Abstract
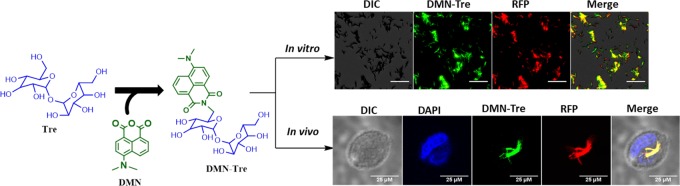
4-N,N-Dimethylamino-1,8-naphthalimide conjugate of trehalose (DMN-Tre) is a fluorogenic dye recently developed as a diagnostic tool for tuberculosis. DMN-Tre selectively labels the mycobacterial cell wall through the Ag85 enzymes. In this work, we disclose a protocol describing the total synthesis of DMN-Tre with more than 99% purity. We further developed a protocol for in vitro and intercellular labeling of various mycobacterial strains. DMN-Tre labeling was found to be a useful tool to study in vitro and intracellular Mycobacterium tuberculosis (Mtb) physiology and as an end-point readout system in high-content image-based screening (HCS) of drug molecules. Such uses of DMN-Tre labeling provide a simple, fast, and cheap alternative to the existing, time-consuming approach that requires Mtb strains to be genetically transformed with fluorescent reporter genes.
Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), is the leading cause of death by an infectious disease. Each year, TB claims the lives of 1.3 million people,1 despite being curable with affordable and accessible antibiotics to treat this infection. However, major drawbacks of current TB therapy are lengthy treatment durations and poor efficacy against multidrug-resistant (MDR) Mtb.2,3 Accordingly, drug development programs focus on developing new agents that are capable of tackling these challenges.
Targeting intracellular mycobacteria is an established approach in current TB drug development4−9 and provides great potential to offer novel TB treatment compounds.10,11 The approach, in general, involves an in-depth study of the intracellular lifestyle of Mtb and development of compounds that target specific metabolic pathways essential for intracellular growth or factors rendering the pathogen less effective in evading the host’s natural ability to clear the pathogen.12 Successful development of such compounds requires an in-depth understanding of the disease pathology, as it helps to identify novel drug targets and a suitable, preferably, image-based high-content drug screening platforms that facilitate active compound identification.
Tracing intracellular mycobacteria typically relies on the expression of fluorescent proteins that facilitate detection of the bacteria by fluorescent microscopy.13−16 This approach became popular in mycobacterial research after Kremer et al. successfully cloned Mtb with a green fluorescent (GFP) reporter gene for the first time.17 However, though proven to be effective, the standard method for producing fluorescent mycobacteria by genetic manipulation has some limitations, including the need to transform each strain of interest with the gene encoding the fluorescent protein. This is especially challenging and time consuming when working with multiple mutant strains or assaying numerous clinical isolates that all require fluorescence. Furthermore, the transformation may alter the pathophysiological properties of the bacterium.18 One strategy to overcome these challenges is to label mycobacteria using chemical-based fluorogenic probes.
The use of chemical fluorogenic dyes for metabolic labeling of mycobacteria is relatively new. The first of such mycobacteria-specific fluorogenic probes was reported by Backus et al.19 This fluorescein-containing trehalose (FITC-Tre) probe is metabolically incorporated into the cell wall of mycobacteria by the Ag85 enzyme complex and was used for both in vitro and ex vivo labeling. Soon after, more trehalose-based labeling probes were reported. These include a series of azide-modified trehaloses (TreAz),20 O- and N-alkylated trehalose monomycolates (O-AlkTMM and N-AlkTMM, respectively),21 a quencher–trehalose–fluorophore (QTF),22 and trehalose conjugated to a dye, 4-N,N-dimethylamino-1,8-naphthalimide (DMN-Tre).23
DMN-Tre (Figure 1) is a solvatochromic dye that exhibits marked enhancement of fluorescence following a transition from an aqueous to a lipophilic environment. Therefore, a fluorescence signal is only detected after mycobacteria take up the dye and metabolically incorporate it into its lipophilic cell wall. Specific incorporation of this dye is achieved by the bacterial Ag85 enzyme complex after mycolation at the 6′-OH position. The dye was shown to rapidly label mycobacteria in vitro and in sputum taken from TB patients, demonstrating its potential for TB diagnosis.23
Figure 1.
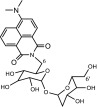
Structure of DMN-Tre.
Labeling of mycobacteria with DMN-Tre offers a number of advantages. First, no postlabeling wash is required, as the excess nonincorporated dye does not fluoresce and therefore does not cause background interference. Second, incorporation of the dye requires that the bacteria be metabolically active and thus the dye can differentiate between viable and nonviable bacterial cells.
A protocol that describes the synthesis of DMN-Tre, from its immediate precursors, 6-aminotrehalose and 4-N,N-dimethylamino-1,8-napthalic anhydride, was previously reported.23 However, the complete synthetic route including the synthesis of the two starting materials was not provided. As both 6-aminotrehalose and 4-N,N-dimethylamino-1,8-napthalic anhydride are not accessible from common commercial sources, a protocol that fully describes the synthesis of DMN-Tre is needed.
The main objective of this work is to synthesize DMN-Tre and adapt it for intracellular labeling of mycobacteria with the aim to develop a tool for end-point detection in high-content image-based screening of active compounds and host–pathogen interaction studies of intracellular mycobacteria.
Results and Discussion
DMN-Tre Synthesis
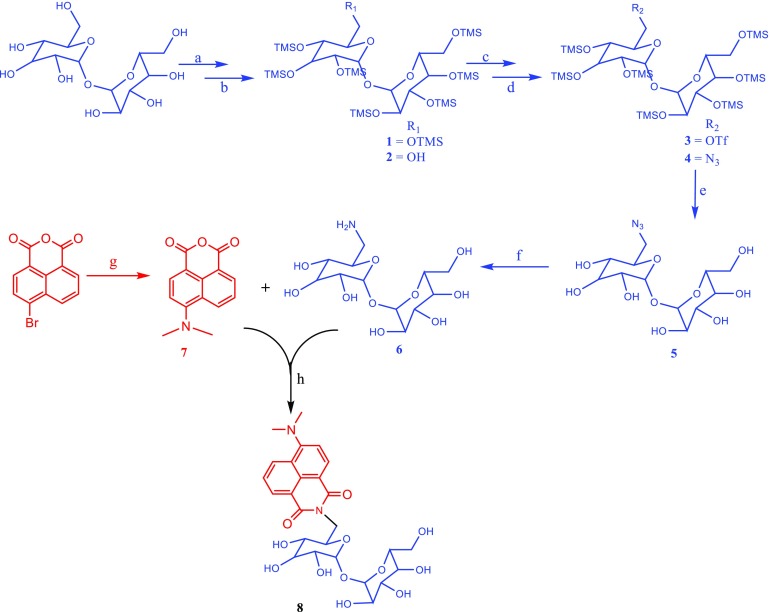
The scheme for the total synthetic route of DMN-Tre is outlined in Figure 2. The first step involved the protection of all free hydroxyl groups of trehalose as trimethylsilyl (TMS) ethers. This protecting group was chosen as it can be easily introduced and selectively removed in the presence of secondary TMS ethers.24 The synthesis was done by treating trehalose with TMS chloride (TMS-Cl) in pyridine solvent as described by Toubiana et al.25 This provided the fully protected 2,3,4,6,2′,3′,4′6′-octa-O-TMS trehalose (1) with a yield of 90% after column purification.
Figure 2.
Total synthetic route for DMN-Tre. Reagents and conditions: (a) TMS-Cl, pyridine, 0 °C to rt, 19 h; (b) 0.2% K2CO3/MeOH, 0 °C, 20 min; (c) Tf2O, pyridine, DCM, 0 °C to rt,2 h; (d) NaN3, dimethylformamide (DMF), 0 °C to rt, 12 h; (e) trifluoroacetic acid (TFA)/dioxane/H2O, rt, 1 h; (f) H2, Pd/C (10%), MeOH, 1 h; (g) 3-dimethylaminopropionitrile, 3-methyl-1-butanol, 132 °C, 12 h; and (h) NaHCO3, EtOH, N2, 85 °C, 8 h.
The next step involved the selective removal of only one of the TMS protection groups and setting the 6-hydroxy group free. This was achieved through controlled alkaline hydrolysis of 1 using a catalytic amount of potassium carbonate (0.2% K2CO3) in methanol26,27 for 20 min. Column purification provided the 2,3,4,2′,3′,4′6′-hepta-O-TMS trehalose-6-ol (2) at 40% 2,3,4,2′,3′,4′-hexa-O-TMS trehalose-6, 6′-diol (30%) and the starting material 1 (30%), which was regenerated and reused.
The free 6-hydroxyl group of 2 was activated to its triflate analogue (3) and then transformed to azide (4) by treatment with sodium azide in DMF.28 Triflation of the primary alcohol of 2 was achieved by treating with triflic anhydride in anhydrous dichloromethane in the presence of pyridine.29 This step was found to be challenging as the use of a little excess triflic anhydride released the very strong triflic acid during the workup and caused a premature deprotection of the remaining TMS protecting groups. For this reason, we opted to use the crude product directly for the next reaction.
After triflate to azide transformation, global deprotection of the remaining TMS protecting groups of 4 was accomplished using a cleaving cocktail made of H2O/TFA/THF (33:8:17).28 Subsequently, the azide functional group of 5 was reduced to its corresponding primary amine group by catalytic hydrogenation using Pd/C (10%) as a catalyst in methanol.30 This provided the first immediate precursor of DMN-Tre, 6-aminotrehalose (6).
The other immediate precursor of the dye, 4-N,N-dimethylaminonaphthalic anhydride (7), was synthesized from 4-bromophthalic anhydride in one step following a previously reported method31 with an isolated yield of 78%.
The final step in the synthesis of DMN-Tre was the coupling of 6 and 7 as previously described by Kamariza et al.23 This reaction gave the target compound, DMN-Tre (8), with an isolated yield of 35% and a purity level greater than 99% after reversed-phase high-pressure liquid chromatography (RP-HPLC) purification. The identity of the product was confirmed by 1H NMR and high-resolution mass spectroscopy (HRMS) (Figure S1).
In Vitro Labeling of Mycobacteria Using DMN-Tre
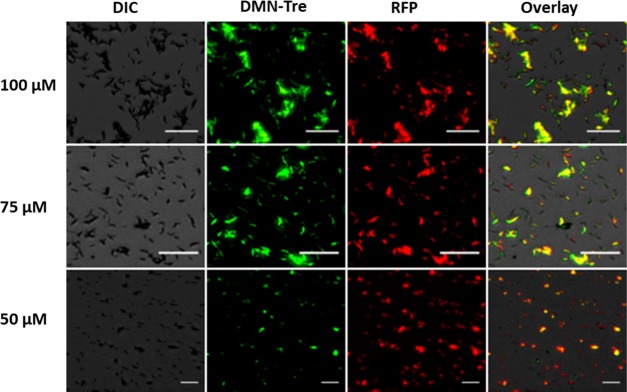
To determine the optimal dose and to assess the suitability of DMN-Tre for labeling different Mtb strains of interest, in vitro labeling was performed. Mtb H37Rv expressing red fluorescent protein (RFP-Mtb H37Rv) was grown in the presence of DMN-Tre at various concentrations for 24 h (Figure 3). Colocalization of the fluorescence signals between GFP (for DMN-Tre) and RFP channels (Figure 3) showed that all of the bacteria that appeared in the RFP channel could also be seen in the GFP channel. This indicates that complete labeling could be achieved at concentrations of 100 μM as previously reported23 and as low as 75 μM. At 50 μM, the labeling efficiency declined and was undetected at 25 μM.
Figure 3.
In vitro labeling of RFP-Mtb H37v with DMN-Tre. Images were collected in the differential interference contrast (DIC), green fluorescent protein (GFP, for DMN-Tre fluorescence), and red fluorescent protein (RFP, for bacterial dTomato fluorescence) channels using the Evos FL auto imaging system fluorescence microscope (Thermo Fisher). Magnifications are 20×. “Overlay” represents merged images from all channels. Scale bars = 25 μm.
We also tested various Mtb mutant strains for their ability to take up the dye. These Mtb mutants were either constructed or grown in our lab to elucidate the mode of action of previously identified anti-TB hit compounds.9 Comparative analysis of Mtb H37Rv-resistant mutants to various anti-TB compounds (Table S1) showed that all of the mutants were labeled (Figures S2–S6). Additionally, we were able to label the Mtb Erdman strain and a Δrv1747 Erdman deletion mutant32 using DMN-Tre, demonstrating the potential of the dye to be used as a viable chemical labeling tool for mycobacterial research.
Intracellular Labeling of Mycobacteria Using DMN-Tre
Macrophages derived from the human monocyte cell line (THP-1) are commonly used to study the intracellular life cycle of Mtb and for screening of anti-TB compounds.9,33 The most common approach for these tasks is to use a genetically modified Mtb strain expressing a fluorescent reporter gene. The use of a chemical probe that can quickly and efficiently label intracellular mycobacteria would greatly facilitate research by reducing the effort and lengthy time needed for genetic transformations. Accordingly, the ability of DMN-Tre to label intracellular Mtb was assessed using this model.
First, we tested whether DMN-Tre was toxic to THP-1 cells. THP-1 cells were exposed to the dye at different concentrations for 4 days, after which cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) to label their nuclei. THP-1 cell viability was assessed based on nuclei count. Tolerability assessment for the dye was then made by comparing the number of viable cell counts of DMN-Tre treated wells vs water control. No statistically significant difference in the cell count between the treated and control groups was observed at all tested concentrations (Figure S7), indicating that DMN-Tre was well tolerated by THP-1 cells.
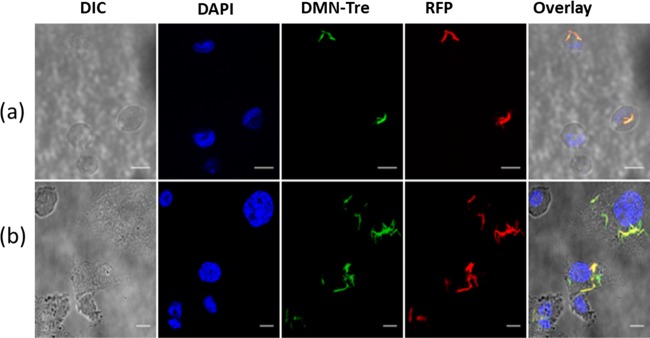
Next, we proceeded to assess if DMN-Tre could be used to label intracellular Mtb within THP-1 cells. Macrophages that had been infected with RFP-Mtb H37v were subsequently incubated in media containing 100 μM DMN-Tre for 24 and 96 h. Microscopy analysis revealed that the DMN-Tre probe was indeed incorporated into intracellular Mtb, indicating that the dye was able to reach the intracellular compartments of the macrophage. The DMN-Tre signal overlapped with the RFP signal, showing that all bacteria were labeled with the dye (Figure 4). Full labeling was observed to be completed as early as 24 h with no difference in intensity when prolonged to 4 days. Additionally, DMN-Tre was also successful in labeling a deletion mutant of Mtb, as well as the vaccine M. bovis BCG strain, intracellularly (Figure S8).
Figure 4.
Intracellular labeling of RFP-Mtb H37v with DMN-Tre. Image was taken after (a) 24 h and (b) 96 h of incubation of the Mtb-infected THP-1 cells with DMN-Tre. Images were collected in the DIC, DAPI (for NucBlue fluorescence of the nuclear stain), GFP (for DMN-Tre fluorescence), and RFP (for bacterial dTomato fluorescence) channels of the Leica SP5 laser scanning confocal microscope. Magnifications are 63× by oil immersion. Overlay represents merged images from all channels. Scale bars = 25 μm.
DMN-Tre Labeling Does Not Alter Intracellular Mtb Growth
The lipid composition of the mycobacterial cell wall contributes to Mtb virulence. Accordingly, modifications to the membrane composition may affect the ability of the bacteria to infect and/or grow within macrophages.12,34 To evaluate the impact of DMN-Tre labeling on Mtb intracellular growth, RFP-Mtb H37v was prelabeled with the dye before macrophage infection. The infected macrophages were then maintained in media containing 100 μM DMN-Tre for the entire incubation period. Macrophages infected with unlabeled RFP-Mtb H37Rv and maintained in media free of DMN-Tre were used as a control.
The impact of the labeling was then assessed by quantifying the intracellular bacterial load immediately after infection and after 4 days of infection. The intracellular Mtb burden was assessed by quantifying the RFP signal at a validated radius around each macrophage’s stained nucleus using the CellInsight CX5 high-content platform. Such techniques for quantifying intracellular Mtb has been previously validated.7,35−37 The intracellular Mtb load was increased by 100% for both prelabeled and nonlabeled bacteria after 4 days of infection (Figure 5), indicating that DMN-Tre does not inhibit the ability of Mtb to infect or grow within the macrophages. A similar finding was also previously reported when assessing the impact of FITC-Tre labeling on the intracellular growth of Mtb.19
Figure 5.
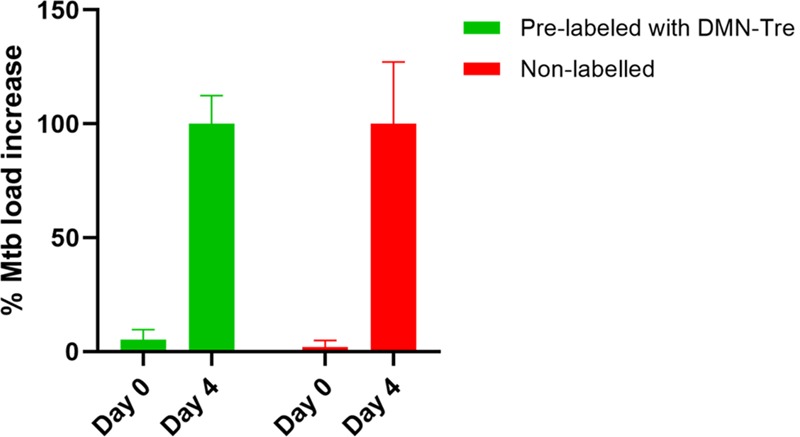
Effect of DMN-Tre labeling on the intracellular growth of RFP-Mtb H37Rv. Macrophages were infected with RFP-expressing Mtb H37Rv with or without prelabeling with DMN-Tre. The intracellular Mtb growth was assessed based on the increase in intensity of the fluorescent signal measured from the infected macrophages on day 0 and day 4. Growth at day 0 was set as 0%. Error bars represent standard error (SE), n = 4.
DMN-Tre as a Tool for Studying Host–Pathogen Interactions
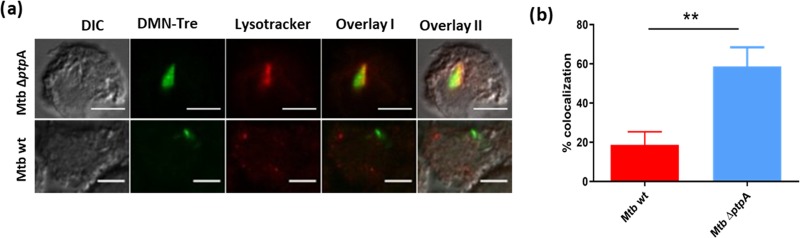
As an intracellular pathogen, Mtb interacts with its host by inhibiting the ability of the macrophage to clear the infection.38 Normally, macrophages destroy invading organisms through a process of phagosome maturation leading to fusion with lysosomes and activation of immune recognition. Mtb secretes a key phosphatase, PtpA, that binds the host vacuolar-H(+)-ATPase, disrupting phagosome acidification, and dephosphorylates VPS33B, involved in tethering the phagosome to the lysosome membranes, thus blocking phagosome–lysosome fusion.39,40 Genetic deletion of ptpA in Mtb (ΔptpA) results in normal macrophage phagosome maturation and colocalization of VPS33B and the V-ATPase subunit H with ΔptpA, but not wild-type (wt) Mtb.40
Here, we tested whether DMN-Tre could be applied to study this colocalization event using fluorescence microscopy. We used DMN-Tre to fluorescently labeled ΔptpA and wt Mtb and lysotracker to label macrophage lysosomal compartments. Figure 6 shows that the ΔptpA mutant colocalized with the lysotracker tag, while the parental wt strain did not, confirming the role of ptpA in controlling this process and validating the use of DMN-Tre.
Figure 6.
DMN-Tre labeling of Mtb allows intracellular tracking of phagolysosome fusion. (a) Colocalization of lysotracker DND 99 (red) with wt or ΔptpA Mtb, labeled with DMN-Tre (green); image was taken by a confocal microscope; magnifications are 63X by oil immersion. “Overlay 1” represents a merge between DMN-Tre signal and lysotracker. “Overlay 2” represents a merge between all channels. (b) Colocalization quantification. Quantification was done using Image J with the Manders coefficient. Error bars represent the standard deviation (SD). **p-value < 0.01.
A similar approach could be followed to prelabel various constructs of Mtb mutants with DMN-Tre and infect macrophages to study other mediators of host–pathogen interaction and monitor Mtb infection and its effect on the host immunophysiology.
DMN-Tre as a Fluorescence End-Point Readout Tool for High-Content Image-Based Screening
High-content screening (HCS) has become a powerful approach for screening new compounds effective against Mtb. However, a prerequisite for HCS is that the bacteria need to be labeled for visualization. Often, there is a need to screen against strains that are not genetically labeled such as clinical isolates and mutant Mtb strains without a priori genetic manipulation. Therefore, a new end-point detection system is needed. The suitability of DMN-Tre as a readout system for HCS was assessed using a 96-well plate format and compared to a traditional RFP readout system. THP-1 cells infected with DMN-Tre-labeled Mtb H37Rv or RFP-Mtb H37Rv were treated with the TB drug, rifampicin (Rif), dissolved in dimethyl sulfoxide (DMSO), or DMSO alone control. Similar effects of the drug were observed using DMN-Tre as readout, albeit with less intensity compared to that of RFP (Figure 7).
Figure 7.
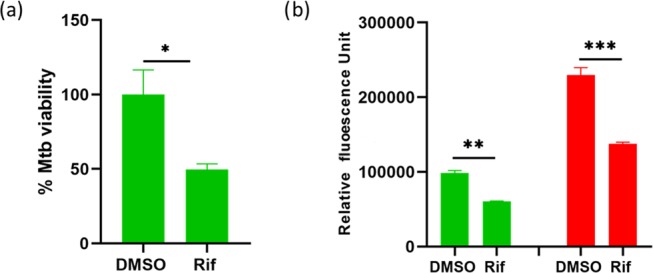
Intracellular Mtb drug susceptibility assay measured by DMN-Tre labeling. (a) Intracellular viability of Mtb H37Rv using DMN-Tre labeling as an end-point readout. THP-1 cells were infected with Mtb H37Rv, treated with or without 5 μM Rif in DMSO and incubated in a media containing DMN-Tre. Mtb viability was calculated as a percentage of the average fluorescence intensity of the DMSO control (set at 100%). (b) Comparison of the fluorescence signal intensities of the dually fluorescent DMN-Tre-labeled RFP-Mtb H37Rv within THP-1 cells. THP-1 cells were infected with RFP-Mtb H37Rv, treated with or without 5 μM Rif in DMSO and incubated in a media containing DMN-Tre. Fluorescence was measured from the GFP channel using DMN-Tre labeling as a readout (green bars) and the RFP channel using red fluorescence protein expression as a readout (red bars). Error bars in (a) and (b) represent the SE. *p-value = 0.0002; ** and ***p-values < 0.0001.
Once the suitability of DMN-Tre for HCS of anti-Mtb control compounds was assessed, a dose–response assay with another TB drug, bedaquiline (BDQ), was performed. The intracellular IC50 values of BDQ obtained from inhibition curves determined using the GFP and RFP channels were similar (Figure 8, 0.12 and 0.14 μM, respectively) and are within the range of the previously reported concentration of 0.13 μM.41 To examine suitability for a screening assay, the Z′ score was calculated for BDQ (3 μM) and determined as 0.6 and 0.4 for DMN-Tre and RFP, respectively. This is a good signal-to-noise ratio, suggesting that DMN-Tre could be used for high-throughput HCS of a larger number of compounds.
Figure 8.
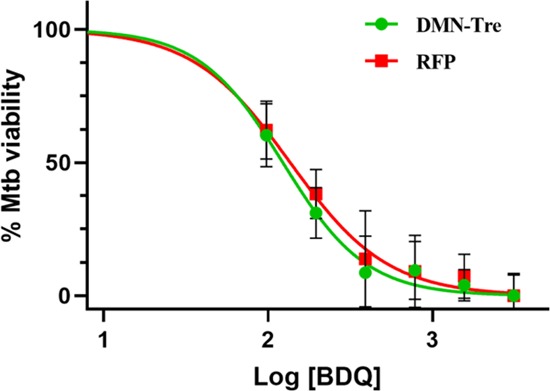
Dose–response plot for BDQ against RFP-Mtb H37v comparing DMN-Tre and RFP. Assay 96-well-plate-containing THP-1 cells infected with RFP-Mtb H37Rv stained or unstained with DMN-Tre were treated with 1% DMSO (six wells) as a negative control corresponding to 100% bacterial growth and 3 μM BDQ (eight wells) as a positive control in which 100% inhibition of bacterial growth was reached. Controls were used to monitor assay quality through the determination of the Z′ score.
Conclusions
In this work, a protocol describing the total synthesis of DMN-Tre at more than 99% purity was developed. The suitability of the dye to label various Mtb mutant strains and other mycobacterial species was assessed, and the optimal dose required for in vitro labeling was determined. Moreover, the dye was shown to be efficient in labeling various intracellular mycobacterial strains in infected macrophages. This intracellular labeling was achieved with limited or no toxicity to the host and without altering virulence characteristics of the bacteria. The finding that DMN-Tre successfully labeled intracellular Mtb was then further applied to study intracellular events of Mtb infection and as an end-point readout tool in image-based HCS of new antitubercular compounds.
Transforming Mtb with fluorescent reporter genes has proven to be a powerful tool to study Mtb physiology and for high-throughput genetic and small molecule screening. However, due to the slow-growing nature of Mtb, this approach is often time consuming, costly, and a tedious process. The applicability of DMN-Tre for in vitro and intracellular labeling of Mtb provides a simple, rapid, and less expensive alternative to the already existing methods. This work underscores the potential of DMN-Tre to be used in drug discovery and basic research in addition to being a useful tool for TB diagnostics as originally described.23
Materials and Methods
Equipment
Fluorescence Microscopes
Leica SP5 laser scanning confocal microscope (Nikon A1R), Evos FL auto cell imaging system fluorescence microscope (Thermo Fisher), and CellInsight CX5 high-content screening platform (Thermo Fisher) were used.
Mycobacterial Strains and Growth Media
Mycobacterial Strains
M. tuberculosis H37Rv, M. tuberculosis H37Rv harboring RFP-expressing pTEC27 plasmid, M. tuberculosis ΔptpA (constructed as previously described42), Mangora bovis BCG, and 14 other Mtb mutants that were grown to elucidate the mode of action of previously identified anti-Mtb compounds (Table S1) were utilized for the experiment.
All of the mycobacterial strains were routinely grown in 7H9 broth (Difco Middlebrook) supplemented with 10% (v/v) OADC (0.05% oleic acid, 5% bovine albumin fraction, 2% dextrose, 0.004% catalase, and 0.8% sodium chloride solution) and 0.05% (v/v) Tween 80 (Sigma-Aldrich) at 37 °C in standing cultures. For Mtb pTEC27, hygromycin B was added to the medium at a final concentration of 50 μg/mL.
THP-1 Cells and Culture Media
The THP-1 cells (ATCC TIB-202) used are derived from human monocytes obtained from a 1 year old male infant with acute monocytic leukemia. The cells were stored in liquid nitrogen.
THP-1 cells were grown in complete RPMI medium. The cells were grown in tissue culture flask with a minimum volume of 20 mL and a maximum volume of 50 mL and were incubated in an atmosphere of 95% air and 5% carbon dioxide (CO2) at a temperature of 37 °C. Cell density was kept between 0.25 and 1 million cells/mL. Every 2 or 3 days, the cells were counted in a microscope and diluted to 0.25 million cells/mL. The cells doubled approximately every 48 h. A culture from liquid nitrogen stock was subcultured for up to 3 months; after this time, a change in morphology and growth behavior was observed and a fresh stock was used.
For culturing of THP-1 cells, RPMI-1640 medium supplemented with 5% fetal bovine serum (FBS), 2% glutamine, 1% nonessential amino acids, and 1% penicillin + streptomycin was used (complete RPMI medium).
For assays, RPMI-1640 medium supplemented with 5% FBS, 1% glutamine, and 1% nonessential amino acids was used (incomplete RPMI medium).
In Vitro Labeling of Mycobacteria with DMN-Tre
Cultures of Mtb and mutants were made by inoculation of a 1 mL frozen glycerol stock into 10 mL of a Middlebrook 7H9 liquid medium supplemented with a 10% (v/v) OADC enrichment and 0.05% (v/v) Tween 80 in a tissue culture flask. Cultures were grown to an optical density at 600 nm (OD600) between 0.4 and 0.6 to begin the experiments. Bacterial cultures were mixed with DMN-Tre at a concentration of 75 μM in 7H9 medium and incubated at 37 °C with shaking for 1 day. Labeled bacterial cells were then harvested by centrifugation (4000° rpm, RT, for 10 min), washed with PBS, and then either used for macrophage infection or fixed in 4% paraformaldehyde (PFA) in PBS (room temperature for 45 min) on coverslips for microscopic analysis.
Tolerability Assay
A cell suspension of THP-1 cells (1 × 106 cells/mL) in RPMI incomplete was seeded in a 96-well plate at a density of 50 000 cells per well and allowed to differentiate overnight by adding PMA (40 ng/mL). Differentiated cells were washed once with warm RPMI, and fresh RPMI incomplete was added to each well with DMN-Tre added at concentrations of 100, 200, and 300 μM to wells in triplicates for each concentration. The vehicle (water) was used as an untreated control. The plates were then sealed by parafilm and incubated in a humidified incubator at 37 °C, 5% CO2 for 4 days. At the end of the incubation period, the media was aspirated, cells were washed twice with PBS, stained with DAPI, and their viability was quantified using a CellInsight CX5 high-content screening (HCS) platform (Thermo Scientific).
Macrophage Infection
Mycobacterial cultures grown to log phase were centrifuged at 4000° rpm, for 10 min at room temperature, washed once in 7H9 media containing 0.05% Tween 80, and resuspended in RPMI medium. Cells were de-clumped using a needle, and OD600 was measured to estimate cell density using the formula (OD600 ≈ 3.3 × 108 CFU/mL). Before infection, bacterial suspension was opsonized by adding a 10% human serum and incubated for 30 min at 37 °C. A cell suspension of THP-1 cells (1 × 106 cells/mL) in RPMI incomplete was incubated with the opsonized Mtb single-cell suspension at a multiplicity of infection (MOI of 2:1) and simultaneously differentiated with 40 ng/mL PMA for 4 h at 37 °C under constant agitation. After infection, the THP-1 cell suspension was centrifuged (750 rpm, RT, 10 min) and washed twice with RPMI. The cell pellet was resuspended in RPMI incomplete media at 1 × 105 cells/mL and dispensed either into 96-well clear, flat bottom plates or 24-well plates containing glass coverslips at the bottom of the wells with a cell density of 1 × 105 cells/mL.
Intracellular Mycobacterial Labeling
Macrophages infected with Mtb, Mtb mutants, or M. bovis BCG (according to the macrophage infection protocol described above) were dispensed in 24-well plates containing glass coverslips at the bottom of the wells. DMN-Tre (100 μM) was added to each well, and the plates were incubated in a humidified incubator at 37 °C, 5% CO2 for 1 or 4 days. At the end of the incubation period, the media was aspirated, coverslips were taken out, and cells were washed twice with PBS buffer, fixed with 4% PFA in PBS buffer for 30 min, stained with NucBlue, and visualized under confocal microscopy.
Impact of DMN-Tre Prelabeling on Infectivity of Mtb
RFP-Mtb H37v was prelabeled with DMN-Tre and used to infect macrophages. The infected macrophages were dispensed in 96-well plates (clear, flat bottom) at a cell density of 5 × 104 cells/well. DMN-Tre (100 μM) was added to wells containing cells infected with prelabeled RFP-Mtb H37v. Similarly, macrophages infected with a nonlabeled RFP-Mtb H37Rv strain were used as control. Plates were sealed with parafilm and incubated in a humidified incubator at 37 °C, 5% CO2 for 1 or 4 days. After incubation, the cells were fixed in 4% PFA for 30 min and stained with NucBlue. Monitoring of the intracellular growth was performed using the CellInsight CX5 high-content platform.
Colocalization Study with Lysotracker
THP-1 cells were infected with Mtb (wild type or ΔptpA) prelabeled, with DMN-Tre (MOI of 5), and dispensed in 24-well plates containing glass coverslips at the bottom of the wells. After 90 min of incubation in a humidified incubator at 37 °C, 5% CO2, the Lysotracker DND 99 was added at 75 nM and the incubation was continued for an additional 30 min. The media was aspirated, and cover glass slips were taken out. Cells were washed twice with PBS, stained with NucBlue Live, fixed in 4% PFA for 45 min, and visualized under confocal microscopy.
DMN-Tre for End-Point Readout System in HCS
Macrophages were infected with RFP-Mtb H37Rv or Mtb H37Rv and seeded (cell density = 50 000 cells/well) to a 96-well plate containing rifampin (5 μM) or different concentrations of bedaquiline (BDQ) in RPMI incomplete media. DMSO (1%) was used as a negative control. DMN-Tre (100 μM) was then added to each well, and the plate was incubated at 37 °C with 5% CO2 for 4 days. On day 4, the media was aspirated and the cells were fixed with 2.5% PFA. Following NucBlue staining of the nuclei, intracellular bacterial loads were quantified using the CellInsight CX5 high-content platform by measuring the intensity of either GFP (green circles for DMN-Tre) or red squares for RFP fluorescence signals.
Statistical Analysis
Data are means ± SEM from at least two independent experiments. Unless otherwise specified, all data were analyzed using the GraphPad Prism software’s analysis of variance (ANOVA) test, as indicated in figure legends. Images obtained from the fluorescent microscopy were analyzed using the Image J program.
Quantification of Intracellular Mtb
Quantification of intracellular Mtb load was done using the CellInsight CX5 high-content platform. First, the macrophage nuclei are identified and counted using the DAPI signal; cell debris and other particles are deducted based on a size filter tool. Following this, a region of interest or “circle” is drawn around each host cell nucleus to encompass where Mtb (spots) are located. Finally, the software identifies, counts, and measures the pixel area of the “spots” within the circle based on the RFP signal. The fluorescent spot area measured within each cell (circle) was then added and quantified for each well and displayed on the data analysis software. The average circle spot area of each well corresponds to the intracellular Mtb burden and was used to quantify the Mtb load.
Acknowledgments
The project is funded by the Canadian Institutes for Health Research (CIHR PJT-148646 and PJT152931) to Y.A.-G. The authors are grateful to Adama Bojang for growing Mtb mutants. The authors also thank Joseph Chao for providing insightful comments and Mary Ko for technical assistance.
Glossary
Abbreviations
- BDQ
bedaquiline
- DAPI
4′,6-diamidino-2-phenylindole
- DIC
differential interference contrast
- DMF
dimethylformamide
- DMN-Tre
4-N,N-dimethylamino-1,8-naphthalimide conjugate of trehalose
- FITC
fluorescein isothiocyanate
- GFP
green fluorescent protein
- HRMS
high-resolution mass spectroscopy
- HCS
high-content screening
- Mtb
Mycobacterium tuberculosis
- RFP
red fluorescent protein
- RP-HPLC
reversed-phase high-pressure liquid chromatography
- Rif
rifampin
- TMS-Cl
trimethylsilyl chloride
- THF
tetrahydrofuran
- TFA
trifluoroacetic acid
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b04173.
Synthesis and characterization of DMN-Tre; NMR spectrum; microscopic images of in vitro and intracellular labeling experiments with DMN-Tre against different mycobacterial species; list of Mtb mutants labeled with DMN-Tre (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- WHO . Global Tuberculosis Report 2018 Executive Summary; WHO, 2018. [Google Scholar]
- Koch A.; Cox H.; Mizrahi V. Drug-Resistant Tuberculosis: Challenges and Opportunities for Diagnosis and Treatment. Curr. Opin. Pharmacol. 2018, 42, 7–15. 10.1016/j.coph.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boogaard J.; Kibiki G. S.; Kisanga E. R.; Boeree M. J.; Aarnoutse R. E. New Drugs against Tuberculosis: Problems, Progress, and Evaluation of Agents in Clinical Development. Antimicrob. Agents Chemother. 2009, 53, 849–862. 10.1128/AAC.00749-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X.; Av-Gay Y.. System for Efficacy and Cytotoxicity Screening of Inhibitors Targeting Intracellular Mycobacterium tuberculosis. J. Vis. Exp. 2017, (), 10.3791/55273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley S. A.; Barczak A. K.; Silvis M. R.; Luo S. S.; Sogi K.; Vokes M.; Bray M. A.; Carpenter A. E.; Moore C. B.; Siddiqi N.; et al. Identification of Host-Targeted Small Molecules That Restrict Intracellular Mycobacterium tuberculosis Growth. PLoS Pathog. 2014, 10, e1003946 10.1371/journal.ppat.1003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song O. R.; Deboosere N.; Delorme V.; Queval C. J.; Deloison G.; Werkmeister E.; Lafont F.; Baulard A.; Iantomasi R.; Brodin P. Phenotypic Assays for Mycobacterium tuberculosis Infection. Cytometry, Part A 2017, 91, 983–994. 10.1002/cyto.a.23129. [DOI] [PubMed] [Google Scholar]
- Christophe T.; Jackson M.; Hee K. J.; Fenistein D.; Contreras-Dominguez M.; Kim J.; Genovesio A.; Carralot J. P.; Ewann F.; Kim E. H.; et al. High Content Screening Identifies Decaprenyl-Phosphoribose 2′ Epimerase as a Target for Intracellular Antimycobacterial Inhibitors. PLoS Pathog. 2009, 5, e1000645 10.1371/journal.ppat.1000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir A.; Rana K.; Arya A.; Kapoor N.; Kumar H.; Siddiqui M. A. Mycobacterium tuberculosis H37Rv: In Silico Drug Targets Identification by Metabolic Pathways Analysis. Int. J. Evol. Biol. 2014, 284170 10.1155/2014/284170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino F.; Del Rio R. G.; Zheng X.; Matilla J. P.; Gomez P. T.; Hoyos M. M.; Herran M. E. P.; Losana A. M.; Av-Gay Y. Development of an Intracellular Screen for New Compounds Able to Inhibit Mycobacterium Tuberculosis Growth in Human Macrophages. Antimicrob. Agents Chemother. 2016, 60, 640–645. 10.1128/AAC.01920-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machelart A.; Song O. R.; Hoffmann E.; Brodin P. Host-Directed Therapies Offer Novel Opportunities for the Fight against Tuberculosis. Drug Discovery Today 2017, 22, 1250–1257. 10.1016/j.drudis.2017.05.005. [DOI] [PubMed] [Google Scholar]
- Kim Y. R.; Yang C. S. Host-Directed Therapeutics as a Novel Approach for Tuberculosis Treatment. J. Microbiol. Biotechnol. 2017, 27, 1549–1558. 10.4014/jmb.1705.05032. [DOI] [PubMed] [Google Scholar]
- Forrellad M. A.; Klepp L. I.; Gioffré A.; Sabio y García J.; Morbidoni H. R.; de la Paz Santangelo M.; Cataldi A. A.; Bigi F. Virulence Factors of the Mycobacterium tuberculosis Complex. Virulence 2013, 4, 3–66. 10.4161/viru.22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y.; Yang D.; Cirillo S. L. G.; Li S.; Akin A.; Francis K. P.; Maloney T.; Cirillo J. D. Application of Fluorescent Protein Expressing Strains to Evaluation of Anti-Tuberculosis Therapeutic Efficacy in Vitro and in Vivo. PLoS One 2016, e0149972 10.1371/journal.pone.0149972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamed D.; Boulle M.; Ganga Y.; Arthur C. M.; Skroch S.; Oom L.; Catinas O.; Pillay K.; Naicker M.; Rampersad S.; et al. Erratum: Intracellular Growth of Mycobacterium tuberculosis after Macrophage Cell Death Leads to Serial Killing of Host Cells. eLife 2017, e22028 10.7554/eLife.28205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins L. A.; Torrero M. N.; Franzblau S. G. Green Fluorescent Protein Reporter Microplate Assay for High-Throughput Screening of Compounds against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 1998, 42, 344–347. 10.1128/AAC.42.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. Y.; Clemens D. L.; Horwitz M. A. The Metabolic Activity of Mycobacterium tuberculosis, Assessed by Use of a Novel Inducible GFP Expression System, Correlates with Its Capacity to Inhibit Phagosomal Maturation and Acidification in Human Macrophages. Mol. Microbiol. 2008, 68, 1047–1060. 10.1111/j.1365-2958.2008.06214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer L.; Baulard A.; Estaquier J.; Poulain-Godefroy O.; Locht C. Green Fluorescent Protein as a New Expression Marker in Mycobacteria. Mol. Microbiol. 1995, 17, 913–922. 10.1111/j.1365-2958.1995.mmi_17050913.x. [DOI] [PubMed] [Google Scholar]
- Sabuquillo P.; Gea A.; Matas I. M.; Ramos C.; Cubero J. The Use of Stable and Unstable Green Fluorescent Proteins for Studies in Two Bacterial Models: Agrobacterium tumefaciens and Xanthomonas campestris Pv. Campestris. Arch. Microbiol. 2017, 199, 581–590. 10.1007/s00203-016-1327-0. [DOI] [PubMed] [Google Scholar]
- Backus K. M.; Boshoff H. I.; Barry C. S.; Boutureira O.; Patel M. K.; D’Hooge F.; Lee S. S.; Via L. E.; Tahlan K.; Barry C. E.; et al. Uptake of Unnatural Trehalose Analogs as a Reporter for Mycobacterium tuberculosis. Nat. Chem. Biol. 2011, 7, 228–235. 10.1038/nchembio.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts B. M.; Holsclaw C. M.; Jewett J. C.; Alber M.; Fox D. M.; Siegrist M. S.; Leary J. A.; Kalscheuer R.; Bertozzi C. R. Probing the Mycobacterial Trehalome with Bioorthogonal Chemistry. J. Am. Chem. Soc. 2012, 134, 16123–16126. 10.1021/ja3062419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley H. N.; Stewart J. A.; Kavunja H. W.; Rundell S. R.; Swarts B. M. Bioorthogonal Chemical Reporters for Selective in Situ Probing of Mycomembrane Components in Mycobacteria. Angew. Chem., Int. Ed. 2016, 55, 2053–2057. 10.1002/anie.201509216. [DOI] [PubMed] [Google Scholar]
- Hodges H. L.; Brown R. A.; Crooks J. A.; Weibel D. B.; Kiessling L. L. Imaging Mycobacterial Growth and Division with a Fluorogenic Probe. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 5271–5276. 10.1073/pnas.1720996115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamariza M.; Shieh P.; Ealand C. S.; Peters J. S.; Chu B.; Rodriguez-Rivera F. P.; Babu Sait M. R.; Treuren W. V.; Martinson N.; Kalscheuer R.; et al. Rapid Detection of Mycobacterium Tuberculosis in Sputum with a Solvatochromic Trehalose Probe. Sci. Transl. Med. 2018, 10, eaam6310 10.1126/scitranslmed.aam6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarpe V. A.; Kulkarni S. S. Synthesis of Maradolipid. J. Org. Chem. 2011, 76, 6866–6870. 10.1021/jo200979n. [DOI] [PubMed] [Google Scholar]
- Toubiana R.; Das B. C.; Defaye J.; Mompon B.; Toubiana M. J. Étude Du Cord-Factor et de Ses Analogues. Partie III. Synthèse Du Cord-Factor (6,6′-Di-O-Mycoloyl-α,α-Tréhalose) et Du 6,6′-Di-O-Palmitoyl-α,α-Tréhalose. Carbohydr. Res. 1975, 44, 308–312. 10.1016/S0008-6215(00)84175-0. [DOI] [PubMed] [Google Scholar]
- Datta A. K.; Takayama K.; Nashed M. A.; Anderson L. An Improved Synthesis of Trehalose 6-Mono and 6,6′-Dicorynomycolates and Related Esters. Carbohydr. Res. 1991, 218, 95–109. 10.1016/0008-6215(91)84089-W. [DOI] [PubMed] [Google Scholar]
- Ahibo-Coffy A.; Aurelle H.; Lacave C.; Prome J. C.; Puzo G.; Savagnac A. Isolation, Structural Studies and Chemical Synthesis of a “palmitone Lipid” from Corynebacterium diphtheriae. Chem. Phys. Lipids 1978, 22, 185–195. 10.1016/0009-3084(78)90025-7. [DOI] [PubMed] [Google Scholar]
- Narouz M. R.; Soliman S. E.; Bassily R. W.; El-Sokkary R. I.; Nasr A. Z.; Nashed M. Improved Synthesis of 6-Azido-6-Deoxy- and 6,6′-Diazido-Dideoxy- α,α-Trehaloses. Synlett 2013, 24, 2271–2273. 10.1055/s-0033-1339843. [DOI] [Google Scholar]
- Ballereau S.; Guédat P.; Poirier S. N.; Guillemette G.; Spiess B.; Schlewer G. Synthesis, Acid-Base Behavior, and Binding Properties of 6-Modified Myo- Inositol 1,4,5-Tris(Phosphate)S. J. Med. Chem. 1999, 42, 4824–4835. 10.1021/jm991084t. [DOI] [PubMed] [Google Scholar]
- Hanessian S.; Lavallee P. Synthesis of 6-Amino-6-Deoxy-α-α-Trehalose: A Positional Isomer of Trehalosamine. J. Antibiot. 1972, 25, 683–684. 10.7164/antibiotics.25.683. [DOI] [PubMed] [Google Scholar]
- Kollár J.; Hrdlovič P.; Chmela Š.; Sarakha M.; Guyot G. Synthesis and Transient Absorption Spectra of Derivatives of 1,8-Naphthalic Anhydrides and Naphthalimides Containing 2,2,6,6-Tetramethylpiperidine; Triplet Route of Deactivation. J. Photochem. Photobiol., A 2005, 170, 151–159. 10.1016/j.jphotochem.2004.07.021. [DOI] [Google Scholar]
- Heinkel F.; Abraham L.; Ko M.; Chao J.; Bach H.; Hui L. T.; Li H.; Zhu M.; Ling Y. M.; Rogalski J. C.; et al. Phase Separation and Clustering of an ABC Transporter in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 2019, 116, 16326–16331. 10.1073/pnas.1820683116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P.; Li W.; Wong D.; Xie J.; Av-Gay Y. Phosphorylation Control of Protein Tyrosine Phosphatase A Activity in Mycobacterium tuberculosis. FEBS Lett. 2015, 589, 326–331. 10.1016/j.febslet.2014.12.015. [DOI] [PubMed] [Google Scholar]
- Ghazaei C. Mycobacterium tuberculosis and Lipids: Insights into Molecular Mechanisms from Persistence to Virulence. J. Res. Med. Sci. 2018, 23, 63 10.4103/jrms.JRMS_904_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollinger J.; Kumar A.; Roberts D. M.; Bailey M. A.; Casey A.; Parish T. A High-Throughput Whole Cell Screen to Identify Inhibitors of Mycobacterium tuberculosis. PLoS One 2019, 14, e0205479 10.1371/journal.pone.0205479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A. J.; Ovechkina Y.; McGillivray A.; Flint L.; Roberts D. M.; Parish T. A High Content Microscopy Assay to Determine Drug Activity against Intracellular Mycobacterium Tuberculosis. Methods 2017, 127, 3–11. 10.1016/j.ymeth.2017.03.022. [DOI] [PubMed] [Google Scholar]
- Love M. S.; Beasley F. C.; Jumani R. S.; Wright T. M.; Chatterjee A. K.; Huston C. D.; Schultz P. G.; McNamara C. W. A High-Throughput Phenotypic Screen Identifies Clofazimine as a Potential Treatment for Cryptosporidiosis. PLoS Negl. Trop. Dis. 2017, 11, e0005373 10.1371/journal.pntd.0005373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier V.; Av-Gay Y. Mycobacterium Tuberculosis Modulators of the Macrophage’s Cellular Events. Microbes Infect. 2012, 14, 1211–1219. 10.1016/j.micinf.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Bach H.; Papavinasasundaram K. G.; Wong D.; Hmama Z.; Av-Gay Y. Mycobacterium Tuberculosis Virulence Is Mediated by PtpA Dephosphorylation of Human Vacuolar Protein Sorting 33B. Cell Host Microbe 2008, 3, 316–322. 10.1016/j.chom.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Wong D.; Bach H.; Sun J.; Hmama Z.; Av-Gay Y. Mycobacterium Tuberculosis Protein Tyrosine Phosphatase (PtpA) Excludes Host Vacuolar-H + -ATPase to Inhibit Phagosome Acidification. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 19371–19376. 10.1073/pnas.1109201108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J.; Kim R.; Woo M.; Jeong J.; Park D. E.; Kim G.; Delorme V. Efflux Attenuates the Antibacterial Activity of Q203 in Mycobacterium Tuberculosis. Antimicrob. Agents Chemother. 2017, 61, e02637 10.1128/AAC.02637-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardarov S.; Bardarov S.; Pavelka M. S.; Sambandamurthy V.; Larsen M.; Tufariello J. A.; Chan J.; Hatfull G.; Jacobs W. R. Specialized Transduction: An Efficient Method for Generating Marked and Unmarked Targeted Gene Disruptions in Mycobacterium Tuberculosis, M. bovis BCG and M. Smegmatis. Microbiology 2002, 148, 3007–3017. 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.