Abstract
Ex vivo differentiation of human hematopoietic stem cells is a widely used model for studying hematopoiesis. The protocol described here is for cytokine induced differentiation of CD34+ hematopoietic stem and progenitor cells to the four myeloid lineage cells. CD34+ cells are isolated from human umbilical cord blood and co-cultured with MS-5 stromal cells in the presence of cytokines. Immunophenotypic characterization of the stem and progenitor cells, and the differentiated myeloid lineage cells are described. Using this protocol, CD34+ cells may be incubated with small molecules or transduced with lentiviruses to express myeloid disease mutations to investigate their impact on myeloid differentiation.
Keywords: CD34+ cells, myeloid differentiation, hematopoietic stem and progenitor cells, granulocyte, monocyte, erythrocyte, megakaryocyte, common myeloid progenitor, granulocyte monocyte progenitor, megakaryocyte erythroid progenitor, flow cytometry
SUMMARY:
Here, we present a protocol for immunophenotypic characterization and cytokine induced differentiation of cord blood derived CD34+ hematopoietic stem and progenitor cells to the four myeloid lineages. The applications of this protocol include investigations on the effect of myeloid disease mutations or small molecules on myeloid differentiation of the CD34+ cells.
INTRODUCTION:
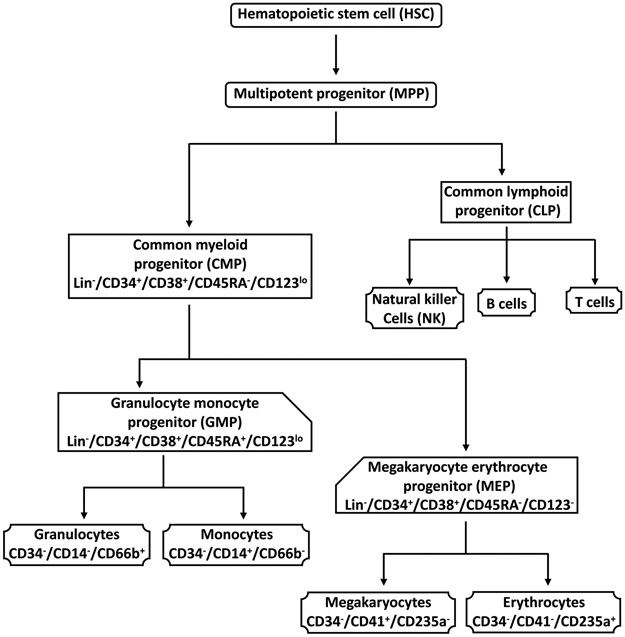
Normal differentiation of hematopoietic stem cells (HSCs) is critical for maintenance of physiological levels of all blood cell lineages. During differentiation, in a coordinated response to extracellular cues including growth factors and cytokines, HSCs first give rise to multipotent progenitor (MPP) cells that have lympho-myeloid potential1-4 (Figure 1). MPPs give rise to common myeloid progenitors (CMPs) and common lymphoid progenitors (CLPs) that are lineage-restricted. CLPs differentiate into the lymphoid lineages comprised of B, T, and natural killer cells. CMPs generate the myeloid lineages through two more restricted progenitor populations, megakaryocyte erythroid progenitors (MEPs), and granulocyte monocyte progenitors (GMPs). MEPs give rise to megakaryocytes and erythrocytes, whereas GMPs give rise to granulocytes and monocytes. In addition to arising through CMPs, megakaryocytes have been reported to also arise directly from HSCs or early MPPs via non-canonical pathways5,6.
Figure 1: Hematopoietic differentiation schematic.
The pluripotent hematopoietic stem cell (HSC) differentiates into the multipotent progenitor (MPP), which gives rise to the common myeloid progenitor (CMP) and the common lymphoid progenitor (CLP) cells. CMPs generate two other myeloid progenitors, the granulocyte monocyte progenitors (GMPs) and the megakaryocyte erythrocyte progenitors (MEPs). Granulocytes and monocytes arise from GMPs, and erythrocytes and megakaryocytes arise from MEPs. CLPs give rise to natural killer, B, and T cells. The cell surface markers used to characterize the cell populations in this protocol are indicated. This figure has been modified from Bapat et al.11.
Hematopoietic stem and progenitor cells (HSPCs) are characterized by the surface marker CD34 and the lack of lineage specific markers (Lin−). Other surface markers that are commonly employed to distinguish HSCs and myeloid progenitor populations include CD38, CD45RA, and CD1232 (Figure 1). HSCs and MPPs are Lin−/CD34+/CD38− and Lin−/CD34+/CD38+, respectively. Myeloid committed progenitor populations are distinguished by the presence or absence of CD45RA and CD123. CMPs are Lin−/CD34+/CD38+/CD45RA−/CD123lo, GMPs are Lin−/CD34+/CD38+/CD45RA+/CD123lo, and MEPs are Lin−/CD34+/CD38+/CD45RA−/CD123−.
The total population of CD34+ stem and progenitor cells can be obtained from human umbilical cord blood (UCB), bone marrow, and peripheral blood. CD34+ cells constitute 0.02% to 1.46% of total mononuclear cells (MNCs) in human UCB, whereas their percentage varies between 0.5% and 5.3% in bone marrow and is much lower at ~0.01% in peripheral blood7-9. The proliferative capacity and differentiation potential of UCB derived CD34+ cells is significantly higher than that of bone marrow or peripheral blood cells1,10, thereby offering a distinct advantage for obtaining sufficient material for molecular analyses in combination with performing immunophenotypic and morphological characterization of the cells during differentiation.
Ex vivo differentiation of umbilical cord blood derived CD34+ HSPCs is a widely applied model for investigating normal hematopoiesis and hematopoietic disease mechanisms. When cultured with the appropriate cytokines, the UCB CD34+ HSPCs can be induced to differentiate along the myeloid or lymphoid lineages11-16. Here, we describe protocols for isolation and immunophenotypic characterization of the CD34+ HSPCs from human UCB, and for their differentiation to myeloid lineage cells. This culture system is based on cytokine-induced differentiation of HSPCs in the presence of MS-5 stromal cells to mimic the microenvironment in bone marrow. The culture conditions cause an initial expansion of the CD34+ cells, followed by their differentiation to cells that express markers for the four myeloid lineage cells, namely granulocytes (CD66b), monocytes (CD14), megakaryocytes (CD41), and erythrocytes (CD235a). Applications of the CD34+ cell differentiation protocol include studies on molecular mechanisms regulating hematopoiesis, and investigations of the impact of myeloid disease associated mutations and small molecules on self-renewal and differentiation of HSPCs.
PROTOCOL:
Ethics Statement: UCB for experimentation was donated by healthy individuals after informed consent to Maricopa Integrated Health Systems (MIHS), Phoenix. The deidentified units were obtained through a Material Transfer Agreement between MIHS and the University of Arizona.
1. Reagents and buffers
NOTE: Prepare all reagents and buffers under sterile conditions in a biological safety cabinet.
1.1. Prepare sterile PBS-BSA-EDTA (PBE) buffer by adding 7.2 mL of sterile 35% bovine serum albumin (BSA; use sterile tissue culture grade 35% BSA) and 5 mL of sterile 0.5 M ethylene diamine tetra acetic acid (EDTA) to 487.8 mL of sterile Dulbecco’s phosphate buffered saline (DPBS). Filter sterilize and de-gas the buffer by leaving the filter attached to vacuum in a biological safety cabinet for at least 2 h. Aliquot the de-gassed buffer in smaller volumes (250 mL) and store at 4 °C.
1.2. Prepare 1x Hanks balanced salt solution (1x HBSS). Dilute 100 mL of 10x HBSS stock in 900 mL of sterile distilled water to make 1 L of 1x HBSS and adjust pH to 7.2. Filter sterilize the 1x HBSS before use.
1.3. Prepare 2% acetic acid solution by adding 2 mL of glacial acetic acid to 98 mL of distilled water.
1.4. Prepare 20 ng/mL recombinant human fibronectin fragment solution in 1x PBS. Make 10 mL per 48-well plate.
1.5. Prepare 2% BSA. To 100 mL 1x PBS, add 2 grams of BSA fraction V. Filter sterilize before use.
1.6. Make complete Dulbecco’s Modified Eagle medium (DMEM) by adding the DMEM powder, 3.7 gm sodium bicarbonate, 100 mL fetal bovine serum (FBS) and 5 mL penicillin-streptomycin (PS) to 800 mL of sterile distilled water. Mix and make up the volume to 1 L, and filter sterilize before use.
1.7. Prepare freezing medium by mixing 90 mL of FBS and 10 mL of sterile dimethyl sulfoxide (DMSO).
1.8. Prepare stimulation medium. To serum free medium, add L-glutamine to 2 mM, and cytokines to a final concentration of 50 ng/mL for stem cell factor (SCF), thrombopoietin (TPO), and Fms-like tyrosine kinase 3 ligand (Flt3L), and 20 ng/mL for interleukin-3 (IL3) (see Table of Materials for preparation of cytokine stock solutions). Make 5 mL per 48-well plate.
Table of Materials
| Name of Material/Reagent | Company | Catalog Number |
Comments/Description |
|---|---|---|---|
| 0.4 % Trypan blue solution | Thermo Fisher Scientific | 15250-061 | Dilute working stock to 0.2 % in sterile 1 x PBS |
| 0.5 M UltraPure Ethylene diamine tetra acetic acid, pH 8.0 | Gibco | 15575–038 | |
| 10x Hanks Balanced Salt Solution (HBSS) | Invitrogen | 14185052 | Dilute to 1x with sterile distilled water & pH to 7.2 |
| 2.5 % Trypsin, no phenol red | Thermo Fisher Scientific | 15090046 | Dilute working stock to 1x with sterile 1 x PBS |
| 30μm Pre-separation filters | Miltenyi biotech | 130-041-407 | |
| 35 % sterile Bovine serum albumin | Sigma-Aldrich | A7979 | |
| 7-AAD | Biolegend | 420404 | Used as a live/dead stain to eliminate dead cells from FACS |
| Anti-human CD10-FITC antibody (Clone HI10a) | Biolegend | 312207 | Use 1:20 dilution |
| Anti-human CD11b-FITC (activated) antibody (Clone CBRM1/5) | Biolegend | 301403 | Use 1:5 dilution |
| Anti-human CD123-APC antibody (Clone 6H6) | Biolegend | 306012 | Use 1:20 dilution |
| Anti-human CD14-PE antibody (Clone M5E2) | Biolegend | 301806 | Use 1:20 dilution |
| Anti-human CD19-FITC antibody (Clone 4G7) | BD Biosciences | 347543 | Use 1:5 dilution |
| Anti-human CD235a-APC antibody (Clone GA-R2 (HIR2)) | BD Biosciences | 551336 | Use 1:20 dilution |
| Anti-human CD235a-FITC antibody (Clone HIR2) | Biolegend | 306609 | Use 1:50 dilution |
| Anti-human CD3-FITC antibody (Clone UCHT1) | Biolegend | 300405 | Use 1:20 dilution |
| Anti-human CD34-APC-Cy7 antibody (Clone 581) | Biolegend | 343514 | Use 1:20 dilution |
| Anti-human CD38-PE antibody (Clone HIT2) | Biolegend | 303506 | Use 1:20 dilution |
| Anti-human CD41a-PerCP-Cy5.5 antibody (Clone HIP8) | Biolegend | 303720 | Use 1:20 dilution |
| Anti-human CD45Ra-PE-Cy7 antibody (Clone HI100) | Biolegend | 304126 | Use 1:20 dilution |
| Anti-human CD66b-PE-Cy7 antibody (Clone G10F5) | Biolegend | 305116 | Use 1:20 dilution |
| Anti-human CD7-FITC antibody (Clone CD7–6B7) | Biolegend | 343103 | Use 1:20 dilution |
| Dimethyl sulfoxide (DMSO) | Fisher Scientific | BP231–100 | Filter sterilize before use |
| Dulbecco’s Modified Eagle Medium (DMEM) powder with L-Glutamine | Gibco | 12100046 | Reconstitute 1 packet to make 1 L of DMEM media with sodium bicarbonate, 10% FBS and 1x |
| Fetal bovine serum, Australian source, heat inactivated | Omega Scientific | FB-22 Lot #609716 | |
| Human CD34 microbead kit | Miltenyi biotech | 130-046-702 | |
| Human Thrombopoietin (TPO), research grade | Miltenyi biotech | 130-094-011 | Make a stock of 100μ/mL in 1x PBS + 0.1 % BSA. Use 50 ng/mL for both myeloid differentiation |
| L-Glutamine | Omega Scientific | GS-60 | 2 mM concentration in stimulation medium |
| LS Columns | Miltenyi biotech | 130-042-401 | |
| MACS Multi stand | Miltenyi biotech | 130-042-303 | |
| MidiMACS magnetic separator | Miltenyi biotech | 130-042-302 | |
| MNC fractionation media (Ficoll-Paque PLUS) | GE Healthcare | 17-1440-03 | |
| MS-5 cells | Gift from the laboratory of Gay | ||
| Paraformaldehyde | Sigma-Aldrich | P6148 | Heat 800 mL of 1x PBS in a glass beaker on a stir plate in a chemical hood to ~65°C. Add 10 g of paraformaldehyde powder. To completely dissolve the paraformaldehyde, raise the pH |
| Penicillin & Streptomycin | Gibco | 15140-122 | |
| Poly-L lysine | Sigma-Aldrich | P2636 | Make a 10 mg/mL stock in 1x |
| Recombinant human erythropoietin-alpha (rHu EPO-α) | BioBasic | RC21-15 | Make a stock of 2000 units/mL in 1x PBS + 0.1 % BSA. Use 4 units/mL for myeloid |
| Recombinant human fibronectin fragment (RetroNectin) | Takara | T100B | Use 20μg/mL diluted in sterile 1x PBS to coat wells prior to stimulation of CD34+ HSCs. |
| Recombinant human Flt-3 ligand (rHu Flt-3L) | BioBasic | RC214-16 | Make a stock of 100μg/mL in 1x PBS + 0.1 % BSA. Use 5 ng/mL for myeloid differentiation and |
| Recombinant human interleukin-3 (rHu IL-3) | BioBasic | RC212-14 | Make a stoc k of 100μg/mL in 1x PBS + 0.1 % BSA. Use 5 ng/mL for myeloid differentiation and |
| Recombinant human stem cell factor (rHu SCF) | BioBasic | RC213-12 | Make a stoc k of 100μg/mL in 1x PBS + 0.1 % BSA. Use 5 ng/mL for myeloid differentiation and |
| Serum free medium (X-Vivo-15) | Lonza | 04-418Q | |
| Sodium bicarbonate | Fisher Scientific | BP328-500 | |
| Wright-Giemsa stain, modified | Sigma-Aldrich | WG16-500 | Use according to manufacturer's instructions |
| Equipment | |||
| BD LSR II flow cytometer | BD | ||
| Centrifuge | Sorvall Legend RT | ||
| Light microscope | Olympus |
1.9. Prepare 1x myeloid differentiation medium. To complete DMEM, add cytokines to a final concentration of 5 ng/mL for SCF, Flt3L, and IL3, 50 ng/mL for TPO, and 4 units/mL for erythropoietin (EPO) (see Table of Materials for preparation of cytokine stock solutions). Make 5 mL per 48-well plate.
1.10. Prepare 2x myeloid differentiation medium. To complete DMEM, add cytokines to a final concentration 10 ng/mL SCF, Flt3L, and IL3, 100 ng/mL for TPO, and 4 units/mL for EPO (see Table of Materials for preparation of cytokine stock solutions). Make 5 mL per 48-well plate.
1.11. Prepare flow cytometry buffer. To 1 L of 1x PBS, add 10 g of BSA fraction V.
1.12. Prepare 1% paraformaldehyde in 1x PBS (see Table of Materials for details).
1.13. Prepare poly-L-lysine solution by adding 500 μL of 10 mg/mL poly-L-lysine to 49.5 mL of 1x PBS.
2. Isolation of mononuclear cells from umbilical cord blood
NOTE: For the protocol described below, cord blood volumes of 90 to 100 mL were used. The isolation of CD34+ cells must be performed under sterile conditions in a biological safety cabinet.
2.1. Spray the bag containing UCB with 70% ethanol to sterilize the outer surface before introducing it into the biological safety cabinet. Transfer the UCB into a sterile 250 mL plastic bottle and dilute it 1:1 by adding an equal volume of room temperature (RT) 1x HBSS.
2.2. Dispense 15 mL of MNC fractionation medium (MFM; see Table of Material) per tube into approximately six sterile 50 mL conical tubes. Tilt the tube with MFM, and gently pour 30 mL of the diluted UCB onto the MFM layer without disturbing it.
2.3. Centrifuge the tubes at 400 x g for 30 min at RT with slow acceleration and no brake.
2.4. After centrifugation, aspirate between 15–20 mL of the plasma above the MNC layer. Gently collect the MNCs with a pipette and transfer to a new 50 mL conical tube.
NOTE: MNCs deposit as a white buffy layer at the interface of plasma and MFM, and the red blood cells (RBCs) sediment at the bottom of the conical tube.
2.5. Pool MNCs collected from 2–3 tubes together. Make up the volume of each tube to 50 mL with cold PBE buffer.
2.6. Centrifuge the tubes at 300 x g for 10 min at 4 °C, with the brake on low.
2.7. Aspirate the supernatant and tap the tube gently to loosen the cell pellet.
2.8. Re-suspend the pelleted cells in 5 mL of ice cold PBE buffer. Use the same 5 mL of re-suspended cells to collect cells from other tubes. Combine cells from 3 to 4 tubes together in one 50 mL conical tube.
2.9. To collect any remaining cells, wash all the tubes with 5 mL of cold PBE buffer and add to the 50 mL conical tube.
2.10. Count the MNCs by diluting 10 μL of cell suspension into 490 μL acetic acid solution.
NOTE: Acetic acid lyses any contaminating RBCs to allow easier counting of MNCs.
2.11. Make up the volume of the cell suspension to 50 mL with ice-cold PBE buffer. Centrifuge the cells at 300 x g for 10 min at 4 °C, with brake on low.
NOTE: The MNC suspension may be processed immediately for isolation of CD34+ cells or frozen for later applications.
2.12. To freeze the MNCs, remove the supernatant and re-suspend at a density of 2 x 107 cells/mL in freezing medium at RT.
2.13. Freeze the cells at −80 °C overnight in a cell freezing container and then transfer them to liquid nitrogen.
3. Isolation of CD34+ cells from mononuclear cells
3.1. If using freshly isolated MNCs proceed directly to step 3.3.
3.2. If using frozen MNCs, quickly thaw the frozen cells in a 37 °C water bath and transfer to a 50 mL conical tube. Collect any remaining cells in the vials with 1 mL of ice-cold PBE buffer and transfer to the 50 mL conical tube.
3.3. Make up the volume of the MNC suspension to 50 mL with cold PBE buffer and centrifuge at 600 x g for 5 min at 4 °C.
3.4. Remove the supernatant and tap the tube gently to loosen the cell pellet. Re-suspend the MNCs to 1 x 108 cells in 300 μL of ice-cold PBE buffer.
3.5. Add 100 μL of FcR blocking reagent from the magnetic-activated cell sorting (MACS) human CD34 microbead kit per 1 x 108 MNCs and mix gently.
NOTE: This blocks nonspecific binding to the CD34 microbeads.
3.6. Add 100 μL of the CD34 microbeads per 1 x 108 cells. Mix gently and incubate at 4 °C for 30 min in a refrigerator. Do not incubate on ice.
3.7. While the cells are incubating, place an LS column and pre-separation filter in the MACS separator magnetic field. Equilibrate the column with ice-cold PBE buffer by adding 2 mL directly into column and 1 mL in the pre-separation filter. Allow the column to empty into a 15 mL conical tube and discard the flow through.
3.8. Remove the MNCs from 4 °C, add ice-cold PBE buffer to make up the volume to 15 mL, and centrifuge the cells at 300 x g for 10 min at 4 °C, with the brake on low.
3.9. Aspirate the supernatant and re-suspend the cells in 1 mL of ice-cold PBE buffer.
3.10. Add the cell suspension to the pre-separation filter placed on the LS column.
NOTE: The pre-separation filter removes any large cell aggregates that may block the column.
3.11. Rinse the tube with 3 mL of ice-cold PBE buffer and add it to pre-separation filter placed on the LS column. After this, discard the pre-separation filter.
3.12. Wash the LS column four times by adding 3 mL of ice-cold PBE buffer, allowing the column to drain each time.
NOTE: The CD34+ cells will remain bound to the column and the unlabeled cells will flow through the column.
3.13. After the final wash, remove the column from the MACS separator magnetic field and place it in a new 15 mL conical tube, away from the magnet.
3.14. Add 5 mL ice-cold PBE buffer to the column and expel the bound CD34+ cells by firmly pushing in the plunger.
3.15. Optionally, pass CD34+ cells isolated from the first column over another LS column with the pre-separation filter as described above (steps 3.9–3.13).
3.16. Count the isolated CD34+ cells with trypan blue (10 μL of cells and 10 μL of trypan blue) to determine the total number of live cells.
NOTE: At this stage, the isolated CD34+ cells may be frozen for later use or may be processed directly for analysis of HSPCs by flow cytometry (section 4) or for differentiation to myeloid lineage cells (section 5).
3.17. Centrifuge the cell suspension at 600 x g for 5 min at RT, with brake on low.
3.18. To freeze the CD34+ cells, aspirate the supernatant and re-suspend up to 5 x 106 cells in 1 mL of freezing medium and freeze as described above (step 2.13). Otherwise, proceed to section 4 or 5.
4. Determination of stem and progenitor populations by flow cytometry
4.1. To determine the fractions of stem and progenitor populations in the freshly isolated CD34+ HSPCs, centrifuge them at 600 x g for 5 min and re-suspend 1 x 106 cells in 50 μL of flow cytometry buffer.
4.2. To 1 x 106 CD34+ cells, add antibodies (see Table of Materials for dilutions) to lineage markers (CD3, CD7, CD10, CD11b, CD19 and CD235a – all FITC labelled), CD34 (APC-Cy7), CD38 (PE), CD45RA (PE-Cy7), CD123 (APC) and 7-aminoactinomycin D (7-AAD; as a live/dead stain) and make up the total volume to 100 μL. Incubate on ice in the dark for 20 min.
NOTE: Fewer than 1 x 106 CD34+ cells may be employed for analysis. If staining fewer cells, the total volume and the volume of antibodies added should be reduced accordingly. For flow cytometry analysis, unstained and single stained MNCs should be used as controls to set positive and negative gates for each fluorophore.
4.3. After incubation, wash the cells with 1 mL of flow cytometry buffer and centrifuge at 600 x g for 5 min.
4.4. Optionally, fix the stained cells in 0.5 mL of 1% paraformaldehyde solution for 10 min in the dark at RT.
4.5. Centrifuge at 600 x g for 5 min and aspirate the supernatant.
4.6. Wash the fixed/stained cells with 1 mL of flow cytometry buffer and centrifuge at 600 x g for 5 min at 4 °C.
4.7. Aspirate the supernatant, re-suspend the cells in 500 μL of flow cytometry buffer, and analyze by a flow cytometer (Table of Materials).
5. Myeloid differentiation of the CD34+ hematopoietic stem and progenitor cells:
NOTE: To differentiate the CD34+ HSPCs to the myeloid lineage, they are first stimulated in recombinant human fibronectin fragment coated plates and then seeded on a layer of MS-5 stromal cells in myeloid differentiation medium. Differentiation may be monitored each week for 3 weeks based on expression of cell surface markers specific to the four myeloid lineages. During stimulation and differentiation, all incubations are performed at 37 °C and 5% CO2 in a humidified chamber. All steps should be performed in a biological safety cabinet.
5.1. Coat the wells of a sterile uncoated 48-well plate by adding 200 μL/well of recombinant human fibronectin solution in 1x PBS for 2 h at RT in the biological safety cabinet.
5.2. After 2 h, carefully remove the recombinant human fibronectin solution and discard.
5.3. Block the wells with 200 μL/well of 2% BSA for 30 min at RT in the biological safety cabinet.
5.4. Aspirate the BSA solution and wash the wells twice with 500 μL of sterile 1x PBS.
NOTE: Fibronectin fragment coated plates may be stored at 4 °C for up to 2 weeks.
5.5. To stimulate the CD34+ HSPCs, re-suspend the freshly isolated cells (from step 3.17) at a density of 1 x 105 cells/200 μL or 5 x 105 cells/mL of warm 1x stimulation medium.
5.6. If using frozen CD34+ cells, quickly thaw the cells and transfer them to a 15 mL conical tube containing warm complete DMEM. Centrifuge at 600 x g for 5 min and re-suspend the cells as described in step 5.5.
5.7. Plate 200 μL of the CD34+ cell suspension per well of the fibronectin fragment coated 48-well plate and incubate for 48 h.
5.8. On the next day, seed 15,000 MS-5 stromal cells in 200 μL of complete DMEM per well of a 48-well tissue culture treated plate and incubate at 37 °C for 24 h.
5.9. After 48 h of stimulation, collect the CD34+ cells by gentle pipetting. Wash the wells with 500 μL of warm DMEM and pool with the cell suspension.
5.10. Count the cells with trypan blue. Centrifuge the suspension at 600 x g for 5 min and re-suspend at a density of 5000 cells/200 μL of 1x myeloid differentiation medium.
5.11. From the 48-well tissue culture plate seeded with MS-5 stromal cells, gently aspirate the medium without disturbing the cells. Layer 200 μL (5000 cells) of the CD34+ cell suspension per well of the plate and incubate at 37 °C.
NOTE: Culture of MS-5 cells can be maintained in complete DMEM. On the day the CD34+ cells are seeded, the MS-5 cells must be in a uniform layer (80–90% confluence). It is recommended that the MS-5 cells are plated at least 24 hours prior to seeding of the CD34+ cells. If MS-5 cells are to be plated 48 h or 72 h prior to seeding the CD34+ cells, the cell density should be adjusted accordingly. It is also recommended that the peripheral wells of the 48-well plate be left blank or filled with 200 μL of sterile water to avoid edge effects.
5.12. Perform half-media change every 3–4 days by gently removing 100 μL of the medium from the top of each well and replacing it with 100 μL of 2x myeloid differentiation medium per well.
5.13. On day 21, harvest the cells for analysis of myeloid lineage marker expression by flow cytometry. Without disturbing the MS-5 layer, gently pipette the media and transfer cells to a 5 mL tube. Wash each well with 500 μL of complete DMEM and add to the 5 mL tube.
NOTE: For staining and flow cytometry analysis, cells from 1–2 wells are sufficient. Differentiation may also be monitored on days 1, 7, and 14. If performing immunophenotyping on multiple days, the numbers of wells needed for analysis should be calculated accordingly.
5.14. Stain 5 x 105 cells with antibodies to CD34 (APC-Cy7), CD66b (PE-Cy7), CD14 (PE), CD41 (PerCP-Cy5.5), and CD235a (APC) in a total volume of 50 μL. Incubate on ice in the dark for 20 min. Include unstained and single stained MNCs as controls for setting positive and negative gates for each fluorophore, and 7-AAD as a live/dead stain to eliminate dead cells from the analysis.
5.15. Wash and fix (optional) the stained cells as described above (steps 4.3–4.6).
5.16. Re-suspend the cells in 500 μL of flow cytometry buffer and analyze by the flow cytometer.
6. Assessment of cellular morphology
6.1. Clean microscope slides by spraying ethanol and wiping until squeaky.
6.2. Immerse clean slides in poly-L-lysine solution for 5 min at RT and dry for 1 h at RT.
6.3. Resuspend the HSPCs from step 3.14 and differentiated cells from step 5.13 in 10 μL of flow cytometry buffer.
6.4. Transfer the cell suspension to the coated slides and leave them to air-dry.
NOTE: Slides from day 1 and day 21 can be stored at RT and stained simultaneously.
6.5. Pour 0.5 mL of the Wright-Giemsa stain on the slide and let stand for 3 min at RT.
6.6. Add equal volume of deionized water and let stand for additional 10 min at RT.
6.7. Wash the slide in deionized water.
6.8. Visualize stained cells at a magnification of 60x or 100x by a microscope.
REPRESENTATIVE RESULTS:
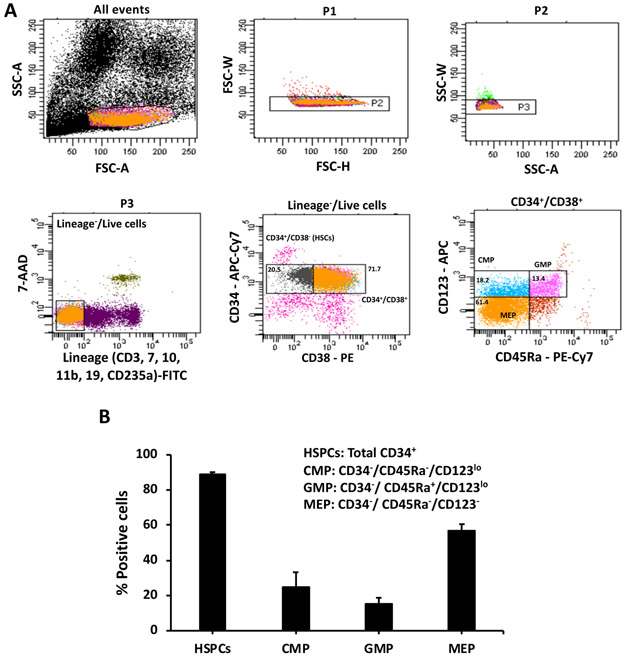
Application of the above protocols yields 5.6 (± 0.5) x 108 MNCs and 1 (± 0.3) x 106 CD34+ cells from a cord blood unit of ~100 mL. The percentage of total CD34+ cells ranges between 80–90% (Figure 2A,B). Immunophenotypic analysis by the scheme described by Manz et al.5 demonstrates that the CD34+ cells typically consist of ~20% HSCs and ~2% MPPs that are Lin− /CD34+/CD38− and Lin−/CD34+/CD38+, respectively (Figure 1 and Figure 2A). In the MPP population, the percentages of CMPs (Lin−/CD34+/CD38+/CD123lo/CD45RA−), GMPs (Lin− /CD34+/CD38+/CD123lo/CD45RA+), and MEPs (Lin−/CD34+/CD38+/CD123−/CD45RA−) are approximately 25%, 15%, and 56%, respectively (Figure 1 and Figure 2B).
Figure 2: Immunophenotypic analysis of the hematopoietic stem and progenitor cells.
(A) Cells were gated on forward and side scatter to select a single cell population. Dead and lineage positive cells were eliminated by staining with 7-AAD and antibodies to CD3, CD7, CD10, CD11b, CD19, and CD235a (all FITC stained). The Lin−/live cells were analyzed with antibodies to CD34 (APC-Cy7), CD38 (PE), CD123 (APC), and CD45RA (PE-Cy7). The progenitors were distinguished from the CD34+/CD38+ cells. CMPs are CD123lo/CD45RA−, GMPs are CD123lo/CD45RA+ and MEPs are CD123−/CD45RA−. Representative scatter plots from an experiment are shown. (B) Percentages of total HSPCs (total CD34+ cells), CMPs, GMPs, and MEPs in cord blood are represented. Data presented are averages with standard error from at least 3 experiments.
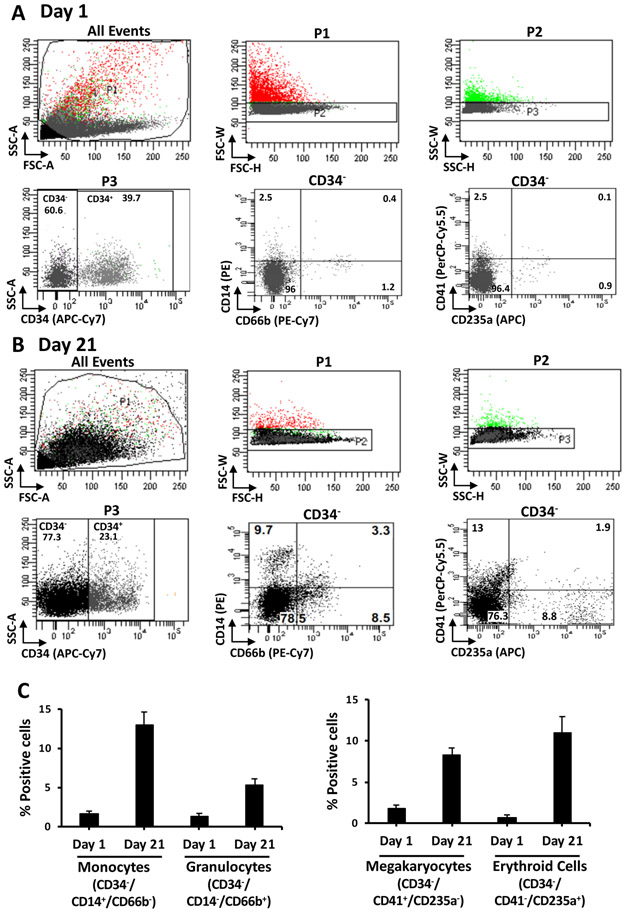
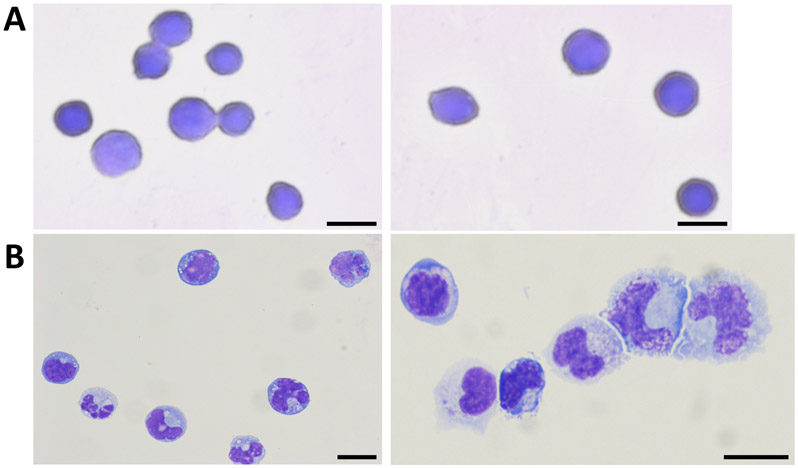
During incubation of the isolated HSPCs in myeloid differentiation conditions, there is progressive loss of the CD34 marker. Immunophenotyping shows that the percentage of CD34+ cells reduces from ~90% at isolation (Figure 2A) to ~23% on day 21 (Figure 3B). Analysis of the CD34− population demonstrates a concomitant rise in the percentage of cells expressing mature myeloid lineage markers. There is an increase in the percentage of cells expressing markers for monocytes (CD34−/CD14+/CD66b−) from an average of 1.7% to 12%, for granulocytes (CD34− /CD14−/CD66b+) from 1.3% to 5.3%, for megakaryocytes (CD34−/CD41+/CD235a−) from 1.8% to 8%, and for erythroid cells (CD34−/CD41−/CD235a+) from 0.7% to 11% (Figure 3C). Examination of Wright-Giemsa stained cells demonstrates that the morphological characteristics of the cells on day 1 and day 21 are distinct. The undifferentiated cells have large round nuclei, and very little cytoplasm. On the other hand, cells from differentiated cultures exhibit characteristics of lineage cells including mature monocytes, granulocytes, and erythroblasts (Figure 4). Thus, the culture conditions described in this protocol promote differentiation of UCB CD34+ cells to the myeloid lineage cells.
Figure 3: Immunophenotypic analysis of myeloid lineage cells.
Single cells were gated based on forward and side scatter. CD34− cells were selected by staining with antibody to CD34 (APC-Cy7). Myeloid lineages in the CD34− population were analyzed with antibodies to CD14 (PE), CD66b (PE-Cy7), CD41 (PerCP-Cy5.5), and CD235a (APC) on day 1 (A) and day 21 (B). Monocytes are CD14+/CD66b−, granulocytes are CD14−/CD66b+, megakaryocytes are CD41+/CD235a− and erythroid cells are CD41−/CD235a+. Representative scatter plots from an experiment are shown. Fractions of monocytes, granulocytes, megakaryocytes, and erythroid cells in the CD34− population on day 1 and day 21 are represented (C). Data presented are averages with standard error from at least 3 experiments. This figure has been modified from Bapat et al.11.
Figure 4: Representative images of HSPCs and differentiated cells.
CD34+ HSPCs (A) and cells from myeloid cultures at day 21 (B) were stained with the Wright-Giemsa stain. Cells corresponding to granulocytes (black arrows), monocytes (blue arrows) and erythroid cells (red arrows) are shown. Scale bars represent 10 μM. This figure has been modified from Bapat et al.11.
DISCUSSION:
The protocol described here is suitable for ex vivo differentiation of UCB derived CD34+ HSPCs to the four myeloid lineages. Initial incubation with a cytokine mix consisting of SCF, TPO, Flt3L and IL3 stimulates the CD34+ cells. Subsequently, differentiation is achieved with a cocktail of SCF, IL3, Flt3L, EPO, and TPO. In this mix, SCF, IL3, and Flt3L are important for survival and proliferation of CD34+ HSCs. EPO and TPO promote differentiation toward erythrocytes and megakaryocytes, respectively, and IL3 promotes differentiation of early granulocyte-monocyte precursors and mature cells13,17,18. One caveat to this method is the observed variation in the percentages of differentiated cells. This could be because of differences in capacities of the CD34+ HSPCs isolated from different donors to give rise to the lineage cells.
The layer of MS-5 cells is critical for efficient differentiation of the CD34+ HSPCs. In our experience, the MS-5 cells need to be plated at least 24 hours prior to seeding of the CD34+ cells. It is important to maintain the confluence of MS-5 cells at 80–90%. Overconfluent MS-5 cells tend to detach, and lower confluency causes reduced differentiation. During incubation, the CD34+ cells expand by ~50-fold. They become confluent by day 14 and must split into new wells containing MS-5 cells on a 48-well plate.
Frequency of media changes is also critical for efficient differentiation and must be performed every 3–4 days to maintain optimal cytokine concentrations. Fewer media changes and lower cytokine concentrations, both cause a reduction in proliferation and differentiation of CD34+ HSPCs. This requirement for large amounts of cytokines can significantly increase costs and thus, is a limitation of this protocol. Another limitation for large scale experiments is the number of CD34+ cells obtained from a unit of cord blood. Typically, a fresh UCB unit of ~100 mL yields about one million CD34+ cells. Storage of the unit beyond 24 hours significantly reduces the yield. Further loss of purity of the CD34+ cells can occur during the column wash steps that are important for removing any unlabeled contaminating cells. If the column clogs during the wash steps, it is recommended that the bound cells from the clogged column be eluted and reloaded on a new column in order to get a pure population of CD34+ cells.
In the literature, different cytokine combinations have been reported that promote differentiation towards either megakaryocytic, erythroid, or granulo-monocyte lineages19-22. An advantage of this protocol is that it allows differentiation along all four myeloid populations, namely monocytes, granulocytes, megakaryocytes, and erythrocytes. Thus, it can be employed for studying normal myeloid differentiation and for investigating the impact of myeloid disease (myelodysplastic syndromes, acute myeloid leukemia, myeloproliferative neoplasms, and others) associated point mutations and chromosomal translocations on molecular and cellular phenotype during proliferation and differentiation of CD34+ cells11,12,23-25. This protocol can also be employed for examining the impact of anti- and pro-inflammatory cytokines, and of potential therapeutic drugs on myeloid differentiation26-28.
ACKNOWLEDGEMENTS:
The authors would like to thank Wendy Barrett, Rachel Caballero, and Gabriella Ruiz of Maricopa Integrated Health Systems for the de-identified and donated cord blood units, Mrinalini Kala for assistance with flow cytometry, and Gay Crooks and Christopher Seet for advice on ex vivo myeloid differentiation. This work was supported by funds to S.S. from the National Institutes of Health (R21CA170786 and R01GM127464) and the American Cancer Society (the Institutional Research Grant 74-001-34-IRG). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
DISCLOSURES:
The authors have nothing to disclose.
REFERENCES:
- 1.Hao QL, Shah AJ, Thiemann FT, Smogorzewska EM & Crooks GM A functional comparison of CD34 + CD38- cells in cord blood and bone marrow. Blood. 86 (10), 3745–3753, (1995). [PubMed] [Google Scholar]
- 2.Manz MG, Miyamoto T, Akashi K & Weissman IL Prospective isolation of human clonogenic common myeloid progenitors. Proc Natl Acad Sci U S A. 99 (18), 11872–11877, (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kondo M, Weissman IL & Akashi K Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91 (5), 661–672, (1997). [DOI] [PubMed] [Google Scholar]
- 4.Seita J & Weissman IL Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med. 2 (6), 640–653, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haas S et al. Inflammation-Induced Emergency Megakaryopoiesis Driven by Hematopoietic Stem Cell-like Megakaryocyte Progenitors. Cell Stem Cell. 17 (4), 422–434, (2015). [DOI] [PubMed] [Google Scholar]
- 6.Sanjuan-Pla A et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature. 502 (7470), 232–236, (2013). [DOI] [PubMed] [Google Scholar]
- 7.Bender JG et al. Phenotypic analysis and characterization of CD34+ cells from normal human bone marrow, cord blood, peripheral blood, and mobilized peripheral blood from patients undergoing autologous stem cell transplantation. Clin Immunol Immunopathol. 70 (1), 10–18, (1994). [DOI] [PubMed] [Google Scholar]
- 8.Fritsch G et al. The composition of CD34 subpopulations differs between bone marrow, blood and cord blood. Bone Marrow Transplant. 17 (2), 169–178, (1996). [PubMed] [Google Scholar]
- 9.Nimgaonkar MT et al. A unique population of CD34+ cells in cord blood. Stem Cells. 13 (2), 158–166, (1995). [DOI] [PubMed] [Google Scholar]
- 10.Hordyjewska A, Popiolek L & Horecka A Characteristics of hematopoietic stem cells of umbilical cord blood. Cytotechnology. 67 (3), 387–396, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bapat A et al. Myeloid Disease Mutations of Splicing Factor SRSF2 Cause G2-M Arrest and Skewed Differentiation of Human Hematopoietic Stem and Progenitor Cells. Stem Cells. 36 1–13, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yip BH et al. The U2AF1S34F mutation induces lineage-specific splicing alterations in myelodysplastic syndromes. J Clin Invest. 127 (6), 2206–2221, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoo ES et al. Myeloid differentiation of human cord blood CD34+ cells during ex vivo expansion using thrombopoietin, flt3-ligand and/or granulocyte-colony stimulating factor. Br J Haematol. 105 (4), 1034–1040, (1999). [DOI] [PubMed] [Google Scholar]
- 14.Hao QL, Smogorzewska EM, Barsky LW & Crooks GM In vitro identification of single CD34+CD38- cells with both lymphoid and myeloid potential. Blood. 91 (11), 4145–4151, (1998). [PubMed] [Google Scholar]
- 15.Moretta F et al. The generation of human innate lymphoid cells is influenced by the source of hematopoietic stem cells and by the use of G-CSF. Eur J Immunol. 46 (5), 1271–1278, (2016). [DOI] [PubMed] [Google Scholar]
- 16.Sanz E et al. Ordering human CD34+CD10-CD19+ pre/pro-B-cell and CD19- common lymphoid progenitor stages in two pro-B-cell development pathways. Proc Natl Acad Sci U S A. 107 (13), 5925–5930, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egeland T et al. Myeloid differentiation of purified CD34+ cells after stimulation with recombinant human granulocyte-monocyte colony-stimulating factor (CSF), granulocyte-CSF, monocyte-CSF, and interleukin-3. Blood. 78 (12), 3192–3199, (1991). [PubMed] [Google Scholar]
- 18.Ogawa M Differentiation and proliferation of hematopoietic stem cells. Blood. 81 (11), 2844–2853, (1993). [PubMed] [Google Scholar]
- 19.Perdomo J, Yan F, Leung HHL & Chong BH Megakaryocyte Differentiation and Platelet Formation from Human Cord Blood-derived CD34+ Cells. J Vis Exp. 10.3791/56420 (130), (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palii CG, Pasha R & Brand M Lentiviral-mediated knockdown during ex vivo erythropoiesis of human hematopoietic stem cells. J Vis Exp. 10.3791/2813 (53), (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies C et al. Silencing of ASXL1 impairs the granulomonocytic lineage potential of human CD34(+) progenitor cells. Br J Haematol. 160 (6), 842–850, (2013). [DOI] [PubMed] [Google Scholar]
- 22.Caceres G et al. TP53 suppression promotes erythropoiesis in del(5q) MDS, suggesting a targeted therapeutic strategy in lenalidomide-resistant patients. Proc Natl Acad Sci U S A. 110 (40), 16127–16132, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi H et al. ASXL1 plays an important role in erythropoiesis. Sci Rep. 6 28789, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazumdar C et al. Leukemia-Associated Cohesin Mutants Dominantly Enforce Stem Cell Programs and Impair Human Hematopoietic Progenitor Differentiation. Cell Stem Cell. 17 (6), 675–688, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung KY et al. Enforced expression of an Flt3 internal tandem duplication in human CD34+ cells confers properties of self-renewal and enhanced erythropoiesis. Blood. 105 (1), 77–84, (2005). [DOI] [PubMed] [Google Scholar]
- 26.Ambrosini P et al. IL-1beta inhibits ILC3 while favoring NK-cell maturation of umbilical cord blood CD34(+) precursors. Eur J Immunol. 45 (7), 2061–2071, (2015). [DOI] [PubMed] [Google Scholar]
- 27.Batard P et al. TGF-(beta)1 maintains hematopoietic immaturity by a reversible negative control of cell cycle and induces CD34 antigen up-modulation. J Cell Sci. 113 ( Pt 3) 383–390, (2000). [DOI] [PubMed] [Google Scholar]
- 28.Huang N, Lou M, Liu H, Avila C & Ma Y Identification of a potent small molecule capable of regulating polyploidization, megakaryocyte maturation, and platelet production. J Hematol Oncol. 9 (1), 136, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]