Summary
Oculocutaneous albinism (OCA) is a heterogeneous autosomal recessive genetic disorder that affects melanin synthesis. OCA results in reduced or absent pigmentation in the hair, skin and eyes. Type 1 OCA (OCA1) is the result of tyrosinase (TYR) gene mutations and is a severe disease type. This study investigated TYR mutations in a Chinese cohort with OCA1. This study included two parts: patient genetic study and prenatal genetic diagnosis. A total of 30 OCA1 patients were subjected to TYR gene mutation analysis. Ten pedigrees were included for prenatal genetic diagnosis. A total of 100 unrelated healthy Chinese individuals were genotyped for controls. The coding sequence and the intron/exon junctions of TYR were analysed by bidirectional DNA sequencing. In this study, 20 mutations were identified, four of which were novel. Of these 30 OCA1 patients, 25 patients were TYR compound heterozygous; two patients carried homozygous TYR mutations; and three were heterozygous. Among the ten prenatally genotyped fetuses, three fetuses carried compound heterozygous mutations and seven carried no mutation or only one mutant allele of TYR and appeared normal at birth. In conclusion, we identified four novel TYR mutations and showed that molecular-based prenatal screening to detect TYR mutations in a fetus at risk for OCA1 provided essential information for genetic counselling of couples at risk.
Introduction
Oculocutaneous albinism (OCA) affects melanin synthesis resulting in reduced or absent pigmentation in the hair, skin and eyes, with severe visual defects being a major effect of this disease (Ray et al., 2007; Montoliu et al., 2014). Although patients with OCA have similar phenotypes the disease actually is due to autosomal recessive mutations in different genes including TYR, OCA2, TRYP1 and SLC45A2. OCA1A is the most common and severe form of albinism in most populations (Oetting & King, 1999; King et al., 2003; Li et al., 2006 a; Wei et al., 2010; Chaki et al., 2011; Preising et al., 2011). One in every 40 000 individuals has OCA1 (type I) albinism (Ray et al., 2007), and OCA1 is the most common type of albinism in Japanese (Suzuki & Tomita, 2008), non-Hispanic Caucasians (Hutton & Spritz, 2008), Danes (Grønskov et al., 2009), a mixed population of Europeans, Asians and Africans (Rooryck et al., 2008), and Chinese (Wei et al., 2010). Other subtypes include OCA type II, which is caused by a mutation in the pink-eyed dilution gene (OCA2; MIM 203200) (Rinchik et al., 1993); OCA2 is the most common type in Africans and African–Americans (Durham-Pierre et al., 1994). OCA type III is caused by a mutation in the tyrosinase-related protein (TYRP1; MIM 203290) (Boissy et al., 1996), and OCA type IV is caused by a mutation in the membrane-associated transport protein (SLC45A2; MIM 606574) (Spritz, 1993; Fernandez et al., 2008; Hutton & Spritz, 2008), and is the only gene affected in OCA4 patients (Ko et al., 2012). Although OCA4 is rare worldwide, OCA4 is the second most common type in Japanese (Suzuki & Tomita, 2008).
TYR encodes a 58 kD (529 amino acids) bifunctional type-1 integral membrane protein that is required for melanin biosynthesis in the melanocytes of hair follicles, skin and eyes (Shibahara et al., 1988; Takeda et al., 1989; Hearing et al., 1992). The enzyme catalyses the conversion of tyrosine to l-dihydroxy-phenylalanine (DOPA) and subsequently oxidizes DOPA to dopaquinone in melanocytes (Hearing et al., 1992). The TYR protein also has 5,6-dihydroxyindole oxidase activity (Hearing et al., 1992). The tyrosinase activity is more stable in the presence of two other factors, tyrosinase-related protein (TRP)-1 and TRP-2 (Hearing et al., 1992). Human TYR is located at 11q14-q21 and contains five exons (Barton et al., 1988; Giebel et al., 1991).
OCA1 can be classified into two subtypes: OCA1A in which there is a complete lack of enzyme activity, and OCA1B in which there remains residual enzyme activity (Tomita et al., 1989; Ray et al., 2007; Hutton & Spritz, 2008; Montoliu et al., 2014). OCA1A is the most severe form of OCA1. OCA1B patients typically have little or no pigmentation at birth but progressive melanization can occur over time. The range of pigmentation can vary from little pigmentation to almost normal pigmentation, and the degree of pigmentation is dependent upon family pigment patterns and ethnicity (Oetting & King, 1999; King et al., 2003; Li et al., 2006 a; Ray et al., 2007; Chaki et al., 2011; Preising et al., 2011). The prevalence of OCA subtypes differs widely among different populations.
Over 273 mutations in TYR have been identified in different ethnic groups (Li et al., 2006 b; Simeonov et al., 2013). Although a distribution of TYR mutations has previously been described in Chinese patients (Wang et al., 2009; Wei et al., 2010), there is still a paucity of information regarding the types, frequency and distribution of mutations. In addition, diagnosis of OCA subtype of a patient based purely on clinical features is challenging. Further characterizing OCA1 mutations may help in the development of molecular tools that could be used for prenatal diagnosis of the disease. Prenatal screening for albinism by molecular analysis or high performance liquid chromatography and sequencing has been performed in Israeli families (Rosenmann et al., 2009) and Taiwanese families (Lin et al., 2006), respectively. There is a need for a comprehensive genetic analysis in a large sample to better characterize mutations in TYR that may cause OCA1 in mainland China. In this study, we characterized TYR mutations associated with OCA1 in 30 affected Chinese patients, and identified four novel mutations that had not previously been described.
Methods
Subjects and methods
This study was performed at the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China) and was approved by the hospital's ethics committee and performed according to the principles of the Declaration of Helsinki. All patients gave their written informed consent.
Study subjects
All subjects analysed in this study were recruited from the Genetic Counseling clinic of the Prenatal Diagnosis Center of the hospital and had albinism type I, type II or type IV. A clinical information form recorded the hair, skin and eye colour at birth, the age of sample submission and fundus examination by an ophthalmologist on the most affected family members. This study focused on OCA1 patients only. Inclusion criteria were symptoms of the eyes (severely poor eyesight, refractive error and photophobia) and skin (white or light yellow skin or hair that did not change with age). Normal pigmentation in a Chinese population is black hair, black eye colour and yellow skin colour. A total of 39 families were screened, including 35 patients with albinism, family members of three patients with albinism who had died and a person receiving genetic counselling. Patients were screened using DNA sequencing of the TYR, OCA2 and SCL45A2 genes. We did not assess patients for type III albinism as this is only found in black patients. We found 30 patients had type 1 albinism due to mutation in the TYR gene, three patients had type II albinism with mutations in the OCA2 gene and two patients had type IV albinism resulting from mutations in the SCL45A2 gene. The parents of the three deceased patients were heterozygous for TYR mutations. The couple receiving genetic counselling were heterozygous for TYR mutations. Only patients who carried the TYR mutation were included in the study. Patients with albinism but without TYR mutation were excluded. In addition, exclusion criteria were abnormalities in the immune system, symptoms in other organs (syndromic albinism) or solely ocular symptoms (ocular albinism). After birth, all surviving babies had a full OCA1 examination including eye examinations.
Collection of samples and genomic DNA extraction
Peripheral blood samples were collected for DNA analysis from study subjects. Genomic DNA was isolated using a TIANamp DNA Kit (Tiangen Biotech, Beijing, China) following the manufacturer's instructions.
PCR and DNA sequencing
All exons and exon/intron junctions of the TYR gene were amplified by PCR. Specific primers were designed according to the UCSC Genome Bioinformatics database (see Supplementary Table 1 for primer sequences; available online). PCR was performed using GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA). The reaction included 20–50 ng of genomic DNA, 1 μl of each primer and 13 μl 2 × Taq PCR MasterMix (containing a cocktail of dNTP, Tris-HCl, taq polymerase, KCl and MgCl2) and ddH2O to a final volume of 25 μl. The PCR amplification was as follows: 96 °C for 5 min, followed by 35 cycles of 96 °C for 30 s, then 58 to 64 °C (depending on the primer) for 40 s, followed by 72 °C for 1 min. Following the last cycle, the reaction products were further extended at 72 °C for 7 min. PCR products were visualized by electrophoresing in 1·5% agarose gel and ethidium bromide staining. Recovered PCR amplicons (50 ng) were bidirectionally sequenced using an ABI PRISM 3130 Genetic Analyzer (Applied Biosystems, Carslbad, CA, USA).
Novel mutation naming and verification
Novel mutations were identified using PubMed and OCA databases (http://www.hgmd.cf.ac.uk/ac/; http://www.ncbi.nlm.nih.gov/SNP/; and http://www.ifpcs.org/albinism/index.html). To confirm that the mutation was new and not a single nucleotide polymorphism, the relevant region of the gene was sequenced using DNA isolated from 100 unrelated healthy individuals and was analysed with PROVEN and Polyphen-2. Novel mutations were named according to Mutalyzer (https://mutalyzer.nl/check)
Prenatal testing
Fetal tissue was obtained from transabdominal chorionic villi isolated during the first trimester or amniocentesis performed during the second trimester. Fetal DNA was also isolated using the TIANamp DNA Kit. Maternal contamination was ruled out from fetal samples and paternity was confirmed using PowerPlex 16 HS System Kit (Promega Corporation, Madison, WI, USA) and GeneMapper ID v3·2 Soft.
Results
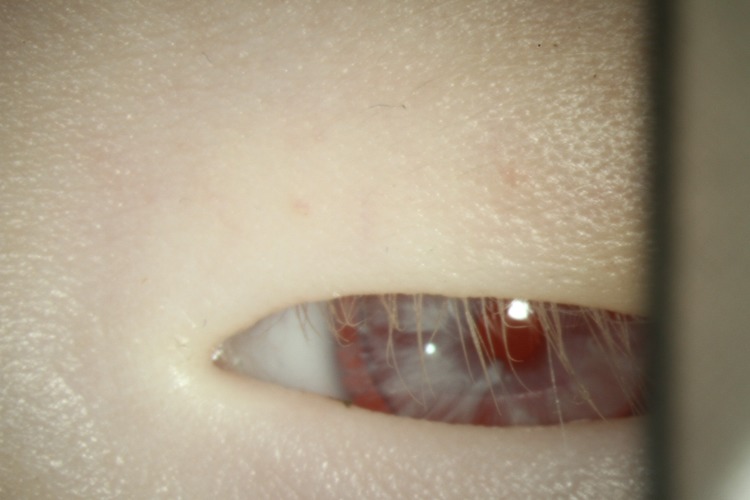
A total of 98 subjects from 34 families were subject to TYR gene mutation analysis (Table 1). Among them, subjects 1–30 were living OCA1 patients and these probands plus both parents were genotyped (n = 90). Subjects 31, 33 and 34 were deceased albinism probands and their genotypes were deduced from the genotypes of their asymptomatic parents (n = 6); the parents each carried heterozygous TYR mutations. Subject 32 was the fetus from a single couple who had poor development of optic disc, displayed pathological photophobia and had normal eyesight without nystagmus; this couple were identified as asymptomatic carriers of OCA1 mutations after genotyping. The parents of OCA1 subjects were asymptomatic and not in consanguineous marriages. A total of 100 unrelated healthy Chinese individuals were genotyped as controls. Most subjects had white hair and skin colour, and were positive for nystagmus (Table 1). Interestingly, Chinese children with typical oculocutaneous albinism usually presented with red irises, rarely blue irises, at birth (Fig. 1).
Table 1.
Clinical characteristics and identified mutations in 34 OCA1 subjects
| Subject genotype | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Subject | Sex | Age (years) | Hair colour at birth | Hair colour at analysis | Skin colour | Iris colour | Nystagmus | OCA1 subtype | Paternal allele | Maternal allele |
| 1 | M | 0·5 | White | White | White | Red–brown | Positive | 1A | c.929_930insC | p.R299H |
| 2 | M | 7 | White | White | White | Red–brown | Positive | 1A | c.929_930insC | — |
| 3 | F | 0·06 | White | White | White | Amber | Positive | 1A | p.R299C | p.R299C |
| 4 | M | 23 | White | Yellow | White | Grey | Negative | 1B | p.R299H | p.R116X |
| 5 | M | 2·5 | White | White | White | Red–brown | Positive | 1A | p.R77Q | p.R278X |
| 6 | F | 6 | White | White | White | Red–brown | Positive | 1A | c.232_233insGGG | p.W400L |
| 7 | F | 22 | White | White | White | Red–brown | Positive | 1A | c.929_930insC | p.R116X |
| 8 | M | 1 | White | Yellow | White | Black | Positive | 1B | IVS2-10delTT-7 T > A | p.E219 K |
| 9 | F | 50 | Unknown | Brown | White | Grey | Negative | 1B | c.232_233insGGG | IVS2-10delTT-7 T > A |
| 10 | F | 0·07 | White | White | White | Red–brown | Positive | 1A | c.560_561ins25 bpa | c.929_930insC |
| 11 | M | 2 | White | White | White | Red–brown | Positive | 1A | IVS2-10delTT-7 T > A | c.929_930insC |
| 12 | F | 0·83 | White | White | White | Red–brown | Positive | 1A | c.232_233insGGG | p.M1 V |
| 13 | F | 13 | White | White | White | Red–brown | Positive | 1A | p.E294 K | p.E294 K |
| 14 | M | 24 | White | White | White | Red–brown | Positive | 1A | p.R299H | — |
| 15 | M | 5·5 | White | Brown | White | Grey | Positive | 1B | p.R77Q | p.R299H |
| 16 | F | 0·58 | White | White | White | Red–brown | Positive | 1A | p.G446Va | p.R299H |
| 17 | F | 32 | White | White | White | Red–brown | Positive | 1A | p.G253E | p.R299H |
| 18 | M | 30 | White | White | White | Red–brown | Positive | 1A | c.929_930insC | p.R299H |
| 19 | F | 0·02 | White | White | White | Red–brown | Positive | 1A | p.W400L | p.R77Q |
| 20 | M | 48 | White | White | White | Red–brown | Positive | 1A | p.R116X | IVS4 + 1delGa |
| 21 | F | 0·17 | White | White | White | Red–brown | Positive | 1A | IVS3 + 2 T > Ca | p.R116X |
| 22 | F | 5 | White | White | White | Red–brown | Positive | 1A | p.R278X | p.R299H |
| 23 | F | 4 | White | White | White | Red–brown | Positive | 1A | p.C24Y | c.929_930insC |
| 24 | F | 7 | White | Yellow | White | Black | Positive | 1B | c.232_233insGGG | IVS2-10delTT-7 T > A |
| 25 | F | 21 | White | White | White | Red–brown | Positive | 1A | — | p.R299H |
| 26 | M | 0·25 | White | White | White | Red–brown | Negative | 1A | c.232_233insGGG | IVS2-10delTT-7 T > A |
| 27 | F | 3 | White | White | White | Red–brown | Positive | 1A | p.K142M | p.R278X |
| 28 | M | 1·4 | White | White | White | Red–brown | Positive | 1A | p.W400L | p.G253E |
| 29 | M | 1·2 | White | White | White | Red–brown | Positive | 1A | p.R299H | p.R116X |
| 30 | F | 7 | White | White | White | Red–brown | Positive | 1A | p.R299H | p.Q399X |
| 31 | F | / | White | White | White | Blue | / | / | p.R299H | c.232_233insGGG |
| 32 | p.R299H | p.R278X | ||||||||
| 33 | M | / | White | White | White | Blue | / | / | p.R278X | p.R116X |
| 34 | M | / | White | White | White | Red–brown | Positive | / | p.W400L | p.R116X |
Previously unknown alleles.
A dash (—) in the genotype column denotes a putative uncharacterized allelic mutation that may not be in another portion of TYR gene and was not identify by the PCR primers used.
, no correlated information; F, female; M, male.
Fig. 1.
Image of iris of Chinese child aged 1 year and 9 months showing the red iris often seen in Chinese children with typical oculocutaneous albinism.
TYR gene mutation analysis
DNA analysis revealed 20 different TYR alleles among which 16 had previously been reported (Tables 1 and 2). The four novel alleles that were not present in healthy controls were an insertion in the coding region (c.560_561ins25 bp), a nucleotide change in the second nucleotide of the third intron (IVS3 + 2 T > C), a deletion in the first nucleotide of the fourth intervening sequence (IVS4 + 1delG) and a non-conservative missense mutation (p.G446 V).
Table 2.
Mutation frequency of TYR gene in Chinese OCA1 patients
| Mutation type | Allele | Location | Mutation frequency |
|---|---|---|---|
| Missense | p.M1 V | Exon 1 | 1/68 |
| p.C24Y | Exon 1 | 1/68 | |
| p.R77Q | Exon 1 | 3/68 | |
| p.K142M | Exon 1 | 1/68 | |
| p.E219 K | Exon 1 | 1/68 | |
| p.G253E | Exon 1 | 2/68 | |
| p.E294 K | Exon 2 | 1/68 | |
| p.R299H | Exon 2 | 12/68 | |
| p.R299C | Exon 2 | 1/68 | |
| p.Q399X | Exon 4 | 1/68 | |
| p.W400L | Exon 4 | 4/68 | |
| p.G446 V | Exon 4 | 1/68 | |
| Nonsense | p.R116X | Exon 1 | 7/68 |
| p.R278X | Exon 2 | 6/68 | |
| Insertion | c.232_233insGGG | Exon 1 | 7/68 |
| c.560_561ins25 bp | Exon 1 | 1/68 | |
| c.929_930insC | Exon 2 | 6/68 | |
| Splice | IVS2-10delTT-7 T > A | Intron 2 | 5/68 |
| IVS3 + 2 T > C | Intron 3 | 1/68 | |
| Deletion | IVS4 + 1delG | Intron 4 | 1/68 |
Seven TYR alleles accounted for 74·6% of the alleles with mutations detected in this study. The most common allele was p.R299H (12/68 mutant alleles analysed) (exon 2), followed by c.232_233insGGG (7/68) (exon 1), p.R116X (7/68) (exon 1), p.R278X (6/68) (exon 2), c.929_930insC (6/68) (exon 2), IVS2-10delTT-7 T > A (5/68) (intron 2) and p.W400L (4/68) (exon 4) (Table 2). All other identified alleles occurred in only one or two of the subjects.
A total of 29 OCA1 subjects were compound heterozygous for mutant alleles of the TYR gene. Two subjects were homozygous (Subject 3 and 13), and three subjects were single heterozygous for mutant TYR alleles((Subjects 2, 14 and 25) (Table 1).
The novel missense mutation p.G446 V changes a glycine at the copper-binding site of tyrosinase, to valine, which likely impacts copper binding and enzyme activity. We tested for pathogenicity of this mutation using PROVEN and Polyphen-2, which indicated the mutation likely affects the structure and function of the protein.
The mutation c.560_561ins25 bp, was a frameshift mutation that caused an abnormality in the amino acid sequence downstream of the mutation (Leu-Leu-Cys-Val-Lys-Leu-Ser-Pro-Thr-Ala-Trp-Gly-Ile-STOP).
Two novel mutations, IVS3 + 2 T > C and IVS4 + 1delG were located in the recognition sequence of the donor splice site of the third and fourth introns, respectively. These nucleotide changes are expected to alter the RNA splicing pattern and frequency, mRNA sequence, levels of mRNA and, subsequently, protein levels and function.
Prenatal genetic diagnoses
Parental genotypes for ten high-risk fetuses suggested that these fetuses had a 25% risk of carrying two mutant alleles of TYR. Prenatal DNA analysis of high-risk fetuses (n = 10) identified three fetuses (Subjects 12, 32 and 34) that carried compound heterozygous alleles (Table 3). Following genetic counselling, the parents of these fetuses chose to terminate the pregnancy. Analysis of the aborted tissue confirmed that these fetuses carried two mutant TYR alleles. Five fetuses (Subjects 5, 6, 8, 11 and 33) were heterozygous carrying one mutant allele and two fetuses (Subjects 30 and 31) did not carry any detectable mutant allele (Table 3). After birth, none of these seven subjects showed symptoms of OCA1. DNA analysis supported the paternity of all fetuses analysed.
Table 3.
Prenatal genetic diagnosis of the ten high-risk fetuses
| Proband genotype | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pedigree | Sex | Age years) | Hair colour (at birth/at analysis) | Skin colour | Iris colour | Nystagmus | OCA1 subtype | Paternal allele | Maternal allele | Fetus genotype | Fetus outcome | Baby phenotype |
| 5 | M | 2·5 | White/white | White | Red–brown | Positive | 1A | p.R278X | p.R77Q | p.R278X | Normal birth | Normal |
| 6 | F | 6 | White/white | White | Red–brown | Positive | 1A | c.232_233insGGG | p.W400L | p.W400L | Normal birth | Normal |
| 8 | M | 1 | White/yellow | White | Black | Positive | 1B | IVS2-10delTT-7 T > A | p.E219 K | p.E219 K | Normal birth | Normal |
| 11 | M | 2 | White/white | White | Red–brown | Positive | 1A | IVS2-10delTT-7 T > A | c.929_930insC | IVS2-10delTT-7 T > A | Normal birth | Normal |
| 12 | F | 0·83 | White/white | White | Red–brown | Positive | 1A | c.232_233insGGG | p.M1 V | p.M1 V | Odinopoeia | / |
| c.232_233insGGG | ||||||||||||
| 30 | F | 7 | White/white | White | Red–brown | Positive | 1A | p.R299H | p.Q399X | No mutation | Normal birth | Normal |
| 31 | F | / | White/white | White | Red–brown | / | / | Deceased | No mutation | Normal birth | Normal | |
| 32 | / | / | / | / | / | / | / | No proband | p.R278X | Odinopoeia | / | |
| p.R299H | ||||||||||||
| 33 | M | / | White/white | White | Red–brown | / | / | Deceased | p.R116X | Normal birth | Normal | |
| 34 | M | / | White/White | White | Red–brown | Positive | / | Deceased | p.R116X | Odinopoeia | / | |
| p.W400L | ||||||||||||
, no correlated information; F, female; M, male.
Discussion
In this study, we assayed TYR for mutations in 30 OCA1 individuals and ten fetuses at risk for the disease. We identified four novel mutations. Two of the novel mutations (p.G446 V and c.560_561ins25 bp) altered the coding sequence and the other two (IVS4 + 1delG and IVS3 + 2 T > C) affected donor splice site sequences. All four mutations were associated with the OCA1A clinical diagnosis, suggesting they are likely null or strong loss-of-function alleles and that the companion mutation on the other chromosome also produced little active enzyme. It is possible that IVS4 + 1delG and IVS3 + 2 T > C disrupt gene function by altering splicing to produce a non-functional protein product or an unstable mRNA. Further studies are required to elucidate how these mutations alter gene function and to determine if these four novel mutations are specific to Han Chinese or are present in other populations.
The most common mutation in our study was p.R299H; 12/68 analysed chromosomes carried this mutation. Codon 299 is highly conserved and is located close to the two copper binding sites. Disrupting copper binding may inhibit enzyme activity. Prior works also found mutations at position 299 (R299H, R299S and R299C) accounted for almost 60% of missense mutant alleles in Chinese OCA1 patients and represent almost 35% of all OCA1 mutations in Chinese (Tsai et al., 1999; Hsieh et al., 2001; Lin et al., 2006). By contrast, mutations at 299 account for <1·0% in white populations (Spritz et al., 1997; Opitz et al., 2004) possibly indicating the difference in founder mutations between populations. The p.W400L, c.232_233insGGG and p.C24Y mutations have only been reported in Chinese OCA patients to date (Wang et al., 2009; Wei et al., 2010). The c929_930insC mutation appears to be common in east Asian populations as it is prevalent in Chinese, Japanese and Korean OCA1 patients (Goto et al., 2004; Suzuki & Tomita, 2008; Wang et al., 2009; Wei et al., 2010; Ko et al., 2012; Park et al., 2012). The other alleles we found at a high frequency (i.e. pR116X, p.R278X and IVS-10delTT-7 T > A) have also been shown to be common in Chinese in previous studies (Wei et al., 2010). On the other hand, p.R278X is the third most common mutation in Japanese and Chinese patients (11·6 and 11·8%, respectively) (Ko et al., 2012).
In these 34 OCA1 patients, 29 patients were confirmed to carry compound heterozygous mutations in the two TYR alleles and two were homozygous for a TYR mutation. For the other three of the patients, only one mutation-carrying allele was detected although we sequenced all five exons and all intron/exon boundaries of the other allele. This may indicate that another mutation(s) outside the region we sequenced, such as in the regulatory regions (both upstream and downstream) or in other intronic sequences, are present that may alter or regulate splicing efficiency and/or accuracy. It is also possible that a mutation in other genes either directly or indirectly affect TYR gene expression or activity. A prior study that genetically analysed OCA1 in Caucasian patients with albinism found 26% of patients with OCA1 did not have two mutations in TYR (Simeonov et al., 2013). They further analysed the TYR gene as well as other genes involved in OCA and found possible reasons for this phenotype in two cases: in one case the patient carried a sequence variant in the SLC24A5 gene and in another case the patient was hemizygote for OCA1 (Simeonov et al., 2013). SLC24A5 protein is required for proper routing of tyrosinase and mutations in this gene result in a similar phenotype as OCA2 mutations. Further studies are required to identify additional mutations within this Han Chinese population and to elucidate the mechanism resulting in the OCA1 phenotype.
We genotyped ten high-risk fetuses for mutations in TYR, and found three fetuses carried mutations on both alleles and were highly likely to have the disease. Five fetuses carried only one mutant allele and the other two carried no TYR mutations, which is similar frequencies to a previous report (Rosenmann et al., 2009). The parents of the three affected fetuses chose to terminate the pregnancy. After birth, the seven other babies showed no signs of OCA1. These data confirm that molecular-based prenatal screening is possible for this disease.
It would be of interest to understand how the different TYR mutations affect gene expression and function. This information may give insight into disease severity and possibly facilitate the development of treatment for the disease. In conclusion, we identified four novel TYR mutations and 16 known TYR mutations in a Chinese OCA1 population. This molecular-based prenatal screening to detect TYR mutations in a fetus at risk for OCA1 provided essential information for genetic counselling of “at risk” couples.
Acknowledgments
Supported by Youth Innovation Fund of First Affiliated Hospital of Zhengzhou University. We are grateful to the albino patients and their family members, the volunteers for donating bloodsamples, and all participants in the study for their kind cooperation.
Declaration of interest
None.
Supplementary material
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0016672314000160.
click here to view supplementary material
References
- Barton D. E., Kwon B. S. & Francke U. (1988). Human tyrosinase gene, mapped to chromosome 11 (q14––q21), defines second region of homology with mouse chromosome 7. Genomics 3, 17–24. [DOI] [PubMed] [Google Scholar]
- Boissy R. E., Zhao H., Oetting W. S., Austin L. M., Wildenberg S. C., Boissy Y. L., Zhao Y., Sturm R. A., Hearing V. J., King R. A. & Nordlund J. J. (1996). Mutation in and lack of expression of tyrosinase-related protein-1 (TRP-1) in melanocytes from an individual with brown oculocutaneous albinism: a new subtype of albinism classified as “OCA3”. American Journal of Human Genetics 58, 1145–1156. [PMC free article] [PubMed] [Google Scholar]
- Chaki M., Sengupta M., Mondal M., Bhattacharya A., Mallick S., Bhadra R., Indian Genome Variation Consortium & Ray K. (2011). Molecular and functional studies of tyrosinase variants among Indian oculocutaneous albinism type 1 patients. The Journal of Investigative Dermatology 131, 260–262. [DOI] [PubMed] [Google Scholar]
- Durham-Pierre D., Gardner J. M., Nakatsu Y., King R. A., Francke U., Ching A., Aquaron R., del Marmol V. & Brilliant M. H. (1994). African origin of an intragenic deletion of the human P gene in tyrosinase positive oculocutaneous albinism. Nature Genetics 7, 176–179. [DOI] [PubMed] [Google Scholar]
- Fernandez L. P., Milne R. L., Pita G., Aviles J. A., Lazaro P., Benitez J. & Ribas G. (2008). SLC45A2: a novel malignant melanoma-associated gene. Human Mutation 29, 1161–1167. [DOI] [PubMed] [Google Scholar]
- Giebel L. B., Strunk K. M. & Spritz R. A. (1991). Organization and nucleotide sequences of the human tyrosinase gene and a truncated tyrosinase-related segment. Genomics 9, 435–445. [DOI] [PubMed] [Google Scholar]
- Goto M., Sato-Matsumura K. C., Sawamura D., Yokota K., Nakamura H. & Shimizu H. (2004). Tyrosinase gene analysis in Japanese patients with oculocutaneous albinism. Journal of Dermatological Science 35, 215–220. [DOI] [PubMed] [Google Scholar]
- Grønskov K., Ek J., Sand A., Scheller R., Bygum A., Brixen K., Brondum-Nielsen K. & Rosenberg T. (2009). Birth prevalence and mutation spectrum in Danish patients with autosomal recessive albinism. Investigative Ophthalmology & Visual Science 50, 1058–1064. [DOI] [PubMed] [Google Scholar]
- Hearing V. J., Tsukamoto K., Urabe K., Kameyama K., Montague P. M. & Jackson I. J. (1992). Functional properties of cloned melanogenic proteins. Pigment Cell Research 5, 264–270. [DOI] [PubMed] [Google Scholar]
- Hsieh Y. Y., Wu J. Y., Chang C. C., Tsai F. J., Lee C. C., Tsai H. D., & Tsai C. H. (2001). Prenatal diagnosis of oculocutaneous albinism two mutations located at the same allele. Prenatal Diagnosis 21, 200–201. [PubMed] [Google Scholar]
- Hutton S. M. & Spritz R. A. (2008). A comprehensive genetic study of autosomal recessive ocular albinism in Caucasian patients. Investigative Ophthalmology & Visual Science 49, 868–872. [DOI] [PubMed] [Google Scholar]
- King R. A., Pietsch J., Fryer J. P., Savage S., Brott M. J., Russell-Eggitt I., Summers C. G. & Oetting W. S. (2003). Tyrosinase gene mutations in oculocutaneous albinism 1 (OCA1): definition of the phenotype. Human Genetics 113, 502–513. [DOI] [PubMed] [Google Scholar]
- Ko J. M., Yang J. A., Jeong S. Y. & Kim H. J. (2012). Mutation spectrum of the TYR and SLC45A2 genes in patients with oculocutaneous albinism. Molecular Medicine Reports 5, 943–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Y., Wu W. I., Zheng H., Duan H. L., Chen Z. & Chen L. M. (2006. a). [Prenatal gene diagnosis of oculocutaneous albinism type I]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 23, 280–282. [PubMed] [Google Scholar]
- Li W., He M., Zhou H., Bourne J. W. & Liang P. (2006. b). Mutational data integration in gene-oriented files of the Hermansky-Pudlak Syndrome database. Human Mutation 27, 402–407. [DOI] [PubMed] [Google Scholar]
- Lin S. Y., Chien S. C., Su Y. N., Lee C. N. & Chen C. P. (2006). Rapid genetic analysis of oculocutaneous albinism (OCA1) using denaturing high performance liquid chromatography (DHPLC) system. Prenatal Diagnosis 26, 466–470. [DOI] [PubMed] [Google Scholar]
- Montoliu L., Grønskov K., Wei A. H., Martinez-Garcia M., Fernandez A., Arveiler B., Morice-Picard F., Riazuddin S., Suzuki T., Ahmed Z. M., Rosenberg T. & Li W. (2014). Increasing the complexity: new genes and new types of albinism. Pigment Cell & Melanoma Research 27, 11–18. [DOI] [PubMed] [Google Scholar]
- Oetting W. S. & King R. A. (1999). Molecular basis of albinism: mutations and polymorphisms of pigmentation genes associated with albinism. Human Mutation 13, 99–115. [DOI] [PubMed] [Google Scholar]
- Opitz S., Kasmann-Kellner B., Kaufmann M., Schwinger E. & Zuhlke C. (2004). Detection of 53 novel DNA variations within the tyrosinase gene and accumulation of mutations in 17 patients with albinism. Human Mutation 23, 630–631. [DOI] [PubMed] [Google Scholar]
- Park S. H., Chae H., Kim Y. & Kim M. (2012). Molecular analysis of Korean patients with oculocutaneous albinism. Japanese Journal of Ophthalmology 56, 98–103. [DOI] [PubMed] [Google Scholar]
- Preising M. N., Forster H., Gonser M. & Lorenz B. (2011). Screening of TYR, OCA2, GPR143, and MC1R in patients with congenital nystagmus, macular hypoplasia, and fundus hypopigmentation indicating albinism. Molecular Vision 17, 939–948. [PMC free article] [PubMed] [Google Scholar]
- Ray K., Chaki M. & Sengupta M. (2007). Tyrosinase and ocular diseases: some novel thoughts on the molecular basis of oculocutaneous albinism type 1. Progress in Retinal and Eye Research 26, 323–358. [DOI] [PubMed] [Google Scholar]
- Rinchik E. M., Bultman S. J., Horsthemke B., Lee S. T., Strunk K. M., Spritz R. A., Avidano K. M., Jong M. T. & Nicholls R. D. (1993). A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature 361, 72–76. [DOI] [PubMed] [Google Scholar]
- Rooryck C., Morice-Picard F., Elcioglu N. H., Lacombe D., Taieb A. & Arveiler B. (2008). Molecular diagnosis of oculocutaneous albinism: new mutations in the OCA1–4 genes and practical aspects. Pigment Cell & Melanoma Research 21, 583–587. [DOI] [PubMed] [Google Scholar]
- Rosenmann A., Bejarano-Achache I., Eli D., Maftsir G., Mizrahi-Meissonnier L. & Blumenfeld A. (2009). Prenatal molecular diagnosis of oculocutaneous albinism (OCA) in a large cohort of Israeli families. Prenatal Diagnosis 29, 939–946. [DOI] [PubMed] [Google Scholar]
- Shibahara S., Tomita Y., Tagami H., Muller R. M. & Cohen T. (1988). Molecular basis for the heterogeneity of human tyrosinase. The Tohoku Journal of Experimental Medicine 156, 403–414. [DOI] [PubMed] [Google Scholar]
- Simeonov D. R., Wang X., Wang C., Sergeev Y., Dolinska M., Bower M., Fischer R., Winer D., Dubrovsky G., Balog J. Z., Huizing M., Hart R., Zein W. M., Gahl W. A., Brooks B. P. & Adams D. R. (2013). DNA variations in oculocutaneous albinism: an updated mutation list and current outstanding issues in molecular diagnostics. Human Mutation 34, 827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spritz R. A. (1993). Molecular genetics of oculocutaneous albinism. Seminars in Dermatology 12, 167–172. [PubMed] [Google Scholar]
- Spritz R. A., Oh J., Fukai K., Holmes S. A., Ho L., Chitayat D., France T. D., Musarella M. A., Orlow S. J., Schnur R. E., Weleber R. G. & Levin A. V. (1997). Novel mutations of the tyrosinase (TYR) gene in type I oculocutaneous albinism (OCA1). Human Mutation 10, 171–174. [DOI] [PubMed] [Google Scholar]
- Suzuki T. & Tomita Y. (2008). Recent advances in genetic analyses of oculocutaneous albinism types 2 and 4. Journal of Dermatological Science 51, 1–9. [DOI] [PubMed] [Google Scholar]
- Takeda A., Tomita Y., Okinaga S., Tagami H. & Shibahara S. (1989). Functional analysis of the cDNA encoding human tyrosinase precursor. Biochemical and Biophysical Research Communications 162, 984–990. [DOI] [PubMed] [Google Scholar]
- Tomita Y., Takeda A., Okinaga S., Tagami H. & Shibahara S. (1989). Human oculocutaneous albinism caused by single base insertion in the tyrosinase gene. Biochemical and Biophysical Research Communications 164, 990–996. [DOI] [PubMed] [Google Scholar]
- Tsai C. H., Tsai F. J., Wu J. Y., Lin S. P., Chang J. G., Yang C. F., Lee C. C. (1999). Insertion/deletion mutations of type I oculocutaneous albinism in chinese patients from Taiwan. Human Mutation 14, 542. [DOI] [PubMed] [Google Scholar]
- Wang Y., Guo X., Li W. & Lian S. (2009). Four novel mutations of TYR gene in Chinese OCA1 patients. Journal of Dermatological Science 53, 80–81. [DOI] [PubMed] [Google Scholar]
- Wei A., Wang Y., Long Y., Guo X., Zhou Z., Zhu W., Zhu W., Liu J., Bian X., Lian S. & Li W. (2010). A comprehensive analysis reveals mutational spectra and common alleles in Chinese patients with oculocutaneous albinism. The Journal of Investigative Dermatology 130, 716–724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0016672314000160.
click here to view supplementary material