Short abstract
Objective
To perform molecular diagnosis and genetic counseling in a young Chinese couple with congenital hearing loss.
Methods
Variant screening analysis was performed by PCR and direct Sanger sequencing or targeted next-generation sequencing of all known hearing loss genes. Novel variants were evaluated by PolyPhen2 and PROVEAN software tools to evaluate possible effects on protein function.
Results
We identified causative variants in the young couple: c.235delC (rs80338943)/c.299-300delAT (rs111033204) compound heterozygous variants of GJB2 in the husband and c.1828G>A (p.Glu610Lys, rs535637788)/c.2825-2827delAGA compound heterozygous variants of LOXHD1 in the wife. The LOXHD1 c.1828G>A variant has only previously been reported in a Mexican-American individual in the 1000 Genomes Project database. Using PolyPhen2 and PROVEAN, we speculated that the LOXHD1 variant c.1828G>A is potentially pathogenic.
Conclusion
We carried out molecular diagnosis in a young couple with congenital hearing loss, and identified different disease-causing genes in the two individuals. The LOXHD1 variant c.1828G>A present in the wife had not previously been reported in individuals with congenital hearing loss. We determined this to be a potential pathogenic variant, and a novel variant associated with hearing loss in a Chinese individual.
Keywords: Hearing loss, LOXHD1, DFNB77, Sanger sequencing, next-generation sequencing, genetic counseling
Introduction
Hearing loss (HL) is the most frequent sensory deficit in humans, with a prevalence of around 1/1000 in newborns.1,2 Approximately 50% to 60% of hearing loss cases are caused by genetic factors.3 The genetic mode of HL inheritance can be autosomal recessive, autosomal dominant, mitochondrial, or X/Y-linked. To date, 121 genes have been reported to be associated with hearing loss (http://hereditaryhearingloss.org/): 45 are autosomal dominant genes,71 are autosomal recessive, and 5 are X-linked. However, most of these genes have only been reported in one or a few families.4 Epidemiological studies showed that variants in GJB2, SLC26A4, and 12S rRNA genes are highly correlated with hereditary HL.2 The most frequent genetic cause of HL is variants in GJB2, and most of these cases occur with non-progressive HL. Variants in SLC26A4, CDH23, and MYO3A were also shown to be associated with naturally occurring progressive HL.5,6
The genetic diagnosis of HL is very important because the findings can be used to aid treatment decisions, and provide prognostic information and genetic counseling for the patient’s family.7 Here, we describe a young couple with HL in whom the husband carried compound heterozygous variants of GJB2, and the wife had an extremely rare form of deafness and compound heterozygous variants of LOXHD1. We provided genetic counseling for this couple and followed them up during their pregnancy.
Patients and methods
Study population
We recruited a young Chinese couple (husband: 27 years old; wife: 25 years old) with congenital HL and 100 healthy controls (aged 25–30 years) from Gansu Provincial Maternal and Child Health Care Hospital. The couple had been married for 6 months and requested pre-pregnancy genetic counseling. The study was in accordance with the tenets of the Declaration of Helsinki and was approved by the Ethics Committee of Gansu Provincial Maternal and Child Health Care Hospital. Written informed consent was obtained from all participants.
Sample collection and genomic DNA preparation
Blood samples (2–3 mL) were collected from the probands and their parents and control individuals. Genomic DNA was extracted using a Tiangen DNA extraction kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions and quantified spectrophotometrically.
Targeted next-generation sequencing (NGS) and Sanger sequencing
First, the coding exon and flanking sequences of GJB2 were screened by PCR and direct sequencing using primers and conditions described in Table 1. If no GJB2 variant was found, targeted capture of candidate disease genes (n = 165, Table 2) was performed using a GenCap custom enrichment kit (MyGenostics, Beijing, China). Briefly, 1 μg of DNA library was mixed with BL buffer and a GenCap hypercholesterolemia probe (MyGenostics) and heated in a PCR cycler at 95°C for 7 minutes then 65°C for 2 minutes. A total of 23 μL HY buffer (pre-warmed to 65°C; MyGenostics) was added and the mixture was incubated at 65°C for 22 hours for hybridization. MyOne beads (50 μL; Thermo Fisher Scientific Inc., Rockford, IL, USA) were washed three times in 500 μL binding buffer (1×) and re-suspended in 80 μL binding buffer (1×). Next, 64 μL binding buffer (2×) was added and the mixture was transferred into a tube containing 80 μL MyOne beads, and spun for 1 hour on a rotator. The beads were then washed once with WB1 buffer at room temperature for 15 minutes and three times with WB3 buffer at 65°C for 15 minutes. Elution buffer was used to elute the bound DNA, which was amplified as follows: 98°C for 30 seconds then 15 cycles of 98°C for 25 seconds, 65°C for 30 seconds, and 72°C for 30 seconds, followed by 72°C for 5 minutes. PCR products were purified using SPRI beads (Beckman Coulter Inc., Brea, CA, USA) following the manufacturer’s protocol. Enrichment libraries were sequenced on an Illumina HiSeq 2000 sequencer (Illumina, San Diego, CA, USA) for 100-bp paired reads.
Table 1.
Primers and PCR conditions for GJB2 and LOXHD1.
| Primer name | Sequence (5′–3′) | Product size (bp) | Amplification reaction conditions |
|---|---|---|---|
| GJB2-F | CATGCTTGCTTACCCAGACTCA | 873 | 95°C for 5 minutes, then 20 cycles of 94°C for 30 seconds, 62°C for 45 seconds, and 72°C for 1 minute95°C for 5 minutes, then 15 cycles of 94°C for 30 seconds, 58°C for 45 seconds, and 72°C for 1 minute. |
| GJB2-R | TAGCGACTGAGCCTTGACAGC | ||
| GJB2-S1 | TGGGTTTTGATCTCCTCGATG | Sequencing primers | |
| GJB2-S2 | GCCTACCGGAGACATGAGAAG | ||
| LOXHD1-E14F | GGTAGTAGGGCTGGGTCTTCC | 355 | |
| LOXHD1-E14R | AGTTGCCTAACCCATCAGCTC | ||
| LOXHD1-E19F | CACCAACTCCACGACAAGTTC | 594 | |
| LOXHD1-E19R | GAGGTGGTGGAAGGATCTGAG |
GJB2-S1 and GJB2-S2 are the sequencing primers for GJB2.
Table 2.
Genes in the hearing loss panel.
| Nuclear genes associated with hereditary HL | ||||||
| ACTG1 | ADGRV1 | ALX3 | BSND | CABP2 | CCDC50 | CDH23 |
| CEACAM16 | CHD7 | CIB2 | CLDN14 | CLPP | CLRN1 | COCH |
| COL11A1 | COL11A2 | COL1A1 | COL1A2 | COL2A1 | COL4A3 | COL4A4 |
| COL4A5 | COL4A6 | COL9A1 | COL9A2 | CRYM | DFNB59 | DIABLO |
| DIAPH1 | DIAPH3 | DSPP | ECM1 | EDN3 | EDNRB | ELMOD3 |
| ESPN | ESRRB | EYA1 | EYA4 | FGF3 | FGF8 | FGFR1 |
| FGFR3 | FLNA | FOXI1 | FREM1 | FXN | GATA3 | GIPC3 |
| GJB1 | GJB2 | GJB3 | GJB6 | GLYAT | GPSM2 | GRHL2 |
| GRXCR1 | GSDME | HARS | HARS2 | HGF | HMX1 | HOXA2 |
| HSD17B4 | IL13 | ILDR1 | KARS | KCNE1 | KCNJ10 | KCNQ1 |
| KCNQ4 | KITLG | KRT9 | LAMA3 | LARS2 | LHFPL5 | LOXHD1 |
| LRTOMT | MARVELD2 | MIR96 | MITF | MPZ | MSRB3 | MYH14 |
| MYH9 | MYO15A | MYO1A | MYO1E | MYO3A | MYO6 | MYO7A |
| NDP | NDRG1 | NEFL | NELL2 | NF2 | OPA1 | OTOA |
| OTOF | OTOG | OTOGL | P2RX2 | PABPN1 | PAX3 | PCDH15 |
| PCDH9 | PDZD7 | PMP22 | PNPT1 | POLR1C | POLR1D | POU3F4 |
| POU4F3 | PROK2 | PROKR2 | PRPS1 | PTPN11 | PTPRQ | PTPRR |
| RDX | RPGR | SALL1 | SALL4 | SEC23A | SEMA3E | SERPINB6 |
| SIX1 | SIX5 | SLC17A8 | SLC19A2 | SLC26A4 | SLC26A5 | SMAD4 |
| SMPX | SNAI2 | SOX10 | STRC | TBC1D24 | TCIRG1 | TCOF1 |
| TECTA | TIMM8A | TJP2 | TMC1 | TMEM126A | TMIE | TMPRSS3 |
| TMPRSS4 | TNC | TPRN | TRIOBP | TRMU | TSPEAR | TYR |
| USH1C | USH1G | USH2A | WFS1 | WHRN | ||
| Mitochondrial gene | ||||||
| MT-RNR1 | RNR-TL1 | MT-CO1 | RNR-TS1 | MT-TK | RNR-TE | |
After sequencing, high-quality reads were retrieved by filtering out adaptors, low-quality reads, and short sequences (<40 bp). Data quality control standards were: 10× > 95%, depth = 200 ± 30. The SOAPaligner program (SOAP v2.21) was used to align clean read sequences to the human reference genome (UCSC Genome Browser hg19). After removing duplicates with Picard software (v1.119), single nucleotide polymorphisms (SNPs) were identified using SOAPsnp v1.03. Subsequently, reads were realigned to the reference genome using the Burrows–Wheeler alignment program (0.7.12-r1044), and insertions or deletions (InDels) were detected by the HaplotypeCaller of GATK software (https://software.broadinstitute.org/gatk/, GATK-3.5) and filtered by VariantFiltration of GATK software. We annotated the identified SNPs and InDels using the Exome-assistant program. Short read alignment and candidate SNP and InDel validation were performed using MagicViewer.
We performed Sanger sequencing for all identified variants in the probands and their parents. PCR primers for Sanger sequencing were designed by Primer 3.0 software (http://bioinfo.ut.ee/primer3-0.4.0/). Primers and PCR conditions for GJB2 and LOXHD1 are shown in Table 1. DNA sequencing was performed on an ABI 3500DX Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Bioinformatics analysis
If a novel variant was found that was not reported in the Human Gene Variant Database (http://www.hgmd.cf.ac.uk/) or ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/), we used PolyPhen2 (http://genetics.bwh.harvard.edu/pph2) and PROVEAN (http://provean.jcvi.org/index.php) tools to predict its possible functional role. To exclude the possibility that the variant was a polymorphism, we also performed direct sequencing in 100 healthy controls.
Results
Variant analysis
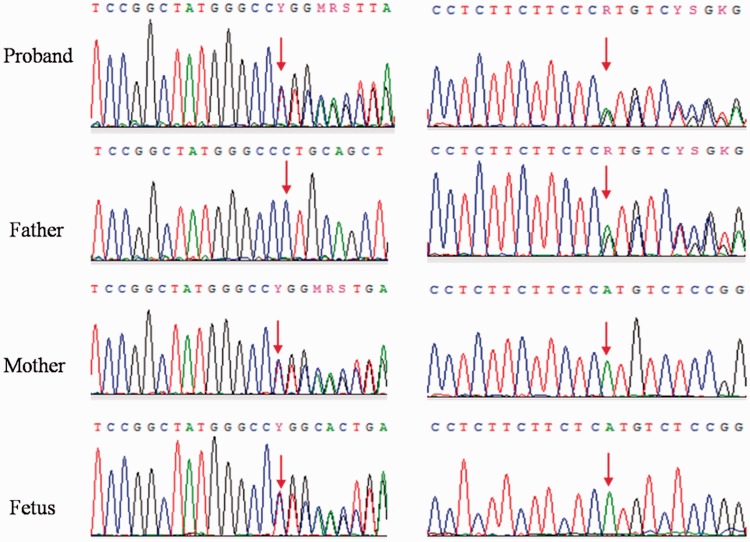
PCR and direct Sanger sequencing identified c.235delC (rs80338943)/c.299-300delAT (rs111033204) compound heterozygous variants of GJB2 in the husband. c.235delC was inherited from his mother and c.299-300delAT was inherited from his father (Figure 1).
Figure 1.
Results of Sanger sequencing. Compound heterozygous variants were detected in the proband.
c.235delC (rs80338943, left)/c.299-300delAT (rs111033204, right) of GJB2. The father carried the heterozygous variant c.299-300delAT, while the mother and fetus carried the heterozygous variant c.235delC.
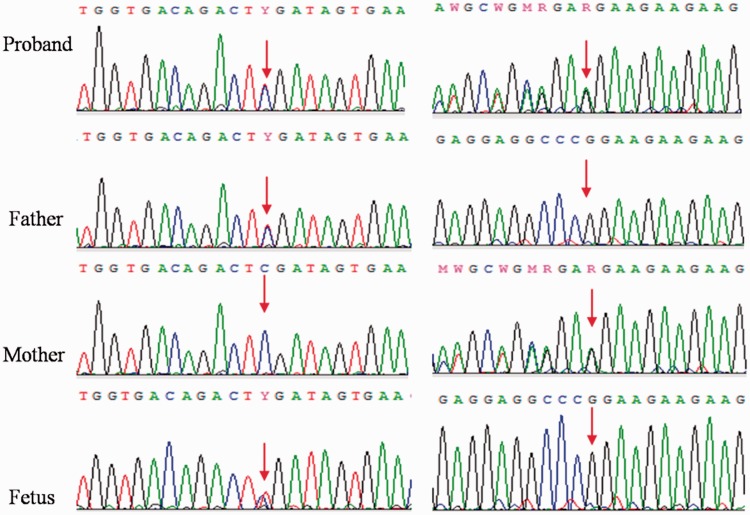
No GJB2 variants were identified in the wife, so targeted NGS was used to search for potential pathogenic variants. She was shown to carry c.1828G>A (p.Glu610Lys, rs535637788)/c.2825-2827delAGA compound heterozygous variants of LOXHD1, with c.2825-2827delAGA inherited from her mother and c.1828G>A from her father (Figure 2). Her hearing loss is an extremely rare form known as DFNB77 (OMIM: 613079). Variant c.2825-2827delAGA has previously been reported to be associated with DFNB77,7 but variant c.1828G>A (p.E610K) was only reported in a Mexican-American individual in the 1000 Genomes database. It has not been reported to be associated with DFNB77. PCR and direct Sanger sequencing did not identify this variant in any of our 100 healthy controls.
Figure 2.
Results of Sanger sequencing. Compound heterozygous variants were detected in the proband.
c.1828G>A(p.Glu610Lys, rs535637788, left)/c.2825-2827delAGA (right) of LOXHD1. The father and fetus carried the heterozygous variant c.1828G>A, while the mother carried the heterozygous variant c.2825-2827delAGA.
Bioinformatics analysis score
PolyPhen2 and PROVEAN tools were used to evaluate the possible functional role of variant c.1828G>A. PolyPhen2 gave a score of 1, suggesting that the site might be a damaging variation. The PROVEAN score was –3.203, and the site was considered “deleterious”.
Pregnancy outcome
Following molecular analysis, we provided genetic counseling to the young couple with HL. We explained that their children were unlikely to have HL because they both carried different genetic variants. During their pregnancy, they underwent regular prenatal checkups which detected the presence of the c.235delC GJB2 variant and the c.1828G>A LOXHD1 variant in the fetus (Figures 1 and 2). The baby was born in June 2018, and both ears passed the hearing screening test.
Discussion
We identified causative variants of HL in both individuals of a young Chinese couple. The variants of the husband were common and the c.2825-2827delAGA LOXHD1 variant of the wife was previously associated with DFNB77; however, the c.1828G>A LOXHD1 variant of the wife has only been reported in a Mexican-American individual in the 1000 Genomes database, and not in the HGMD database or elsewhere. PolyPhen2 and PROVEAN tools suggested that it is a likely pathogenic variant.
LOXHD1 is located on chromosome 18q12-q21 and contains at least 43 exons.8 It encodes lipoxygenase homology domain 1-containing protein 1 which has 15 PLAT domains4 that are involved in targeting proteins to the plasma membrane and mediating protein interactions.9–12 Mouse studies showed that the Loxhd1 product is localized to the stereocilia of sensory hair cells, and that Loxhd1 variants can induce deafness with defects in the stereocilia followed by hair cell degeneration.13 This indicates that LOXHD1 plays an important role in maintaining normal hair cell function.
Although DFNB77 has previously been associated with LOXHD1 variants, it is a highly heterogeneous disease both phenotypically and genetically. More than 23 probands with DFNB77 have been reported worldwide on PubMed, and 37 different disease-causing variants have been identified (Table 3).5,7–21 Most of these probands come from Asia, suggesting that it has a high incidence of DFNB77. They show different auditory characteristics and audiometric phenotypes, varying from mild to profound and from stable to progressive sensorineural HL.5,7–21 Animal studies revealed that homozygous missense variants of Loxhd1 induced profound deafness while homozygous nonsense variants caused progressive HL.13 However, Wesdorp et al.4 found that the type of variant (nonsense or missense) did not associate with HL severity, and that the combination of a nonsense and missense variant could cause different audiometric phenotypes. Such research is limited, so correlations between LOXHD1 variants and phenotypic characteristics of HL remain unclear.
Table 3.
Known LOXHD1 variants causative of hearing loss.
| No. | Nucleotide or amino acid change | Variant type | Zygosity | Type of HL | Progressiveness | Population |
|---|---|---|---|---|---|---|
| 15 | c.879 + 1G>A | Splice | Hom | Profound | Non-progressive | Japanese |
| 25 | c.5869G>T | Nonsense | Het | Moderate-severe | Non-progressive | Japanese |
| 35 | c.4480C>T | Nonsense | Het | Moderate-severe | Non-progressive | Japanese |
| 47 | c.884C>T | Missense | Het | Moderate-severe | Progressive | – |
| 57 | c.2825_2827delAGA | Frameshift | Het | Moderate-severe | Progressive | – |
| 67 | c.2797C>T | Nonsense | Het | Profound | Non-progressive | – |
| 77 | c.1730T>G | Frameshift | Het | Profound | Non-progressive | – |
| 87 | c.2722G>A | Missense | Het | Profound | Non-progressive | – |
| 97 | c.3015_3017delCTT | Frameshift | Het | Profound | Non-progressive | – |
| 107 | c.766G>T | Nonsense | Het | Profound | Non-progressive | – |
| 117 | c.3596T>C | Missense | Het | – | – | – |
| 127 | c.2696G>C | Missense | Het | – | – | – |
| 137 | c.4526G>A | Missense | Hom | Profound | Progressive | – |
| 147 | c.4480C>T | Nonsense | Hom | Profound | Progressive | – |
| 157 | c.3206G>A | Missense | Het | Moderate-severe | Non-progressive | – |
| 167 | c.894T>G | Nonsense | Het | Moderate-severe | Non-progressive | – |
| 177 | c.1501delG | Frameshift | Het | Profound | Progressive | – |
| 187 | c.1193G>A | Missense | Het | Profound | Progressive | – |
| 197 | c.1147C>T | Nonsense | Het | Profound | Progressive | – |
| 208 | c.4714C>T | Nonsense | Hom | Profound | Non-progressive | Jewish |
| 219 | c.5674G>T | Missense | Het | Moderate-severe | Non-progressive | Japanese |
| 229 | c.4212 + 1G>A | Splice | Het | Moderate-severe | Non-progressive | Japanese |
| 3313 | c.2008C>T | Nonsense | Hom | Moderate-severe | Progressive | Iranian |
| 2414 | p.Gly398Glu | Missense | Het | Profound | Progressive | American |
| 2514 | p.Arg383X | Nonsense | Het | Profound | Progressive | American |
| 2615 | c.2863G>T | Nonsense | Hom | – | – | Turkey |
| 2715 | c.4480C>T | Nonsense | Hom | – | – | Turkey |
| 2816 | c.1588C>T | Nonsense | Hom | Profound | Progressive | Qatar |
| 2917 | c.71delT | Frameshift | Hom | – | – | Turkish |
| 3018 | c.3371G>A | Missense | Het | Profound | Non-progressive | Cameroonian |
| 3118 | c.3979T>A | Missense | Het | Profound | Non-progressive | Cameroonian |
| 3219 | c.1751C>T | Missense | Het | Moderate-severe | Progressive | Chinese |
| 3319 | c.5815G>A | Missense | Het | Moderate-severe | Progressive | Chinese |
| 3420 | p.A1406V | Missense | Het | – | – | – |
| 3520 | p.K148* | Nonsense | Het | – | – | – |
| 3621 | c.797 G > A | Missense | Het | – | – | Chinese |
| 37* | c.1828G>A | Missense | Het | Profound | Non-progressive | Chinese |
References shown as superscript numbers in first column; * This study
Hom, homozygous; het, heterozygous
LOXHD1 variants have not only been linked to HL but are also associated with late-onset Fuchs corneal dystrophy (FCD), a genetic disorder of the corneal endothelium.22 A case–control study by Stehouwer et al.23 reported a significant association between FCD and hearing disorders, but this should be investigated in larger sample sizes.22 We believe that it is important to check for ophthalmology disorders in patients with HL caused by LOXHD1 variants; however, we found no FCD phenotype in the current proband with LOXHD1 variants.
In conclusion, we carried out molecular diagnosis in a young couple with congenital HL and identified different disease-causing variants in the two individuals. The husband had compound heterozygous variants of GJB2, while the wife had the extremely rare HL known as DFNB77 and compound heterozygous variants of LOXHD1. We followed up the pregnancy outcome of this couple, and report that both ears of their baby passed the hearing screening test. To the best of our knowledge, this is the third case reported in Chinese individuals and the first in the northwest of the country. PCR and direct Sanger sequencing cannot provide effective detection of diseases caused by such rare variants. However, with the development of molecular diagnostic technology, the cost of tests is decreasing and NGS will become a more effective way of providing accurate molecular diagnosis and genetic counseling for rare diseases.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This work was supported by the Gansu Natural Science Foundation (Grant Nos.: 18JR3RA036, 1606RJZA151), the Lanzhou Talent Innovation and Entrepreneurship Project (Grant No.: 2018-RC-95), and the National Population and Reproductive Health Science Data Center of the People’s Republic of China (Grant No.: 2005DKA32408).
References
- 1.Fortnum HM, Summerfield AQ, Marshall DH, et al. Prevalence of permanent childhood hearing impairment in the United Kingdom and implications for universal neonatal hearing screening: questionnaire based ascertainment study. BMJ 2001; 323: 536–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du W, Wang Q, Zhu Y, et al. Associations between GJB2, mitochondrial 12S rRNA, SLC26A4 variants, and hearing loss among three ethnicities. Biomed Res Int 2014; 2014: 746838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nance WE, Lim BG, Dodson KM. Importance of congenital cytomegalovirus infections as a cause for pre-lingual hearing loss. J Clin Virol 2006; 35: 221–225. [DOI] [PubMed] [Google Scholar]
- 4.Wesdorp M, Schreur V, Beynon AJ, et al. Further audiovestibular characterization of DFNB77, caused by deleterious variants in LOXHD1, and investigation into the involvement of Fuchs corneal dystrophy. Clin Genet 2018; 94: 221–231. [DOI] [PubMed] [Google Scholar]
- 5.Mori K, Moteki H, Kobayashi Y, et al. Variants in LOXHD1 gene cause various types and severities of hearing loss. Ann Otol Rhinol Laryngol 2015; 124: 135S–141S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hilgert N, Smith R, Van Camp G. Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutat Res 2009; 681: 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sloan-Heggen CM, Bierer AO, Shearer AE, et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet 2016; 135: 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edvardson S, Jalas C, Shaag A, et al. A deleterious variant in the LOXHD1 gene causes autosomal recessive hearing loss in Ashkenazi Jews. Am J Med Genet A 2011; 155A: 1170–1172. [DOI] [PubMed] [Google Scholar]
- 9.Minami SB, Mutai H, Namba K, et al. Clinical characteristics of a Japanese family with hearing loss accompanied by compound heterozygous variants in LOXHD1. Auris Nasus Larynx 2016; 43: 609–613. [DOI] [PubMed] [Google Scholar]
- 10.Bateman A, Sandford R. The PLAT domain: a new piece in the PKD1 puzzle. Curr Biol 1999; 9: R588–R590. [DOI] [PubMed] [Google Scholar]
- 11.Hu J, Barr MM. ATP-2 interacts with the PLAT domain of LOV-1 and is involved in Caenorhabditis elegans polycystin signaling. Mol Biol Cell 2005; 16: 458–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aleem AM, Jankun J, Dignam JD, et al. Human platelet 12-lipoxygenase, new findings about its activity, membrane binding and low-resolution structure. J Mol Biol 2008; 376: 193–209. [DOI] [PubMed] [Google Scholar]
- 13.Grillet N, Schwander M, Hildebrand MS, et al. Variants in LOXHD1, an evolutionarily conserved stereociliary protein, disrupt hair cell function in mice and cause progressive hearing loss in humans. Am J Hum Genet 2009; 85: 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eppsteiner RW, Shearer AE, Hildebrand MS, et al. Prediction of cochlear implant performance by genetic variant: the spiral ganglion hypothesis. Hear Res 2012; 292: 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diaz-Horta O, Duman D, Foster J, et al. Whole-exome sequencing efficiently detects rare variants in autosomal recessive nonsyndromic hearing loss. PLoS One 2012; 7: e50628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vozzi D, Morgan A, Vuckovic D, et al. Hereditary hearing loss: a 96 gene targeted sequencing protocol reveals novel alleles in a series of Italian and Qatari patients. Gene 2014; 542: 209–216. [DOI] [PubMed] [Google Scholar]
- 17.Atik T, Onay H, Aykut A, et al. Comprehensive analysis of deafness genes in families with autosomal recessive nonsyndromic hearing loss. PLoS One 2015; 10: e0142154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lebeko K, Sloan-Heggen CM, Noubiap JJ, et al. Targeted genomic enrichment and massively parallel sequencing identifies novel nonsyndromic hearing impairment pathogenic variants in Cameroonian families. Clin Genet 2016; 90: 288–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu S, Sun F, Zhang J, et al. Genetic etiology study of ten Chinese families with nonsyndromic hearing loss. Neural Plast 2018; 2018: 4920980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Posey JE, Harel T, Liu P, et al. Resolution of disease phenotypes resulting from multilocus genomic variation. N Engl J Med 2017; 376: 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He L, Pang X, Chen P, et al. Carrier re-sequencing reveals rare but benign variants in recessive deafness genes. Sci Rep 2017; 7: 11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riazuddin SA, Parker DS, McGlumphy EJ, et al. Variants in LOXHD1, a recessive-deafness locus, cause dominant late-onset Fuchs corneal dystrophy. Am J Hum Genet 2012; 90: 533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stehouwer M, Bijlsma WR, Van der Lelij A. . Hearing disability in patients with Fuchs’ endothelial corneal dystrophy: unrecognized co-pathology? Clin Ophthalmol 2011; 5: 1297–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]