Abstract
Background:
FOLFOX treatment is a method used widely to reduce tumor size in low-set rectal cancer, with variable clinical results. FOLFOX agents comprise a mixture of oxaliplatin and 5-fluorouracil, the efficacy of which might be modulated by the gut microbiome in humans. This study aimed to determine whether the bowel microbiota is a factor that influences FOLFOX treatment.
Methods:
To investigate the role of gut microbiota during FOLFOX treatment, we carried out comprehensive metagenomic and metabolomic analyses on 62 fecal samples collected from 37 low-set rectal cancer patients. A set of 31 samples was collected before the patients underwent treatment; another 31 samples were obtained after the treatment was completed. Among these samples, 50 were paired samples collected before and after FOLFOX treatment. The patients were divided into responder and nonresponder groups according to the treatment outcome. Metagenomic sequencing was performed on these fecal samples. Diverse bacterial taxa were identified by MetaGeneMark, Soapaligner, and DIAMOND; microbiotal data analyses were carried out in the R environment. Differences in microbial taxa and metagenomic linkage groups were observed in multiple comparative analyses.
Results:
The gut microbiota was altered after treatment. Compared with before treatment, the changes in bacterial diversity and microbiotal composition after treatment were more apparent in the responder group than in the nonresponder group. Bacterial species analysis revealed a group of gut bacteria in multiple comparisons, with a group of eight specific species being associated with the outcome of FOLFOX treatment. Responders and nonresponders before treatment were clearly separated based on this bacterial subset. Finally, the metagenomic linkage group network and metabolomic analyses based on the genomic data confirmed a more significant change in the gut microbiota during FOLFOX treatment in the responder group than in the nonresponder group.
Conclusions:
Overall, our results describe a dynamic process of gut microbiotal changes from the start to the end of FOLFOX treatment, and verified a close relationship between microbiota and treatment outcome. Recognition of the significance of microbiotal intervention before FOLFOX treatment for low-set rectal cancer may improve the effects of these agents.
Keywords: chemotherapy, FOLFOX treatment, gut microbiota, low-set rectal cancer, metagenomic sequencing analysis
Introduction
Colorectal cancer (CRC) is one of the most prevalent malignant tumors worldwide. During the initiation and progression of CRC, the gut microbiota affects the host bowel environment to interfere with tumor growth, and engages in cross-talk with the host immune system.1–3 Fusobacterium nucleatum is a carcinogenic bacterium that exhibits increasing abundance in the polyp-to-tumor transformation.4,5 Another group has recently shown that a combination of F. nucleatum, Bacteroides clarus, Roseburia intestinalis, Clostridium hathewayi, and one undefined species (labeled as m7) might serve as a novel noninvasive diagnostic biomarker for CRC.6
The most effective strategy for CRC treatment, which is widely approved for both clinical and general settings, is surgical excision and chemotherapy before metastasis. However, for low-set rectal cancer, which is located in the lower part of the bowel at 6 cm (2 inches) from the anus,7 the likely consequence of surgical treatment is problem with bowel function over the remaining lifespan of the patient. The clinical use of FOLFOX, a type of neoadjuvant chemotherapy (NC) recommended by the National Comprehensive Cancer Network (NCCN) Guidelines (2018),8 has been beneficial for these patients, who otherwise might have to undergo anus removal. In contrast to adjuvant chemotherapy, FOLFOX involves drug treatment before surgical excision of a tumor.
In clinical settings, the FOLFOX agents used for rectal cancer are oxaliplatin (Oxp) and 5-fluorouracil (5-FU).8 The efficacy of these two agents has been confirmed to be modulated by the gut microbiome.9,10 Oxp, which is a platinum-based chemical therapy, is widely used to treat several malignancies in clinical settings. Studies have verified that the anticarcinogenic effects of Oxp rely on gram-negative (G−) bacteria in the host intestinal tract. Lipopolysaccharide (LPS), the main component of G− bacterial cell walls, induces myeloid cells to produce high levels of reactive oxygen species (ROS). Under oxidative stress, the DNA damage caused by Oxp is enhanced in the tumor, triggering cancer cell death.9 5-FU, the other component of FOLFOX, has been defined as a first-line drug for NC, and has been used extensively in CRC.11,12 As an analogue of uracil, 5-FU and its prodrugs block nucleotide biosynthesis and induce cell division, in addition to being converted to thymidine-5’-monophosphoric acid by thymidylate synthase.13 Based on clinical feedback and previous studies, there is a clear difference in drug potency among patients.14–16 The observed variation in 5-FU efficacy may be due to the different genetic backgrounds of each patient,17 though it is difficult to explain the widespread 5-FU tolerance among different races.18 Such variation suggests that the bowel microflora environment is a key factor in the action of NC.
To better understand the role of the microbiome and its interaction with chemical therapy, we collected 62 microbiome samples from 37 low-set rectal cancer patients undergoing NC treatment, including 21 responders with 35 stool samples, and 16 nonresponders with 27 stool samples.19 In each group, patient characteristics were monitored during the treatment process, and stool samples were collected both before and after treatment. We performed deep metagenomics sequencing of all stool samples, and metabolomic analyses of their metabolic profiles; we further built a classification system for the responders and nonresponders based on gut microbiota and metabolites. Our results provide evidence that chemical therapy may affect the composition of the gut microbiota. Additionally, the gut microbiota might be a factor used to improve the effects of FOLFOX agents before treatment for low-set rectal cancer.
Material and methods
Sample collection and DNA preparation
We recruited, from the China-Japan Friendship Hospital, 37 low-set rectal cancer patients without tumor metastases who were diagnosed on the basis of clinical criteria according to a previous study (Supplementary Table S1).7 The participants, who were not treated with any antineoplastic agent or antibiotic treatment before NC, all underwent FOLFOX therapy. The dose was based on body surface area. After treatment, the patients were divided into responder and nonresponder groups according to NCCN criteria.
In total, 62 fecal samples, including 50 paired samples collected before and after FOLFOX treatment (Table 1), were available for metagenomic analysis. Based on the group information, the fecal samples, which were collected before therapy, were classified into subgroups labeled responders before treatment (RB, n = 16) and nonresponders before treatment (NRB, n = 19). Correspondingly, the samples collected at the end of FOLFOX treatment were labelled responders after treatment (RA, n = 15) and nonresponders after treatment (NRA, n = 12). This study was approved by local ethics committees (Institute of Microbiology, Chinese Academy of Sciences, IRB No. APIMCAS2016003), and informed consent was obtained from all participants.
Table 1.
Sample collection table.
The number presents the number of samples.
The number in brackets represent the number of paired samples.
The number after the slash represents the number of patients.
Stool samples were collected from each patient at the hospital before colonoscopy, and immediately frozen at −80°C until analysis. Total DNA extraction was performed according to the manufacturer’s guidelines for bacterial DNA enrichment, with minor modifications, using QIAamp® FAST DNA Stool Mini Kit (cat. No.51604, Qiagen, Hilden, Germany). To improve the efficiency of gram-positive bacteria cell disruption, fecal samples were disrupted in InhibitEX Buffer, and homogenized with 100 mg of zirconium beads (0.1 mm) using a Mini-Beadbeater-1 (Biospec Products Inc., Bartlesville, OK, USA) at a rate of 4800 rpm/min four times for 30 s at room temperature (15–25°C), with a 10 s interval each time. The mixture was heated at 95°C for 5 min in a metal bath to further increase the amount of total bacterial DNA extracted. Ensuing steps were carried out according to the manufacturer’s recommendations.
Metagenomic sequencing and gene catalogue construction
The Illumina HiSeq platform (insert size 250–450 bp, read length 150 bp) was used for the analysis of all fecal samples. Raw reads were quality controlled to remove certain sequences, including bases with low quality, with more than 10 bp N or with more than a 15 bp overlap with the adapter, and were aligned to the human genome (alignment with SOAP2, parameters: identity ⩾90%, −l 30, −v 10, −M 4, −m 200, −x 400). The remaining clean data (Supplementary Table S2) were used to assemble scaffolds with SOAP denovo (Version 2.04, parameters: −d 1, −M 3, −R, −u, −F). For each sample, the scaffolds were cut off at N bases. Fragments without N bases were called scaftigs. The clean data agonist scaftigs were mapped using SOAP2 (parameters: −u, −2, −m 200), and we obtained the unused reads of each sample. All unused reads were assembled as scaftigs using the same method and parameters as mentioned above.
Scaftigs longer than 500 bp were used to predict open reading frames (ORFs) in MetaGeneMark (prokaryotic GeneMark.hmm version 2.10), and scaftigs with a length of less than 100 nt were deleted. After redundancies were removed by CD-HIT, a primitive gene catalogue was constructed (parameters: identity ⩾95%, converge ⩾90%, −c 0.95, −G 0, −aS 0.9, −g 1, −d 0). Finally, only genes with at least two mapped reads were used to calculate abundance using SoapAligner (parameters: identity ⩾95%, −m 200, −x 400).
Generating gene profiles, constructing metagenomic linkage groups (MLGs), creating a Kyoto Encyclopedia of Genes and Genomes (KEGG) ortholog, module, and pathway, were all accomplished according to previously published methods.20 Reads were quality controlled and assembled in the same way as the scaftigs used to predict ORFs. Finally, each ORFs was aligned to the nonresponder database of NCBI and distinguished taxonomic groups.
Raw Illumina read data for all samples were deposited in the National Center for Biotechnology Information GenBank Sequence Read Archive under BioProject accession PRJNA484031.
α diversity and rarefaction curve
Within a sample, α diversity (within-sample diversity) was used to represent genus richness (Supplementary Table S3): the higher is the richness of a sample, the greater the diversity. The α diversity value was calculated according to the Chao 1 index as described previously.21,22
To assess the gene richness in all groups, we performed sampling 100 times randomly in the cohort, and counted the total number of genes that could be classified from these samples in the R environment.
Taxonomic annotation and abundance calculation
According to the gene sequence information we obtained based on the above analyses, each gene was aligned to the NR database of NCBI using DIAMOND (default parameter: e-value ⩽10−5). For each gene, only reads with an e-value ⩽10 × minimal e-value were used to distinguish taxonomic groups. The lowest common ancestor-based algorithm (LCA) was employed to define the gene’s taxonomical level in the MEGAN environment. The relative abundance of a taxonomic group was summed up by the abundance of its matching genes.
Metagenome-wide association study and co-occurrence network of MLGs
According to the gene clustering results, we constructed a gene set that included 1,048,574 genes. In total, 201,136 genes showed statistically significant differences (Wilcoxon rank sum test, p < 0.05) in multiple relative abundance comparisons and were considered marker genes, including 50,886 genes for RB versus NRB, 49,553 genes for RB versus RA, 15,177 genes for NRB versus NRA, and 93,718 genes for RA versus NRA.
To determine the relationships between these marker genes, we created MLGs to cluster genes according to their variation in abundance, as previously described.20 The MLGs that included less than 50 genes were removed. The remaining 237 MLGs were assigned to taxonomic groups, and abundance was profiled according to the relative abundance of the constituent genes, as previously described.20 Briefly, assignment to a species required more than 90% of the genes in an MLG to be aligned with a species’ genome at 95% identity and 70% overlap with the query sequence. Similarly, assignment to a genus required at least 80% of the genes in an MLG to align to the genome at 85% identity for both DNA and protein sequences.
According to the MLG abundance, we calculated Spearman’s correlations for all samples to further cluster the MLGs. Enrichment of MLGs was identified according to the OR value, which was calculated according to the abundance of the compared samples of each group as previously described.23 The computing formula of an MLG, k, was calculated as OR(k)= [∑s=group 1 Ask/∑s=group 1 (∑i≠k Asi)]/ [∑s=group 2 Ask/∑s=group 2 (∑i≠k Asi)], where Ask is the abundance of MLG k in sample s. For the comparative analysis between two groups, MLGs were classified according to the OR values, where OR > 2 belongs to group 1, and OR < 0.5 to another group. The co-occurrence network of MLGs was visualized with Cytoscape 3.5.1.
Functional annotation
All genes in our catalogue were translated into putative amino acid sequences, and then aligned against the proteins or domains in KEGG databases (release 59.0, without animal and plant genes) using DIAMOND software (version 0.7.9.58, default parameter except that −k 50 –sensitive −e 0.00001). According to the highest scoring annotated hit(s), each protein was assigned to a KEGG orthology group in which at least one of the high-scoring segment pair (HSP) scores was over 60 bits.24 The abundance of KEGG orthology groups was the sum of the annotated gene abundance.
Statistical analyses
The Shannon index, Chao 1 index, and Bray-Curtis distance were all calculated in R (Version 3.4.0 by vegan package). Principal component analysis (PCA) and principal coordinate analysis PCoA were performed and displayed by the cluster package, clusterSim package, and ade4 package. Spearman’s correlation was calculated for all samples using R (Version 3.4.0, psych package), as previously described.25,26 The ggplot2 package was applied to construct plots. Significant differences in the relative abundance of gut microbes or MLGs between two compared groups were identified by two-tailed Wilcoxon rank sum tests with p < 0.05. Enrichment in any group was determined according to the higher rank sum
Results
Bacterial diversity is influenced by FOLFOX treatment
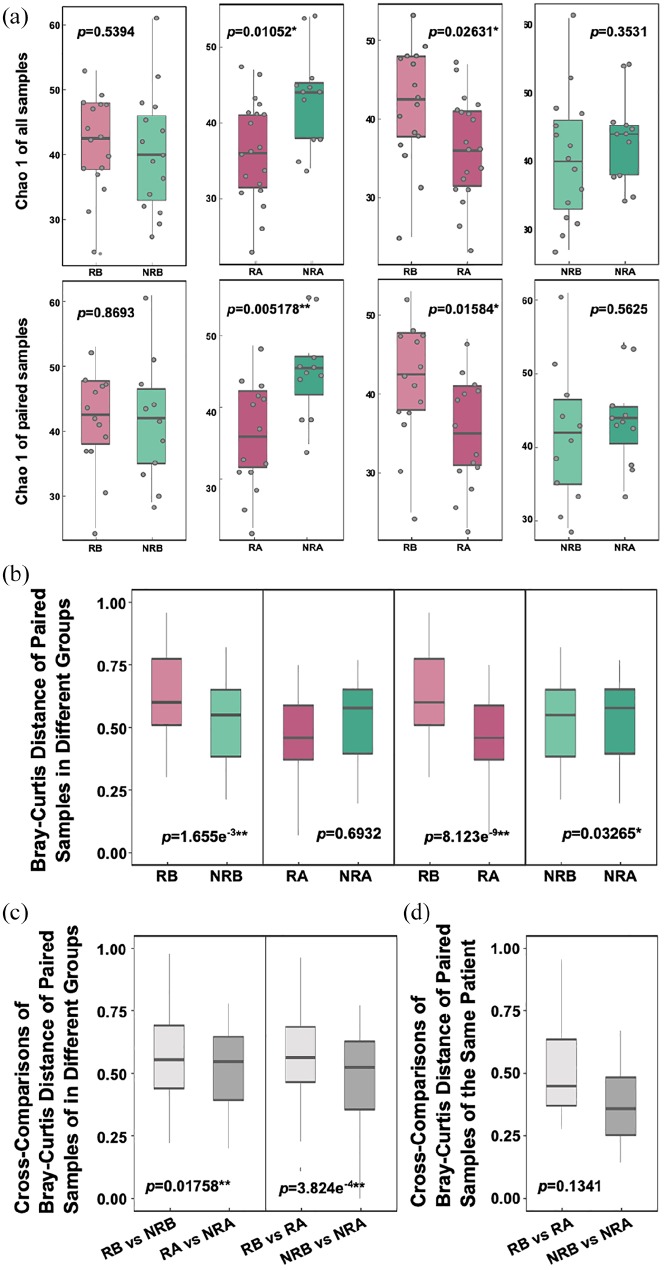
To identify whether gut microbial diversity changes are associated with FOLFOX treatment, we performed shotgun metagenomics sequencing of stool samples from our cohort, and used the Chao 1 index27 to reveal the bacterial diversity present in the samples. First, we compared Chao 1 values in different groups according to clinical outcomes. Before treatment, bacterial diversity did not differ significantly between responders and nonresponders (p = 0.5394), but a significant difference was found (p = 0.01052) after the groups received identical NC (Figure 1a). Next, we compared Chao 1 values to display the dynamic change in bacterial diversity with treatment. After treatment, Chao 1 values decreased significantly (p = 0.02631) in the responder group, but there was no significant difference in the nonresponder group (p = 0.3531), with an increasing trend in all samples and paired samples (Figure 1a). The results suggest that the composition of gut microbes changed to a considerably greater degree in the responder group than in the nonresponder group.
Figure 1.
Change in diversity and sample distance under FOLFOX treatment. (a) Comparison of the microbial gene count and α diversity (as assessed by the Chao 1 index) by cross-comparisons at the genus level. The first line represents the comparison without considering the paired samples; the second line presents the comparison considering the paired samples. (b) Distance between two paired samples (as assessed by the Bray-Counts distance index) by cross-comparisons. (c) Count of the distances of two different group samples and then their cross-comparison assessed by the Bray-Counts distance index. (d) Count of the distances before and after treatment of the paired samples using the Bray-Counts distance index.
NRA, nonresponders after treatment; NRB, nonresponders before treatment; RA, responders after treatment; RB, responders before treatment.
To verify the extent of the change to the gut microbiota, we employed Bray-Curtis distances,28 which indicate the different degree of bacterial composition between two random samples. We calculated distances within two samples of the same group. When comparing Bray-Curtis distances between responders before treatment (RB) and nonresponders before treatment (NRB), the difference was very significant, both in paired samples (p = 1.655e−3, Figure 1) and in all samples (p = 2.68e−3, Figure S1b). However, no significant differences between responders after treatment (RA) and nonresponders after treatment (NRA) were observed (p = 0.6932, Figure 1b; p = 0.1758, Figure S1). Next, we examined the dynamic change in Bray-Curtis distances in each FOLFOX outcome group. In the responder group, the distance between two samples dropped sharply after treatment (p = 8.123e−9, Figure 1b; p = 0.06675, Figure S1); in the nonresponder group, although the p value was less than 0.05, the trend was flatter (p = 0.03265, Figure 1b). In a comparison of the total NRB and NRA samples, the p value was almost 1 (p = 0.9633, Figure S1). These results suggest that NC increased the degree of change in gut microbial composition only in the responder group.
To further explain the differences in gut microbial composition, we again employed the Bray-Curtis distance to cross-compare samples in different groups. We calculated the distance between every two samples: one was RB, and the other was NRB, labelled as RB versus NRB. The same calculation method was applied between other groups with similar labels. When we then performed multiple comparisons of the RB versus NRB and RA versus NRA separately, the distance clearly decreased (p = 0.01758, Figure 1c); when multiple comparisons of RB versus RA and NRB versus NRA were performed separately, the distance decreased more significantly (p = 3.824e−4, Figure 1c; p = 5.767e−5, Figure S1). These results support the view that, during FOLFOX treatment, the gut microbiotal composition of responders underwent a sharper change than that of nonresponders.
Finally, we calculated the distance of paired samples with cross-comparison before and after treatment. There was a clear trend towards a higher average distance in the responder group, though the difference was not significant (p = 0.1341, Figure 1d).
This finding indicated that the difference in composition of the low-set rectal cancer patients’ gut microbes was present before chemotherapy, and that this difference might influence the effectiveness of NC. Moreover, gut microbial composition changed during FOLFOX treatment, and the range of variation was shown to be closely associated with treatment outcome.
Changes in relative abundance of taxa post-FOLFOX treatment
To describe the features of FOLFOX treatment-associated gut microbiota, genes were aligned as scaftigs, which were added to gene catalogues by MetaGeneMark. The gene catalogues were used to annotate and calculate the abundance of taxonomic groups by MetaPhlAn2 (Supplementary Table S3). When comparing genus abundances before and after treatment, only seven genera showed significant changes. Porphyromonas, Peptostreptococcus, and Veillonella decreased after therapy, whereas Gemella, Parvimonas, Solobacterium, and Pyramidobacter increased (Supplementary Table S3). Next, we compared genus groups by multiple comparison and labelled them according to comparison groups, including RB versus NRB, RA versus NRA, RB versus RA, and NRB versus NRA. Based on these multiple comparisons, we found a key sensitive genus in the different comparison groups.
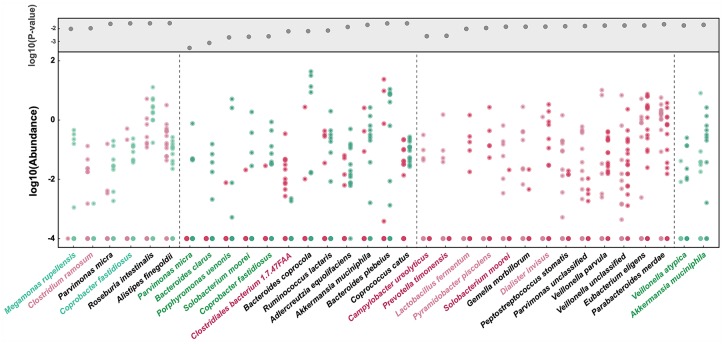
To further explore FOLFOX treatment-associated gut microbes, we assessed species abundance difference in each comparison group. The species’ relative abundances were more than 10−4, as shown in Figure 2 (Wilcoxon rank sum test, p < 0.05). When comparing differences in abundance between the responder and nonresponder groups before and after treatment, only Clostridium species were significantly more abundant in the responder group both before and after treatment. As shown in Figure 2, before NC, Clostridium ramosum (labelled with light purple) was detected in the responder group but could not be found in more than 90% of nonresponders. After treatment, Clostridiales bacterium 1.7.47 FAA (labelled with dark purple) was at a higher level in the responder group. In contrast, Megamonas rupellensis and Coprobacter fastidiosus (labelled with light green) were enriched in NRB, and Parvimonas micra, Bacteroides clarus, Porphyromonas uenonis, Solobacterium moorei, and Coprobacter fastidiosus (labelled with dark green) were enriched in NRA.
Figure 2.
Species are strikingly different across groups. Species with an average relative abundance greater than 10e−5 are displayed based on p values (top grey area) <0.05 according to the Wilcoxon rank sum test. The points show the relative abundance of each sample. The colors represent the four groups: responders before treatment (light purple), responders after treatment (dark purple), nonresponders before treatment (light green), and nonresponders of after treatment (dark green). The species with corresponding group color are mentioned in the text.
When comparing the changes in dynamic abundance in the responder and nonresponder groups, the statistical results verified our hypothesis, with the differences between before and after chemotherapy being larger in the former than in the latter (Figure 2). After treatment, Lactobacillus fermentum, Pyramidobacter piscolens, and Dialister invisus (labelled with light purple), which were almost undetectable before chemotherapy in the responder group, were detected. Conversely, Campylobacter ureolyticus, Prevotella timonensis, and Solobacterium moorei (labelled with dark purple) nearly disappeared in the responder group after NC. However, in the nonresponder group, only Veillonella atypica and Akkermansia muciniphila (labelled with dark green) showed a change in abundance after FOLFOX treatment.
Bacterial subset as marker for outcome of FOLFOX treatment
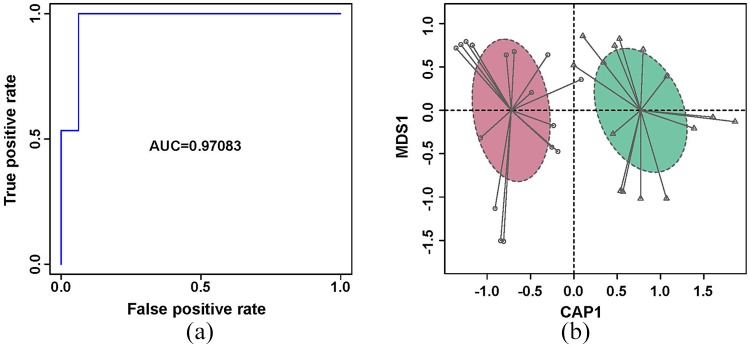
To illustrate the microbial signature of responders and nonresponders before FOLFOX treatment, and to further exploit the potential biomarker in the gut microbiome, a random forest disease classifier was employed to explain the variables of the two groups. We defined the different species of RB versus NRB as a set that contained eight species (Supplementary Table S4); we then extracted two to eight species from the set randomly as a subset, with 247 subsets in total. For each subset, we calculated the area under the receiver operating curve (AUC) (Supplementary Table S4). The subset including Coprobacter fastidiosus, Alistipes finegoldii, Gemella unclassified, Granulicatella adiacens, Parvimonas micra, and Clostridium ramosum had the highest AUC of 0.9708 (Figure 3a, Supplementary Table S4). Based on this subset, samples from RB and NRB were obviously separated by constrained PCoA (Figure 3b). This subset might be used as a biomarker to predict the results of NC.
Figure 3.
ROC curve and constrained PCoA. (a) ROC of the random forest classifier using species relative abundance of the highest AUC subset based on the identified bacteria between responder and nonresponder groups before FOLFOX treatment; and (b) Constrained PCoA of the responder (purple) and nonresponder (green) groups based on this subset.
AUC, area under the curve; PCoA, principal coordinate analysis; ROC, receiver operating characteristic.
Metagenomic linkage groups enriched in responder and nonresponder groups before and after treatment
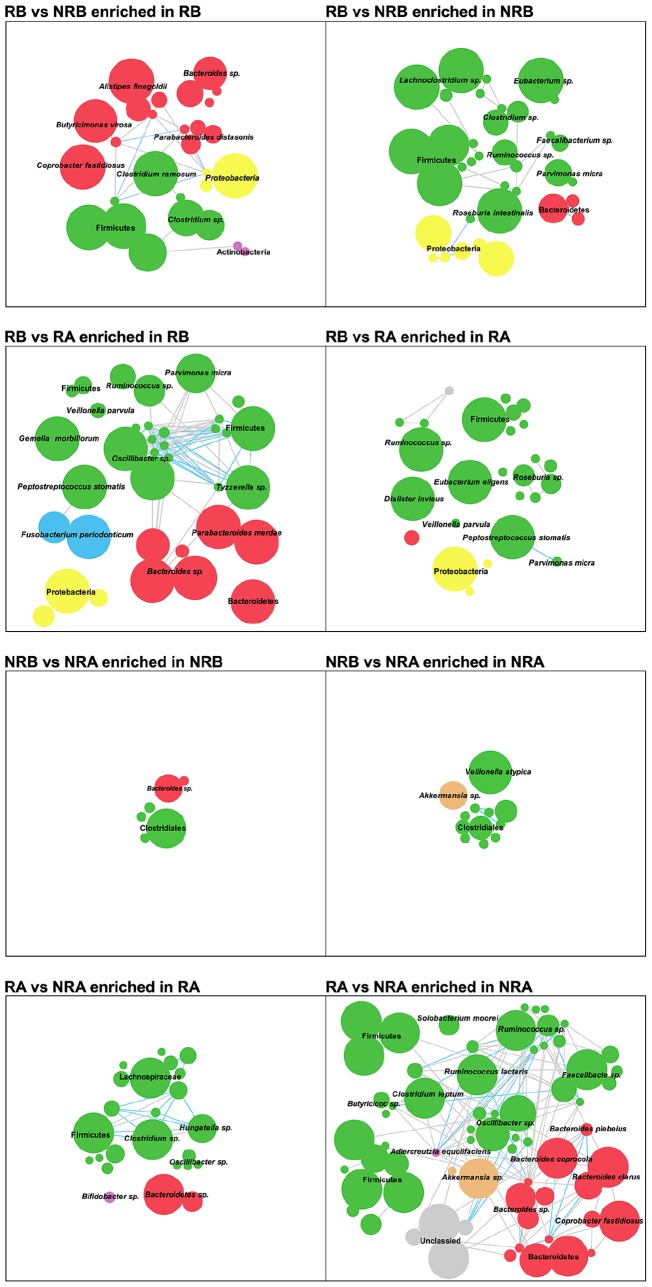
To describe the features of chemical-therapy-associated gut microbiota, we compared genes using multiple comparisons, including RB versus NRB, RA versus NRA, RB versus RA, and NRB versus NRA. In total, we identified 201,136 gene markers that were differentially enriched in the multiple comparisons (Wilcoxon rank sum test, Supplementary Table S5); there were 50,886, 93,718, 49,553, and 15,177 gene markers in each comparison (Supplementary Table S5). These genes were then separately clustered into metagenomic linkage groups (MLGs) based on correlated abundance variation among samples. MLGs containing more than 50 genes, 235 MLGs in total, were used for network analysis. There were 64, 91, 62, and 18 MLGs in each comparison, and the Simpson correlation value, more than 0.6, was 51, 136, 97, and 9, respectively. Among all comparison groups, the most complex network was RA versus NRA, and the simplest was NRB versus NRA. This result suggests that the degree of difference in these comparison groups can be estimated by the complexity of the network.
Moreover, the parameters of the networks showed a similar tendency as the gut microbe analysis, in that the numbers of nodes and edges in the RA versus NRA network were much larger than those in the NRB versus NRA network. Furthermore, enrichment of the species exhibiting a significant difference showed similar trends. For example, in the microbe composition analysis, enrichment of Alistipes finegoldii in RB, and Parvimonas micra in NRB showed the same result in the network study (Figure 4).
Figure 4.
Comparative analysis of GM enrichment across groups of MLGs. Each MLG involves at least 50 linked genes, and the correlation network of MLGs differently enriched in each group is based on the abundance based on Spearman’s correlation. MLGs are colored by the taxonomic assignment at the phylum level, including Bacteroidetes (red), Firmicutes (green), Proteobacteria (yellow), Actinobacteria (purple), Fusobacteria (blue), Verrucomicrobia (orange) and others (grey). The node size is scaled to the number of genes within the MLG. Edges connected with nodes denote Spearman correlation >0.8 (blue) or between 0.7 and 0.8 (grey).
GM, gut microbiota; MLG, metagenomic linkage group.
Functional alteration in responder and nonresponder groups before and after FOLFOX treatment
To understand gut microbial function during the course of FOLFOX treatment in our study cohort, we used the KEGG and Carbohydrate-Active EnZymes (CAZy) databases to align all genes and to assign KEGG orthology and CAZy families as proteins (Supplementary Table S6). Based on KEGG orthology, PCA revealed striking differences in microbial functions. A set of 31 (n = 31) KEGG modules were differentially enriched in different groups in the cross-comparisons (adjusted p < 0.05, Wilcoxon rank sum test, Figure 5, Supplementary Table S6). With regard to FOLFOX treatment, the degree of functional alteration was similar to the microbiotal composition and MLGs, and the number of different modules was greatest in the RB and RA comparison. During this process, the modules involving essential amino acid and polyamine biosynthesis increased, and modules of some nonessential amino acids and fucose decreased; only the pyruvate fermentation function was increased in NRB. In previous studies, metabolic functional alterations in CRC patient fecal samples included iron, phosphate, and amino acid transport, short-chain acid biosynthesis, and methanogenesis.29–31 However, in our study, these microbiota pathways did not change during the NC treatment. Moreover, the result was unexpected when comparing the different modules between the responder and nonresponder groups before and after treatment. There were 21 (n = 21) differential modules when RB was compared with NRB, but only 4 (n = 4) when RA was compared with NRA. When comparing the microbial composition and MLGs, the range of divergence was consistent. Nonetheless, the functional analytical results were conflicting. Before NC treatment, the modules involving aromatic amino acid and deoxyribonucleotide biosynthesis were enriched in NRB, whereas tetrahydrofolate, ornithine, and ubiquinol biosynthesis and degradation pathways, which might produce short-chain fatty acids, were enriched in RB. After NC treatment, only four (n = 4) modules displayed differences, and these involved in petroselinate biosynthesis, glycolysis, and ester metabolism. Considering the contribution rate of the microbes (Supplementary Table S7), although there were fewer different microbes between RB and NRB, they are polyfunctional during metabolism; in contrast, microbes that displayed abundance differences between RA and NRA have a similar metabolic function.
Figure 5.
Microbial gene function annotation in each group. The heat map shows the abundance of KEGG modules differentially enriched in each sample gut microbiome. The criterion used was a p value <0.05 determined by the Wilcoxon rank sum test. The bars on the top of each heat map represent the four groups: responders before treatment (light purple), responders after treatment (dark purple), nonresponders before treatment (light green), and nonresponders after treatment (dark green).
KEGG, Kyoto Encyclopedia of Genes and Genomes.
Discussion
In recent years, many studies have focused on the modulating role of gut microbiota in drug efficacy,9,32–34 and a series of gut microbes have been confirmed to increase or decrease the efficacy of some agents.32,33,35,36 To explain the relationship between gut microbes and NC agents, stool samples were collected, and the composition of microbes was analyzed. In almost all studies, stool samples were collected at the end of the treatment when the outcomes were known. According to these outcomes, the patients were separated into responder and nonresponder groups.36–38 However, in this approach, the gut microbial composition at the beginning of therapy remained unknown. Many changes, especially in the immune system and gut microbiota, occur during the use of antineoplastics because of the effects of these drugs within the body.9,32–34 To address these limitations, we assembled a cohort of 37 Chinese low-set rectal cancer patients among patients at the China-Japan Friendship Hospital in Beijing, China, and carried out genomic sequencing technology. Based on the genetic information, we applied metagenomic and metabolomic analyses, and observed a dynamic process of the gut microbiota during FOLFOX treatment.
Metasequencing detected bacterial signals from fecal samples, combined with the patients’ clinical outcomes and analysis of α diversity and Bray-Curtis distances. In our analysis, α diversity was reduced after FOLFOX treatment, which was consistent with previous studies.39 Additionally, after treatment, α diversity was lower in responders than in nonresponders. This result was different from our general awareness that microbial diversity loss is always associated with chronic health conditions,40–42 including cancer,3,43,44 and low diversity was usually related to poor outcomes of some therapeutic regimen, such as stem cell transplant and antiprogrammed cell death protein 1 (PD-1) immunotherapy.36,45 However, some authors have also reported that stool microbial diversity was higher in CRC patients than in healthy controls.39 In murine studies, reduced gut microbiome diversity using antibiotics was able to reduce ischemic brain injury.46 As few results were similar to ours, there may not be a unified tendency for gut flora diversity, which is unique in some cases.
We next employed Bray-Curtis distances to describe the degree of change in the gut bacterial composition. The results revealed that the gut microbiota changed during FOLFOX treatment, to a higher degree in the responder group than in the nonresponder group. Although we did not verify this result, it suggests that the gut microbiotal composition is closely associated with chemical agent usage, and might be linked to treatment outcomes.
In our analysis of gut microbial relative abundance, a group of species displayed significant differences in their comparison groups. Clostridium ramosum was the only species with a higher abundance in RB than in NRB (Figure 2). C. ramosum participates in the metabolism of glucose and insulin,47 with the ability to increase insulin sensitivity via transfer of lean donor fecal samples in animal models.7 CRC has a high correlation with fat and red meat intake, and glucose metabolism may benefit from the avoidance of these foods. In our function analysis, there was a higher level of degradation of these molecules in RB than in NRB.
Megamonas rupellensis, Parvimonas micra, and Coprobacter fastidiosus were enriched in the NRB. Previous studies rarely report on the relationship between M. rupellensis or C. fastidiosus and CRC. In our work, these microbes were scarce in the responder group. Conversely, several studies supported the notion that P. micra has a close association with CRC,35,48–50 and is involved in tumor growth.51 Moreover, the high prevalence of P. micra in NRA suggests that this species might be an important element affecting NC results. This hypothesis should be verified in the future.
Bacteroides clarus, Porphyromonas uenonis, Solobacterium moorei, and Coprobacter fastidiosus are other species that were enriched in NRA. B. clarus is another candidate microbe that might be used to identify CRC. According to a large cohort network research study, the combination of Fusobacterium nucleatum, B. clarus, Roseburia intestinalis, and Clostridium hathewayi was verified as being a sensitive and specific marker for CRC.6 In our data, a significant difference for R. intestinalis was found between RB and NRB, whereby abundance was higher in the former than in the latter. A significant difference in B. clarus was only found after NC. Conversely, differences in F. nucleatum and C. hathewayi were observed in our study, which might have been caused by the sample size in this analysis. S. moorei, an anaerobic gram-positive bacterium first isolated from human feces,52 and has a strong relationship with gut microbiotal disturbance diseases, such as CRC, adenoma, and acute proctitis.53–55 S. moorei also colonizes wounds or lesion tissues to induce thrombophlebitis and septic pulmonary embolism,56,57 and affects the intestinal microbiota and immune development.58 P. uenonis was originally isolated from patients with serious infection, and from stool samples from children,59 but there are few reports on the role of this species in CRC. There are also few reports on the relationship between C. fastidiosus and CRC outcomes.
After NC, only Veillonella atypica and Akkermansia muciniphila displayed differences in the nonresponder group. V. atypica is a gastrointestinal bacterium with a high detection rate in dysbacteriosis and samples from patients with celiac diseases.60 It also has a high detection rate in human oral, gastric juice and gastric mucosa samples.61 Moreover, V. atypica can multiply on oral cancer lesion surfaces and constitutes the oral mucosal core bacteriome of oral cell carcinoma.62,63 This suggests that V. atypica has a strong association with intestinal mucosa or tissue lesions, which might worsen the outcomes of tumor treatment. A. muciniphila is an intestinal mucin-degrading bacterium (phylum Verrucomicrobia) that is widespread in humans and animals.64 Recent studies have shown its beneficial roles in re-establishing gut barrier function, maintaining gut microbial balance, reducing obesity, and relieving metabolic disturbance.65–67 This bacterium may also enhance the effects of antiprogrammed cell death protein/ligand 1 (anti-PD-1/anti-PD-L1) during treatment for epithelial tumors.38 Nonetheless, some studies have reported that this bacterium has a higher abundance in mucosal samples from CRC patients than from healthy controls.68,69 In animal studies, A. muciniphila colonization was found to exacerbate intestinal inflammation, which is induced by Salmonella typhimurium, or increase the tumor burden in Apc gene-mutation mouse models.64,70 This suggests that A. muciniphila may have a dual effect on host health that is dependent on the host and environmental conditions. In our study, intestinal mucosa lesions were worse in the nonresponder group after treatment, and the level of disturbance in the gut was more severe. This condition might accelerate the growth of A. muciniphila.
Regarding biomarkers, our result applies a new standard to predicting the result of the FOLFOX. Some members of the biomarker group, such as Granulicatella adiacens, were in low abundance, which may be difficult to detect, and Gemella unclassified could not be identified at the species level. Although further investigation is needed to verify these findings, the results will inspirit the development of a novel noninvasive diagnostic method for patients before chemotherapy and help them to avoid unnecessary treatment.
Assessment of the differences in species indicated that most of bacteria were enriched in the nonresponder group both before and after treatment. Similar outcomes were observed in the MLG analysis. As in previous studies,71–73 we built a co-occurrence network to investigate the concordance of microbes between each comparison group, and distinguishing the co-occurrence networks according to their components and topographies was very successful. The MLG assembly method was different from taxonomic annotation, but the most notable differential bacteria were also significantly different in the relative abundance analysis. According to these results, consistency of gut microbiotal variation occurred not only across the FOLFOX but also between different outcomes in bacterial relative abundance and in their interactive relationship. At the end of our study, we performed functional alteration analysis according to the microbiotal pathways, with unexpected results. When checked for their contribution, some of the bacteria were found to be involved in more than one pathway, which was always a significantly different species in the comparison group. Perhaps in future research, these species will help in our understanding of the ways in which drugs modulate the gut microbiome.
In addition to the results reported here, we performed an analysis of enterotypes,74 which represent a classification of the human gut microbiome based on the relative abundances of different microbial groups in the individual’s gut microbiota at the genus level. In our cohort, 62 samples were clustered into two distinct enterotypes,75 with Bacteroides and Prevotella as dominate bacteria (Figure S2, Supplementary Table S7). After FOLFOX treatment, eight of the paired samples’ enterotypes shifted, including five responders and three nonresponders. Notably, in the responder group, the change occurred consistently from enterotype II to enterotype I. Moreover, according to clinical records, three patients showed significant treatment efficacy, as their tumors had almost disappeared and their carcinoembryonic antigen (CEA) levels were almost normal at 5–10 ng/ml after the entire course of treatment (Supplementary Table S1). Nevertheless, it was difficult to identify the underlying principle with regard to these three nonresponder group samples, with some changes from enterotype I to enterotype II and some in the other direction. Although this result is not very convincing, it suggests a relationship between enterotype and NC efficacy as well as the significance of gut microbes in the course of chemical therapy (Figure S3).
In this study, we unfortunately could not recruit as many patients with FOLFOX as intended. Most patients choose surgery as the first treatment option, though some patients accepted NC as their first treatment. However, during the 2 months of therapy, patients experienced serious side effects due to the chemical agents, and some could not tolerate the therapy and opted to receive excision surgery. Therefore, it is very difficult to build a large cohort to obtain more information on the association between the gut microbiota and agent efficacy or to construct a model to predict treatment results. Although our cohort size was limited, we were able to demonstrate dynamic trends in the gut microbiota during the therapeutic process. Regardless, we could not explain the role of these bacteria in the efficacy of FOLFOX in this study, which did not report on the relationship between bacteria and CRC or gut tumors.
Conclusion
Taken together, our results clearly described the profiles of the gut microbiota in low-set rectal cancer patients before and after FOLFOX treatment in responder and nonresponder groups. We also revealed a dynamic process of gut bacterial composition change during treatment, and established a relationship between gut bacterial and FOLFOX outcomes. Although we could not explain the role of the differential bacteria before FOLFOX, our findings highlight an opportunity to improve the effects of chemical therapy with bacterial intervention before treatment of low-set rectal cancer.
Supplemental Material
Supplemental material, Authorship-Change-Request-Form-P1 for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Authorship-Change-Request-Form-P2 for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Authorship-Change-Request-Form-P3 for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Supplementary_Figure_S1.Bray-Curitis_distance_between_all_samples.svg for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Supplementary_Figure_S2.Shift_of_gut_enterotypes_in_patients_with_FOLFOX_treatment for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Supplementary_Figure_S3._Genus_construction_of_enterotype_changed_patients_during_FOLFOX_treatment for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Table_S1_Patients_clinical_background_information for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Table_S2_Data_information_of_HiSeq_sequencing for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Table_S3__Relative_abundance_profile_at_each_taxonomy_level for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Table_S4__AUC_of_each_combination for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Table_S5._Detailed_information_for_gene_markers for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Table_S6__Pathway_information for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Table_S7_Enterotype_information for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Acknowledgments
Authors Jing Li and Jingtao Li are joint first authors.
Footnotes
Authors’ Note: Xiaofeng Song is also affiliated with State Key Laboratory of Infectious Disease Prevention and Control, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China.
Yingdian Wang is also affiliated with College of Life sciences, Qinghai Normal University, Xining, Qinghai, China.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and publication of this article: This study was partially supported by the China-Japan Friendship Hospital Research Funding (2016-1-MS-5), Beijing Municipal Natural Science Foundation (5174037), Major State Basic Research Development Program (2015CB554200), and National High Technology Research and Development Program (2014AA020801).
Conflict of interest statement: The authors declare that there is no conflict of interest.
ORCID iD: Cheng Xiao  https://orcid.org/0000-0002-5601-9670
https://orcid.org/0000-0002-5601-9670
Supplementary material: Supplemental material for this article is available online.
Contributor Information
Jing Li, College of Life Sciences, Beijing Normal University, Beijing, China; CAS Key Laboratory of Pathogenic Microbiology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.
Jingtao Li, Department of Gastroenterology, China-Japan Friendship Hospital, Beijing, 100029, China.
Na Lyu, CAS Key Laboratory of Pathogenic Microbiology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.
Yue Ma, CAS Key Laboratory of Pathogenic Microbiology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.
Fei Liu, CAS Key Laboratory of Pathogenic Microbiology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.
Yuqing Feng, CAS Key Laboratory of Pathogenic Microbiology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.
Li Yao, Department of Surgery, China-Japan Friendship Hospital, Beijing, China.
Zhiyong Hou, Department of Surgery, China-Japan Friendship Hospital, Beijing, China.
Xiaofeng Song, CAS Key Laboratory of Pathogenic Microbiology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.
Hongchuan Zhao, Department of Gastroenterology, China-Japan Friendship Hospital, Beijing, China.
Xiaoya Li, Institute of Clinical Medicine, China-Japan Friendship Hospital, Beijing, China; Graduate School of Peking Union Medical College, Chinese Academy of Medical Sciences/Peking Union Medical College, Beijing, China.
Yingdian Wang, College of Life Sciences, Beijing Normal University, Beijing, China.
Cheng Xiao, Institute of Clinical Medicine, China-Japan Friendship Hospital, Beijing, 100029, China; Graduate School of Peking Union Medical College, Chinese Academy of Medical Sciences/Peking Union Medical College, Beijing, China.
Baoli Zhu, CAS Key Laboratory of Pathogenic Microbiology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China; Savaid Medical School, University of Chinese Academy of Sciences, Beijing, China; Beijing Key Laboratory of Antimicrobial Resistance and Pathogen Genomics, Beijing, China; Department of Pathogenic Biology, School of Basic Medical Sciences, Southwest Medical University, Luzhou, Sichuan, China.
References
- 1. Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012; 338: 120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer 2013; 13: 800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garrett WS. Cancer and the microbiota. Science 2015; 348: 80–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 2012; 22: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 2012; 22: 292–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liang Q, Chiu J, Chen Y, et al. Fecal bacteria act as novel biomarkers for non-invasive diagnosis of colorectal cancer. Clin Cancer Res 2017; 23: 2061–2070. [DOI] [PubMed] [Google Scholar]
- 7. Huang MJ, Wang XD, Hu YJ, et al. Short-course neoadjuvant chemoradiotherapy and surgery are beneficial in Chinese patients: a retrospective study. Medicine (Baltimore) 2017; 96: e9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. National Comprehensive Cancer Network. Rectal cancer (version 1.2018), https://www.nccn.org/store/login/login.aspx?ReturnURL=https://www.nccn.org/professionals/physician_gls/pdf/rectal.pdf. (accessed 14 March 2018).
- 9. Iida N, Dzutsev A, Stewart CA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013; 342: 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scott TA, Quintaneiro LM, Norvaisas P, et al. Host-microbe co-metabolism dictates cancer drug efficacy in C. elegans. Cell 2017; 169: 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koehne CH, Midgley R, Seymour M, et al. Advanced colorectal cancer: which regimes should we recommend? Ann Oncol 1999; 10: 877–882. [DOI] [PubMed] [Google Scholar]
- 12. IMPACT. Efficacy of adjuvant fluorouracil and folinic acid in colon cancer. International multicantre pooled analysis of colon cancer trials (IMPACT) investigators. Lancet 1995; 345: 939–944. [PubMed] [Google Scholar]
- 13. Kozovska Z, Gabrisova V, Kucerova L. Colon cancer: cancer stem cells markers, drug resistance and treatment. Biomed Pharmacother 2014; 68: 911–916. [DOI] [PubMed] [Google Scholar]
- 14. Johnston PG, Kaye S. Capecitabine: a novel agent for the treatment of solid tumors. Anticancer Drugs 2001; 12: 639–646. [DOI] [PubMed] [Google Scholar]
- 15. Giacchetti S, Perpoint B, Zidani R, et al. Phase III multicenter randomized trial of oxaliplatin added to chronomodulated fluorouracil-leucovorin as first-line treatment of metastatic colorectal cancer. J Clin Oncol 2000; 18: 136–147. [DOI] [PubMed] [Google Scholar]
- 16. Douillard JY, Cunningham D, Roth AD, et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. Lancet 2000; 355: 1041–1047. [DOI] [PubMed] [Google Scholar]
- 17. Offer SM, Diasio RB. Is it finally time for a personalized medicine approach for fluorouracil-based therapies? J Clin Oncol 2016; 34: 205–207. [DOI] [PubMed] [Google Scholar]
- 18. O’Donnell PH, Dolan ME. Cancer pharmacoethnicity: ethnic differences in susceptibility of the effects of chemotherapy. Clin Cancer Res 2009; 15: 4806–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pham TT, Liney G, Wong K, et al. Study protocol: multi-parametric magnetic resonance imaging for therapeutic response prediction in rectal cancer. BMC Cancer 2017; 17: 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012; 490: 55–60. [DOI] [PubMed] [Google Scholar]
- 21. Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature 2012; 486: 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rehman A, Rausch P, Wang J, et al. Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut 2016; 65: 238–248. [DOI] [PubMed] [Google Scholar]
- 23. Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci U S A 2012; 109: 594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bäckhed F, Roswall J, Peng Y, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015; 17: 690–703. [DOI] [PubMed] [Google Scholar]
- 25. Feng Q, Liang S, Jia H, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun 2015; 6: 6528. [DOI] [PubMed] [Google Scholar]
- 26. Karisson FH, Tremaroli V, Nookaew I, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013; 498: 99–103. [DOI] [PubMed] [Google Scholar]
- 27. Chao A. Nonparametric estimation of the number of classes in a population. Scand J Statis 1984; 11: 265–270. [Google Scholar]
- 28. Bray JR, Curtis JT. An ordination of upland forest communities of southern Wisconsin. Ecol Monogr 1957; 27: 325–349. [Google Scholar]
- 29. Brown DG, Rao S, Weir TL, et al. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab 2016; 4: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin Y, Ma C, Liu C, et al. NMR-based fecal metabolomics fingerprinting as predictors of earlier diagnosis in patients with colorectal cancer. Oncotarget 2016; 7: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kong C, Gao R, Yan X, et al. Research progression of blood and fecal metabolites in colorectal cancer. Int J Surg Oncol 2018; 3: e51. [Google Scholar]
- 32. Viaud S, Saccheri F, Mignot G, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013; 342: 971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-Li efficacy. Science 2015; 350: 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015; 350: 1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017; 170: 548–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018; 359: 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018; 359: 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Routy B, Emmanuelle LC, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018; 395: 91–97. [DOI] [PubMed] [Google Scholar]
- 39. Deng X, Li Z, Li B, et al. Comparison of microbiota in patients treated by surgery or chemotherapy by 16S rRNA sequencing reveals potential biomarkers for colorectal cancer therapy. Front Microbiol 2018; 9: 1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012; 486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Turnbaugh PJ, Bäckhed F, Fulton L, et al. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008; 3: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010; 464: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Segre JA. Microbial growth dynamics and human disease. Science 2015; 349: 1058–1059. [DOI] [PubMed] [Google Scholar]
- 44. Drewes JL, Housseau F, Sears CL. Sporadic colorectal cancer: microbial contributors to disease prevention, development and therapy. Br J Cancer 2016; 115: 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taur Y, Jenq RR, Perales MA, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 2014; 124: 1174–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Winek K, Engel O, Koduah, et al. Depletion of cultivatable gut microbiota by broad-spectrum antibiotic pretreatment worsens outcome after murine stroke. Stroke 2016; 47: 1354–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vrieze A, Van Nood E, Holleman F, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012; 143: 913–916. [DOI] [PubMed] [Google Scholar]
- 48. Coker OO, Dai Z, Nie Y, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018; 67: 1024–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 2018; 67: 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shah MS, DeSantis TZ, Weinmaier T, et al. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut 2018; 67: 882–891. [DOI] [PubMed] [Google Scholar]
- 51. Nakatsu G, Li X, Zhou H, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun 2015; 6: 8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kageyama A, Benno Y. Phylogenic and phenotypic characterization of some Eubacterium-like isolates from human feces: descrition of Solobacterium moorei Gen. Nov., Sp. Nov. Microbiol Immunol 2000; 44: 223–227. [DOI] [PubMed] [Google Scholar]
- 53. Lau SK, Teng JL, Leung KW, et al. Bacteremia caused by Solobacterium moorei in a patient with acute proctitis and carcinoma of the Cervix. J Clin Microbiol 2006; 44: 3031–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pedersen RM, Holt HM, Justesen US. Solobacterium moorei bacteremia: identification, antimicrobial susceptibility, and clinical characteristics. J Clin Microbiol 2011; 49: 2766–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jobin C. Human intestinal microbiota and colorectal cancer: moving beyond associative studies. Gastroenterology 2017; 153: 1475–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Martin CA, Wijesurendra RS, Borland CDR, et al. Femoral vein thrombophlebitis and septic pulmonary embolism due to a mixed anaerobic infection including Solobacterium moorei: a case report. J Med Case Rep 2007; 1: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zheng G, Summanen PH, Talan D, et al. Phenotypic and molecular characterization of Solobacterium moorei isolates from patients with wound infection. J Clin Microbiol 2010; 48: 873–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Andrade-Navarro MA, Schokker D, Zhang J, et al. Early-life environmental variation affects intestinal microbiota and immune development in new-born piglets. PLoS One 2014; 9: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Finegold SM, Vaisanen ML, Rautio M, et al. Porphyromonas uenonis sp. nov., a pathogen for humans distinct from P. asaccharolytica and P. endodontalis. J Clin Microbiol 2004; 42: 5298–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Herran AR, Perez-Andres J, Caminero A, et al. Gluten-degrading bacteria are present in the human small intestine of healthy volunteers and celiac patients. Res Microbiol 2017; 168: 673–684. [DOI] [PubMed] [Google Scholar]
- 61. Liu J, Xue Y, Zhou L. Detection of gastritis-associated pathogens by culturing of gastric juice and mucosa. Int J Exper Pathol 2018; 11: 2214–2220. [PMC free article] [PubMed] [Google Scholar]
- 62. Fujii R, Saito Y, Tokura Y, et al. Characterization of bacterial flora in persistent apical periodontitis lesions. Oral Microbiol Immunol 2009; 24: 502–505. [DOI] [PubMed] [Google Scholar]
- 63. Zhao H, Chu M, Huang Z, et al. Variations in oral microbiota associated with oral cancer. Sci Rep 2017; 7: 11773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dingemanse C, Belzer C, van Hijum SA, et al. Akkermansia muciniphila and Helicobacter typhlonius modulate intestinal tumor development in mice. Carcinogenesis 2015; 36: 1388–1396. [DOI] [PubMed] [Google Scholar]
- 65. Png CW, Lindén SK, Gilshenan KS, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 2010; 105: 2420–2428. [DOI] [PubMed] [Google Scholar]
- 66. Santacruz A, Collado MC, García-Valdés L, et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br J Nutr 2010; 104: 83–92. [DOI] [PubMed] [Google Scholar]
- 67. Everard A, Lazarevic V, Derrien M, et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes 2011; 60: 2775–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Weir TL, Manter DK, Sheflin AM, et al. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One 2013; 8: e70803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mira-Pascual L, Cabrera-Rubio R, Ocon S, et al. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol 2014;50: 167–179. [DOI] [PubMed] [Google Scholar]
- 70. Ryffel B, Ganesh BP, Klopfleisch R, et al. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella typhimurium-infected gnotobiotic mice. PLoS One 2013; 8: e74963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang JF, Zheng JY, Shi WY, et al. Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut 2018; 67: 1614–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang X, Zhang DY, Jia HJ, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med 2015; 21: 895–906. [DOI] [PubMed] [Google Scholar]
- 73. Li J, Zhao FQ, Wang YD, et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017; 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Caliński T, Harabasz J. A dendrite method for cluster analysis. Commun Stat Theory Methods 1974; 3: 1–27. [Google Scholar]
- 75. Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 1987; 20: 53–65. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Authorship-Change-Request-Form-P1 for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Authorship-Change-Request-Form-P2 for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Authorship-Change-Request-Form-P3 for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Supplementary_Figure_S1.Bray-Curitis_distance_between_all_samples.svg for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Supplementary_Figure_S2.Shift_of_gut_enterotypes_in_patients_with_FOLFOX_treatment for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Supplementary_Figure_S3._Genus_construction_of_enterotype_changed_patients_during_FOLFOX_treatment for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Table_S1_Patients_clinical_background_information for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Table_S2_Data_information_of_HiSeq_sequencing for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Table_S3__Relative_abundance_profile_at_each_taxonomy_level for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Table_S4__AUC_of_each_combination for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Table_S5._Detailed_information_for_gene_markers for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Table_S6__Pathway_information for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease
Supplemental material, Table_S7_Enterotype_information for Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX by Jing Li, Jingtao Li, Na Lyu, Yue Ma, Fei Liu, Yuqing Feng, Li Yao, Zhiyong Hou, Xiaofeng Song, Hongchuan Zhao, Xiaoya Li, Yingdian Wang, Cheng Xiao and Baoli Zhu in Therapeutic Advances in Chronic Disease