Abstract
Vertebrate protein SAMHD1 (sterile-α-motif and HD domain containing protein 1) regulates the cellular dNTP (2′-deoxynucleoside-5′-triphosphate) pool by catalysing the hydrolysis of dNTP into 2′-deoxynucleoside and triphosphate products. As an important regulator of cell proliferation and a key player in dNTP homeostasis, mutations to SAMHD1 are implicated in hypermutated cancers, and germline mutations are associated with Chronic Lymphocytic Leukaemia and the inflammatory disorder Aicardi–Goutières Syndrome. By limiting the supply of dNTPs for viral DNA synthesis, SAMHD1 also restricts the replication of several retroviruses, such as HIV-1, and some DNA viruses in dendritic and myeloid lineage cells and resting T-cells. SAMHD1 activity is regulated throughout the cell cycle, both at the level of protein expression and post-translationally, through phosphorylation. In addition, allosteric regulation further fine-tunes the catalytic activity of SAMHD1, with a nucleotide-activated homotetramer as the catalytically active form of the protein. In cells, GTP and dATP are the likely physiological activators of two adjacent allosteric sites, AL1 (GTP) and AL2 (dATP), that bridge monomer–monomer interfaces to stabilise the protein homotetramer. This review summarises the extensive X-ray crystallographic, biophysical and molecular dynamics experiments that have elucidated important features of allosteric regulation in SAMHD1. We present a comprehensive mechanism detailing the structural and protein dynamics components of the allosteric coupling between nucleotide-induced tetramerization and the catalysis of dNTP hydrolysis by SAMHD1.
Keywords: allosteric regulation, HD domain, HIV, hydrolase, oligomerization, SAMHD1
Introduction
Sterile-α-motif and HD domain containing protein 1 (SAMHD1) is a vertebrate, cellular protein that catalyses the hydrolysis of 2′-deoxynucleoside-5′-triphosphate (dNTP) into 2′-deoxynucleoside and triphosphate (Figure 1A) [1–6]. The replication of some DNA viruses and several retroviruses, such as HIV-1, is restricted by the catalytic activity of SAMHD1, which depletes the cellular dNTPs required for viral DNA synthesis [7–19].
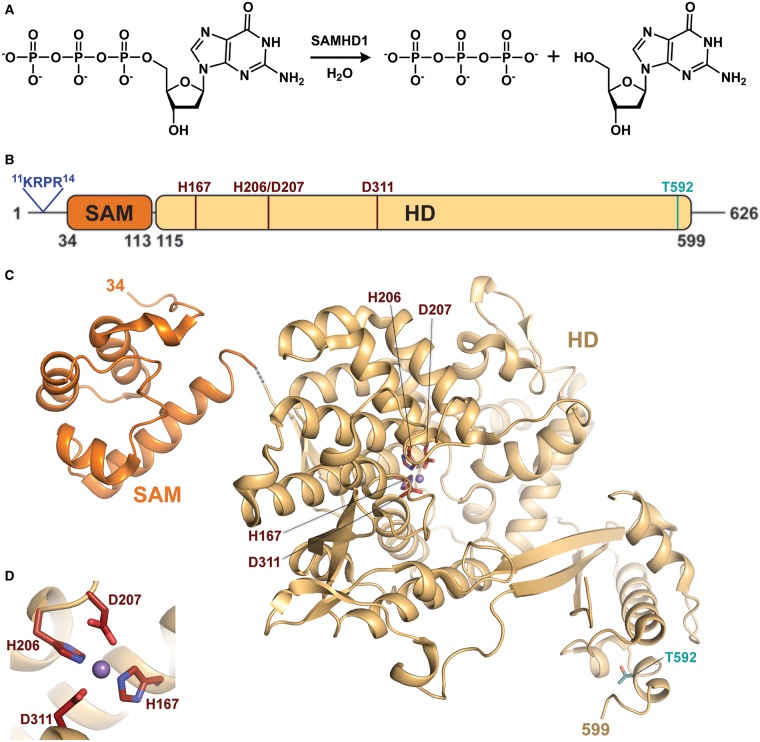
Figure 1. SAMHD1 structure and catalytic activity.
(A) The dNTP triphosphohydrolysis reaction catalysed by SAMHD1, dGTP is shown as the example substrate. (B) Domain organisation of human SAMHD1, showing the nuclear localisation signal in blue, the SAM domain in dark orange, the HD domain in light orange, the HD motif residues in maroon and phosphorylated residue Thr592 in teal. (C) Left: NMR structure of the human SAMHD1 SAM domain (PDB code: 2E8O residues 34–113). Right: X-ray crystal structure of WT human SAMHD1 HD domain (PDB code: 4BZC, monomer A, residues 115–599) [85]. The structures of the two domains are connected by a short, dotted and grey line. The HD motif-co-ordinated manganese ion is shown as a purple sphere, and the HD motif residues are shown as maroon sticks. (D) A close-up view of the HD motif-co-ordinated manganese ion in the catalytic site. PyMOL was used to prepare the structural figures.
Tight regulation of the cellular dNTP concentration, and of the balance among the individual dNTPs (dATP, dGTP, dCTP and TTP), is required to prevent genomic instability and tumourigenesis [20,21]. SAMHD1 mutations have been shown to alter cellular dNTP concentrations [22,23] and are associated with the development of certain cancers [23–29] (reviewed comprehensively by Mauney and Hollis [30]). Furthermore, SAMHD1 catalytic activity modulates the efficacy of anti-cancer and anti-viral nucleoside analogues [31–40].
In addition to its dNTP triphosphohydrolase activity, it was reported that SAMHD1 possesses nuclease activity against DNA and RNA substrates [41–45]. However, the observed nuclease activity could not be replicated in other studies [46–48] and has been associated with a contaminant co-purified with recombinantly expressed SAMHD1 [46]. Despite lacking intrinsic nuclease activity, SAMHD1 binds nucleic acids [41,46,48–51], with selectivity for single-stranded over double-stranded oligonucleotides [46,50], and for RNA over DNA [46,49].
SAMHD1 may utilise dNTP hydrolase activity, nucleic acid-binding or a scaffolding activity in other cellular roles such as in DNA replication [52], DNA repair [53] and inhibiting LINE-1 retrotransposition [54–57]. Germline mutation to SAMHD1 or any of six other genes (TREX1, RNaseH2A, RNaseH2B and RNaseH2C, ADAR1 and IFIH1) can cause the auto-inflammatory condition Aicardi–Goutières Syndrome [30,58–65]. More broadly, SAMHD1 appears to repress immune responses to viral infection and other inflammatory stimuli [52,60,66–68]. However, these mechanisms remain to be elucidated.
Regulation of cellular dNTP levels
The dNTP hydrolase activity of SAMHD1 is essential for both cellular dNTP homeostasis and regulating cell proliferation [24,29,69]. While SAMHD1 catalyses dNTP hydrolysis to reduce the cellular dNTP pool, several enzymes act antagonistically to increase the dNTP pool by catalysing dNTP synthesis either de novo or via salvage pathways. dNTPs are continually synthesised and degraded throughout the cell cycle, with the highest rates of dNTP flux occurring in S-phase [70]. dNTP levels in mammalian cells are ∼10- to 18-fold higher in S-phase than G0/G1 [70–72], and dNTP synthesis must continue during S-phase to complete chromosomal replication [72–75].
The catalytic activity of SAMHD1 is tightly controlled throughout the cell cycle through a mechanism of phosphorylation and dephosphorylation [3]. In S-phase, SAMHD1 appears to be phosphorylated at residue Thr592 by cyclin-dependent kinase 1 or 2 and cyclin A (CDK1/2-cyclinA) to lower the rate of dNTP hydrolysis [76–79]. At the end of M-phase, SAMHD1 catalytic activity is recovered due to dephosphorylation by phosphatase PP2A-B55α [80]. SAMHD1 expression levels may also vary throughout the cell cycle to further regulate dNTP hydrolase catalytic activity [3,4,30].
Allostery
In addition to post-translational regulation, SAMHD1 is subject to allosteric regulation by nucleotides to fine-tune its catalytic activity. Allosteric regulation occurs when the binding of a ligand at one site affects the affinity of ligand binding or catalysis at a second site in the same protein. Allosteric effects can be positive, termed ‘allosteric activation’, or negative, ‘allosteric inhibition’. Allostery is observed in many proteins, from single-domains to large multimeric complexes [81–84]. SAMHD1 is allosterically activated by nucleotide binding in two allosteric sites, AL1 and AL2 [5,6,85–88]. However, there is little evidence to suggest allosteric site coordination modifies catalytic site selectivity in SAMHD1 [87]. This review focuses on the mechanism by which allostery regulates human SAMHD1 catalysis and so the experiments described refer to human SAMHD1 studies, unless explicitly stated otherwise.
SAMHD1 domain organisation
Human SAMHD1 is a 626-residue protein that contains an N-terminal nuclear localisation signal, 11KRPR14 [89], a sterile-α-motif (SAM) domain (residues 34–113) and an HD catalytic domain (residues 115–599; Figure 1B–D). The SAM domain is an α-helical domain with an unknown function in SAMHD1 [90,91]. The HD catalytic domain is a phosphohydrolase domain that is named after the two pairs of Histidine–Aspartate residues (His167, His206, Asp207 and Asp311) that co-ordinate a metal ion at the catalytic site (Figure 1D) [92]. The HD domain of SAMHD1 is itself sufficient to catalyse the hydrolysis of dNTPs into 2′-deoxynucleoside and triphosphate [3,5,6,51,85,86,93].
Structural studies on human SAMHD1 have primarily focussed on the HD catalytic domain, as the flexibility of the linker connecting the SAM and HD domains has hindered structural studies on full-length human SAMHD1. More recently, X-ray crystal structures have been determined for mouse SAMHD1 containing both SAM and HD domains [94], providing some insight into how the human SAM and HD domains may interact with one another.
Nucleotide-dependent tetramer assembly
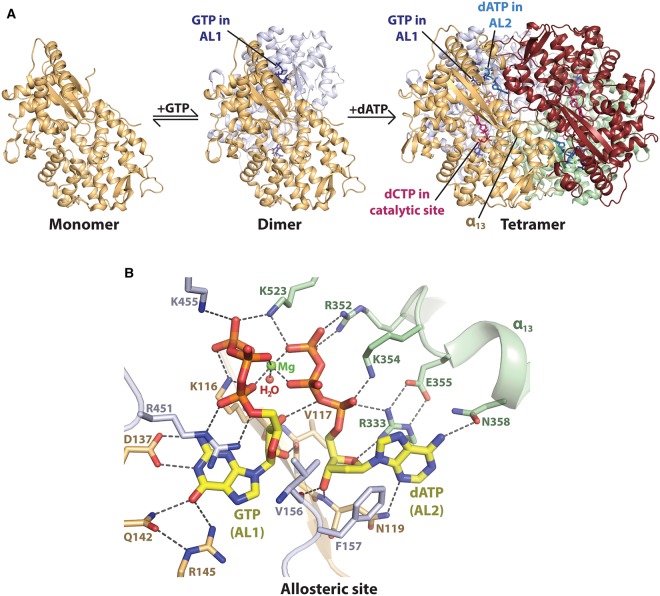
SAMHD1 is catalytically active as a homotetramer [93]. The HD domain tetramerises in a nucleotide-dependent manner (Figure 2A), with crystal structures revealing how nucleotides bind at two allosteric sites, AL1 and AL2, per protein monomer (Figure 2B) [85–88,95]. AL1 is specific for guanine-based nucleotides [85,87,96], which stabilise HD domain dimerisation [97]. The second allosteric site, AL2, lies adjacently to AL1 and is specific for a dNTP, which supports the dimer–dimer interactions required for tetramerization [85–88,95,97]. A magnesium ion co-ordinates the phosphoester oxygens of the nucleotides in AL1 and AL2 to further stabilise the tetramer [85], which explains the observation that SAMHD1 catalytic activity is magnesium-dependent [6]. Overall, four pairs of AL1–AL2 activators, each bridged by a magnesium ion, stabilise the SAMHD1 tetramer.
Figure 2. Nucleotide-dependent tetramerization of SAMHD1 HD domain.
(A) Ordered pathway for tetramer assembly, proposed by Hansen et al. [97], and elucidated structurally using X-ray crystallography [77,78,85–88,94,95,101]. The four protein monomers of SAMHD1 are shown in light orange, grey, maroon and light green. From left to right: The apo SAMHD1 HD domain is in a monomer–dimer equilibrium with GTP (dark blue), which stabilises dimerisation by coordinating AL1 (PDB code: 4RXO) [95]. dATP (light blue) co-ordinates AL2, and a magnesium ion (green sphere) bridges adjacent AL1–AL2 allosteric sites to stabilise the HD domain tetramer (PDB code: 4RXP) [95]. dCTP in the catalytic site is shown in pink. (B) GTP–dATP coordination at adjacent allosteric sites, AL1–AL2, of an HD domain tetramer (PDB code: 4TO0) [87]. GTP in AL1 and dATP in AL2 are shown in stick representation, with Cα atoms shown in yellow. Hydrogen-bonding interactions are represented by dashed lines.
GTP is the likely physiological activator of AL1 [87,97–99], as the cellular concentration of GTP is greater than that of the other guanine-based nucleotides [100], and, unlike dGTP, GTP is not a substrate of SAMHD1 [5]. In AL1, the guanine base of a co-ordinated nucleotide is recognised through a hydrogen-bonding network involving residues Asp137, Gln142 and Arg145 [85,87,95]. AL1 has a preference for GTP ≥ dGTP > ddGTP [77,97,98], due to hydrogen bonds formed between the GTP ribose and the backbone carbonyl of Val117 and the adjacent AL2-co-ordinated dNTP [87,88,95]. The preference in AL1 for GTP > GDP >> GMP [98] is due to extensive salt-bridges formed between the GTP triphosphate, Lys116 of one SAMHD1 monomer, and Arg451 and Lys455 of a second monomer [85,86], explaining how AL1-coordination stabilises dimerisation.
In AL2, the bulky side chains of Val156 and Phe157 prevent binding by nucleotides functionalised at the 2′-ribose position, such as ribonucleoside-5′-triphosphates (NTPs), although fluoro-substitution at the 2′-proS position is tolerated [38,87,101]. AL2 selects for dNTPs over 2′,3′-dideoxynucleoside triphosphates (ddNTPs) due to two hydrogen bonds formed between the 3′-hydroxyl of a dNTP and the protein backbone of Asn119 and Val156 [85,86]. Salt-bridges between the dNTP triphosphate and residues Arg333, Arg352 and Lys354 further stabilise nucleotide coordination in AL2. Additionally, Lys523 forms salt-bridges with the γ-phosphates of each AL1–AL2 pair to select for triphosphorylated nucleotides in AL1 and AL2.
All four canonical dNTPs can co-ordinate AL2, with a preference for dATP > dGTP > TTP > dCTP [6,87,88]. The polar side chains of residues Asn119 and Asn358 and several water molecules adapt their hydrogen-bonding network to accommodate all four bases in AL2 [87]. The preference in AL2 for the purine nucleotides dATP and dGTP, over the pyrimidines TTP and dCTP, is due to more extensive cation-π stacking of the larger purine bases with the guanidino side chain of Arg333 [87]. Ji et al. [87] proposed that dATP is the primary activator of AL2, as they observed a stronger salt-bridge formed between the side chains of Arg333 and Glu355 only when dATP is bound in AL2. In contrast, dCTP is a poor AL2-activator of SAMHD1 [88], likely due to the inability of the cytosine base to form a direct hydrogen bond with Asn358.
SAMHD1 tetramerization is essential for catalysis
Extensive experimental evidence supports the hypothesis that SAMHD1 tetramerization is essential for catalysing dNTP hydrolysis [85,86,88,93,97]. Firstly, dGTP, which can co-ordinate AL1, AL2 and the catalytic site, is hydrolysed by SAMHD1 in vitro in the absence of additional nucleotides [5,6,86,93], whereas the three other canonical dNTPs (dATP, dCTP and TTP) are only hydrolysed by SAMHD1 in the presence of AL1-activating GTP or dGTP [5,6,98]. Secondly, point mutations to key residues in AL1 (D137A, Q142A, R145A and R451E), AL2 (R333E) and the dimer–dimer interface (D361K, H364K and R372D) reduced SAMHD1 tetramerization and dNTP hydrolysis in vitro [77,85,86,88,93,95,102]. Finally, N- and C-terminal truncations in constructs 120–626 (Δ115–119) and 115–583 (Δ584–626), respectively, abolished tetramerization and reduced catalytic activity in comparison with constructs 1–626 and 115–626 [5,51,77], demonstrating the importance of tetramerization for catalysis.
Long-lived, activated state of SAMHD1 corresponds to the homotetramer
Hansen et al. [97] observed that GTP and dNTPs, or dGTP alone, generate a long-lived, activated state of SAMHD1 that corresponds to the SAMHD1 homotetramer. Furthermore, the activated, homotetrameric state of SAMHD1 is not in equilibrium with free GTP or dNTP activators in solution. Strikingly, the SAMHD1 homotetramer persisted in vitro for hours without further exchange of nucleotides in AL1 or AL2 [77,97]. It is proposed that this slow rate of tetramer dissociation, despite activator depletion, enables SAMHD1 to deplete cellular dNTP concentrations to the nanomolar concentrations observed in macrophages and resting CD4+ T-cells.
Thr592 phosphorylation destabilises the tetramer
Phosphorylation of human SAMHD1 residue Thr592 by CDK1/2-cyclinA regulates catalytic activity throughout the cell cycle [76–78]. Phosphorylation of residue Thr592 in vitro or introducing the phosphomimetic mutation T592E reduced tetramerization and catalytic activity [77,78]. The mutation T592E also eliminated the ability of SAMHD1 to restrict HIV-1 infection in macrophage-like PMA-differentiated U937 cells [77]. Similarly, another phosphomimetic mutation, T592D, also impaired the ability of SAMHD1 to block the lytic replication of the Epstein–Barr herpesvirus in producer Akata cells [103].
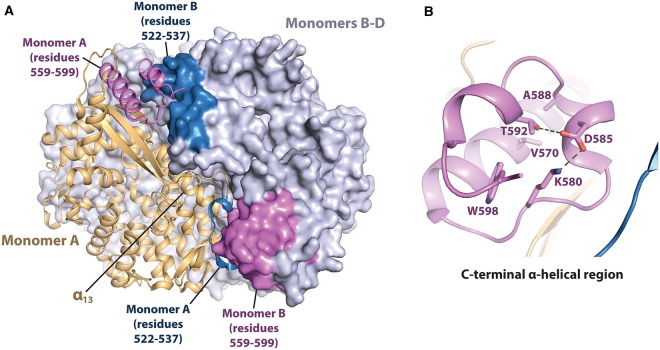
SAMHD1 residue Thr592 is buried in an α-helical region spanning residues 559–599, and this region interacts with residues 522–537 of an adjacent monomer at the dimer–dimer interface in the SAMHD1 tetramer (Figure 3A). From crystal structures, it appears that phospho-T592 would experience electrostatic repulsion with the adjacent acidic residue Asp585 and may sterically clash with hydrophobic residues Val570 and Trp598 (Figure 3B) [77,78]. Correspondingly, crystal structures of phospho-T592 or T592E SAMHD1 reveal that phosphorylating Thr592 or making the phosphomimetic mutation T592E disrupts local protein folding, causing the C-terminal residues 585–599 to become disordered in the crystal lattice [77,78].
Figure 3. Thr592 phosphorylation modulates SAMHD1 tetramer stability.
(A) X-ray crystal structure of SAMHD1 HD domain tetramer (PDB code: 4BZC) [85]. Monomer A is shown in cartoon representation in light orange. Monomers B–D are shown in surface representation in grey. Residues 559–599 are in pink and residues 522–537 are in blue. (B) C-terminal, the α-helical region between residues 559–599, comprising phosphorylated residue Thr592.
Molecular dynamics (MD) simulations by Patra et al. [104] showed that mutation T592E caused minor local perturbations to residues 585–595, but did not affect the integrity of the allosteric or catalytic sites on the timescale modelled. Further analysis of correlated motions across the SAMHD1 tetramer in the MD simulations revealed that the mutation T592E decoupled a signalling pathway between residue Thr592 and the allosteric sites, and increased the dynamic coupling between Thr592 and α-helix 13 (α13; residues 352–375) at the dimer–dimer interface [105]. The authors concluded that phosphorylation of Thr592 may trigger a loosening of the HD domain tetramer.
Catalytic site selectivity
The SAMHD1 catalytic site can accommodate all four canonical dNTP substrates (dATP, dGTP, dCTP and TTP), as well as dUTP and a variety of dNTP analogues (Figure 4). In each case, SAMHD1 catalyses their hydrolysis into triphosphate and 2′-deoxynucleoside products. As all four canonical dNTPs can be accommodated in the catalytic site, they also act as competitive inhibitors of one another's hydrolysis in mixed dNTP pools [87,97,98]. Chromatography-based dNTP hydrolysis experiments, which were performed in the presence of GTP and all four dNTPs to minimise allosteric effects on catalysis, demonstrated the following rank order for dNTP hydrolysis rate: dGTP > dCTP > TTP > dATP [5,6,87]. Co-crystallization experiments by Ji et al. [87] supported a catalytic site-binding preference of: dCTP > dGTP ≈ TTP > dATP.
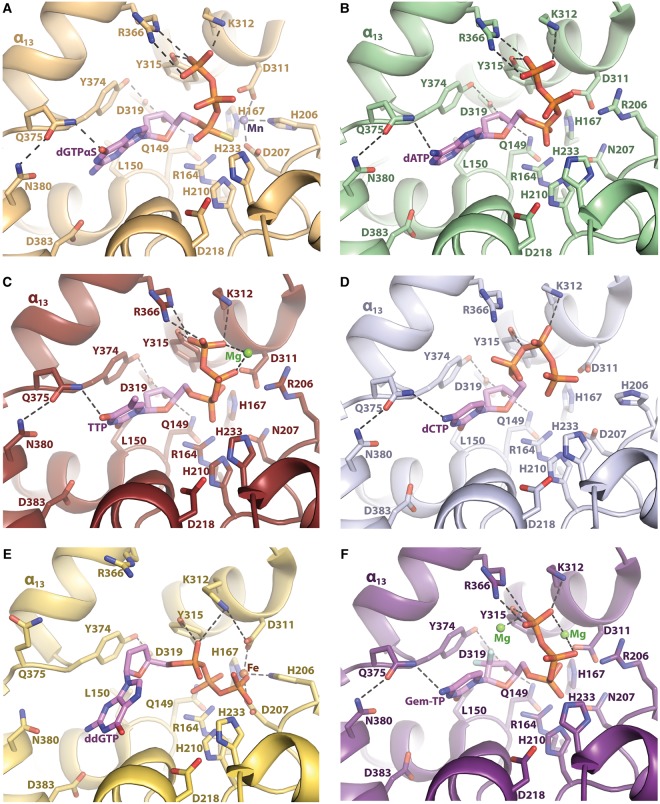
Figure 4. SAMHD1 catalytic site accommodates dNTPs and dNTP analogues.
(A–F) Nucleotide coordination in the catalytic site of the human SAMHD1 HD domain. (A) WT SAMHD1 with dGTPαS (PDB: 4BZC) [85]. (B) H206R/D207N SAMHD1 with dATP (PDB: 4QG1) [88]. (C) H206R/D207N SAMHD1 with TTP (PDB: 4TNZ) [87]. (D) WT SAMHD1 with dCTP (PDB: 4RXR) [95]. (E) WT SAMHD1 with ddGTP (PDB: 5AO1) [77]. (F) H206R/D207N SAMHD1 with gemcitabine-TP (Gem-TP) (PDB: 6DW5) [101]. Nucleotides are shown in stick representation with Cα atoms in pink. Manganese (purple), magnesium (green) and iron (brown) metal ions are represented as spheres.
Base selectivity in the catalytic site is achieved through subtle differences in the hydrogen bonding between a dNTP base, its network of hydrating water molecules and residues Leu150, Tyr374, Gln375, Asn380 and Asp383, which line the catalytic pocket [87,88]. The side chains of residues Leu150, Tyr315 and Tyr374 form a tight-binding pocket around the base and 2′-deoxyribose moieties of a dNTP substrate [38,85,101]. The 3′-hydroxyl group on a dNTP substrate is hydrogen bonded by the polar side chains of residues Gln149 and Asp319 [86,88]. Nucleotide binding in the catalytic site is further stabilised by salt-bridges between the triphosphate and the basic side chains of Arg164, Lys312 and Arg366 [85,88].
In addition to canonical dNTPs and dUTP, the SAMHD1 catalytic site can co-ordinate and hydrolyse particular dNTP analogues. The poorly hydrolysed analogue ddGTP co-ordinates the SAMHD1 catalytic site, as revealed through crystal structures (Figure 4E) [77]. Knecht et al. [101] solved co-crystal structures in which the catalytic site was occupied by the triphosphorylated forms of anti-cancer drugs cladribine, clofarabine, fludarabine, cytarabine and gemcitabine (Figure 4F), and the anti-viral agent vidarabine. The authors’ structural and biophysical studies revealed that the catalytic site could tolerate fluoro- and chloro-substitutions at the carbon-2 position on an adenine base, fluoro- and hydroxyl-substitutions at the 2′-proS ribose position, and a fluoro-group at the 2′-proR position. Such substitutions at the 2′-proS position are tolerated by a compensatory rotation of the ribose moiety of these analogues within the catalytic pocket. Leu150 and Tyr374 side chains prevent bulkier functionalisation at the 2′-proR position, while Tyr315 prevents functionalisation to the 3′-proS position [38,85,101].
dNTP geometry in the catalytic site
Numerous crystal structures have been solved of the SAMHD1 HD domain with dNTPs or dNTP analogues in the catalytic site [77,78,85–88,94,95,101]. Frequently, the inactivating double mutation H206R/D207N has been employed in these studies [78,85,87,88,101]. The H206R/D207N mutation to the HD motif (His167, His206, Asp207 and Asp311) prevents coordination of a metal ion at the HD motif and eliminates catalytic activity in SAMHD1 [85,93]. Metal ion coordination at the HD motif is likely important for catalysis, as a further HD motif mutant, D311A, is also catalytically inactive [5,44,106].
While it is possible that dNTP or dNTP analogue coordination may be perturbed in crystal structures of SAMHD1 mutant H206R/D207N (Figure 4B,C,F), a similar binding mode is observed for the analogue dGTPαS in a wild-type (WT) catalytic site (Figure 4A) [85]. The consensus between independently reported H206R/D207N-dNTP and WT-dGTPαS structures (Figure 4A–C) [85,87,88] suggests there may be a physiological basis for this nucleotide-binding mode in the catalytic site. Therefore, it could be postulated that these non-catalytically competent SAMHD1-nucleotide structures represent enzyme–substrate complexes prior to catalysis.
In comparison, a different triphosphate geometry is modelled in the crystal structures of catalytically competent WT–dNTP complexes (Figure 4D) [86,95]. The base and 2′-deoxyribose portions of the dNTP ligands superimpose with those of non-catalytically competent H206R/D207N-dNTP structures. However, the triphosphate moiety is modelled in different configurations. Thus, the WT–dNTP structures may represent intermediate- or product-like states during catalysis.
Structures of WT SAMHD1 with the poorly hydrolysed analogue ddGTP reveal a further binding mode for the nucleotide in the catalytic site [77], with ddGTP less well buried within the catalytic pocket (Figure 4E). The WT–ddGTP crystal structures reveal a unique substrate-binding mode that may be required for ddGTP hydrolysis, but importantly could represent a nucleotide-bound state along the dNTP substrate-binding pathway of SAMHD1. Further SAMHD1-nucleotide structural studies may be required to elucidate nucleotide-binding modes at various stages of catalysis, including substrate binding, hydrolysis and product release.
Catalytic mechanism
The chemical reaction catalysed by SAMHD1 was initially identified through chromatography-based experiments in which dNTP substrates were demonstrated to be hydrolysed directly into 2′-deoxynucleoside and triphosphate products (Figure 1A), rather than by sequential monophosphate cleavages via 2′-deoxynucleoside-5′-diphosphate (dNDP) and 2′-deoxynucleoside-5′-monophosphate (dNMP) intermediates [5,6]. The catalytic mechanism was further investigated using mass spectrometry experiments that determined oxygen from bulk water is incorporated into the triphosphate product, rather than the 2′-deoxynucleoside product, supporting a mechanism of nucleophilic attack on the α-phosphorous that results in cleavage of the α-phosphorous-to-5′-oxygen covalent bond [107].
In addition to residues in the HD motif, residues His210, Asp218 and His233 have been proposed to be important for catalysis, based on the observation that mutations H210A and H233A disrupt catalysis [88], and on the conservation of these three residues across HD phosphohydrolase domains, including in the homologous protein EF1143 from the bacterium Enterococcus faecalis [6,85,108]. Furthermore, a crystal structure of mutant H210A was found to lack nucleotide coordination in the catalytic site, supporting a function for residue His210 in substrate dNTP coordination [88].
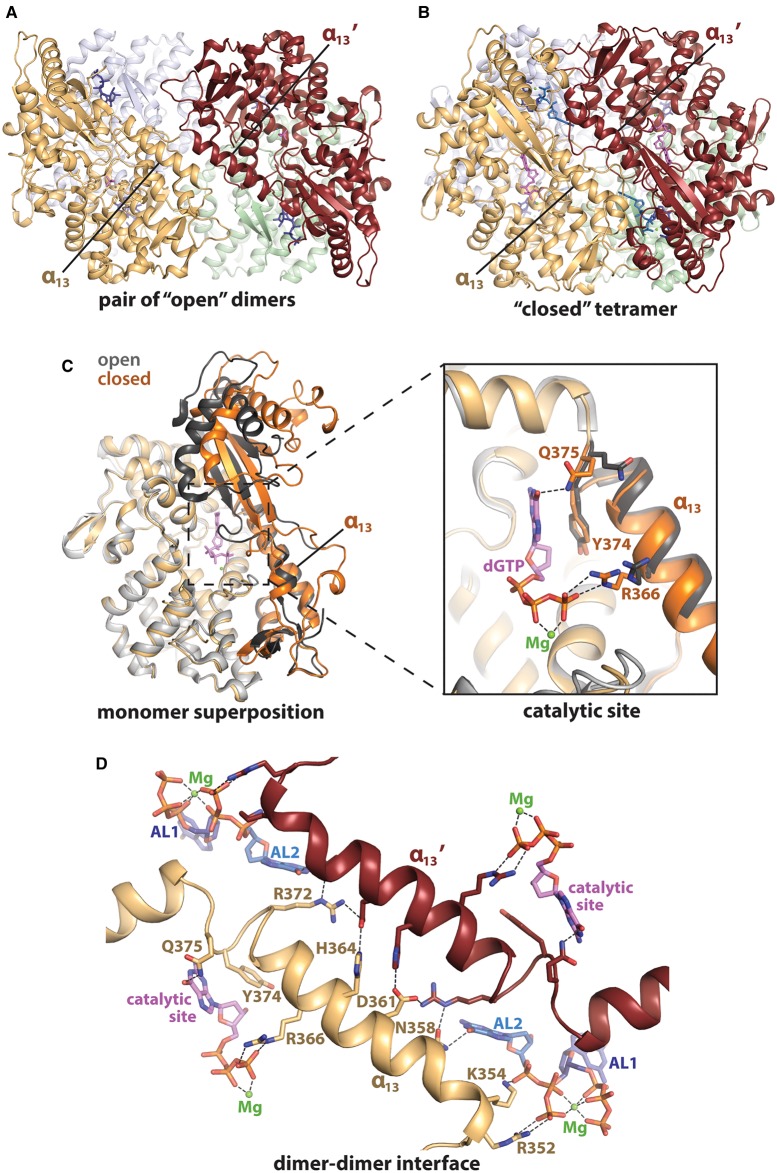
‘Open’ and ‘closed’ HD domain conformations
Crystal structures of human SAMHD1, either apo or with nucleotides co-ordinated, reveal that the HD domain contains intrinsic conformational flexibility [5,77,85,86,95,102], adopting two distinct conformations in crystal structures, which we term the ‘open’ and ‘closed’ conformations (Figure 5A–C). In the absence of co-ordinated nucleotides, or with only GTP bound in AL1, the SAMHD1 HD domain adopts an ‘open’ conformation (Figure 5A), with a more expanded catalytic site pocket, and is disordered between residues 278–283, 507–546 and 583–599 [5,77,95,102]. In the crystal lattices of ‘open’ structures, the HD domains are arranged in dimeric repeating units. These dimeric units likely correspond to the SAMHD1 dimer of the monomer–dimer equilibrium that is present in solution in the absence of nucleotides or in the presence of only GTP [93,97].
Figure 5. ‘Open’ and ‘closed’ conformations of HD domain.
(A) X-ray crystal structure of the HD domain in ‘open’ conformation co-ordinated to GTP (dark blue) in AL1 (PDB: 4RXO) [95]. The tetramer comprises two dimers, with one formed from the orange and grey monomers, and the second from the maroon and light green monomers. (B) X-ray crystal structure of HD domain in ‘closed’ tetrameric conformation, with dGTP co-ordinated in AL1, AL2 and the catalytic site (PDB: 4BZB) [85]. dGTP is shown in dark blue (AL1), pale blue (AL2) or pink (catalytic site) and magnesium ions are shown as green spheres. (C) Superposition of an HD domain monomer in ‘open’ (grey) and ‘closed’ (orange) conformations. Residues 326–375 and 454–599 differ in their conformation between the ‘open’ and ‘closed’ states and are represented by darker shades of grey and orange, respectively. The dGTP- and magnesium-coordination observed in the catalytic site of the ‘closed’ state is displayed, while the manganese- and phosphate-coordination observed in the ‘open’ state is not shown. (D) Interactions formed by the α13 helix at the dimer–dimer interface (residues Asn358, Asp361, His364 and Arg372), in the catalytic site (Arg366, Tyr374 and Gln375) and in AL2 (Arg352, Lys354 and Asn358). One monomer is shown in pale orange, and its neighbouring monomer at the dimer–dimer interface is shown in maroon. dGTP in AL1 (dark blue), AL2 (pale blue) and the catalytic site (pink) is shown in stick representation. Magnesium ions are represented by green spheres. Hydrogen bonds are shown as dashed lines.
SAMHD1 HD domain crystal structures with nucleotides simultaneously bound in AL1, AL2 and the catalytic site adopt a so-called ‘closed’ conformation (Figure 5B) that is more compact about the catalytic site, and ordered to a greater extent, with density observed for the HD domain backbone for all residues between positions 115–599, except for a short loop between residues 278–283 [85]. In ‘closed’ structures, the HD domains assemble in the crystal lattice into homotetramers that contain D2 dihedral symmetry, whereby the four monomers are related to one another by three 2-fold symmetry axes.
Structural comparisons suggest that the HD domain must undergo a change in conformation during the dimer-to-tetramer transition to accommodate dNTPs into AL2 and the catalytic site. Secondary structural elements are conserved between dimeric (‘open’) and tetrameric (‘closed’) conformations of the SAMHD1 HD domain. However, between dimeric and tetrameric structures, motions of up to ∼5 Å affect tertiary packing in two regions of the protein, between residues 326–375, and 454–599 (Figure 5C) [85,86]. Several residues in these two regions are important for nucleotide coordination in AL1, AL2 and the catalytic site. Therefore, it is likely that these regions have important functions in allosteric regulation in SAMHD1.
Linkage between allosteric and catalytic sites
As described above, the HD domain conformation varies between dimeric (apo or AL1-occupied) and tetrameric (AL1-, AL2- and catalytic site-occupied) states of SAMHD1. While the majority of residues across the catalytic site do not appear to be significantly structurally perturbed during tetramer assembly, tertiary structural changes alter the positioning of catalytic site residues Arg366 and Gln375 (Figure 5C), which lie on one face of α13 [85,86]. Residues Arg366 and Gln375 are involved in dNTP coordination in the catalytic site of closed, tetrameric SAMHD1 structures, but appear too distal for substrate coordination in open, dimeric structures that lack dNTPs in AL2 and the catalytic site.
Helix α13, which spans residues 352–375, bridges the catalytic and allosteric sites, and makes important interactions at the dimer–dimer interface (Figure 5D) [86]. Catalytic site residues Arg366 and Gln375 are at the C-terminal end of α13, while residues Arg352, Lys354 and Asn358 at the N-terminal end of α13 are involved in dNTP coordination in AL2. At the dimer–dimer interface, the α13 helix of one monomer interacts with the neighbouring monomer's helix, α13', through a network of salt-bridges and hydrogen bonds involving residues Asn358, Asp361, His364 and Arg372 from both α13 and α13' elements. Thus, α13 appears to be crucial for allosteric regulation, by communicating allosteric site occupancy and tetramerization to the catalytic site, with residues Arg366 and Gln375 supporting substrate dNTP binding once AL2 is occupied and the protein has tetramerized.
In addition to structural changes in the catalytic site, HD domain tetramerization likely alters protein dynamics. Patra et al. [104,105] explored mechanisms for cross-talk between allosteric and catalytic sites in SAMHD1 using correlation analysis of MD simulations. The authors observed that correlated motions between allosteric and catalytic sites were reciprocated across the HD domain tetramer, revealing both short-range and long-range allosteric signal transduction in SAMHD1. Furthermore, removing dATP from one AL2 site in the SAMHD1 HD domain tetramer significantly reduced the rigidity of the protein around the dATP-occupied catalytic site. In separate MD simulations, Cardamone et al. [109] observed that removing all nucleotides and magnesium ions from the protein tetramer weakened α13–α13' interactions at the dimer–dimer interface, and the AL1 mutation R145E accelerated the destabilisation of the tetramer [110]. Overall, biophysical and computational experiments demonstrate that interactions at the dimer–dimer interface and nucleotide occupancy at the allosteric sites modulate the catalytic site structure and dynamics in order to regulate catalysis.
Summary
Allosteric site occupancy and HD domain tetramerization control both the structural integrity and the rigidity of the SAMHD1 catalytic site [85,86,105,109,110]. In the absence of nucleotide coordination in AL1, AL2 and the catalytic site, SAMHD1 exists in a monomer–dimer equilibrium [93,97]. GTP or dGTP binding in AL1 increases the proportion of dimeric SAMHD1 [97] and is necessary for subsequent dNTP binding in AL2 [97]. Changes in tertiary structure and protein dynamics result from the dNTP-induced dimer-to-tetramer transition, including structural perturbations to residues 326–375 and 454–599 [85,86], and changes in the dynamics of catalytic site residues, including His206, Tyr374 and Gln375 [104]. The catalytic site becomes more rigid upon nucleotide-induced tetramerization [104,105], and there appears to be an energetic coupling between nucleotide binding in AL2 and the catalytic site [97]. Subsequent to catalysis, the reaction products, 2′-deoxynucleoside and triphosphate, dissociate from SAMHD1 and the catalytic site of a closed tetramer appears sufficiently accessible for nucleotide exchange to occur without tetramer disassembly. Kinetic experiments demonstrate that the AL1- and AL2-co-ordinated tetramer is a long-lived, activated state, in which the AL1- and AL2-co-ordinated nucleotides are not in exchange with free nucleotides [97]. This is relevant to a cellular environment in which the dNTP pool has been largely depleted. Stable, active SAMHD1 tetramers persist [77,97] and hydrolyse dNTPs to drive the cellular dNTP pool to nanomolar concentrations that are observed in resting cells and are required for the restriction of HIV-1 replication.
Perspectives
SAMHD1 has important anti-viral, anti-cancer and anti-inflammation functions in the cell. SAMHD1 restricts HIV-1 replication in dendritic and myeloid lineage cells. Mutations to SAMHD1 have been identified in hypermutated cancers, and germline mutation to SAMHD1 can cause Chronic Lymphocytic Leukaemia and auto-immune condition Aicardi–Goutières Syndrome.
The dNTP triphosphohydrolase catalytic function of SAMHD1 is essential for HIV-1 restriction but also for cellular dNTP homeostasis. The catalytic domain of SAMHD1 tetramerizes in a nucleotide-dependent manner, with GTP and dATP coordinating two allosteric sites (AL1 and AL2) per SAMHD1 monomer to stimulate dNTP hydrolysis in the catalytic site. This review combines the results of biophysical, structural and MD studies to present a unified mechanism for the allosteric regulation of catalysis by SAMHD1.
X-ray crystallographic studies have revealed how nucleotide coordination in AL1 and AL2 stabilises catalytic domain tetramerization. However, it remains unclear how a substrate dNTP is co-ordinated in the WT catalytic site of SAMHD1 prior to catalysis and how SAMHD1 catalyses dNTP triphosphohydrolysis. Further studies are required to elucidate the catalytic mechanism of SAMHD1.
Acknowledgements
The Authors thank Dr Sarah J. Caswell, Dr Joshua D. Wright, Dr Matthew A. Cottee, Dr Paula Ordonez and Dr Chloe Ming-Han Tsai for their comments on the manuscript.
Abbreviations
- AL1
allosteric site 1 of SAMHD1
- AL2
allosteric site 2 of SAMHD1
- CDK1/2-cyclinA
cyclin-dependent kinase 1 or 2 and cyclin A
- ddNTP
2′,3′-dideoxynucleoside-5′-triphosphate
- dNDP
2′-deoxynucleoside-5′-diphosphate
- dNMP
2′-deoxynucleoside-5′-monophosphate
- dNTP
2′-deoxynucleoside-5′-triphosphate
- Gem-TP
gemcitabine-triphosphate
- HD
Histidine–Aspartate motif or domain
- MD
molecular dynamics
- NTP
ribonucleoside-5′-triphosphate
- PDB
Protein Data Bank
- SAM
sterile-α-motif domain
- SAMHD1
sterile-α-motif and HD domain containing protein 1
- WT
wild-type
- α13
α-helix 13
Author Contribution
E.R.M. wrote the manuscript. E.R.M. and I.A.T. edited the manuscript.
Funding
This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001178), the UK Medical Research Council (FC001178) and the Wellcome Trust (FC001178), and by a Wellcome Trust Senior Research Fellowship to I.A.T. (108014/Z/15/Z).
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Li N., Zhang W. and Cao X. (2000) Identification of human homologue of mouse IFN-gamma induced protein from human dendritic cells. Immunol. Lett. 74, 221–224 10.1016/S0165-2478(00)00276-5 [DOI] [PubMed] [Google Scholar]
- 2.Descours B., Cribier A., Chable-Bessia C., Ayinde D., Rice G., Crow Y. et al. (2012) SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology 9, 87 10.1186/1742-4690-9-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franzolin E., Pontarin G., Rampazzo C., Miazzi C., Ferraro P., Palumbo E. et al. (2013) The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc. Natl Acad. Sci. U.S.A. 110, 14272–14277 10.1073/pnas.1312033110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt S., Schenkova K., Adam T., Erikson E., Lehmann-Koch J., Sertel S. et al. (2015) SAMHD1's protein expression profile in humans. J. Leukoc. Biol. 98, 5–14 10.1189/jlb.4HI0714-338RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstone D.C., Ennis-Adeniran V., Hedden J.J., Groom H.C.T., Rice G.I., Christodoulou E. et al. (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 10.1038/nature10623 [DOI] [PubMed] [Google Scholar]
- 6.Powell R.D., Holland P.J., Hollis T. and Perrino F.W. (2011) Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286, 43596–43600 10.1074/jbc.C111.317628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lahouassa H., Daddacha W., Hofmann H., Ayinde D., Logue E.C., Dragin L. et al. (2012) SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228 10.1038/ni.2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sze A., Belgnaoui S.M., Olagnier D., Lin R., Hiscott J., van Grevenynghe J. et al. (2013) Host restriction factor SAMHD1 limits human T cell leukemia virus type 1 infection of monocytes via STING-mediated apoptosis. Cell Host Microbe 14, 422–434 10.1016/j.chom.2013.09.009 [DOI] [PubMed] [Google Scholar]
- 9.Gramberg T., Kahle T., Bloch N., Wittmann S., Müllers E., Daddacha W. et al. (2013) Restriction of diverse retroviruses by SAMHD1. Retrovirology 10, 26 10.1186/1742-4690-10-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollenbaugh J.A., Gee P., Baker J., Daly M.B., Amie S.M., Tate J. et al. (2013) Host factor SAMHD1 restricts DNA viruses in non-dividing myeloid cells. PLoS Pathog. 9, e1003481 10.1371/journal.ppat.1003481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z., Zhu M., Pan X., Zhu Y., Yan H., Jiang T. et al. (2014) Inhibition of Hepatitis B virus replication by SAMHD1. Biochem. Biophys. Res. Commun. 450, 1462–1468 10.1016/j.bbrc.2014.07.023 [DOI] [PubMed] [Google Scholar]
- 12.Jeong G.U., Park I.-H., Ahn K. and Ahn B.-Y. (2016) Inhibition of hepatitis B virus replication by a dNTPase-dependent function of the host restriction factor SAMHD1. Virology 495, 71–78 10.1016/j.virol.2016.05.001 [DOI] [PubMed] [Google Scholar]
- 13.Antonucci J.M., St. Gelais C., de Silva S., Yount J.S., Tang C., Ji X. et al. (2016) SAMHD1-mediated HIV-1 restriction in cells does not involve ribonuclease activity. Nat. Med. 22, 1072–1074 10.1038/nm.4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim B., Nguyen L.A., Daddacha W. and Hollenbaugh J.A. (2012) Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J. Biol. Chem. 287, 21570–21574 10.1074/jbc.C112.374843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.St. Gelais S., de Silva S., Amie S.M., Coleman C.M., Hoy H., Hollenbaugh J.A. et al. (2012) SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 9, 105 10.1186/1742-4690-9-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E. et al. (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 10.1038/nature10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hrecka K., Hao C., Gierszewska M., Swanson S.K., Kesik-Brodacka M., Srivastava S. et al. (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 10.1038/nature10195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baldauf H.M., Pan X., Erikson E., Schmidt S., Daddacha W., Burggraf M. et al. (2012) SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat. Med. 18, 1682–1687 10.1038/nm.2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berger A., Sommer A.F.R., Zwarg J., Hamdorf M., Welzel K., Esly N. et al. (2011) SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 7, e1002425 10.1371/journal.ppat.1002425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathews C.K. (2006) DNA precursor metabolism and genomic stability. FASEB J. 20, 1300–1314 10.1096/fj.06-5730rev [DOI] [PubMed] [Google Scholar]
- 21.Bester A.C., Roniger M., Oren Y.S., Im M.M., Sarni D., Chaoat M. et al. (2011) Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145, 435–446 10.1016/j.cell.2011.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansson P., Klein-Hitpass L., Choidas A., Habenberger P., Mahboubi B., Kim B. et al. (2018) SAMHD1 is recurrently mutated in T-cell prolymphocytic leukemia. Blood Cancer J. 8, 11 10.1038/s41408-017-0036-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rentoft M., Lindell K., Tran P., Chabes A.L., Buckland R.J., Watt D.L. et al. (2016) Heterozygous colon cancer-associated mutations of SAMHD1 have functional significance. Proc. Natl Acad. Sci. U.S.A. 113, 4723–4728 10.1073/pnas.1519128113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clifford R., Louis T., Robbe P., Ackroyd S., Burns A., Timbs A.T. et al. (2014) SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood 123, 1021–1031 10.1182/blood-2013-04-490847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landau D.A., Carter S.L., Stojanov P., McKenna A., Stevenson K., Lawrence M.S. et al. (2013) Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152, 714–726 10.1016/j.cell.2013.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuh A., Becq J., Humphray S., Alexa A., Burns A., Clifford R. et al. (2012) Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood 120, 4191–4196 10.1182/blood-2012-05-433540 [DOI] [PubMed] [Google Scholar]
- 27.Landau D.A., Tausch E., Taylor-Weiner A.N., Stewart C., Reiter J.G., Bahlo J. et al. (2015) Mutations driving CLL and their evolution in progression and relapse. Nature 526, 525–530 10.1038/nature15395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merati M., Buethe D.J., Cooper K.D., Honda K.S., Wang H. and Gerstenblith M.R. (2015) Aggressive CD8(+) epidermotropic cutaneous T-cell lymphoma associated with homozygous mutation in SAMHD1. JAAD Case Rep. 1, 227–229 10.1016/j.jdcr.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J.L., Lu F.Z., Shen X.-Y., Wu Y. and Zhao L.T. (2014) SAMHD1 is down regulated in lung cancer by methylation and inhibits tumor cell proliferation. Biochem. Biophys. Res. Commun. 455, 229–233 10.1016/j.bbrc.2014.10.153 [DOI] [PubMed] [Google Scholar]
- 30.Mauney C.H. and Hollis T. (2018) SAMHD1: recurring roles in cell cycle, viral restriction, cancer, and innate immunity. Autoimmunity 51, 96–110 10.1080/08916934.2018.1454912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ordonez P., Kunzelmann S., Groom H.C.T., Yap M.W., Weising S., Meier C. et al. (2017) SAMHD1 enhances nucleoside-analogue efficacy against HIV-1 in myeloid cells. Sci. Rep. 7, 42824 10.1038/srep42824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amie S.M., Daly M.B., Noble E., Schinazi R.F., Bambara R.A. and Kim B. (2013) Anti-HIV host factor SAMHD1 regulates viral sensitivity to nucleoside reverse transcriptase inhibitors via modulation of cellular deoxyribonucleoside triphosphate (dNTP) levels. J. Biol. Chem. 288, 20683–20691 10.1074/jbc.M113.472159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ballana E., Badia R., Terradas G., Torres-Torronteras J., Ruiz A., Pauls E. et al. (2014) SAMHD1 specifically affects the antiviral potency of thymidine analog HIV reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 58, 4804–4813 10.1128/AAC.03145-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider C., Oellerich T., Baldauf H.-M., Schwarz S.-M., Thomas D., Flick R. et al. (2017) SAMHD1 is a biomarker for cytarabine response and a therapeutic target in acute myeloid leukemia. Nat. Med. 23, 250–255 10.1038/nm.4255 [DOI] [PubMed] [Google Scholar]
- 35.Herold N., Rudd S.G., Ljungblad L., Sanjiv K., Myrberg I.H., Paulin C.B.J. et al. (2017) Targeting SAMHD1 with the Vpx protein to improve cytarabine therapy for hematological malignancies. Nat. Med. 23, 256–263 10.1038/nm.4265 [DOI] [PubMed] [Google Scholar]
- 36.Herold N., Rudd S.G., Sanjiv K., Kutzner J., Bladh J., Paulin C.B.J. et al. (2017) SAMHD1 protects cancer cells from various nucleoside-based antimetabolites. Cell Cycle 16, 1029–1038 10.1080/15384101.2017.1314407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rassidakis G.Z., Herold N., Myrberg I.H., Tsesmetzis N., Rudd S.G., Henter J.-I. et al. (2018) Low-level expression of SAMHD1 in acute myeloid leukemia (AML) blasts correlates with improved outcome upon consolidation chemotherapy with high-dose cytarabine-based regimens. Blood Cancer J. 8, 98 10.1038/s41408-018-0134-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hollenbaugh J.A., Shelton J., Tao S., Amiralaei S., Liu P., Lu X. et al. (2017) Substrates and inhibitors of SAMHD1. PLoS ONE 12, e0169052 10.1371/journal.pone.0169052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y.T., Yuan B., Chen H.-D., Xu L., Tian Y.-N., Zhang A. et al. (2018) Acquired resistance of phosphatase and tensin homolog-deficient cells to poly(ADP-ribose) polymerase inhibitor and Ara-C mediated by 53BP1 loss and SAMHD1 overexpression. Cancer Sci. 109, 821–831 10.1111/cas.13477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huber A.D., Michailidis E., Schultz M.L., Ong Y.T., Bloch N., Puray-Chavez M.N. et al. (2014) SAMHD1 has differential impact on the efficacies of HIV nucleoside reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 58, 4915–4919 10.1128/AAC.02745-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beloglazova N., Flick R., Tchigvintsev A., Brown G., Popovic A., Nocek B. et al. (2013) Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J. Biol. Chem. 288, 8101–8110 10.1074/jbc.M112.431148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryoo J., Hwang S.-Y., Choi J., Oh C. and Ahn K. (2016) SAMHD1, the Aicardi-Goutieres syndrome gene and retroviral restriction factor, is a phosphorolytic ribonuclease rather than a hydrolytic ribonuclease. Biochem. Biophys. Res. Commun. 477, 977–981 10.1016/j.bbrc.2016.07.013 [DOI] [PubMed] [Google Scholar]
- 43.Ryoo J., Choi J., Oh C., Kim S., Seo M., Kim S.-Y. et al. (2014) The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat. Med. 20, 936–941 10.1038/nm.3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi J., Ryoo J., Oh C., Hwang S. and Ahn K. (2015) SAMHD1 specifically restricts retroviruses through its RNase activity. Retrovirology 12, 46 10.1186/s12977-015-0174-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brandariz-Nuñez A., Valle-Casuso J., White T.E., Nguyen L., Bhattacharya A., Wang Z. et al. (2013) Contribution of oligomerization to the anti-HIV-1 properties of SAMHD1. Retrovirology 10, 131 10.1186/1742-4690-10-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seamon K.J., Sun Z., Shlyakhtenko L.S., Lyubchenko Y.L. and Stivers J.T. (2015) SAMHD1 is a single-stranded nucleic acid binding protein with no active site-associated nuclease activity. Nucleic Acids Res. 43, 6486–6499 10.1093/nar/gkv633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Welbourn S. and Strebel K. (2016) Low dNTP levels are necessary but may not be sufficient for lentiviral restriction by SAMHD1. Virology 488, 271–277 10.1016/j.virol.2015.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seamon K.J., Bumpus N.N. and Stivers J.T. (2016) Single-stranded nucleic acids bind to the tetramer interface of SAMHD1 and prevent formation of the catalytic homotetramer. Biochemistry 55, 6087–6099 10.1021/acs.biochem.6b00986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goncalves A., Karayel E., Rice G.I., Bennett K.L., Crow Y.J., Superti-Furga G. et al. (2012) SAMHD1 is a nucleic-acid binding protein that is mislocalized due to aicardi-goutieres syndrome-associated mutations. Hum. Mutat. 33, 1116–1122 10.1002/humu.22087 [DOI] [PubMed] [Google Scholar]
- 50.Tüngler V., Staroske W., Kind B., Dobrick M., Kretschmer S., Schmidt F. et al. (2013) Single-stranded nucleic acids promote SAMHD1 complex formation. J. Mol. Med. 91, 759–770 10.1007/s00109-013-0995-3 [DOI] [PubMed] [Google Scholar]
- 51.White T.E., Brandariz-Nuñez A., Valle-Casuso J.C., Amie S., Nguyen L., Kim B. et al. (2013) Contribution of SAM and HD domains to retroviral restriction mediated by human SAMHD1. Virology 436, 81–90 10.1016/j.virol.2012.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coquel F., Silva M.-J., Técher H., Zadorozhny K., Sharma S., Nieminuszczy J. et al. (2018) SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 557, 57–61 10.1038/s41586-018-0050-1 [DOI] [PubMed] [Google Scholar]
- 53.Daddacha W., Koyen A.E., Bastien A.J., Head P.E., Dhere V.R., Nabeta G.N. et al. (2017) SAMHD1 promotes DNA end resection to facilitate DNA repair by homologous recombination. Cell Rep. 20, 1921–1935 10.1016/j.celrep.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao K., Du J., Han X., Goodier J.L., Li P., Zhou X. et al. (2013) Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutieres syndrome-related SAMHD1. Cell Rep. 4, 1108–1115 10.1016/j.celrep.2013.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu S., Li J., Xu F., Mei S., Le Duff Y., Yin L. et al. (2015) SAMHD1 inhibits LINE-1 retrotransposition by promoting stress granule formation. PLOS Genet. 11, e1005367 10.1371/journal.pgen.1005367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herrmann A., Wittmann S., Thomas D., Shepard C.N., Kim B., Ferreirós N. et al. (2018) The SAMHD1-mediated block of LINE-1 retroelements is regulated by phosphorylation. Mob. DNA 9, 11 10.1186/s13100-018-0116-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du J., Peng Y., Wang S., Hou J., Wang Y., Sun T. et al. (2019) Nucleocytoplasmic shuttling of SAMHD1 is important for LINE-1 suppression. Biochem. Biophys. Res. Commun. 510, 551–557 10.1016/j.bbrc.2019.02.009 [DOI] [PubMed] [Google Scholar]
- 58.Crow Y.J., Leitch A., Hayward B.E., Garner A., Parmar R., Griffith E. et al. (2006) Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat. Genet. 38, 910–916 10.1038/ng1842 [DOI] [PubMed] [Google Scholar]
- 59.Crow Y.J., Hayward B.E., Parmar R., Robins P., Leitch A., Ali M. et al. (2006) Mutations in the gene encoding the 3′–5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat. Genet. 38, 917–920 10.1038/ng1845 [DOI] [PubMed] [Google Scholar]
- 60.Rice G.I., Bond J., Asipu A., Brunette R.L., Manfield I.W., Carr I.M. et al. (2009) Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 41, 829–832 10.1038/ng.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xin B., Jones S., Puffenberger E.G. Hinze C., Bright A., Tan H. et al. (2011) Homozygous mutation in SAMHD1 gene causes cerebral vasculopathy and early onset stroke. Proc. Natl Acad. Sci. U.S.A. 108, 5372–5377 10.1073/pnas.1014265108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rice G.I., Kasher P.R., Forte G.M.A., Mannion N.M., Greenwood S.M., Szynkiewicz M. et al. (2012) Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat. Genet. 44, 1243–1248 10.1038/ng.2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rice G.I., del Toro Duany Y., Jenkinson E.M., Forte G.M.A., Anderson B.H., Ariaudo G. et al. (2014) Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat. Genet. 46, 503–509 10.1038/ng.2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crow Y.J., Chase D.S., Lowenstein Schmidt J., Szynkiewicz M., Forte G.M.A., Gornall H.L. et al. (2015) Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am. J. Med. Genet. A 167A, 296–312 10.1002/ajmg.a.36887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oda, et al. (2014) Am. J. Hum. Genet. 95 10.1016/j.ajhg.2014.06.007 [DOI] [Google Scholar]
- 66.Chen S., Bonifati S., Qin Z., St. Gelais C., Kodigepalli K.M., Barrett B.S. et al. (2018) SAMHD1 suppresses innate immune responses to viral infections and inflammatory stimuli by inhibiting the NF-kappaB and interferon pathways. Proc. Natl Acad. Sci. U.S.A. 115, E3798–E3807 10.1073/pnas.1801213115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White T.E., Brandariz-Nuñez A., Martinez-Lopez A., Knowlton C., Lenzi G., Kim B. et al. (2017) A SAMHD1 mutation associated with Aicardi-Goutieres syndrome uncouples the ability of SAMHD1 to restrict HIV-1 from its ability to downmodulate type I interferon in humans. Hum. Mutat. 38, 658–668 10.1002/humu.23201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maelfait J., Bridgeman A., Benlahrech A., Cursi C. and Rehwinkel J. (2016) Restriction by SAMHD1 limits cGAS/STING-dependent innate and adaptive immune responses to HIV-1. Cell Rep. 16, 1492–1501 10.1016/j.celrep.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bonifati S., Daly M.B., St. Gelais C., Kim S.H., Hollenbaugh J.A., Shepard C. et al. (2016) SAMHD1 controls cell cycle status, apoptosis and HIV-1 infection in monocytic THP-1 cells. Virology 495, 92–100 10.1016/j.virol.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rampazzo C., Miazzi C., Franzolin E., Pontarin G., Ferraro P., Frangini M. et al. (2010) Regulation by degradation, a cellular defense against deoxyribonucleotide pool imbalances. Mutat. Res. 703, 2–10 10.1016/j.mrgentox.2010.06.002 [DOI] [PubMed] [Google Scholar]
- 71.Hakansson P., Hofer A. and Thelander L. (2006) Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J. Biol. Chem. 281, 7834–7841 10.1074/jbc.M512894200 [DOI] [PubMed] [Google Scholar]
- 72.Reichard P. (1988) Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 57, 349–374 10.1146/annurev.bi.57.070188.002025 [DOI] [PubMed] [Google Scholar]
- 73.Walters R.A., Tobey R.A. and Ratliff R.L. (1973) Cell-cycle-dependent variations of deoxyribonucleoside triphosphate pools in Chinese hamster cells. Biochim. Biophys. Acta 319, 336–347 10.1016/0005-2787(73)90173-1 [DOI] [PubMed] [Google Scholar]
- 74.Skoog K.L., Nordenskjold B.A. and Bjursell K.G. (1973) Deoxyribonucleoside-triphosphate pools and DNA synthesis in synchronized hamster cells. Eur. J. Biochem. 33, 428–432 10.1111/j.1432-1033.1973.tb02699.x [DOI] [PubMed] [Google Scholar]
- 75.Kohalmi S.E., Glattke M., McIntosh E.M. and Kunz B.A. (1991) Mutational specificity of DNA precursor pool imbalances in yeast arising from deoxycytidylate deaminase deficiency or treatment with thymidylate. J. Mol. Biol. 220, 933–946 10.1016/0022-2836(91)90364-C [DOI] [PubMed] [Google Scholar]
- 76.Cribier A., Descours B., Valadão A.L.C., Laguette N. and Benkirane M. (2013) Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 3, 1036–1043 10.1016/j.celrep.2013.03.017 [DOI] [PubMed] [Google Scholar]
- 77.Arnold L.H., Groom H.C.T., Kunzelmann S., Schwefel D., Caswell S.J., Ordonez P. et al. (2015) Phospho-dependent regulation of SAMHD1 oligomerisation couples catalysis and restriction. PLoS Pathog. 11, e1005194 10.1371/journal.ppat.1005194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tang C., Ji X., Wu L. and Xiong Y. (2015) Impaired dNTPase activity of SAMHD1 by phosphomimetic mutation of Thr-592. J. Biol. Chem. 290, 26352–26359 10.1074/jbc.M115.677435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.White T.E., Brandariz-Nuñez A., Valle-Casuso J.C., Amie S., Nguyen L.A., Kim B. et al. (2013) The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13, 441–451 10.1016/j.chom.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schott K., Fuchs N.V., Derua R., Mahboubi B., Schnellbächer E., Seifried J. et al. (2018) Dephosphorylation of the HIV-1 restriction factor SAMHD1 is mediated by PP2A-B55alpha holoenzymes during mitotic exit. Nat. Commun. 9, 2227 10.1038/s41467-018-04671-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Monod J., Changeux J.P. and Jacob F. (1963) Allosteric proteins and cellular control systems. J. Mol. Biol. 6, 306–329 10.1016/S0022-2836(63)80091-1 [DOI] [PubMed] [Google Scholar]
- 82.Perutz M.F. (1989) Mechanisms of cooperativity and allosteric regulation in proteins. Q. Rev. Biophys. 22, 139–237 10.1017/S0033583500003826 [DOI] [PubMed] [Google Scholar]
- 83.Nussinov R. and Tsai C.J. (2013) Allostery in disease and in drug discovery. Cell 153, 293–305 10.1016/j.cell.2013.03.034 [DOI] [PubMed] [Google Scholar]
- 84.Volkman B.F., Lipson D., Wemmer D.E. and Kern D. (2001) Two-state allosteric behavior in a single-domain signaling protein. Science 291, 2429–2433 10.1126/science.291.5512.2429 [DOI] [PubMed] [Google Scholar]
- 85.Ji X., Wu Y., Yan J., Mehrens J., Yang H., DeLucia M. et al. (2013) Mechanism of allosteric activation of SAMHD1 by dGTP. Nat. Struct. Mol. Biol. 20, 1304–1309 10.1038/nsmb.2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu C., Gao W., Zhao K., Qin X., Zhang Y., Peng X. et al. (2013) Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase. Nat. Commun. 4, 2722 10.1038/ncomms3722 [DOI] [PubMed] [Google Scholar]
- 87.Ji X., Tang C., Zhao Q., Wang W. and Xiong Y. (2014) Structural basis of cellular dNTP regulation by SAMHD1. Proc. Natl Acad. Sci. U.S.A. 111, E4305–E4314 10.1073/pnas.1412289111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koharudin L.M., Wu Y., DeLucia M., Mehrens J., Gronenborn A.M. and Ahn J. (2014) Structural basis of allosteric activation of sterile alpha motif and histidine-aspartate domain-containing protein 1 (SAMHD1) by nucleoside triphosphates. J. Biol. Chem. 289, 32617–32627 10.1074/jbc.M114.591958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brandariz-Nuñez A., Valle-Casuso J., White T.E., Laguette N., Benkirane M., Brojatsch J. et al. (2012) Role of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac. Retrovirology 9, 49 10.1186/1742-4690-9-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim C.A. and Bowie J.U. (2003) SAM domains: uniform structure, diversity of function. Trends Biochem. Sci. 28, 625–628 10.1016/j.tibs.2003.11.001 [DOI] [PubMed] [Google Scholar]
- 91.Qiao F. and Bowie J.U. (2005) The many faces of SAM. Sci. STKE 2005, re7 10.1126/stke.2862005re7 [DOI] [PubMed] [Google Scholar]
- 92.Aravind L. and Koonin E.V. (1998) The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23, 469–472 10.1016/S0968-0004(98)01293-6 [DOI] [PubMed] [Google Scholar]
- 93.Yan J., Kaur S., DeLucia M., Hao C., Mehrens J., Wang C. et al. (2013) Tetramerization of SAMHD1 is required for biological activity and inhibition of HIV infection. J. Biol. Chem. 288, 10406–10417 10.1074/jbc.M112.443796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Buzovetsky O., Tang C., Knecht K.M., Antonucci J.M., Wu L., Ji X. et al. (2018) The SAM domain of mouse SAMHD1 is critical for its activation and regulation. Nat. Commun. 9, 411 10.1038/s41467-017-02783-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhu C.F., Wei W., Peng X., Dong Y.-H., Gong Y. and Yu X.-F. (2015) The mechanism of substrate-controlled allosteric regulation of SAMHD1 activated by GTP. Acta Crystallogr. D Biol. Crystallogr. 71, 516–524 10.1107/S1399004714027527 [DOI] [PubMed] [Google Scholar]
- 96.Brown N.C. and Reichard P. (1969) Ribonucleoside diphosphate reductase. Formation of active and inactive complexes of proteins B1 and B2. J. Mol. Biol. 46, 25–38 10.1016/0022-2836(69)90055-2 [DOI] [PubMed] [Google Scholar]
- 97.Hansen E.C., Seamon K.J., Cravens S.L. and Stivers J.T. (2014) GTP activator and dNTP substrates of HIV-1 restriction factor SAMHD1 generate a long-lived activated state. Proc. Natl Acad. Sci. U.S.A. 111, E1843–E1851 10.1073/pnas.1401706111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Amie S.M., Bambara R.A. and Kim B. (2013) GTP is the primary activator of the anti-HIV restriction factor SAMHD1. J. Biol. Chem. 288, 25001–25006 10.1074/jbc.C113.493619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Miazzi C., Ferraro P., Pontarin G., Rampazzo C., Reichard P. and Bianchi V. (2014) Allosteric regulation of the human and mouse deoxyribonucleotide triphosphohydrolase sterile alpha-motif/histidine-aspartate domain-containing protein 1 (SAMHD1). J. Biol. Chem. 289, 18339–18346 10.1074/jbc.M114.571091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Traut T.W. (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell Biochem. 140, 1–22 10.1007/BF00928361 [DOI] [PubMed] [Google Scholar]
- 101.Knecht K.M., Buzovetsky O., Schneider C., Thomas D., Srikanth V., Kaderali L. et al. (2018) The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1. Proc. Natl Acad. Sci. U.S.A. 115, E10022–E10031 10.1073/pnas.1805593115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li Y., Kong J., Peng X., Hou W., Qin X. and Yu X.-F. (2015) Structural insights into the high-efficiency catalytic mechanism of the sterile α-motif/histidine-aspartate domain-containing protein. J. Biol. Chem. 290, 29428–29437 10.1074/jbc.M115.663658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang K., Lv D.-W. and Li R. (2018) Conserved herpesvirus protein kinases target SAMHD1 to facilitate virus replication. bioRxiv 10.1101/399063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Patra K.K., Bhattacharya A. and Bhattacharya S. (2017) Uncovering allostery and regulation in SAMHD1 through molecular dynamics simulations. Proteins 85, 1266–1275 10.1002/prot.25287 [DOI] [PubMed] [Google Scholar]
- 105.Patra K.K., Bhattacharya A. and Bhattacharya S. (2017) Allosteric signal transduction in HIV-1 restriction factor SAMHD1 proceeds via reciprocal handshake across monomers. J. Chem. Inf. Model. 57, 2523–2538 10.1021/acs.jcim.7b00279 [DOI] [PubMed] [Google Scholar]
- 106.Bhattacharya A., Wang Z., White T., Buffone C., Nguyen L.A., Shepard C.N. et al. (2016) Effects of T592 phosphomimetic mutations on tetramer stability and dNTPase activity of SAMHD1 cannot explain the retroviral restriction defect. Sci. Rep. 6, 31353 10.1038/srep31353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Seamon K.J., Hansen E.C., Kadina A.P., Kashemirov B.A., McKenna C.E., Bumpus N.N. et al. (2014) Small molecule inhibition of SAMHD1 dNTPase by tetramer destabilization. J. Am. Chem. Soc. 136, 9822–9825 10.1021/ja5035717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vorontsov I.I., Minasov G., Kiryukhina O., Brunzelle J.S., Shuvalova L. and Anderson W.F. (2011) Characterization of the deoxynucleotide triphosphate triphosphohydrolase (dNTPase) activity of the EF1143 protein from Enterococcus faecalis and crystal structure of the activator-substrate complex. J. Biol. Chem. 286, 33158–33166 10.1074/jbc.M111.250456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cardamone F., Iacovelli F., Chillemi G., Falconi M. and Desideri A. (2017) A molecular dynamics simulation study decodes the early stage of the disassembly process abolishing the human SAMHD1 function. J. Comput. Aided Mol. Des. 31, 497–505 10.1007/s10822-017-0014-9 [DOI] [PubMed] [Google Scholar]
- 110.Cardamone F., Falconi M. and Desideri A. (2018) Molecular dynamics characterization of the SAMHD1 Aicardi-Goutieres Arg145Gln mutant: structural determinants for the impaired tetramerization. J. Comput. Aided Mol. Des. 32, 623–632 10.1007/s10822-018-0115-0 [DOI] [PubMed] [Google Scholar]