote its transcriptional activity. In our study, the expression of KLF6 is transcriptionally downregulated by CBX4. Combinative treatment of HDACi and CBX4 knockdown should be more powerful to reduce the ccRCC cell growth. Since HDACis have potential in clinical management of human cancers [41, 42], targeting CBX4 represents an adjuvant strategy to improve the efficacy of chemotherapy in ccRCC. Collectively, our study suggests CBX4 function as an oncogene and serve as a promising prognostic factor and a potential therapeutic target in ccRCC.
Declaration of Competing Interest
All authors declare no conflict of interest.
Acknowledgments
Acknowledgement
We thank Dr. Yun Cao from the Sun Yat-sen University Cancer Center for his valuable suggestions on the writing of this paper.
Funding sources
This study is supported by National Natural Science Foundation of China (No. 81872266).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102692.
Contributor Information
Chris Zhiyi Zhang, Email: zhangzy@jnu.edu.cn.
Huimin Shen, Email: huimin_shen@126.com.
Appendix. Supplementary materials
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Capitanio U., Bensalah K., Bex A., Boorjian S.A., Bray F., Coleman J. Epidemiology of renal cell carcinoma. Eur Urol. 2019;75(1):74–84. doi: 10.1016/j.eururo.2018.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barata P.C., Rini B.I. Treatment of renal cell carcinoma: current status and future directions. CA Cancer J Clin. 2017;67(6):507–524. doi: 10.3322/caac.21411. [DOI] [PubMed] [Google Scholar]
- 5.Mozgova I., Hennig L. The polycomb group protein regulatory network. Annu Rev Plant Biol. 2015;66:269–296. doi: 10.1146/annurev-arplant-043014-115627. [DOI] [PubMed] [Google Scholar]
- 6.Ma R.G., Zhang Y., Sun T.T., Cheng B. Epigenetic regulation by polycomb group complexes: focus on roles of CBX proteins. J Zhejiang Univ Sci B. 2014;15(5):412–428. doi: 10.1631/jzus.B1400077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mardaryev A.N., Liu B., Rapisarda V., Poterlowicz K., Malashchuk I., Rudolf J. Cbx4 maintains the epithelial lineage identity and cell proliferation in the developing stratified epithelium. J Cell Biol. 2016;212(1):77–89. doi: 10.1083/jcb.201506065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li B., Zhou J., Liu P., Hu J., Jin H., Shimono Y. Polycomb protein Cbx4 promotes SUMO modification of de novo DNA methyltransferase Dnmt3a. Biochem. J. 2007;405(2):369–378. doi: 10.1042/BJ20061873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q., Huang L., Pan D., Zhu L.J., Wang Y.X. Cbx4 sumoylates prdm16 to regulate adipose tissue thermogenesis. Cell Rep. 2018;22(11):2860–2872. doi: 10.1016/j.celrep.2018.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ismail I.H., Gagne J.P., Caron M.C., McDonald D., Xu Z., Masson J.Y. CBX4-mediated SUMO modification regulates BMI1 recruitment at sites of DNA damage. Nucleic Acids Res. 2012;40(12):5497–5510. doi: 10.1093/nar/gks222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng J.S., Zhang Z.D., Pei L., Bai Z.Z., Yang Y., Yang H. CBX4 exhibits oncogenic activities in breast cancer via Notch1 signaling. Int J Biochem Cell Biol. 2018;95:1–8. doi: 10.1016/j.biocel.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Yang J., Cheng D., Zhu B., Zhou S., Ying T., Yang Q. Chromobox homolog 4 is positively correlated to tumor growth, survival and activation of HIF-1alpha signaling in human osteosarcoma under normoxic condition. J Cancer. 2016;7(4):427–435. doi: 10.7150/jca.13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J., Xu Y., Long X.D., Wang W., Jiao H.K., Mei Z. Cbx4 governs HIF-1alpha to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell. 2014;25(1):118–131. doi: 10.1016/j.ccr.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Wang X., Li L., Wu Y., Zhang R., Zhang M., Liao D. CBX4 suppresses metastasis via recruitment of HDAC3 to the Runx2 promoter in colorectal carcinoma. Cancer Res. 2016;76(24):7277–7289. doi: 10.1158/0008-5472.CAN-16-2100. [DOI] [PubMed] [Google Scholar]
- 15.Gao Y., Li H., Ma X., Fan Y., Ni D., Zhang Y. KLF6 suppresses metastasis of clear cell renal cell carcinoma via transcriptional repression of E2F1. Cancer Res. 2017;77(2):330–342. doi: 10.1158/0008-5472.CAN-16-0348. [DOI] [PubMed] [Google Scholar]
- 16.Jing X., Sui W.H., Wang S., Xu X.F., Yuan R.R., Chen X.R. HDAC7 ubiquitination by the E3 ligase CBX4 is involved in contextual fear conditioning memory formation. J Neurosci Off J Soc Neurosci. 2017;37(14):3848–3863. doi: 10.1523/JNEUROSCI.2773-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Touma S.E., Goldberg J.S., Moench P., Guo X., Tickoo S.K., Gudas L.J. Retinoic acid and the histone deacetylase inhibitor trichostatin a inhibit the proliferation of human renal cell carcinoma in a xenograft tumor model. Clin Cancer Res. 2005;11(9):3558–3566. doi: 10.1158/1078-0432.CCR-04-1155. [DOI] [PubMed] [Google Scholar]
- 18.Yamagishi M., Uchimaru K. Targeting EZH2 in cancer therapy. Curr Opin Oncol. 2017;29(5):375–381. doi: 10.1097/CCO.0000000000000390. [DOI] [PubMed] [Google Scholar]
- 19.Ren H., Du P., Ge Z., Jin Y., Ding D., Liu X. TWIST1 and BMI1 in cancer metastasis and chemoresistance. J Cancer. 2016;7(9):1074–1080. doi: 10.7150/jca.14031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Costanzo A., Del Gaudio N., Conte L., Dell'Aversana C., Vermeulen M., de The H. The HDAC inhibitor SAHA regulates CBX2 stability via a SUMO-triggered ubiquitin-mediated pathway in leukemia. Oncogene. 2018;37(19):2559–2572. doi: 10.1038/s41388-018-0143-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wheeler L.J., Watson Z.L., Qamar L., Yamamoto T.M., Post M.D., Berning A.A. CBX2 identified as driver of anoikis escape and dissemination in high grade serous ovarian cancer. Oncogenesis. 2018;7(11):92. doi: 10.1038/s41389-018-0103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clermont P.L., Crea F., Chiang Y.T., Lin D., Zhang A., Wang J.Z. Identification of the epigenetic reader CBX2 as a potential drug target in advanced prostate cancer. Clin Epigenetics. 2016;8:16. doi: 10.1186/s13148-016-0182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng H., Jiang W.H., Tian T., Tan H.S., Chen Y., Qiao G.L. CBX6 overexpression contributes to tumor progression and is predictive of a poor prognosis in hepatocellular carcinoma. Oncotarget. 2017;8(12):18872–18884. doi: 10.18632/oncotarget.14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang Y.K., Lin H.Y., Chen C.F., Zeng prognostic values of distinct CBX family members in breast cancer. Oncotarget. 2017;8(54):92375–92387. doi: 10.18632/oncotarget.21325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meseure D., Vacher S., Alsibai K.D., Nicolas A., Chemlali W., Caly M. Expression of ANRIL-Polycomb complexes-CDKN2A/B/ARF genes in breast tumors: identification of a two-gene (EZH2/CBX7) signature with independent prognostic value. Mol Cancer Res. 2016;14(7):623–633. doi: 10.1158/1541-7786.MCR-15-0418. [DOI] [PubMed] [Google Scholar]
- 26.Pallante P., Terracciano L., Carafa V., Schneider S., Zlobec I., Lugli A. The loss of the CBX7 gene expression represents an adverse prognostic marker for survival of colon carcinoma patients. Eur J Cancer. 2010;46(12):2304–2313. doi: 10.1016/j.ejca.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 27.Zhang C.Z., Chen S.L., Wang C.H., He Y.F., Yang X., Xie D. CBX8 exhibits oncogenic activity via AKT/beta-Catenin activation in hepatocellular carcinoma. Cancer Res. 2018;78(1):51–63. doi: 10.1158/0008-5472.CAN-17-0700. [DOI] [PubMed] [Google Scholar]
- 28.Chung C.Y., Sun Z., Mullokandov G., Bosch A., Qadeer Z.A., Cihan E. Cbx8 acts non-canonically with WDR5 to promote mammary tumorigenesis. Cell Rep. 2016;16(2):472–486. doi: 10.1016/j.celrep.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luis N.M., Morey L., Mejetta S., Pascual G., Janich P., Kuebler B. Regulation of human epidermal stem cell proliferation and senescence requires polycomb- dependent and -independent functions of CBX4. Cell Stem Cell. 2011;9(3):233–246. doi: 10.1016/j.stem.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Forzati F., Federico A., Pallante P., Abbate A., Esposito F., Malapelle U. CBX7 is a tumor suppressor in mice and humans. J Clin Invest. 2012;122(2):612–623. doi: 10.1172/JCI58620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adelman E.R., Huang H.T., Roisman A., Olsson A., Colaprico A., Qin T. Aging human hematopoietic stem cells manifest profound epigenetic reprogramming of enhancers that may predispose to leukemia. Cancer Discov. 2019;9(8):1080–1101. doi: 10.1158/2159-8290.CD-18-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kremer-Tal S., Reeves H.L., Narla G., Thung S.N., Schwartz M., Difeo A. Frequent inactivation of the tumor suppressor Kruppel-like factor 6 (KLF6) in hepatocellular carcinoma. Hepatology. 2004;40(5):1047–1052. doi: 10.1002/hep.20460. [DOI] [PubMed] [Google Scholar]
- 33.Syafruddin S.E., Rodrigues P., Vojtasova E., Patel S.A., Zaini M.N., Burge J. A KLF6-driven transcriptional network links lipid homeostasis and tumour growth in renal carcinoma. Nat Commun. 2019;10(1):1152. doi: 10.1038/s41467-019-09116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masilamani A.P., Ferrarese R., Kling E., Thudi N.K., Kim H., Scholtens D.M. KLF6 depletion promotes NF-KappaB signaling in glioblastoma. Oncogene. 2017;36(25):3562–3575. doi: 10.1038/onc.2016.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimmelman A.C., Qiao R.F., Narla G., Banno A., Lau N., Bos P.D. Suppression of glioblastoma tumorigenicity by the Kruppel-like transcription factor KLF6. Oncogene. 2004;23(29):5077–5083. doi: 10.1038/sj.onc.1207662. [DOI] [PubMed] [Google Scholar]
- 36.Narla G., DiFeo A., Fernandez Y., Dhanasekaran S., Huang F., Sangodkar J. KLF6-SV1 overexpression accelerates human and mouse prostate cancer progression and metastasis. J Clin Invest. 2008;118(8):2711–2721. doi: 10.1172/JCI34780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatami R., Sieuwerts A.M., Izadmehr S., Yao Z., Qiao R.F., Papa L. KLF6-SV1 drives breast cancer metastasis and is associated with poor survival. Sci Transl Med. 2013;5(169) doi: 10.1126/scitranslmed.3004688. 169ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen C.H., Huang P.H., Chu P.C., Chen M.C., Chou C.C., Wang D. Energy restriction-mimetic agents induce apoptosis in prostate cancer cells in part through epigenetic activation of KLF6 tumor suppressor gene expression. J Biol Chem. 2011;286(12):9968–9976. doi: 10.1074/jbc.M110.203240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li D., Yea S., Dolios G., Martignetti J.A., Narla G., Wang R. Regulation of Kruppel-like factor 6 tumor suppressor activity by acetylation. Cancer Res. 2005;65(20):9216–9225. doi: 10.1158/0008-5472.CAN-05-1040. [DOI] [PubMed] [Google Scholar]
- 40.Camolotto S.A., Racca A.C., Ridano M.E., Genti-Raimondi S., Panzetta-Dutari G.M. PSG gene expression is up-regulated by lysine acetylation involving histone and nonhistone proteins. PLoS ONE. 2013;8(2):e55992. doi: 10.1371/journal.pone.0055992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang C.Z., Zhang H.T., Chen G.G., Lai P.B. Trichostatin a sensitizes HBx-expressing liver cancer cells to etoposide treatment. Apoptosis. 2011;16(7):683–695. doi: 10.1007/s10495-011-0597-x. [DOI] [PubMed] [Google Scholar]
- 42.Mishra V.K., Wegwitz F., Kosinsky R.L., Sen M., Baumgartner R., Wulff T. Histone deacetylase class-I inhibition promotes epithelial gene expression in pancreatic cancer cells in a BRD4- and MYC-dependent manner. Nucleic Acids Res. 2017;45(11):6334–6349. doi: 10.1093/nar/gkx212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.