Abstract
Nanopores bear great potential as single-molecule tools for bioanalytical sensing and sequencing, due to their exceptional sensing capabilities, high-throughput, and low cost. The detection principle relies on detecting small differences in the ionic current as biomolecules traverse the nanopore. A major bottleneck for the further progress of this technology is the noise that is present in the ionic current recordings, because it limits the signal-to-noise ratio (SNR) and thereby the effective time resolution of the experiment. Here, we review the main types of noise at low and high frequencies and discuss the underlying physics. Moreover, we compare biological and solid-state nanopores in terms of the SNR, the important figure of merit, by measuring translocations of a short ssDNA through a selected set of nanopores under typical experimental conditions. We find that SiNx solid-state nanopores provide the highest SNR, due to the large currents at which they can be operated and the relatively low noise at high frequencies. However, the real game-changer for many applications is a controlled slowdown of the translocation speed, which for MspA was shown to increase the SNR > 160-fold. Finally, we discuss practical approaches for lowering the noise for optimal experimental performance and further development of the nanopore technology.
Keywords: biological nanopores, solid-state nanopores, translocation, ion current noise, signal-to-noise ratio, single-molecule detection, biosensors, DNA sequencing
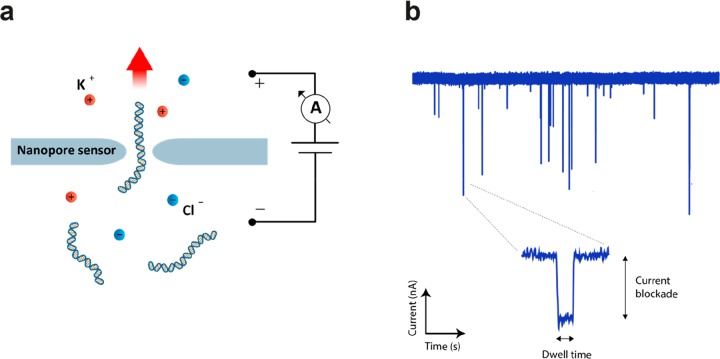
Nanopores are promising tools for biosensing applications and sequencing of DNA and proteins, as they can resolve single analyte molecules, resolve structural modifications of molecules, and even discriminate between nucleotide sequences.1−10 The detection mechanism is simple: While passing through the pore, a (part of a) molecule transiently blocks the ionic current, thereby inducing a small dip in the current signal, which is detectable by the electronics (Figure 1). The electrical read-out is carried out by an amplifier, which senses and amplifies the current signal, followed by a digitizer that performs the analog-to-digital conversion (ADC) of the data. Digital low-pass (LP) filtering is typically used to reduce the high-frequency noise and thus improve the signal-to-noise ratio (SNR). Such a gain in SNR comes, however, at the expense of a lower time resolution, thereby imposing an inherent trade-off.
Figure 1.
Fundamental principle of nanopore sensing. (a) A nanopore separates two aqueous compartments filled with electrolyte solution (e.g., potassium chloride), and small molecules (e.g., DNA) are electrokinetically pulled through the pore by an applied potential. (b) While passing through the nanopore, the molecule temporarily induces a partial current blockade which is detected by an amplifier. The signature of a single-molecule translocation event is generally characterized by the amplitude of the current blockade, which is proportional to the volume of the molecule in the nanopore, and by the dwell time, which depends on the electrophoretic driving force and transient interactions between the passing molecule and the pore surface.
The detection of analytes with nanopores thus is, on the one hand, limited by the ionic current noise which requires LP filtering that sets a finite operating bandwidth,11,12 but on the other hand, by the fast speed (typically submilliseconds) at which molecules translocate through the pore, which conversely requires a high time resolution for accurate sampling. Various approaches have been investigated in order to slow down the molecular translocation. For biological nanopores, a DNA-translocating motor protein (such as a helicase or polymerase) has been used to slowly feed a ssDNA strand into a protein pore for DNA sequencing.13−15 For solid-state nanopores fabricated in thin SiNx membranes16−18 or two-dimensional (2D) materials (graphene,19−21 boron nitride,22−24 molybdenum disulfide25−27), various efforts have been made to either increase time resolution16,17,28−31 or slow down the translocation process32 by the use of ionic liquids,27 pore surface engineering,33 mechanical manipulation with a double pore system,34 optical trapping,35 and sequential DNA unzipping.36 Nevertheless, while fingerprinting approaches have been developed to detect individual portions of a DNA sequence using dCas9,7,37 streptavidin,38 DNA hairpins,39 or DNA-origami as probes,40 the SNR has not yet allowed de novo DNA sequencing with solid-state pores. An understanding of the noise sources that affect nanopore systems and how these govern the SNR is key for achieving signals wherein molecular structures can be resolved fast and reliably. Noise characteristics of nanopores have been reported in various isolated reports, but a systematic overview and comparison between biological and solid-state nanopores is lacking.
In this review, we first describe the typical noise sources that affect the ionic current recordings of biological and solid-state nanopores, both at low and high frequencies. Next, we compare their respective performances of various nanopores using ssDNA poly(dT) translocations as a test system. We assess the SNR under typical experimental conditions for different protein pores Mycobacterium smegmatis porin A (the M2 mutant with a neutral constriction and positively charged vestibule, subsequently referred to as MspA),41Staphylococcus aureus alpha-hemolysin (α-HL),42,43Fragaceatoxin C (the mutant of FraC with a positively charged constriction, referred to as ReFraC),44,45 and SiNx29 and MoS246 solid-state nanopores. We find that biological pores generally exhibit lower noise (Figure 2a). Nevertheless, solid-state nanopores achieve the best SNR, largely because of the higher voltages and bandwidths that such devices can operate at, as compared to biological nanopores. Finally, we discuss approaches for lowering the ionic current noise and improving the SNR in biological and solid-state nanopores.
Figure 2.
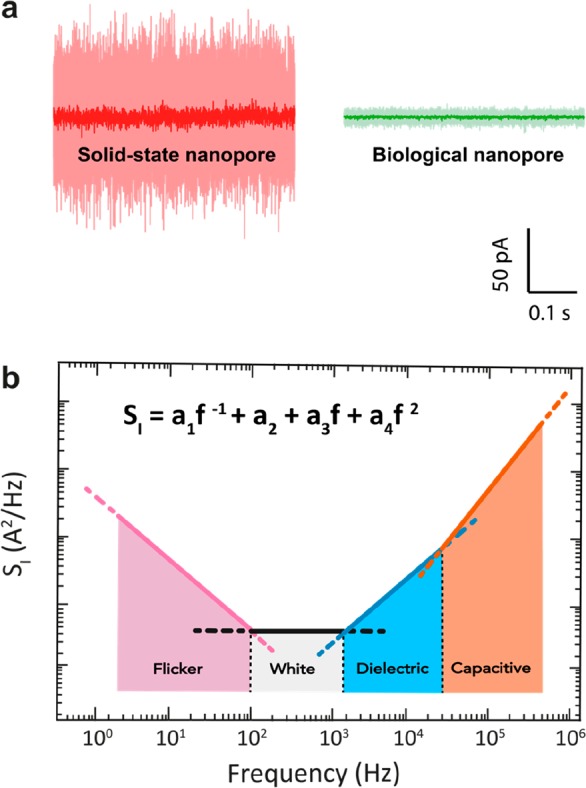
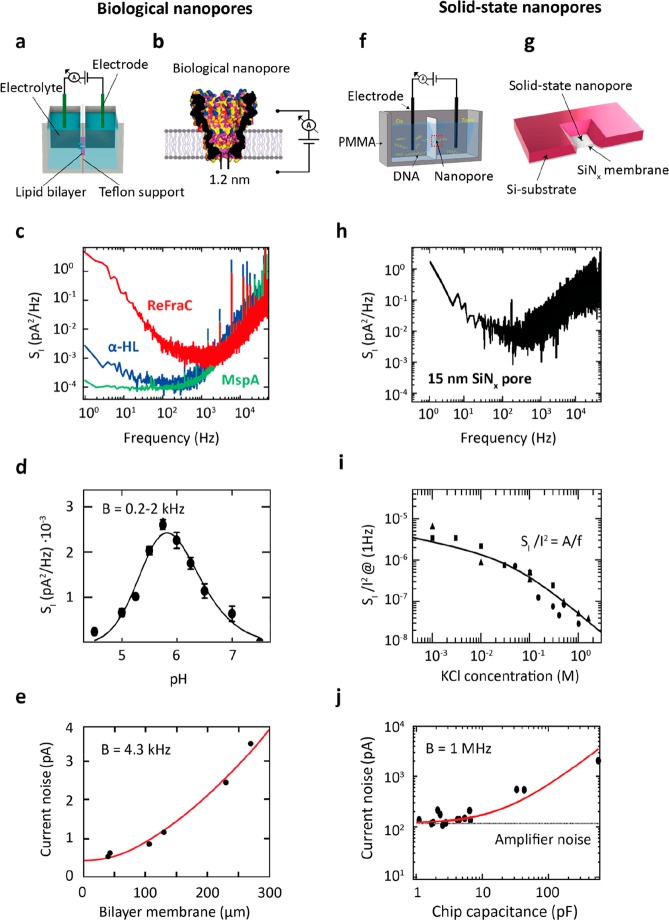
Ionic current noise in nanopores. (a) Example current traces for a 1.3 nm diameter solid-state SiNx nanopore (red) and a 1.4 nm diameter biological α-HL pore (green), performed at a constant applied bias of 100 mV in 1 M KCl buffer at pH 7 at a bandwidth of 10 kHz (light) and 1 kHz (dark). α-HL pore was measured using the typical Montal–Muller approach,47 with a bilayer diameter of ∼100 μm, as described by Maglia et al.3 The solid-state pore was fabricated on a Si-supported 20 nm-thick SiNx freestanding membrane using transmission electron microscopy. Currents through both pores were amplified with Axopatch 200B. (b) Schematic of the current PSD for a typical nanopore. Common types of noise are highlighted in the various frequency ranges.
Noise Sources in Nanopores
Noise refers to any statistical fluctuation of a signal. It can be characterized by the standard deviation σ or root-mean-square (rms) variation around the average value as measured over the full bandwidth B of the signal and by its power spectral density (PSD). Generally, noise is undesirable, as it can distort or even completely mask the actual signal. Nanopores typically operate by measuring a through-pore ionic current that is driven by a constant applied bias voltage. For the open-pore current measurement, where no analyte molecules are present, any deviation from the baseline current can be regarded as noise (Figure 2a).
Understanding the origins of noise is fundamental for optimizing signal detection. Nanopore systems exhibit a range of different noise sources.48,49 In Figure 2b, we illustrate the major current noise sources that affect nanopore systems at different frequencies. Generally, these can be divided in (i) low-frequency (≲100 Hz) 1/f noise and protonation noise; (ii) shot noise and thermal current noise (∼0.1–2 kHz), which are both white noise sources (i.e., frequency-independent); (iii) high-frequency dielectric (∼1–10 kHz); and (iv) capacitive (>10 kHz) noise.
In the low-frequency range, 1/f noise (also referred to as “flicker” or “pink” noise) typically is the dominant source of noise. Its power decreases with frequency f following a 1/fβ scaling, with β ≈ 1. While this type of noise is found in many biological and physical systems, a fundamental understanding of it is still missing.50 Based on phenomenological evidence, 1/f noise in nanopores has been associated with physical processes such as slow fluctuations in the number and mobility of the charge carriers,51−54 nanometer-sized bubbles in the pore channel,55 noise arising from the electrodes,56 mechanical fluctuations of the freestanding membrane (e.g., for 2D materials),23,57,58 and conformational changes in the case of biological nanopores.59,60 Smeets et al.61 found that Hooge’s phenomenological formula54 could effectively describe the 1/f noise in solid-state56,61−64 nanopores:
| 1 |
where Hooge’s constant, αH, is an empirical parameter that quantifies the magnitude of 1/f noise fluctuations, I the ionic current, and Nc the number of charge carriers in the pore volume, which was further validated by follow-up studies.56,62−64 As discussed below, solid-state nanopores typically feature a relatively pronounced 1/f noise, whose microscopic origin often remains unresolved. For biological pores, the low-frequency noise is typically dominated by protonation noise, which is generated by protonation/deprotonation of ionizable sites within the protein channel.65−67 It can be described by fluctuations between two different current levels with mean lifetimes τ1 and τ2 for the protonated and deprotonated states, respectively, yielding a Lorentzian-shaped component in the frequency spectrum (for a complete derivation see Machlup et al.):68
| 2 |
where Δi is the difference in current between the two levels, and τ is the characteristic relaxation time that can be expressed as τ = τ1τ2/(τ1 + τ2). For α-HL, for example,66 τ was found to be 3.1 × 10–5 s. A distribution of multiple Lorentzian processes such as in eq 2 can lead to 1/f noise.51 Temporal conformational changes of the pore channel can also generate conductance fluctuations resulting in 1/f noise. Such a phenomenon, also known as “channel breathing”, was reported to affect protein pores such as bacterial porin channels.59,60
In the midfrequency range (typically ∼0.1–2 kHz), a frequency-independent white noise is observed that derives from thermal noise (also known as Johnson–Nyquist noise) and shot noise. Thermal current noise is fundamental to any dissipative element69,70 and adds to the current noise as
| 3 |
where kB is the Boltzmann constant, T is temperature, and R the equivalent resistance of the nanopore. Shot noise, on the other hand, is due to the quantization of charge and is generated when charge carriers flow across a potential barrier.71,72 Its current-dependent contribution to the noise can be expressed as
| 4 |
where q is the charge of a single carrier. In practice, shot noise and thermal noise are comparable in magnitude for the conditions that are typically used in nanopore experiments.
Another contribution to the nanopore noise originates from the loss conductance of the membrane and chip support.48,49 Such dissipation, resulting from dipolar relaxation and charge carrier migration (details can be found in Chen et al.),73 generates thermal energy causing thermal noise, also known as dielectric noise.74,75 As this loss conductance scales linearly with frequency, this noise can be described by
| 5 |
where Cchip is the parasitic capacitance, and D a dissipation factor of the dielectric materials constituting the membrane and support chip. This source of noise typically dominates in the 2–10 kHz frequency range. To estimate Cchip, one can simply use the expression for a parallel plate capacitor C = εA/d, where ε is the dielectric constant of the membrane material, and A and d are the area and the thickness of the membrane, respectively. For f > 10 kHz, the current noise is determined by the input-referred thermal voltage noise vn across the total capacitance Ctot at the amplifier input:48,49
| 6 |
where vn is the input voltage noise (3 nV/Hz–1 for the commonly used amplifier Axopatch 200B,76 Molecular Devices, San Jose, USA). Ctot is the total capacitance including the membrane and support chip capacitance Cchip, the capacitance Camp at the input of the amplifier, and the capacitance Cw of the wiring between the electronics and the pore. Notably, SI, capacitance has an even stronger, f2, frequency dependence than SI, dielectric. The total current noise of a nanopore system over its full bandwidth is the sum of all contributions (Figure 2b), i.e., the sum of eqs 1–6.
Noise in Biological Nanopores
Biological nanopores are formed by the spontaneous insertion of membrane proteins into a lipid bilayer, which creates nanopores with typical diameters ranging from ∼1–4 nm,77 although larger pores with diameters up to ∼40 nm, e.g., the nuclear pore complex,78 are also found in nature. Figure 3a shows a schematic of a standard setup for measuring the ionic current through such a protein pore. Briefly, a thick (tens of micrometers) insulating film of amorphous polytetrafluoro-ethylene (PTFE, or Teflon) separates two liquid compartments and contains a ∼50–100 μm-sized hole where the lipid bilayer is assembled.3,79 Teflon is the preferred support material due to the relatively low high-frequency noise and ease of fabrication.80 Insertion of a protein pore (Figure 3b) short-circuits the insulating bilayer membrane and an ionic current between the two reservoirs can be measured by a pair of Ag/AgCl electrodes. The current signal is amplified by a transimpedance amplifier (e.g., Axopatch 200B) and digitized by an analog-to-digital converter (ADC, e.g., Axon Digidata, same supplier). To shield from external radiative electric noise, the flow-cell and the amplifier headstage are enclosed in a metallic Faraday cage.3 For biological nanopores, ionic conductances are typically on the order of 0.1–2 nS.
Figure 3.
Noise in biological and solid-state nanopores. (a) Standard setup used for measuring the ionic current through a biological nanopore embedded within a lipid membrane. (b) Sketch of a biological MspA nanopore. Adapted with permission from ref (14). Copyright 2010 National Academy of Sciences. (c) Typical current PSD for three biological nanopores, ReFraC (D10R/K159E mutant of FraC)44 (red), α-HL (blue), and the D90N/D91N/D93N/D118R/E139 K/D134R mutant of MspA (green), measured in the same setup at 50 mV applied voltage, 1 M KCl salt, pH 7. (d) Low-frequency protonation noise of α-HL as a function of pH. Adapted with permission from ref (67). Copyright 1995 The Biophysical Society. (e) Current noise Irms measured at a 4.3 kHz bandwidth of a lipid bilayer setup (where no pore was inserted) vs the size of the bilayer membrane. Adapted with permission from ref (80). Copyright 2003 The Biophysical Society. (f) Schematic of a typical flow cell for measuring the ionic current through a solid-state nanopore. Adapted with permission from ref (4). Copyright 2015 Elsevier. (g) Sketch of a solid-state nanopore fabricated onto a Si-supported SiNx membrane. (h) Current PSD for a 15.6 nm SiNx solid-state nanopore. Data were measured at 100 mV applied voltage for 1 M KCl salt.61 (i) Relative low-frequency noise SI/I2 at 1 Hz versus salt concentration. Solid line shows a fit to the data using Hooge’s relation, cf. eq 1. (h) and (i) were adapted with permission from ref (61). Copyright 2008 National Academy of Sciences. (j) Current noise Irms measured at a 1 MHz bandwidth vs capacitance of the nanopore chip. Adapted with permission from ref (16). Copyright 2014 American Chemical Society.
Characteristic examples of the current PSD for three biological nanopores (α-HL,42 MspA, and ReFraC44) are shown in Figure 3c, as measured at 1 M KCl, pH 7.5, under 50 mV applied bias. Noticeably, both α-HL and MspA exhibit a noise plateau at low frequencies (<1 kHz) which is due to protonation noise, cf. eq 2 for f ≪ 1/τ. The associated PSD is ∼10–4 to 10–3 pA2/Hz, which is higher than the corresponding white noise of ∼10–5 pA2/Hz, set by the sum of thermal and shot noise, eqs 3 and 4. In the context of single-molecule sensing, protonation noise in biological nanopores was first investigated by Bezrukov and Kasianowicz in the mid 1990s.66,67 Spectral analysis of the current noise of α-HL pores revealed the presence of a Lorentzian-shaped component at low-frequencies (0.2–2 kHz). Given the strong dependence on pH (Figure 3d), this noise source was associated with the reversible protonation of ionizable residues occurring in the α-HL constriction. This notion was further established in a later work by Nestorovich et al.,65 where the bacterial porin, OmpF, was shown to produce a similar pH-dependence of the protonation noise.
ReFraC instead shows a pronounced 1/f noise with a PSD of ∼10–1 pA2/Hz at 1 Hz, which is almost three times more than for α-HL and MspA. 1/f noise in biological nanopores was first studied by Benz and co-workers59,81 and described using Hooge’s model, eq 1. The low-frequency fluctuations observed in a family of bacterial porins were associated with a number of possible phenomena, e.g., gating of the pore channel.59 In later work by Bezrukov and Winterhalter,60 conformational changes of the protein pore channel, termed “channel breathing”,82 were discussed as the main cause for the observed 1/f noise.
At higher frequencies (>1 kHz), the noise in biological nanopores is dominated by dielectric noise arising from the loss conductance of the lipid membrane. In fact, since the dielectric loss and dielectric constant of the Teflon are relatively low (D = (0.8–2) × 10–4 and εr = 1.89–1.93, respectively), the major contribution to the dielectric noise is set by the capacitance of the thin lipid bilayer membrane. This can be attenuated by reducing the area of the Teflon hole (Figure 3e).80,83 A noise characterization at even higher frequencies (MHz-GHz; above the experimentally accessible frequency range) was performed using molecular dynamics simulations based on a comprehensive model of MspA.84
Noise in Solid-State Nanopores
Solid-state nanopores are generally fabricated in a freestanding membrane of a solid-state material such as silicon nitride (SiNx),85 graphene,19 hexagonal boron nitride (h-BN),86 or molybdenum disulfide (MoS2),46 with thicknesses ranging from ∼0.3 to 30 nm. In common nanopore chips (Figure 3g), such a membrane is structurally supported by a ∼200–500 μm-thick substrate material, typically silicon(Si),85 glass (SiO2),16 or Pyrex.87,88 Nanopores can be drilled into the membrane in a variety of ways, e.g., by using a transmission electron microscope (TEM),89,90 focused ion beam milling (FIB),91,92 reactive ion etching (RIE),93 laser-etching,94,95 or by dielectric breakdown,96,97 resulting in pore diameters from sub-1 nm to tens of nanometers. In a standard solid-state nanopore experiment, the chip is sandwiched between two rubber O-rings that seal two compartments containing the electrolyte solution (Figure 3f). Alternatively, solid-state pores of ∼5–50 nm size can be made by mechanical pulling of hollow glass (SiO2) pipettes,98−100 which are immersed in electrolyte during the measurement. Current sensing, amplification, and recording is the same as for biological nanopores.
Figure 3h displays a typical current PSD measured for a 15 nm diameter SiNx solid-state nanopore61 in a 20 nm-thick membrane. Characteristic of solid-state nanopores is the pronounced 1/f noise that dominates the low-frequency part of the spectrum (<100 Hz). It can originate from a range of physical processes, see eq 1 and associated discussion. Smeets et al.55 showed that poor wettability of the pore surface, associated with the formation of nanobubbles, resulted in high 1/f noise in SiNx. Tabard-Cossa et al.101 discussed that high 1/f noise in SiNx pores correlates with surface contamination: inhomogeneities of the pore surface resulted in fluctuations of the number and mobility of charge carriers due to trapping at the pore surface,63,101 analogous to 1/f noise found in semiconductors.102 As shown by Smeets et al.,61,62 such low-frequency noise in SiNx pores obeys Hooge’s relation, eq 1, which describes an inverse proportionality between the 1/f current noise power and the number of charge carriers present within the nanopore volume (Figure 3i).54 For nanopores made in 2D materials, the 1/f noise depends strongly on the size of the freestanding area,22,57,58,103 indicating that mechanical fluctuations of the ultrathin 2D membrane (thickness <1 nm) are the main source. The high-frequency noise in solid-state nanopores is dominated by dielectric (∼2–10 kHz) and capacitive noise (>10 kHz),16,104 see Figure 3j. The PSD of these noise sources depends mostly on the capacitance of the chip, cf. eq 5 and 6, which in turn is set by the membrane and substrate size, thickness, and dielectric constant. Additionally, parasitic capacitances from the amplifier and the interconnects between nanopore and amplifier contribute to the total capacitance at the amplifier input.
Comparing the Performance of Biological and Solid-State Nanopores
So far, we provided a general overview of the typical noise sources in biological and solid-state nanopores. We now turn to a mutual comparison between these two classes of nanopores. We compare their performances in terms of the SNR, a more relevant figure of merit than the mere magnitude of the current noise. We define the SNR as the ratio between the signal modulation ΔI produced by the translocation of a ssDNA molecule and the baseline current rms (Irms) measured at the operating bandwidth (Figure 4a). Although other definitions of SNR are found in the literature, e.g., as the ratio between open pore current and baseline current noise Io/Irms105 or the capability to discern current levels when sequencing DNA,13,41 we find this definition the most appropriate for the diverse nanopore systems compared in our study.
Figure 4.
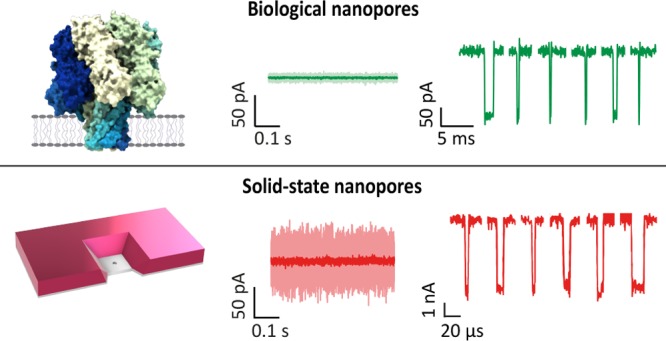
Detection of DNA homopolymer poly(dT) with protein and solid-state nanopores. (a) Example of a translocation event, illustrating the SNR. (b) Schematic comparing the relative sizes of MspA (green), α-HL (red), ReFraC (blue), MoS2 (black), and solid-state SiNx (purple). Adapted with permission from ref (2). Copyright 2015 IOP Publishing Ltd. (c) Example of translocation events of poly(dT) molecules through MspA14 channel (green), α-HL pore (red), ReFraC pore (blue), 1.4 nm MoS2 pore (black), and 1.4 nm SiNx pore (purple, Adapted with permission from ref (18). Copyright 2013 American Chemical Society) all in a 1 M KCl solution at transmembrane voltages of 180 mV, 180 mV, 180 mV, 300 mV, and 1 V and at bandwidths of 30 kHz, 10 kHz, 10 kHz, 10 kHz, and 500 kHz, respectively. Experiments for biological pores were done using an Axopatch 200B amplifier, a Teflon-supported lipid membrane (∼50–100 μm wide; DPhPC lipids), 10–30 kHz bandwidth, 1 M KCl, pH 7.5, and a forward bias voltage of 180 mV, as in ref (106). The solid-state SiNx pore was built on a glass chip and measured with the VC100 high-bandwidth, low-noise voltage-clamp amplifier (Chimera Instruments, New York, NY, USA) which allowed for low-noise measurements at high bandwidth. A broad bandwidth of 500 kHz was required in order to fully resolve the fast translocations (∼22 μs)18 of poly(dT)30 through the solid-state SiNx pore. Notably, the positively charged constriction of ReFraC causes the negatively charged poly(dT)50 to translocate with much slower (491 ± 114 μs) translocation times compared to MspA (17.7 ± 1.1 μs), which permitted to filter out more high-frequency noise. (d) Comparison of various figures of merit for different nanopore systems under typical experimental conditions. Io indicates the open pore ionic current at the applied bias V.
Given that the experimental conditions reported in the literature differ considerably, we carried out a dedicated comparative study by complementing reported data with data that were, to the extent possible, obtained in our lab under the same experimental conditions. The bandwidth was chosen such as to fully resolve the current blockade ΔI generated by the poly(dT) substrate (avoiding a reduced ΔI due to a too narrow bandwidth). The translocation time, in turn, is determined by a combination of electrophoresis, electro-osmosis, and interactions between the passing molecule and the pore surface, which will depend on each individual nanopore system. The applied bias was chosen as to maximize the current signal and is limited by experimental conditions, as will be discussed below. We selected five popular nanopore systems, MspA, a-HL, ReFraC, MoS2, and SiNx, that are commonly used and that were shown to possess good spatiotemporal resolution, allowing for accurate discrimination of short homopolymers.13,27,29,44,107 All pores considered had a similar diameter of ∼1.3 nm. Figure 4b illustrates the relative sizes of the different pores.
Nanopore experiments probing the translocation of poly(dT)50 were carried out in-house using three biological pores, MspA, α-HL, and ReFraC. We compared these data to experimental results on two types of solid-state nanopores, SiNx29 and MoS2,27 that were measured at the same electrolyte conditions. Translocation data of poly(dT)80 through a 1.4 nm MoS2 pore were kindly shared by the Radenovic lab,46 whereas poly(dT)30 data for a 1.4 nm SiNx pore with ∼5 nm length were taken from the literature.29Figure 4c shows examples of single-molecule poly(dT) translocations for the 5 pores. A range of SNR values are observed, with, at face value, a better performance for SiNx and ReFraC than for MoS2, α -HL, and MspA.
Figure 4d quantitatively compares the data for the different nanopore systems. For the biological nanopores, ReFraC gives the best SNR of 15, while MspA resulted in a much lower SNR of 4. This is mainly due to the faster translocations of poly(dT) through MspA, which required a higher bandwidth (30 kHz), and hence larger noise, in order to resolve the translocation events. Conversely, translocations through the positively charged constriction of ReFraC were significantly slower, thus permitting to employ a lower bandwidth (10 kHz). Among the solid-state nanopores, SiNx showed the best SNR: an impressive value of 37, which was higher than the SNR of 5 obtained for MoS2, as well as higher than the values for all biological nanopores. The greater SNR for SiNx results from the very high voltage applied (1000 mV vs 300 mV for MoS2), producing a particularly large current signal ΔI. The applied voltage for MoS2 pores was limited by the degradation of the 2D membrane and pore growth under high bias voltages, which typically limited the applied bias to <400 mV. In biological nanopores, the range of bias voltages is limited by the membrane stability, affected by electroporation and rupture around 200–300 mV.108,109 Note furthermore that the SiNx nanopore system was operated at a much higher bandwidth (500 kHz vs 10 kHz for MoS2), the regime where dielectric and capacitive noise dominate. This is advantageous for high-voltage sensing, since these noise sources do not scale with voltage, cf. eqs 5 and 6. As a result, the high bias voltage improves the signal (ΔI), while it does not affect the noise. Lastly, we note that, while MoS2 has a lower SNR than SiNx, it features a better spatial resolution along the molecule, given its 0.7 nm pore length, as compared to the ∼5 nm of SiNx.
Finally, it is important to point out that the above comparison was carried out for voltage-driven translocation of DNA through nanopores. A controlled slowdown of the translocation speed can change these numbers dramatically. Indeed, despite the fact that Figure 4d shows that the best SNR was obtained for the solid-state SiNx nanopores, with values exceeding those of biological nanopores, todays commercialized nanopore-based DNA sequencers employ protein pores to read off DNA bases with (sub)-nucleotide resolution over very long reads.15,110 Using a helicase to slow down ssDNA molecules through MspA, allowed Laszlo et al.41 to use a very low LP filter frequency of ∼200 Hz and fully resolve the stepwise DNA translocation at half-base resolution. By comparing the noise at a 200 Hz bandwidth with the signal obtained for voltage-driven poly(dT) translocations in our experiments, we find an exquisite SNR of ∼650 for MspA – 2 orders of magnitude higher than the SNR = 4 noted above. Applying the same reasoning to α -HL and ReFraC increases their SNR to ∼270 and ∼220, respectively, i.e., somewhat lower values, consistent with their higher low-frequency noise compared to MspA (Figure 3c). Thus, in the context of DNA sequencing, the real game-changer lies in the enzymatic control over the translocation speed by use of an additional motor protein.13,15,41,107,111 For solid-state nanopores, despite the progresses in slowing down the DNA translocation,27,32−36 time control has so far remained a challenge, and accordingly, DNA sequencing has not yet been realized with such nanopores. Furthermore, mechanical instability of solid-state nanopores over time, which particularly affects smaller pores, should be minimized in order to achieve sufficiently long observation times.
Approaches To Overcome Noise Limitations
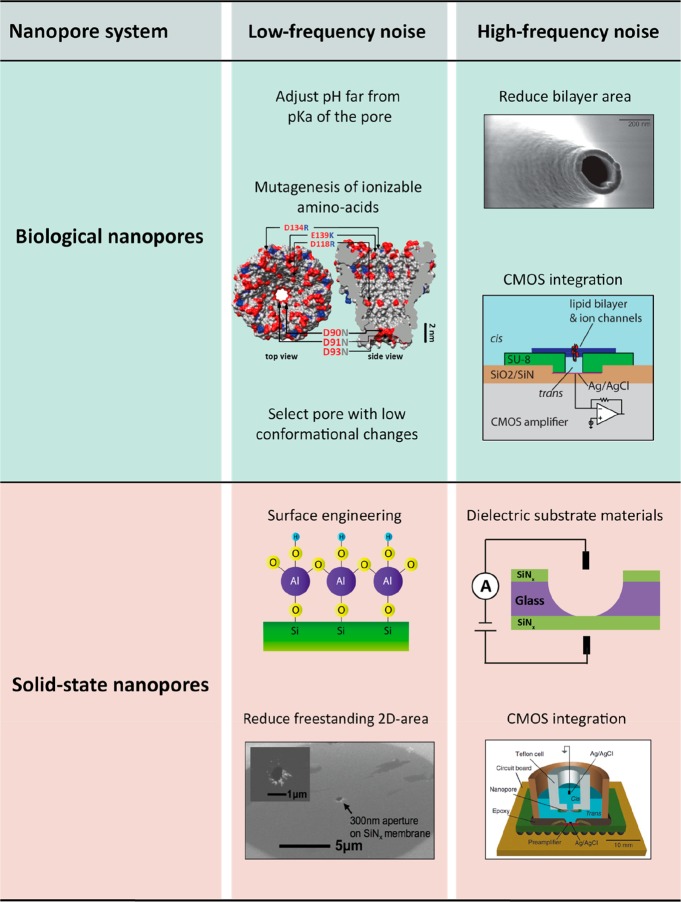
Figure 5 shows important approaches to lower the ionic current noise in nanopores. We first describe efforts to reduce the low-frequency noise. As protonation noise is the main source of low-frequency noise in biological nanopores, it is advantageous to choose a pH value that is far away from the pKa of the ionizable amino acids to attenuate the noise. Another way to reduce it, is to remove charged amino acids near the constriction site, which is expected to yield lower noise levels. Furthermore, increasing the conformational stiffness of biological pores can help to reduce conductance fluctuations associated with channel breathing.
Figure 5.
Approaches to reduce the noise in nanopore systems. For biological nanopores, low-frequency protonation noise can be minimized by adjusting the pH far from pKa of the amino acids in the pore constriction, as reported in ref. (66) or by mutating the ionizable amino acids (Arg, Lys, Asp, Glu) to neutral ones (e.g., Asn), as was done for MspA. Reprinted with permission from ref (106). Copyright 2008 National Academy of Sciences. Low-frequency 1/f noise instead can only be avoided by selecting a pore that is mechanically stable under an applied bias, e.g. MspA or α-HL. High-frequency noise can be minimized by reducing the size of the freestanding lipid membrane by, e.g., employing a nanocapillary as a support (reprinted with permission from ref (123). Copyright 2011 American Chemical Society) and by reducing the capacitance of the interconnects by smart CMOS integration (reprinted with permission from ref (105). Copyright 2013 American Chemical Society). For solid-state nanopores, low-frequency 1/f noise can be reduced by coating the surface with a hydrophilic, homogeneous material, e.g., Al2O3, as reported in ref (115). For 2D-materials, 1/f noise can be suppressed by lowering the area of the freestanding 2D membrane. Adapted with permission from ref (17). Copyright 2015 Springer Nature. High-frequency noise can be minimized by employing dielectric chip substrate materials, e.g. glass or by tight integration of the amplifier and nanopore chip (adapted with permission from ref (28). Copyright 2012 Springer Nature).
For solid-state nanopores, the low-frequency 1/f noise can be efficiently suppressed by surface functionalization of the SiNx nanopore with a hydrophilic surface layer, such as Al2O3 or SiO2.112−115 In principle, any surface treatment that reduces the amount of contaminants and improves hydrophilicity of the pore surface will lower the 1/f noise. Indeed, Tabard-Cossa et al.101 showed that piranha treatment (30% H2O2/H2SO4, 1:3) substantially reduced the 1/f noise by up to 3 orders of magnitude. Beamish et al.116 demonstrated that cyclic application of high electric fields to the nanopore also suppressed this noise source. Similar to protein pores, work from Wen et al.56 showed that the 1/f noise could be minimized by choosing a pH that is far from the isoelectric point of the nanopore material (∼5 for Si3N4).117,118 Nanopores built with 2D materials suffer from pronounced 1/f noise that was found to correlate with the area and thickness of the freestanding 2D membrane.47,48 A decrease of the freestanding area was shown to reduce the 1/f noise, while employing multilayer membranes was also helpful for obtaining less noise, though that approach is less desirable due to a loss of spatial resolution.23,57,58 Use of freestanding 2D membranes that are directly grown on a SiNx-supporting membrane was also shown to lower the 1/f noise for both graphene119 and MoS2120 pores, as compared to transferred 2D membranes.19,57
The noise at higher frequencies, constituted by dielectric and capacitive noise, has a well-characterized physical origin, namely the thermal voltage noise in conjunction with the loss conductance of the membrane and substrate materials as well as the amplifier input capacitance. Suppression of dielectric noise is generally achieved by minimizing the capacitance Cchip and dielectric loss D of the chip, cf. eq 5. To effectively decrease capacitive noise, the total input capacitance Ctot needs to be reduced, see eq 6 and related discussion. In biological nanopores, the high-frequency noise can be reduced by decreasing the area of the lipid bilayer. Mayer et al.80 fabricated Teflon holes of only ∼25 μm in diameter with soft lithography using SU-8 resist as master mold, providing a Cchip of 10–28 pF. By using a U-shaped Teflon patch tube as the support, Akeson and co-workers83,106 built horizontal bilayers <20 μm in diameter. Lipid bilayers with a comparable size were also created with the droplet-interface-bilayer (DIB) technique.121 Kitta et al.122 reported on the fabrication of yet smaller bilayers, with sizes down to 2–3 μm in diameter, by using a heated tungsten tip to create a microhole across the Teflon film.
Similarly sized 1–3 μm bilayers can be obtained by inserting protein pores into giant unilamellar vescicles (GUVs) and using patch-clamp pipets to measure the conductance of the pores.124,125 More recently, Gornall et al.123 showed that borosilicate glass nanopipets with diameters as low as 230 nm could be fabricated and used for current recordings on an OmpF protein channel. Hartel et al.126 achieved high-bandwidth (>500 kHz) recordings with biological pores with complementary metal-oxide-semiconductor (CMOS)-suspended membranes that were built directly over a ∼30 μm well on top of a CMOS-amplifier chip. This offered a reduction of the total input capacitance Ctot to <4 pF and provided a bandwidth as high as 1 MHz and a SNR > 8 at 500 kHz, for detecting the gating of a RyR1 pore (type 1 ryanodine receptor).126 Combined with extended β distribution data analysis127 (which exploits the characteristics of the excess current noise to reconstruct the true current signal), it was possible to achieve a time resolution of 35 ns.126
For reducing the high-frequency noise in SiNx solid-state nanopores, an established method, first reported by Tabard-Cossa et al.,101 is to lower Cchip by coating the area of the chip around the pore with a dielectric, e.g. PDMS, thereby providing additional thickness to the chip membrane surrounding the pore and thus a low series capacitance. Similarly, a substantial reduction of Cchip was achieved by employing a dielectric, e.g., amorphous glass16,17 or Pyrex23,88 as substrate material instead of the commonly used crystalline silicon which is intrinsically conductive. In work by Balan et al.,17 glass chips were shown to reduce Cchip to <1 pF, compared to >300 pF for standard silicon chips.61 Similarly to biological nanopores, the highest working bandwidths were so far achieved by integrating a low Cchip nanopore device with an on-chip CMOS-amplifier,28,30 which lowered the total input capacitance to Ctot ≈ 4 pF. In this way, ssDNA molecules were recorded using ultrathin (<4 nm) sub-2 nm pores yielding a SNR > 10 at 5 MHz.30 In 2D nanopores, the high-frequency noise can be addressed in similar ways to SiNx pores. The use of glass as substrate material, combined with a small ∼300 nm freestanding 2D-membrane of graphene or MoS2, resulted in a Cchip < 2 pF.17
Conclusions
In this paper, we illustrated the main sources of noise affecting various nanopore systems, with a particular emphasis on comparing biological and solid-state nanopores, and we discussed practical approaches to lower the noise. We compared the SNR of poly(dT) translocations through a representative set of biological and solid-state pores and found that silicon nitride nanopores gave the highest SNR. This can be attributed to the higher currents (i.e., larger signals) that solid-state systems offer and to the relatively low high-frequency noise. Despite these good noise characteristics, prominent applications such as DNA or protein sequencing have so far remained out of reach for solid-state nanopores, because the fast translocation speed provides only a short observation time per single molecule. There are two ways to improve this: One can either shift the sampling rate into even higher frequencies (≫MHz) or alternatively slow down the translocation of the molecule. The latter strategy has led to the successful commercialization of DNA sequencers based on protein nanopores that are coupled with an enzymatic stepping motor. In our comparison, we found that the SNR of MspA increased >160-fold by such speed control, mainly due to the decoupling of the signal from the high-frequency noise. Additionally, the motor protein provides a ratcheting mechanism that translocates the substrate with a constant discrete step size. Since the sensing region of the pore is typically larger than the individual monomer size (nucleotide or amino acid), such a mechanism is indispensable to reproducibly resolve and identify the sequence. Future improvements of the solid-state nanopore system could thus be directed toward either a further increase of the temporal resolution, e.g., by reducing even more the overall parasitic capacitances, or by creating an efficient slowdown mechanism, similar to biological nanopores. In general, the understanding of noise sources, associated time scales, and techniques to lower the noise at both low and high frequencies are greatly beneficial to maximize the sensitivity of nanopore detection and thereby extend the range of its applications.
Acknowledgments
We would like to thank Aleksandra Radenovic, Michael Graf, and Thakur Mukeshchand (EPFL, Switzerland) for sharing and discussing current traces measured on MoS2 nanopores, Marija Drndic and Siddharth Shekar (University of Pennsylvania, USA) for sharing current measurement performed on SiNx nanopores, Hagan Bayley and Nicholas Bell (Oxford University, UK) for sharing current traces measured through α-HL pores, and Giovanni Maglia and Gang Huang (University of Groningen, The Netherlands) for FraC mutants. The alpha-hemolysin was a kind gift of Jingyue Ju and Sergey Kalachikov (Columbia University, USA). MspA was a kind gift of Jens Gundlach and Andrew H. Laszlo (University of Washington, USA). We thank Meng-yue Wu for technical assistance on TEM and Wayne Yang, Stephanie Heerema, Laura Restrepo Perez, Sergii Pud, Daniel Verschueren (TU Delft, The Netherlands) for fruitful discussions. This work was supported by ERC Advanced Grant SynDiv (no. 669598) and the NanoFront and BaSyC programs. S.S. acknowledges the Postdoc. Mobility fellowship no. P400PB_180889 by the Swiss National Science Foundation.
Glossary
Vocabulary
- Nanopore
nanometer-sized hole in a thin freestanding membrane
- single-molecule detection
the capability to sense individual molecules one by one
- root-mean-square current noise (rms)
square root of the average squared value of the current fluctuations from the mean current
- power spectral density (PSD)
current noise power per unit of bandwidth
- signal-to-noise ratio
ratio between the ion current signal that is produced by a molecule that translocates through a nanopore, and the ion current noise of the baseline
The authors declare no competing financial interest.
References
- Dekker C. Solid-State Nanopores. Nat. Nanotechnol. 2007, 2, 209–216. 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- Carson S.; Wanunu M. Challenges in DNA Motion Control and Sequence Readout Using Nanopore Devices. Nanotechnology 2015, 26, 074004. 10.1088/0957-4484/26/7/074004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglia G.; Heron A. J. ; Stoddart D.; Japrung D.; Bayley H. Walter G. N.. Analysis of Single Nucleic Acid Molecules with Protein Nanopores. In Methods in Enzymology; Elsevier Inc.: San Diego, 2010; Vol. 475, pp 591–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.; Zhang Y.; Ying C.; Wang D.; Du C. Nanopore-Based Fourth-Generation DNA Sequencing Technology. Genomics, Proteomics Bioinf. 2015, 13, 4–16. 10.1016/j.gpb.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.; Ying Y. L.; Long Y. T. Nanopore Confinement for Electrochemical Sensing at the Single-Molecule Level. Curr. Opin. Electrochem. 2018, 7, 172–178. 10.1016/j.coelec.2017.12.002. [DOI] [Google Scholar]
- Atas E.; Singer A.; Meller A. DNA Sequencing and Bar-Coding Using Solid-State Nanopores. Electrophoresis 2012, 33, 3437–3447. 10.1002/elps.201200266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.; Birnie A.; Restrepo-Pérez L.; van der Torre J.; Bengtson M.; Heerema S. J.; Dekker C. Detection of CRISPR-DCas9 on DNA with Solid-State Nanopores. Nano Lett. 2018, 18, 6469–6474. 10.1021/acs.nanolett.8b02968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires A. H.; Hersey J. S.; Grinstaff M. W.; Meller A. A Nanopore-Nanofiber Mesh Biosensor to Control DNA Translocation. J. Am. Chem. Soc. 2013, 135, 16304–16307. 10.1021/ja408685x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles B. N.; Ivanov A. P.; Wilson K. A.; Doğan F.; Japrung D.; Edel J. B. Single Molecule Sensing with Solid-State Nanopores: Novel Materials, Methods, and Applications. Chem. Soc. Rev. 2013, 42, 15–28. 10.1039/C2CS35286A. [DOI] [PubMed] [Google Scholar]
- Wu D.; Bi S.; Zhang L.; Yang J. Single-Molecule Study of Proteins By Biological Nanopore Sensors. Sensors 2014, 14, 18211–18222. 10.3390/s141018211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm A. J.; Storm C.; Chen J.; Zandbergen H.; Joanny J. F.; Dekker C. Fast DNA Translocation through a Solid-State Nanopore. Nano Lett. 2005, 5, 1193–1197. 10.1021/nl048030d. [DOI] [PubMed] [Google Scholar]
- Plesa C.; Kowalczyk S. W.; Zinsmeester R.; Grosberg A. Y.; Rabin Y.; Dekker C. Fast Translocation of Proteins through Solid State Nanopores. Nano Lett. 2013, 13, 658–663. 10.1021/nl3042678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrao E. A.; Derrington I. M.; Laszlo A. H.; Langford K. W.; Hopper M. K.; Gillgren N.; Pavlenok M.; Niederweis M.; Gundlach J. H. Reading DNA at Single-Nucleotide Resolution with a Mutant MspA Nanopore and Phi29 DNA Polymerase. Nat. Biotechnol. 2012, 30, 349–353. 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrington I. M.; Butler T. Z.; Collins M. D.; Manrao E.; Pavlenok M.; Niederweis M.; Gundlach J. H. Nanopore DNA Sequencing with MspA. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 16060–16065. 10.1073/pnas.1001831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter J.-M.; Hussain S. Robust Long-Read Native DNA Sequencing Using the ONT CsgG Nanopore System. Wellcome Open Res. 2017, 2, 23. 10.12688/wellcomeopenres.11246.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balan A.; Machielse B.; Niedzwiecki D.; Lin J.; Ong P.; Engelke R.; Shepard K. L.; Drndić M. Improving Signal-to-Noise Performance for DNA Translocation in Solid-State Nanopores at MHz Bandwidths. Nano Lett. 2014, 14, 7215–7220. 10.1021/nl504345y. [DOI] [PubMed] [Google Scholar]
- Balan A.; Chien C. C.; Engelke R.; Drndic M. Suspended Solid-State Membranes on Glass Chips with Sub 1-PF Capacitance for Biomolecule Sensing Applications. Sci. Rep. 2016, 5, 17775. 10.1038/srep17775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venta K.; Shemer G.; Puster M.; Rodríguez-Manzo J. A.; Balan A.; Rosenstein J. K.; Shepard K.; Drndić M. Differentiation of Short, Single-Stranded DNA Homopolymers in Solid-State Nanopores. ACS Nano 2013, 7, 4629–4636. 10.1021/nn4014388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant A. C.; Healy K.; Wanunu M.; Ray V.; Peterman N.; Bartel J.; Fischbein M. D.; Venta K.; Luo Z.; Johnson A. T. C.; Drndic M. DNA Translocation through Graphene Nanopores. Nano Lett. 2010, 10, 2915–2921. 10.1021/nl101046t. [DOI] [PubMed] [Google Scholar]
- Schneider G. F.; Xu Q.; Hage S.; Luik S.; Spoor J. N. H.; Malladi S.; Zandbergen H.; Dekker C. Tailoring the Hydrophobicity of Graphene for Its Use as Nanopores for DNA Translocation. Nat. Commun. 2013, 4, 2619. 10.1038/ncomms3619. [DOI] [PubMed] [Google Scholar]
- Schneider G. F.; Kowalczyk S. W.; Calado V. E.; Pandraud G.; Zandbergen H. W.; Vandersypen L. M. K.; Dekker C. DNA Translocation through Graphene Nanopores. Nano Lett. 2010, 10, 3163–3167. 10.1021/nl102069z. [DOI] [PubMed] [Google Scholar]
- Zhou Z.; Hu Y.; Wang H.; Xu Z.; Wang W.; Bai X.; Shan X.; Lu X. DNA Translocation through Hydrophilic Nanopore in Hexagonal Boron Nitride. Sci. Rep. 2013, 3, 3287. 10.1038/srep03287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K. B.; Kim H. J.; Kim H. M.; Han S. A.; Lee K. H.; Kim S. W.; Kim K. B. Noise and Sensitivity Characteristics of Solid-State Nanopores with a Boron Nitride 2-D Membrane on a Pyrex Substrate. Nanoscale 2016, 8, 5755–5763. 10.1039/C5NR09085G. [DOI] [PubMed] [Google Scholar]
- Liu K.; Lihter M.; Sarathy A.; Caneva S.; Qiu H.; Deiana D.; Tileli V.; Alexander D. T. L.; Hofmann S.; Dumcenco D.; Kis A.; Leburton J.; Radenovic A. Geometrical Effect in 2D Nanopores. Nano Lett. 2017, 17, 4223–4230. 10.1021/acs.nanolett.7b01091. [DOI] [PubMed] [Google Scholar]
- Graf M.; Lihter M.; Thakur M.; Georgiou V.; Topolancik J.; Ilic B. R.; Liu K.; Feng J.; Astier Y.; Radenovic A. Fabrication and Practical Applications of Molybdenum Disulfide Nanopores. Nat. Protoc. 2019, 14, 1130–1168. 10.1038/s41596-019-0131-0. [DOI] [PubMed] [Google Scholar]
- Liu K.; Feng J.; Kis A.; Radenovic A. Atomically Thin Molybdenum Disulfide Nanopores with High Sensitivity for DNA Translocation. ACS Nano 2014, 8, 2504–2511. 10.1021/nn406102h. [DOI] [PubMed] [Google Scholar]
- Feng J.; Liu K.; Bulushev R. D.; Khlybov S.; Dumcenco D.; Kis A.; Radenovic A. Identification of Single Nucleotides in MoS 2 Nanopores. Nat. Nanotechnol. 2015, 10, 1070–1076. 10.1038/nnano.2015.219. [DOI] [PubMed] [Google Scholar]
- Rosenstein J. K.; Wanunu M.; Merchant C. A.; Drndic M.; Shepard K. L. Integrated Nanopore Sensing Platform with Sub-Microsecond Temporal Resolution. Nat. Methods 2012, 9, 487–492. 10.1038/nmeth.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venta K.; Shemer G.; Puster M.; Rodríguez-Manzo J. A.; Balan A.; Rosenstein J. K.; Shepard K.; Drndić M. Differentiation of Short, Single-Stranded DNA Homopolymers in Solid-State Nanopores. ACS Nano 2013, 7, 4629–4636. 10.1021/nn4014388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekar S.; Niedzwiecki D. J.; Chien C. C.; Ong P.; Fleischer D. A.; Lin J.; Rosenstein J. K.; Drndić M.; Shepard K. L. Measurement of DNA Translocation Dynamics in a Solid-State Nanopore at 100 Ns Temporal Resolution. Nano Lett. 2016, 16, 4483–4489. 10.1021/acs.nanolett.6b01661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel G.; Shepard K. L.; Hartel A. J. W.; Shekar S.; Schroeder I.; Ong P. High Bandwidth Approaches in Nanopore and Ion Channel Recordings – A Tutorial Review. Anal. Chim. Acta 2019, 1061, 13–27. 10.1016/j.aca.2019.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyser U. F. Controlling Molecular Transport through Nanopores. J. R. Soc., Interface 2011, 8, 1369–1378. 10.1098/rsif.2011.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanunu M.; Meller A. Chemically Modified Solid-State Nanopores. Nano Lett. 2007, 7, 1580–1585. 10.1021/nl070462b. [DOI] [PubMed] [Google Scholar]
- Pud S.; Chao S. H.; Belkin M.; Verschueren D.; Huijben T.; Van Engelenburg C.; Dekker C.; Aksimentiev A. Mechanical Trapping of DNA in a Double-Nanopore System. Nano Lett. 2016, 16, 8021–8028. 10.1021/acs.nanolett.6b04642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa T.; Meller A. Optical Sensing and Analyte Manipulation in Solid-State Nanopores. Analyst 2015, 140, 4733–4747. 10.1039/C4AN02388A. [DOI] [PubMed] [Google Scholar]
- Yamazaki H.; Hu R.; Zhao Q.; Wanunu M. Photothermally Assisted Thinning of Silicon Nitride Membranes for Ultrathin Asymmetric Nanopores. ACS Nano 2018, 12, 12472–12481. 10.1021/acsnano.8b06805. [DOI] [PubMed] [Google Scholar]
- Weckman N. E.; Ermann N.; Gutierrez R.; Chen K.; Graham J.; Tivony R.; Heron A.; Keyser U. F. Multiplexed DNA Identification Using Site Specific DCas9 Barcodes and Nanopore Sensing. ACS Sensors 2019, 4, 2065–2072. 10.1021/acssensors.9b00686. [DOI] [PubMed] [Google Scholar]
- Chen K.; Juhasz M.; Gularek F.; Weinhold E.; Tian Y.; Keyser U. F.; Bell N. A. W. Ionic Current-Based Mapping of Short Sequence Motifs in Single DNA Molecules Using Solid-State Nanopores. Nano Lett. 2017, 17, 5199–5205. 10.1021/acs.nanolett.7b01009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.; Kong J.; Zhu J.; Ermann N.; Predki P.; Keyser U. F. Digital Data Storage Using DNA Nanostructures and Solid-State Nanopores. Nano Lett. 2019, 19, 1210. 10.1021/acs.nanolett.8b04715. [DOI] [PubMed] [Google Scholar]
- Bell N. A. W.; Keyser U. F. Digitally Encoded DNA Nanostructures for Multiplexed, Single-Molecule Protein Sensing with Nanopores. Nat. Nanotechnol. 2016, 11, 645–651. 10.1038/nnano.2016.50. [DOI] [PubMed] [Google Scholar]
- Laszlo A. H.; Derrington I. M.; Gundlach J. H. MspA Nanopore as a Single-Molecule Tool: From Sequencing to SPRNT. Methods 2016, 105, 75–89. 10.1016/j.ymeth.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L.; Hobaugh M. R.; Shustak C.; Cheley S.; Bayley H.; Gouaux J. E. Structure of Staphylococcal α-Hemolysin, a Heptameric Transmembrane Pore. Science 1996, 274, 1859–1866. 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- Menestrina G. Ionic Channels Formed By Staphylococcus Aureus Alpha-Toxin: Voltage-Dependent Inhibition By Divalent and Trivalent Cations. J. Membr. Biol. 1986, 90, 177–190. 10.1007/BF01869935. [DOI] [PubMed] [Google Scholar]
- Wloka C.; Mutter N. L.; Soskine M.; Maglia G. Alpha-Helical Fragaceatoxin C Nanopore Engineered for Double-Stranded and Single-Stranded Nucleic Acid Analysis. Angew. Chem., Int. Ed. 2016, 55, 12494–12498. 10.1002/anie.201606742. [DOI] [PubMed] [Google Scholar]
- Huang G.; Willems K.; Soskine M.; Wloka C.; Maglia G. Electro-Osmotic Capture and Ionic Discrimination of Peptide and Protein Biomarkers with FraC Nanopores. Nat. Commun. 2017, 8, 935. 10.1038/s41467-017-01006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf M.; Lihter M.; Thakur M.; Georgiou V.; Topolancik J.; Ilic B. R.; Liu K.; Feng J.; Astier Y.; Radenovic A. Fabrication and Practical Applications of Molybdenum Disulfide Nanopores. Nat. Protoc. 2019, 14, 1130–1168. 10.1038/s41596-019-0131-0. [DOI] [PubMed] [Google Scholar]
- Montal M.; Mueller P. Formation of Bimolecular Membranes from Lipid Monolayers and a Study of Their Electrical Properties. Proc. Natl. Acad. Sci. U. S. A. 1972, 69, 3561–3566. 10.1073/pnas.69.12.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakmann B.; Neher E.. Single-Channel Recording; Kluwer Academic/Plenum Publishers: New York, 2009. [Google Scholar]
- Tabard-Cossa V.; Joshua B. E.; Tim A.. Instrumentation for Low-Noise High-Bandwidth Nanopore Recording. In Engineered Nanopores for Bioanalytical Applications: A Volume in Micro and Nano Technologies; Elsevier Inc., Waltham, 2013; pp 59–93. [Google Scholar]
- Milotti E.1/F Noise: A Pedagogical Review. arXiv 0204033. https://arxiv.org/abs/physics/0204033 (accessed April 12, 2002).
- Dutta P.; Horn P. M. Low-Frequency Fluctuations in Solids: 1/F Noise. Rev. Mod. Phys. 1981, 53, 497–516. 10.1103/RevModPhys.53.497. [DOI] [Google Scholar]
- Zhang D.; Solomon P.; Zhang S. L.; Zhang Z. An Impedance Model for the Low-Frequency Noise Originating from the Dynamic Hydrogen Ion Reactivity at the Solid/Liquid Interface. Sens. Actuators, B 2018, 254, 363–369. 10.1016/j.snb.2017.07.054. [DOI] [Google Scholar]
- Jindal R. P.; Van Der Ziel A. Model for Mobility Fluctuation 1/F Noise. Appl. Phys. Lett. 1981, 38, 290–291. 10.1063/1.92310. [DOI] [Google Scholar]
- Hooge F. N. 1/f noise. Physica B+C (Amsterdam) 1976, 83, 14–23. 10.1016/0378-4363(76)90089-9. [DOI] [Google Scholar]
- Smeets R. M. M.; Keyser U. F.; Wu M. Y.; Dekker N. H.; Dekker C. Nanobubbles in Solid-State Nanopores. Phys. Rev. Lett. 2006, 97, 088101. 10.1103/PhysRevLett.97.088101. [DOI] [PubMed] [Google Scholar]
- Wen C.; Zeng S.; Arstila K.; Sajavaara T.; Zhu Y.; Zhang Z.; Zhang S. L. Generalized Noise Study of Solid-State Nanopores at Low Frequencies. ACS Sensors 2017, 2, 300–307. 10.1021/acssensors.6b00826. [DOI] [PubMed] [Google Scholar]
- Heerema S. J.; Schneider G. F.; Rozemuller M.; Vicarelli L.; Zandbergen H. W.; Dekker C. 1/F Noise in Graphene Nanopores. Nanotechnology 2015, 26, 074001 10.1088/0957-4484/26/7/074001. [DOI] [PubMed] [Google Scholar]
- Zhang Z.-Y.; Deng Y.-S.; Tian H.-B.; Yan H.; Cui H.-L.; Wang D.-Q. Noise Analysis of Monolayer Graphene Nanopores. Int. J. Mol. Sci. 2018, 19, 2639. 10.3390/ijms19092639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohnsland F.; Benz R. 1/F-Noise of Open Bacterial Porin Channels. J. Membr. Biol. 1997, 158, 77–85. 10.1007/s002329900245. [DOI] [PubMed] [Google Scholar]
- Bezrukov S. M.; Winterhalter M. Examining Noise Sources at the Single-Molecule Level: 1/F Noise of an Open Maltoporin Channel. Phys. Rev. Lett. 2000, 85, 202–205. 10.1103/PhysRevLett.85.202. [DOI] [PubMed] [Google Scholar]
- Smeets R. M. M.; Keyser U. F.; Dekker N. H.; Dekker C. Noise in Solid-State Nanopores. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 417–421. 10.1073/pnas.0705349105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets R. M. M.; Dekker N. H.; Dekker C. Low-Frequency Noise in Solid-State Nanopores. Nanotechnology 2009, 20, 095501 10.1088/0957-4484/20/9/095501. [DOI] [PubMed] [Google Scholar]
- Fragasso A.; Pud S.; Dekker C. 1/F Noise in Solid-State Nanopores Is Governed By Access and Surface Regions. Nanotechnology 2019, 30, 395202. 10.1088/1361-6528/ab2d35. [DOI] [PubMed] [Google Scholar]
- Tasserit C.; Koutsioubas A.; Lairez D.; Zalczer G.; Clochard M. C. Pink Noise of Ionic Conductance through Single Artificial Nanopores Revisited. Phys. Rev. Lett. 2010, 105, 260602. 10.1103/PhysRevLett.105.260602. [DOI] [PubMed] [Google Scholar]
- Nestorovich E. M.; Rostovtseva T. K.; Bezrukov S. M. Residue Ionization and Ion Transport through OmpF Channels. Biophys. J. 2003, 85, 3718–3729. 10.1016/S0006-3495(03)74788-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasianowicz J. J.; Bezrukov S. M. Current Noise Reveals Protonation Kinetics and Number of Ionizable Sites in an Open Protein Ion Channel. Phys. Rev. Lett. 1993, 70, 2352–2355. 10.1103/PhysRevLett.70.2352. [DOI] [PubMed] [Google Scholar]
- Kasianowicz J. J.; Bezrukov S. M. Protonation Dynamics of the Alpha-Toxin Ion Channel from Spectral Analysis of PH-Dependent Current Fluctuations. Biophys. J. 1995, 69, 94–105. 10.1016/S0006-3495(95)79879-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlup S. Noise in Semiconductors: Spectrum of a Two-Parameter Random Signal. J. Appl. Phys. 1954, 25, 341–343. 10.1063/1.1721637. [DOI] [Google Scholar]
- Johnson J. B. Thermal Agitation of Electricity in Conductors. Phys. Rev. 1928, 32, 97–109. 10.1103/PhysRev.32.97. [DOI] [Google Scholar]
- Nyquist H. Thermal Agitation of Electric Charge in Conductors. Phys. Rev. 1928, 32, 110–113. 10.1103/PhysRev.32.110. [DOI] [Google Scholar]
- Blanter Y. M.; Büttiker M. Shot Noise in Mesoscopic Conductors. Phys. Rep. 2000, 336, 1–166. 10.1016/S0370-1573(99)00123-4. [DOI] [Google Scholar]
- Schottky W. Über Spontane Stromschwankungen in Verschiedenen Elektrizitätsleitern. Ann. Phys. 1918, 362, 541–567. 10.1002/andp.19183622304. [DOI] [Google Scholar]
- Chen L. F.; Ong C. K.; Neo C. P.; Varadan V. V.; Varadan V. K.. Microwave Electronics: Measurement and Materials Characterization; John Wiley & Sons Ltd: Chichester, 2004. [Google Scholar]
- Levis R. A.; Rae J. L. The Use of Quartz Patch Pipettes for Low Noise Single Channel Recording. Biophys. J. 1993, 65, 1666–1677. 10.1016/S0006-3495(93)81224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uram J. D.; Ke K.; Mayer M. L. Noise and Bandwidth of Current Recordings from Submicrometer Pores and Nanopores. ACS Nano 2008, 2, 857–872. 10.1021/nn700322m. [DOI] [PubMed] [Google Scholar]
- Sherman-Gold R.The Axon CNS Guide to Electrophysiology and Biophysics Laboratory Techniques; Molecular Devices, LLC: Sunnyvale, 2012. [Google Scholar]
- Ayub M.; Bayley H. Engineered Transmembrane Pores. Curr. Opin. Chem. Biol. 2016, 34, 117–126. 10.1016/j.cbpa.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z.; Jarrold M. F.; Williams R.; Ludtke S. J.; Echeverria I.; Slaughter B. D.; Chemmama I. E.; Unruh J. R.; Mironska R.; de la Cruz M. J.; Hogan J. A.; Shivaraju M.; Chaudhury A. S.; Raveh B.; Wang J.; Pellarin R.; Upla P.; Kim S. J.; Herricks T.; Nudelman I.; et al. Integrative Structure and Functional Anatomy of a Nuclear Pore Complex. Nature 2018, 555, 475–482. 10.1038/nature26003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J. W. F; Joshua B. E.; Tim A.. Biological Pores on Lipid Bilayers. In Engineered Nanopores for Bioanalytical Applications: A Vol. in Micro and Nano Technologies; Elsevier Inc.: Waltham, 2013. [Google Scholar]
- Mayer M.; Kriebel J. K.; Tosteson M. T.; Whitesides G. M. Microfabricated Teflon Membranes for Low-Noise Recordings of Ion Channels in Planar Lipid Bilayers. Biophys. J. 2003, 85, 2684–2695. 10.1016/S0006-3495(03)74691-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekolla S.; Andersen C.; Benz R. Noise Analysis of Ion Current through the Open and the Sugar-Induced Closed State of the LamB Channel of Escherichia Coli Outer Membrane: Evaluation of the Sugar Binding Kinetics to the Channel Interior. Biophys. J. 1994, 66, 1388–1397. 10.1016/S0006-3495(94)80929-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Läuger P. Structural Fluctuations and Current Noise of Ionic Channels. Biophys. J. 1985, 48, 369–373. 10.1016/S0006-3495(85)83793-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akeson M.; Brandin E.; Branton D.; Deamer D. W.; Kasianowicz J. J. Microsecond Time-Scale Discrimination Among Polycytidylic Acid, Polyadenylic Acid, and Polyuridylic Acid as Homopolymers or as Segments Within Single RNA Molecules. Biophys. J. 1999, 77, 3227–3233. 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S.; Yoo J.; Aksimentiev A. Water Mediates Recognition of DNA Sequence via Ionic Current Blockade in a Biological Nanopore. ACS Nano 2016, 10, 4644–4651. 10.1021/acsnano.6b00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb T.; Ayub M.. Solid-State Nanopore Fabrication. Engineered Nanopores for Bioanalytical Applications: A Vol. in Micro and Nano Technologies; Elsevier Inc.: Waltham, 2013; pp 121–140. [Google Scholar]
- Gilbert S. M.; Dunn G.; Azizi A.; Pham T.; Shevitski B.; Dimitrov E.; Liu S.; Aloni S.; Zettl A. Fabrication of Subnanometer-Precision Nanopores in Hexagonal Boron Nitride. Sci. Rep. 2017, 7, 15096. 10.1038/s41598-017-12684-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H.; Kumar A.; Park K. B.; Cho S. Y.; Kim H. M.; Lim M. C.; Kim Y. R.; Kim K. B. A Low-Noise Solid-State Nanopore Platform Based on a Highly Insulating Substrate. Sci. Rep. 2015, 4, 7448. 10.1038/srep07448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitchford W. H.; Kim H. J.; Ivanov A. P.; Kim H. M.; Yu J. S.; Leatherbarrow R. J.; Albrecht T.; Kim K. B.; Edel J. B. Synchronized Optical and Electronic Detection of Biomolecules Using a Low Noise Nanopore Platform. ACS Nano 2015, 9, 1740–1748. 10.1021/nn506572r. [DOI] [PubMed] [Google Scholar]
- Storm A. J.; Chen J. H.; Ling X. S.; Zandbergen H. W.; Dekker C. Fabrication of Solid-State Nanopores with Single-Nanometre Precision. Nat. Mater. 2003, 2, 537–540. 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- Van Den Hout M.; Hall A. R.; Wu M. Y.; Zandbergen H. W.; Dekker C.; Dekker N. H. Controlling Nanopore Size, Shape and Stability. Nanotechnology 2010, 21, 115304. 10.1088/0957-4484/21/11/115304. [DOI] [PubMed] [Google Scholar]
- Lanyon Y. H.; De Marzi G.; Watson Y. E.; Quinn A. J.; Gleeson J. P.; Redmond G.; Arrigan D. W. M. Fabrication of Nanopore Array Electrodes By Focused Ion Beam Milling. Anal. Chem. 2007, 79, 3048–3055. 10.1021/ac061878x. [DOI] [PubMed] [Google Scholar]
- Schiedt B.; Auvray L.; Bacri L.; Oukhaled G.; Madouri A.; Bourhis E.; Patriarche G.; Pelta J.; Jede R.; Gierak J. Direct FIB Fabrication and Integration of “Single Nanopore Devices” for the Manipulation of Macromolecules. Microelectron. Eng. 2010, 87, 1300–1303. 10.1016/j.mee.2009.12.073. [DOI] [Google Scholar]
- Verschueren D. V.; Yang W.; Dekker C. Lithography-Based Fabrication of Nanopore Arrays in Freestanding SiN and Graphene Membranes. Nanotechnology 2018, 29, 145302. 10.1088/1361-6528/aaabce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa T.; Zvuloni E.; Zrehen A.; Squires A. H.; Meller A. Automated, Ultra-Fast Laser-Drilling of Nanometer Scale Pores and Nanopore Arrays in Aqueous Solutions. Adv. Funct. Mater. 2019, 1900642, 1900642. 10.1002/adfm.201900642. [DOI] [Google Scholar]
- Gilboa T.; Zrehen A.; Girsault A.; Meller A. Optically-Monitored Nanopore Fabrication Using a Focused Laser Beam. Sci. Rep. 2018, 8, 9765. 10.1038/s41598-018-28136-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pud S.; Verschueren D.; Vukovic N.; Plesa C.; Jonsson M. P.; Dekker C. Self-Aligned Plasmonic Nanopores By Optically Controlled Dielectric Breakdown. Nano Lett. 2015, 15, 7112–7117. 10.1021/acs.nanolett.5b03239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok H.; Briggs K.; Tabard-Cossa V. Nanopore Fabrication By Controlled Dielectric Breakdown. PLoS One 2014, 9, e92880 10.1371/journal.pone.0092880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper J. D.; Clarke R. W.; Korchev Y. E.; Ying L.; Klenerman D. A Renewable Nanosensor Based on a Glass Nanopipette. J. Am. Chem. Soc. 2006, 128, 16462–16463. 10.1021/ja0650899. [DOI] [PubMed] [Google Scholar]
- Xu X.; Li C.; Zhou Y.; Jin Y. Controllable Shrinking of Glass Capillary Nanopores Down to Sub-10 Nm By Wet-Chemical Silanization for Signal-Enhanced DNA Translocation. ACS Sensors 2017, 2, 1452–1457. 10.1021/acssensors.7b00385. [DOI] [PubMed] [Google Scholar]
- Bafna J. A.; Soni G. V. Fabrication of Low Noise Borosilicate Glass Nanopores for Single Molecule Sensing. PLoS One 2016, 11, e0157399 10.1371/journal.pone.0157399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabard-Cossa V.; Trivedi D.; Wiggin M.; Jetha N. N.; Marziali A. Noise Analysis and Reduction in Solid-State Nanopores. Nanotechnology 2007, 18, 305505. 10.1088/0957-4484/18/30/305505. [DOI] [Google Scholar]
- Vandamme L.K.J.; Xiaosong Li; Rigaud D. 1/F Noise in MOS Devices, Mobility or Number Fluctuations?. IEEE Trans. Electron Devices 1994, 41, 1936–1945. 10.1109/16.333809. [DOI] [Google Scholar]
- Garaj S.; Liu S.; Golovchenko J. A.; Branton D. Molecule-Hugging Graphene Nanopores. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 12192–12196. 10.1073/pnas.1220012110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelen Z.; Bustamante J. A.; Carlsen A.; Baker-Murray A.; Tabard-Cossa V. Instrumentation for Low Noise Nanopore-Based Ionic Current Recording under Laser Illumination. Rev. Sci. Instrum. 2018, 89, 015007 10.1063/1.5006262. [DOI] [PubMed] [Google Scholar]
- Rosenstein J. K.; Ramakrishnan S.; Roseman J.; Shepard K. L. Single Ion Channel Recordings with CMOS-Anchored Lipid Membranes. Nano Lett. 2013, 13, 2682–2686. 10.1021/nl400822r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler T. Z.; Pavlenok M.; Derrington I. M.; Niederweis M.; Gundlach J. H. Single-Molecule DNA Detection with an Engineered MspA Protein Nanopore. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 20647–20652. 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrao E.; Butler T. Z.; Pavlenok M.; Collins M. D.; Niederweis M.; Gundlach J. H.; Derrington I. M. Nanopore DNA Sequencing with MspA. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 16060–16065. 10.1073/pnas.1001831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlin M.; Kotnik T.; Miklavčič D.; Kramar P.; Maček Lebar A.; Iglič A.. Electroporation of Planar Lipid Bilayers and Membranes. In Advances in Planar Lipid Bilayers and Liposomes; Elsevier Inc.: Oxford, 2012; Vol. 6, pp 165–226. [Google Scholar]
- Tarek M. Membrane Electroporation: A Molecular Dynamics Simulation. Biophys. J. 2005, 88, 4045–4053. 10.1529/biophysj.104.050617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M.; Koren S.; Miga K. H.; Quick J.; Rand A. C.; Sasani T. A.; Tyson J. R.; Beggs A. D.; Dilthey A. T.; Fiddes I. T.; Malla S.; Marriott H.; Nieto T.; O’Grady J.; Olsen H. E.; Pedersen B. S.; Rhie A.; Richardson H.; Quinlan A. R.; Snutch T. P.; et al Nanopore Sequencing and Assembly of a Human Genome with Ultra-Long Reads. Nat. Biotechnol. 2018, 36, 338–345. 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M.; Olsen H. E.; Paten B.; Akeson M. The Oxford Nanopore MinION: Delivery of Nanopore Sequencing to the Genomics Community. Genome Biol. 2016, 17, 256. 10.1186/s13059-016-1122-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. M.; Kong D. L.; Chen Q.; Xue J. M. Surface Engineering of Synthetic Nanopores By Atomic Layer Deposition and Their Applications. Front. Mater. Sci. 2013, 7, 335–349. 10.1007/s11706-013-0218-4. [DOI] [Google Scholar]
- Nilsson J.; Lee J. R. I.; Ratto T. V.; Létant S. E. Localized Functionalization of Single Nanopores. Adv. Mater. 2006, 18, 427–431. 10.1002/adma.200501991. [DOI] [Google Scholar]
- Danelon C.; Santschi C.; Brugger J.; Vogel H. Fabrication and Functionalization of Nanochannels By Electron-Beam-Induced Silicon Oxide Deposition. Langmuir 2006, 22, 10711–10715. 10.1021/la061321c. [DOI] [PubMed] [Google Scholar]
- Chen P.; Mitsui T.; Farmer D. B.; Golovchenko J.; Gordon R. G.; Branton D. Atomic Layer Deposition to Fine-Tune the Surface Properties and Diameters of Fabricated Nanopores. Nano Lett. 2004, 4, 1333–1337. 10.1021/nl0494001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beamish E.; Kwok H.; Tabard-Cossa V.; Godin M. Precise Control of the Size and Noise of Solid-State Nanopores Using High Electric Fields. Nanotechnology 2012, 23, 405301. 10.1088/0957-4484/23/40/405301. [DOI] [PubMed] [Google Scholar]
- Kosmulski M. The PH-Dependent Surface Charging and the Points of Zero Charge. J. Colloid Interface Sci. 2002, 253, 77–87. 10.1006/jcis.2002.8490. [DOI] [PubMed] [Google Scholar]
- Firnkes M.; Pedone D.; Knezevic J.; Döblinger M.; Rant U. Electrically Facilitated Translocations of Proteins through Silicon Nitride Nanopores: Conjoint and Competitive Action of Diffusion, Electrophoresis, and Electroosmosis. Nano Lett. 2010, 10, 2162–2167. 10.1021/nl100861c. [DOI] [PubMed] [Google Scholar]
- Waduge P.; Larkin J.; Upmanyu M.; Kar S.; Wanunu M. Programmed Synthesis of Freestanding Graphene Nanomembrane Arrays. Small 2015, 11, 597–603. 10.1002/smll.201402230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waduge P.; Bilgin I.; Larkin J.; Henley R. Y.; Goodfellow K.; Graham A. C.; Bell D. C.; Vamivakas N.; Kar S.; Wanunu M. Direct and Scalable Deposition of Atomically Thin Low-Noise MoS2Membranes on Apertures. ACS Nano 2015, 9, 7352–7359. 10.1021/acsnano.5b02369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley H.; Cronin B.; Heron A.; Holden M. A.; Hwang W. L.; Syeda R.; Thompson J.; Wallace M. Droplet Interface Bilayers RID B-8725–2008. Mol. BioSyst. 2008, 4, 1191–1208. 10.1039/b808893d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitta M.; Tanaka H.; Kawai T. Rapid Fabrication of Teflon Micropores for Artificial Lipid Bilayer Formation. Biosens. Bioelectron. 2009, 25, 931–934. 10.1016/j.bios.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Gornall J. L.; Mahendran K. R.; Pambos O. J.; Steinbock L. J.; Otto O.; Chimerel C.; Winterhalter M.; Keyser U. F. Simple Reconstitution of Protein Pores in Nano Lipid Bilayers. Nano Lett. 2011, 11, 3334–3340. 10.1021/nl201707d. [DOI] [PubMed] [Google Scholar]
- Criado M.; Keller B. U. A Membrane Fusion Strategy for Single-Channel Recordings of Membranes Usually Non-Accessible to Patch-Clamp Pipette Electrodes. FEBS Lett. 1987, 224, 172–176. 10.1016/0014-5793(87)80442-8. [DOI] [PubMed] [Google Scholar]
- Riquelme G.; Lopez E.; Garcia-segura L. M.; Ferragut J. A.; Gonzalez-ros J. M. Giant Liposomes: A Model System in Which To Obtain Patch-Clamp Recordings. Biochemistry 1990, 29, 11215–11222. 10.1021/bi00503a009. [DOI] [PubMed] [Google Scholar]
- Hartel A. J. W.; Ong P.; Schroeder I.; Giese M. H.; Shekar S.; Clarke O. B.; Zalk R.; Marks A. R.; Hendrickson W. A.; Shepard K. L. Single-Channel Recordings of RyR1 at Microsecond Resolution in CMOS-Suspended Membranes. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, E1789–E1798. 10.1073/pnas.1712313115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder I. How to Resolve Microsecond Current Fluctuations in Single Ion Channels: The Power of Beta Distributions. Channels 2015, 9, 262–280. 10.1080/19336950.2015.1083660. [DOI] [PMC free article] [PubMed] [Google Scholar]