Abstract
Plant nucleotide-binding leucine-rich repeat (NLR) immune receptors activate cell death and confer disease resistance by unknown mechanisms. We demonstrate that plant Toll/interleukin-1 receptor (TIR) domains of NLRs are enzymes capable of degrading nicotinamide adenine dinucleotide in its oxidized form (NAD+). Both cell death induction and NAD+ cleavage activity of plant TIR domains require known self-association interfaces and a putative catalytic glutamic acid that is conserved in both bacterial TIR NAD+-cleaving enzymes (NADases) and the mammalian SARM1 (sterile alpha and TIR motif containing 1) NADase. We identify a variant of cyclic adenosine diphosphate ribose as a biomarker of TIR enzymatic activity. TIR enzymatic activity is induced by pathogen recognition and functions upstream of the genes enhanced disease susceptibility 1 (EDS1) and N requirement gene 1 (NRG1), which encode regulators required for TIR immune function. Thus, plant TIR-NLR receptors require NADase function to transduce recognition of pathogens into a cell death response.
Plants rely on an innate immune system to detect potential pathogens. Molecular identification of disease resistance genes has revealed that resistance to pathogens from all kingdoms is conferred by nucleotide-binding leucine-rich repeat (NLR) immune receptors (1). Despite their agronomic value and over 25 years of molecular study, mechanistic explanations for how NLR receptors trigger cell death and disease resistance remain opaque (2, 3).
Toll/interleukin-1 receptor (TIR) domains are found in both intracellular plant TIR-NLR receptors and the animal cell surface Toll-like receptor system (3). The noncanonical animal TIR protein SARM1 (sterile alpha and TIR motif containing 1) is required for Wallerian degeneration in axons after nerve injury (4, 5). Although other animal TIRs function as oligomeric signaling scaffolds, SARM1 has a distinct mechanism: SARM1’s TIR domain is the founding member of a class of enzymes that cleave nicotinamide adenine dinucleotide (oxidized form; NAD+) (fig. S1) (6). SARM1-triggered cell death is proposed to result from severe depletion of NAD+ (4, 6). Prokaryotic TIR domains also metabolize NAD+ and NADP+ (7). NAD+ and NADP+ are essential metabolic components, and their cleavage is implicated in diverse interspecies interactions, including toxin-triggered cell death (8), effector-mediated immunosuppression (9), antiphage defenses (10), and bacterial competition (11).
Plant TIR-NLRs typically trigger hypersensitive cell death in response to pathogens. There is no evidence that plant TIRs function as scaffolds, as canonical animal TIRs do, and transient expression of some plant TIRs is sufficient to cause ectopic cell death. We sought to determine if plant TIR domains are mechanistically similar to SARM1. A structural homology search with SARM1-TIR returned plant TIR domains with high confidence (table S1). Homology models suggest that SARM1’s putative catalytic glutamic acid at position 642 (E642) (6) and neighboring residues are positionally conserved in plants (Fig. 1A). Of 146 Arabidopsis TIR domains from the reference genome, 131 contain this conserved glutamic acid (Fig. 1B and table S2). TIR domains lacking the glutamic acid have genomic signatures reminiscent of the cell death–inactive “sensor” NLR RRS1 (table S2 and fig. S2) (12, 13). Thus, the putative catalytic residue of SARM1 is present in potentially active plant TIR domains but absent from TIR-NLRs likely to lack autonomous activity.
Fig. 1. Plant TIR proteins require a conserved putative catalytic residue for cell death activity in planta.
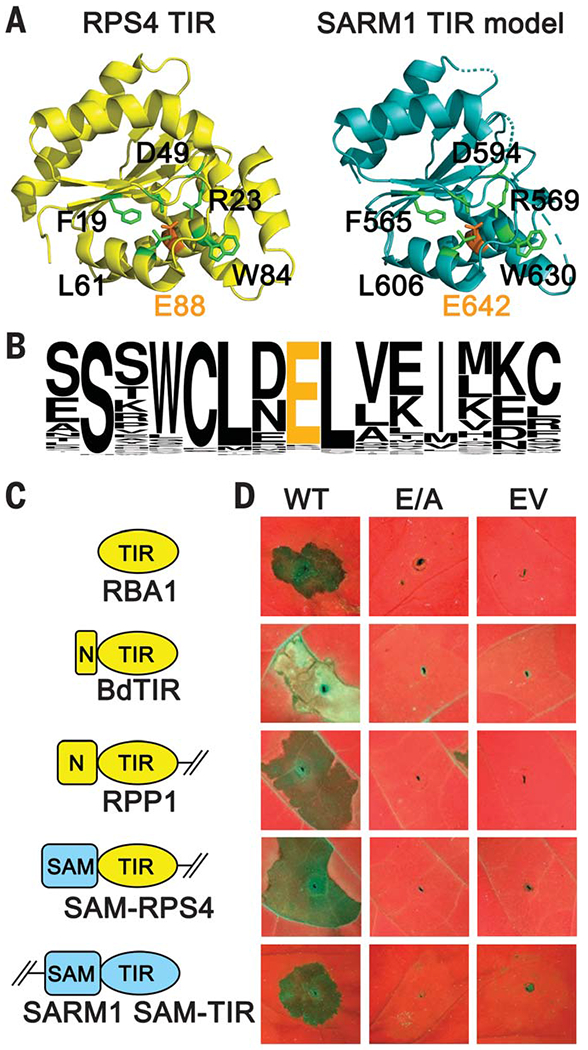
(A) A homology model of SARM1-TIR (right, blue) indicates that its putative catalytic residue (E642, orange) is positioned similarly to E88 in the plant TIR RPS4 (PDB 4C6R; left, yellow). Neighboring conserved residues are green (with black labels). (B) Sequence pileup of the 146 TIR domains from the Arabidopsis Col-0 genome. (C) Proteins expressed in planta. Plant TIR proteins are shown in yellow, and human SARM1 domains are shown in blue. The NBS-LRR autoinhibitory domains are deleted in the RPP1NdA and RPS4 truncations. N, N-terminal extension; SAM, oligomerization domain from human SARM1. (D) In planta phenotypes of TIR proteins. RBA1 and autoactive SARM1 were expressed in Nicotiana benthamiana, and other TIRs were expressed in Nicotiana tabacum. Putative catalytic mutants (E/A) lose autoactive cell death capabilities. WT, wild type; EV, empty vector. Full images and protein accumulation data are shown in fig. S3.
To determine if the putative catalytic residue is functionally required, we generated glutamic acid–to–alanine mutations in the Arabidopsis TIR-only immune receptor RBA1 and the TIR domains of the Arabidopsis TIR-NLR immune receptors RPS4 and RPP1NdA (13–15). We also tested a TIR-only protein (BdTIR) of unknown function from the monocot Brachypodium distachyon. Monocot genomes have no full-length TNL immune receptors (16). All four TIRs triggered glutamic acid–dependent cell death in Nicotiana species (Fig. 1, C and D). In our study, the RPS4 TIR required fusion to an orthologous oligomerization domain [the sterile alpha motif (SAM) domain of SARM1] to trigger cell death (Fig. 1, C and D, and fig. S4). Consistent with our transient-expression assay results, mutants disrupting the conserved glutamic acid were identified in genetic screens as suppressors of TIR and TIR-NLR function (table S3). Expression of an autoactive fragment of SARM1 also triggered glutamic acid–dependent cell death in planta (Fig. 1C). Thus, the putative SARM1 catalytic residue is conserved and functionally required in plant TIRs.
Similar to SARM1-TIR (6, 7), expression of plant TIR domains in Escherichia coli resulted in strong NAD+ depletion, as measured by high-performance liquid chromatography (HPLC) (Fig. 2, A to D, and fig. S5). As with SARM1 (6, 7), activity was dependent on the conserved glutamic acid (Fig. 2, A to D, and fig. S5). We observed accumulation of potential breakdown products accompanying NAD+ depletion (Fig. 2, E to H). To characterize reaction products and demonstrate the intrinsic enzymatic activity of plant TIR domains, we purified RBA1 and BdTIR after in vitro transcription-translation and tested their ability to degrade NAD+. Both HPLC and mass spectrometry analysis revealed that, like SARM1-TIR, purified RBA1 and BdTIR proteins degrade NAD+ into nicotinamide (Fig. 2, I to K, and fig. S6, A and B). Purified BdTIR additionally produced adenosine diphosphate ribose (ADP-ribose) from the NAD+ cleavage reaction (Fig. 2K and fig. S6C). The NAD+ cleavage activity of RBA1 and BdTIR was dependent on the putative catalytic glutamic acid residue (Fig. 2I). Unlike SARM1, plant TIRs also generated an NAD+ cleavage product that appears identical to an NAD+ cleavage product from the archaeal TIR protein TcpO (fig. S7) (7). This product, which derives from the same parent ion as cyclic ADP-ribose (cADPR) but has a distinct chromatography retention time, is suggestive of a cyclization variant of cADPR, termed v-cADPR (7). v-cADPR serves as a proxy for plant TIR NAD cleavage (NADase) activity, as its accumulation is dependent on the putative catalytic glutamic acid (Fig. 2, E to H). SARM1-TIR cleaves NAD+ in vitro to produce cADPR rather than v-cADPR (fig. S5F) (6) and appears to have higher activity than plant TIRs in this assay (fig. S5, E and G, and Fig. 2I), indicating mechanistic differences. We observed NAD+ depletion by RPS4 and RPP1 TIR domains in the E. coli assay but were unable to confirm NAD+-cleaving enzyme (NADase) activity with these TIR domains as in vitro–purified proteins. We speculate that this assay fails to recreate functionally relevant higher-order structures that are generated during expression in E. coli. SARM1-TIR can degrade NADP+ but not another NAD+-related molecule, nicotinic acid adenine dinucleotide (NaAD) (7). Similar to SARM1-TIR, RBA1 and BdTIR proteins degraded NADP+ (Fig. 2L) into nicotinamide and ADP-ribose phosphate (fig. S6, D to G) but did not degrade NaAD (Fig. 2L and fig. S1). Altogether, these results support our hypothesis that plant TIR domains are enzymes.
Fig. 2. Plant TIR proteins can degrade NAD+ and NADP+.
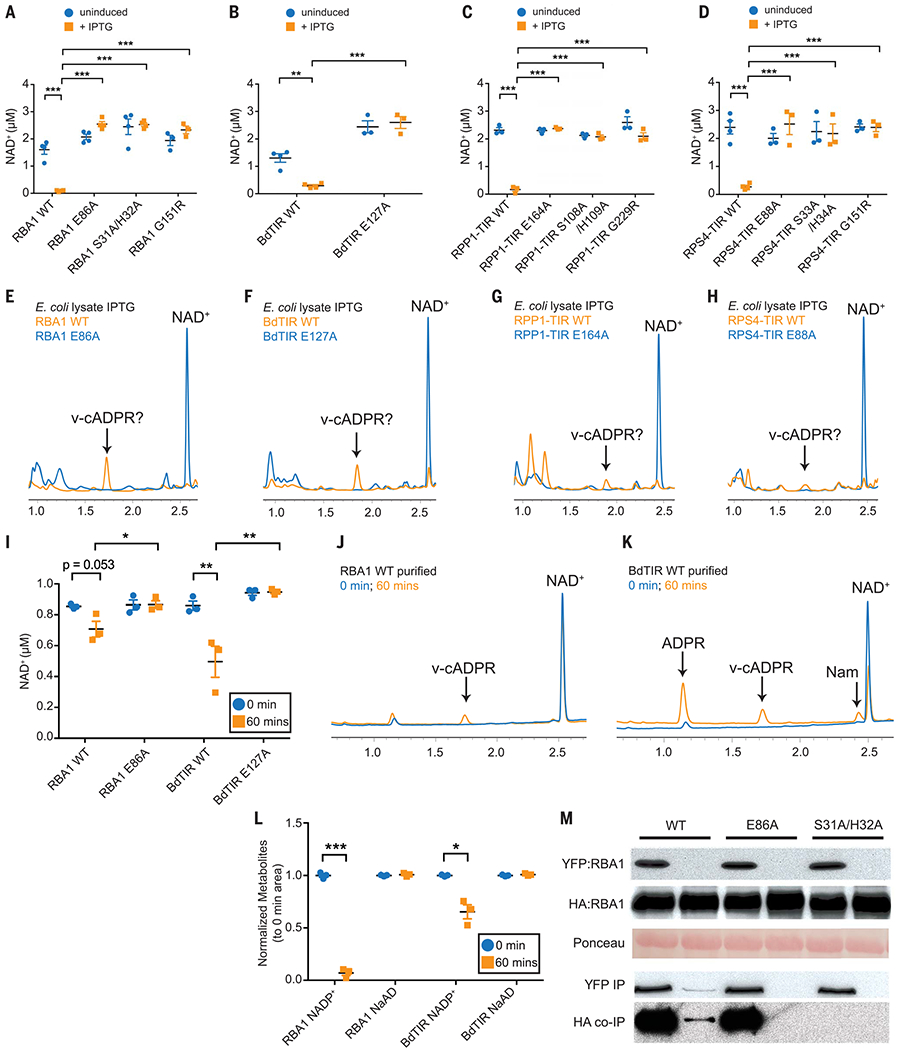
(A to D) Plant TIR proteins drive glutamic acid–dependent NAD+ depletion in E. coli after a 2-hour induction with isopropyl-β-d-thiogalactopyranoside (IPTG). Lysates were assayed using HPLC. Error bars represent SEM. Asterisks indicate statistical significance [one-way analysis of variance (ANOVA) with Tukey’s test: *P < 0.05; **P < 0.01; ***P < 0.001]. (E to H) HPLC results with E. coli lysates [from assays shown in (A) to (D)]. (I) In vitro transcription-translation–generated TIR-only proteins lowered NAD+ levels relative to putative catalytic mutants. Statistical significance levels, calculated separately for each gene and indicated by asterisks, are the same as for (A) to (D). (J) HPLC traces corresponding to (I) indicate RBA1-generated v-cADPR. (K) HPLC traces corresponding to (I) show that BdTIR decreases NAD+ and leads to accumulation of nicotinamide, ADPR, and v-cADPR. (L) Plant TIR-only proteins generated by in vitro transcription-translation degrade NADP+, but not the related molecule NaAD, after 60 min. Error bars represent SEM. Statistical significance was assessed using Welch’s t test: *P < 0.05; ***P < 0.001. (M) Mutation of RBA1’s putative catalytic residue (E86A) does not affect coimmunoprecipitation (co-IP) of hemagglutinin-tagged RBA1 (HA:RBA1) with yellow fluorescent protein-tagged RBA1 (YFP:RBA1) from N. benthamiana. WT, wild type. Amino acid abbreviations: A, alanine; E, glutamic acid; G, glycine; H, histidine; R, arginine; S, serine. The mutant protein E86A indicates a change at residue 86 from glutamic acid to alanine; the same nomenclature is used for other mutant proteins.
Plant TIR domains require two self-association interfaces for immune function (fig. S8) (3). To determine if self-association is required for NADase activity, we generated TIR domains containing demonstrated, or proposed, loss-of-self-association mutations (13, 14). Mutation of either AE- or DE-type self-association interfaces abolished TIR cell death in planta (figs. S3, S4, and S8B) and NADase activity in E. coli (Fig. 2, A, C, and D). The in planta self-association requirements were retained in the human HsSAMRPS4 TIR fusion (fig. S4, C and D), despite fusion to the SAM oligomerization domain, demonstrating that specific TIR-TIR interaction is required for function. We asked if mutation of the putative catalytic residue had an impact on self-association as measured by coimmunoprecipitation. RBA1SH/AA lost self-association (Fig. 2M) (14), but the putative catalytically dead RBA1E86A mutant retained self-association (Fig. 2M). This result is consistent with the hypothesis that TIR enzymatic activity is downstream of, and promoted by, self-association, thus explaining why plant TIR self-association is a common requirement for function.
Activation of SARM1 in neurons leads to strong NAD+ depletion and subsequent axonal demise (4). In planta, the autoactive SARM1 truncation strongly depletes NAD+ (fig. S9A). Consistent with an apparent weaker activity of plant TIRs in vitro, we have been unable to detect depletion of NAD+ after transient expression of TIRs in planta (fig. S9, A to C). As noted above (Fig. 2), NAD+ cleavage by plant TIR domains was accompanied by production of nicotinamide, ADPR, and v-cADPR. NAD+ cleavage is thus a plausible signaling event, as ADPR and cADPR trigger calcium influx to the cytoplasm (17, 18), and this is correlated with plant NLR function (19).
To validate that plant TIRs are indeed NADases in planta, we measured TIR-dependent accumulation of v-cADPR in planta. v-cADPR accumulated in planta in response to plant TIR transient expression (Fig. 3, A to D). Similar to our E. coli assay (Fig. 2, E to H), the levels of v-cADPR produced in planta by the TIR-NLR TIRs were lower than those induced by the TIR-only proteins. TIR-dependent accumulation of v-cADPR in planta was dependent upon the putative catalytic glutamic acid (Fig. 3, A to D) and on self-association (Fig. 3, A, C, and D). We measured v-cAPDR accumulation in a biologically relevant system. Delivery of the type III effector HopBA1 by Pseudomonas fluorescens activates RBA1-dependent cell death in Arabidopsis (14). HopBA1 induced v-cAPDR accumulation in Arabidopsis, whereas bacteria containing a loss-of-function allele, HopBA1H56F (14), did not (Fig. 3E). Cell death triggered by non-TIR immune function did not induce v-cADPR accumulation (fig. S9, E and F). In summary, v-cADPR is an in planta biomarker of plant TIR enzymatic function. The lack of in planta NAD+ depletion by plant TIRs suggests that plant TIRs function through an NADase-dependent signaling mechanism distinct from that for animal SARM1 NAD+ depletion.
Fig. 3. TIRs produce v-cADPR in planta.
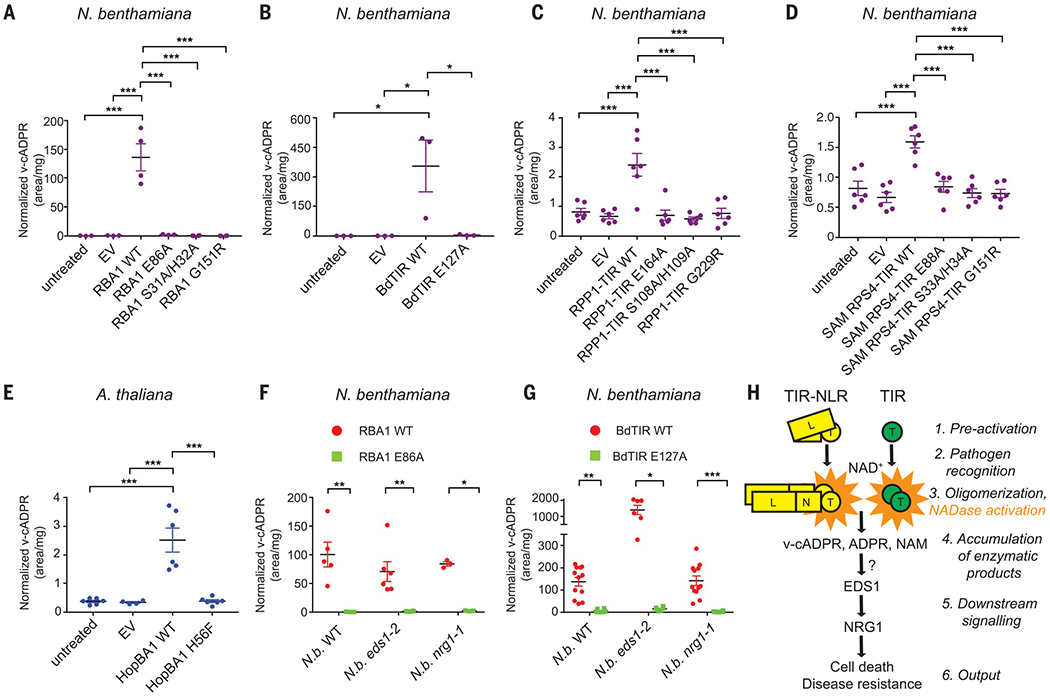
(A to D) v-cADPR accumulates after transient expression of plant TIRs. v-cADPR does not accumulate in plants expressing empty vector (EV) or TIR loss-of-function mutants. Experiments shown in (C) and (D) were performed together and share untreated and EV data. Error bars represent SEM. Asterisks indicate statistical significance (one-way ANOVA with Tukey’s test: *P < 0.05; ***P < 0.001). (E) v-cADPR accumulates in Arabidopsis (Ag-0) 24 hours after inoculation with P. fluorescens (HopBA1). HopBA1H56F is encoded by a loss-of-function mutant [with histidine (H) at residue 56 changed to phenylalanine (F)] (14). (F and G) v-cADPR accumulates upon RBA1 and BdTIR expression in N. benthamiana and eds1-2 and nrg1-1 mutants. Error bars represent SEM. Asterisks indicate statistical significance [one-way ANOVA with Tukey’s test for (F) and Kruskal-Wallis with Dunn’s test for (G): *P < 0.05; **P < 0.01; ***P < 0.001]. (H) Hypothetical pathway wherein TIR enzymatic function is induced by effector recognition and activation by TIR-NLR (left) or by TIR only (right). This leads to accumulation of signaling products derived from NAD+ that accumulate upstream of EDS1/NRG1.T, TIR; N, NBD; L, LRR. Definitions of amino acid abbreviations and mutation designations are provided in the Fig. 2 legend.
All tested TIR-NLR phenotypes require the catalytically inactive lipase ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) (20, 21). Consistent with this requirement, autoactivity triggered by the four tested plant TIR domains was EDS1 dependent (fig. S10). In contrast, autoactive SARM1 cell death in planta was EDS1 independent (fig. S10), again suggesting that plant and animal TIR NADases act via different mechanisms to drive cell death. The functional mechanism of EDS1 is unknown, but this lipase can be found in complex with TIR-NLR proteins (22). Thus, EDS1 could be required for TIR function directly or be required for downstream signaling. Similarly, protein encoded by N-required gene 1 (NRG1), a coiled-coil “helper” NLR, functions downstream of EDS1 (23). v-cADPR accumulates in both eds1 and nrg1 mutants upon expression of RBA1 or BdTIR, despite a lack of cell death (Fig. 3, F and G, and fig. S11). Thus, TIR enzymatic activity, via the generation of metabolites such as v-cADPR, may signal downstream through EDS1 and then NRG1 to promote cell death (Fig. 3H).
NAD+ is a signaling substrate that influences myriad aspects of biology, including DNA repair, aging, and transcriptional regulation, in addition to its metabolic function (24). Our results establish plant TIR domains as enzymes that can cleave NAD+ and NADP+. Direct cell death by NAD+ depletion would not require downstream genetic components, suggesting that SARM1 and plant TIRs trigger cell death by distinct mechanisms and that the latter generate a signal via NADase activity that activates EDS1 and NRG1. Our data present a framework for understanding plant TIRs as enzymes that cleave NAD+ to activate immunity.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Grant and M. Redinbo for careful reading and discussion of the manuscript. We thank B. Staskawicz for sharing N. benthamiana eds1 and nrg1 mutants. We thank B. Kobe for sharing the sequence of the SARM1 SAM domain pentamutant. We thank K. Simburger and T. Fahrner for technical assistance. We thank C. Argueso and D. Bush for use of facilities. We thank B. Staskawicz, K. H. Sohn, and F. El Kasmi for plasmids.
Funding: This work was supported by the National Science Foundation (grant IOS-1758400 to J.L.D. and M.T.N.) and National Institutes of Health (grants GM107444 to J.L.D., RF1AG013730 to J.M., and R01NS087632 to J.M. and A.D.). J.L.D. is a Howard Hughes Medical Institute (HHMI) Investigator. M.T.N. was supported by startup funds from Colorado State University. R.G.A. was supported by an NIH Ruth L. Kirschstein NRSA fellowship (F32GM108226). K.E. was an HHMI Medical Research Fellow. F.M. is supported by a grant from the Gordon and Betty Moore Foundation (GBMF4725) to the Two Blades Foundation.
Footnotes
Competing interests: J.M. and Y.S. may derive income from licensing technology to ChromaDex. J.M. and A.D. are cofounders and scientific advisors of Disarm Therapeutics. K.E., Y.S., A.D., and J.M. may derive income from licensing of technology to Disarm Therapeutics.
Data and materials availability: All data are available in the manuscript or the supplementary materials.
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Jones JDG, Dangl JL, Nature 444, 323–329 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Jones JDG, Vance RE, Dangl JL, Science 354, aaf6395 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, Dodds PN, Bernoux M, Annu. Rev. Phytopathol 55, 205–229 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Gerdts J, Brace EJ, Sasaki Y, DiAntonio A, Milbrandt J, Science 348, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerdts J, Summers DW, Sasaki Y, DiAntonio A, Milbrandt J, J. Neurosci 33, 13569–13580 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Essuman K et al. , Neuron 93, 1334–1343.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Essuman K et al. , Curr. Biol 28, 421–430.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pajuelo D et al. , Cell Rep. 24, 429–440 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shidore T et al. , PLOS Pathog. 13, e1006442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doron S et al. , Science 359, eaar4120 (2018).29371424 [Google Scholar]
- 11.Tang JY, Bullen NP, Ahmad S, Whitney JC, J. Biol. Chem 293, 1504–1514 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narusaka M et al. , Plant J. 60, 218–226 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Williams SJ et al. , Science 344, 299–303 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Nishimura MT et al. , Proc. Natl. Acad. Sci. U.S.A 114, E2053–E2062 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schreiber KJ, Bentham A, Williams SJ, Kobe B, Staskawicz BJ, PLOS Pathog. 12, e1005769 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao ZQ et al. , Plant Physiol. 170, 2095–2109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fliegert R, Gasser A, Guse AH, Biochem. Soc. Trans 35, 109–114 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Wu Y et al. , Science 278, 2126–2130 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Grant M et al. , Plant J. 23, 441–450 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Aarts N et al. , Proc. Natl. Acad. Sci. U.S.A 95, 10306–10311 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner S et al. , Cell Host Microbe 14, 619–630 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Huh SU et al. , PLOS Pathog. 13, e1006376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi T et al. , Proc. Natl. Acad. Sci. U.S.A 115, E10979–E10987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demarest TG et al. , Annu. Rev. Cancer Biol 3, 105–130 (2019). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


