Abstract
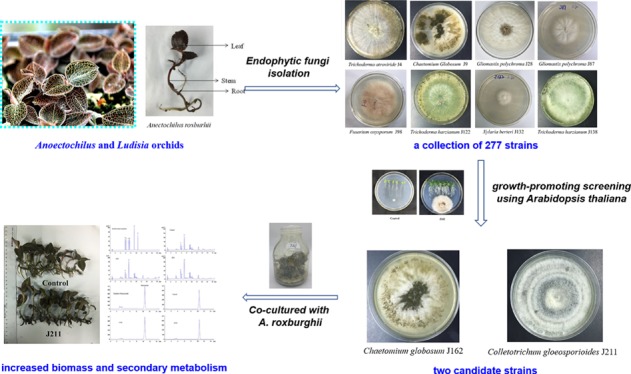
Endophytic fungi possess favorable effects on their host plants, including disease-resistance improvement, secondary metabolite induction, and growth promotion. It is therefore a promising and sustainable strategy to utilize endophytic fungi for the quality improvement of medicinal herbs or important crops. In our study, a collection of 277 strains of endophytic fungi were isolated from Anoectochilus and Ludisia orchids. Two strains J162 and J211 can be symbiotically cocultured with the tissue culture seedlings of Anoectochilus roxburghii, a popular medicinal and edible plant in southern China. Both strains can significantly enhance the biomass of A. roxburghii and induce the biosynthesis and accumulation of its active ingredients, including flavonoids, kinsenoside, and polysaccharides. J162 and J211 were further identified as Chaetomium globosum and Colletotrichum gloeosporioides based on multilocus phylogenetic analysis. Immunocytochemical staining indicated that J162 and J211 mainly colonized the intercellular gap of xylem parenchyma cells of A. roxburghii roots without obvious harm. In addition, quantitative real-time polymerase chain reaction showed that the expression of three growth-related genes, namely, uracil phosphoribosyl transferase, amino acid transmembrane transporter, and maturase K, were significantly altered in A. roxburghii plants when treated with J162 and J211. In conclusion, the two strains are highly beneficial microbial resources for the growth and accumulation of active ingredients of A. roxburghii in agricultural cultivation.
Introduction
There are more than 15 Anoectochilus species (Orchidaceae family), widely distributed in Fujian, Taiwan, Jiangxi, Yunnan, and Guangxi, from the south to the east and west of China.1,2Anoectochilus roxburghii (Wall.) Lindl. is the most well-known medicinal and edible species and the only officially designated origin of a valuable herb named “Jinxianlian” in the “Standards of Chinese Medicinal Materials of Fujian Province in China” (2006 Edition).3A. roxburghii, also known as “Jewel Orchid”, is a reputed folk medicine in southern China with high efficacy for the treatment of hypertension, diabetes, and liver diseases,3,4 which also has nourishing functions and widely used as tonic diet and tea in Fujian.5 Because of the lack of wild resources, many Anoectochilus, Goodyera, and Ludisia plants have also been used as substitutes of A. roxburghii or even adulterants in local herbal markets and inevitably confused with A. roxburghii because of their morphological similarities.6 Environmental deterioration and excessive human consumption aggravated the wild resource destruction of A. roxburghii, which subsequently led to a low yield and short supply. Thus, it is important to find some promising ways in facilitating plant growth and enhancing the yield of bioactive metabolites to improve the total quality of A. roxburghii.
Endophytes are microorganisms that inhabit living plants without causing apparent disease symptoms to the host. Previous studies have also shown that endophytic fungi could benefit their hosts in different ways, such as promoting germination and shoot growth and enhancing host plant tolerance to biotic or abiotic stresses, as well as inducing bioactive metabolite accumulation in host plants.7,8 In addition, endophytes were widely used in agricultural cultivation, such as Oryza sativa L. and Oncidium hybridum.9,10
Previous studies have revealed that orchid plants need to establish a symbiotic relationship with mycorrhizal fungi to maintain their normal growth and development.11 Two unidentified strains of mycorrhizal fungi, ASF-15 and ASF-18, were reported to significantly enhance the biomass of the tissue culture seedlings of A. roxburghii.12 Two strains of Epulorhiza mycorrhizal fungi, MF-15 and MF-18, were found to promote the growth and polysaccharide accumulation of A. roxburghii, while two strains of Mycena fungi, MF-23 and MF-24, only enhanced the content of polysaccharides in A. roxburghii.13 The abovementioned six strains of mycorrhizal fungi were not isolated from A. roxburghii but from other Orchidaceae plants. To date, most reported beneficial fungal strains for A. roxburghii are exogenous mycorrhizal fungi rather than endogenous strains, and little is known about the profiles and functions of the endophytes in Anoectochilus and Ludisia species. What is more, favorable evidence regarding the effects of endophytic fungi on the accumulation of secondary metabolites in A. roxburghii is still limited and their functional significances remain unclear. Therefore, in our study, the endophytic fungi harbored in healthy Anoectochilus and Ludisia plants were systematically isolated. All isolated strains were further screened for their ability to promote the growth of Arabidopsis thaliana and A. roxburghii. The effects of these bioactive strains on the accumulation of secondary metabolites in A. roxburghii were further studied, and the underlying mechanism was also investigated to better understand the role of endophytic fungi in host–microbe interactions.
Materials and Methods
Sample Collection
A total of ten batches of samples (including five Anoectochilus and one Ludisia species) were collected from Guangxi, Jiangxi, Yunnan, and Fujiang in China (Table S1). All samples were authenticated by Dr. Zehao Huang, School of Pharmacy, Fujian University of Traditional Chinese Medicine. All voucher specimens were deposited in the Herbarium of Fujian University of Traditional Chinese Medicine, Fuzhou, China. The collected plants were placed in plastic bags, transferred to the laboratory within 24 h, and stored at 4 °C until the fungal isolation procedure was carried out.
Isolation of Endophytic Fungi
Plant surface sterilization and fungal isolation were carried out according to the procedures described by Jia.14 Briefly, all plant samples were washed thoroughly with running tap water to remove soil particles and then rinsed several times with distilled water. The separated parts (roots, stems, leaves, and flowers; Figure S1) were successively immersed in 75% ethanol solution for 4 min, then in 2.5% sodium hypochlorite solution for 4 min, followed by immersion in 75% ethanol solution for 30 s. These surface-sterilized tissues were then rinsed four times with sterile distilled water and subsequently dried with sterile filter paper. Next, 0.5–1.0 cm pieces were cut from each tissue and evenly placed on Petri dishes containing potato dextrose agar (PDA) with 50 mg/L penicillin to inhibit bacterial growth. To validate the effectiveness of surface sterilization, an aliquot of 0.3 mL of the last rinsed water was also inoculated onto the PDA plates as a control. The Petri dishes were incubated at 26 ± 2 °C for 7–14 days, and fungal growth was monitored daily. Each colony was subcultured on new penicillin-free PDA medium for further purification. For long-term storage, the fungal colonies were stored at −80 °C in slant tubes containing PDA.
Initial Screening of Growth-Promoting Strains Using A. thaliana
The sterilization of A. thaliana seeds, which were obtained from Prof. Meili Guo, School of Pharmacy, Second Military Medical University, was carried out as follows. Briefly, A. thaliana seeds were immersed in 75% ethanol for 5 min and subsequently washed five times with sterile deionized water. The seeds were then inoculated onto the 1/2 Murashige and Skoog (MS) medium. An aliquot of 0.3 mL of the last rinsed water was inoculated onto the 1/2 MS medium as a control to validate the effectiveness of sterilization. After three days of seed vernalization at 4 °C, the plates were incubated in a plant growth chamber under controlled conditions (22 ± 2 °C, 16 h light/8 h dark cycle and a light intensity of 2000 lux) for germination and growth.
To estimate the effects of fungal inoculation on the plant growth of A. thaliana, a vertical agar plate (VAP) assay was performed with six plants per plate, and each experiment was performed in triplicate. On the 4th day after germination, A. thaliana seedlings were inoculated with a 0.5 cm square fungal plug punched from fungus-containing PDA or a sterile PDA piece as a control. The fungal or PDA plug was placed at a 5 cm distance from the primary root tip of A. thaliana. The plates were incubated vertically in the same plant growth chamber as previously mentioned. After ten days of coculture, the root microstructure was observed by using a biological microscope (Leica DM2500) at a magnification of 50×, and the fresh weight, number of lateral roots, and root length of A. thaliana seedlings were also calculated based on a manual measurement. Subsequently, A. thaliana seedlings were dried at 50 °C to a constant weight to determine the dry weight.
Identification of Growth-Promoting Strains in the Initial Screening
Pure fungal colonies were selected for DNA extraction. DNA was extracted from 100 mg mycelia chilled in liquid nitrogen by using a Hi-DNA Secure Plant Kit (TIANGEN Biotech, Beijing, China) according to the manufacturer’s protocol. The internal transcribed spacer (ITS) region and the β-tubulin (TUB2), elongation factor 1-α (EF-1α) were amplified with the primer pairs ITS4/ITS5, Bt2a/Bt2b, and EFa/EFb which were synthesized by Sangon Biotech (Shanghai, China). The polymerase chain reaction (PCR) was performed in a 50 μL reaction mixture containing 2 μL template DNA, 1 μL forward primer (10 μM), 1 μL reverse primer (10 μM), 25 μL 2× TransTap HiFi PCR SuperMix, and 21 μL double distilled water (Table S4). The PCR cycling protocol consisted of initial denaturation at 94 °C for 4 min, followed by 30 cycles at 94 °C for 50 s, 54 °C for 50 s, and 72 °C for 90 s, and a final elongation step at 72 °C for 7 min (Table S5). The PCR-amplified products without further purification were sequenced by Sangon Biotech (Shanghai, China).
For strain identification, combined with morphological observation, the ITS and the TUB2, EF-1α sequences were compared with those submitted in the NCBI database (http://www.ncbi.nlm.nih.gov/) using the BLAST search program. A phylogenetic tree was constructed according to the maximum parsimony method using MEGA version 5.0 software (http://www.megasoftware.net).15Postia caesia 12974 and Amesia nigricolor CBS 600.66 were used as outgroup sequences, respectively.
Immunocytochemical Staining of Fungal Infection in A. roxburghii
The immunocytochemical staining of the plant tissue was performed as described by Zhai.16 Briefly, a portion of the fresh root tissues of A. roxburghii were fixed in 4% paraformaldehyde for more than 24 h. Then, paraffin sections of 4 μm were prepared according to the following procedures. The slices were sequentially placed into dimethylbenzene xylene I for 20 min, dimethylbenzene xylene II for 20 min, anhydrous ethanol I for 10 min, anhydrous ethanol II for 10 min, 95% alcohol for 5 min, 90% alcohol for 5 min, 80% alcohol for 5 min, 70% alcohol for 5 min, and distilled water for rinsing. The plant tissue sections were then placed in a repair box filled with EDTA antigen repair buffer (pH 8.0) (Servicebio, Wuhan, China) for antigen repair in a microwave oven with medium boiling for 5 min. After natural cooling, the slice was placed into the PBS (pH 7.4) and washed for three times (5 min each time) on the decoloring table. After the slices were spin-dried, a circle was drawn on the center of the slice with a Pap Pen to prevent the antibody from running off the slide. Then, 3% BSA was dropped into the circle to cover the tissue evenly and hermetically stored the sample at room temperature for 30 min. After shaking off the blocking solution gently, the primary antibody [1:100 concanavalin A (GenBank ID, 72333; Sigma) labeled with FITC fluorescence] was added, and the slice was subsequently placed flat on the wet box and incubated at 4 °C for 24 h. Then, the slice was transferred into PBS for decoloration again. Subsequently, DAPI (Servicebio, Wuhan, China) was added to the slice and coincubated in the dark for 10 min at room temperature. Finally, the slice was decolored three times in PBS and sealed with Antifade Mounting Medium (Servicebio, Wuhan, China), and their images were collected by using a fluorescent inverted microscope (Nikon, Japan).
Ultra Performance Liquid Chromatography and High Performance Liquid Chromatography Analysis of Bioactive Constituents in A. roxburghii
The tissue culture plants of A. roxburghii were desiccated at 60 °C in the oven and then ground into a powder. A total of 0.5 g of plant powder was accurately weighed and placed in 50 mL centrifuge tubes. Subsequently, 25 mL of 70% methanol was added into the centrifugal tube, and the gross weight was recorded. After ultrasonic extraction for 45 min, the centrifuge tube was weighed again, and the weight loss was made up with methanol. The sample solution was filtered with a 0.22 μm syringe filter and subsequently used for ultra performance liquid chromatography (UPLC) analysis of flavonoid contents.
The UPLC analysis was performed on an Agilent-1290 UPLC system equipped with a Phenomenex Kinelex C18 100A chromatographic column (100 mm × 2.10 mm, 1.7 μm) at 35 °C. The mobile phase consisted of acetonitrile (A) and 0.1% formic acid in water (B), and a gradient program (Table S6) was used. The injection volume was 5 μL with a flow rate of 0.3 mL/min.
In HPLC–ELSD analysis of kinsenoside content, 0.5 g of plant powder was weighed and extracted with 100 mL methanol, and other procedures in sample preparation were the same as those described in UPLC analysis. The HPLC–ELSD system was equipped with a Phenomenex NH2 column (250 × 4.6 mm, 5 μm). An isocratic elution system of acetonitrile–water (92:8, v/v) was adopted as the mobile phase. The flow rate was 0.8 mL/min, the column temperature was 35 °C, the ELSD spray chamber temperature was 70 °C, and the nitrogen flow rate was 1.5 L/min.
Determination of Polysaccharide Content in A. roxburghii
The polysaccharide content in A. roxburghii was quantified by the phenol–sulfuric acid method. A standard curve was established as described by Chen et al.13 Briefly, 10 mg of glucose was accurately weighed and dissolved in 100 mL distilled water as the standard solution. Then, 0.0, 0.2, 0.4, 0.6, 0.8, 1.0, 1.2, 1.4, 1.6, 1.8, and 2.0 mL of standard solutions were drawn to prepare different concentrations of glucose solutions in volumetric flasks with a final volume of 2.0 mL. Subsequently, these solutions were transferred to test tubes, and 1.0 mL of 6% phenol solution was added to each tube. Then, the mixture was shaken after the quick addition of 5.0 mL of concentrated sulfuric acid. The solution was then allowed to stand for 20 min and subsequently react in a boiling water bath for 10 min, after which the solution was quickly removed into the iced water bath for 5 min and finally maintained at room temperature for 30 min. The absorbance of the mixture was determined at 490 nm in a 96-well plate using an absorbance microplate reader (BioTek Co., USA). All treatments were performed in triplicate.
A total of 120 mg of A. roxburghii powder was accurately weighed and extracted with 20 mL distilled water in an 85 °C water bath. After filtration, 1.0 mL filtrate was drawn, and 4.0 mL of anhydrous ethanol was added, which was then stored at 4 °C for 24 h. The precipitation of polysaccharide was collected and washed with a small amount of 80% ethanol and dissolved in 2.0 mL of distilled water. To determine the polysaccharide content of the tested samples, 1.0 mL of the sample solution from each group was drawn, and then the phenol solution and sulfuric acid were added as described before with a final volume of 2.0 mL. Standard solutions of glucose and distilled water were regarded as the reference and blank control, respectively. The results were calculated based on the calibration curve and expressed as μg/mL.
Determination of the Activities of Three Enzymes in A. roxburghii
Plant samples were homogenized in precooled mortars using extracting solution according to the protocol of the β-1,3-glucanase Activity Assay Kit (Solarbio, Beijing, China), the Chitinase Activity Detection Kit, and the T-SOD Activity Assay Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). After centrifugation (12,000g for 10 min, 12,000g for 20 min, and 4,000g for 10 min, for β-1,3-glucanase, chitinase, and T-SOD, respectively), the supernatants of the treated samples were collected and used as enzyme extracts.
The enzyme extract was mixed with the corresponding kit reagent to initiate the reaction, and then the absorptions of different samples were measured on a microplate reader (BioTek Co., USA). All assays were performed in triplicate. β-1,3-glucanase activity was assayed by measuring the rate of reducing sugar release from laminarin as a substrate at 540 nm, and the enzyme activity was expressed as U/g fresh weight. One unit of glucanase activity was defined as the amount of enzyme that released 1.0 μM reducing sugar equivalent, expressed as glucose, per hour. Chitinase activity was assayed by a modified method recorded at 570 nm following the kit instruction, and the enzyme activity was also expressed as U/g fresh weight. In addition, the activity of superoxide dismutase (SOD) was determined by measuring its ability to inhibit the photochemical reduction of NBT. One unit of SOD activity is the amount of enzyme that produces 50% inhibition on the rate of NBT reduction.
RNA Isolation and Real-Time Quantitative PCR Analysis
The total RNA of A. roxburghii from different groups was extracted using the TRIzol Total RNA Isolation Kit (Keygene Biotech, NanJing, China) according to the manufacturer’s instructions. RNA yield and quality were verified by an average optical density (OD) OD260/OD280 nm absorption ratio range from 1.8 to 2.0, with a BioMate 3S UV–Visible spectrophotometer (Thermo Fisher Scientific, USA). Once the integrity of the total RNA was detected to be good, the cDNA was then obtained by the PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian, China). All cDNA samples were stored at −20 °C before further analysis by quantitative real-time PCR (qRT-PCR). The reference gene was 18S rRNA, and the specific primers used in qRT-PCR are listed in Table S7. All primers were synthesized by Sangon Biotech (Shanghai, China). qRT-PCR was conducted on an ABI 7500 Real-time PCR System (Applied Biosystems, Foster City, CA, USA) using a Fast Start Universal SYBR Green Master Mix (Rox) (Roche, China), and the procedure was set as follows: initial degradation at 94 °C for 10 min, followed by 40 cycles at 94 °C for 15 s, 60 °C for 15 s, 72 °C for 30 s, and a final dissociation cycle for melting curve analysis at 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 30 s. Melting curves were generated for each reaction to ensure specific amplification. All reactions were performed in triplicate. We establish a standard curve and determine the PCR efficiency, a good efficiency was verified with a slope close to −3.32 (100% theoretical efficiency). Cycle threshold (CT) values were generated from the ABI PRISM 7500 SDS Software version 1.4 (Applied Biosystems, Foster City, CA, USA). The relative gene expression was normalized by 18S rRNA and quantified using the comparative CT method and the data were analyzed as follows
| 1 |
| 2 |
Equation 2 represents the full form of eq 1, which is used to compare the gene expression in two different samples (sample A: treated sample and sample B: untreated control), with the 18S rRNA gene as the internal control.
Statistical Analysis
All data were expressed as mean ± SD (standard deviation), and the error bars in the figures represent the standard deviation of biological triplicates. Significant differences between two groups were analyzed by one-way analysis of variance (ANOVA), based on by the Dunnett test in SPSS software. Differences were considered significant when p < 0.05.
Results
Effects of Endophytic Fungi on the Plant Growth of A. thaliana
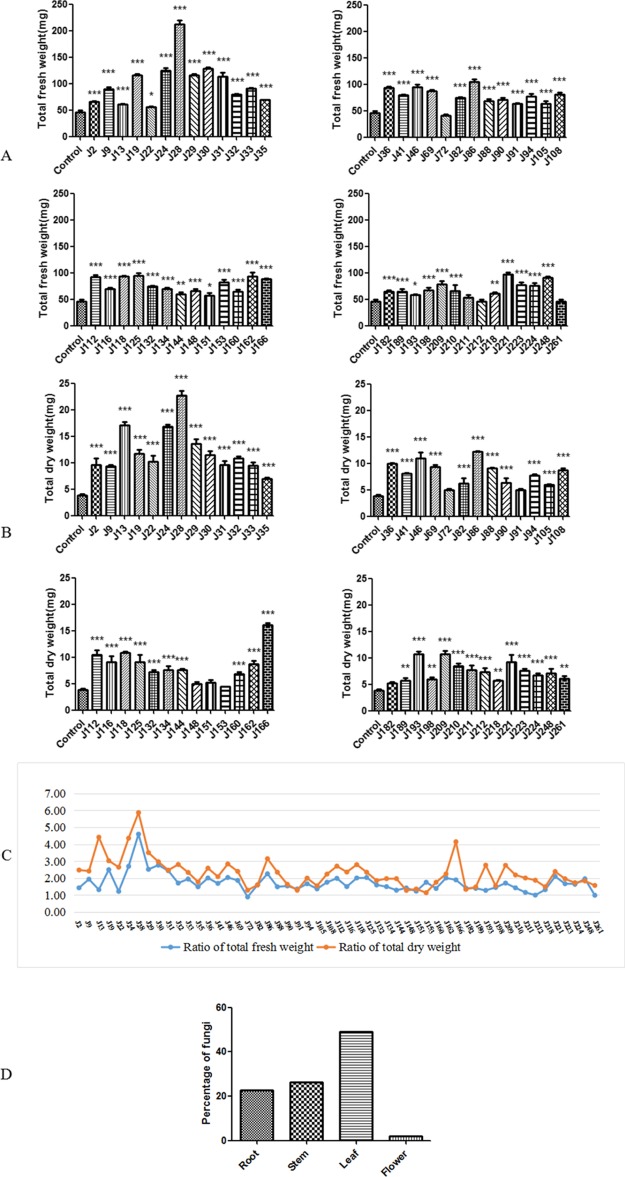
A total of 277 strains of endophytic fungi were isolated from ten batches of samples (including five Anoectochilus and one Ludisia species) that were collected from Guangxi, Jiangxi, Yunnan, and Fujiang in China (Table S1), with 112 strains isolated from leaves, 91 from stems, 72 from roots, and only two from flowers (Table S2) (Figure S1). To evaluate the growth-promoting effects of these isolated endophytes, A. thaliana seedlings were cocultured with individual fungi in VAPs. The total fresh weight and total dry weight of A. thaliana were measured after ten days of coculture with the tested fungus. Our results revealed that many isolated fungal strains significantly altered the growth phenotype of A. thaliana (Figure S2). Among the 277 strains, a total of 53 endophytes significantly enhanced the total fresh weight or total dry weight of A. thaliana (Figure 1), with the ratio of the total fresh weight between the fungus-treated group and the control group ranging from 0.89 (J72) to 4.60 (J28) (Figure 1A) and the ratio of the total dry weight between the fungus-treated group and the control group ranging from 1.14 (J153) to 5.87 (J28) (Figure 1B). The changes in the total dry weight after fungal inoculation were not completely consistent with those of the total fresh weight. Some strains did not show effects on the fresh weight but significantly enhanced the dry weight (Figure 1C). Based on further detailed analyses of the quantity of identified endophytes from different tissues, we found that most growth-promoting fungi were isolated from the leaves of these plants (Figure 1D).
Figure 1.
Biomass of A. thaliana seedlings at 10 days of coculture and the percentage of growth-promoting fungi from different tissues. The values are presented as the mean ± SD, n = 3, *P < 0.05; **P < 0.01; ***P < 0.001 vs the control group. [(A) Total fresh weight; (B) total dry weight; (C) comparison of total fresh weight and total dry weight between different groups; and (D) percentage of growth promoting fungi from different tissues]. (Ratio = the total fresh weight or total dry weight of the treatment group/control).
The root length and lateral root number of A. thaliana seedlings were also measured on the tenth day after coculture with each of the abovementioned 53 fungi. Our results showed that a total of 31 fungal strains inhibited the root length of A. thaliana seedlings, with the ratio between the fungal treatment group and the control group ranging from 0.54 (J261) to 0.99 (J69) (Figure S2A), whereas the other 22 strains increased the root length (Figure S2B) with the ratio between the fungal treatment group and the control group ranging from 1.04 (J9) to 1.64 (J198). Interestingly, as shown in Figures S2C and S3D, all 53 strains of growth promoting fungi significantly enhanced the lateral root germination of Arabidopsis seedlings. A total of 19 fungal strains notably increased the lateral root number, which was more than threefold that of the control group (Figure S2D). In addition, the microscopic characteristics of the Arabidopsis roots inoculated with the growth-promoting fungi were observed at a magnification of 50× using a biological microscope on the tenth day of cocultivation. All the strains with growth promoting effects significantly changed the root architecture of Arabidopsis seedlings. The microscopic characteristics of the root hair distribution on the root meristem of Arabidopsis seedlings are presented in Figure S2E. The dramatic changes in the seedlings inoculated with the growth promoting fungus (J162 and J211) showed evidently increased lateral root numbers compared to control seedlings. Furthermore, there was also a notable increase in the density of root hairs observed in J162-and J211-treated seedlings (Figure S2E), whereas the root hair of control seedlings was sparse. Our results indicated that these growth-promoting fungi mainly induced the differentiation of the root hair and promoted the development of lateral root formation in Arabidopsis, thus increasing nutrient uptake and plant biomass.
Identification and Analysis of Growth-Promoting Fungi for A. thaliana
Our results indicated that a total of 53 strains could be cocultivated long-term with A. thaliana, with promoted plant growth and enhanced biomass (Figures S2 and 1).
According to our detailed phylogenetic analysis (Table S3), except for three unknown strains, the other 50 strains were classified into two phyla (Basidiomycota and Ascomycota), eight orders (Polyporales, Auriculariales, Pleosporales, Glomerellales, Hypocreales, Xylariales, Diaporthales, and Sordariales) and 13 genera (Gliomastix, Bjerkandera, Auricularia, Helminthosporium, Colletotrichum, Acremonium, Bionectria, Fusarium, Hypoxylon, Xylariaceae, Diaporthe, Phomopsis, and Chaetomium). These fungi mainly belong to three genera, Gliomastix, Colletotrichum, and Fusarium, which occupied 22.6, 17.0, and 13.2% of all the growth promoting fungi, respectively.
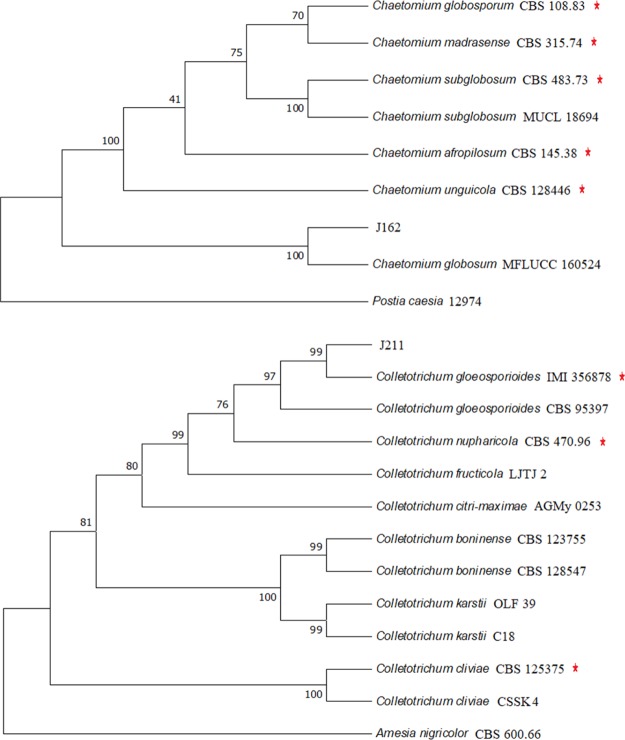
The phylogenetic tree (Figure 2) revealed that strain J162 was most closely related to Chaetomium globosum MFLUCC with 99.06% similarity and was therefore gathered into the same cluster. Thus, strain J162 was identified as C. globosum. In addition, strain J211 shared 99.38% similarity with Colletotrichum gloeosporioides IMI 356878, which suggested that endophyte J211 could be identified as a member of the species C. gloeosporioides.
Figure 2.
Phylogenetic trees showing the relationships of strains to closer species. The tree was based on maximum parsimony analysis of the ITS, EF-1α, and β-tubulin gene sequences of the endophytic fungi (Only 2 of 53 growth promoting fungi were shown). Bootstrap analysis was done with 1000 cycles. red star: Ex-type or ex-epitype. Postia caesia 12974 and Amesia nigricolor CBS 600.66 were used as outgroup sequences, respectively.
Effects of the Selected Fungi on Plant Growth and Secondary Metabolism in A. roxburghii
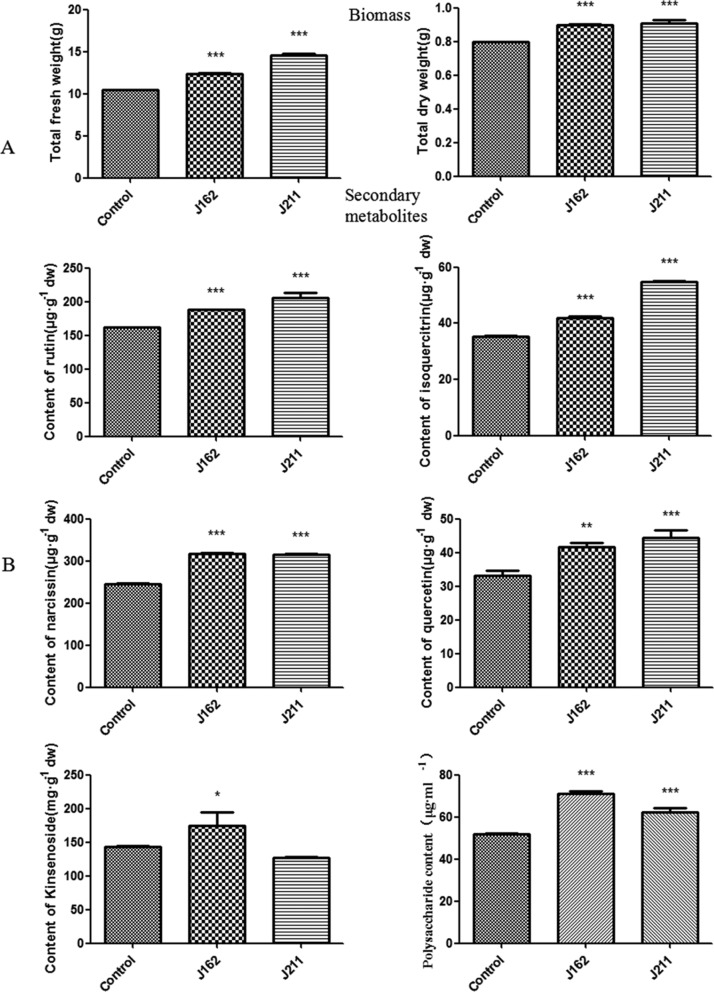
We further evaluated the effects of 53 candidate fungi on plant growth and secondary metabolism in A. roxburghii tissue culture seedlings. The biomass of A. roxburghii was determined after symbiosis with the tested fungi for 30 days. As a result, two strains, J162 and J211, showed not only long-term coculture with A. roxburghii without causing disease symptoms but also significantly enhanced its biomass. As shown in Figure 3A, both C. globosum J162 and C. gloeosporioides J211 improved the total fresh weight and dry weight of A. roxburghii plants, with total dry weights 12.50 and 13.75% higher than that of the control plants (Figure 3A), respectively.
Figure 3.
Effects of endophytic fungi J162 and J211 on the biomass (A) and secondary metabolism (B) in A. roxburghii seedlings (values are presented as means ± SD, n = 3, *P < 0.05; **P < 0.01; ***P < 0.001 vs the control group).
The contents of secondary metabolites (flavonoids, kinsenoside, and polysaccharides) in A. roxburghii after fungal infection were also determined. In our experiment, the accumulation of rutin in A. roxburghii was enhanced by both J162 and J211. The largest increase in rutin content was observed in the J211 group, which was 26.90% higher than that of the control samples (Figure 3B). Significant increases in the content of isoquercitrin were also observed in J211- and J162-treated plants, which reached 54.84 and 41.88 μg/g, respectively, compared to 35.34 μg/g in control plants (Figure 3B). These two endophytic fungi also improved the content of narcissin in A. roxburghii, which was 29.29 and 28.49% higher than that of the control samples, respectively. In addition, the content of quercetin reached the highest level in J211-treated plants, which was 1.33-fold of that in the control group. Our results showed that both J162 and J211 can be used as potential flavonoid-stimulating fungi for A. roxburghii.
As shown in Figure 3B, only J162 significantly stimulated the accumulation of kinsenoside in A. roxburghii with contents 21.35% higher than that in the control plants, while the content of kinsenoside in the J211 group was comparable with that in the control group. In addition, both J162 and J211 significantly enhanced the polysaccharide content, which was 37.16 and 20.37% higher than that in control plants, respectively. These data revealed that the two selected strains had promoted effects on secondary metabolite accumulation in A. roxburghii.
Immunocytochemical Staining of Fungal Infection in A. roxburghii
Observation under light microscopy showed that, after 30 days of coculture with the selected fungi, A. roxburghii seedlings were successfully infected with J162 and J211. As shown in Figure 4, both J162 and J211 were mainly located in the intercellular space or cell junction (red triangle), while a minority invaded the tissue cells of A. roxburghii seedlings. Therefore, the results suggested that these two fungi could noninvasively and stably interact with the host mainly in the intercellular space and thus form a nonaggressive symbiosis pattern with A. roxburghii seedlings.
Figure 4.
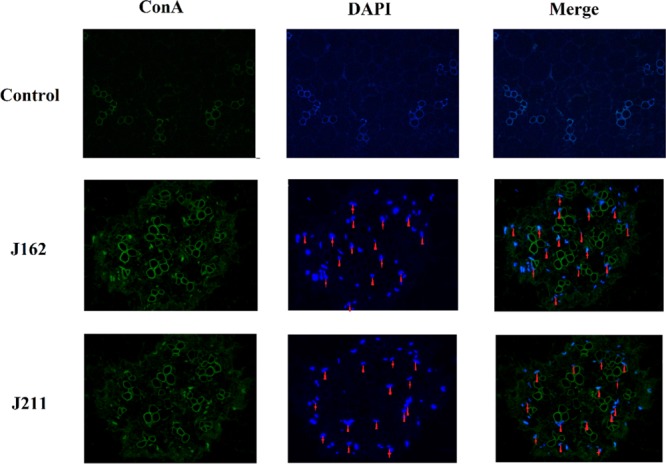
Transection of endophytic fungi J162 and J211 localization in the rhizome of A. roxburghii seedlings after immunocytochemical staining. Green: ConA-FITC; blue: DAPI. Red triangle indicates endophytic fungi J162 and J211 located in the rhizome intercellular space, while the red shuriken indicates endophytic fungi J162 and J211 located in root cells (magnification at 400×).
Effects of the Fungi on the Activities of Three Enzymes
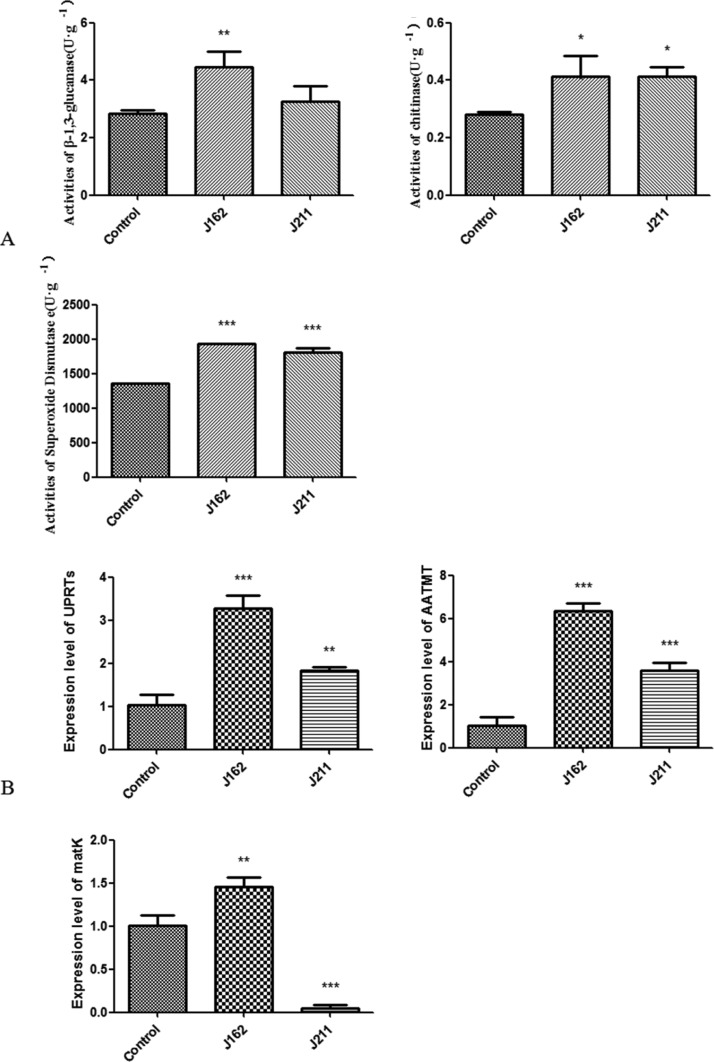
After 30 days of coculture, the activities of three enzymes, including β-1,3-glucanase, chitinase, and superoxide dismutase, in J162- and J211-treated plants were all significantly increased (Figure 5A). J162 strongly enhanced the activity of β-1,3-glucanase in A. roxburghii, which was 56.67% higher than that of the control plants. In addition, the activities of chitinase in J162- and J211-treated plants were approximately 0.50-fold higher than that of the control group. Moreover, the activities of superoxide dismutase were also highly increased in J162- and J211-inoculated plants, which were 42.09 and 33.14% higher than that in control samples, respectively. Our results showed that these two endophytes, particularly J162, could significantly improve the activities of favorable enzymes involved in plant growth.
Figure 5.
Effects of endophytic fungi J162 and J211 on the activities of three enzymes (A) and the expression of three growth-related genes (B) in A. roxburghii (UPRT, uracil phosphoribosyl transferase; AATMT, amino acid transmembrane transporter; and matK, maturase K. The values are presented as the mean ± SD, n = 3, *P < 0.05; **P < 0.01; ***P < 0.001 vs the control group).
Expression of Three Growth-Related Genes in A. roxburghii
Increased growth and secondary metabolism in plants are commonly associated with the expression levels of growth-related genes. To determine whether our fungi acted as inducers of plant growth in A. roxburghii, 18S rRNA gene was used as the reference gene, and the transcription levels of three growth-related genes in A. roxburghii were determined by real-time quantitative PCR.
As shown in Figure 5B, the expression of uracil phosphoribosyl transferase (UPRT) in A. roxburghii seedlings was strongly increased by both J162 and J211. The UPRT gene was upregulated by J162 to the highest level at 3.20-fold that of the control plants. In addition, the expression levels of the amino acid transmembrane transporter (AATMT) and maturase K (matK) in A. roxburghii were significantly enhanced by J162 to 6.10- and 1.45-fold of that in the control group, respectively. The administration of J211 also significantly increased the expression of AATMT in plants, which was 1.50-fold that of the control plants. However, matK was significantly downregulated by J211. These data indicated that both J162 and J211 could promote the plant growth of A. roxburghii by affecting the expression of growth-related genes but most likely in different ways.
Discussion
Endophytes have been reported as the promising and sustainable microbial resources with favorable plant growth promoting activity because they can serve their host effectively and efficiently under a wide range of environmental conditions.17 Plant growth-promoting (PGP) endophytes may become a biological alternative to traditional chemical fertilizers and be utilized to increase the yields of certain valuable crops or medicinal plants. These endophytes can be safely used as biofertilizers or disease-preventing agents because they do not exert ecological pressures and are environmentally friendly.18
In this study, we reported the isolation of 277 strains of endophytic fungi from different tissues of five Anoectochilus and one Ludisia plants collected from four provinces in China. The isolated strains were initially screened for their growth-promoting activity on A. thaliana seedlings. As a result, a total of 53 strains displayed significant promoting effects on the plant growth of A. thaliana and were identified by both morphological and molecular characterization, except for three unknown species. Our study revealed that three genera, Gliomastix, Colletotrichum, and Fusarium, were the dominant community members among the 53 strains of plant growth promoting fungi. Colletotrichum sp. and Fusarium sp. are common phytopathogens in many plants.19,20 In particular, Colletotrichum sp. are among the most important plant pathogens worldwide and are responsible for anthracnose, an economically important disease in a wide range of hosts, including cereals, legumes, vegetables, and tree fruits.21 However, in our study, several Colletotrichum sp. exhibited favorable effects on the growth of A. thaliana. A recent interesting study revealed that Colletotrichum tofieldiae, an endophytic fungus of the A. thaliana root, can transfer the macronutrient phosphorus to A. thaliana shoots, which confers plant fitness benefits upon an intact phosphate starvation response.22 This finding indicated that Colletotrichum sp. may also exert beneficial effects on their host under environmental stress, which merits further studies regarding the underlying actions and mechanisms.
Our results revealed that all 53 strains of endophytes significantly increased the number of lateral roots and enhanced the density of root hairs because of their possible effects on stimulating the germination of lateral roots and differentiation of root hairs. Therefore, we deduced that these endophytes could improve the capacity of A. thaliana seedlings in nutrient and water uptake by increasing the surface area of the plant root system, thus enhancing the plant biomass.
However, the in vitro detection of PGP traits of natural endophytes does not implicate a general and predictable improvement of the growth and health of all types of host plants in vivo.23 Therefore, further investigations were conducted to assess whether these PGP strains for Arabidopsis seedlings, could produce beneficial effects on A. roxburghii plant growth and secondary metabolism. In our study, only two strains, J162 and J211, could be long-term cocultured with A. roxburghii tissue culture seedlings without causing obvious disease symptoms and significantly increased the biomass of A. roxburghii. The increased plant biomass in the fungus-treated group may be attributed to the enhanced plant nutrient uptake of the host plant stimulated by these two fungi.24 Our findings were consistent with those of the previous reports on the promotion of plant growth by fungal endophytes.25,26
Flavonoids in A. roxburghii were reported to be responsible for its versatile pharmacological activities.27,28 In our experiment, the contents of four flavonoids were also enhanced by J162 and J211 inoculation compared with that of the control plants, which indicated that these endophytic fungi may activate the biosynthetic pathway of flavonoids in the whole A. roxburghii plant. Kinsenoside, a characteristic component of Anoectochilus plants, is one of the main active ingredients and is widely distributed in A. roxburghii, Anoectochilus formosanus, and Anoectochilus chapaensis, which is commonly used for the treatment of diabetes, hepatitis, liver disorders, osteoporosis, and hyperliposis.3−6 To the best of our knowledge, there are no reports of the promoting effects of fungal infection on the content of kinsenoside in A. roxburghii. Our study suggested that the content of kinsenoside in A. roxburghii seedlings was significantly increased by J162, and J211 did not affect the accumulation of kinsenoside. In addition, polysaccharide is also the main constituent of A. roxburghii, possessing antioxidant, hepatoprotective, immunostimulatory, and antitumor effects.5 Thus, we also investigated the changes in polysaccharide content after fungal treatments. As a result, both J162 and J211 promoted the accumulation of polysaccharides in A. roxburghii. Our data showed that these two fungi could notably activate the secondary metabolism in the whole A. roxburghii plants, especially J162, which possesses great potential in application and may lead to a practical breakthrough in the facilitation of A. roxburghii cultivation.
J162 and J211 were further molecularly identified as C. globosum and C. gloeosporioides, respectively. C. globosum has been recognized as an effective biocontrol fungus, especially in agriculture,29 which could enhance seedling tolerance to stress.30 Recently, we obtained the endophyte D38 from Salvia miltiorrhiza, also identified as C. globosum, which significantly promoted the accumulation of bioactive constituents and root production in S. miltiorrhiza.16 Therefore, we suggest that C. globosum may be a universal, beneficial and crucial endophytic fungus for the quality of many valuable crops and medicinal plants. However, Colletotrichum sp. is among the most important plant pathogens worldwide, causing anthracnose in a wide range of hosts.21 Our study revealed that J211, identified as C. gloeosporioides, could also produce favorable effects on A. roxburghii. This finding coincided with an interesting report published in Cell, suggesting that C. tofieldiae, an endophytic fungus of Arabidopsis root, confers plant fitness benefits upon an intact phosphate starvation response,22 which indicated that Colletotrichum sp. may also exert beneficial effects on their host under biotic and abiotic stresses.
Pathogens, as well as endophytic fungi, have evolved different strategies to overcome various barriers that they encounter during the infection of their hosts.31 A previous study reported that the fungal hyphae could directly enter root epidermal cells and produce intracellular hyphae, and the fungus establishes a biotrophic interaction with the living host cells and invades roots via junctions between epidermal cells to form intercellular hyphae. Furthermore, the fungi could enter the aboveground parts of the plant via the central cylinder.22,32 Our results showed that both the candidate fungi J162 and J211 mainly colonized the intercellular gap of the xylem parenchyma cells of A. roxburghii roots during long-term cocultivation, which suggested that these fungi may penetrate into tissue cells through the intercellular space and form a nonaggressive symbiosis pattern with the host. However, more precise investigations at the ultrastructural level are urgently needed in future studies.
Previous studies showed that stress-related enzymes, such as peroxidases, polyphenoloxides, and SOD, were induced and increased when the plants were subjected to pathogenic attack.33,34 Furthermore, chitinase and glucanase are also important enzymes for the catalytic hydrolysis of pathogenic fungus cell walls, which contribute to increasing the disease resistance of the host plant.35,36 Therefore, we hypothesized that beneficial fungi may induce stress-related enzymes to resist disadvantages and thus improve plant growth. In our study, the activities of β-1,3-glucanase, chitinase, and SOD were significantly increased in plants treated with the tested fungi J162 and J211, which indicated that these two strains enhanced the activities of stress-related enzymes to protect the plantlets and thus influence the growth of A. roxburghii.
Additionally, to explore the possible mechanism of the endophytic effects of J162 and J211 on plant growth and metabolism, the expression of three growth-related genes in A. roxburghii was further investigated. A previous study found that three clones were differentially expressed in A. roxburghii during symbiosis with a growth-promoting mycorrhizal fungus Epulorhiza sp., including two genes encoding UPRT and AATMT, respectively, and a matK pseudogene expressed only in seedlings after fungal treatment.37 UPRT is involved in salvaging pyrimidines by catalyzing the formation of uridine monophosphate from uracil and phosphoribosylpyrophosphate.38 Uracil salvage is of major importance for plant development,39 which, in our study, was markedly upregulated by J162 and J211 inoculation, and the expression of AATMT was also increased by both fungal treatments. Amino acids are known to play fundamental roles in multiple processes in plants, including hormone metabolism, cell growth, production of metabolic energy, nitrogen metabolism, and urea biosynthesis.40 Furthermore, matK was significantly increased in J162-inoculated plants but notably downregulated in J211-treated plants, thus indicating different mechanisms in the plant growth regulation of these two fungi. These data suggested that the endophytes J162 and J211 may regulate the gene expression of UPRT, AATMT, and matK to facilitate nutrient uptake, transportation, and utilization, thus promoting the plant growth and secondary metabolism of A. roxburghii. However, it is still unknown how fungi stimulate these genes through signal transduction, which requires further study to understand the detailed mechanism of action.
In conclusion, we isolated a collection of 277 strains of endophytic fungi from five Anoectochilus and one Ludisia species, among which C. globosum J162 and C. gloeosporioides J211 significantly promoted the growth and secondary metabolism of A. roxburghii. Our results suggest that endophytic fungi from the Anoectochilus and Ludisia species are highly beneficial microbial resources, especially J162 and J211, which can be utilized to improve the yield and quality of Chinese folk medicine “Jinxianlian” in agricultural cultivation.
Acknowledgments
This work was supported by the Program of Shanghai Health System Subject Chief Scientist (2017BR004), Shanghai Rising Star Program (18QA1405200), the National High Technology Research and Development Program of China (no. 2014AA 020508), the “Chen Guang” project supported by Shanghai Municipal Education Commission, the Shanghai Education Development Foundation (13CG40), the National Nature Science Foundation of China (nos. 81473301, 81603230, and 81773846), and the Program for Distinguished Young Research Talents in Fujian Province University.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b03789.
Other relevant data and detailed experimental procedure that supports the findings of this study (PDF)
Author Contributions
B.Y., Y.W., and X.Z. contributed equally to this work. C.Z. and T.H. conceived and designed the experiments; B.Y. and Y.W. performed the experiments; B.Y., X.Z., and C.Z. analyzed the data; B.Y. and C.Z. wrote the manuscript; and K.R. and L.Q. provided technical assistance to B.Y. and Y.W.
The authors declare no competing financial interest.
Supplementary Material
References
- Tian H.-Z.; Liu Q.-X.; Cheng Z.-Q.; Hu A.-Q.; Jiang H. Anoectochilus longilobus (Orchidoideae: Orchidaceae), a new species from Yunnan, China. Phytotaxa 2014, 164, 276–280. 10.11646/phytotaxa.164.4.6. [DOI] [Google Scholar]
- Qu X.-C.; Huang Y.-F.; Feng H.-Z.; Hu R.-C. Anoectochilus nandanensis, sp. nov. (Orchidaceae) from northern Guangxi, China. Nord. J. Bot. 2015, 33, 572–575. 10.1111/njb.00847. [DOI] [Google Scholar]
- Huang Y. L.Standards of Chinese Medicinal Materials in Fujian Province of China; Haifeng Press, 2006; p 154. [Google Scholar]
- Zeng B.; Su M.; Chen Q.; Chang Q.; Wang W.; Li H. Protective effect of a polysaccharide from Anoectochilus roxburghii against carbon tetrachloride-induced acute liver injury in mice. J. Ethnopharmacol. 2017, 200, 124–135. 10.1016/j.jep.2017.02.018. [DOI] [PubMed] [Google Scholar]
- Ye S. Y.A Preliminary Study on the Yield and Quality Control of Anoectochilus roxburghii; Zhejiang Agriculture & Forestry University, 2018. [Google Scholar]
- Yin Z. N.; Xu K. X.; Fan J. J.; Ma Z. Q.; Lin R. C. Research progress on chemical constituents of Anoectochilus and pharmacological activities. Global Tradit. Chin. Med. 2016, 9, 1153–1160. [Google Scholar]
- Johnson L. J.; de Bonth A. C. M.; Briggs L. R.; Caradus J. R.; Finch S. C.; Fleetwood D. J.; Fletcher L. R.; Hume D. E.; Johnson R. D.; Popay A. J.; Tapper B. A.; Simpson W. R.; Voisey C. R.; Card S. D. The exploitation of epichloae endophytes for agricultural benefit. Fungal Divers. 2013, 60, 171–188. 10.1007/s13225-013-0239-4. [DOI] [Google Scholar]
- Mei C.; Flinn B. The use of beneficial microbial endophytes for plant biomass and stress tolerance improvement. Recent Pat. Biotechnol. 2010, 4, 81–95. 10.2174/187220810790069523. [DOI] [PubMed] [Google Scholar]
- Li X.; Wang C.; Ren C. G.; Dai C. C. Effect of endophytic fungus B3 and different amounts of nitrogen applied on growth and yield in rice (Oryza sativa L.). Jiangsu J. Agric. Sci. 2009, 25, 1207–1212. [Google Scholar]
- Jin Y. L.; Song X. Q.; Lin M. G.; Hu M. J.; Yang F. S. Effects of endophytic fungi on development of dancing-lady-orchid (Oncidium) seedling at S2 stage in protected cultivation. Guangdong Agric. Sci. 2012, 15, 28–32. [Google Scholar]
- Zhou D. P.; Wu S. H.; Jiang Z. F.; Zhang X. The function and application prospect of endophytic fungi in Orchidaceae. J. Shanghai Agric. Sci. 2005, 21, 110–113. [Google Scholar]
- Wang Y. J.; Meng Z. X.; Yu X. M.; Wang C. L.; Guo S. X. Screening of Endophytic Fungi Promoting and Development of Anoectochilus roxburghii. J. Chin. Pharm. Sci. 2009, 44, 976–979. [Google Scholar]
- Chen X. M.; Guo S. X.; Wang C. L. Effects of four endophytic fungi on the growth and polysaccharide content of Anectochilus roxburhii (Wall.) Lindl. Chin. Pharm. J. 2005, 40, 13–16. [Google Scholar]
- Jia M.Biodiversity of Endophytic Fungi from Adlay (Coix lacrymajobi L.var. mayuen.) and their Correlations with the Crud Drug Quality of the Host Plants; The Second Military Medical University, 2014. [Google Scholar]
- Tamura K.; Peterson D.; Peterson N.; Stecher G.; Nei M.; Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai X.; Luo D.; Li X. Q.; Han T.; Jia M.; Kong Z. Y.; Ji J. C.; Rahman K.; Qin L. P.; Zheng C. J. Endophyte Chaetomium globosum D38 Promotes Bioactive Constituents Accumulation and Root Production in Salvia miltiorrhiza. Front. Microbiol. 2018, 8, 2694. 10.3389/fmicb.2017.02694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compant S.; Clément C.; Sessitsch A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. 10.1016/j.soilbio.2009.11.024. [DOI] [Google Scholar]
- Pavlo A.; Leonid O.; Iryna Z.; Natalia K.; Maria P. A. Endophytic bacteria enhancing growth and disease resistance of potato. Biolcontrol 2011, 56, 43–49. 10.1016/j.biocontrol.2010.09.014. [DOI] [Google Scholar]
- Than P. P.; Jeewon R.; Hyde K. D.; Pongsupasamit S.; Mongkolporn O.; Taylor P. W. J. Characterization and pathogenicity of Colletotrichum species associated with anthracnose on chilli (Capsicum spp.) in Thailand. Plant Pathol. 2008, 57, 562–572. 10.1111/j.1365-3059.2007.01782.x. [DOI] [Google Scholar]
- Munawar M.; Khan S. A.; Javed N.; Ul Haq I.; Gondal A. S. Biological control of wilt disease complex on tomato crop caused by Meloidogyne javanica and Fusarium oxysporum f. sp. lycopersici by Verticillium leptobactrum. Nematology 2015, 17, 479–485. 10.1163/15685411-00002882. [DOI] [PubMed] [Google Scholar]
- Bailey J. A.; Jeger M. J. Colletotrichum: Biology, Pathology and Control. Mycologica 1993, 85, 879. 10.2307/3760628. [DOI] [Google Scholar]
- Hiruma K.; Gerlach N.; Sacristán S.; Nakano R. T.; Hacquard S.; Kracher B.; Neumann U.; Ramírez D.; Bucher M.; O’Connell R. J.; Schulze-Lefert P. Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 2016, 165, 464–474. 10.1016/j.cell.2016.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H. H.; Schmidt D. D.; Baldwin I. T. Native bacterial endophytes promote host growth in a species-specifc manner; phytohormone manipulations do not result in common growth responses. PLoS One 2008, 3, e2702 10.1371/journal.pone.0002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukumarasamy R.; Revathi G.; Loganathan P. Effect of inorganic N on the population in vitro colonization and morphology of Acetobacter diazotrophicus (syn.Gluconacetobacter diazotrophicus). Plant Soil 2002, 243, 91–102. 10.1023/a:1019963928947. [DOI] [Google Scholar]
- Waqas M.; Khan A. L.; Hamayun M.; Shahzad R.; Kang S.-M.; Kim J.-G.; Lee I.-J. Endophytic fungi promote plant growth and mitigate the adverse effects of stem rot: an example of Penicillium citrinum and Aspergillus terreus. J. Plant Interact. 2015, 10, 280–287. 10.1080/17429145.2015.1079743. [DOI] [Google Scholar]
- Hamayun M.; Khan S. A.; Iqbal I.; Ahmad B.; Lee I.-J. Isolation of a gibberellin-producing fungus (Penicillium sp. MH7) and growth promotion of crown daisy (Chrysanthemum coronarium). J. Microbiol. Biotechnol. 2010, 20, 202–207. 10.4014/jmb.0905.05040. [DOI] [PubMed] [Google Scholar]
- He C.-N.; Wang C.-L.; Guo S.-X.; Yang J.-S.; Xiao P.-G. A novel flavonoid glucoside from Anoectochilus roxburghii (Wall.) Lindl. J. Integr. Plant Biol. 2006, 48, 359–363. 10.1111/j.1744-7909.2006.00179.x. [DOI] [Google Scholar]
- Wu Y.; Zhang C.; Zhang X. C.; Wu J. G.; Yi J.; Wu J. Z.; Zheng C. J. Determination of the total flavonoids content of Jinxianlian from different sources and its in vitro antioxidant and hypoglycemic activities. Pharm. Care Res. 2017, 17, 206–209. 10.5428/pcar20170314. [DOI] [Google Scholar]
- Reissinger A.; Winter S.; Steckelbroeck S.; Hartung W.; Sikora R. A. Infection of barley roots by Chaetomium globosum: evidence for a protective role of the exodermis. Mycol. Res. 2003, 107, 1094–1102. 10.1017/s0953756203008189. [DOI] [PubMed] [Google Scholar]
- Abou Alhamed M. F.; Shebany Y. M. Endophytic Chaetomium globosum enhances maize seedling copper stress tolerance. Plant Biol. 2012, 14, 859–863. 10.1111/j.1438-8677.2012.00608.x. [DOI] [PubMed] [Google Scholar]
- Mendgen K.; Hahn M.; Deising H. Morphogenesis and mechanisms of penetration by plant pathogenic fungi. Annu. Rev. Phytopathol. 1996, 34, 367–386. 10.1146/annurev.phyto.34.1.367. [DOI] [PubMed] [Google Scholar]
- Sukno S. A.; Garcia V. M.; Shaw B. D.; Thon M. R. Root infection and systemic colonization of maize by Colletotrichum graminicola. Appl. Environ. Microbiol. 2008, 74, 823–832. 10.1128/aem.01165-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H.-c.; Zhao H.; Wang B.-c.; Wang J.-b. Effect of local stress induction on resistance-related enzymes in cucumber seeding. Colloids Surf., B 2005, 43, 37–42. 10.1016/j.colsurfb.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Gechev T.; Willekens H.; Van Montagu M.; Inzé D.; Van Camp W. i. m.; Toneva V.; Minkov I. Different responses of tobacco antioxidant enzymes to light and chilling stress. J. Plant Physiol. 2003, 160, 509–515. 10.1078/0176-1617-00753. [DOI] [PubMed] [Google Scholar]
- Wang J.; Cao S.; Wang L.; Wang X. L.; Jin P.; Zheng Y. H. Effect of β-aminobutyric acid on disease resistance against Rhizopus rot in harvested peaches. Front. Microbiol. 2018, 9, 1505. 10.3389/fmicb.2018.01505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas-Gaudot E.; Slezack S.; Dassi B.; Pozo M. J.; Gianinazzi-Pearson V.; Gianinazzi S. Plant hydrolytic enzymes (chitinases and β-l,3-glucanases) in root reactions to pathogenic and symbiotic microorganisms. Plant Soil 1996, 185, 211–221. 10.1007/bf02257526. [DOI] [Google Scholar]
- Li B.; Tang M.; Tang K.; Zhao L.; Guo S. Screening for differentially expressed genes in Anoectochilus roxburghii (Orchidaceae) during symbiosis with the mycorrhizal fungus Epulorhiza sp. Life Sci. 2012, 55, 164–171. 10.1007/s11427-012-4284-0. [DOI] [PubMed] [Google Scholar]
- Islam M. R.; Kim H.; Kang S.-W.; Kim J.-S.; Jeong Y.-M.; Hwang H.-J.; Lee S.-Y.; Woo J.-C.; Kim S.-G. Functional characterization of a gene encoding a dual domain for uridine kinase and uracil phosphoribosyltransferase in Arabidopsis thaliana. Plant Mol. Biol. 2007, 63, 465–477. 10.1007/s11103-006-9101-3. [DOI] [PubMed] [Google Scholar]
- Mainguet S. E.; Gakière B.; Majira A.; Pelletier S.; Bringel F.; Guérard F.; Caboche M.; Berthomé R.; Renou J. P. Uracil salvage is necessary for early Arabidopsis development. Plant J. 2009, 60, 280–291. 10.1111/j.1365-313x.2009.03963.x. [DOI] [PubMed] [Google Scholar]
- Wipf D.; Ludewig U.; Tegeder M.; Rentsch D.; Koch W.; Frommer W. B. Conservation of amino acid transporters in fungi, plants and animals. Trends Biochem. Sci. 2002, 27, 139–147. 10.1016/s0968-0004(01)02054-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.