Abstract
Background
The CDKN2A/B locus contains crucial tumor suppressors and a lncRNA gene ANRIL. However, the mechanisms that coordinately regulate their expression levels are not clear.
Methods
Novel RNAs transcribed from the CDKN2A gene were screened by CDKN2A-specific RNA capture deep-sequencing and confirmed by Northern blotting and clone-sequencing. Long non-coding RNA (lncRNA) binding proteins were characterized by RNA pull-down combined with mass spectrometry and RNA immunoprecipitation. LncRNA functions in human cells were studied using a set of biological assays in vitro and in vivo.
Results
We characterized a novel lncRNA, P14AS with its promoter in the antisense strand of the fragment near CDKN2A exon 1b in human cells. The mature P14AS is a three-exon linear cytoplasmic lncRNA (1043-nt), including an AU-rich element (ARE) in exon 1. P14AS decreases AUF1-ANRIL/P16 RNA interaction and then increases ANRIL/P16 expression by competitively binding to AUF1 P37 and P40 isoforms. Interestingly, P14AS significantly promoted the proliferation of cancer cells and tumor formation in NOD-SCID mice in a P16-independent pattern. Moreover, in human colon cancer tissues, the expression levels of P14AS and ANRIL lncRNAs were significantly upregulated compared with the paired normal tissues.
Conclusion
A novel lncRNA, P14AS, transcribed from the antisense strand of the CDKN2A/P14 gene, promotes colon cancer development by cis upregulating the expression of oncogenic ANRIL.
Keywords: lncRNA, CDKN2A, P14AS, ANRIL, P16, AUF1, Colon cancer
Background
Three important tumor suppressor genes (P16INK4A, P14ARF and P15INK4B) and the oncogenic lncRNA ANRIL (CDKN2B-AS1) resides in the human CDKN2A/B locus at chromosome 9p21 [1]. P16 and P15 proteins target CDK4/6 through the CDK4/6-RB-E2F pathway, and inactivation of P15 and/or P16 allows cells to escape cell cycle arrest in G1 while P14 protein binds to MDM2 and results in P53 activation. While ANRIL was reported to downregulate P15 and P16 expression by interacting with components of polycomb repressive complex-1/-2 [2–5], ANRIL was also found to be coordinately transcribed with P16 in cancer cells and transcriptionally repressed by P16 DNA methylation [6]. This gene locus is frequently inactivated in cancer genome by somatic copy-number deletion and DNA methylation, leading to familial pancreatic cancer and melanoma.
AUF1 is an essential RNA binding protein that promotes the decay of many cancer-related RNAs, including P16, c-MYC, NEAT1 [7–10]. In this study, we designed a probe set to capture all possible transcripts from the CDKN2A locus and performed an extra-deep sequencing (CDKN2A RNACap-Seq) to identify given and novel RNAs from this gene and flanking regions. We characterized, for the first time, a novel lncRNA called P14AS (NCBI GenBank MK574077) transcribed from the antisense strand of the fragment around exon 1β of the CDKN2A gene. We found that AUF1 binds to P14AS and the AUF1 binding of the ARE (AU-rich element) in exon 1 of the P14AS gene increases ANRIL/P16 level. Furthermore, a significantly higher level of P14AS exists in colon cancers compared to paired normal tissues and that P14AS could markedly promote the proliferation of cancer cells and tumor formation in a P16-independent pattern in vitro and in vivo.
Methods
Cell culture and tissue samples
The human cell line HEK293T was kindly provided by Professor Yasuhito Yuasa at Tokyo Medical and Dental University. HCT116 and SW480 cells were kindly provided by Professor Yuanjia Chen at Peking Union Medical College Hospital. The MCF7 cell line was kindly provided by Professor Yuntao Xie; BGC823, MGC803 and SGC7901 cells were kindly provided by Professor Yang Ke; A549 and HEK293FT cells were kindly provided by Professor Zhiqian Zhang; the HepG2 cell line was kindly provided by Professor Qingyun Zhang; the AGS cell line was kindly provided by Professor Chengchao Shou at Peking University Cancer Hospital and Institute.
The colon cancer (CC) tissues, paired normal tissues from the surgical margin (SM, > 5 cm from cancer lesion) from CC patients (N = 172, including 75 cases in the pilot study and 97 additional cases in the clinical association analysis), and normal colon mucosa biopsy samples from noncancer patients (N = 50) were collected and stored at − 70 °C at Peking University Cancer Hospital from 2004 to 2011. Research protocols were approved by the Institutional Review Board of the Peking University Cancer Hospital and Institute, China. Clinical and histopathological data for each patient were obtained according to approved institutional guidelines.
CDKN2A-RNACap-Seq
Total RNA was used to synthesize complementary DNA (cDNA). In total, 3 μg of cDNA was fragmented by nebulization. The fragmented DNA was repaired, and an ‘A’ was ligated to the 3′ end. Illumina adapters were then ligated to the fragments, and the sample was size-selected with a 350–400-bp product. The size-selected product was amplified by PCR. Each sample was tagged with a unique index during this procedure, and the final product was validated using a Bioanalyzer (Agilent, USA). The amplified DNA (1 μg) was mixed with a CDKN2A-specific biotinylated probe panel to tile along chr9: 21,959,171-21,999,170 (30-kb; hg19; excluding repeat sequences; MyGenostics GenCap Enrichment Technologies, Beijing, China; Fig. 1a), held at 65 °C with the PCR lid heat on for 22 h for hybridization. The hybridized DNA was heated at 95 °C for 7 min and 65 °C for 2 min in the PCR machine. Then, 23 μL of the 65 °C prewarmed Buffer HY (MyGenostics, Beijing, China) was added to the mix. 50 μL of MyOne beads (Life Technology, USA) was washed in 500 μL 1× binding buffer 3 times and resuspended in 80 μL 1× binding buffer. Then, 64 μL 2× binding buffer was added to the hybrid mix and transferred to the tube with 80 μL MyOne beads. The mixture was rotated for 1 h on a rotator. The beads were then washed once with WB1 buffer at room temperature for 15 min and three times WB3 buffer at 65 °C for 15 min. The bound DNA was then eluted with Buffer Elute. The eluted DNA was finally amplified for 15 cycles using the following program: 98 °C for 30 s (1 cycle); 98 °C for 25 s, 65 °C for 30 s, 72 °C for 30 s (15 cycles); and 72 °C for 5 min (1 cycle). The PCR product was purified using SPRI beads (Beckman Coulter) according to the manufacturer’s protocol. The enrichment libraries were sequenced on an Illumina HiSeq 2000 sequencer for paired reads of 100-bp.
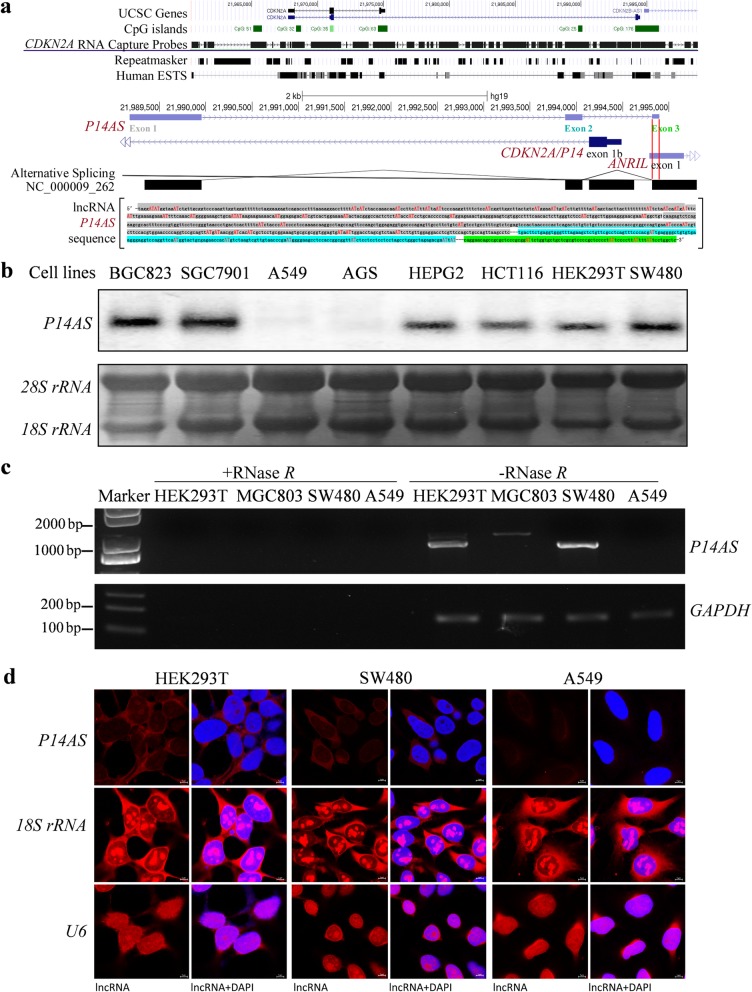
Fig. 1.
Characterization of lncRNA P14AS in cancer cells. a Illustration of the location of CDKN2A RNA capture probes (30 k) and lncRNA P14AS transcribed from the antisense strand of the sequence around exon 1β of the CDKN2A gene (chr9: 21,989,178-21,994,898 in the human genome GRCh37/hg19). Sequences for the three exons are highlighted with different colors: exon 1 in gray, exon 2 in blue, and exon 3 in green. AT (AU in RNA) sites are marked with red letters. b The expression status of P14AS in various human cell lines by Northern blotting. An 80-nt P14AS-specific probe was labeled with 32P for Northern blot analysis. The 18S rRNA and 28S rRNA were used as RNA controls. c cDNA synthesized from total RNA with or without RNase R digestion was used as the PCR template. d Cytoplasm P14AS (red) was visualized by RNA-FISH in HEK293T and SW480 cells, but not in A459 cells without the CDKN2A/B locus. P14AS-specific probes were labeled with Cy3 dye. The 18S rRNA and U6 RNA were used as the cytoplasm and nucleus RNA controls, respectively. Scale bar, 5 μm
Illumina clean reads were aligned to each human reference genome using the BWA program, and quality scores were recalibrated and realigned to references using the GATK software package. Duplicated reads were removed using Sequence Alignment/Map tools (SAMtools), and only uniquely mapping reads were used for variation detection. The RNA-seq datasets were uploaded into the NCBI website (GES128205).
Northern blotting
Total RNA was electrophoresed in a formaldehyde denaturing agarose gel after extracting from cells with the Ultrapure RNA kit (Beijing Com Win Biotech Co., Ltd., China). The RNA (10 μg) was transferred to a positively charged NC film with 20× SSC buffer (3.0 M NaCl and 0.3 M sodium citrate, pH 7.0). The membrane was UV-cross-linked and incubated with 32P-labeled RNA probes (Table 1) generated by the Rediprime II Random Prime Labelling System according to the manufacturer’s instruction. The membrane was washed 2–3 times using 2× SSC/0.2% SDS under the condition of 65 °C for 15 min. The membrane was put into the phosphorus screen and pressed for several hours to overnight according to the signal strength.
Table 1.
Sequences of oligo probes and primers
| Assay | Oligo name | Oligo sequence (5′-3′) | Temp. | |
|---|---|---|---|---|
| Northern blotting | P14AS probe | acgctgcggatcccagaggcttaactggcagctggaacgaggtcctccaacaagaatttagacgctaggtccaattatca | ||
| RNA pull-down | P14AS probe #1 | accaacttgggaccgcaacagatttaccata | (1–200 nt) | |
| P14AS probe #2 | gtccttttaaagggtctgactcttcctagaa | (1–200 nt) | ||
| P14AS probe #3 | ggtgcaggatggtatagagagtggcccgtag | (201–400 nt) | ||
| P14AS probe #4 | ctgggtcacctctccagcttggaactggcta | (401–600 nt) | ||
| P14AS probe #5 | gcaggagcgatgtgatccgttatcataactg | (601–777 nt) | ||
| P14AS probe #6 | aacgaggtcctccaacaagaatttagacgct | (601–777 nt) | ||
| Control #1 | ttaacgcctcgaatcagcaa | |||
| Control #2 | gatcttccagataactgccg | |||
| RNA EMSA | probe#1 | tttcatttgaaaagaaaattttcaaacatggggaaagctgcaatataagaagaaaacaatggagagacatcgtcactgga | ||
| probe#2 | aatactacgggccactctctataccatcctgcacccccagatggagaaactgagggaagtcgtggcctttcaacactctt | |||
| P14AS CRISPR/Cas9 | gRNA #1 | catgacagtaagccaaccgatgg | (HEK293T) | |
| gRNA #2 | gttagtggactcgagacgaaagg | (HEK293T) | ||
| gRNA #3 | tgttgcggtcccaagttggtggg | (HCT116) | ||
| gRNA #4 | gttagtggactcgagacgaaagg | (HCT116) | ||
| P14AS promoter | gRNA#5 | acaattagatgttcaactggggg | (HEK293T) | |
| CRISPR/Cas9 | gRNA#6 | cgtatcttatatagcttatgtgg | ||
| qRT-PCR primers | P14AS-F1 | aacggatcacatcgctcctg | 254 bp | 58 °C |
| P14AS-R1 | tccccattcgggttacaacg | |||
| ALU-F | gaggctgaggcaggagaatcg | 87 bp | 60 °C | |
| ALU-R | gtcgcccaggctggagtg | |||
| GAPDH-F | gagatggtgatgggatttc | 224 bp | 62 °C | |
| GAPDH-R | gaaggtgaaggtcggagt | |||
| P16-F | gctgcccaacgcaccgaata | 180 bp | 60 °C | |
| P16-R | accaccagcgtgtccaggaa | |||
| P15-F | agtcaaccgtttcgggaggcg | 168 bp | 62 °C | |
| P15-R | accaccagcgtgtccaggaag | |||
| (q)RT-PCR primers | ANRIL-F | cagcagaaggtgggcagcagat | 145 bp | 64 °C |
| ANRIL-R | ttcctcgacagggcaggcaggt | |||
| 18S rRNA-F | gcttaatttgactcaacacggga | 69 bp | 58 °C | |
| 18S rRNA-R | agctatcaatctgtcaatcctgtc | |||
| RT-PCR primers | P14AS-F2 | taggatatggtaaatctgttgcggt | 1043 bp | 58 °C |
| P14AS-R2 | gagccaggaataaaataaggggaat | |||
| P14AS promoter primers | P14AS-F3 | ccatgtgatttaggaagaaagtttc | 1281 bp/628 bp | 58 °C |
| P14AS-R3 | ttaacaacagcattattacctgggc | |||
| PCR Out-primers | P14AS-F4 | taggatatggtaaatctgttgcggt | 1234 bp | 58 °C |
| P14AS-R4 | tagggagggaggaaagacaaggaa | |||
| PCR In-primers | P14AS-F5 | caaacatggggaaagctgcaa | 164 bp | 58 °C |
| P14AS-R5 | cccttccaagccagatggag | |||
| siRNA | P14AS-S1F | ggaagagucagacccuuuatt | ||
| P14AS-S1R | uaaagggucugacucuucctt | |||
| P14AS-S2F | gcuuacugucauggaaauutt | |||
| P14AS-S2R | aauuuccaugacaguaagctt | |||
| P14AS-S3F | agccugggcuagagacgaatt | |||
| P14AS-S3R | uucgucucuagcccaggcutt | |||
| AUF1-S1F | cguggguucugcuuuauuatt | |||
| AUF1-S1R | uaauaaagcagaacccacgtt | |||
| AUF1-S2F | gccaugucgaaggaacaautt | |||
| AUF1-S2R | auuguuccuucgacauggctt | |||
| AUF1-S3F | cuacuauggauauggugautt | |||
| AUF1-S3R | aucaccauauccauaguagtt | |||
| Scramble-F | uucuccgaacgugucacgutt | |||
| Scramble-R | acgugugacacguucggagaatt |
Plasmid construction and transfection
The 1043-nt P14AS lentiviral vector was constructed in PCDH-CMV-EF1a-copGFP-T2A- Puro lentiviral vector by Syngentech Co., Ltd. (Beijing, China). The lentiviruses for the empty control and P14AS expression vector were generated with the lentiviral packaging kit (BG20401S, Beijing Syngentech Co., Ltd., China) according to the manufacturer’s manual. Briefly, HEK293FT cells were seeded in 6 cm diameter plates, and transfected with vectors at 40% confluence. The medium was collected after 48 h, and filtered with a 0.45 μm filter. The stably transfected HCT116, SW480 and MGC803 cells were selected for 3 days after infection of these viruses with 1 μg/mL puromycin (Sigma, St. Louis, MO, USA).
The four AUF1 isoforms (p37, p40, p42 and p45 KD) in pcDNA3.1 vector were kindly provided by Professor Xiaotian Zhang at Beijing Normal University [11]. To construct the pGEX-4 T-1-AUF1 expression vectors, the full-length coding region of four AUF1 isoforms was amplified by PCR using the primers (Table 1) and then inserted between the EcoRI and XhoI sites of the pGEX-4 T-1 vector. These proteins were purified with GST-tag from bacteria [12].
RNA sequencing
The AUF1 gene was knocked down in stably P14AS-overexpressed HCT116 cells for 72 h. The transcriptomes of these cells were sequenced, as described in the supplemental method (Additional file 1). The data sets were deposited in the Gene Expression Omnibus database with the accession number GSE127905. Function annotation for differentially expressed genes was performed using the David 6.8 online tools at the website (https://david.ncifcrf.gov/tools.jsp) [13].
Knockout of AU-rich element (ARE) in exon 1 of P14AS and its promoter by CRISPR/Cas9
A dual gRNA approach was used to knock out the ARE-containing sequence in exon 1 of the P14AS gene and its promoter by the CRISPR/Cas9 system, respectively. The oligonucleotides used for sgRNA construction were individually designed upstream and downstream of the target fragment (Table 1) and were synthesized by Thermo Scientific, Inc. (Rockford, IL, USA). The sgRNAs were cloned into the Lenti-CRISPR-V2 vector expressing Cas9 (Plasmid #52961, Addgene, Inc.). Then, the lentiviral plasmid expressing both gRNAs and Cas9 was introduced into HEK293FT cells. The viral supernatants were collected 48 h after transfection and were used to infect HEK293T and HCT116 cells. One week later, the infected cells were subjected to puromycin selection, and surviving cells were seeded into 96-well plates to select the monoclonal cells. Initial identification of knockout cell clones was carried out by genomic PCR, the sequences of out- and in-primers are listed in Table 1. P14AS ARE−/promoter-KO-negative clones were pooled and served as a wildtype (WT) control.
RNA pull-down assay
Biotin-labeled targeted P14AS probe #1-#6, and control probe #1-#2 (Escherichia coli strain genome) (Table 1) were in vitro synthesized by the Beijing Genomics Institute using PierceTM RNA 3′ End Desthiobiotinylation Kit (20,163, Thermo Scientific, Rockford, IL, USA) These probes were incubated with separated lysates from HEK293T cells, as described in the supplemental methods (Additional file 1).
RNA immunoprecipitation assay (RIP)
The RIP assay was carried out using the RNA-Binding Protein Immunoprecipitation Kit (Cat# 17–701, EZ-Magna, Millipore, USA) according to the manufacturer’s instructions. Total AUF1-binding RNAs were immunoprecipitated and extracted using AUF1 antibody (ab61193, Abcam, UK). cDNA was synthesized from the RIP-RNAs using random primers, and gene-specific quantitative PCR was then performed using the primer P14AS-F1 and -R1 set to amplify the 254-bp fragment within the 1043-nt P14AS gene (Table 1).
Animal experiments
HCT116 cells stably transfected with the control, P14AS expression vector or P14AS knockout vector were harvested by trypsinization, washed twice with PBS, and then subcutaneously injected into the bilateral inguinal area of female NOD/SCID mice (body weight, 18–20 g; age, 6 weeks, Beijing Huafukang Bioscience Co. Inc., 2 × 106 cells per injection). On the 17th posttransplantation day, mice were sacrificed. For the experiment on tumor growth from P14AS−/− and P14AS+/+ HCT116 cells (2 × 106 cells per injection), the mice were sacrificed on the 19th posttransplantation day. Tumors were fixed in 4% paraformaldehyde, sectioned, and stained with hematoxylin and eosin (H&E).
Statistical analyses
All statistical analyses were performed using SPSS 18.0 software. The Kolmogorov–Smirnov test was used to estimate the normality of distributions. The Mann–Whitney U-test was conducted for non-normally distributed data. Student’s t-test was conducted for normally distributed data. All statistical tests were two-sided. Statistical significance was assigned at P < 0.05 (*) or P < 0.01 (**).
Other methods
Other used methods, including cell culture, Western blotting, RT-PCR, RNA sequencing, RNA-FISH, RNA pull-down, cell proliferation and migration assays, siRNA downregulation, electrophoretic mobility shift assay (EMSA), and induction of methylation of P16 CpG islands, were listed in the supplemental method (Additional file 1).
Results
Characterization of endogenous lncRNA P14AS in cancer cells
An RNACap-Seq technology was established and used to screen novel CDKN2A-specific RNA transcripts in HEK293T and MCF7 cells (Fig. 1a). A three-exon 1043-nt lncRNA transcribed from the antisense strand of the fragment around exon 1β of the CDKN2A locus was detected from the RNACap-Seq readouts (chr9: 21,989,178-21,994,898 in the human genome GRCh37/hg19; (Additional file 2: Fig. S1A). The results of Northern blot analysis confirmed the existence of the endogenous lncRNA P14AS in 6 cell lines (BGC823, SGC7901, HepG2, HCT116, HEK293T, and SW480 cells; Fig. 1b). P14AS was not detected in the negative control cell lines A549 (without CDKN2A/B allele) and AGS (containing fully methylated P16 alleles).
LncRNAs, including ANRIL, are often processed into circular RNAs. To study whether P14AS is a circular RNA, the linear RNAs were digested by RNase R before cDNA synthesis by reverse transcription (RT). No RT-PCR product was amplified from the RNase R-digested samples, indicating that P14AS is a linear lncRNA (Fig. 1c). The PhyloCSF analysis results showed that P14AS has no coding capacity (Additional file 2: Figure S1B). RNA-FISH analysis revealed that endogenous P14AS was mainly distributed in the cytoplasm of HEK293T and SW480 cells (Fig. 1d).
It is well-known that P14 and P16 genes are transcribed from different transcription start sites (TSSs) in the human CDKN2A locus. Also, P14 and P16 mRNAs share the same CDKN2A exon 2 with different translation reading frames (Additional file 2: Figure S1a). Similarly, P14AS exon 3 (79-nt) completely overlaps with the 5′-sequence in exon 1 of ANRIL (Fig. 1a and S1a). Therefore, ANRIL and P14AS might be spliced from the same primary transcript, and P14AS might be an isoform of ANRIL. Alternatively, ANRIL and P14AS may be transcribed from the CDKN2A gene using different TSS and partially shared exon 1 of ANRIL.
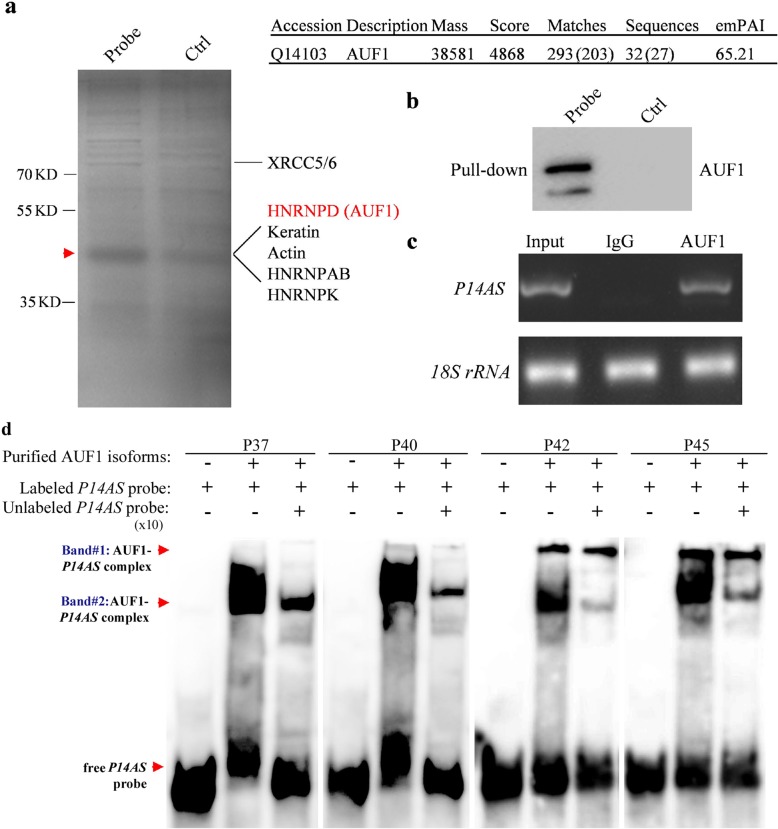
Direct lncRNA-protein interaction between P14AS and AUF1
We performed a biotin-labeled RNA pull-down and mass spectrometry assay to identify potential P14AS-binding proteins from HEK293T cell lysates. We found that endogenous AUF1 proteins were the top proteins in the P14AS pull-down complexes (Additional file 3: Table S1, Fig. 2a). Western blot analysis confirmed the interaction between AUF1 and P14AS (Fig. 2b). To determine whether P14AS naturally binds to AUF1 protein in cells (without gene overexpression), we performed the RNA-Immunoprecipitation (RIP) assay and found that P14AS was significantly enriched in the AUF1 antibody-RIP complexes compared with the IgG control (Fig. 2c), indicating the occurrence of endogenous P14AS-AUF1 binding in cells. The results of the RNA-EMSA assay further confirmed that P14AS could directly bind to four AUF1 protein isoforms (Fig. 2d). Unlabeled P14AS probe (× 10) could inhibit most of the P14AS-binding to AUF1 P37 and P40. In addition, there were two P14AS-binding bands for the AUF1 P42 and P45 complexes and unlabeled P14AS probe could only inhibit the P14AS-binding to one of these AUF1 bands. These results suggest specific bindings between P14AS and AUF1, at least for P37 and P40 isoforms.
Fig. 2.
P14AS interacts with AUF1 through the AU-rich element in exon 1. a Biotin-labeled P14AS pull-down complexes from HEK293T cell lysate, following mass spectrometry; (b) Western blot analyses; (c) Interaction of AUF1 protein with P14AS in RIP-PCR; (d) Interaction of four purified AUF1 protein isoforms with biotin-labeled P14AS probe in the RNA-electrophoretic mobility shift assay (RNA-EMSA). The interaction of Band #2-AUF1 P42 isoform with biotin-labeled P14AS probe was almost completely inhibited by the unlabeled probe (×10). However, the interaction of Band #1-AUF1 P42 and P45 isoforms with the P14AS was not affected by the unlabeled probe
P14AS-AUF1 interaction upregulates ANRIL expression
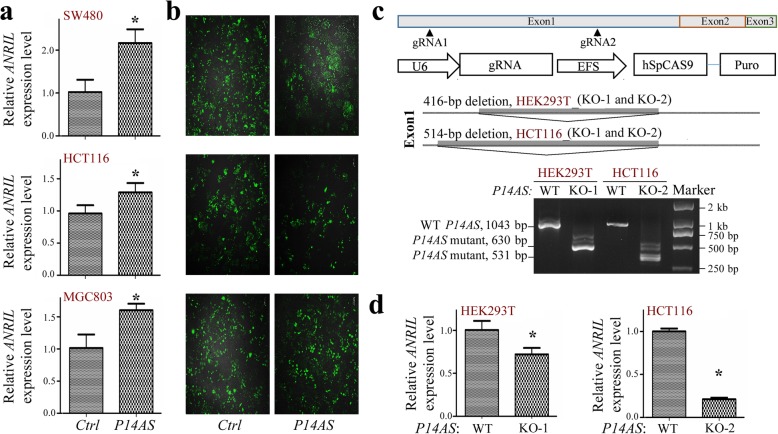
As lncRNAs often act as cis regulators of nearby genes, we determined whether P14AS could affect the expression of ANRIL and other neighboring genes. The qRT-PCR results revealed that stable P14AS overexpression consistently increased the expression level of ANRIL in SW480, HCT116, and MGC803 cells (Fig. 3a-3b). Similar results were also observed for P16, P15, and P14 genes hosted by the same 9p21.3 locus (Additional file 4: Figure S2a-S2b).
Fig. 3.
P14AS affects the expression level of the ANRIL gene. qRT-PCR data are normalized to GAPDH mRNA levels and shown as the means ± SD. a The expression levels of ANRIL in the P14AS vector stably transfected colon cancer cells (HCT116 and SW480), and gastric cancer cells (MGC803) were analyzed by qRT-PCR. b GFP-expression efficiency in the P14AS expression and control vector in the stably transfected cells. c Knockout (KO) of the ARE-containing element within P14AS exon 1 by CRISPR/Cas9. A 416-bp (chr9: 21,989,309-21,989,724) or 514-bp (chr9: 21,989,211-21,989,724) fragment deletion in P14AS exon 1 was detected by RT-PCR (bottom chart) in HEK293T or HCT116 P14AS-KO clones. d The expression change of ANRIL in HCT116 and HEK293T cells whose ARE-containing elements in P14AS exon 1 were homogenously deleted in the qRT-PCR analysis. Pooled P14AS ARE-KO-negative clones were used as the wild-type (WT) control
We then hypothesized that P14AS-AUF1 interaction could protect ANRIL and other RNAs from degradation. To test this hypothesis, the ARE element in P14AS exon 1 (Fig. 1a) was homogenously knocked out (KO) in HCT116 and HEK293T cells using CRISPR/Cas9 technology (Fig. 3c). PCR-sequencing and RT-PCR analyses confirmed the homogenous deletion of P14AS exon 1 (416-bp and 514-bp deletions) in the HEK293T (KO-1 and -2) and HCT116 clones (KO-1 and -2) (Additional file 4: Figure S2c). The expression levels of ANRIL were significantly decreased in the HCT116 and HEK293T P14AS-KO cells in the qRT-PCR analyses (Fig. 3d). Similar decreases of P15, P14, and P16 expression levels by P14AS knockout were also observed in these clones (Additional file 4: Figure S2c-S2d).
We next determined if P14AS impacts ANRIL/P16 RNA stability through interaction with the AUF1 protein. As expected, ANRIL and P16 mRNA indeed existed in the AUF1-RIP RNA complexes in HCT116 cells (Additional file 5: Figure S3a), suggesting that both ANRIL and P16 mRNA are AUF1-binding RNAs. The amount of the AUF1-RIP ANRIL and P16 mRNA was significantly decreased in HCT116 cells with P14AS overexpression compared with the control cells (Additional file 5: Figure S3b), suggesting a competitive AUF1-binding between P14AS and ANRIL/P16 RNAs (Additional file 5: Figure S3c).
We further transiently transfected scramble control and AUF1 siRNA in P14AS-overexpressed stable HCT116 cells, and performed RNA sequencing at 72 h after transfection. The results showed that ANRIL was one of the P14AS-upregulated lncRNAs (fold change > 2.0) while AUF1 knockdown upregulated many lncRNA genes (N = 341). However, after the AUF1 knockdown, P14AS could not upregulate ANRIL expression (Additional file 6: Figure S4). In addition, after the AUF1 knockdown, P14AS downregulated many lncRNA genes (N = 433). No significant changes in P15 and P16 mRNA levels were observed. These results imply that P14AS could protect the decay of lncRNAs, including ANRIL, in an AUF1-dependent pattern. The results of David functional annotation analyses showed that about half of P14AS-upregulated and -downregulated genes were significantly enriched with sequences containing signal peptide feature or related to glycoproteins (Additional file 7: Table S2 and Additional file 8: Table S3). However, 155 of 377 (41%) upregulated genes and 246 of 437 (56%) downregulated genes were not included in the David 6.8 functional annotation analyses. Thus, more functions of P14AS-affected RNAs were expected.
Interestingly, a positive relationship between the expression levels of ANRIL and AUF1 mRNA was also observed using the transcript databases for 1037 cell lines in the cancer cell line encyclopedia (CCLE) project [R = 0.23 (or 0.30 for 224 cell lines with relative CDKN2A copy number > 0), P < 0.001; Additional file 9: Figure S5]. Again, the AUF1 expression did not correlate with that of P16 and inversely correlated with P15 expression in these cell lines. Overall, these results suggest that P14AS could upregulate the expression of ANRIL (and other lncRNAs), probably in an AUF1 binding-dependent pattern.
Promotion of cell proliferation and tumor formation by P14AS
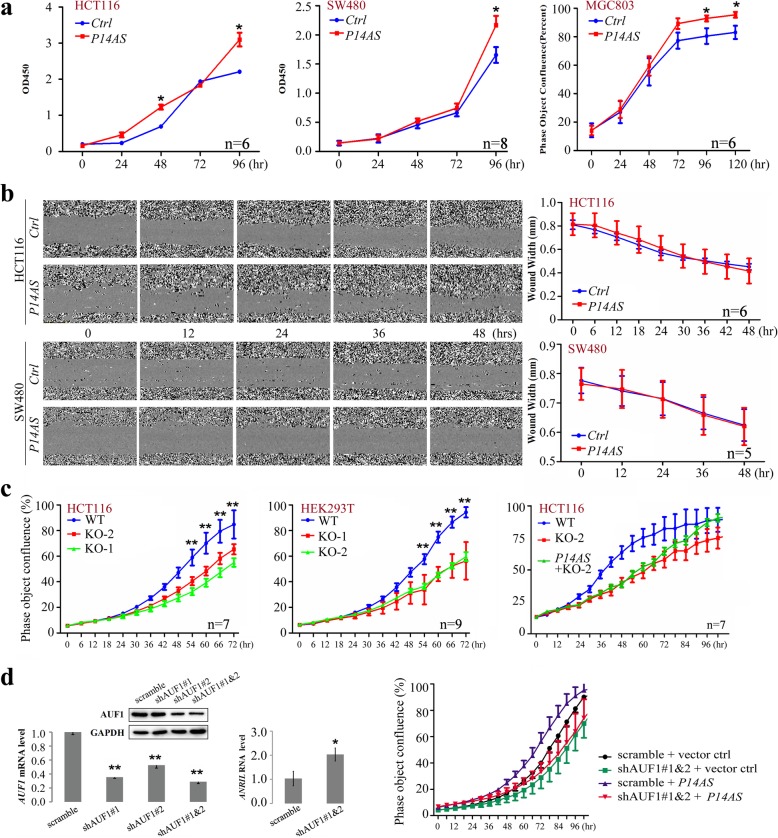
We further studied the effects of P14AS overexpression on cancer cell behavior in vitro and in vivo. Both human colon cell lines SW480 and HCT116 and gastric cancer cell line MGC803 were stably transfected with the P14AS expression vector. The results of CCK-8 and long-term dynamic observation assays revealed that P14AS overexpression promoted cell proliferation but did not promote cell migration (Fig. 4a and b).
Fig. 4.
P14AS promoted cancer cell proliferation. aP14AS overexpression promoted cell proliferation in the CCK-8 assays and IncuCyte long-term dynamic observation. b Wound width curve for HCT116 and SW480 cells with P14AS overexpression in the long-term observation. c Effect of P14AS knockout (KO) by CRISPR/Ccas9 and P14AS re-expression in the proliferation of HCT116 and HEK293T cells. d Effect of shRNA-knockdown of AUF1 and/or transient P14AS overexpression on the proliferation of HCT116 cells (Right chart). The levels of AUF1 and ANRIL expression in HCT116 cells with stable shAFU1 transfection for four weeks was tested with qRT-PCR (Left and Right charts, respectively) and Western blotting (top chart) before these cells were transiently transfected with the P14AS expression vector. Student’s t-test: *, P < 0.05; **, P < 0.01
Moreover, the proliferation of both HCT116 and HEK293T cells was significantly inhibited by the P14AS-KO compared to the wild-type (WT) controls (Fig. 4c, left and middle, respectively). The results of rescue experiments showed that enforced P14AS re-expression could completely recover the P14AS-KO-inhibited proliferation of HCT116 cells after 96 h, though the proliferation difference was still observed at the beginning between the P14AS-KO cells and P14AS re-expressing cells after cell seeding (Fig. 4c, right). Notably, stably AUF1 shRNA-knockdown completely abolished the P14AS-induced enhancement of the proliferation of HCT116 cells (Fig. 4d). As expected, the level of ANRIL expression was significantly increased in these cells with stably AUF1 knockdown. Since the P16 gene is inactivated in HCT116 cells, endogenous lncRNA P14AS may serve as a sponge to protect other AUF1 targets from decay and promote cancer cell proliferation. These phenomena suggest that the enhancement of cell proliferation by P14AS may depend on the ARE element in P14AS exon 1.
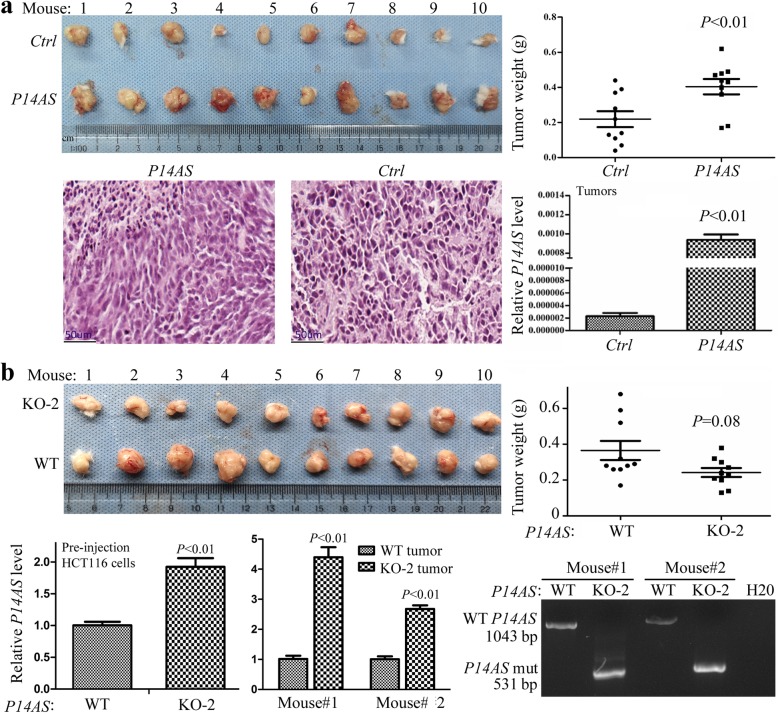
To validate the in vitro results, NOD-SCID mice (N = 10) were injected with HCT116 cells stably transfected with P14AS into the left inguinal area and the empty control vector into the right inguinal area for each mouse. On the 17th posttransplantation day, the number and weight of tumors in the P14AS group were significantly higher than those in the control group (P < 0.01, Fig. 5a). The qRT-PCR results indicate that P14AS remained to be actively transcribed in these tumors. In contrast, knockout of the ARE-containing element in P14AS exon 1 decreased the growth of HCT116 cells (P = 0.08, Fig. 5b). The qRT-PCR results illustrated that the level of KO-truncated P14AS lncRNA was higher in the P14AS-KO cells than that of wild-type P14AS in the pre-injection P14AS-KO cells and derived tumors. Thus, knockout of the exon 1 ARE-containing element may lead to compensatory upregulation of the truncated P14AS gene residue. Collectively, these results indicate that P14AS promotes cancer cell proliferation in vitro and tumor formation in vivo.
Fig. 5.
Effect of P14AS overexpression and knockout on tumor growth from HCT116 cells in NOD SCID mice. (a) Tumors derived from HCT116 cells stably transfected with the P14AS and control vectors on the 17th posttransplantation day. Hematoxylin and eosin (H&E) staining images were attached. (b) Tumors derived from P14AS-knockout HCT116 KO-2 clone on the 19th posttransplantation day. The tumor weight and the levels of wild-type P14AS (WT) and its KO-truncated counterpart were displayed. Ten mice per group
Coordinate overexpression of P14AS, ANRIL, and AUF1 in colon cancer tissues
To determine whether P14AS, ANRIL, and P16 expression were coordinately upregulated in cancer development, we examined their expression status in colon cancer (CC) and their corresponding surgical margin (SM) tissue samples from 172 patients, and normal colon mucosa biopsy samples from 50 noncancer patients. The results of qRT-PCR detection (254-bp) revealed that the average P14AS level in CC tissues, with and without ANRIL expression, was significantly higher than that in the paired SM tissues and normal biopsies (Additional file 10: Figure S6a-b). The average P14AS level in SM samples was also markedly higher than that in normal colon biopsies. More P14AS was detected in CCs with advanced local invasion (trend-test, P = 0.034; Additional file 11: Table S4). These phenomena suggest that P14AS upregulation is an early event in CC development and correlates with CC invasion.
In addition, the positive rate of ANRIL (by RT-PCR) was significantly higher in CCs than SMs (90/167 [53.9%] vs. 52/167 [31.1%], P < 0.001; Additional file 13: Table S5). Similarly, the average P16 mRNA level was also significantly higher in CCs than SMs (Additional file 10: Figure S6a). In contrast, the average P15 mRNA level in CCs was considerably lower than that in SM tissues. The level of AUF1 mRNA (by qRT-PCR) was significantly higher in CCs than SMs (median, 22.02 vs. 1.77, P < 0.001; Additional file 14: Table S6). While no significant assoication between clinicopathological characteristics and the level of AUF1 mRNA was detected for CCs, the level of AUF1 mRNA was higher for SMs from patients with stage I&II CC than those with stage III&IV (Mann–Whitney U-test, P = 0.014).
The P16 mRNA level (by qRT-PCR) was also significantly higher in the P14AS-positive colon CC and SM tissues (by RT-PCR; 1043-bp) than in the P14AS-negative tissues (nonparametric tests, P = 0.039; Additional file 10: Figure S6c). Once again, no significant correlations were observed between the levels of P14AS and P15 mRNA (Additional file 10: Figure S6d). Thus, P14AS expression is coordinately overexpressed with ANRIL and P16 in colon tissues.
In addition, the level of AUF1 mRNA was positively associated with P14AS expression in CC or SM samples (Pearson_r = 0.27, P = 0.002; Additional file 12: Figure S7a). The level of AUF1 mRNA was significantly higher in ANRIL-positive SMs than ANRIL-negative SMs (P = 0.047). A similar difference was also observed between CCs with and without ANRIL expression (P = 0.061; Additional file 12: Figure S7b).
Combined analysis showed that the frequency of P14AS and ANRIL coexpression was significantly higher in distant metastatic colon cancers than non-metastatic colon cancers (53.3% vs. 30.7%, P = 0.018; Additional file 15: Table S7). Such an association could not be observed when P14AS and ANRIL were individually analyzed.
Characterization of the P14AS promoter
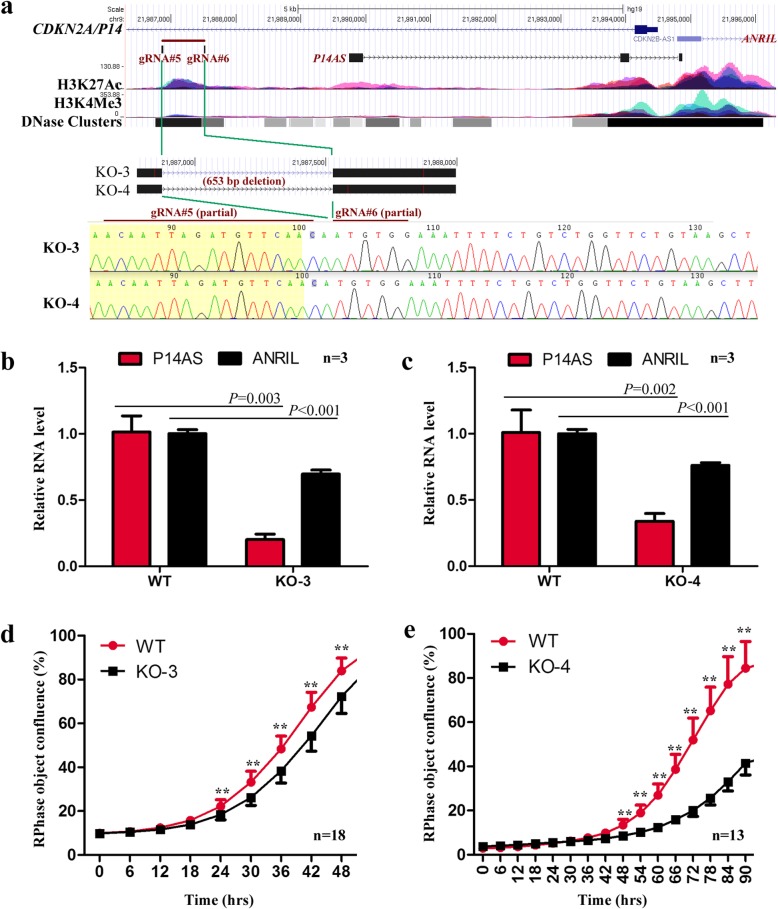
According to the ENCODE datasets for 7 cell lines, there is a promoter-like sequence 2-kb upstream of P14AS exon 1. This region is enriched with active H3K27Ac and H3K4Me3 modifications (Additional file 16: Figure S8a-S8b), where RNA polymerase II (POLR2A)-binding was detected (Additional file 16: Figure S8c, highlighted in red lines). This promoter-like sequence is conserved among vertebrates (Additional file 16: Figure S8d) and implies its biological significance. To study whether this sequence is the P14AS promoter, we knocked out of this sequence (653-bp) with CRISPR/Cas9 and found that the level of P14AS transcription was dramatically decreased in two HEK293T subclones (KO-3 and KO-4; Fig. 6a-c). In addition, the level of ANRIL expression (by qRT-PCR) was significantly reduced (Fig. 6d). The results of long-term dynamic observation assays showed that the proliferation of these P14AS promoter-KO cells was also repressed (Fig. 6e). These data strongly suggest that this sequence may be the P14AS promoter.
Fig. 6.
Effects of the knockout of the P14AS promoter on the P14AS and ANRIL transcription and the proliferation of HEK293T cells. (a) Remaining sequences of the P14AS promoter with a 653 bp deletion (chr9:21,986,875-21,987,527; hg19) by CRISPR/Cas9 in two HEK293T subclones (KO-3 and KO-4). Locations of the wildtype P14AS promoter, two guide RNAs, and the 653 bp knockout (KO) fragment are also illustrated; (b and c) The levels of P14AS and ANRIL expression in wildtype and KO-3 or KO-4 cells by qRT-PCR. (d and e) Effect of the promoter deletion on KO-3 and KO-4 cell proliferation in the IncuCyte long-term dynamic observation. Pooled P14AS promoter-KO-negative clones were used as a wild-type (WT) control. Error bars, S.D. **, P < 0.01
Discussion
Accumulating evidence has revealed that lncRNAs function in multiple cellular processes, including transcription, intracellular trafficking, chromosome remodeling, and disease development. Deregulation of lncRNAs was found in various cancers [14–16]. The 9p21 locus is the most frequently deleted allele in cancer genomes. This locus contains several important tumor suppressor genes (CDKN2A/B encodes the P16, P14, and P15 proteins). It also hosts the oncogenic antisense lncRNA gene ANRIL, which is implicated in cancers of the colon, breast, lung, and bladder [17–22]. In the present study, we characterized a new oncogenic antisense lncRNA, P14AS, in this locus. Our systemic studies show that P14AS cis upregulates ANRIL and promotes cancer cell proliferation.
We found that P14AS was frequently overexpressed in CC tissues, and that P14AS overexpression promoted the proliferation of cancer cells. In contrast, P14AS knockdown by siRNA and knockout of the ARE-containing element in P14AS exon 1 by CRISPR/Cas9 could repress cancer cell proliferation. The effect of P14AS on cancer cell proliferation was confirmed in a mouse model. These results suggest that P14AS may be an oncogene. We also analyzed the expression status of P14AS in gastric carcinoma tissues (N = 40), P14AS expression was not detected (data not shown). This result warrants studying its roles in cancer development in other organs.
In addition, although P14AS could upregulate P16 expression in cell lines and P14AS expression is positively correlated with P16 expression in colon tissues, P14AS overexpression or knockout remains to affect the proliferation of the P16-inactive cell lines (HCT116 and SW480), suggesting that P14AS may function as an oncogene in a P16-independent way. P16 is a weakly expressed nucleic protein in normal cells. However, the P16 protein is sequestered in the cytoplasm without tumor suppressor function in many cancer tissues, including CCs [6]. Therefore, the upregulation of P16 expression by P14AS in cancers may not affect cancer development. Instead, dysfunctions of P16, P14, and P15 cause abnormal G1-S shift in the cell cycle that plays crucial role in the malignant transformation of human cells due to increased phosphorylation of RB1 [2].
AUF1 is a member of the family of RNA binding proteins that is termed ARE/poly (U)-binding/ degradation factor 1. The canonical functions of AUF1 are to regulate mRNA decay and translation via recognition of specific sequence elements in mRNA 3′ untranslated regions (3’UTR) [23]. For example, AUF1 is involved in the regulation of c-MYC and P16 mRNA stability [7, 8]. Previous studies have identified AUF1 as an important destabilizer for P16 mRNA, thereby influencing cell senescence [24]. Moreover, AUF1 can interact with lncRNAs, including NEAT1 (10) and linc-ROR [9]. Interestingly, we found that there is an ARE element in P14AS exon 1 that binds to AUF1. Notably, knockout of the ARE-containing element not only abolishes the function of P14AS to upregulate ANRIL and P16/P14/P15 expression but also eliminates the effect of P14AS on cancer cell proliferation. Because re-expression of the wild-type P14AS could restore the effect of P14AS-KO on cell proliferation and stable knockdown of AUF1 expression could abolish the effect of P14AS on cell proliferation, P14AS-AUF1 binding is highly considered to be the main pathway for the function of P14AS.
It has been reported that AUF1 isoforms (P37, P40, P42, And P45) could form dimers, which could yield oligomeric AUF1 complexes [25, 26]. In our RNA-EMSA assay, most P14AS-binding to the AUF1 P37 and P40 band was inhibited by the unlabeled P14AS probe (× 10), suggesting high binding specificity between these AUF1 isoforms to P14AS.
Moreover, we found that P14AS overexpression and knockout/knockdown significantly increased and decreased ANRIL expression in many cell lines, respectively. The results of our RNA-seq analyses confirmed the upregulation (fold change > 2.5) of ANRIL expression by P14AS in HCT116 cells. Both P14AS and ANRIL were upregulated in CC tissues, and more P14AS was detected in the ANRIL-positive tissues than the ANRIL-negative tissues. In addition, the results of our AUF1 RIP-PCR experiment indicate that ANRIL lncRNA is an AUF1-binding RNA. Analysis of the public CCLE data reveals that the AUF1 mRNA level was positively and significantly correlated with the ANRIL lncRNA level in 1037 human cancer cell lines. The level of AUF1 mRNA was positively and significantly associated with the P14AS level in CC tissues. These findings support that P14AS may function as an oncogene through the cis upregulation of ANRIL expression in cancer cells through AUF1-P14AS binding.
Consistent with the reported functions of AUF1 in degrading RNAs, AUF1 knockdown by siRNA could upregulate the expression of many lncRNAs in HCT116 cells in our RNA-seq analysis. However, in cells with AUF1 knockdown, P14AS downregulated the expression level of many lncRNAs (fold change <− 2.0), which was not observed in cells with P14AS overexpression alone. This implies that AUF1 may be a dominant factor in regulating P14AS functions.
It is well-recognized that DNA demethylation of CpG islands around TSS is needed for gene transcription [27] and that methylation of CpG islands in the gene body is needed for active genes to repress abnormal transcription from cryptic TSSs [28]. Surprisingly, it was reported that the ANRIL expression level was positively associated with the methylation level of the P14 promoter CpG islands in tissues [29], implying that the P14 promoter CpG islands may not be the true ANRIL promoter but the ANRIL gene body CpG islands. We also reported that ANRIL and P16 are coordinately transcribed in CCLE cell lines, and P16 methylation repressed both ANRIL and P16 expression and did not affect P14/P15 expression [6]. Furthermore, we found that P16 methylation repressed P14AS expression in BGC 823 cells (Additional file 17: Figure S9), suggesting a co-repression of P14AS, ANRIL, and P16 expression by DNA methylation of the P16 CpG island around TSS. Chromosome conformation capture (3C) analysis showed that the P16 promoter could play a key role in formation of compact chromatin loops [30]. P16 promoter methylation may lead to focal chromatin condensing and downregulation of these genes. We also found that the 79-nt sequence of P14AS exon 3 was completely overlapped with the 5′-sequence of ANRIL exon 1. To test the probability that P14AS and ANRIL might be spliced from the same primary transcript, we knocked out the promoter-like sequence 2-kb upstream of P14AS exon 1 with CRISPR/Cas9 and found a dramatically decreased P14AS transcription and slightly decreased ANRIL expression in two subclones, providing strong evidence to support the conserved sequence is the P14AS promoter. The fact that KO of the P14AS promoter only weakly decreased ANRIL transcription suggests that the P14AS promoter is likely not the ANRIL promoter or the ANRIL gene could be transcribed using multiple TSSs.
Unlike mature mRNAs that may contain many exons, approximately 42% of mature lncRNAs contain only two exons [31]. In melanoma cells, preliminary ANRIL transcripts could be spliced into diverse mature forms, including linear ANRILs mainly located in the nucleus and circular ANRILs located in the cytoplasm, which imply function difference between linear and circular ANRILs [32]. The specific processing patterns for lncRNAs make the characterization of lncRNA genes much more complicated than the identification of mRNAs. This processing might account for misidentification of TSSs for lncRNA genes.
It was reported that ANRIL knockdown inhibited the proliferation migration of hepatoma cells [33, 34]. However, we found that P14AS overexpression or knockout only increased or decreased the proliferation of cell lines HCT116, SW480, and HEK293T, but did not affect the migration of these cells. As we observed in RNA-sequencing data, P14AS could target to both ANRIL and other ncRNAs in AUF1-dependent manner. This may account for the difference of biological functions between ANRIL and P14AS. It is worth studying whether linear or circular ANRIL transcription and degradation are regulated by P14AS.
Conclusions
The antisense strand of the CDKN2A locus hosts the P14AS gene, which might be connected with a true TSS for the ANRIL gene. P14AS is an AUF1-binding lncRNA that upregulates the expression of AUF1 target genes, including ANRIL. P14AS maybe an oncogenic lncRNA involved in the development of colon cancer.
Supplementary information
Additional file 1. Supplementary methods
Additional file 2 Figure S1. Characterization of P14AS in the CDKN2A/B locus. (A) Sashimi view for transcripts detected by CDKN2A-specific probe-captured RNA (RNACap)-Seq in HEK293T cells containing two wild-type CDKN2A/B alleles and in MCF7 cells with the homogenous P16 deletion. (B) Graphic view of the protein-coding potential for P14AS, CDKN2A/P14, and ANRIL genes (adapted from the UCSC website).
Additional file 3 Table S1. List of proteins interacted with P14AS characterized in the HEK293T cells in RNA pull-down mass spectrum analysis
Additional file 4 Figure S2.P14AS affects the expression level of the neighboring genes at 9p21.3. qRT-PCR data are normalized to GAPDH mRNA levels and shown as the means ± SD. (a) The expression levels of P14, P16, and P15 genes in the P14AS vector stably transfected colon cancer cells (HCT116 and SW480), and gastric cancer cells (MGC803) were analyzed by qRT-PCR. (b) Detection of P16/P15/P14 proteins in MGC803 cells in Western blot analyses. (c) A fragment deletion in P14AS exon 1 was detected by PCR (top chart) in HEK293T or HCT116 P14AS-KO clones (KO). The expression changes of P14, P16, and P15 genes in HCT116 and HEK293T cells whose ARE-containing elements in P14AS exon-1 were homogenously deleted in the qRT-PCR analysis (bottom chart). Pooled P14AS ARE-KO-negative subclones were used as a wild-type (WT) control. (d) Detection of the P16, P15, and P14 proteins in HEK293T cells in Western blot analyses.
Additional file 5 Figure S3. P14AS expression decreased P16 mRNA-AUF1 binding. (a) AUF1 directly bound to ANRIL and P16 mRNA in HCT116 cells in the AUF1-RIP-PCR. (b) P14AS overexpression decreased ANRIL and P16 mRNA-AUF1 interaction in by the AUF1-RIP-qPCR. (c) An illustration of how the competitive AUF1-P14AS binding protects ANRIL and P16 mRNA from the decay. AUF1 complexes were drawn as dimers based on the reports that AUF1 isoforms (p37, p40, p42, and p45) could form functional dimers [25, 26].
Additional file 6 Figure S4. Genome-wide analyses of transcriptome by RNA sequencing for HCT116 cells with and without P14AS overexpression and/or AUF1 downregulation. The HCT116 cells with stable P14AS overexpression were transfected with AUF1 siRNAs (siAUF1) for 72 h, and then harvested for RNA sequencing. The number of genes with > 2 fold changes (UP, upregulated; Down, downregulated) for different types of RNAs were labeled. Western blot analysis for the determination of AUF1 downregulation by siRNAs was inserted into the top chart. Two samples were sequenced for each group.
Additional file 7 Table S2. Function annotations for P14AS-upregulated genes (n = 241) with fold change > 2 in HCT116 cells with the David 6.8: Functional Annotation Tools at the website http://david.ncifcrf.gov/tools.jsp [13]
Additional file 8 Table S3. Function annotations for P14AS-downregulated genes (n = 299) with fold change > 2 in HCT116 cells with the David 6.8: Functional Annotation Tools at the website http://david.ncifcrf.gov/tools.jsp [13]
Additional file 9 Figure S5. Association analyses between the expression levels of AUF1 and ANRIL, P16, or P15 using the publicly available transcriptome databases for human cancer cell lines in the CCLE project. (A) All 1037 cell lines; (B) 224 cell lines without the CDKN2A allele deletion (relative copy number > 0).
Additional file 10 Figure S6. Comparison of the levels of P14AS, P16, and P15 expression in colon tissues from cancer and noncancer patients. (a) The expression status of P14AS, P16, and P15 in colon cancer (CC), paired surgical margin (SM), and normal colon biopsy (Normal) tissues from noncancer patients by qRT-PCR. (b) The level of P14AS expression in ANRIL-positive and -negative (by RT-PCR) colon CC and SM tissues. (c and d) Comparison of the expression levels of P16 and P15 mRNA (by qRT-PCR) in P14AS-positive and -negative (by RT-PCR; 1043 bp) colon CC and SM tissues. Error bars, S.E.M. *, P < 0.05; **, P < 0.01; N.S. no significance.
Additional file 11 Table S4. Comparison of the P14AS expression level (by qRT-PCR) in colon cancer (CC) and surgical margin (SM) tissue samples from patients with different clinicopathological characteristics
Additional file 12 Figure S7. Comparisons of the levels of AUF1 mRNA (by qRT-PCR) with those of P14AS and ANRIL lncRNA in colon cancer tissues (CCs).
Additional file 13 Table S5. Comparison of the ANRIL expression level (by RT-PCR) in colon cancer (CC) and surgical margin (SM) tissue samples from patients with different clinicopathological characteristics
Additional file 14 Table S6. Comparison of the AUF1 mRNA level (by qRT-PCR) in colon cancer (CC) and surgical margin (SM) tissue samples from patients with different clinicopathological characteristics
Additional file 15 Table S7. Comparison of P14AS and ANRIL coexpression in colon cancer (CC) and surgical margin (SM) tissue samples from patients with different clinicopathological characteristics
Additional file 16 Figure S8. Graph of the P14AS gene in the CDKN2A/B locus. (A) CpG islands within the CDKN2A/P14 gene. (B) The transcription and active histone modification status in the chromatin upstream of the P14AS gene in 7 cell lines from ENCODE. (C) Transcription factors binding to various fragments around the P14AS gene from ENCODE. The RNA polymerase II (POLR2A) is highlighted in red lines. (D) The conservation status of various fragments among vertebrates (adapted from the UCSC website).
Additional file 17 Figure S9. Repression of P16, P14AS, and ANRIL expression in gastric cancer cells by engineered P16-specific DNA methyltransferase (P16-Dnmt). The gene expression level was detected using qRT-PCR assay. The detailed processes for the construction of P16-Dnmt and transfection experiments are previously described [27]
Acknowledgements
We appreciated Dr. Shengyan Xiang, Florida, USA, for English language editing.
Abbreviations
- ARE
AU-rich element
- CC
colon cancer
- KO
knockout
- lncRNA
long non-coding RNA
- P16-Dnmt
engineered P16-specific zinc-protein-based DNA methyltransferease
- qRT-PCR
quantitative reverse transcription-PCR
- RIP
RNA immunoprecipitation
- RNACap-Seq
probe captured-RNA extra-deep sequencing
- RNA-EMSA
RNA-electrophoretic mobility shift assay
- SM
surgical margin
Authors’ contributions
WM and JQ performed the data analyses and wrote the draft of the manuscript. WM, JQ, JZ and LG performed the experiments. DD designed the study, reviewed the manuscript. All of the authors approved the study.
Funding
This work is supported by the National Natural Science Foundation of China to D.D. (Grant no.91640108).
Availability of data and materials
The data and materials of the study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
All patients in this study provided written informed consent. Animal experiment in this study was approved by the animal ethics committee, Peking University Cancer Hospital and Institute (Approval #AE-2012-06).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wanru Ma and Juanli Qiao contributed equally to this work.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12943-020-01150-4.
References
- 1.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 2.Lukas J, Parry D, Aagaard L, Mann DJ, Bartkova J, Strauss M, Peters G, Bartek J. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 3.Pasmant E, Laurendeau I, Héron D, Vidaud M, Vidaud D, Bièche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 4.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg A, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montes M, Nielsen MM, Maglieri G, Jacobsen A, Højfeldt J, Agrawal-Singh S, Hansen K, Helin K, van de Werken HJG, Pedersen JS, Lund AH. The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nat Commun. 2015;6:6967. doi: 10.1038/ncomms7967. [DOI] [PubMed] [Google Scholar]
- 6.Gan Y, Ma W, Wang X, Qiao J, Zhang B, Cui C, Liu Z, Deng D. Coordinate transcription of ANRIL and P16 genes silenced by DNA methylation. Chin J Cancer Res. 2018;30:93–103. doi: 10.21147/j.issn.1000-9604.2018.01.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones TR, Cole MD. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3′ untranslated sequences. Mol Cell Biol. 1987;7:4513–4521. doi: 10.1128/MCB.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang W, Martindale JL, Yang X, Chrest FJ, Gorospe M. Increased stability of the p16 mRNA with replicative senescence. EMBO Rep. 2005;6:158–164. doi: 10.1038/sj.embor.7400346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J, Zhang A, Ho TT, Zhang Z, Zhou N, Ding X, Zhang X, Xu M, Mo YY. Linc-RoR promotes c-Myc expression through hnRNP I and AUF1. Nucleic Acids Res. 2016;44:3059–3069. doi: 10.1093/nar/gkv1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoon JH, De S, Srikantan S, Abdelmohsen K, Grammatikakis I, Kim J, Kim KM, Noh JH, White EJ, Martindale JL, et al. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat Commun. 2014;5:5248. doi: 10.1038/ncomms6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun S, Zhang X, Lyu L, Li X, Yao S, Zhang J. Autotaxin expression is regulated at the post-transcriptional level by the RNA-binding proteins HuR and AUF1. J Biol Chem. 2016;291:25823–25836. doi: 10.1074/jbc.M116.756908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schäfer F, Seip N, Maertens B, Block H, Kubicek J. Purification of GST-tagged proteins. Methods Enzymol. 2015;559:127–139. doi: 10.1016/bs.mie.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Huang Da Wei, Sherman Brad T, Lempicki Richard A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2008;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 14.Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet. 2014;48:13. doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt AM, Chang HY. Long noncoding RNAs in Cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolha L, Ravnik-Glavač M, Glavač D. Long noncoding RNAs as biomarkers in Cancer. Dis Markers. 2017;2017:7243968. doi: 10.1155/2017/7243968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khorshidi HR, Taheri M, Noroozi R, Sarrafzadeh S, Sayad A, Ghafouri-Fard S. Genetic variants in Iranian breast Cancer patients. Cell J. 2017;19:72–78. doi: 10.22074/cellj.2017.4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meseure D, Vacher S, Alsibai KD, Nicolas A, Chemlali W, Caly M, Lidereau R, Pasmant E, Callens C, Bieche I. Expression of ANRIL-Polycomb complexes-CDKN2A/B/ARF genes in breast tumors: identification of a two-gene (EZH2/CBX7) signature with independent prognostic value. Mol Cancer Res. 2016;14:623–633. doi: 10.1158/1541-7786.MCR-15-0418. [DOI] [PubMed] [Google Scholar]
- 19.Naemura M, Tsunoda T, Inoue Y, Okamoto H, Shirasawa S, Kotake Y. ANRIL regulates the proliferation of human colorectal cancer cells in both two- and three-dimensional culture. Mol Cell Biochem. 2016;412:141–146. doi: 10.1007/s11010-015-2618-5. [DOI] [PubMed] [Google Scholar]
- 20.Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, Liu YW, Liu XH, Zhang EB, Lu KH, Shu YQ. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14:268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 21.Sun Z, Ou C, Ren W, Xie X, Li X, Li G. Downregulation of long non-coding RNA ANRIL suppresses lymphangiogenesis and lymphatic metastasis in colorectal cancer. Oncotarget. 2016;7:47536–47555. doi: 10.18632/oncotarget.9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu H, Li X, Song Y, Zhang P, Xiao Y, Xing Y. Long non-coding RNA ANRIL is up-regulated in bladder cancer and regulates bladder cancer cell proliferation and apoptosis through the intrinsic pathway. Biochem Biophys Res Commun. 2015;467:223–228. doi: 10.1016/j.bbrc.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 23.White Elizabeth J.F., Matsangos Aerielle E., Wilson Gerald M. AUF1 regulation of coding and noncoding RNA. Wiley Interdisciplinary Reviews: RNA. 2016;8(2):e1393. doi: 10.1002/wrna.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang N, Yi J, Guo G, Liu X, Shang Y, Tong T, Cui Q, Zhan M, Gorospe M, Wang W. HuR uses AUF1 as a cofactor to promote p16INK4 mRNA decay. Mol Cell Biol. 2010;30:3875–3886. doi: 10.1128/MCB.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson GM, Sun Y, Lu H, Brewer G. Assembly of AUF1 oligomers on U-rich RNA targets by sequential dimer association. J Biol Chem. 1999;274:33374–33381. doi: 10.1074/jbc.274.47.33374. [DOI] [PubMed] [Google Scholar]
- 26.Zucconi BE, Ballin JD, Brewer BY, Ross CR, Huang J, Toth EA, Wilson GM. Alternatively expressed domains of AU-rich element RNA-binding protein 1 (AUF1) regulate RNA-binding affinity, RNA-induced protein oligomerization, and the local conformation of bound RNA ligands. J Biol Chem. 2010;285:39127–39139. doi: 10.1074/jbc.M110.180182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui C, Gan Y, Gu L, Wilson J, Liu Z, Zhang B, Deng D. P16-specific DNA methylation by engineered zinc finger methyltransferase inactivates gene transcription and promotes cancer metastasis. Genome Biol. 2015;16:252. doi: 10.1186/s13059-015-0819-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brocks D, Schmidt CR, Daskalakis M, Jang HS, Shah NM, Li D, Li J, Zhang B, Hou Y, Laudato S, et al. DNMT and HDAC inhibitors induce cryptic transcription start sites encoded in long terminal repeats. Nat Genet. 2017;49:1052–1060. doi: 10.1038/ng.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lillycrop K, Murray R, Cheong C, Teh AL, Clarke-Harris R, Barton S, Costello P, Garratt E, Cook E, Titcombe P, et al. ANRIL promoter DNA methylation: a perinatal marker for later adiposity. EBioMedicine. 2017;19:60–72. doi: 10.1016/j.ebiom.2017.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirosue A, Ishihara K, Tokunaga K, Watanabe T, Saitoh N, Nakamoto M, Chandra T, Narita M, Shinohara M, Nakao M. Quantitative assessment of higher-order chromatin structure of the INK4/ARF locus in human senescent cells. Aging Cell. 2012;11:553–556. doi: 10.1111/j.1474-9726.2012.00809.x. [DOI] [PubMed] [Google Scholar]
- 31.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarkar D, Oghabian A, Bodiyabadu PK, Joseph WR, Leung EY, Finlay GJ, Baguley BC, Askarian-Amiri ME. Multiple isoforms of ANRIL in melanoma cells: structural complexity suggests variations in processing. Int J Mol Sci. 2017;18:e1378. doi: 10.3390/ijms18071378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, Xiang B, Liu Y, Wang Y, Kan H. LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer Lett. 2018;437:56–66. doi: 10.1016/j.canlet.2018.08.024. [DOI] [PubMed] [Google Scholar]
- 34.Huang D, Bi C, Zhao Q, Ding X, Bian C, Wang H, Wang T, Liu H. Knockdown long non-coding RNA ANRIL inhibits proliferation, migration and invasion of HepG2 cells by down-regulation of miR-191. BMC Cancer. 2018;18:919. doi: 10.1186/s12885-018-4831-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary methods
Additional file 2 Figure S1. Characterization of P14AS in the CDKN2A/B locus. (A) Sashimi view for transcripts detected by CDKN2A-specific probe-captured RNA (RNACap)-Seq in HEK293T cells containing two wild-type CDKN2A/B alleles and in MCF7 cells with the homogenous P16 deletion. (B) Graphic view of the protein-coding potential for P14AS, CDKN2A/P14, and ANRIL genes (adapted from the UCSC website).
Additional file 3 Table S1. List of proteins interacted with P14AS characterized in the HEK293T cells in RNA pull-down mass spectrum analysis
Additional file 4 Figure S2.P14AS affects the expression level of the neighboring genes at 9p21.3. qRT-PCR data are normalized to GAPDH mRNA levels and shown as the means ± SD. (a) The expression levels of P14, P16, and P15 genes in the P14AS vector stably transfected colon cancer cells (HCT116 and SW480), and gastric cancer cells (MGC803) were analyzed by qRT-PCR. (b) Detection of P16/P15/P14 proteins in MGC803 cells in Western blot analyses. (c) A fragment deletion in P14AS exon 1 was detected by PCR (top chart) in HEK293T or HCT116 P14AS-KO clones (KO). The expression changes of P14, P16, and P15 genes in HCT116 and HEK293T cells whose ARE-containing elements in P14AS exon-1 were homogenously deleted in the qRT-PCR analysis (bottom chart). Pooled P14AS ARE-KO-negative subclones were used as a wild-type (WT) control. (d) Detection of the P16, P15, and P14 proteins in HEK293T cells in Western blot analyses.
Additional file 5 Figure S3. P14AS expression decreased P16 mRNA-AUF1 binding. (a) AUF1 directly bound to ANRIL and P16 mRNA in HCT116 cells in the AUF1-RIP-PCR. (b) P14AS overexpression decreased ANRIL and P16 mRNA-AUF1 interaction in by the AUF1-RIP-qPCR. (c) An illustration of how the competitive AUF1-P14AS binding protects ANRIL and P16 mRNA from the decay. AUF1 complexes were drawn as dimers based on the reports that AUF1 isoforms (p37, p40, p42, and p45) could form functional dimers [25, 26].
Additional file 6 Figure S4. Genome-wide analyses of transcriptome by RNA sequencing for HCT116 cells with and without P14AS overexpression and/or AUF1 downregulation. The HCT116 cells with stable P14AS overexpression were transfected with AUF1 siRNAs (siAUF1) for 72 h, and then harvested for RNA sequencing. The number of genes with > 2 fold changes (UP, upregulated; Down, downregulated) for different types of RNAs were labeled. Western blot analysis for the determination of AUF1 downregulation by siRNAs was inserted into the top chart. Two samples were sequenced for each group.
Additional file 7 Table S2. Function annotations for P14AS-upregulated genes (n = 241) with fold change > 2 in HCT116 cells with the David 6.8: Functional Annotation Tools at the website http://david.ncifcrf.gov/tools.jsp [13]
Additional file 8 Table S3. Function annotations for P14AS-downregulated genes (n = 299) with fold change > 2 in HCT116 cells with the David 6.8: Functional Annotation Tools at the website http://david.ncifcrf.gov/tools.jsp [13]
Additional file 9 Figure S5. Association analyses between the expression levels of AUF1 and ANRIL, P16, or P15 using the publicly available transcriptome databases for human cancer cell lines in the CCLE project. (A) All 1037 cell lines; (B) 224 cell lines without the CDKN2A allele deletion (relative copy number > 0).
Additional file 10 Figure S6. Comparison of the levels of P14AS, P16, and P15 expression in colon tissues from cancer and noncancer patients. (a) The expression status of P14AS, P16, and P15 in colon cancer (CC), paired surgical margin (SM), and normal colon biopsy (Normal) tissues from noncancer patients by qRT-PCR. (b) The level of P14AS expression in ANRIL-positive and -negative (by RT-PCR) colon CC and SM tissues. (c and d) Comparison of the expression levels of P16 and P15 mRNA (by qRT-PCR) in P14AS-positive and -negative (by RT-PCR; 1043 bp) colon CC and SM tissues. Error bars, S.E.M. *, P < 0.05; **, P < 0.01; N.S. no significance.
Additional file 11 Table S4. Comparison of the P14AS expression level (by qRT-PCR) in colon cancer (CC) and surgical margin (SM) tissue samples from patients with different clinicopathological characteristics
Additional file 12 Figure S7. Comparisons of the levels of AUF1 mRNA (by qRT-PCR) with those of P14AS and ANRIL lncRNA in colon cancer tissues (CCs).
Additional file 13 Table S5. Comparison of the ANRIL expression level (by RT-PCR) in colon cancer (CC) and surgical margin (SM) tissue samples from patients with different clinicopathological characteristics
Additional file 14 Table S6. Comparison of the AUF1 mRNA level (by qRT-PCR) in colon cancer (CC) and surgical margin (SM) tissue samples from patients with different clinicopathological characteristics
Additional file 15 Table S7. Comparison of P14AS and ANRIL coexpression in colon cancer (CC) and surgical margin (SM) tissue samples from patients with different clinicopathological characteristics
Additional file 16 Figure S8. Graph of the P14AS gene in the CDKN2A/B locus. (A) CpG islands within the CDKN2A/P14 gene. (B) The transcription and active histone modification status in the chromatin upstream of the P14AS gene in 7 cell lines from ENCODE. (C) Transcription factors binding to various fragments around the P14AS gene from ENCODE. The RNA polymerase II (POLR2A) is highlighted in red lines. (D) The conservation status of various fragments among vertebrates (adapted from the UCSC website).
Additional file 17 Figure S9. Repression of P16, P14AS, and ANRIL expression in gastric cancer cells by engineered P16-specific DNA methyltransferase (P16-Dnmt). The gene expression level was detected using qRT-PCR assay. The detailed processes for the construction of P16-Dnmt and transfection experiments are previously described [27]
Data Availability Statement
The data and materials of the study are available from the corresponding author upon reasonable request.