Abstract
Introduction:
Enterococci are now recognized as the second most cause of nosocomial infections worldwide. The emergence of multidrug-resistant strains in the organism has given rise to alternative strategies such as phage therapy. In this study, an Enterococcus faecalis infecting phage was isolated and its efficiency against biofilms formed by drug-resistant enterococci obtained from chronic periodontitis was evaluated.
Materials and Methods:
Bacteriophage against E. faecalis was isolated from sewage sample. The phage was propagated and identified using transmission electron microscope (TEM). In vitro biofilm formation was assessed.
Results:
TEM microscopy showed that the phage belonged to Siphoviridae family. In the presence of the novel phage, the metabolic activity of enterococci biofilm was reduced at 48 h of contact. A difference of at least 5 log CFU/ml was seen in the live cells of the control biofilm, and the phage treated biofilm of enterococci isolates.
Conclusion:
The study shows that the novel phage inhibits biofilm production in oral enterococci isolates from periodontitis patients but has a narrow host range. The role of bacteriophages as strong biotechnological and natural therapeutic agents for E. faecalis in chronic periodontitis can be considered.
Keywords: Bacteriophage, biofilm, enterococci, periodontitis
INTRODUCTION
Enterococci are Gram-positive facultative anaerobic bacteria initially regarded as nonvirulent but are now recognized as one of the major causes of nosocomial infection.[1,2] The two most common enterococci species are Enterococcus faecalis and Enterococcus faecium. Both the species readily produce biofilm, which is a population of cells surrounded by a matrix of macromolecules such as polysaccharides, proteins, lipids, and extracellular DNA.[3]
Periodontitis is one of the infections induced by oral biofilms. It is a biofilm mediated disease, which is refractory to antibiotic agents and host defenses.[4] Enhanced biofilm resistance to antibiotic includes decreased antibiotic penetration, trapping of antibiotics, and presence of persister cells.[5,6,7]E. faecalis has been recovered from periodontal pockets in 1%–51.8% of chronic periodontitis patients.[8,9,10] Enterococci infections are difficult to treat as many strains have been found to be antibiotic resistant.[11,12] The surviving ability of E. faecalis in the starvation phase and the biofilm formation capability in alkaline environment make it a difficult organism to eradicate.[13] Newly acquired vancomycin-resistant genes producing resistant phenotypes have been reported in E. faecalis and E. faecium.[14,15]
Phage therapy has recently begun to prove superior to conventional chemotherapy.[16,17,18] Phages also have no harmful effect on the nontarget microbiome.[19] Biofilm destruction by phages is much more efficient as compared to antibiotics as phages can replicate and penetrate the inner layers of the biofilm.[20] Oral isolates of E. faecalis have been found to be infected by lytic and temperate phages.[21,22] Phage therapy trial using E. faecalis infecting phage in a human root canal model has been attempted and a substantial reduction in bacterial cell viability seen.[23] The efficacy of a genetically engineered phage on E. faecalis biofilm has also been seen in a recent study. The biomass of vancomycin sensitive and vancomycin-resistant E. faecalis biofilms is markedly reduced following infection with the genetically engineered phage.[24] Passage of phage through the E. faecalis strain JH2-2 harboring a defective prophage produced a new strain with more antimicrobial efficacy.[25] Using bacteriophages singly, in cocktail or genetically engineering them, could be a promising approach for treating biofilms formed by bacteria. However, these are a few attempts which have been made to treat E. faecalis infection using phage therapy and no such study has been reported for treating enterococci infection by phages in chronic periodontitis.
In this study, an E. faecalis infecting phage was isolated from sewage, and its efficacy against biofilms formed by enterococci isolates obtained from chronic periodontitis patients was evaluated. The drug-resistant oral enterococci isolates of chronic periodontitis patients were from our previous study.[26] This cross-sectional study had observed a highly significant association of oral drug-resistant enterococci with biofilm formation.[26]
MATERIALS AND METHODS
This study was conducted at the Department of Microbiology in Dr. Harvansh Singh Judge Institute of Dental Sciences and Hospital, Panjab University, Chandigarh, from June 2016 to January 2017. In our previous study entitled “Biofilm formation by drug-resistant enterococci isolates obtained from chronic periodontitis patients” 46 enterococci were isolated from 100 patients (52 males and 48 females). The study was approved by the Ethics Committee of the Panjab University. The patients were informed of the study protocol and written consent was obtained as per the Indian Council of Medical Education and Research guidelines. Briefly, the bacterial sample was collected from the subgingival sites and two sterile paper points were inserted into each periodontal pocket for 15–30 s. The paper points were pooled and transferred immediately in a sterile transport medium (glucose azide broth, HiMedia Laboratories, Mumbai) and taken to the laboratory for microbiological analysis. The samples were inoculated onto the 5% defibrinated sheep blood agar (HiMedia Laboratories, Mumbai) and incubated in microaerophilic condition overnight. Every growth showing Gram-positive cocci, positive bile esculin, positive 6.5% NaCl test, catalase-negative was processed. Enterococci speciation was done by motility tests, pigmentation (on tryptic soy agar) and 1% carbohydrate fermentation tests (mannitol, sorbitol, arabinose, lactose, and glucose) as per standard microbiological guidelines.[27] Standard antibiotic discs of amoxicillin (10 μg), ciprofloxacin (10 μg), erythromycin (15 μg), vancomycin (30 μg), gentamycin (10 μg), and teicoplanin (30 μg) were obtained from HiMedia Laboratories, Mumbai, India. The antibiotic susceptibility of the test strains to different antibiotics was determined by Kirby-Bauer disc diffusion susceptibility testing technique of the Clinical and laboratory standards Institute.[28] The quality control strain used was E. faecalis (ATCC 29212). The evaluation of biofilm formation was done by standard protocol using microtiter plate method.[29] The isolates were then cultured in nutrient agar stabs and stored at 4°C for this study. Bacterial strain-E. faecalis (ATCC29212) was grown in brain heart infusion (BHI) broth at 37°C under aerobic conditions. The nutrient agar stabs of bacterial strains stored at 4°C were revived in BHI broth and further used for screening by the phage.
Isolation of the phage
The phage was isolated from the sewage effluent of Panjab University, Chandigarh, by the method of Smith and Huggins.[30] The sewage water (50 ml) was collected in a sterile conical flask and treated with few drops of chloroform. To this, an equal volume of the lactic phage broth (HiMedia Laboratories, Mumbai) and 1 ml of the 24 h old broth culture of E. faecalis ATCC29212 culture was added and incubated at 37°C for 12–24 h in a shaker water bath.[30] After 12–24 h, the lysate was shaken with a few drops of chloroform for about 10 min, centrifuged at 10,000 rpm for 10 min. The supernatant was filtered through a 0.22 um pore size Acrodisc membrane filters (HiMedia Laboratories, Mumbai) to remove the bacteria and subjected to the plaque-forming unit (PFU) assay using the standard double-layer agar method.[31] Briefly, the lysate obtained was diluted in 5 ml BHI broth, plated using soft agar (0.6%) and overlaid with 0.5 ml test (109 CFU/ml) overnight culture and incubated at 37°C for 24 h. Plaque morphology was observed the next day. For the recovery of the phage, a single well-isolated plaque was picked and repeated twice to ensure the isolation of the phage-type. The phage was isolated. The lysate was then stored in BHI with chloroform (40 ml/L) at 4°C–6°C.
Propagation of phage
The concentration of PFU was determined according to the standard method.[31] Lysate was diluted in 10 fold dilutions in 5 ml of BHI soft agar (0.6%). 100 ul of an overnight culture of test strain was added to the tube and put on BHI agar plate. The number of plaques was counted and the initial concentration of 109 plaque-forming particles was obtained.
Transmission electron microscopy visualization
The enterococci phage solution was filtered with the Acrodisc filter to remove soluble biological macromolecule fragments of host bacteria. After washing three times with 0.1M ammonium acetate solution (pH 7.0), the retained phage solution was used directly for negative staining with 2% phosphotungstic acid. A transmission electron microscope (TEM) (FEI Tecnai, G2 F20, Netherlands-analyzed at 120 KV) was used to capture the images. The classification of the phage was done according to the guidelines of the International Committee on Taxonomy of viruses.
Biofilm assays
Microtiter plate assay was performed for analyzing biofilm formation.[29] BHI broth containing 0.25% glucose was inoculated with test strains and incubated at 37°C overnight. The overnight BHI broth cultures supplemented with glucose were diluted 1:20 in fresh BHI broth with glucose. Diluted strain (200 ul) was dispensed into triplicate wells in a single column of a sterile 96 well flat bottom plate and incubated at 37°C for48 h. Microtiter plate was gently tapped, and planktonic cells were removed. Phage was added to the wells at 109 PFU/well, and the plate was incubated for 24 h at 37°C. The microtiter plates were then washed three times with sterile phosphorus buffer saline (PBS). The plates were inverted and dried for 1 h at room temperature. 0.5% aqueous crystal violet solution (200 ul) was added to each well for 15 min. The wells were then washed thrice with sterile PBS to wash off the excess crystal violet. 80:20 (vol/vol) mixtures of ethyl alcohol and acetone (200 ul) was added to solubilized bound crystal violet. The absorbance of the extracted crystal violet was read at 550 nm automatic microplate reader (APW). A biofilm-forming bacteria Staphylococcus epidermidis was taken as positive control and nonbiofilm forming bacteria Salmonella typhi was taken as a negative control. All biofilm assays were repeated three times. The cutoff value (ODc) was established. The final optical density (OD) value of a tested strain was expressed as average OD value of the strain reduced by ODc value of the triplicate assays.[29]
Statistical analysis
SPSS software version 22(IBM corp, Armonk, NY, USA) was used for statistical analysis. T-test and Chi-square test were performed for data analysis. For comparison of biofilm formation with phage and biofilm formation without phage (data were normally distributed) paired t-test was carried out. P < 0.05 was considered to be statistically significant.
RESULTS
Isolation and determination of phage efficacy
E. faecalis phage was isolated from sewage water. The phage had lytic activity, which was seen by clear plaque formation on double-layer agar [Figure 1]. TEM microscopy showed that the phage has a hexagonal head about 73 nm in diameter and a 100 nm long-tail thus being morphologically similar to phages belonging to Siphoviridae family [Figure 2].
Figure 1.
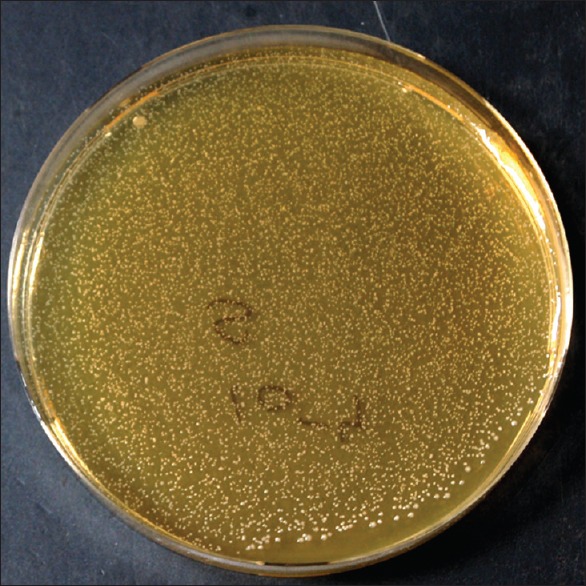
Plaque of phage on double-layered agar
Figure 2.
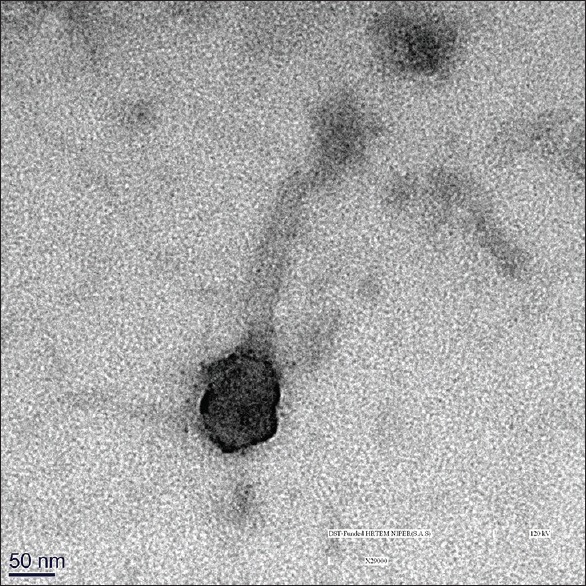
Transmission electron microscopy of the phage
Determination of phage efficacy against biofilms of enterococci
Enterococci phage was tested against 48 h attached enterococci cells in 96 well plates. After phage treatment with an initial dose concentration equal to or higher than 102 PFU/well, phage significantly reduced the biofilm activity within 24 h of contact (P < 0.05) [Figure 3a and b]. A difference of at least 5 log CFU/ml is noticeable in the control biofilm, and the phage treated biofilm. The one-step curve shows a linear line in control biofilm that is no reduction in the biofilm biomass whereas the one step curve of phage treated biofilm shows a significant reduction in the biofilm mass (P < 0.05) whereas biofilm formed by isolates of E. faecium shows no significant reduction in the presence of phage.
Figure 3.
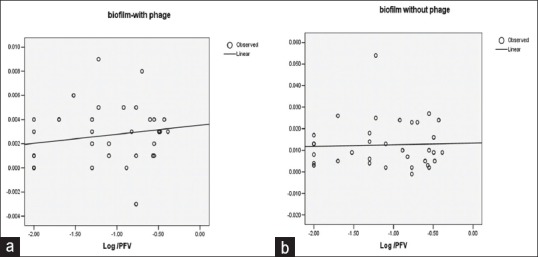
One step growth curve (a) Biofilms of enterococci isolates treated. with phage. (b) Biofilms of enterococci isolates not treated with phage
DISCUSSION
Enterococci are able to colonize the oral cavity, particularly in patients with periodontitis or root canal infections associated with oral mucosal lesions and in immunocompromised patients.[32] It has been reported that patients with periodontitis have a more diverse combination of species as compared to healthy persons.[33]
The phage was isolated from the wastewater. It has been seen that wastewater is a rich source of phage isolation against multidrug-resistant bacteria. A recent study shows that the application of the wastewater isolated phage has promising results in controlling vancomycin-resistant E. faecium in the milk samples.[34] TEM of phage revealed an icosahedral head around 73 nm in diameter and more than 100 nm long tail similar to phages belonging to Siphoviridae family.[35] These phages are lytic infecting Gram-positive bacteria and seem to be a promising candidate against Gram-positive pathogens.[36,37] The host range of phage was limited to E. faecalis and less to E. faecium. This high host specificity is common among phages and is beneficial that the phage does not harm the natural microbiome of the host as antibiotics do. In our previous cross-sectional study 46 enterococci isolates were obtained from subgingival samples collected from seventy periodontitis patients. No enterococci isolate was obtained from control group which consisted of 30 subjects. Based on the identification, the species of enterococci were E. faecalis 39 (84.78%) followed by E. faecium 7 (15.21%).[26] The quantitative microtitre assay for biofilm formation was positive in 39/46 (84.78%) isolates. The remaining isolates were nonbiofilm producers considered as negative. E. faecalis strain positive for biofilm production was (29/39) 71.8% and E. faecium was 25.6% (10/39). Among the enterococci isolates tested for antimicrobial susceptibility by Kirby Bauer disc diffusion method resistance to erythromycin was 3/46 (6.5%), ciprofloxacin 4/46 (8.7%), tiecoplanin 6/46 (13.0%), amoxicillin 2/46 (4.3%), gentamycin 4/46 (8.7%), and vancomycin 6/46 (13.3%).[26]
In this study, the ability of the phage to reduce enterococcal biofilm and growth was tested. The phage efficiently reduced the growth of enterococci by at least 5 log CFU/ml in laboratory broth. Biofilm formation is indirect evidence of adhesiveness and microtiter plate assay is the indirect way to measure adhesion of Enterococci. Survival advantages conferred by the biofilm community include resistance to phagocytosis and antimicrobial agents.[38]
The use of phages could help in reducing the colonization of teeth surface by E. faecalis, particularly the ability of the phage to reduce the biofilm after a minimum 24 h of contact. The reduction of the E. faecalis growth and biofilm formation has been successfully tested by the use of other antimicrobial agents[39,40] probiotic bacteria[41] and a genetic engineered bacteriophage.[42] This could overcome the narrow host range of the phage and thus result in more effective prevention of enterococci biofilm and periodontitis. A recent finding on bacteriophage availability for human application has shown that bacteriophages are more virulent to bacteria in human cells and less in bacterial cultures.[24] This should lead to further clinical research and application of bacteriophages as therapeutic agents in human applications.
The results support that bacteriophage is a promising strategy to conventional antibiotic treatment, particularly in the case of biofilm and multidrug-resistant strains. The phage was active on all enterococci tested, including the vancomycin, ciprofloxacin, erythromycin, teicoplanin, and gentamycin-resistant strains.
CONCLUSION
The narrow host range of enterococci phage is a disadvantage, but it has the ability to reduce enterococci growth and biofilm formation. This study strengthens the view that phage therapy is effective on multidrug-resistant enterococci. If the incidence of vancomycin-resistant enterococci increases, it could be a stand-alone therapy for infections fully resistant to antibiotics like vancomycin.
Research quality and ethics statement
The authors of this manuscript declare that this scientific work complies with reporting quality, formatting, and reproducibility guidelines set forth by the EQUATOR Network. The authors also attest that this clinical investigation was determined to require the Institutional Review Board/Ethics Committee Review, and the corresponding approval number is (PU/IEC/116/8). We also certify that we have not plagiarized the contents in this submission and have done a plagiarism check.
Financial support and sponsorship
The funds for this project were sanctioned by the Department of Science and Technology (DST Letter No S&T/Sanc/08/2016/934-940), India vide grant.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
The funds for this project were sanctioned by the Department of Science and Technology (DST Letter No S&T/Sanc/08/2016/934-940), India, vide grant. The technical help of Ms. Arti is duly acknowledged.
REFERENCES
- 1.Marra A, Dib-Hajj F, Lamb L, Kaczmarek F, Shang W, Beckius G, et al. Enterococcal virulence determinants may be involved in resistance to clinical therapy. Diagn Microbiol Infect Dis. 2007;58:59–65. doi: 10.1016/j.diagmicrobio.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infect Control Hosp Epidemiol. 2013;34:1–4. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 3.Jakubovics NS, Burgess JG. Extracellular DNA in oral microbial biofilms. Microbes Infect. 2015;17:531–7. doi: 10.1016/j.micinf.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Schaudinn C, Gorur A, Keller D, Sedghizadeh PP, Costerton JW. Periodontitis: An archetypical biofilm disease. J Am Dent Assoc. 2009;140:978–86. doi: 10.14219/jada.archive.2009.0307. [DOI] [PubMed] [Google Scholar]
- 5.Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–10. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 6.Lewis K. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol. 2008;322:107–31. doi: 10.1007/978-3-540-75418-3_6. [DOI] [PubMed] [Google Scholar]
- 7.Dale JL, Nilson JL, Barnes AMT, Dunny GM. Restructuring of Enterococcus faecalis biofilm architecture in response to antibiotic-induced stress. NPJ Biofilms Microbiomes. 2017;3:15. doi: 10.1038/s41522-017-0023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soothill JS. Treatment of experimental infections of mice with bacteriophages. J Med Microbiol. 1992;37:258–61. doi: 10.1099/00222615-37-4-258. [DOI] [PubMed] [Google Scholar]
- 9.Balaei-Gajan E, Shirmohammadi A, Abashov R, Agazadeh M, Faramarzie M. Detection of Enterococcus faecalis in subgingival biofilm of patients with chronic refractory periodontitis. Med Oral Patol Oral Cir Bucal. 2010;15:e667–70. doi: 10.4317/medoral.15.e667. [DOI] [PubMed] [Google Scholar]
- 10.Sun J, Song X. Assesment of antimicrobial susceptibility of E. faecalis isolated from chronic periodontitis biofilms vs planktonic phase. J Periodontol. 2011;82:513–655. doi: 10.1902/jop.2010.100378. [DOI] [PubMed] [Google Scholar]
- 11.Miller WR, Murray BE, Rice LB, Arias CA. Vancomycin-Resistant Enterococci: therapeutic challenges in the 21st century. Infect Dis Clin North Am. 2016;30:415–39. doi: 10.1016/j.idc.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Liu H, Wei X, Ling J, Wang W, Huang X. Biofilm formation capability of Enterococcus faecalis cells in starvation phase and its susceptibility to sodium hypochlorite. J Endod. 2010;36:630–5. doi: 10.1016/j.joen.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 13.Palmer SM, Rybak MJ. Vancomycin-resistant enterococci. Pharmacotherapy. 1996;16:819–29. [PubMed] [Google Scholar]
- 14.Talebi M, Asghari Moghadam N, Mamooii Z, Enayati M, Saifi M, Pourshafie MR. Antibiotic resistance and biofilm formation of Enterococcus faecalis in patient and environmental samples. Jundishapur J Microbiol. 2015;8:e23349. doi: 10.5812/jjm.23349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox JL. The business of developing antibacterials. Nat Biotechnol. 2006;24:1521–8. doi: 10.1038/nbt1206-1521. [DOI] [PubMed] [Google Scholar]
- 16.Shlezinger M, Khalifa L, Houri-Haddad Y, Coppenhagen-Glazer S, Resch G, Que YA, et al. Phage therapy: A new horizon in the antibacterial treatment of oral pathogens. Curr Top Med Chem. 2017;17:1199–211. doi: 10.2174/1568026616666160930145649. [DOI] [PubMed] [Google Scholar]
- 17.Marks T, Sharp R. Bacteriophages and biotechnology: A review. J Chem Technol Biotechnol. 2000;75:6–17. [Google Scholar]
- 18.Szafrański SP, Winkel A, Stiesch M. The use of bacteriophages to biocontrol oral biofilms. J Biotechnol. 2017;250:29–44. doi: 10.1016/j.jbiotec.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM. Phage treatment of human infections. Bacteriophage. 2011;1:66–85. doi: 10.4161/bact.1.2.15845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutiérrez D, Vandenheuvel D, Martínez B, Rodríguez A, Lavigne R, García P. Two Phages, phiIPLA-RODI and phiIPLA-C1C, Lyse mono- and dual-Species Staphylococcal Biofilms. Appl Environ Microbiol. 2015;81:3336–48. doi: 10.1128/AEM.03560-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachrach G, Leizerovici-Zigmond M, Zlotkin A, Naor R, Steinberg D. Bacteriophage isolation from human saliva. Lett Appl Microbiol. 2003;36:50–3. doi: 10.1046/j.1472-765x.2003.01262.x. [DOI] [PubMed] [Google Scholar]
- 22.Stevens RH, Porras OD, Delisle AL. Bacteriophages induced from lysogenic root canal isolates of Enterococcus faecalis. Oral Microbiol Immunol. 2009;24:278–84. doi: 10.1111/j.1399-302X.2009.00506.x. [DOI] [PubMed] [Google Scholar]
- 23.Khalifa L, Brosh Y, Gelman D, Coppenhagen-Glazer S, Beyth S, Poradosu-Cohen R, et al. Targeting Enterococcus faecalis biofilms with phage therapy. Appl Environ Microbiol. 2015;81:2696–705. doi: 10.1128/AEM.00096-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tinoco JM, Buttaro B, Zhang H, Liss N, Sassone L, Stevens R. Effect of a genetically engineered bacteriophage on Enterococcus faecalis biofilms. Arch Oral Biol. 2016;71:80–6. doi: 10.1016/j.archoralbio.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Fouts DE, DePew J, Stevens RH. Genetic modifications to temperate Enterococcus faecalis phage Ef11 that abolish the establishment of lysogeny and sensitivity to repressor, and increase host range and productivity of lytic infection. Microbiology. 2013;159:1023–35. doi: 10.1099/mic.0.067116-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhardwaj SB, Mehta M, Sood S, Sharma J. Biofilm Formation by Drug Resistant Enterococci Isolates Obtained from Chronic Periodontitis Patients. J Clin Diagn Res. 2017;11:DC01–3. doi: 10.7860/JCDR/2017/24472.9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teixeira LM, Facklam RR. Enterococcus. In: Murray PR, Baron EJ, Jorgensen JH, PFaller MA, Yolken RH, editors. Manual of Clinical Microbiology. 8th ed. Washington: ASM Press; 2003. pp. 422–433. [Google Scholar]
- 28.Performance Standards for Antimicrobial Susceptibility Testing: M100-S23. Wayne, Pennsylvania, USA: Clinical and Laboratory Standards Institute; 2013. Clinical and Laboratory Standards Institute. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, et al. The enterococcal surface protein, ESP, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol. 2001;67:4538–45. doi: 10.1128/AEM.67.10.4538-4545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith HW, Huggins MB. Successful treatment of experimental Escherichia coli infections in mice using phage: Its general superiority over antibiotics. J Gen Microbiol. 1982;128:307–18. doi: 10.1099/00221287-128-2-307. [DOI] [PubMed] [Google Scholar]
- 31.Clokie MR, Kropinski AM. Bacteriophages: Methods and Protocols. New York, NY: Humana Press; 2009. [Google Scholar]
- 32.Pinheiro ET, Anderson MJ, Gomes BP, Drucker DB. Phenotypic and genotypic identification of enterococci isolated from canals of root-filled teeth with periapical lesions. Oral Microbiol Immunol. 2006;21:137–44. doi: 10.1111/j.1399-302X.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- 33.Raza T, Saadia A, Ullah SR, Jamal M, Mehmood K, Ali M. Isolation and characterization of a phage to control vancomycin resistant E. faecium. Open Life Sci. 2018;13:553–60. doi: 10.1515/biol-2018-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee HJ, Kim JK, Cho JY, Lee JM, Hong SH. Quantification of subgingival bacterial pathogens at different stages of periodontal diseases. Curr Microbiol. 2012;65:22–7. doi: 10.1007/s00284-012-0121-8. [DOI] [PubMed] [Google Scholar]
- 35.King AM, Adams MJ, Carstens EB, Lefkowitz EJ. Virus Taxonomy. Ninth report of the International Committee on Taxonomy of Viruses. 2012. pp. 86–98. [Google Scholar]
- 36.Uchiyama J, Takemura I, Hayashi I, Matsuzaki S, Satoh M, Ujihara T, et al. Characterization of lytic enzyme open reading frame 9 (ORF 9) derived from E. faecalis bacteriophage phage EF 24 C with a spontaneous point mutation. PloS ONE. 2011;6:E26648. doi: 10.1371/journal.pone.0026648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Łobocka M, Hejnowicz MS, Dąbrowski K, Gozdek A, Kosakowski J, Witkowska M, et al. Genomics of staphylococcal Twort-like phages, potential therapeutics of the post-antibiotic era. Adv Virus Res. 2012;83:143–216. doi: 10.1016/B978-0-12-394438-2.00005-0. [DOI] [PubMed] [Google Scholar]
- 38.Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–25. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathur T, Singhal S, Khan S, Upadhyay DJ, Fatma T, Rattan A. Detection of biofilm formation among the clinical isolates of Staphylococci: An evaluation of three different screening methods. Indian J Med Microbiol. 2006;24:25–9. doi: 10.4103/0255-0857.19890. [DOI] [PubMed] [Google Scholar]
- 40.de la Fuente-Núñez C, Reffuveille F, Fernández L, Hancock RE. Bacterial biofilm development as a multicellular adaptation: Antibiotic resistance and new therapeutic strategies. Curr Opin Microbiol. 2013;16:580–9. doi: 10.1016/j.mib.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 41.Chhibber S, Nag D, Bansal S. Inhibiting biofilm formation by Klebsiella pneumoniae B5055 using an iron antagonizing molecule and a bacteriophage. BMC Microbiol. 2013;13:174. doi: 10.1186/1471-2180-13-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montecinos FE, Jofre FM, Amendola I, Goncalves CR, Leao MV, Santos SS. Relationship between the probiotic Lactobacillus rhamnosus and E. faecalis during the biofilm formation. Afri J Micro Res. 2016;31:1182–86. [Google Scholar]


