Trypanosoma brucei causes human African trypanosomiasis and regularly switches its major surface antigen, VSG, to evade the host immune response. VSGs are expressed from subtelomeres in a monoallelic fashion. TbRAP1, a telomere protein, is essential for cell viability and VSG monoallelic expression and suppresses VSG switching. Although TbRAP1 has conserved functional domains in common with its orthologs from yeasts to mammals, the domain functions are unknown. RAP1 orthologs have pleiotropic functions, and interaction with different partners is an important means by which RAP1 executes its different roles. We have established a Cre-loxP-mediated conditional knockout system for TbRAP1 and examined the roles of various functional domains in protein expression, nuclear localization, and protein-protein interactions. This system enables further studies of TbRAP1 point mutation phenotypes. We have also determined functional domains of TbRAP1 that are required for several different protein interactions, shedding light on the underlying mechanisms of TbRAP1-mediated VSG silencing.
KEYWORDS: RAP1, telomere, Trypanosoma brucei, protein-protein interaction, RAP1
ABSTRACT
RAP1 is a telomere protein that is well conserved from protozoa to mammals. It plays important roles in chromosome end protection, telomere length control, and gene expression/silencing at both telomeric and nontelomeric loci. Interaction with different partners is an important mechanism by which RAP1 executes its different functions in yeast. The RAP1 ortholog in Trypanosoma brucei is essential for variant surface glycoprotein (VSG) monoallelic expression, an important aspect of antigenic variation, where T. brucei regularly switches its major surface antigen, VSG, to evade the host immune response. Like other RAP1 orthologs, T. brucei RAP1 (TbRAP1) has conserved functional domains, including BRCA1 C terminus (BRCT), Myb, MybLike, and RAP1 C terminus (RCT). To study functions of various TbRAP1 domains, we established a strain in which one endogenous allele of TbRAP1 is flanked by loxP repeats, enabling its conditional deletion by Cre-mediated recombination. We replaced the other TbRAP1 allele with various mutant alleles lacking individual functional domains and examined their nuclear localization and protein interaction abilities. The N terminus, BRCT, and RCT of TbRAP1 are required for normal protein levels, while the Myb and MybLike domains are essential for normal cell growth. Additionally, the Myb domain of TbRAP1 is required for its interaction with T. brucei TTAGGG repeat-binding factor (TbTRF), while the BRCT domain is required for its self-interaction. Furthermore, the TbRAP1 MybLike domain contains a bipartite nuclear localization signal that is required for its interaction with importin α and its nuclear localization. Interestingly, RAP1’s self-interaction and the interaction between RAP1 and TRF are conserved from kinetoplastids to mammals. However, details of the interaction interfaces have changed throughout evolution.
IMPORTANCE Trypanosoma brucei causes human African trypanosomiasis and regularly switches its major surface antigen, VSG, to evade the host immune response. VSGs are expressed from subtelomeres in a monoallelic fashion. TbRAP1, a telomere protein, is essential for cell viability and VSG monoallelic expression and suppresses VSG switching. Although TbRAP1 has conserved functional domains in common with its orthologs from yeasts to mammals, the domain functions are unknown. RAP1 orthologs have pleiotropic functions, and interaction with different partners is an important means by which RAP1 executes its different roles. We have established a Cre-loxP-mediated conditional knockout system for TbRAP1 and examined the roles of various functional domains in protein expression, nuclear localization, and protein-protein interactions. This system enables further studies of TbRAP1 point mutation phenotypes. We have also determined functional domains of TbRAP1 that are required for several different protein interactions, shedding light on the underlying mechanisms of TbRAP1-mediated VSG silencing.
INTRODUCTION
Telomeres are nucleoprotein complexes at linear chromosome ends. They are essential for genome integrity and chromosome stability (1). Telomere DNA in most eukaryotic cells consists of TG-rich tandem repeats (2), and proteins that associate with the telomere chromatin play critical roles in all aspects of telomere biology, including telomere length regulation (3) and protection of the natural chromosome ends from nucleolytic degradation and illegitimate DNA damage repair processes (1).
Among the core telomere protein components, RAP1 orthologs have been identified in many eukaryotes, including vertebrates (4–7), yeasts (8–15), and kinetoplastids (16). RAP1 orthologs have similar functions in chromosome end protection (17–27), telomere length control (4, 11, 17, 28–37), and telomeric silencing (12, 16, 34, 38–44) — where the expression of genes located at subtelomeric regions are suppressed by the heterochromatic telomere structure (45). Strikingly, RAP1 orthologs also have nontelomeric functions, including both transcription activation and repression activities (8, 46–55). In budding yeast, Saccharomyces cerevisiae RAP1 (ScRAP1) achieves different goals through interactions with various protein partners (50). For example, the C-terminal domain of ScRAP1 interacts with Sir3 and Sir4, which helps maintain telomeric silencing (39, 56, 57), and the same region of ScRAP1 also interacts with Rif1 and Rif2 to regulate the telomere length (58, 59). Interestingly, RAP1 orthologs have several conserved protein-protein interaction domains. All known RAP1s also have a BRCA1 C terminus (BRCT) domain (4, 11, 12, 16) that is found in many proteins involved in DNA damage repair and replication (60, 61) and that frequently interacts with phosphorylated peptides (62–64). Human and yeast RAP1 orthologs also have a C-terminal conserved domain termed RAP1 C terminus (RCT) (4, 65), which is mainly involved in protein-protein interactions (39, 56–59, 66–68). All known RAP1 domains have a central Myb domain (4, 11–13, 16, 69). Although ScRAP1 uses its Myb and MybLike domains to bind duplex DNA directly (69), the human RAP1 Myb domain does not have any DNA binding activity, as its third helix has a negatively charged surface that is not suitable for DNA recognition (70).
A RAP1 ortholog has been identified in Trypanosoma brucei (16), a protozoan parasite that causes human African trypanosomiasis. T. brucei proliferates in extracellular spaces of its mammalian host and is directly exposed to the host immune surveillance. However, the parasite regularly switches its major surface antigen, variant surface glycoprotein (VSG), thereby effectively evading the host immune response (71). The T. brucei genome has >2,500 VSG genes and pseudogenes (72), which are all located at subtelomeres (72–74). VSGs are expressed exclusively from VSG expression sites (ESs), which are subtelomeric polycistronic transcription units transcribed by RNA polymerase I (RNA Pol I) (75, 76). VSG is the last gene in any ES, located within 2 kb of the telomere repeats, while the ES promoter is 40 to 60 kb upstream (73). There are 13 different ESs in the Lister 427 strain (74), all with the same gene organization and with ∼90% sequence identity (73). However, at any given moment, only one ES is fully transcribed, presenting a single type of VSG on the cell surface (77). Monoallelic VSG expression ensures the effectiveness of VSG switching by avoiding presentation of a previously active VSG on the cell surface after a VSG switch, which helps the parasite to establish long-term infections. Many factors have been shown to regulate monoallelic VSG expression, including chromatin structure, transcription elongation, inositol phosphate pathway, and nuclear lamina (78, 79); a subtelomere and VSG-associated VEX complex (80, 81); and telomeric silencing (16, 44). VSG switching has two major pathways (82, 83). In an in situ switch, the originally active ES is silenced while a different one becomes fully active (82, 83). In recombination-mediated switches, either a silent VSG gene exchanges places with the originally active VSG without any loss of genetic information or a silent VSG gene is duplicated into the active ES to replace the originally active VSG gene (84). Many factors important for homologous recombination, DNA damage repair, and DNA replication influence VSG switching frequencies (84). Several telomere proteins also suppress VSG switching (85–88).
T. brucei RAP1 (TbRAP1) was identified as a factor interacting with T. brucei TTAGGG repeat-binding factor (TbTRF) (16), which binds the duplex telomere DNA directly (89). TbRAP1 is essential for cell proliferation, and depletion of TbRAP1 leads to a dramatic derepression of all ES-linked VSG genes up to >1,000-fold (16, 44). Transient depletion of TbRAP1 also results in an increased VSG switching frequency (88). Additionally, depletion of TbRAP1 leads to an increased level of the telomeric transcript (TERRA), an increased amount of telomeric RNA:DNA hybrids, and an elevated amount of telomeric/subtelomeric DNA damage (88). We showed that TbRAP1 has a BRCT domain located toward its N terminus, a central Myb domain, and a weak MybLike domain toward its C terminus (16). Here, we report that TbRAP1 also has a C-terminal RCT domain. However, functions of TbRAP1 domains are poorly understood. The interaction interface between TbRAP1 and TbTRF is unknown. Whether this interaction is required for the nuclear localization of TbRAP1 is unclear. These limitations have hindered further investigation of how TbRAP1 regulates VSG silencing and switching.
In this study, we established several strains in which one endogenous TbRAP1 allele is flanked by two loxP repeats so that it can be conditionally deleted by inducing Cre expression. Using this system, we determined that TbRAP1 Myb is necessary for TbTRF interaction and VSG silencing. Using TbRAP1 MybLike as bait in a yeast 2-hybrid screen, we have determined that importin α interacts with TbRAP1’s nuclear localization signal (NLS) residing in the MybLike domain. We found that TbRAP1 interacts with itself through the BRCT domain. In addition, the N terminus, BRCT, and RCT of TbRAP1 are required for normal TbRAP1 protein levels, while Myb and MybLike are essential for normal cell growth. These results not only provide further evidence of conserved essential functions of RAP1 orthologs throughout evolution but also pave the way for a better understanding of the mechanisms explaining how TbRAP1 silences subtelomeric VSG genes and helps maintain telomere stability and integrity.
RESULTS
Establishing a T. brucei strain with a floxed TbRAP1 allele.
TbRAP1 is an essential protein (16), making it difficult to study phenotypes of various small deletions or point mutations using RNA interference (RNAi) (90). To better characterize functions of TbRAP1, we established a strain in which one TbRAP1 allele was flanked by two loxP repeats so that it was able to be conditionally deleted through Cre-mediated recombination (Fig. 1A). The other TbRAP1 allele can be replaced by various TbRAP1 mutants. Upon induction of Cre by the use of doxycycline, we are able to examine the phenotypes of the mutant TbRAP1, even if they are lethal.
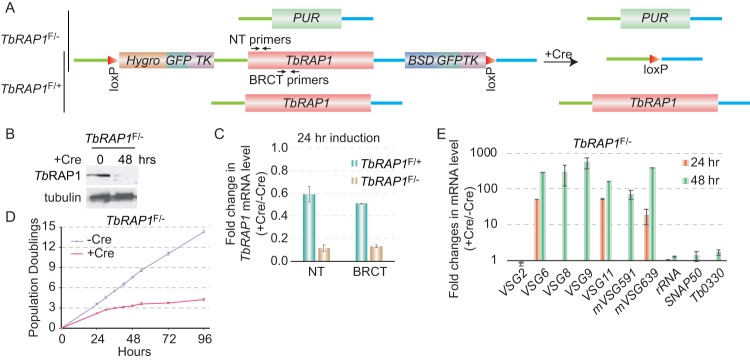
FIG 1.
Conditional knockout of T. brucei RAP1 (TbRAP1) led to cell growth arrest and VSG derepression. (A) A diagram showing three different TbRAP1 alleles, including deleted (top), floxed (middle), and WT (bottom) alleles, in TbRAP1F/− (top two) and TbRAP1F/+ (bottom two) cells before (left) and after (right) the Cre induction. NT, N terminus. (B) Western analysis of cell lysates prepared from TbRAP1F/− cells before and after the Cre induction. A rabbit antibody (16) was used to detect TbRAP1 (top). In this and other figures, TAT-1 (107) was used to detect tubulin (as a loading control). (C) Quantitative RT-PCR was performed using TbRAP1 primers that anneal to the N terminus or the BRCT domain of TbRAP1 (marked in panel A) to estimate the change in the TbRAP1 mRNA level in TbRAP1F/+ and TbRAP1F/− strains. (D) Growth curves of TbRAP1F/− cells with and without the doxycycline-induced Cre expression. (E) Quantitative RT-PCR to estimate the changes in mRNA level of a number of ES-linked VSG genes and several control genes. Average values were calculated from three independent inductions. In this and following figures, error bars represent standard deviations.
A loxP site with the hygromycin resistance gene (HYG) and a loxP site with the blasticidin resistance gene (BSD) were targeted to locations upstream and downstream, respectively, of a given TbRAP1 allele (Fig. 1A). Both selectable markers were fused with the thymidine kinase gene (TK) from the herpes simplex virus, allowing selection for cells that had lost the floxed TbRAP1 allele (denoted as “F”) by the use of ganciclovir (GCV), as expression of the TK gene in T. brucei renders the parasite sensitive to GCV (91). To enable selection of cells that had lost the floxed allele by the use of fluorescence-activated cell sorting (FACS) analysis, we also fused the green fluorescent protein gene (GFP) with both selectable markers (Fig. 1A). Therefore, cells carrying a floxed TbRAP1 allele expressed HYG-GFP-TK and BSD-GFP-TK fusion proteins, and this was confirmed by Western analysis using a rabbit anti-GFP antibody (Life Technologies) in two TbRAP1F/+ clones (without the Cre expression construct Cre-EP1 [91]) (see Fig. S1A in the supplemental material). Southern blotting also confirmed the genotype of these clones (Fig. S1B).
Validation of Cre-loxP-mediated conditional deletion of TbRAP1. (A) Western blotting using a rabbit GFP antibody (Life Technologies) to detect HYG-GFP-TK and BSD-GFP-TK expression in TbRAP1F/+ (-Cre-Ep1) cells before and after transient transfection of Cre-EP1 (91). (B) Southern blotting of BamHI/XbaI-digested genomic DNA isolated from TbRAP1F/+ (-Cre-Ep1) cells before and after transient transfection of Cre-EP1 (91). A DNA fragment containing the full-length TbRAP1 gene was used as the probe. The WT TbRAP1 allele gave a 7.9-kb band, while the floxed TbRAP1 allele gave a 4.9-kb band. (C) Growth curves of two clones of TbRAP1F/+ cells (with Cre-EP1 targeted to an rDNA spacer region) before and after induction of Cre by doxycycline. Cells were selected with different combinations of antibiotics as indicated. The Cre-EP1 plasmid has a phleomycin resistance (BLE) marker (91). Download FIG S1, EPS file, 1.9 MB (1.9MB, eps) .
Copyright © 2020 Afrin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To validate that the floxed TbRAP1 allele can be excised by Cre efficiently, we transiently transfected the Cre-EP1 plasmid into TbRAP1F/+ (-Cre-EP1) cells. Pools of cells and individual clones were selected with GCV. Western analysis performed with the GFP antibody showed that HYG-GFP-TK and BSD-GFP-TK were no longer expressed in either the pool or the clones (Fig. S1A). Southern analysis further confirmed that the floxed TbRAP1 allele was lost in the selected pool and clones (Fig. S1B).
To further increase the feasibility of conditional deletion of the floxed TbRAP1 allele, we integrated Cre-EP1 into a ribosomal DNA (rDNA) spacer region, whose expression can be induced by doxycycline (91). The Cre-EP1-integrated TbRAP1F/+ cells grew normally in the presence of phleomycin, hygromycin, and blasticidin (Fig. S1C). Upon addition of doxycycline, these cells still grew normally in the presence of phleomycin only but did not survive in the presence of all three antibiotics (phleomycin, hygromycin, and blasticidin) (Fig. S1C), confirming that the doxycycline-induced Cre had excised the floxed TbRAP1 allele together with the HYG-GFP-TK and BSD-GFP-TK markers. In the subsequent studies, the Cre-EP1 integrated TbRAP1F/+ strain was used.
Conditional deletion of TbRAP1 leads to cell growth arrest and VSG derepression.
In TbRAP1F/+ cells, we replaced the unfloxed TbRAP1 allele with a puromycin resistance marker (PUR) (Fig. 1A). In the resulting TbRAP1F/− cells, addition of doxycycline led to the loss of the TbRAP1 protein in 48 h (Fig. 1B), confirming the efficient deletion of the floxed TbRAP1 allele. The TbRAP1 mRNA level was estimated by quantitative reverse transcription PCR (RT-PCR), and two sets of TbRAP1 primers were used: one annealed specifically to the N-terminal region and another annealed to the BRCT domain (Fig. 1A). After 24 h of Cre induction, the TbRAP1 mRNA level dropped to 10% of the wild-type (WT) level in TbRAP1F/− cells (Fig. 1C), indicating that the sole TbRAP1 allele in these cells was excised by Cre. In contrast, the TbRAP1 mRNA level dropped to ∼50% of the WT level in TbRAP1F/+ cells (Fig. 1C), as only the floxed TbRAP1 allele had been deleted, leaving the other WT allele intact.
We previously showed that depletion of TbRAP1 by RNAi led to cell growth arrest and VSG derepression (16, 44). Induction of Cre in TbRAP1F/− cells also led to a severe growth defect (Fig. 1D). Quantitative RT-PCR analysis showed that ES-linked VSG6, VSG8, VSG9, VSG11, VSG591, and VSG639 (all originally silent) were derepressed by several 10-fold orders after induction of Cre for 24 h and by several 100-fold orders after 48 h (Fig. 1E). As a control, the rRNA levels and mRNA levels of two chromosome-internal genes, SNAP50 and Tb11.0330, did not increase significantly (Fig. 1E). The mRNA level of the originally active VSG2 gene was decreased ∼20% (Fig. 1E). Therefore, conditional deletion of TbRAP1 by Cre-loxP exhibited the same phenotypes as depletion of TbRAP1 by RNAi (16), indicating that the Cre-loxP-mediated conditional deletion is feasible and efficient.
The TbRAP1 Myb domain is essential for normal cell growth and VSG silencing.
We previously showed that TbRAP1 has a BRCT domain, a Myb domain, and a MybLike domain (Fig. 2, top) (16). With a careful sequence analysis, we found that the C terminus of TbRAP1 has recognizable similarities to the RCT domains of other RAP1 orthologs (Fig. 2, bottom). The level of sequence identity between TbRAP1 and other RAP1 orthologs in this domain was 11.7%, which is approximately the same as that in the BRCT domain (16). We named this region “RCT.” Therefore, TbRAP1 has several conserved domains like other known RAP1 orthologs (Fig. 2, top).
FIG 2.
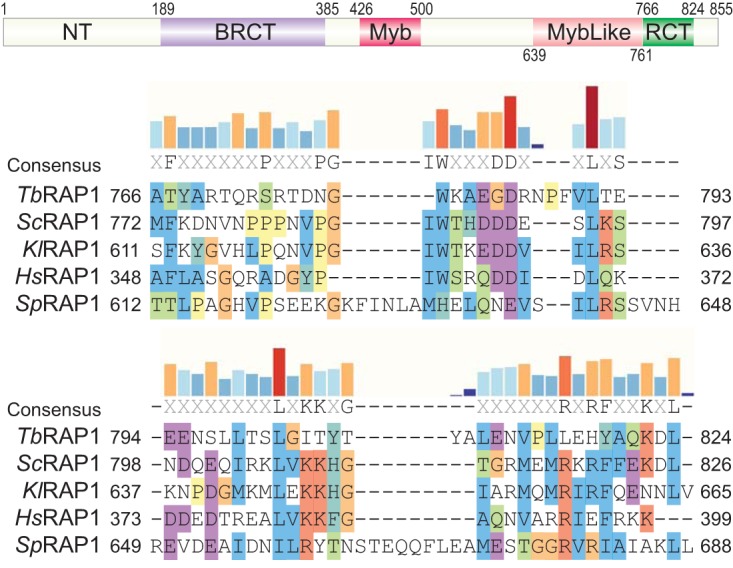
TbRAP1 has an RCT domain. (Top) TbRAP1 domain structure. The positions of each domain are labeled. NT, N terminus; BRCT, BRCA1 C terminus; RCT, RAP1 C terminus. (Bottom) Sequence alignment of the TbRAP1 RCT domain and the RCT domains in Saccharomyces cerevisiae (Sc) RAP1, Kluyveromyces lactis (Kl) RAP1, Homo sapiens (Hs) RAP1, and Schizosaccharomyces pombe (Sp) RAP1 performed using Clustal X. Alignment is visualized in SnapGene. Amino acid positions are indicated. Amino acids are highlighted based on their properties and level of conservation (Clustal X). Specifically, nonpolar amino acids are highlighted in blue, positively charged amino acids in red, negatively charged amino acids in magenta, G amino acid (lacking of bonding characteristics) in orange, H and Y amino acids (mixture of nonpolar and polar ends to the side chain) in dark turquoise, P amino acid (between polar and nonpolar) in yellow, and S and T amino acids (mostly nonpolar) in green. Levels of conservation at each position (percent identical residues among the five RAP1 orthologs) are shown as bar graphs on top of the aligned sequences.
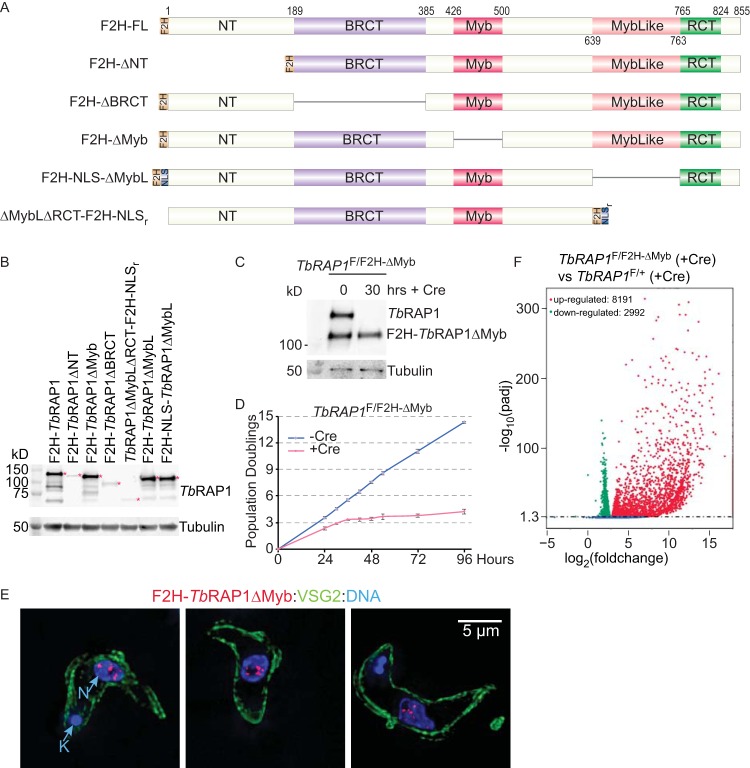
The TbRAP1F/+ strain would be a good choice to examine phenotypes of various TbRAP1 mutants lacking individual domains. As a proof of principle, in the TbRAP1F/+ strain, we replaced the WT TbRAP1 allele with a mutant that lacks the Myb domain. Because the C-terminal FLAG-hemagglutinin-hemagglutinin (FLAG-HA-HA [F2H])-tagged TbRAP1ΔMyb did not express well, we replaced the WT TbRAP1 allele with a TbRAP1F2H-ΔMyb mutant (Fig. 3A). Western analysis using the HA probe antibody (Santa Cruz Biotechnologies) showed that F2H-TbRAP1ΔMyb was expressed at the same level as F2H-TbRAP1 from TbRAP1−/F2H+ cells (Fig. 3B; see also Table S1 in the supplemental material). Southern blotting confirmed the genotype of TbRAP1F/F2H-ΔMyb (Fig. S2A). Using a TbRAP1 rabbit antibody (16) that recognizes a recombinant TbRAP1 fragment containing only the MybLike domain (Fig. S2B), both WT TbRAP1 and F2H-TbRAP1ΔMyb were observed by Western analysis at the same level in TbRAP1F/F2H-ΔMyb cells (Fig. 3C). Upon induction of Cre, WT TbRAP1 was depleted whereas the expression of F2H-TbRAP1ΔMyb remained the same (Fig. 3C). F2H-TbRAP1ΔMyb did not support normal cell growth as TbRAP1F/F2H-ΔMyb cells exhibited a severe growth defect after Cre induction (Fig. 3D), even though the mutant is located in the nucleus in immunofluorescence (IF) analysis (Fig. 3E).
FIG 3.
The Myb domain of TbRAP1 is essential for normal cell growth and VSG silencing. (A) Diagrams of FLAG-HA-HA (F2H)-tagged WT and mutant TbRAP1 used in this study. NLS, simian virus 40 (SV40) large T nuclear localization signal. NLSr, the second half of TbRAP1 nuclear localization signal (aa 737 to 742). (B) Whole-cell lysates from TbRAP1F/F2H-mut cells (after induction of Cre for 30 h) and TbRAP1−/F2H+ cells (as a positive control) were analyzed by Western blotting. HA probe monoclonal antibody (Santa Cruz Biotechnologies) was used to detect the F2H-tagged TbRAP1. Red asterisks indicate the TbRAP1 fragments. F2H-TbRAP1ΔNT ran at a much lower rate than expected. (C) Western blotting of cell lysates prepared from TbRAP1F/F2H-ΔMyb cells before and after induction of Cre. An anti-TbRAP1 rabbit antibody (16) was used to detect TbRAP1 and F2H-TbRAP1ΔMyb. To differentiate WT and mutant TbRAP1, proteins were separated on a 7.5% Tris polyacrylamide gel for 7 h 40 min. (D) Growth curves of TbRAP1F/F2H-ΔMyb cells with and without the Cre induction. (E) F2H-TbRAP1ΔMyb is located in the nucleus. The monoclonal HA antibody 12CA5 (Memorial Sloan Kettering Cancer Center [MSKCC] monoclonal antibody core) was used to detect F2H-TbRAP1ΔMyb. An anti-VSG2 rabbit antibody (16) was used to show the outline of the cell body. DAPI was used to stain DNA. The small DAPI-positive circle represents the kinetoplast (K), and the large DAPI-positive circle represents the nucleus (N). Three different cells are shown in three panels. In this and other figures, IF images are in the same scale and a size bar is shown in one of the panels. (F) Differential gene expression in the TbRAP1ΔMyb mutant was summarized in a volcano plot. TbRAP1F/F2H-ΔMyb and TbRAP1F/+ cells were induced for Cre expression for 30 h and analyzed by RNA-seq. Compared to TbRAP1F/+ cells, more than 8,000 genes were upregulated and nearly 3,000 genes were downregulated in the Cre-induced TbRAP1F/F2H-ΔMyb cells. A log10(adjusted P [padj]) value of 1.3 or higher is considered to be significant.
(A) Southern analysis confirmed the TbRAP1F/F2H-ΔMyb genotype. Genomic DNA from TbRAP1F/+ and a number of candidates of the TbRAP1F/F2H-ΔMyb strain was digested with EcoRI and BamHI and probed with a DNA fragment containing the region of the TbRAP1 gene before the BRCT domain. The three different TbRAP1 alleles are labeled. C1, B1, and B3 were positive clones, while the other three did not have the intended genotype. (B) The rabbit TbRAP1 antibody (16) used in this study recognizes an epitope(s) inside the MybLike domain. (Left) Polyacrylamide gel electrophoresis of the partially purified TRX-His6-TbRAP1-MybLike fragment. (Right) Western blotting of the same recombinant fragment using the rabbit TbRAP1 antibody. (C and D) Classification of genes affected in Cre-induced TbRAP1F/F2H-ΔMyb cells. Distributions (percent of total affected genes) of different types of genes whose mRNA levels were upregulated (C) or downregulated (D) are shown. The number of genes affected in each category is listed in parentheses. Download FIG S2, EPS file, 2.7 MB (2.7MB, eps) .
Copyright © 2020 Afrin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
T. brucei strains used in the current study. Download Table S1, DOCX file, 0.01 MB (15.3KB, docx) .
Copyright © 2020 Afrin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ScRAP1 has both transcription activation and repression functions (8, 50). To examine whether the Myb domain is required for VSG silencing and affects expression of other genes, we performed transcriptome sequencing (RNA-seq) analysis. Both TbRAP1F/+ and TbRAP1F/F2H-ΔMyb cells were induced for Cre expression for 30 h, after which the total RNA was isolated. Poly(A) RNA was purified and used for library construction at Novogene followed by paired-end high-throughput sequencing using Illumina (Materials and Methods). Differential gene expression analysis showed that more than 8,000 genes were upregulated and nearly 3,000 genes were downregulated in Cre-induced TbRAP1F/F2H-ΔMyb cells compared to the results seen with TbRAP1F/+ (Fig. 3F). However, the fold change in mRNA levels was much greater for upregulated genes than for downregulated ones (Fig. 3F), and upregulated genes were present in much greater numbers than downregulated ones, suggesting that TbRAP1 has a major role in gene silencing and a minor role in gene activation. Sequence read coverage in all VSG bloodstream-form (BF) expression sites (BESs) (74) showed that all silent BES-linked VSG genes and some BES-linked expression site-associated genes (ESAG genes) were upregulated, but other BES-linked ESAG genes were not affected or were even downregulated (Fig. S3). Based on available annotation of the affected genes, a total of more than 2,700 VSG genes and pseudogenes were upregulated (Fig. S2C), which included nearly all reported VSG genes/pseudogenes in the Lister 427 genome (72). Therefore, the TbRAP1 Myb domain is essential for the functions of TbRAP1 in normal cell growth and VSG silencing. Interestingly, the mRNA levels of some ribosomal protein genes were decreased in the mutant, although at only up to 60% of the normal level (Fig. S2D). It is possible that TbRAP1 may also participate in transcription activation of ribosomal protein genes, as was seen previously with ScRAP1 (8), although further investigation is necessary to validate this.
RNA sequence read coverage in all VSG bloodstream-form expression sites (BESs) for Cre-induced TbRAP1F/+ and TbRAP1F/F2H-ΔMyb cells. RNA samples were isolated from three independently induced cells in both strains. RNA sequence reads were mapped to the T. brucei Lister 427 genome TriTrypDB-45_TbruceiLister427_2018_Genome.fasta, which was obtained from TriTrypDB. Read coverage for each sample was viewed in IGV. Each BES is labeled, and the genes inside each BES are listed at the bottom. The whole BES region is shown with the promoter located on the left side. The y axis data represent the number of RNA reads. The range is indicated on the left of each trace. BES1 was fully active in the cells analyzed. Download FIG S3, PDF file, 1.2 MB (1.2MB, pdf) .
Copyright © 2020 Afrin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The nuclear localization signal of TbRAP1 is required for its interaction with Importin α and nuclear localization.
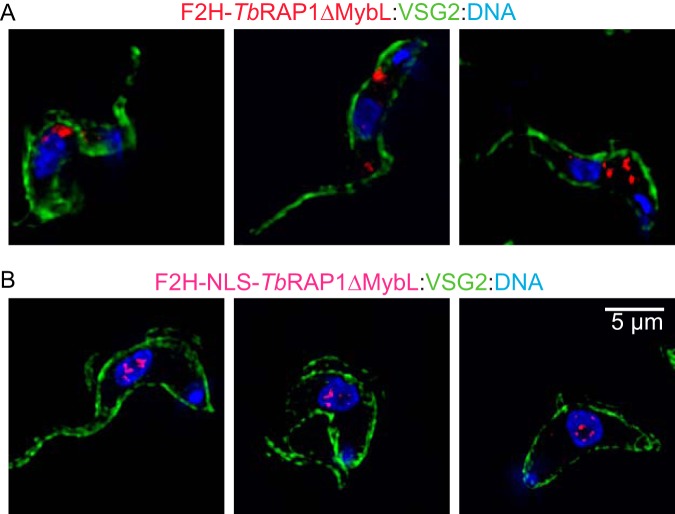
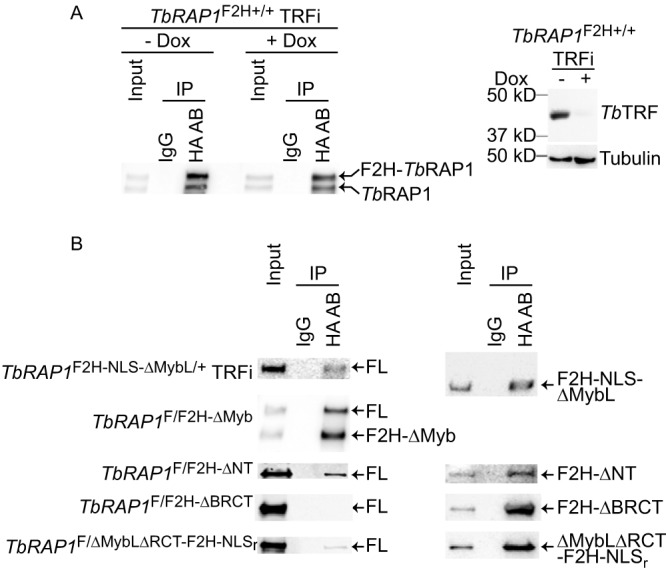
Using the same approach, we replaced the WT TbRAP1 allele with an F2H-tagged TbRAP1 mutant lacking the MybLike domain (TbRAP1ΔMybL). Southern blotting confirmed its genotype (Fig. S4A). Western blotting showed that F2H-TbRAP1ΔMybL was expressed (Fig. S4B) and that the expression was at the same level as F2H-TbRAP1 (Fig. 3B). However, IF showed that this mutant was localized in the cytoplasm (Fig. 4A). Using Motif Scan analysis (https://myhits.isb-sib.ch/cgi-bin/motif_scan) (92), we found that the sequence consisting of amino acids (aa) 727 to 741 of TbRAP1 represents a bipartite nuclear localization signal (NLS). F2H-TbRAP1ΔMybL lacks this NLS, which is likely why this mutant is localized in the cytoplasm. To confirm this, we added the simian virus 40 (SV40) large T NLS to the N terminus of TbRAP1ΔMybL (Fig. 3A). Southern and Western analyses confirmed the genotype of this strain and that the expression of F2H-NLS-TbRAP1ΔMybL was at the WT level (Fig. 3B; see also Fig. S4C and D). IF analysis showed that F2H-NLS-TbRAP1ΔMybL was indeed localized in the nucleus (Fig. 4B).
FIG 4.
Immunofluorescent analysis of TbRAP1F/F2H-ΔMybL TRFi and TbRAP1F/F2H-NLS-ΔMybL TRFi cells. F2H-TbRAP1ΔMybL and F2H-NLS-TbRAP1ΔMybL were stained with 12CA5 HA antibody. A rabbit VSG2 antibody was used to show the outline of the cell body. DNA was stained with DAPI. In each strain, three different cells are shown in three panels.
Southern analyses confirmed the genotypes of TbRAP1F/F2H-ΔMybL (A) and TbRAP1F/F2H-NLS-ΔMybL (C) cells. Genomic DNA was digested with EcoRI and probed with a DNA fragment amplified by PCR with the forward primer (5′-GCGCCTCGAGACCGACGTTGACGGTG-3′) and the backward primer (5′-GCGCTCTAGAACACGAAGAAATCGGGACGTG-3′) using TbRAP1 as the template. Western analyses showed the expression levels of F2H-TbRAP1ΔMybL (B) and F2H-NLS-TbRAP1ΔMybL (D) in the same clones as those indicated in panels A and C, respectively. Download FIG S4, EPS file, 1.1 MB (1.1MB, eps) .
Copyright © 2020 Afrin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To further explore how TbRAP1 is imported into the nucleus, we screened a normalized yeast 2-hybrid library generated from T. brucei cDNA using the TbRAP1 MybLike domain as bait. In this screen, 16.5 million yeast primary transformants were obtained, and a total of 711 clones were positive in the initial screen. The majority of the candidates represented the same gene, Tb427.06.2640, which is annotated as encoding the importin α subunit in TriTrypDB (93, 94). The canonical function of importin α is to bind the NLS of nuclear proteins, form a complex with importin β, and transport the protein into the nucleus through the nuclear pore (95). Once inside the nucleus, importin α releases its cargo and exits the nucleus to transport the next cargo (95). TbRAP1 MybLike contains the predicted bipartite NLS. Therefore, we expected that importin α would interact with the TbRAP1 NLS and this interaction would be essential for transporting TbRAP1 into the nucleus.
To confirm the interaction between importin α and TbRAP1, we inserted a C-terminal myc13 (13 repeats of myc) epitope at one endogenous importin α allele. PCR analysis confirmed correct targeting in three different TbRAP1 backgrounds: both alleles were WT, one of the two WT alleles had an N-terminal F2H tag, and one of the alleles was replaced with the TbRAP1F2H-ΔMybL mutant (Fig. S5). The latter two strains also carried an inducible TbTRF RNAi cassette inserted into an rDNA spacer, although the RNAi was not induced for the analysis of TbRAP1-importin α interaction. The expression of importin α-myc13 was confirmed by Western blotting (Fig. 5A). Subsequently, we performed coimmunoprecipitation (co-IP) experiments in these three strains. In the TbRAP1+/+ background, IP experiments were performed using a rabbit TbRAP1 antibody (16) or the myc monoclonal antibody 9E10 (Memorial Sloan Kettering Cancer Center [MSKCC] monoclonal antibody core). In TbRAP1F2H+/+ TRF RNAi (TRFi) and TbRAP1F2H-ΔMybL/+ TRFi cells, IP experiments were performed using HA monoclonal antibody 12CA5 (MSKCC monoclonal antibody core) or 9E10. In cells carrying untagged or F2H-tagged WT TbRAP1, both importin α-myc13 and TbRAP1 were present in the IP products (Fig. 5B, top two rows). However, F2H-TbRAP1ΔMybL and importin α-myc13 were not in the same IP product (Fig. 5B, bottom row). Therefore, the MybLike domain of TbRAP1 (and most likely the NLS in this domain) is necessary for TbRAP1’s interaction with importin α and its nuclear localization.
FIG 5.
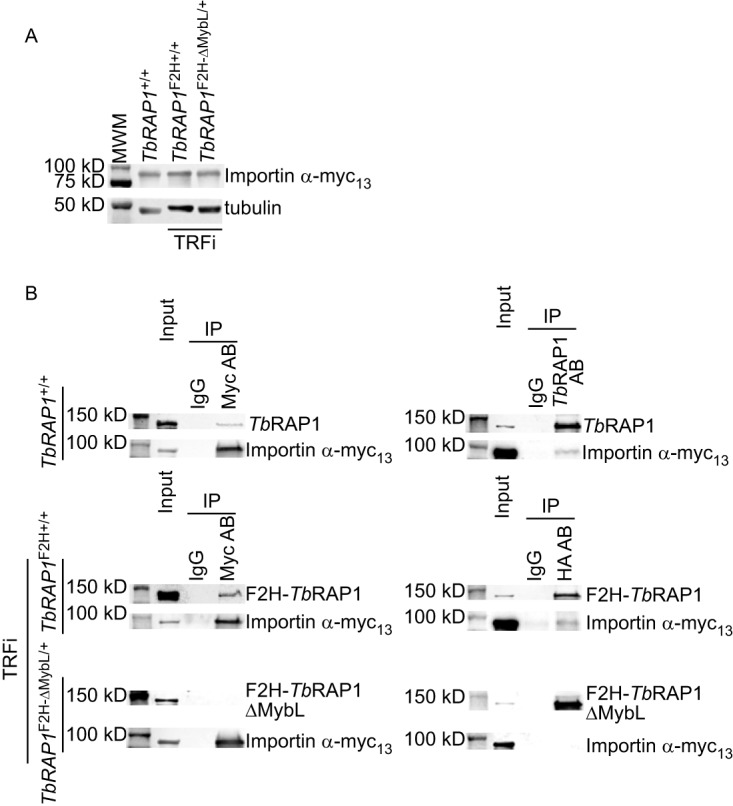
The TbRAP1 MybLike domain is required for interaction with importin α. (A) Expression of importin α-myc13 in TbRAP1+/+, TbRAP1F2H+/+ TRFi, and TbRAP1F2H-ΔMybL/+ TRFi cells. Myc13-tagged proteins were detected by a myc monoclonal antibody, 9E10 (MSKCC monoclonal antibody core). (B) Co-IP of TbRAP1 and importin α. The myc antibody 9E10, an anti-TbRAP1 rabbit antibody (16), and IgG (as a negative control) were used for IP in TbRAP1+/+ cells. Western analysis was performed using the antibodies mentioned above to detect importin α-myc13 and TbRAP1. In TbRAP1F2H+/+ TRFi and TbRAP1F2H-ΔMybL/+ TRFi cells, the 9E10 myc antibody, the 12CA5 HA antibody, and IgG (as a negative control) were used for IP, and Western blotting was performed to detect importin α-myc13 (by 9E10) and F2H-tagged WT and mutant TbRAP1 (by 12CA5 in the left panels and HA probe in the right panels). In this and other figures, input samples represent 1% of the materials used for IP.
PCR confirmed the endogenous tagging of importin α with a C-terminal myc13 tag. Genomic DNAs from the parent TbRAP1+/+ strain (as a negative control) and three transfected cells in the TbRAP1+/+, TbRAP1F2H+/+ TRFi, and TbRAP1F2H-ΔMybL/+ TRFi background were used as the templates. Forward primer (5′-CACTTTGTCTCCTCGAGACA-3′) and backward primer (5′-CGATCAGAAACTTCTCGACAG-3′) were used in the PCR amplification. A product of 1.26 kb is expected for correct targeting of the myc13 tag. Download FIG S5, EPS file, 1.2 MB (1.2MB, eps) .
Copyright © 2020 Afrin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The TbRAP1 Myb domain interacts with TbTRF.
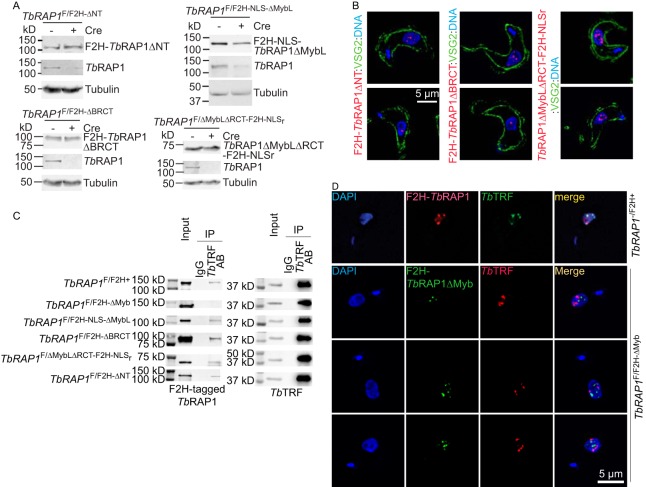
TbRAP1 was originally identified as a TbTRF-interacting factor in a yeast 2-hybrid screen (16). The N-terminal part of TbRAP1 (including the N terminus, BRCT, and Myb) is sufficient to interact with TbTRF in yeast 2-hybrid analysis (16), and WT TbRAP1 and TbTRF co-IP in vivo (16), although both assays showed a weak interaction between the two proteins. To examine which TbRAP1 functional domain(s) is essential for the TbRAP1-TbTRF interaction, in the TbRAP1F/+ cells, we either targeted an F2H tag to the N terminus of the WT TbRAP1 allele (to generate the TbRAP1F/F2H+ strain) or replaced the WT allele with an F2H-tagged TbRAP1 mutant lacking various functional domains (to generate the TbRAP1F/F2H-mut strains) (Fig. 3A). Southern analyses confirmed the replacement of the WT TbRAP1 allele by the TbRAP1F2H-ΔNT, TbRAP1F2H-ΔBRCT, or TbRAP1ΔMybLΔRCT-F2H-NLSr allele in the corresponding strains (Fig. S6A to C). Here, we added the second half (aa 736 to 742) of the endogenous TbRAP1 NLS (labeled as NLSr) at the C terminus of TbRAP1ΔMybLΔRCT. These TbRAP1 mutants were expressed (Fig. 6A) but at much lower levels than F2H-TbRAP1 (Fig. 3B). F2H-TbRAP1ΔNT and F2H-TbRAP1ΔBRCT still contained the TbRAP1 NLS, and they were localized in the nucleus as expected (Fig. 6B, left and middle). Interestingly, TbRAP1ΔMybLΔRCT-F2H-NLSr was also localized in the nucleus (Fig. 6B, right), indicating that the sequence consisting of aa 736 to 742 contains a minimum nuclear localization signal and that its presence is sufficient to target TbRAP1 to the nucleus. This observation further validated the function of TbRAP1 NLS.
FIG 6.
The TbRAP1 Myb domain is required for interaction with TbTRF. (A) Western analyses showed the expression levels of the F2H-tagged TbRAP1 mutants (using the HA probe antibody) and WT TbRAP1 (using a rabbit TbRAP1 antibody [16]) before and after the Cre induction in TbRAP1F/F2H-ΔNT, TbRAP1F/F2H-ΔBRCT, TbRAP1F/F2H-NLS-ΔMybL, and TbRAP1F/ΔMybLΔRCT-F2H-NLSr cells. (B) IF analysis showed that F2H-TbRAP1ΔNT, F2H-TbRAP1ΔBRCT, and TbRAP1ΔMybLΔRCT-F2H-NLSr were localized in the nucleus. 12CA5 was used to detect the F2H-tagged TbRAP1 mutants. A rabbit anti-VSG2 antibody was used to show the outline of the cell body. DNA was stained by DAPI. In each strain, two different cells are shown in two panels. (C) Co-IP experiments were performed in a series of TbRAP1F/F2H-mut cells (as labeled on the left). An anti-TbTRF rabbit antibody (89) and IgG (as a negative control) were used for IP. The antibodies used for Western analyses of the IP products were the 12CA5 HA antibody (except for TbRAP1F/F2H-ΔNT, where the HA probe was used) and an anti-TbTRF chicken antibody (16). (D) (Top) F2H-TbRAP1 (detected by 12CA5) partially colocalized with TbTRF (detected by a rabbit TbTRF antibody [89]) in TbRAP1−/F2H+ cells. (Bottom) F2H-TbRAP1ΔMyb (detected by 12CA5) was not colocalized with TbTRF (detected by a chicken TbTRF antibody [16]). DNA was stained by DAPI. Three different TbRAP1F/F2H-∆Myb cells are shown.
The TbRAP1F/F2H-ΔNT, TbRAP1F/F2H-ΔBRCT, TbRAP1F/F2H-NLS-ΔMybL, and TbRAP1F/ΔMybLΔRCT-F2H-NLSr cells exhibited a growth arrest phenotype after induction of Cre. (A to C) Southern analyses confirmed the genotype of TbRAP1F/F2H-ΔNT (A), TbRAP1F/F2H-ΔBRCT (B), and TbRAP1F/ΔMybLΔRCT-F2H-NLSr (C) cells. Genomic DNAs were digested with XbaI and BamHI in the experiments represented in panels A and C and with EcoRI in the experiments represented in panel B. A DNA fragment containing the TbRAP1 sequence between the BRCT domain and the Myb domain was used as the probe in the experiments represented in panels A and C. A DNA fragment containing the N-terminal region of TbRAP1 was used as the probe used in the experiments represented in panel B. (D to G) Growth curves of TbRAP1F/F2H-NLS-ΔMybL (D), TbRAP1F/F2H-ΔNT (E), TbRAP1F/F2H-ΔBRCT (F), and TbRAP1F/ΔMybLΔRCT-F2H-NLSr (G) cells before and after the induction of Cre expression are shown. Download FIG S6, EPS file, 1.9 MB (1.9MB, eps) .
Copyright © 2020 Afrin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Since human RAP1 interacts with itself (4), it is possible that TbRAP1 may also interact with itself (see below). To avoid detecting possible indirect interactions between TbTRF and mutant TbRAP1 mediated by the WT TbRAP1, we induced Cre in these TbRAP1F/F2H-mut strains for 30 h to ensure the depletion of the WT TbRAP1 protein (Fig. 6A). Depletion of TbRAP1 by RNAi for up to 36 h still allows complete cell growth recovery 24 h after removal of the RNAi induction (88). In addition, upon Cre induction, the number of TbRAP1F/− and TbRAP1F/F2H-mut cells did not decrease for several days (Fig. 1D) (Fig. 3D; see also Fig. S6D to G). Therefore, inducing TbRAP1F/F2H-mut for 30 h caused only cell growth arrest rather than cell death. Subsequently, co-IPs were performed using a rabbit TbTRF antibody (89). In all cases, TbTRF was detected in the IP products by Western analysis using a chicken TbTRF antibody (16) (Fig. 6C, right). Although F2H-TbRAP1ΔNT, F2H-TbRAP1ΔBRCT, and TbRAP1ΔMybLΔRCT-F2H-NLSr were expressed at very low levels (Fig. 3B), these mutants still interacted with TbTRF, as they were detected in the IP products in the same manner as F2H-TbRAP1 and F2H-NLS-TbRAP1ΔMybL (Fig. 6C, left). However, F2H-TbRAP1ΔMyb was not detected in the IP product (Fig. 6C, left). In addition, IF analysis showed that F2H-TbRAP1ΔMyb was not colocalized with TbTRF even though these proteins were in the nucleus (Fig. 6D). As a control, WT TbRAP1 was partially colocalized with TbTRF (Fig. 6D), as we have shown previously (16). Therefore, the Myb domain of TbRAP1 is required for its interaction with TbTRF. Interestingly, TbRAP1F/F2H-NLS-ΔMybL cells exhibited a growth arrest phenotype after induction of Cre for 30 h (Fig. S6D), even though F2H-NLS-TbRAP1ΔMybL was expressed at the WT level (Fig. 3B) and was localized in the nucleus (Fig. 4B), indicating that the MybLike domain has essential functions other than transporting TbRAP1 into the nucleus. On the other hand, F2H-TbRAP1ΔNT, F2H-TbRAP1ΔBRCT, and TbRAP1ΔMybLΔRCT-F2H-NLSr were expressed at much lower levels than WT TbRAP1 (Fig. 3B), indicating that the N terminus and BRCT and RCT domain of TbRAP1 are required for normal TbRAP1 protein levels, which is likely the reason why TbRAP1F/F2H-ΔNT, TbRAP1F/F2H-ΔBRCT, and TbRAP1F/ΔMybLΔRCT-F2H-NLSr cells also showed a growth arrest phenotype after induction of Cre (Fig. S6E to G).
The BRCT domain is required for TbRAP1 self-interaction.
To test whether TbRAP1 has any self-interaction ability, we performed co-IP experiments in cells expressing an F2H-TbRAP1 from one of its endogenous alleles. Since TbRAP1 interacts with TbTRF, these co-IP experiments were performed in TbTRF RNAi cells. Both before and after depletion of TbTRF by RNAi (Fig. 7A, right), we detected WT TbRAP1 in the IP product when IP was performed using the 12CA5 HA antibody (Fig. 7A, left). Therefore, TbRAP1 interacts with itself, and this interaction is independent of TbTRF.
FIG 7.
TbRAP1 interacts with itself through the BRCT domain. (A) TbRAP1’s self-interaction activity is independent of TbTRF. Left, co-IP experiments were done using the 12CA5 HA antibody and IgG (as a negative control) in TbRAP1F2H+/+ TRFi cells before and after induction of TbTRF RNAi. IP products were analyzed by Western blotting using a rabbit TbRAP1 antibody (16). To differentiate F2H-TbRAP1 and the untagged TbRAP1, proteins were separated on a 7.5% Tris polyacrylamide gel for 7 h. Right, Western analysis showed that TbTRF was efficiently depleted by RNAi after addition of doxycycline (Dox) for 30 h. A rabbit antibody was used to detect TbTRF (89). (B) The BRCT domain of TbRAP1 is critical for its self-interaction. Co-IP experiments were performed in various TbRAP1F/F2H-mut strains (indicated on the left) without Cre induction and in TbRAP1F2H-NLS-ΔMybL/+ TRFi cells. IP experiments were done using the 12CA5 HA antibody and IgG (as a negative control). To analyze the IP products, WT TbRAP1 and F2H-TbRAP1ΔMyb were detected with an anti-TbRAP1 rabbit antibody (16), and other F2H-tagged TbRAP1 mutants were detected by the HA probe antibody in Western blotting. To differentiate TbRAP1 and F2H-TbRAP1ΔMyb, proteins were separated on a 7.5% Tris polyacrylamide gel for 7 h 40 min.
To further examine which domain of TbRAP1 is required for its self-interaction, we tested whether any F2H-tagged TbRAP1 domain deletion mutants showed co-IP with the WT TbRAP1. In TbRAP1F/F2H-mut cells (without Cre induction) and in TbRAP1F2H-NLS-ΔMybL/+ TRFi cells, IP experiments were performed using the 12CA5 HA antibody, and the IP products were examined by Western blotting using both the HA probe antibody (Fig. 7B, right) and the rabbit TbRAP1 antibody (16) (Fig. 7B, left). In all cells except TbRAP1F/F2H-ΔBRCT, WT TbRAP1 was detected in the IP products (Fig. 7B, left), indicating that BRCT is essential for TbRAP1 self-interaction. Although F2H-TbRAP1ΔBRCT was expressed at a lower-than-WT level, it was expressed at a higher level than F2H-TbRAP1ΔNT and TbRAP1ΔMybLΔRCT-F2H-NLSr (Fig. 3B), while the latter two mutants interacted with the WT protein (Fig. 7B, left). Therefore, the lack of interaction between F2H-TbRAP1ΔBRCT and WT TbRAP1 is unlikely to have been due to the low level of expression of the mutant.
DISCUSSION
RAP1 orthologs are conserved from protozoa to mammals (4–16), and they have similar domain structures (4, 12, 16, 65) and essential telomeric and nontelomeric functions (96). TbRAP1 also has the BRCT, Myb, MybLike, and RCT functional domains, like other RAP1 orthologs (16). TbRAP1 is essential for VSG silencing and telomere/subtelomere integrity and stability (16, 44, 88). However, whether TbRAP1’s domains are required for these functions was unknown. Study of TbRAP1 domain functions was partly limited by the fact that TbRAP1 is essential for cell proliferation (16). Previous studies of TbRAP1 functions were heavily dependent on the use of conditional RNAi to deplete TbRAP1 (16, 44, 88). Although expressing double-stranded RNA (dsRNA) of the TbRAP1 full-length gene (16, 88), the BRCT fragment, or the RCT fragment (44) can efficiently deplete TbRAP1, expressing dsRNA of the 3′ untranslated region (3′UTR) of TbRAP1 cannot. Therefore, the RNAi approach is not suitable for studying phenotypes of all domain deletion mutants or point mutations of TbRAP1. In this study, we took advantage of the Cre-loxP system (91) and established a series of strains in which one endogenous TbRAP1 allele is flanked by two repeats of loxP, allowing its conditional deletion upon Cre induction. We confirmed that this conditional deletion was able to efficiently deplete the TbRAP1 protein and mRNA. Most importantly, we are now able to examine the phenotypes of a series of TbRAP1 mutants that lack individual functional domains or carry point mutations, even if the mutants are lethal.
In this study, we found that none of the domain deletion mutants of TbRAP1 could support normal cell growth. F2H-TbRAP1ΔNT, F2H-TbRAP1ΔBRCT, and TbRAP1ΔMybLΔRCT-F2H-NLSr were expressed at much lower levels than the WT TbRAP1, while F2H-TbRAP1ΔMyb, F2H-TbRAP1ΔMybL, and F2H-NLS-TbRAP1ΔMybL were expressed at the same level as WT TbRAP1. Therefore, the N terminus, BRCT, and RCT are required for normal TbRAP1 protein level, and the low level of protein expression is most likely the reason why these mutants do not support normal cell growth. F2H-TbRAP1ΔMyb and F2H-NLS-TbRAP1ΔMybL were localized in the nucleus and expressed at the same level as the WT protein, but these mutants still had a severe growth defect, indicating that Myb and MybLike domains are essential for normal cell growth. Only the DNA binding domains of ScRAP1 (Myb and MybLike) are essential for cell viability, and its RCT domain, which is important for telomere length regulation and telomeric silencing, is not essential for cell survival (66). This leads us to speculate that the TbRAP1 Myb and/or MybLike domains may have DNA binding activities. Although Myb domains are frequently involved in DNA binding (97), the human RAP1 Myb domain does not seem to have any DNA binding activity due to its negative surface charge on the third helix, which is typically involved in DNA recognition (70). In addition, the ScRAP1 MybLike domain was revealed to fold into a DNA binding motif only after its crystal structure was solved (69). It will be interesting to investigate whether TbRAP1 Myb and MybLike domains have any DNA binding activities, but further protein structural analysis may be necessary.
Myb domains can also mediate protein-protein interactions (98, 99). We found that TbRAP1 Myb interacts with TbTRF, providing another piece of evidence that the Myb domain can have an important function in protein-protein interaction. Nevertheless, whether the interaction between TbRAP1 and TbTRF is essential for cell survival and/or VSG silencing is still unknown. Further investigation will be necessary to identify key residues in TbRAP1 Myb that are critical for TbTRF interaction, which will help address this issue. Human RAP1 uses its C-terminal RCT domain to interact with TRF2’s linker region (4, 68). Similarly, Schizosaccharomyces pombe RAP1 also uses its C-terminal RCT domain to interact with Taz1, a functional homologue of mammalian TRF1/2 (65, 68), indicating that this interaction interface is conserved from yeast to mammals. However, we found that the TbRAP1 Myb domain is critical for TbTRF interaction. It is interesting that the RAP1-TRF interaction is preserved from kinetoplastids to mammals and yet the functional domains that accomplish this goal have changed.
The transcription profile of more than 10,000 genes was found to have changed in TbRAP1F/F2H-ΔMyb cells after induction of Cre. In particular, ∼2,700 VSG genes were upregulated. Since the VSGnome identified more than 2,500 VSG genes and pseudogenes in the Lister 427 genome (72), this means nearly all of the VSG genes were upregulated, further validating that TbRAP1 has an essential function in silencing VSG genes and that Myb is essential for this function. Interestingly, RNA-seq also identified nearly 3,000 genes that are downregulated in TbRAP1F/F2H-ΔMyb cells, although the fold change in mRNA levels was much lower than that seen with the upregulated genes. ScRAP1 has been well known for both its transcription activation and its repression functions (50), and ScRAP1 is required for ribosomal protein gene activation (8). Our observation suggests that TbRAP1 may have functions similar to those of ScRAP1, although TbRAP1’s transcription activation effect appears to be much weaker than its repressive effect. Some of the downregulated genes encode ribosomal proteins, suggesting that TbRAP1 may also participate in ribosomal protein gene activation. However, further validation is necessary to confirm TbRAP1’s role as a transcription activator.
Importing >45-kDa nuclear proteins frequently depends on the presence of NLS, which can be recognized by importin α/β proteins (100). After the importin-cargo complex is transported through the nuclear pore complexes, binding of RanGTP to the importin β dissociates the complex to release cargo into the nucleoplasm (100). However, nuclear proteins can also be transported into the nucleus through NLS-independent mechanisms, such as interacting with a protein partner that contains a NLS (101). TbRAP1 is a nuclear protein (16). However, how TbRAP1 is imported into the nucleus was unknown. Here, we show that importin α directly interacts with TbRAP1 and that this interaction depends on the MybLike domain, which contains a predicted bipartite NLS. Additionally, F2H-TbRAP1ΔMybL is not localized in the nucleus, while TbRAP1ΔMybLΔRCT-F2H-NLSr with the second half of the TbRAP1 NLS, is. Therefore, TbRAP1’s nuclear localization depends on the NLS in the MybLike domain and requires its recognition by importin α. Addition of an SV40 large T NLS to the N terminus of TbRAP1ΔMybL can target the mutant to the nucleus, indicating that the NLS and importin α are well conserved with other known classical NLS and importin α proteins, respectively. Interestingly, F2H-NLS-TbRAP1ΔMybL still interacts with TbTRF. However, without any NLS, F2H-TbRAP1ΔMybL is localized in the cytoplasm, indicating that the interaction between TbRAP1 and TbTRF is not sufficient to bring TbRAP1 into the nucleus.
Human RAP1 interacts with itself through the RCT domain (4), although the function of this self-interaction is unknown. Human RAP1 interacts tightly with TRF2 with equal stoichiometry (102), and TRF2 homodimerizes (103). It is possible that homodimerization of human RAP1 allows a better interaction with TRF2. The TbTRF-TbRAP1 interaction is much weaker than the TbTRF-TbTIF2 interaction (4, 86), and whether TbTRF interacts with TbRAP1 with equal stoichiometry is unknown. Therefore, whether TbRAP1 self-interaction contributes to its interaction with TbTRF is not clear. We found that TbRAP1’s self-interaction is independent of TbTRF, indicating that this self-interaction is direct and not mediated by the TbRAP1-TbTRF interaction. Additionally, TbRAP1 self-interaction and TbRAP1-TbTRF interaction require different functional domains. The BRCT domain is also important for a normal level of TbRAP1 protein. It is possible that TbRAP1 is better stabilized with self-interaction, possibly by preventing TbRAP1 degradation by proteases. Further analysis with point mutations in the BRCT domain that specifically abolish TbRAP1 self-interaction will be useful to reveal its function and help understand the mechanism of protein stabilization.
The fact that the functional interactions of RAP1 with other telomere proteins, such as TRF, or with itself are conserved from kinetoplastids to mammals suggests that these functions of RAP1 orthologs are critical for essential cellular processes. However, throughout evolution, different organisms have used different approaches to achieve the same goal. Hence, the detailed protein-protein interaction interfaces have been changed even though the consequential protein complex is still preserved. We have observed a similar scenario in interactions of TbTRF and TbTIF2 (86). Although TbTRF interacts with TbTIF2 (86) and mammalian TRF1 and TRF2 interact with TIN2 (104, 105), the two protein pairs interact with different interfaces (86). Therefore, conserved protein-protein interactions without a conserved interaction interface can be common among many telomere protein homologues.
TbRAP1, as with its orthologs in other organisms, has multiple protein-protein interaction domains. A common theme appears to pertain for most RAP1 orthologs: RAP1 interacts with different protein partners to perform different cellular tasks (50). Our results further suggest that TbRAP1 interacts with different partners through different functional domains to achieve various goals. With our established Cre-loxP system, we will be able to investigate details of the functions of each TbRAP1 domain in the future.
MATERIALS AND METHODS
T. brucei strains and plasmids.
All T. brucei strains used in this study (listed in Table S1 in the supplemental material) were derived from bloodstream-form Lister 427 cells that express VSG2 as well as a T7 polymerase and the Tet repressor (also known as the single marker or SM strain) (106). All BF T. brucei cells were cultured in HMI-9 medium supplemented with 10% fetal bovine serum (FBS) and appropriate antibiotics.
To establish the TbRAP1F/+ strain, the NotI/XhoI-digested TbRAP1-5′UTR-loxP-HYGGFPTK plasmid and NotI/XhoI-digested TbRAP1-3′UTR-BSDGFPTK-loxP plasmid were sequentially transfected into SM cells. The A1 and A3 clones of TbRAP1F/+ (-Cre-EP1) were confirmed by Western and Southern analyses (see Fig. S1A and B in the supplemental material). The conditional Cre-expressing Cre-EP1 plasmid with the phleomycin resistance (BLE) marker (91) was subsequently inserted into an rDNA spacer region to generate the final TbRAP1F/+ strain. Clones A1 and B1 were verified by their sensitivity to hygromycin and blasticidin after induction of Cre expression by the use of doxycycline (Fig. S1C).
To establish the TbRAP1F/− strain, the NotI/XhoI-digested pSK-TbRAP1-ko-PUR plasmid was transfected into TbRAP1F/+ cells. The pSK-TbRAP1-ko-PUR plasmid contains PUR flanked by sequences upstream and downstream of the TbRAP1 open reading frame (ORF), respectively.
All TbRAP1F/F2H-mut strains were established using the same strategy. N-terminal F2H-tagged TbRAP1ΔNT, TbRAP1ΔBRCT, TbRAP1ΔMyb, TbRAP1ΔMybL, and NLS-TbRAP1ΔMybL mutants flanked by sequences upstream and downstream of the TbRAP1 gene, together with a PUR marker, were cloned into pSK to generate respective targeting constructs. The TbRAP1ΔMybLΔRCT-F2H-NLSr mutant flanked by sequences upstream and downstream of the TbRAP1 gene was also cloned into pSK with a PUR marker to generate the mutant targeting construct. All mutant targeting plasmids were digested with SacII (or with PvuII in the case of ΔMybLΔRCT) before transfection of the TbRAP1F/+ cells was performed to generate the corresponding TbRAP1F/F2H-mut strains. All mutant strains were confirmed by Southern analyses.
For examination of TbRAP1 self-interaction in the presence and absence of TbTRF, a TbTRF RNAi (TRFi) strain was first established by transfection of the NotI-digested pZJMβ-TbTRF-Mid1 RNAi construct (89) into SM cells. Subsequently, one endogenous TbRAP1 allele was tagged with an N-terminal F2H tag by transfection of a SacII-digested pSK-PUR-F2H-TbRAP1-tar2 construct into the TRFi cells.
Insertion of 13 C-terminal repeats of myc into one endogenous allele of importin α was performed in TbRAP1+/+, TbRAP1F2H+/+ TRFi, and TbRAP1F2H-ΔMybL/+ TRFi cells. TbRAP1F2H-ΔMybL/+ TRFi was obtained by replacing one endogenous TbRAP1 allele with the F2H-TbRAP1ΔMybL mutant in TRFi cells as described above. (The TbRAP1F2H-NLS-ΔMybL/+ TRFi strain used for TbRAP1 co-IP analysis was established in a similar way.) The importin α C-terminal myc13 tagging fragment was amplified by PCR with primers OBL-TbIMPORT-CT-MYC-FW (5′-GAGGGGGCACCTCAGCAGTTTGAGTTGGGGATGGATCATGGAGATCCAAATGGACAGCCACCACAGGGCCAGTTCGATCTTATCCCCGGGTTAATTAACG-3′) and OBL-TbIMPORT-CT-MYC-BW (5′-GTATATATATATACACGTGCCCTCCTTCTTCACCATTTTCATCCCTTTCATCTCGATCTCATTATATTTATCAACTTTTCTCATCTAGATTCCTTTGCCC-3′) by the use of a plasmid containing myc13 and a HYG marker as the template. The PCR product was then purified and transfected into TbRAP1+/+, TbRAP1F2H+/+ TRFi, and TbRAP1F2H-ΔMybL/+ TRFi cells.
Coimmunoprecipitation.
A total of 200 million T. brucei cells in log-phase growth were used for each IP using appropriate antibody or IgG (as a negative control). IP products were pulled down by the use of Dynabeads protein G (Life Technologies) and split equally for two Western blotting analyses using appropriate antibodies. A 1% volume of input sample was loaded as a control. Since F2H-TbRAP1ΔNT, F2H-TbRAP1ΔBRCT, and TbRAP1ΔMybLΔRCT-F2H-NLSr express at low levels, 500 million mutant cells were used for each IP. A 1% volume of input was loaded in Western analysis as a control.
Immunofluorescence analyses.
IF experiments were performed as described previously (86). Specifically, cells were fixed with 2% formaldehyde at room temperature for 10 min, permeabilized in 0.2% NP-40–1× phosphate-buffered saline (PBS) at room temperature for 5 min and blocked by the use of 1× PBS–0.2% cold fish gelatin–0.5% bovine serum albumin (BSA) at room temperature twice for 10 min each time, followed by incubation with the primary antibody at room temperature for 2 h and the secondary antibody at room temperature for 1 h. Cells were then washed with 1× PBS–0.2% cold fish gelatin–0.5% BSA and 1× PBS followed by staining with 0.5 μg/ml DAPI (4′,6-diamidino-2-phenylindole) and by mounting of coverslips on slides. Images were taken by a DeltaVision Elite deconvolution microscope. Images were deconvolved using SoftWoRx.
T. brucei cDNA library.
A normalized T. brucei cDNA library was prepared by Bio S&T. Briefly, using a modified SMART cDNA synthesis method, the total RNA from WT T. brucei cells was used for synthesis of cDNA with either oligo(dT) or a random primer. The cDNA was normalized and amplified, after which it was inserted into a modified pGAD T7 yeast expression vector.
Yeast 2-hybrid screen.
The TbRAP1 MybLike domain was cloned into the pBTM116 vector and transformed into the yeast strain L41 [MATα his 3D200 trp1-901 leu2-3,112 ade2 LYS::(lexAop)4-HIS3 URA3::(lexAop)8–lacZ gal4 gal80]. The resulting cells were transformed with the normalized T. brucei cDNA library. A total of 16.5 million primary transformants were plated onto synthetic drop-out (SD) plates without tryptophan, leucine, or histidine. A total of 711 clones were obtained from this initial screening. Subsequently, these clones were tested by the use of filter lift assays, and 508 candidates were verified to express the reporter lacZ gene. The pGAD T7 candidate plasmids were isolated from these yeast transformants and T. brucei gene insertions were analyzed by restriction digestion, PCR, and sequencing.
RNA-seq.
Cre expression was induced by the use of doxycycline in TbRAP1F/F2H-ΔMyb and TbRAP1F/+ cells for 30 h, after which total RNA was isolated and purified through RNeasy columns (Qiagen). Three independent inductions were performed as biological replicates. RNA samples were run on a BioAnalyzer 2100 system (Agilent Technologies) using an Agilent RNA 6000 Nano kit to verify the RNA quality and then sent to Novogene for library preparation and RNA high-throughput sequencing.
The following processes were performed at Novogene.
(i) RNA quantification and qualification.
RNA degradation and contamination were monitored on 1% agarose gels. RNA purity was checked using a NanoPhotometer spectrophotometer (Implen). RNA integrity and quantitation were assessed using an RNA Nano 6000 assay kit on a Bioanalyzer 2100 system (Agilent Technologies).
(ii) Library preparation for transcriptome sequencing.
Sequencing libraries were generated using a NEBNext Ultra RNA library prep kit for Illumina (NEB, USA) using 1 μg poly(A) RNA according to the manufacturer’s protocol, and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly(T) oligonucleotide-attached magnetic beads. Fragmentation was carried out using divalent cations under conditions of elevated temperature in NEBNext first-strand synthesis reaction buffer (5×). First-strand cDNA was synthesized using random hexamer primers and Moloney murine leukemia virus (M-MuLV) reverse transcriptase (RNase H-). Second-strand cDNA synthesis was performed using DNA polymerase I and RNase H. The remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with a hairpin loop structure was ligated to prepare for hybridization. In order to select cDNA fragments preferentially of length 150 to 200 bp, the library fragments were purified with an AMPure XP system (Beckman Coulter). A 3-μl volume of USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C followed by PCR, which was performed with Phusion high-fidelity DNA polymerase, universal PCR primers, and an Index (X) primer. PCR products were purified (AMPure XP system), and library quality was assessed on an Agilent Bioanalyzer 2100 system.
(iii) Clustering and sequencing (Novogene Experimental Department).
Clustering of the index-coded samples was performed on a cBot cluster generation system using a cBot-HiSeq (HS) paired-end (PE) cluster kit (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina platform and 125-bp and 150-bp paired-end reads were generated.
RNA-seq data analysis.
The RNA-seq data were analyzed by Novogene as follows.
(i) Quality control.
Raw reads of fastq format were first processed through the use of Novogene perl scripts. In this step, clean reads were obtained by removing reads containing adapters, reads containing poly(N), and low-quality reads. At the same time, the levels of Q20, Q30, and GC content of the clean reads were calculated. All downstream analyses were performed on the basis of the clean reads with high quality.
(ii) Mapping of reads to the reference genome.
The T. brucei Lister 427 genome TriTrypDB-45_TbruceiLister427_2018_Genome.fasta and its annotation TriTrypDB-45_TbruceiLister427_2018.gff were downloaded from TriTrypDB and used as the references. The index of the reference genome was built using hisat2 2.1.0, and paired-end clean reads were aligned to the reference genome using HISAT2.
(iii) Quantification of gene expression levels.
HTSeq v0.6.1 was used to calculate the number of reads mapped to each gene. The number of fragments per kilobase per million (FPKM) was calculated for each gene on the basis of the length of the gene and the number of reads mapped to the gene.
(iv) Differential expression analysis.
Differential expression analysis of two conditions/group (three biological replicates per condition) was performed using the DESeq R package (1.18.0). The DESeq R package provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P values were adjusted using the Benjamini and Hochberg approach for controlling the false-discovery rate. Genes with an adjusted P value of <0.05 found by DESeq were assigned as differentially expressed.
Data availability.
The RNA-seq data set determined in this work has been submitted to NCBI GEO under accession number GSE143456.
ACKNOWLEDGMENTS
We thank G. A. M. Cross for providing us the Cre-EP1 plasmid. We thank Keith Gull for providing us the TAT-1 antibody. We thank Amit Gaurav, Arpita Saha, Maiko Tonini, and Brittny Schnur for comments and suggestions with respect to the manuscript.
This work was supported by NIH R01 grant AI066095 (Principal Investigator [PI], B.L.) and NIH S10 grant S10OD025252 (PI, B.L.). Construction of the normalized T. brucei cDNA library was supported by a Cleveland State University (CSU) 2010 Faculty Research and Development award (PI, B.L.). The publication cost was partly met by support from the Center for Gene Regulation in Health and Disease (GRHD) at CSU.
REFERENCES
- 1.Maciejowski J, de Lange T. 2017. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol 18:175–186. doi: 10.1038/nrm.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen JJ. 2008. The telomerase database. Nucleic Acids Res 36:D339–D343. doi: 10.1093/nar/gkm700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martínez P, Blasco MA. 2015. Replicating through telomeres: a means to an end. Trends Biochem Sci 40:504–515. doi: 10.1016/j.tibs.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Li B, Oestreich S, de Lange T. 2000. Identification of human Rap1: implications for telomere evolution. Cell 101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 5.Scherthan H, Jerratsch M, Li B, Smith S, Hulten M, Lock T, de Lange T. 2000. Mammalian meiotic telomeres: protein composition and redistribution in relation to nuclear pores. Mol Biol Cell 11:4189–4203. doi: 10.1091/mbc.11.12.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan M, Wei C, Price CM. 2003. The telomeric protein Rap1 is conserved in vertebrates and is expressed from a bidirectional promoter positioned between the Rap1 and KARS genes. Gene 323:1–10. doi: 10.1016/j.gene.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 7.Viera A, Parra MT, Page J, Santos JL, Rufas JS, Suja JA. 2003. Dynamic relocation of telomere complexes in mouse meiotic chromosomes. Chromosome Res 11:797–807. doi: 10.1023/B:CHRO.0000005781.71466.da. [DOI] [PubMed] [Google Scholar]
- 8.Shore D, Nasmyth K. 1987. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell 51:721–732. doi: 10.1016/0092-8674(87)90095-X. [DOI] [PubMed] [Google Scholar]
- 9.Longtine MS, Wilson NM, Petracek ME, Berman J. 1989. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAP1. Curr Genet 16:225–239. doi: 10.1007/bf00422108. [DOI] [PubMed] [Google Scholar]
- 10.Chikashige Y, Hiraoka Y. 2001. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol 11:1618–1623. doi: 10.1016/s0960-9822(01)00457-2. [DOI] [PubMed] [Google Scholar]
- 11.Kanoh J, Ishikawa F. 2001. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 11:1624–1630. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- 12.Park MJ, Jang YK, Choi ES, Kim HS, Park SD. 2002. Fission yeast Rap1 homolog is a telomere-specific silencing factor and interacts with Taz1p. Mol Cells 13:327–333. [PubMed] [Google Scholar]
- 13.Yu EY, Yen WF, Steinberg-Neifach O, Lue NF. 2010. Rap1 in Candida albicans: an unusual structural organization and a critical function in suppressing telomere recombination. Mol Cell Biol 30:1254–1268. doi: 10.1128/MCB.00986-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoekstra R, Groeneveld P, Van Verseveld HW, Stouthamer AH, Planta RJ. 1994. Transcription regulation of ribosomal protein genes at different growth rates in continuous cultures of Kluyveromyces yeasts. Yeast 10:637–651. doi: 10.1002/yea.320100508. [DOI] [PubMed] [Google Scholar]
- 15.Haw R, Yarragudi AD, Uemura H. 2001. Isolation of a Candida glabrata homologue of RAP1, a regulator of transcription and telomere function in Saccharomyces cerevisiae. Yeast 18:1277–1284. doi: 10.1002/yea.775. [DOI] [PubMed] [Google Scholar]
- 16.Yang X, Figueiredo LM, Espinal A, Okubo E, Li B. 2009. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell 137:99–109. doi: 10.1016/j.cell.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller KM, Ferreira MG, Cooper JP. 2005. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J 24:3128–3135. doi: 10.1038/sj.emboj.7600779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bae NS, Baumann P. 2007. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol Cell 26:323–334. doi: 10.1016/j.molcel.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 19.Sarthy J, Bae NS, Scrafford J, Baumann P. 2009. Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J 28:3390–3399. doi: 10.1038/emboj.2009.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bombarde O, Boby C, Gomez D, Frit P, Giraud-Panis MJ, Gilson E, Salles B, Calsou P. 2010. TRF2/RAP1 and DNA-PK mediate a double protection against joining at telomeric ends. EMBO J 29:1573–1584. doi: 10.1038/emboj.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gustafsson C, Rhodin Edsö J, Cohn M. 2011. Rap1 binds single-stranded DNA at telomeric double- and single-stranded junctions and competes with Cdc13 protein. J Biol Chem 286:45174–45185. doi: 10.1074/jbc.M111.300517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rai R, Chen Y, Lei M, Chang S. 2016. TRF2-RAP1 is required to protect telomeres from engaging in homologous recombination-mediated deletions and fusions. Nat Commun 7:10881. doi: 10.1038/ncomms10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Runnberg R, Narayanan S, Cohn M. 2017. Rap1 and Cdc13 have complementary roles in preventing exonucleolytic degradation of telomere 5’ ends. Sci Rep 7:8729. doi: 10.1038/s41598-017-08663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pardo B, Marcand S. 2005. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J 24:3117–3127. doi: 10.1038/sj.emboj.7600778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vodenicharov MD, Laterreur N, Wellinger RJ. 2010. Telomere capping in non-dividing yeast cells requires Yku and Rap1. EMBO J 29:3007–3019. doi: 10.1038/emboj.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang CW, Tseng SF, Yu CJ, Chung CY, Chang CY, Pobiega S, Teng SC. 2017. Telomere shortening triggers a feedback loop to enhance end protection. Nucleic Acids Res 45:8314–8328. doi: 10.1093/nar/gkx503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. 2010. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 327:1657–1661. doi: 10.1126/science.1185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lustig AJ, Kurtz S, Shore D. 1990. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science 250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- 29.Kyrion G, Boakye KA, Lustig AJ. 1992. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol Cell Biol 12:5159–5173. doi: 10.1128/mcb.12.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krauskopf A, Blackburn EH. 1996. Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature 383:354–357. doi: 10.1038/383354a0. [DOI] [PubMed] [Google Scholar]
- 31.Marcand S, Wotton D, Gilson E, Shore D. 1997. Rap1p and telomere length regulation in yeast. Ciba Found Symp 211:76–93. doi: 10.1002/9780470515433.ch6. [DOI] [PubMed] [Google Scholar]
- 32.Marcand S, Gilson E, Shore D. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 33.Krauskopf A, Blackburn EH. 1998. Rap1 protein regulates telomere turnover in yeast. Proc Natl Acad Sci U S A 95:12486–12491. doi: 10.1073/pnas.95.21.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gurevich R, Smolikov S, Maddar H, Krauskopf A. 2003. Mutant telomeres inhibit transcriptional silencing at native telomeres of the yeast Kluyveromyces lactis. Mol Genet Genomics 268:729–738. doi: 10.1007/s00438-002-0788-9. [DOI] [PubMed] [Google Scholar]
- 35.Li B, de Lange T. 2003. Rap1 affects the length and heterogeneity of human telomeres. Mol Biol Cell 14:5060–5068. doi: 10.1091/mbc.e03-06-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martínez P, Gómez-López G, Pisano DG, Flores JM, Blasco MA. 2016. A genetic interaction between RAP1 and telomerase reveals an unanticipated role for RAP1 in telomere maintenance. Aging Cell 15:1113–1125. doi: 10.1111/acel.12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Liu Z, Liu X, Wang S, Zhang Y, He X, Sun S, Ma S, Shyh-Chang N, Liu F, Wang Q, Wang X, Liu L, Zhang W, Song M, Liu GH, Qu J. 2019. Telomere-dependent and telomere-independent roles of RAP1 in regulating human stem cell homeostasis. Protein Cell 10:649–667. doi: 10.1007/s13238-019-0610-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kyrion G, Liu K, Liu C, Lustig AJ. 1993. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev 7:1146–1159. doi: 10.1101/gad.7.7a.1146. [DOI] [PubMed] [Google Scholar]
- 39.Liu C, Mao X, Lustig AJ. 1994. Mutational analysis defines a C-terminal tail domain of RAP1 essential for telomeric silencing in Saccharomyces cerevisiae. Genetics 138:1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buck SW, Shore D. 1995. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev 9:370–384. doi: 10.1101/gad.9.3.370. [DOI] [PubMed] [Google Scholar]
- 41.Liu C, Lustig AJ. 1996. Genetic analysis of Rap1p/Sir3p interactions in telomeric and HML silencing in Saccharomyces cerevisiae. Genetics 143:81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo K, Vega-Palas MA, Grunstein M. 2002. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev 16:1528–1539. doi: 10.1101/gad.988802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gallegos-García V, Pan S-J, Juárez-Cepeda J, Ramírez-Zavaleta CY, Martin-del-Campo MB, Martínez-Jiménez V, Castaño I, Cormack B, De Las Peñas A. 2012. A novel downstream regulatory element cooperates with the silencing machinery to repress EPA1 expression in Candida glabrata. Genetics 190:1285–1297. doi: 10.1534/genetics.111.138099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pandya UM, Sandhu R, Li B. 2013. Silencing subtelomeric VSGs by Trypanosoma brucei RAP1 at the insect stage involves chromatin structure changes. Nucleic Acids Res 41:7673–7682. doi: 10.1093/nar/gkt562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. 1990. Position effect at S. cerevisiae telomeres: reversible repression of pol II transcription. Cell 63:751–762. doi: 10.1016/0092-8674(90)90141-Z. [DOI] [PubMed] [Google Scholar]
- 46.Brindle PK, Holland JP, Willett CE, Innis MA, Holland MJ. 1990. Multiple factors bind the upstream activation sites of the yeast enolase genes ENO1 and ENO2: ABFI protein, like repressor activator protein RAP1, binds cis-acting sequences which modulate repression or activation of transcription. Mol Cell Biol 10:4872–4885. doi: 10.1128/mcb.10.9.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sussel L, Shore D. 1991. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc Natl Acad Sci U S A 88:7749–7753. doi: 10.1073/pnas.88.17.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mizuta K, Tsujii R, Warner JR, Nishiyama M. 1998. The C-terminal silencing domain of Rap1p is essential for the repression of ribosomal protein genes in response to a defect in the secretory pathway. Nucleic Acids Res 26:1063–1069. doi: 10.1093/nar/26.4.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lieb JD, Liu X, Botstein D, Brown PO. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet 28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 50.Piña B, Fernández-Larrea J, García-Reyero N, Idrissi F-Z. 2003. The different (sur)faces of Rap1p. Mol Genet Genomics 268:791–798. doi: 10.1007/s00438-002-0801-3. [DOI] [PubMed] [Google Scholar]
- 51.Garbett KA, Tripathi MK, Cencki B, Layer JH, Weil PA. 2007. Yeast TFIID serves as a coactivator for Rap1p by direct protein-protein interaction. Mol Cell Biol 27:297–311. doi: 10.1128/MCB.01558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bendjennat M, Weil PA. 2008. The transcriptional repressor activator protein Rap1p is a direct regulator of TATA-binding protein. J Biol Chem 283:8699–8710. doi: 10.1074/jbc.M709436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang D, Xiong Y, Kim H, He Q, Li Y, Chen R, Songyang Z. 2011. Human telomeric proteins occupy selective interstitial sites. Cell Res 21:1013–1027. doi: 10.1038/cr.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martinez P, Thanasoula M, Carlos AR, Gomez-Lopez G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA. 2010. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol 12:768–780. doi: 10.1038/ncb2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez P, Gomez-Lopez G, Garcia F, Mercken E, Mitchell S, Flores JM, de Cabo R, Blasco MA. 2013. RAP1 protects from obesity through its extratelomeric role regulating gene expression. Cell Rep 3:2059–2074. doi: 10.1016/j.celrep.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moretti P, Freeman K, Coodly L, Shore D. 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev 8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 57.Moretti P, Shore D. 2001. Multiple interactions in sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol Cell Biol 21:8082–8094. doi: 10.1128/MCB.21.23.8082-8094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hardy CF, Sussel L, Shore D. 1992. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev 6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 59.Wotton D, Shore D. 1997. Novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev 11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 60.Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. 1997. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J 11:68–76. doi: 10.1096/fasebj.11.1.9034168. [DOI] [PubMed] [Google Scholar]
- 61.Callebaut I, Mornon JP. 1997. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett 400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- 62.Zhang X, Morera S, Bates PA, Whitehead PC, Coffer AI, Hainbucher K, Nash RA, Sternberg MJ, Lindahl T, Freemont PS. 1998. Structure of an XRCC1 BRCT domain: a new protein-protein interaction module. EMBO J 17:6404–6411. doi: 10.1093/emboj/17.21.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu X, Chini CC, He M, Mer G, Chen J. 2003. The BRCT domain is a phospho-protein binding domain. Science 302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 64.Rodriguez M, Yu X, Chen J, Songyang Z. 2003. Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains. J Biol Chem 278:52914–52918. doi: 10.1074/jbc.C300407200. [DOI] [PubMed] [Google Scholar]
- 65.Fujita I, Tanaka M, Kanoh J. 2012. Identification of the functional domains of the telomere protein Rap1 in Schizosaccharomyces pombe. PLoS One 7:e49151. doi: 10.1371/journal.pone.0049151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Graham IR, Haw RA, Spink KG, Halden KA, Chambers A. 1999. In vivo analysis of functional regions within yeast Rap1p. Mol Cell Biol 19:7481–7490. doi: 10.1128/mcb.19.11.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feeser EA, Wolberger C. 2008. Structural and functional studies of the Rap1 C-terminus reveal novel separation-of-function mutants. J Mol Biol 380:520–531. doi: 10.1016/j.jmb.2008.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Y, Rai R, Zhou ZR, Kanoh J, Ribeyre C, Yang Y, Zheng H, Damay P, Wang F, Tsujii H, Hiraoka Y, Shore D, Hu HY, Chang S, Lei M. 2011. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat Struct Mol Biol 18:213–221. doi: 10.1038/nsmb.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Konig P, Giraldo R, Chapman L, Rhodes D. 1996. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell 85:125–136. doi: 10.1016/s0092-8674(00)81088-0. [DOI] [PubMed] [Google Scholar]
- 70.Hanaoka S, Nagadoi A, Yoshimura S, Aimoto S, Li B, de Lange T, Nishimura Y. 2001. NMR structure of the hRap1 Myb motif reveals a canonical three-helix bundle lacking the positive surface charge typical of Myb DNA-binding domains. J Mol Biol 312:167–175. doi: 10.1006/jmbi.2001.4924. [DOI] [PubMed] [Google Scholar]
- 71.Barry JD, McCulloch R. 2001. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv Parasitol 49:1–70. doi: 10.1016/s0065-308x(01)49037-3. [DOI] [PubMed] [Google Scholar]
- 72.Cross GAM, Kim HS, Wickstead B. 2014. Capturing the variant surface glycoprotein repertoire (the VSGnome) of Trypanosoma brucei Lister 427. Mol Biochem Parasitol 195:59–73. doi: 10.1016/j.molbiopara.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 73.Hertz-Fowler C, Figueiredo LM, Quail MA, Becker M, Jackson A, Bason N, Brooks K, Churcher C, Fahkro S, Goodhead I, Heath P, Kartvelishvili M, Mungall K, Harris D, Hauser H, Sanders M, Saunders D, Seeger K, Sharp S, Taylor JE, Walker D, White B, Young R, Cross GAM, Rudenko G, Barry JD, Louis EJ, Berriman M. 2008. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One 3:e3527. doi: 10.1371/journal.pone.0003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Müller LSM, Cosentino RO, Förstner KU, Guizetti J, Wedel C, Kaplan N, Janzen CJ, Arampatzi P, Vogel J, Steinbiss S, Otto TD, Saliba AE, Sebra RP, Siegel TN. 2018. Genome organization and DNA accessibility control antigenic variation in trypanosomes. Nature 563:121–125. doi: 10.1038/s41586-018-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gunzl A, Bruderer T, Laufer G, Schimanski B, Tu LC, Chung HM, Lee PT, Lee MG. 2003. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot Cell 2:542–551. doi: 10.1128/ec.2.3.542-551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kolev NG, Günzl A, Tschudi C. 2017. Metacyclic VSG expression site promoters are recognized by the same general transcription factor that is required for RNA polymerase I transcription of bloodstream expression sites. Mol Biochem Parasitol 216:52–55. doi: 10.1016/j.molbiopara.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cross G. 1975. Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology 71:393–417. doi: 10.1017/s003118200004717x. [DOI] [PubMed] [Google Scholar]
- 78.Cestari I, Stuart K. 2018. Transcriptional regulation of telomeric expression sites and antigenic variation in trypanosomes. Curr Genomics 19:119–132. doi: 10.2174/1389202918666170911161831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gunzl A, Kirkham JK, Nguyen TN, Badjatia N, Park SH. 2015. Mono-allelic VSG expression by RNA polymerase I in Trypanosoma brucei: expression site control from both ends? Gene 556:68–73. doi: 10.1016/j.gene.2014.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Faria J, Glover L, Hutchinson S, Boehm C, Field MC, Horn D. 2019. Monoallelic expression and epigenetic inheritance sustained by a Trypanosoma brucei variant surface glycoprotein exclusion complex. Nat Commun 10:3023. doi: 10.1038/s41467-019-10823-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Glover L, Hutchinson S, Alsford S, Horn D. 2016. VEX1 controls the allelic exclusion required for antigenic variation in trypanosomes. Proc Natl Acad Sci U S A 113:7225–7230. doi: 10.1073/pnas.1600344113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Myler PJ, Allison J, Agabian N, Stuart K. 1984. Antigenic variation in African trypanosomes by gene replacement or activation of alternative telomeres. Cell 39:203–211. doi: 10.1016/0092-8674(84)90206-X. [DOI] [PubMed] [Google Scholar]
- 83.Myler P, Nelson RG, Agabian N, Stuart K. 1984. Two mechanisms of expression of a variant antigen gene of Trypanosoma brucei. Nature 309:282–284. doi: 10.1038/309282a0. [DOI] [PubMed] [Google Scholar]
- 84.McCulloch R, Morrison LJ, Hall JPJ. 2015. DNA recombination strategies during antigenic variation in the African trypanosome. Microbiol Spectr doi: 10.1128/microbiolspec.MDNA3-0016-2014. [DOI] [PubMed] [Google Scholar]
- 85.Jehi SE, Li X, Sandhu R, Ye F, Benmerzouga I, Zhang M, Zhao Y, Li B. 2014. Suppression of subtelomeric VSG switching by Trypanosoma brucei TRF requires its TTAGGG repeat-binding activity. Nucleic Acids Res 42:12899–12911. doi: 10.1093/nar/gku942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jehi SE, Wu F, Li B. 2014. Trypanosoma brucei TIF2 suppresses VSG switching by maintaining subtelomere integrity. Cell Res 24:870–885. doi: 10.1038/cr.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jehi SE, Nanavaty V, Li B. 2016. Trypanosoma brucei TIF2 and TRF suppress VSG switching using overlapping and independent mechanisms. PLoS One 11:e0156746. doi: 10.1371/journal.pone.0156746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nanavaty V, Sandhu R, Jehi SE, Pandya UM, Li B. 2017. Trypanosoma brucei RAP1 maintains telomere and subtelomere integrity by suppressing TERRA and telomeric RNA:DNA hybrids. Nucleic Acids Res 45:5785–5796. doi: 10.1093/nar/gkx184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li B, Espinal A, Cross G. 2005. Trypanosome telomeres are protected by a homologue of mammalian TRF2. Mol Cell Biol 25:5011–5021. doi: 10.1128/MCB.25.12.5011-5021.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ngo H, Tschudi C, Gull K, Ullu E. 1998. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci U S A 95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Scahill MD, Pastar I, Cross G. 2008. CRE recombinase-based positive-negative selection systems for genetic manipulation in Trypanosoma brucei. Mol Biochem Parasitol 157:73–82. doi: 10.1016/j.molbiopara.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. 2010. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res 38:D161–D166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Bohme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth TJ, Churcher C, Clark LN, Corton CH, Cronin A, Davies RM, Doggett J, Djikeng A, Feldblyum T, Field MC, Fraser A, Goodhead I, Hance Z, Harper D, Harris BR, Hauser H, Hostetler J, Ivens A, Jagels K, Johnson D, Johnson J, Jones K, Kerhornou AX, Koo H, Larke N, et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 94.Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, Gardner MJ, Gingle A, Grant G, Harb OS, Heiges M, Hertz-Fowler C, Houston R, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Logan FJ, Miller JA, Mitra S, Myler PJ, Nayak V, Pennington C, Phan I, Pinney DF, Ramasamy G, Rogers MB, Roos DS, Ross C, Sivam D, Smith DF, Srinivasamoorthy G, Stoeckert CJJ, Subramanian S, Thibodeau R, Tivey A, Treatman C, Velarde G, Wang H. 2010. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 38:D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oka M, Yoneda Y. 2018. Importin α: functions as a nuclear transport factor and beyond. Proc Jpn Acad Ser B Phys Biol Sci 94:259–274. doi: 10.2183/pjab.94.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cai Y, Kandula V, Kosuru R, Ye X, Irwin MG, Xia Z. 2017. Decoding telomere protein Rap1: its telomeric and nontelomeric functions and potential implications in diabetic cardiomyopathy. Cell Cycle 16:1765–1773. doi: 10.1080/15384101.2017.1371886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. 1994. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- 98.Cutler G, Perry KM, Tjian R. 1998. Adf-1 is a nonmodular transcription factor that contains a TAF-binding Myb-like motif. Mol Cell Biol 18:2252–2261. doi: 10.1128/mcb.18.4.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Berger SL, Piña B, Silverman N, Marcus GA, Agapite J, Regier JL, Triezenberg SJ, Guarente L. 1992. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70:251–265. doi: 10.1016/0092-8674(92)90100-Q. [DOI] [PubMed] [Google Scholar]
- 100.Freitas N, Cunha C. 2009. Mechanisms and signals for the nuclear import of proteins. Curr Genomics 10:550–557. doi: 10.2174/138920209789503941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Song H, Kim W, Kim SH, Kim KT. 2016. VRK3-mediated nuclear localization of HSP70 prevents glutamate excitotoxicity-induced apoptosis and Aβ accumulation via enhancement of ERK phosphatase VHR activity. Sci Rep 6:38452. doi: 10.1038/srep38452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhu XD, Kuster B, Mann M, Petrini JH, Lange T. 2000. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet 25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- 103.Broccoli D, Smogorzewska A, Chong L, de Lange T. 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet 17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 104.Kim SH, Kaminker P, Campisi J. 1999. TIN2, a new regulator of telomere length in human cells. Nat Genet 23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ye JZ, Donigian JR, van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, de Lange T. 2004. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem 279:47264–47271. doi: 10.1074/jbc.M409047200. [DOI] [PubMed] [Google Scholar]
- 106.Wirtz E, Leal S, Ochatt C, Cross G. 1999. A tightly regulated inducible expression system for dominant negative approaches in Trypanosoma brucei. Mol Biochem Parasitol 99:89–101. doi: 10.1016/S0166-6851(99)00002-X. [DOI] [PubMed] [Google Scholar]
- 107.Woods A, Sherwin T, Sasse R, MacRae TH, Baines AJ, Gull K. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J Cell Sci 93:491–500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Validation of Cre-loxP-mediated conditional deletion of TbRAP1. (A) Western blotting using a rabbit GFP antibody (Life Technologies) to detect HYG-GFP-TK and BSD-GFP-TK expression in TbRAP1F/+ (-Cre-Ep1) cells before and after transient transfection of Cre-EP1 (91). (B) Southern blotting of BamHI/XbaI-digested genomic DNA isolated from TbRAP1F/+ (-Cre-Ep1) cells before and after transient transfection of Cre-EP1 (91). A DNA fragment containing the full-length TbRAP1 gene was used as the probe. The WT TbRAP1 allele gave a 7.9-kb band, while the floxed TbRAP1 allele gave a 4.9-kb band. (C) Growth curves of two clones of TbRAP1F/+ cells (with Cre-EP1 targeted to an rDNA spacer region) before and after induction of Cre by doxycycline. Cells were selected with different combinations of antibiotics as indicated. The Cre-EP1 plasmid has a phleomycin resistance (BLE) marker (91). Download FIG S1, EPS file, 1.9 MB (1.9MB, eps) .
Copyright © 2020 Afrin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Southern analysis confirmed the TbRAP1F/F2H-ΔMyb genotype. Genomic DNA from TbRAP1F/+ and a number of candidates of the TbRAP1F/F2H-ΔMyb strain was digested with EcoRI and BamHI and probed with a DNA fragment containing the region of the TbRAP1 gene before the BRCT domain. The three different TbRAP1 alleles are labeled. C1, B1, and B3 were positive clones, while the other three did not have the intended genotype. (B) The rabbit TbRAP1 antibody (16) used in this study recognizes an epitope(s) inside the MybLike domain. (Left) Polyacrylamide gel electrophoresis of the partially purified TRX-His6-TbRAP1-MybLike fragment. (Right) Western blotting of the same recombinant fragment using the rabbit TbRAP1 antibody. (C and D) Classification of genes affected in Cre-induced TbRAP1F/F2H-ΔMyb cells. Distributions (percent of total affected genes) of different types of genes whose mRNA levels were upregulated (C) or downregulated (D) are shown. The number of genes affected in each category is listed in parentheses. Download FIG S2, EPS file, 2.7 MB (2.7MB, eps) .
Copyright © 2020 Afrin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
T. brucei strains used in the current study. Download Table S1, DOCX file, 0.01 MB (15.3KB, docx) .
Copyright © 2020 Afrin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA sequence read coverage in all VSG bloodstream-form expression sites (BESs) for Cre-induced TbRAP1F/+ and TbRAP1F/F2H-ΔMyb cells. RNA samples were isolated from three independently induced cells in both strains. RNA sequence reads were mapped to the T. brucei Lister 427 genome TriTrypDB-45_TbruceiLister427_2018_Genome.fasta, which was obtained from TriTrypDB. Read coverage for each sample was viewed in IGV. Each BES is labeled, and the genes inside each BES are listed at the bottom. The whole BES region is shown with the promoter located on the left side. The y axis data represent the number of RNA reads. The range is indicated on the left of each trace. BES1 was fully active in the cells analyzed. Download FIG S3, PDF file, 1.2 MB (1.2MB, pdf) .
Copyright © 2020 Afrin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Southern analyses confirmed the genotypes of TbRAP1F/F2H-ΔMybL (A) and TbRAP1F/F2H-NLS-ΔMybL (C) cells. Genomic DNA was digested with EcoRI and probed with a DNA fragment amplified by PCR with the forward primer (5′-GCGCCTCGAGACCGACGTTGACGGTG-3′) and the backward primer (5′-GCGCTCTAGAACACGAAGAAATCGGGACGTG-3′) using TbRAP1 as the template. Western analyses showed the expression levels of F2H-TbRAP1ΔMybL (B) and F2H-NLS-TbRAP1ΔMybL (D) in the same clones as those indicated in panels A and C, respectively. Download FIG S4, EPS file, 1.1 MB (1.1MB, eps) .
Copyright © 2020 Afrin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PCR confirmed the endogenous tagging of importin α with a C-terminal myc13 tag. Genomic DNAs from the parent TbRAP1+/+ strain (as a negative control) and three transfected cells in the TbRAP1+/+, TbRAP1F2H+/+ TRFi, and TbRAP1F2H-ΔMybL/+ TRFi background were used as the templates. Forward primer (5′-CACTTTGTCTCCTCGAGACA-3′) and backward primer (5′-CGATCAGAAACTTCTCGACAG-3′) were used in the PCR amplification. A product of 1.26 kb is expected for correct targeting of the myc13 tag. Download FIG S5, EPS file, 1.2 MB (1.2MB, eps) .
Copyright © 2020 Afrin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The TbRAP1F/F2H-ΔNT, TbRAP1F/F2H-ΔBRCT, TbRAP1F/F2H-NLS-ΔMybL, and TbRAP1F/ΔMybLΔRCT-F2H-NLSr cells exhibited a growth arrest phenotype after induction of Cre. (A to C) Southern analyses confirmed the genotype of TbRAP1F/F2H-ΔNT (A), TbRAP1F/F2H-ΔBRCT (B), and TbRAP1F/ΔMybLΔRCT-F2H-NLSr (C) cells. Genomic DNAs were digested with XbaI and BamHI in the experiments represented in panels A and C and with EcoRI in the experiments represented in panel B. A DNA fragment containing the TbRAP1 sequence between the BRCT domain and the Myb domain was used as the probe in the experiments represented in panels A and C. A DNA fragment containing the N-terminal region of TbRAP1 was used as the probe used in the experiments represented in panel B. (D to G) Growth curves of TbRAP1F/F2H-NLS-ΔMybL (D), TbRAP1F/F2H-ΔNT (E), TbRAP1F/F2H-ΔBRCT (F), and TbRAP1F/ΔMybLΔRCT-F2H-NLSr (G) cells before and after the induction of Cre expression are shown. Download FIG S6, EPS file, 1.9 MB (1.9MB, eps) .
Copyright © 2020 Afrin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The RNA-seq data set determined in this work has been submitted to NCBI GEO under accession number GSE143456.