Abstract
Human induced pluripotent stem cells (iPSCs) that express stable and robust fluorescent proteins have proven to be indispensable in basic and translational research. These reporter iPSC lines can greatly facilitate cell imaging, sorting, and tracking in vitro and in vivo studies. Here, we document two reporter human iPSC lines generated by gene-editing technologies that precisely integrated one-copy of a tdTomato transgene driven by strong CAG promoter into the AAVS1 human safe harbor locus.
1. Resource utility
These fluorescent reporter human induced pluripotent stem cell (iPSC) lines created by introducing a tdTomato transgene driven by a constitutive CAG promoter into the established ND2.0 iPSC cell line are useful for cell tracking and sorting in vitro and in vivo studies concerning human developmental research and disease-modeling.
2. Resource details
The NHLBIi003-A-1 and NHLBIi003-A-2 iPSC reporter cell lines were created by integrating a tdTomato transgene into the AAVS1 safe harbor locus of the established ND2.0 iPSC line with CRISPR/Cas9 (Chen et al., 2011; Cerbini et al., 2015). tdTomato is about three times as bright as the widely used green fluorescent protein (GFP), making it the brightest fluorescent protein used in research. Its long emission wavelength and low light absorption by animal tissues also make tdTomato a better candidate than GFP for in vivo deep-tissue imaging applications (Deliolanis et al., 2008). Furthermore, in our construct, tdTomato expression is under control of the constitutively active CAG promoter (Supplementary Fig. S1B), which is one of strongest promoters reported in iPSC and iPSC-derived cells. These advantages coupled with the transgene’s stable expression within the safe harbor locus make these tdTomato reporter iPSC lines useful for tracking and sorting iPSCs as well as iPSC-derived cell types grown in co-culture in vitro. They will also prove useful for tracking the cells in vivo transplantation applications (Table 1).
Table 1.
Summary of lines.
| iPSC line names | Abbreviation in figures | Gender | Age | Ethnicity | Genotype of locus | Disease |
|---|---|---|---|---|---|---|
| NHLBIi003-A-1 | ND2-tdTom1 | Male | Newborn | Unknown | CAG-tdTomato | N/A |
| NHLBIi003-A-2 | ND2-tdTom4 | Male | Newborn | Unknown | CAG-tdTomato | N/A |
These tdTomato reporter iPSC lines underwent a thorough evaluation after confirming targeted integration of one copy of the CAG-tdTomato transgene at the AAVS1 safe harbor locus with a Southern Blot assay. Targeted integration was confirmed by the expected 3.6 kb band and the remaining wild-type allele was indicated by the presence of the 5.5 kb band (Supplemental Fig. S1B). There was no evidence of additional random integrations of the transgene. The iPSC lines maintained stable and robust tdTomato expression and human embryonic stem cell (ESC)-like morphology over extended cell culture. Their undifferentiated state was characterized by immunofluorescent staining and flow cytometry analysis of several common human ESC/iPSC markers including SOX2, NANOG, OCT4, SSEA4, and TRA-1-60 (Fig. 1A and B). FITC or Alexa Fluor 488 conjugated isotype control antibodies were used to stain iPSCs separately for proper gating and measuring the percentages of positive iPSCs stained by pluripotent stem cell marker-specific antibodies (Table 3). The percentage of tdTomato positive cells were quantified by flow cytometry, using parental ND2.0 iPSCs as negative control, to confirm that nearly 100% of reporter iPSCs express tdTomato (Fig. 1B). In addition, G-banding karyotyping at passage 42 indicated a normal karyotype (46, XY) (Supplemental Fig. S1A) and short tandem repeat (STR) DNA profiling analysis at 15 loci showed the genotypes of these two iPSC lines did match that of the parental ND2.0 line (available with the authors). The cell lines’ mycoplasma status was also confirmed to be negative by quantitative PCR (qPCR) (Supplemental Fig. S1C). Lastly, pluripotency was demonstrated by a teratoma formation assay in which the cells successfully differentiated into all three germ layers (ectoderm, neural tube; mesoderm, cartilage; endoderm, gut) in vivo (Fig. 1C).
Fig. 1.
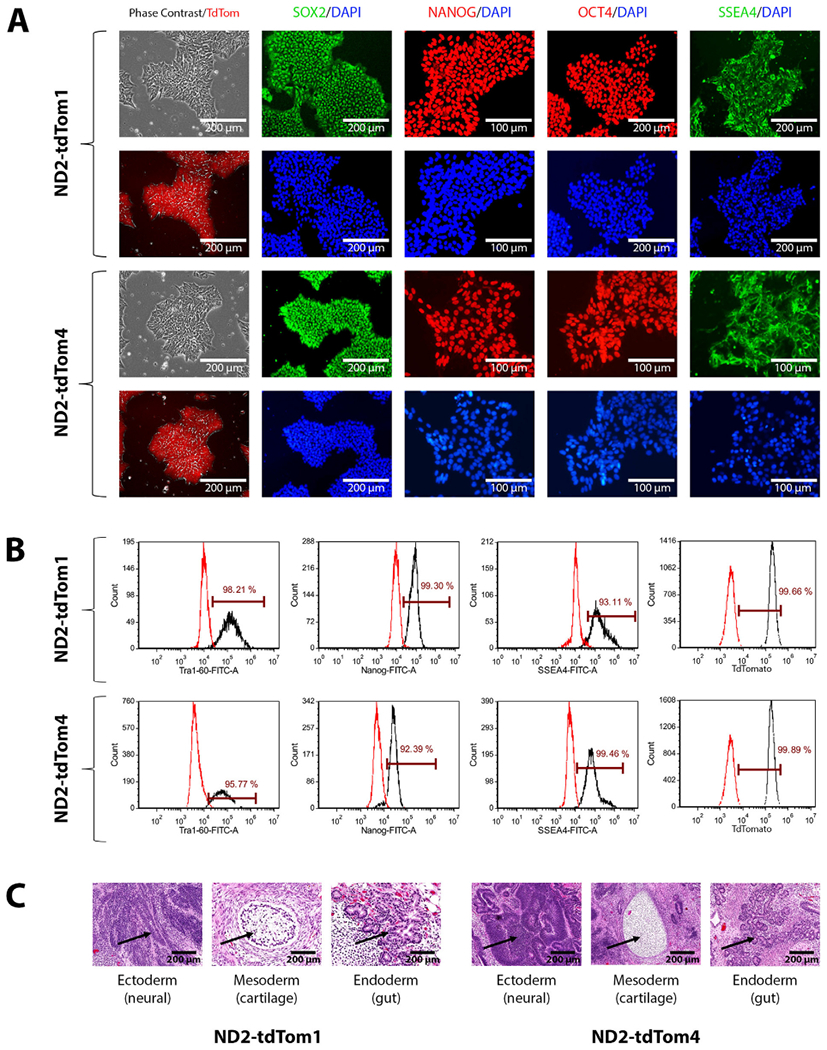
(A) Images of phase contrast and flurorescence microscopy showing the expression of tdTomato and pluripotency markers by ND2-tdTom1 and ND2-tdTom4 iPSCs. (B) Flow cytometry analysis of pluripotency markers of ND2-tdTom1 and ND2-tdTom4 iPSCs. (C) Teratoma formation assay shows ND2-tdTom1 and ND2-tdTom4 iPSCs can generate three germ layers in vivo.
Table 3.
Reagents details. RRID Requirement for antibodies: use http://antibodyregistry.org/ to retrieve RRID for antibodies and include ID in table as shown in examples.
| Antibodies used for immunocytochemistry/flow-cytometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency Markers | Mouse anti-SOX2 | 1:250 | BioLegend, Cat# 656102, RRID: AB_2,562246 |
| Pluripotency Markers | Rabbit anti-NANOG | 1:400 | Cell Signaling Technology, Cat# 4903, RRID: AB_10559205 |
| Pluripotency Markers | Rabbit anti-OCT4 | 1:400 | Thermo Fisher, Cat# 701756, RRID: AB_2633031 |
| Pluripotency Markers | Mouse anti-SSEA4 | 1:1000 | Cell Signaling Technology, Cat# 4755, RRID: AB_1264259 |
| Secondary Antibodies | Donkey anti-Mouse IgG (Alexa Fluor 488) | 1:400 | Thermo Fischer, Cat# A21202, RRID: AB_141607 |
| Secondary Antibodies | Donkey anti-Rabbit IgG (Alexa Fluor 594) | 1:400 | Thermo Fischer, Cat# A21207, RRID: AB_141637 |
| Flow Cytometry Antibodies | Anti-Tra-1-60-DyLight 488 | 1:50 | Thermo Fischer, Cat# MA1–023-D488X, RRID: AB_2536700 |
| Flow Cytometry Antibodies | Anti-Nanog-Alexa Fluor 488 | 1:50 | Millipore, Cat# FCABS352A4, RRID: AB_10807973 |
| Flow Cytometry Antibodies | Anti-SSEA-4-Alexa Fluor 488 | 1:50 | Thermo Fischer, Cat# 53-8843-41, RRID: AB_10597752 |
| Flow Cytometry Antibodies | Mouse-IgM-DyLight 488 | 1:50 | Thermo Fischer, Cat# MA1-194-D488, RRID: AB_2536969 |
| Flow Cytometry Antibodies | Rabbit IgG-Alexa Fluor 488 | 1:50 | Cell Signaling Technology, Cat# 4340S, RRID: AB_10694568 |
| Flow Cytometry Antibodies | Mouse IgG3-FITC | 1:50 | Thermo Fischer, Cat# 11-4742-42, RRID: AB_2043894 |
| Primers | |||
| Target | Forward/Reverse primer (5′ – 3′) | ||
| Mycoplasma detection primers (qPCR) | GPO-1_MGSO/724bp | 5′-ACGGCCCAGACTCCTACGGGAGGCAGCAGTA | |
| 5′-CCATGCACCATCTGTCACTCTGTTAACCTC | |||
| House-keeping gene primers (qPCR) | GAPDH-3/488 bp | 5′-GGGAGCCAAAAGGGTCATCA | |
| 5′-TGATGGCATGGACTGTGGTC | |||
3. Materials and methods
3.1. CRISPR/Cas9-mediated targeted integration of the tdTomato transgene in human iPSCs
All iPSCs were maintained in cell culture incubators at 37 °C with 5% CO2 and 20% O2. ND2.0 iPSCs were maintained in a 6-well plate in E8 medium (A1517001, Thermo Fisher) with 1:10 passaging every 3–4 days using the EDTA method (Beers et al., 2012). The iPSCs were dissociated with TrypLE (12563029, Thermo Fisher) once they reached 70–90% confluency. 300,000 cells were then re-plated onto one 12-well coated with Matrigel (Corning, 354277) in E8 medium with 10 μl RevitaCell (A2644501, Thermo Fisher). The ND2.0 iPSCs were transfected after they were seeded in the morning and attached 4–6 h later in the afternoon, using Lipofectamine 3000 Transfection Reagent according to the manufacturer’s protocol (L3000015, Thermo Fisher). We transfected 1.5 μg of the plasmid pCAG-SpCas9-GFP-U6-gRNA (79144, Addgene) containing the Cas9 protein sequence and the sgRNA targeting the 5′-GGGGCCACTAGGGACAGGAT sequence in the AAVS1 safe harbor locus along with 1.5 μg of the plasmid pAAVS1p-iCAG.copGFP (66577, Addgene) with the cloned tdTomato cassette (Supplementary Fig. S1B). E8 medium was added the next day and the cells were passaged after 48 h if confluent. After 2–3 days the iPSCs underwent selection with 0.25 μg/ml puromycin in E8 medium. The medium was changed every day for 7–12 days or until selection was complete and only targeted colonies remained. Colonies were then picked and expanded in E8 medium without puromycin. tdTomato1 and tdTomato4 iPSC clones were selected from these colonies for further characterization.
3.2. Southern blot
A Southern blot assay was performed by Lofstrand Labs Limited (Rockville, MD) using a 32P labelled PCR probe recognizing the left homology arm as described previously, except that EcoRV and HindIII restriction enzymes were used to digest 10ug genomic DNA (Cerbini et al., 2015). The probe can be used to detect wild-type, targeted integration, and random integration alleles. The wild-type allele is expected to show a 5.5 kb band and the targeted integration allele is expected to show a 3.6 kb band.
3.3. Immunocytochemistry
NHLBIi003-A-1 and NHLBIi003-A-2 iPSCs were fixed and stained as previously described, though we blocked the cells and diluted the primary antibodies with a 10 mg/ml Bovine Serum Albumin (BSA) in DPBS solution (Hong et al., 2019). Cell nuclei were stained with DAPI and the cells were imaged with an EVOS® FL Cell Imaging System (Thermo Fisher) and a 10 or 20 × objective lens with Texas Red, FITC, and DAPI filters.
3.4. Flow cytometry analysis
iPS cells were dissociated from the plate with TrypLE (12563029, Thermo Fisher) and were prepared for flow cytometry as previously described (Beers et al., 2012), except that a different permeabilization buffer (2%FBS and 0.2% Tween 20 in DPBS) was used. We used fluorophore conjugated antibodies as listed in Table 2. The cells were analyzed with an AccuriC6 Flow Cytometry system (BD Biosciences) and FCS Express 5 software.
Table 2.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | Normal | Fig. 1A |
| Phenotype | Qualitative analysis: Immunocytochemistry | Positive for SOX2, OCT4, NANOG, SSEA-4 | Fig. 1A |
| Quantitative analysis: Flow Cytometry (tdTom1, tdTom4) | TRA-1-60 (98.21%, 95.77%); NANOG (99.30%, 92.39%); SSEA-4 (93.11%, 99.46%); tdTomato (99.66%, 99.89%) | Fig. 1B | |
| Genotype | Karyotype (G-banding) and resolution | 46XY; Resolution 425–500 | Supplementary Fig. S1A |
| Identity | Microsatellite PCR (mPCR) OR STR analysis | Not performed | N/A |
| 15 loci plus amelogenin (Promega PowerPlex 16) tested, all matched | Available with the authors | ||
| Mutation analysis (IF APPLICABLE) | Sequencing | N/A | N/A |
| Southern Blot | Monoallelic targeted integration without random integration | Supplementary Fig. S1B | |
| Microbiology and virology | Mycoplasma testing by qPCR | Negative | Supplementary Fig. S1C |
| Differentiation potential | Teratoma formation | Teratoma formed with three germ layers: Ectoderm, Mesoderm, and Endoderm | Fig. 1C |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A |
3.5. G-banding karyotyping
G-banding karyotyping was performed at passage 42 by WiCell Cytogenetics lab (Madison, WI) using twenty randomly selected metaphases.
3.6. Short tandem repeat (STR) analysis
STR analysis was performed by WiCell Cytogenetics lab using a Powerplex® 16 System (Promega) and genomic DNA extracted from the iPSCs with DNeasy Blood and Tissue Kit (Qiagen).
3.7. Mycoplasma detection
2 ml of medium from the iPSC culture was spun down at > 20,000 g for 20 min to collect a small pellet of cells. After removing all medium, the pellet was lysed by 0.5x Phusion HF Buffer (NEB, #B0518S) with 8 U/ml Proteinase K (NEB, #P8107S) at 55 °C for 1–3 h followed by heat-inactivation at 95 °C for 10 min. Quantitative PCR (qPCR) detection of mycoplasma was carried out using the primer pair GPO-1_MGSO with the SsoAdvanced™Universal SYBR Green Supermix (Bio-Rad Laboratories) for 40 cycles. The RFU values at the end of the PCR were used to compare samples with positive (a known contaminated sample) and negative (sterile water) controls to evaluate the presence of mycoplasma contamination. A pair of GAPDH primers (GAPDH-3) that amplify in human samples was used to ensure cell material was present.
3.8. Teratoma assay
NHLBIi003-A-1 and NHLBIi003-A-2 iPSCs were removed from 6-well plates when ~90% confluent using the EDTA dissociation method. 1 × 107 cells per clone were resuspended in E8 medium and kept on ice. The suspension was mixed with a 50% volume of cold Matrigel (Corning, 354277) and 150 μl of the resulting mixture was injected subcutaneously into NSG mice (JAX No. 005557) at two sites. Tumors were visible after 6–8 weeks at which point they were removed and fixed in 10% Neutral Buffer Formalin. They were then embedded in paraffin and stained with hematoxylin and eosin.
Supplementary Material
Resource Table:
| Unique stem cell lines identifier | NHLBIi003-A-1 |
| NHLBIi003-A-2 | |
| Alternative names of stem cell lines | ND2-tdTom1 (NHLBIi003-A-1) |
| ND2-tdTom4 (NHLBIi003-A-2) | |
| Institution | National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health, Bethesda, USA |
| Contact information of distributor | Dr. Jizhong Zou jizhong.zou@nih.gov |
| Type of cell lines | iPSC |
| Origin | Human |
| Cell Source | Fibroblast |
| Clonality | Clonal |
| Method of reprogramming | Episomal vectors |
| Multiline rationale | Stable and bright fluorescent protein reporter iPSC lines generated from a previously published wild-type iPSC line |
| Gene modification | Yes |
| Type of modification | Transgene expression (fluorescent reporter and drug-resistance genes) by targeted integration |
| Associated disease | N.A. |
| Gene/locus | AAVS1/PPP1R12C |
| Method of modification | CRISPR/Cas9 |
| Name of transgene or resistance | tdTomato and Puromycin |
| Inducible/constitutive system | Constitutive |
| Date archived/stock date | September 2019 |
| Cell line repository/bank |
https://hpscreg.eu/cell-line/NHLBIi003-A-1
https://hpscreg.eu/cell-line/NHLBIi003-A-2 |
| Ethical approval | The original fibroblast CCD-1079sk (ATCC® CRL-2097) was obtained from ATCC (https://www.atcc.org/products/all/CRL-2097.aspx#specifications) |
Acknowledgment
We would like to thank Dr. Zu-xi Yu of the Pathology Core and Dr. Chengyu Liu of the Transgenic Core of National Heart, Lung, and Blood Institute, NIH for performing teratoma assay. We would also like to thank WiCell Cytogenetics lab for performing karyotyping and STR assays. This work was supported by the Intramural Research Program of National Heart, Lung, and Blood Institute at NIH (grant number ZIC HL006145-08).
Footnotes
Declaration of Competing Interest
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.scr.2019.101673.
References
- Beers J, Gulbranson DR, George N, Siniscalchi LI, Jones J, Thomson JA, Chen G, 2012. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat. Protoc. 7, 2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerbini T, Funahashi R, Luo Y, Liu C, Park K, Rao M, Malik N, Zou J, 2015. Transcription activator-like effector nuclease (TALEN)-mediated CLYBL targeting enables enhanced transgene expression and one-step generation of dual reporter human induced pluripotent stem cell (iPSC) and neural stem cell (NSC) lines. PLoS One 10, e0116032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, Wagner R, Lee GO, Antosiewicz-Bourget J, Teng JM, Thomson JA, 2011. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods 8, 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deliolanis NC, Kasmieh R, Wurdinger T, Tannous BA, Shah K, Ntziachristos V, 2008. Performance of the red-shifted fluorescent proteins in deep-tissue molecular imaging applications. J. Biomed. Opt. 13, 044008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J, Xu M, Li R, Cheng YS, Kouznetsova J, Beers J, Liu C, Zou J, Zheng W, 2019. Generation of an induced pluripotent stem cell line (TRNDi008-A) from a Hunter syndrome patient carrying a hemizygous 208insC mutation in the IDS gene. Stem Cell Res. 37, 101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


