Abstract
Objectives
Carbapenem resistance is a serious clinical and public health threat. Carbapenemase can confer carbapenem resistance, and most carbapenemase genes are plasmid encoded so resistance can easily spread. In this study, we aimed to develop a novel system based on the TaqMan platform for the rapid detection of 6 clinically prevalent carbapenemase genes: Klebsiella pneumoniae carbapenemase, New Delhi metallo-β-lactamase, oxacillinase, imipenem-hydrolyzing, Verona integron-encoded metallo-β-lactamase, and Guiana extended-spectrum β-lactamase.
Methods
The triplex assay was verified by testing genomic DNA of 6 carbapenemase-producing Klebsiella pneumoniae. It was validated with a blinded panel of 310 Enterobacteriaceae isolates, including 225 carbapenemase-producers and 85 non-producers, by direct colony triplex real-time polymerase chain reaction (PCR). The real-time PCR was performed using the ABI 7500 fast instrument (Applied Biosystems, CA, USA) and specific primers for each carbapenemase target were designed to include modified peptide-nucleic acid oligonucleotides.
Results
No amplification was detected among the negative samples. The result showed 100% concordance with the genotypes previously identified. The entire assay, including DNA extraction and real-time PCR, was completed within 2 hours.
Conclusion
The newly developed triplex real-time PCR assay was useful for the rapid, accurate and simultaneous detection of 6 carbapenemase genes in Enterobacteriaceae, suggesting its potential to allow an early decision on the appropriate treatment, management, and prevention of the spread of resistant infections in hospitals.
Keywords: carbapenemase, Enterobacteriaceae, real-time PCR, triplex PCR,
Introduction
Enterobacteriaceae is the largest family of gram-negative bacteria that are significant causes of community- and healthcare-associated human infections. In particular, the considerable increase of multidrug-resistant Enterobacteriaceae with extended spectrum β-lactamase (ESBL) poses a serious clinical challenge as these infections are increasingly difficult to control in many hospitals [1–3]. Carbapenems, given their broad spectrum of antibacterial activity against hydrolysis by most β-lactamases, are agents currently regarded as the last-line of defense for the treatment of infections by ESBL producers [4–6]. The increased use of carbapenems has led to an emergence of carbapenem-resistant Enterobacteriaceae (CRE) and/or carbapenemase-producing Enterobacteriaceae (CPE) [7–9]. CPEs are classified as A, B, and D, based on their amino acid sequence according to the Ambler system. Class A includes the active-site serine β-lactamases such as Klebsiella pneumoniae carbapenemase (KPC) and Guiana extended-spectrum β-lactamase (GES), class B includes the metallo-b-lactamases such as New Delhi metallo-β-lactamase (NDM), imipenem-hydrolyzing (IMP), and Verona integron-encoded metallo-β-lactamase (VIM) and class D is the oxacillinase (OXA-48 like)[10,11]. The rapid spread of carbapenem resistance is mainly attributed to the horizontal transfer of carbapenemase genes between organisms, since they are located on mobile plasmids [12,13].
Since the first imported case of CPE infection in 2010, the Korea Centers for Disease Control and Prevention has implemented a national laboratory surveillance system of carbapenem-non-susceptible Enterobacteriaceae (CNSE). Based on these data, we recently identified a remarkable increase of CPE infections from 2011 to 2016 in Korea [14,15].
The seriousness of this situation resulted in the Korean government designating the CRE/CPE infection as a notifiable disease, and expanded the monitoring system for strengthening strict control measures of the spread of these bacteria within individual hospitals.
Many clinicians have emphasized the need for improved laboratory capacity to facilitate the rapid and accurate detection of CPE to identify patients at high risk, and initiate appropriate treatments as soon as possible [16,17]. Indeed, early intervention, by administering the most appropriate antimicrobials is the most important factor to prevent death, and prevent transmission of infection to other patients in the hospital. Traditional culture-based confirmation of carbapenemase producers requires supplementation with additional, laborious phenotypic tests such as the modified Hodge test, which can take up to 18 hours to obtain results [18,19]. As an alternative, polymerase chain reaction (PCR) based-molecular detection is a promising tool for the rapid identification of a specific carbapenemase gene in Enterobacteriaceae. Most carbapenemase kits available on the market can identify one or a few genes at a time, and kits that can cover all 6 genes of carbapenemase are not common [20–24]. As target genes, the 6 most prevalent types of carbapenemases identified in the CNSE surveillance of Korea were selected: KPC, NDM, OXA-48 like, IMP, VIM, and GES [14]. In particular, the dissemination of GES-producing Enterobacteriaceae was considered of significant importance to include in this CPE screening system [25–27].
In this study, two triplex real-time PCR assays for the simultaneous detection of 6 carbapenemase genes were developed, reducing the turn-around time of screening carbapenemase producers in hospital laboratories, and in large-scale surveillance. To improve the specificity of this assay, the modified peptide-nucleic acid (PNA) oligonucleotides technique was adopted. PNA is an artificially synthesized nucleic acid analogue in which the phosphate-ribose backbone of DNA is replaced with a peptide-like amide backbone, and therefore exhibits a stronger binding affinity and a higher specificity to the target DNA for molecular diagnosis [28,29].
The assays were optimized by using direct colony PCR, without complex sample preparation, and the results and performance were compared to the conventional PCR method. This developed triplex real-time format assay is widely accessible to clinical diagnosis for rapid detection of the carbapenemase enzyme in Enterobacteriaceae.
Materials and Methods
1. Bacterial strains
Six reference strains of K. pneumoniae were used to verify the assay: CKPN20100021 with blaKPC, CKPN20160166 with blaNDM, CKPN20140654 with blaOXA-48 like, CKPN20130418 with blaIMP, CKPN20140712 with blaVIM, and CKPN20150958 with blaGES.
For the assessment of specificity and sensitivity, 310 Enterobacteriaceae isolates from the National Laboratory Surveillance for CRE from 2010 to 2017 were tested, which were previously characterized using conventional PCR and/or sequencing analysis. Among them, 201 strains carried only 1 type of carbapenemase gene, and 24 strains carried several combinations of one or more carbapenemase genes. The remaining 85 strains had none of the 6 carbapenemase genes. These strains included 26 Enterobacteriaceae species (Table 1) and variants of KPC (blaKPC-2, 3, 4, 6, 12, 19), NDM (blaNDM-1, 3, 4, 5, 6, 7), OXA-48 like (blaOXA-48, 181, 232), IMP (blaIMP-1, 4, 6), VIM (blaVIM-1, 2), and GES (blaGES-5). All CPE isolates were confirmed by both the phenotypic method, and an in-house conventional PCR method, as previously described [14,30].
Table 1.
Distribution of CPE types and species by carbapenemase gene.
| Carbapenemase | CPE type (n) | Species (n) | |
|---|---|---|---|
| Single-producing | blaKPC | KPC-2 (46) / KPC-3 (5) / KPC-4 (17) / KPC-12 (1) / KPC-19 (4) | C. murliniae (1) / C. freundii (2) / C. koseri (1) / E. coli (6) / E. cloacae (5) / E. asburiae (1) / E. aerogenes (1) / E. hormaechei (1) / K. pneumoniae (49) / K. oxytoca (1) / K. variicola (1) / R. ornithinolytica (2) / S. marcescens (1) / H. alvei (1) |
|
|
|||
| blaNDM | NDM-1 (23) / NDM-3 (1) / NDM-4 (3) / NDM-5 (6) / NDM-6 (1) / NDM-7 (3) | C. freundii (2) / C. amalonaticus (1) / K. pneumoniae (14) / E. coli (11) / E. cloacae (5) / E. aerogenes (1) / K. oxytoca (1) / M. morganii (1) / R. ornithinolytica (1) | |
|
|
|||
| blaOXA-48 like | OXA-48 (3) / OXA-181 (2) / OXA-232 (21) | E. coli (5) / K. ascorbata (1) / K. oxytoca (1) / K. pneumoniae (19) | |
|
|
|||
| blaIMP | IMP-1 (20) / IMP-4 (4) / IMP-6 (1) | E. cloacae (7) / K. oxytoca (2) / K. pneumoniae (12) / Pantoea spp. (3) / S. marcescens (1) | |
|
|
|||
| blaVIM | VIM-1 (5) / VIM-2 (22) | C. freundii (6) / E. coli (1) / E. hormaechei (1) / E. cloacae (3) / E. asburiae (2) / E. aerogenes (1) / K. oxytoca (2) / K. pneumoniae (9) / K. variicola (1) / S. marcescens (1) | |
|
|
|||
| blaGES | GES-5 (13) | C. freundii (1) / E. aerogenes (2) / E. cloacae (2) / K. oxytoca (2) / K. pneumoniae (3) / S. marcescens (3) | |
|
| |||
| Co-producing | ≤ 2 * | KPC-2, NDM-1 (5) / KPC-2, VIM-1 (2) / KPC-4, NDM-1 (2) / KPC-6, NDM-1 (1)/ NDM-1, OXA-232 (2) / NDM-1, VIM-2 (1) / NDM-1, GES-5 (2) / NDM-5, OXA-181 (5) / IMP-1, VIM-2 (2) / VIM-2, GES-5 (1) / KPC-4, VIM-2, NDM-1 (1) | C. freundii (5) / K. pneumoniae (8) / E. asburiae (1) / E. cloacae (3) / K. oxytoca (1) / R. planticola (1) / R. ornithinolytica (2) |
|
| |||
| non-CPE | - | - | C. freundii (7) / C. koseri (2) / E. aerogenes (8) / E. cloacae (11) / E. coli (9) / E. hormaechei (2) / E. asburiae (3) / K. oxytoca (2) / K. pneumoniae (12) / M. morganii (5) / P. mirabilis (5) / P. vulgaris (2) / P. penneri (1) / P. hauseri (1) / P. Rettgeri (6) / P. favisporus (1) / P. Stuartii (2) / S. marcescens (5) / R. planticola (1) |
Producing ≥ 2 carbapenemase genes.
CPE = carbapenemase-producing Enterobacteriaceae; KPC = Klebsiella pneumoniae carbapenemase; NDM = New Delhi metallo-β-lactamase; OXA-48 like = oxacillinase; IMP = imipenem-hydrolyzing; VIM = Verona integron-encoded metallo-β-lactamase; GES = Guiana extended-spectrum β-lactamase.
2. DNA preparation
All bacteria were grown on tryptic soy agar at 37°C overnight. Genomic DNA from the reference strains used for triplex RT-PCR assay optimization was extracted using the Qiagen DNeasy Blood & Tissue Kit (Qiagen GmbH, Hilden, Germany) according to manufacturer’s instructions. The DNA concentration was determined on a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Dreieich, Germany). Serial dilutions of genomic DNA were prepared, calculated, and adjusted to cover the range of 105 to 101 to evaluate the limit of detection of each triplex real-time PCR. In addition, specificity was determined as the detection of a single carbapenemase gene from the pooled sample of 6 different genetic types of carbapenemase.
For direct-colony PCR, DNA was extracted directly from the bacterial colonies by boiling. In brief, 1–2 colonies of bacteria were picked in a 1 uL loop and suspended in 100 uL of sterile distilled water, and heated at 100°C for 10 minutes. The culture supernatant (2 uL) was then used for real-time PCR. The total turn-around time of DNA preparation was approximately 15 minutes.
3. Design of primer and probe sets for real-time PCR
There were two sets of triplex real-time PCR assays developed: one (Rx1) targeting KPC, NDM, and OXA-48 like, and the other (Rx2) targeting IMP, VIM, and GES. Primers and probes were designed to amplify the targeted region of each carbapenemase gene based on the conserved nucleotide sequence to cover all known, and published subtypes from the NCBI database (http://www.ncbi.nlm.nih.gov). Detailed information of primer and probe sets is given in Table 2. The PNA probes with superior binding affinity to a short base sequence, were manufactured by the PNA synthesizer (PANAGENE Inc., Daejeon, Korea), and labelled with different fluorophores such as FAM (6-carboxyfluorescein), HEX (4,4,7,2′,4′,5′,7′-hexachloro-6-carboxyfluorescein), and ROX (carboxy-X-rhodamine) dye for the respective target attached to the 3′-end.
Table 2.
Primers and probes for detecting the carbapenemase genes blaKPC, blaNDM, blaOXA-48 like, blaIMP, blaVIM, and blaGES.
| Reaction | bla target | Primer and Probe | Sequence (5′-3′) | Conc. of oligo in Rxn (nM) | Amplicon (bp) | Condition |
|---|---|---|---|---|---|---|
| Rx1 | KPC | KPC F | TGACAACAGGCATGACGG | 40 | 121 | 95°C 15 min + 40× (95°C 15 sec + 58°C 45 sec + 72°C 15 sec) |
| KPC R | GCGCATGAAGGCCGTC | 600 | ||||
| KPC probe | Dabcyl-GCAATACAGTGAT-FAM | 5 | ||||
|
| ||||||
| NDM | NDM F | GATCAAGCAGGAGATCAAC | 120 | 111 | ||
| NDM R | GGCATAAGTCGCAATCC | 400 | ||||
| NDM probe | Dabcyl-CATCAGGACAAG-HEX | 10 | ||||
|
| ||||||
| OXA-48 like | OXA-48 F | CGTAGTTGTGCTCTGGAATGA | 160 | 163 | ||
| OXA-48 R | CATCCCACTTAAAGACTTGGTG | 640 | ||||
| OXA-48 probe | Dabcyl-AATAGCTTGATC-ROX | 10 | ||||
|
| ||||||
| Rx2 | IMP | IMP F | CATAGCGACAGCACRGG | 160 | 124 | 95°C 15 min + 40× (95°C 15 sec + 58°C 45 sec + 72°C 15 sec) |
| IMP R | CCGCTAAATGAATTTKTRGCTTG | 1000 | ||||
| IMP probe | Dabcyl-AATAGCTTGATC-FAM | 20 | ||||
|
| ||||||
| VIM | VIM F | GTCTCATTGTCCGTGATG | 40 | 141 | ||
| VIM R | TCGTCATGAAAGTGCGTGGA | 400 | ||||
| VIM probe | Dabcyl-GATTGATACAGC-HEX | 5 | ||||
|
| ||||||
| GES | GES F | TGGCTAAAGTCCTCTATGG | 120 | 110 | ||
| GES R | TAGGAAAACCCGCTCGTAGTG | 600 | ||||
| GES probe | Dabcyl-GTGGCTGATCGG-ROX | 10 | ||||
KPC = Klebsiella pneumoniae carbapenemase; NDM = New Delhi metallo-β-lactamase; OXA-48 like = oxacillinase; IMP = imipenem-hydrolyzing; VIM = Verona integron-encoded metallo-β-lactamase; GES = Guiana extended-spectrum β-lactamase.
4. Optimization of two triplex real-time PCR assays
The assays (Rx1 and Rx2) were performed on the ABI7500 fast real-time PCR system (Applied Biosystems, Inc., Foster City, CA, USA) using the Panagene triplex real-time PCR Mix (PANAGENE Inc., Daejeon, Korea). The following parameters were reviewed and optimized to ensure valid results of the triplex real-time PCR: primer and probe concentration, input DNA concentration, and reaction cycle conditions (Table 2). The assays were carried out in a single tube with a 25 uL reaction volume containing the optimum concentration of each component: 9 uL of 2.5× concentrated master mix, 0.5 uL of each of 3 up- and downstream primer pairs, 0.25 uL of each of 3 probes, and 2 primers of DNA per reaction. Concentrations of the primers and probes are listed in Table 2. The real-time PCR involved the following steps: 15 minutes of initial denaturation at 95°C, followed by 40 cycles of denaturation at 95°C for 15 seconds, annealing at 58°C for 45 seconds, and extension at 72°C for 15 seconds. The intensity of the fluorescent signal was measured, and the cycle threshold (Ct) was determined as the point at which the fluorescence reached the threshold value in each reaction. The limit of detection of all target genes was calculated as the mean Ct values using 10-fold serial dilutions of DNA from each corresponding reference strains throughout three independent PCR experiments.
Results
Confirmation that the single primer pair, and designed probes produced the expected signal individually using monoplex real-time PCR for each target gene alongside positive-and negative-control DNA was performed.
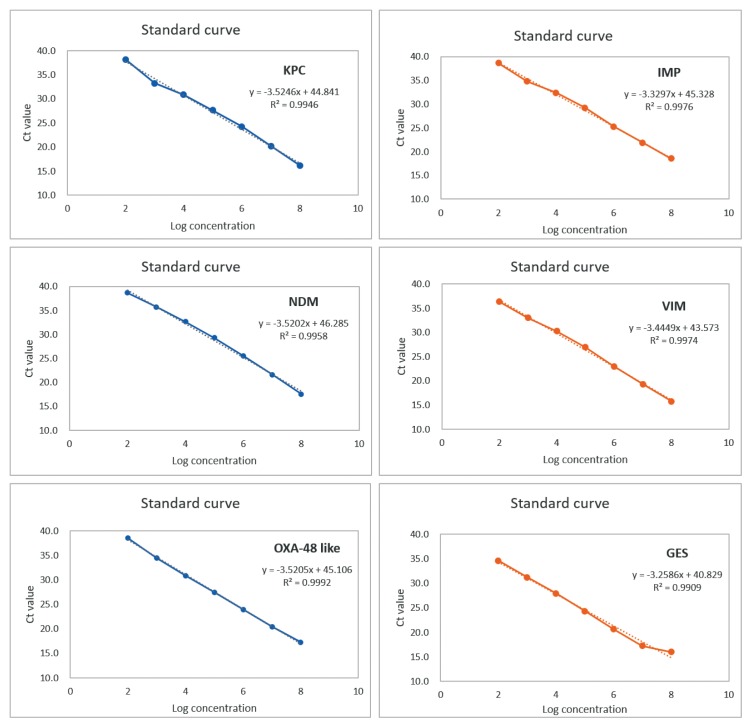
Spiked serial-dilution PCRs for each of the 6 carbapenemase genes were linear over a correlated well with KPC (R2 = 0.995), NDM (R2 = 0.996), OXA-48 like (R2 = 0.999), IMP (R2 = 0.997), VIM (R2 = 0.997), and GES (R2 = 0.991) over the entire range, with efficiencies of at least 92.2%, 92.3%, 92.3%. 99.6%, 95.1%, and 102.9%, respectively (Figure 1).
Figure 1.
Triplex real-time PCR efficiencies of 6 carbapenemases.
KPC = Klebsiella pneumoniae carbapenemase; NDM = New Delhi metallo-β-lactamase; IMP = imipenem-hydrolyzing; VIM = Verona integron-encoded metallo-β-lactamase; GES = Guiana extended-spectrum β-lactamase; PCR = polymerase chain reaction.
Three pairs of primers and probes were then pooled for the individual triplex PCRs (Rx1 and Rx2), and the detailed parameters were adjusted until the program that was effective for the simultaneous differential amplification of the three targets in a single reaction tube was found. Under these final optimized conditions, all six K. pneumoniae reference strains carrying only one type of carbapenemase gene, showed the corresponding positive results for each of the 6 carbapenemase genes tested by the two triplex real-time PCR assays, via direct colony PCR (data not shown). Moreover, the newly developed assays did not amplify DNA from isolates that either lacked carbapenemase genes, or that harbored other types of carbapenemase genes not included as targets of the primers and probes. These results confirmed the simultaneous amplification and accurate identification of the three targets in each single reaction tube.
For specificity of evaluation, the test collection of 310 Enterobacteriaceae clinical isolates were applied to the two new assays, which identified the corresponding target genes in all 225 positive isolates. None of the 85 negative isolates generated a positive signal. Detection of the 6 carbapenemase genes (blaKPC, blaNDM, blaOXA-48 like, blaOXA-48 like, blaVIM, and blaGES) showed 100% concordance between the previous conventional PCR, and newly developed triplex real-time PCR methods based on direct colony real-time PCR.
In all cases, the 6 target genes were detected with Ct values ranging from 10 to 35 (Supplementary Table S1). There was no significant difference in the Ct value range of triplex real-time PCR between single gene-producing and co-producing isolates. The interpretation of detection for each of the 6 carbapenemases was determined as a Ct value of 35 for the two triplex real-time PCR assays. The sensitivity test for the 10-fold serial dilutions of the bacterial suspensions experimentally demonstrated that these triplex real-time PCR assays can detect approximately 2 × 102 colony forming units carrying the carbapenemase genes.
Discussion
Since the first report of non-metalloenzyme carbapenemase in 1993, CPE have been identified in many countries [31]. However, until recently, CPE isolates were rarely detected in the Korean antimicrobial resistance surveillance system (Kor-GLASS), with a rate of less than 0.1% estimated in 2017 [30]. However, increasing awareness of the sharp increase of CPE isolates reported from health settings prompted the need to intensify the strict preparedness of control strategies for the potential risks of the amplification and transmission of CPE within individual institutions in Korea.
Above all, early screening, and confirmation of carbapenemase producers cannot be overestimated for appropriate infection control, and treatment in acute hospital settings [32]. There is no perfect single laboratory diagnostic method available for rapidly detecting all known carbapenemases. The routine culture-based susceptibility tests to detect carbapenemase producers generally require additional confirmatory tests, and careful interpretation owing to varied levels of carbapenemase expression, and the poor specificity of some antibiotic combinations [33,34].
In addition, some exceptional cases cannot be detected with phenotypic testing methods, such as CPE with low-level carbapenem resistance or susceptibility to carbapenems (e.g., KPC, GES enzymes) [35,36]. Moreover, the main drawback of the phenotypic testing method is the prolonged turnaround time of at least 24–72 hours. To overcome these limitations, various molecular-based diagnostic methods have been developed recently, as a first-line screening strategy of CPE infection, which are now being employed in hospital settings [37].
In this study the applicability of a triplex real-time PCR assay was assessed in the routine diagnosis or surveillance of CPE, to shorten the turnaround time of the previous conventional PCR. Indeed, the triplex real-time PCR assay capable of easily identifying more positive samples than the previous conventional PCR method was optimized in this study. Several factors were examined and adjusted to allow for the simultaneous amplification of triple targets in a single reaction tube. Consequently, the two triplex real-time PCR assays showed similar sensitivities to those determined with monoplex real-time PCR for each target gene. Importantly, these assays also allowed for the definitive identification of the most common 6 types of carbapenemase genes in Korea and their variants, showing good concordance with previously determined characteristics of clinical Enterobacteriaceae isolates. Overall, these results indicate that the newly developed triplex real-time PCR assay is an attractive tool owing to the rapid turn-around time (approximately 2 hours), and high sensitivity for CPE detection.
A key drawback of any PCR-based method targeting limited types of carbapenemases is that other carbapenemase types cannot be detected, including novel types [38,39]. Although the distribution of carbapenemase types differs widely among countries, even within regions, the representative 6 types of carbapenemases targeted in this study accounted for more than 97% of the total number of CPE strains collected by the national laboratory surveillance system of CNSE in Korea [14]. Accordingly, these two assays are expected to cover a very high proportion of CPE in the country. Nevertheless, further research is needed to uncover novel carbapenemases or rare enzyme types that were not included in these assays.
In addition, we intend to examine whether these assays are applicable for the detection of carbapenemase genes in other gram-negative bacteria such as Acinetobacter spp. and Pseudomonas spp., and the feasibility of the assays in direct testing of clinical specimens or environmental swabs for screening of CPE carriage and/or patients in hospital settings.
Importantly, the entire assay flow, including DNA extraction, and real-time PCR, can be completed within only 2 hours. This fast turnaround time will markedly shorten the time to derive results, representing a significant advantage for initiating timely measures at a hospital and/or intensive care unit, and primary settings for outbreaks of CPE reported in Korea.
Moreover, our proposed broad triplex real-time PCR screening strategy would be suitable for epidemiologic surveys in an outbreak situation.
Acknowledgments
This research was supported by Research of Korea Centers for Disease Control and Prevention (2016-NI44001-00).
Footnotes
Supplementary Materials
Supplementary materials are available at https://www.kcdcphrp.org.
Conflicts of Interest
The authors have no conflicts of interest declared.
References
- 1.Pitout JDD, Laupland KB. Extended-spectrum β-lactamase-producing Enterobacteriaceae: An emerging public-health concern. Lancet Infect Dis. 2008;8(3):159–66. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 2.Falagas ME, Bliziotis IA. Pandrug-resistant Gram-negative bacteria: The dawn of the post-antibiotic era? Int J Antimicrob Agents. 2007;29(6):630–6. doi: 10.1016/j.ijantimicag.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 3.Davies SC, Fowler T, Watson J, et al. Annual Report of the Chief Medical Officer: Infection and the rise of antimicrobial resistance. Lancet. 2013;381(9878):1606–9. doi: 10.1016/S0140-6736(13)60604-2. [DOI] [PubMed] [Google Scholar]
- 4.Shaikh S, Fatima J, Shakil S, et al. Antibiotic resistance, and extended spectrum beta-lactamases: Types, epidemiology, and treatment. Saudi J Biol Sci. 2015;22(1):90–101. doi: 10.1016/j.sjbs.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez F, Van Duin D. Carbapenem-resistant Enterobacteriaceae: A menace to our most vulnerable patients. Cleve Clin J Med. 2013;80(4):225–33. doi: 10.3949/ccjm.80a.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomson KS. Extended-spectrum-β-lactamase, AmpC, and carbapenemase issues. J Clin Microbiol. 2010;48(4):1019–25. doi: 10.1128/JCM.00219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yanling X, Bing G, Mao H, et al. Epidemiology of carbapenem resistant Enterobacteriaceae (CRE) during 2000–2012 in Asia. J Thorac Dis. 2015;7(3):376–85. doi: 10.3978/j.issn.2072-1439.2014.12.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantón R, Akóva M, Carmeli Y, et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect. 2012;18(5):413–31. doi: 10.1111/j.1469-0691.2012.03821.x. [DOI] [PubMed] [Google Scholar]
- 9.Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect. 2014;20(9):821–30. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 10.Lee CR, Lee JH, Park KS, et al. Global Dissemination of Carbapenemase-Producing Klebsiella pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front Microbiol. 2016;7:895. doi: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tzouvelekis LS, Markogiannakis A, Psichogiou M, et al. Carbapenemases in Klebsiella pneumoniae and Other Enterobacteriaceae: An Evolving Crisis of Global Dimensions. Clin Microbiol Rev. 2012;25(4):682–707. doi: 10.1128/CMR.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blair JM, Webber MA, Baylay AJ, et al. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13(1):42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 13.Munoz-Price LS, Quinn JP. The Spread of Klebsiella pneumoniae Carbapenemases: A Tale of Strains, Plasmids, and Transposons. Clin Infect Dis. 2009;49(11):1739–41. doi: 10.1086/648078. [DOI] [PubMed] [Google Scholar]
- 14.Yoon EJ, Yang JW, Kim JO, et al. Carbapenemase-producing Enterobacteriaceae in South Korea: a report from the National Laboratory Surveillance System. Future Microbiol. 2018;13:771–83. doi: 10.2217/fmb-2018-0022. [DOI] [PubMed] [Google Scholar]
- 15.Lee EJ, Lee SJ, Bahk HJ, et al. Analysis of carbapenemase-producing Enterobacteriaceae (CPE) surveillance results for 2017 in Korea: Comparison with the surveillance results of the previous 5 years 2012–2016. Cheongju (Korea): Korean Centers for Disease Control & Prevention; 2018. [Internet] Available from: https://www.cdc.go.kr/board/board.es?mid=a20602010000&bid=0034&list_no=141909&act=view#. [Google Scholar]
- 16.Gupta N, Limbago BM, Patel JB, et al. Carbapenem-resistant Enterobacteriaceae: Epidemiology and prevention. Clin Infect Dis. 2011;53(1):60–7. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 17.Logan LK, Renschler JP, Gandra S, et al. Carbapenem-resistant Enterobacteriaceae in children, United States, 1999–2012. Emerg Infect Dis. 2015;21(11):2014–21. doi: 10.3201/eid2111.150548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girlich D, Poirel L, Nordmann P. Value of the Modified Hodge Test for Detection of Emerging Carbapenemases in Enterobacteriaceae. J Clin Microbiol. 2012;50(2):477–9. doi: 10.1128/JCM.05247-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nordmann P, Gniadkowski M, Giske CG, et al. Identification and screening of carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect. 2012;18(5):432–8. doi: 10.1111/j.1469-0691.2012.03815.x. [DOI] [PubMed] [Google Scholar]
- 20.Kaase M, Szabados F, Wassill L, et al. Detection of carbapenemases in Enterobacteriaceae by a commercial multiplex PCR. J Clin Microbiol. 2012;50(9):3115–8. doi: 10.1128/JCM.00991-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tracy DL, Kathleen A, Alan M, et al. Rapid Detection of KPC, NDM, and OXA-48-Like Carbapenemases by Real-Time PCR from Rectal Swab Surveillance Samples. J Clin Microbiol. 2015;53(8):2731–3. doi: 10.1128/JCM.01237-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ewa K, Tomasz D, Grażyna M. Rapid Detection of NDM, VIM, KPC and IMP Carbapenemases by Real-Time PCR. J Bacteriol Parasitol. 2016;7(6):1000299. [Google Scholar]
- 23.Yusuf E, Van Der Meeren S, Schallier A, et al. Comparison of the Carba NP test with the Rapid CARB Screen Kit for the detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Eur J Clin Microb Infect Dis. 2014;33(12):2237–40. doi: 10.1007/s10096-014-2199-3. [DOI] [PubMed] [Google Scholar]
- 24.Laurent D, Mathieu F, Thierry N. Improvement of the Xpert Carba-R Kit for the Detection of Carbapenemase-Producing Enterobacteriaceae. Antimicrob Agents Chemother. 2016;60(6):3832–7. doi: 10.1128/AAC.00517-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cuzon G, Bogaerts P, Bauraing C, et al. Spread of Plasmids Carrying Multiple GES Variants. Antimicrob Agents Chemother. 2016;60(8):5040–3. doi: 10.1128/AAC.00360-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonnin RA, Jousset AB, Urvoy N, et al. Detection of GES-5 Carbapenemase in Klebsiella pneumoniae, a Newcomer in France. Antimicrob Agents Chemother. 2017;61(3):e02263–16. doi: 10.1128/AAC.02263-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedersen T, Sekyere JO, Govinden U, et al. Spread of Plasmid-Encoded NDM-1 and GES-5 Carbapenemases among Extensively Drug-Resistant and Pandrug-Resistant Clinical Enterobacteriaceae in Durban, South Africa. Antimicrob Agents Chemother. 2018;62(5):e02178–17. doi: 10.1128/AAC.02178-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ray A, Nordén B. Peptide nucleic acid (PNA): Its medical and biotechnical applications and promise for the future. FASEB J. 2000;14(9):1041–60. doi: 10.1096/fasebj.14.9.1041. [DOI] [PubMed] [Google Scholar]
- 29.Guo S, Du D, Tang L, et al. PNA-assembled graphene oxide for sensitive and selective detection of DNA. Analyst. 2013;138(11):3216–20. doi: 10.1039/c3an00266g. [DOI] [PubMed] [Google Scholar]
- 30.Lee H, Yoon EJ, Kim D, et al. Antimicrobial resistance of major clinical pathogens in South Korea, May 2016 to April 2017: First one-year report from Kor-GLASS. Euro Surveill. 2018;23(42):1800047. doi: 10.2807/1560-7917.ES.2018.23.42.1800047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791–8. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okeke IN, Peeling RW, Goossens H, et al. Diagnostics as essential tools for containing antibacterial resistance. Drug Resist Updat. 2011;14(2):95–106. doi: 10.1016/j.drup.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Azimi L, Talebi M, Owlia P, et al. Tracing of false negative results in phenotypic methods for identification of carbapenemase by real-time PCR. Gene. 2016;576(1 Pt 1):166–70. doi: 10.1016/j.gene.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 34.Swayne R, Ellington MJ, Curran MD, et al. Utility of a novel multiplex TaqMan PCR assay for metallo-beta-lactamase genes plus other TaqMan assays in detecting genes encoding serine carbapenemases and clinically significant extended-spectrum beta-lactamases. Int J Antimicrob Agents. 2013;42(4):3523–56. doi: 10.1016/j.ijantimicag.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 35.Banerjee R, Humphries R. Clinical and laboratory considerations for the rapid detection of carbapenem-resistant Enterobacteriaceae. Virulence. 2017;8(4):427–39. doi: 10.1080/21505594.2016.1185577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitchel B, Rasheed JK, Endimiani A, et al. Genetic Factors Associated with Elevated Carbapenem Resistance in KPC-Producing Klebsiella pneumoniae. Antimicrob Agents Chemother. 2010;54(10):4201–7. doi: 10.1128/AAC.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee HJ, Lee DG. Carbapenem-resistant Enterobacteriaceae: Recent updates and treatment strategies. J Korean Med Assoc. 2018;61(4):281–9. doi: 10.5124/jkma.2018.61.4.281. [DOI] [Google Scholar]
- 38.Landman D, Bratu S, Quale J. Contribution of OmpK36 to carbapenem susceptibility in KPC-producing Klebsiella pneumoniae. J Med Microbiol. 2009;58(Pt 10):1303–8. doi: 10.1099/jmm.0.012575-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wozniak A, Villagra NA, Undabarrena A, et al. Porin alterations present in non-carbapenemase-producing Enterobacteriaceae with high and intermediate levels of carbapenem resistance in Chile. J Med Microbiol. 2012;61(Pt9):1270–9. doi: 10.1099/jmm.0.045799-0. [DOI] [PubMed] [Google Scholar]