Abstract
Wolbachia is a widely distributed intracellular bacterial endosymbiont among invertebrates. The wStriCN, the Wolbachia strain that naturally infects an agricultural pest Laodelphax striatellus, has a “Jekyll and Hyde” mode of infection pattern with positive and negative effects: It not only kills many offspring by inducing cytoplasmic incompatibility (CI) but also significantly increases host fecundity. In this study, we assembled the draft genome of wStriCN and compared it with other Wolbachia genomes to look for clues to its Jekyll and Hyde characteristics. The assembled wStriCN draft genome is 1.79 Mb in size, which is the largest Wolbachia genome in supergroup B. Phylogenomic analysis showed that wStriCN is closest to Wolbachia from Asian citrus psyllid Diaphorina citri. These strains formed a monophylogentic clade within supergroup B. Compared with other Wolbachia genomes, wStriCN contains the most diverse insertion sequence families, the largest amount of prophage sequences, and the most ankyrin domain protein coding genes. The wStriCN genome encodes components of multiple secretion systems, including Types I, II, IV, VI, Sec, and Tac. We detected three pairs of homologs for CI factors CifA and CifB. These proteins harbor the catalytic domains responsible for CI phenotypes but are phylogenetically and structurally distinct from all known Cif proteins. The genome retains pathways for synthesizing biotin and riboflavin, which may explain the beneficial roles of wStriCN in its host planthoppers, which feed on nutrient-poor plant sap. Altogether, the genomic sequencing of wStriCN provides insight into understanding the phylogeny and biology of Wolbachia.
Keywords: Wolbachia, Laodelphax striatellus, cytoplasmic incompatibility, mutualistic, parasitic
Introduction
Wolbachia is a genus of Gram-negative intracellular endosymbiotic bacteria belonging to the family Rickettsiaceae (order Alphaproteobacteria). Wolbachia is one of the most widespread endosymbionts in animals, existing in about 40% of arthropod species (Zug and Hammerstein 2012), as well as some nematodes (Lefoulon et al. 2016). Wolbachia is the abbreviation for Wolbachia pipientis (Lo et al. 2007), which contains 16 supergroups (A–Q, except for G, which is a combination of A and B) (Baldo, Dunning Hotopp, et al. 2006; Lo et al. 2007; Haegeman et al. 2009; Ros et al. 2009; Augustinos et al. 2011; Bing et al. 2014; Glowska et al. 2015). In insects, the most common supergroups are A and B (Lo et al. 2007).
As a facultative endosymbiont in arthropods, Wolbachia is famous for manipulating host reproduction. Wolbachia-mediated reproductive manipulation includes producing infected females without males (parthenogenesis-inducing), feminizing genetic males (feminization-inducing), killing infected male progenies (male-killing), and inducing cytoplasmic incompatibility (CI) between infected males and uninfected (or differently infected) females (Werren et al. 2008). These phenotypes directly or indirectly result in a better survival environment for Wolbachia-infected females. In other words, they enhance Wolbachia’s maternal transmission and thus increase its ability to spread in populations. Wolbachia has also been shown to be beneficial in insects by protecting them from pathogenic viral infection and increasing host fitness and fecundity (Nikoh et al. 2014; Moriyama et al. 2015; Zug and Hammerstein 2015; Guo et al. 2018). In nematodes, Wolbachia functions as an obligatory mutualist endosymbiont (Comandatore et al. 2013).
The diverse effects of Wolbachia strains on their hosts are reflected in their genetic diversity. The diversity of genome features has elucidated insights of Wolbachia’s multiple functions in many insects, such as the mutualistic roles of wCle (supergroup F, an obligatory Wolbachia from the bedbug Cimex lectularius) (Nikoh et al. 2014), CI induced by wMel (supergroup A, Wolbachia from Drosophila melanogaster) (LePage et al. 2017), and male-killing from wBol1 (supergroup B, Wolbachia from the butterfly Hypolimnas bolina) (Duplouy et al. 2013). Genomic analyses showed that Wolbachia strains have reduced genome size (Sinha et al. 2019), many pseudogenes, bacteriophages, and transposable mobile elements (Bordenstein and Wernegreen 2004; Wu et al. 2004; Gavotte et al. 2007; Klasson et al. 2009; Bordenstein and Bordenstein 2016), and genetic recombination events (Baldo, Bordenstein, et al. 2006; Zhang et al. 2013), which have contributed significantly to diversifying Wolbachia genomes.
The small brown planthopper Laodelphax striatellus (Fallén) (Hemiptera: Delphacidae) is one of the most destructive pests of rice Oryza sativa Japonica, the primary food source for half of the world’s population. Laodelphaxstriatellus feed on plant sap exclusively and lay eggs in plant tissues, causing direct physiological damage to crops. In addition, L. striatellus can migrate long distances and transmit many rice virus diseases (such as rice stripe virus) (Nault 1994). Previous field investigations have shown that L. striatellus is highly infected by Wolbachia (more than 90%) (Hoshizaki and Shimada 1995; Zhang et al. 2013). Microbiota analysis of field planthoppers showed that Wolbachia accounts for more than 80% of the identified operational taxonomic units (OTUs), indicating its dominance in L. striatellus (Bing et al., 2020, under revision). Wolbachia has been reported to increase host fecundity (Guo et al. 2018) and induce strong CI (Noda 1984a, 1984b) in L. striatellus, a classic type of “Jekyll and Hyde” mode of infection pattern (Jiggins and Hurst 2011; Moriyama et al. 2015; Zug and Hammerstein 2015). In addition, Wolbachia isolated from L. striatellus was recently shown to block the growth of several positive-sense RNA viruses (Dengue virus, Chikungunya virus, Zika virus, and yellow fever virus) by more than 99.9% in mosquito Aedes albopictus cells (Schultz et al. 2018), which increased its potential to block arbovirus infections in vectors. By having both mutualistic and deleterious effects in the same host species, the Wolbachia strain in L. striatellus is a good model for exploring the mechanisms of Wolbachia–host interactions.
We recently analyzed the genome sequence of Wolbachia in L. striatellus (wStriCN) to understand the molecular mechanisms underlying its beneficial effects (Ju et al. 2019). In this article, we provide a more detailed description of the wStriCN genome, including how it differs from other Wolbachia genomes, and unique features that may explain Wolbachia’s expansion, the separation of Wolbachia into supergroups A and B, the functions of Wolbachia in insect hosts, and how Wolbachia and the host communicate.
Materials and Methods
Insect Preparation
Laodelphax striatellus planthoppers were collected at a rice farm field in Nanjing, China, in July 2014 and then were maintained under laboratory conditions on rice seedlings in a climate-controlled room (25 °C, 60% RH, and an 8-h photoperiod). The Wolbachia infection statuses of individual L. striatellus were checked with polymerase chain reaction (PCR) and were confirmed with Sanger sequencing as previously described before (Zhang et al. 2013).
Wolbachia Genome Sequencing, Assembly, and Annotation
The process of Wolbachia genome sequencing and assembly has been described in detail in Ju et al. (2019). The genome size and GC content of assembled Wolbachia genome were calculated with bioawk (https://github.com/lh3/bioawk/, last accessed January 19, 2020). The structural and functional annotation was predicted with Prokka 1.13.3 (Seemann 2014). The insertion sequences (IS) were identified based on ISfinder database (Siguier et al. 2006). Prophage sequences (WO) were annotated with PHASTER (Arndt et al. 2016). Wolbachia genomes were functionally characterized and compared by assigning predicted proteins to the following databases: Clusters of Orthologous Groups (COG) of proteins (Galperin et al. 2015), KEGG Ortholog database (Kanehisa and Goto 2000), and eggNOG (Huerta-Cepas et al. 2016). The signal peptides and transmembrane domains of proteins were identified with SignalP 4.0 (Petersen et al. 2011) and TMHMM 2.0 (Krogh et al. 2001), respectively.
Genome Completeness Assessment
The completenesses of Wolbachia genomes were estimated with the BUSCO pipeline 3.0.2 (Waterhouse et al. 2018), which was performed against 221 highly conserved, single-copy orthologs (BUSCO groups) obtained from 1,520 proteobacterial species (proteobacteria_odb9) (Loetscher et al. 2016). To compare genomic features of various Wolbachia strains, all 53 available insect-associated Wolbachia genomes (by the time of April, 2019, supplementary table S2, Supplementary Material online) were downloaded from NCBI RefSeq or GenBank assembly database (Benson et al. 2018; Marchler-Bauer et al. 2018). Nematode-associated Wolbachia were excluded from this study because they differ distinctly in biology and phylogeny. As a low BUSCO score may indicate a poorly assembled Wolbachia genome and the complete Wolbachia genomes have BUSCO score >80% (Lindsey et al. 2016; Sinha et al. 2019), we used a complete BUSCO score of 80% as the criterion for genome quality control. The wStriCN and 32 other genomes were selected for further comparative genomic analyses. The 33 genomes were annotated all with the same methods mentioned above. wStriCN-specific genes were predicted with OrthoFinder version 2.2.6 (Emms and Kelly 2015). Synteny between wStriCN and other Wolbachia genomes was analyzed with NUCmer and visualized with mummerplot on Mumer 4.0.0beta2 (Kurtz et al. 2004). Average nucleotide identities between Wolbachia genomes were calculated using OrthoANI 1.40 (Yoon et al. 2017).
Phylogenetic and Phylogenomic Analyses
For molecular phylogenetic analysis, we inferred Wolbachia phylogeny using 52 ribosomal protein sequences (twenty-one 30S ribosomal proteins and thirty-one 50S ribosomal proteins), and the Wolbachia ortholog groups generated by OrthoFinder 2.2.6 (Emms and Kelly 2015). The protein sequences were concatenated with SeqKit 0.9.2 (Shen et al. 2016), aligned with MAFFT 7.402 (Katoh and Standley 2013), and trimmed with Gblocks 0.91b (Castresana 2000). The best-fitting nucleotide substitution model was calculated with ModelFinder (Kalyaanamoorthy et al. 2017). The maximum likelihood (ML) phylogenetic tree was constructed with IQ-TREE 1.6.5 (Nguyen et al. 2015). The node support was calculated with 1,000 ultrafast bootstraps. The phylogenetic tree was visualized with ggtree (Yu et al. 2017) and was annotated in Inkscape (https://inkscape.org/).
Analysis of Genome Content among Wolbachia Strains
COG category annotations were used to compare genome contents across the 33 Wolbachia strains. The number of genes in each category for each genome was subjected to principle component analysis (PCA) using R 3.5 (R Core Team 2018). The Wilcoxon rank-sum test was used to test for a significant difference in abundance of genes between supergroups A and B.
Identification of Cif Proteins in wStriCN
Candidate CifA and CifB proteins in wStriCN were identified by searching similar proteins of known Cif proteins with Blast 2.7.1+ (Camacho et al. 2009) and OrthoFinder 2.2.6 (Emms and Kelly 2015). The phylogeny of Cif protein sequences was constructed as described above. Cif protein structures were predicted with HHpred (Söding et al. 2005) following Lindsey’s method (Lindsey et al. 2018).
Results
Genome Assemblies and Annotation of wStriCN
To obtain the genome of wStriCN, we generated two Illumina paired-end libraries (one with PCR and one without PCR) and one mate pair library on the Illumina HiSeq2000 platform. These libraries yielded 210-Mb (117× coverage of assembled Wolbachia), 211-Mb (118× coverage), and 457.6-Mb (280× coverage) data, respectively. In addition, we also generated a 454-pyrosequencing library that yielded 385.3-Mb (216× coverage) data on the Roche 454 GS FLX Titanium platform (supplementary table S1, Supplementary Material online). The draft genome of wStriCN was assembled with de novo assembly of Illumina reads with SOAPdenovo (Li et al. 2010) and closing gaps with 454 reads and Sanger sequencing results. The assembled wStriCN draft genome contains two scaffolds, composed of 42 contigs. The N50 of the contigs was 114,888 bp, about 142 times the average gene length (table 1). The total length of the wStriCN genome is 1,786,382 bp, which is one of the largest insect-associated Wolbachia genomes so far (table 1). The average GC content of the wStriCN genome is 33.72%, which is within the typical range of Wolbachia genomes (31.7% ∼ 38.3%) (supplementary table S2, Supplementary Material online).
Table 1.
Genome Statistics of wStriCN
| wStriCN | |
|---|---|
| Host | Laodelphax striatellus |
| Phenotypes | Cytoplasmic incompatibility, increase fecundity |
| Number of scaffolds | 2 |
| Number of contigs | 42 |
| Total nucleotides | 1,786,382 |
| N50 scaffolds | 1,600,254 |
| N50 contigs | 114,888 |
| GC content (%) | 33.72 |
| Number of CDS | 1,747 |
| Number of tRNAs | 34 |
| Number of rRNAs | 3 |
| Number of tmRNA | 1 |
| Average gene length | 808.7 |
The Prokka pipeline annotation results (Seemann 2014) revealed that the wStriCN genome contained 1,747 protein coding sequences (CDSs) (92.8% of all wStriCN genes), 34 tRNA genes that could transfer all 20 amino acids, 3 rRNA genes (5S, 16S, and 23S rRNA), and 1 tmRNA gene (table 1). The 1,747 CDS-encoded proteins encompassed 182 complete and single-copy, 1 complete and duplicated, 5 fragmented and 33 missing highly conserved, Benchmarking Universal Single-Copy Orthologs (BUSCO groups), resulting in 82.81% BUSCO completeness score (supplementary fig. S1, Supplementary Material online). That score falls within the range of scores for published complete Wolbachia genomes (supplementary table S2, Supplementary Material online) (Sinha et al. 2019), which indicates the wStriCN genome is sufficiently reliable for further comparative genomic analyses.
Taxonomy and Synteny of wStriCN
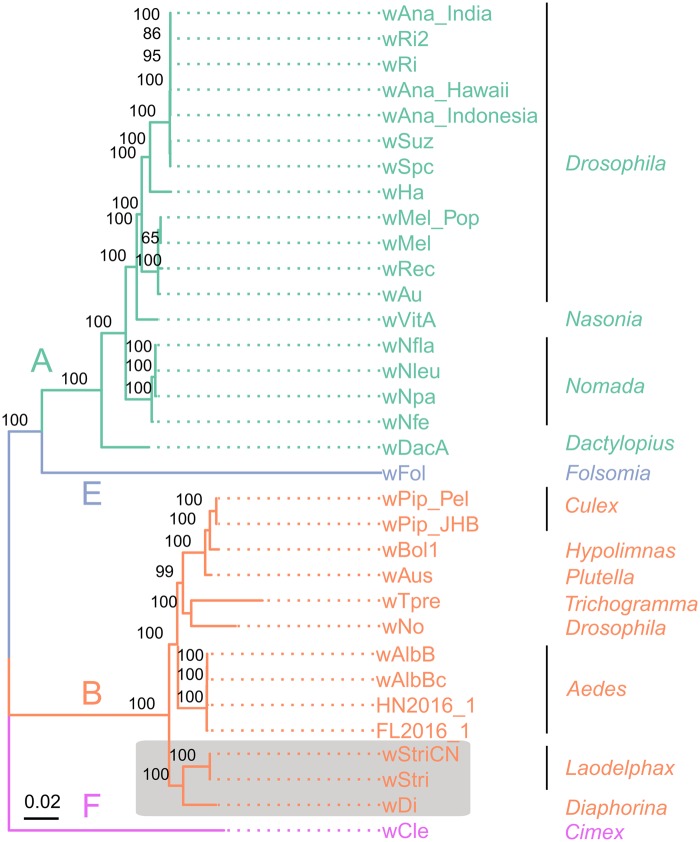
It has been argued that the classic multilocus sequence typing (MLST) loci for Wolbachia (Baldo, Dunning Hotopp, et al. 2006) are problematic and may not reflect the properties of a Wolbachia strain very well (Bleidorn and Gerth 2017). Therefore, we performed two genome-wide phylogenomic analysis using 52 ribosomal protein coding genes (22,386 bp) and 367 single-copy orthologs of insect-associated Wolbachia genomes (215,584 aa), respectively. Both phylogenetic trees allocated wStriCN to supergroup B (fig. 1 and supplementary fig. S2, Supplementary Material online). In addition, wStriCN, wStri (the Wolbachia from a Korean L. striatellus population), and wDi (the Wolbachia isolated from the Asian citrus psyllid Diaphorina citri), clustered together and formed a strongly supported monophyletic group that was distinct from the other supergroup B Wolbachia members (fig. 1), suggesting they are close relatives. The average nucleotide identity (ANI) between wStriCN and wStri/wDi genomes is 97.96%, which is much higher than the ANI between wStriCN and the other Wolbachia (88.88%) (supplementary table S3, Supplementary Material online). The above results suggest that these Wolbachia strains shared the same ancestor.
Fig. 1.
—Phylogenetic relationship of Wolbachia strains. The ML tree was calculated with a concatenated protein sequence of 367 single-copy protein sequences (215,584 amino acids) using an HIVw + F + R8 substitution model. Wolbachia supergroups are color coded as shown on the branches. The host genera of the Wolbachia strains are shown on the side. Bootstrap values are indicated at the respective node (only values >50% are shown). The scale bar represents the average number of substitutions per site.
The colinear regions of MUMmer dot plots between pairs of wStriCN and other supergroup B Wolbachia genomes are larger than those between pairs of Wolbachia genomes of other supergroups (supplementary fig. S3, Supplementary Material online). In agreement with the finding of a high level of genome rearrangement between multiple Wolbachia genomes (Klasson et al. 2008; Duplouy et al. 2013), many inversions were found between wStriCN and other Wolbachia genomes (supplementary fig. S3, Supplementary Material online). It is noteworthy that the wStriCN sequences are more colinear with draft genomes than complete genomes of supergroup B Wolbachia. That may be because the directions of many scaffolds were artificially rearranged during the Mummer analysis. Therefore, it is necessary to be cautious when comparing rearrangements between draft genomes.
Insertion Sequences
Wolbachia associated with arthropods are known to carry a large number of mobile elements, such as IS and WO (Kent and Bordenstein 2010; Bouchon et al. 2011), which may explain the expansion in Wolbachia genomes. As the largest genome in B supergroup so far, wStriCN genome was expected to be enriched in these features.
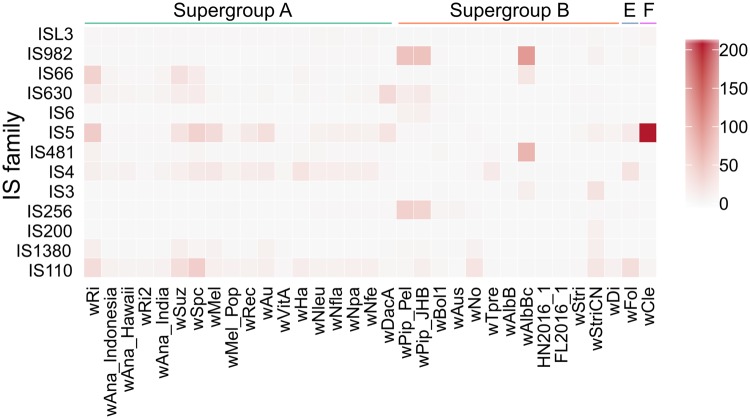
The ISfinder database identified 78 IS elements of 10 IS families from the wStriCN genome (supplementary table S5, Supplementary Material online). The most abundant IS families in wStriCN were the IS3 and IS110 families, containing 21 and 18 genes respectively. The total size of IS elements is 64224 bp, accounting for 3.6% of the wStriCN genome, which is much smaller than the IS elements in Wolbachia wFol and wPip (Klasson et al. 2008; Kampfraath et al. 2019). The top three IS families in abundance in all 33 invertebrate-associated Wolbachia genomes are IS5, IS110, and IS982 (supplementary table S5, Supplementary Material online). The wCle genome harbors the highest number of IS elements, 96% (208/217) of which belong to IS5 (supplementary table S5, Supplementary Material online). Significant differences were found in the distribution of IS families among various Wolbachia genomes (fig. 2, chi-square test, χ2 = 3481.7, df = 384, P value < 0.0001). Within the same Wolbachia supergroup, the numbers of IS elements in different IS families also differ substantially (fig. 2). This is particularly striking for the wAlbB and wTpre genomes, which are all supergroup B members (Lindsey et al. 2016; Sinha et al. 2019). It should be noted that the number of IS elements may be underestimated for Wolbachia genomes that are incomplete.
Fig. 2.
—Comparison of IS families from different Wolbachia supergroups. Abundance of IS elements in different IS families in different Wolbachia supergroups.
Bacteriophage Genes
The PHASTER server predicted that the wStriCN genome has nine prophage regions (WOStriCN1-9), with a combined size of 233.9 kb (supplementary table S5, Supplementary Material online). These regions account for 13.1% of the wStriCN genome, which is a major contributing factor to the large genome size of wStriCN. Blasting prophage region sequences (WOStriCNs) against COG database revealed they encode modules for essential functions such as head or tail formation. For instance, WOStriCN5 contains genes encoding capsid proteins, tail formation proteins, tail sheath proteins, tail tube proteins, baseplate assembly protein, and phage terminase (supplementary fig. S4, Supplementary Material online). In addition, modules for the assembly of baseplate (wStriCN_00262, wStriCN_00303) and tail (wStriCN_00255, wStriCN_00299) are homologous to the P2 phage modules. The regions between the phage modules are mostly IS elements (transposases and other related proteins), ankyrin repeat (ANK) containing proteins and genes of unknown function (supplementary fig. S4, Supplementary Material online). In the WO regions of the wStriCN genome, the most significantly enriched elements other than phage-related genes were IS elements and ANK genes. Seventeen of the 78 IS elements in the genome and 32 of the 113 ANK genes in the genome were in the WO regions, indicating significant enrichment in the WO regions (binomial tests, P = 0.029 and <0.0001, respectively). Other genes that were significantly enriched in WO regions include site-specific DNA recombinase genes (10 out of 13, binomial test, P value < 0.0001), periplasmic serine protease of ClpP class (7 out of 9, binomial test, P value < 0.0001), and proteins with Zn-binding Pro-Ala-Ala-Arg (PAAR) domain, which were involved in Type VI secretion (8 of 8, binomial test, P value < 0.0001). The latter genes may have been horizontally shuttered by phage from Wolbachia. The closest matches of these genes in the NCBI NR databases were other Wolbachia genes.
Functional Categories of wStriCN Genes
The representation of functional categories in Wolbachia genomes was analyzed by assigning the CDSs to COGs and KEGG databases. Of the 1,747 CDS in wStriCN genome, 1,339 CDSs (76.6%) were annotated to COG and 722 CDSs (41.3%) were annotated to KEGG pathways by the KAAS annotation server. KEGG analysis showed wStriCN contained complete pathways for the tricarboxylic acid cycle (map0020), fatty acid biosynthesis (map0061), oxidative phosphorylation (map00190), and lipoic acid metabolism (map00785), which are for essential energy metabolism. It also contained genes for biotin synthesis (map00780), riboflavin synthesis (map00740), and peptidoglycan biosynthesis (map00550). COG analysis showed that mobilome-related genes, such as prophages and transposons (category X, n = 307), were the most abundant genes in wStriCN, in agreement with the large number of WO and IS elements described above. In the 33 Wolbachia genomes, the COG categories with the most variation in gene number per genome were mobilome, prophages, and transposons (X) and signal transduction mechanisms (T). The categories that showed little variance were secondary metabolites biosynthesis, transport and catabolism (Q), cell motility (N), and inorganic ion transport and metabolism (P) (supplementary table S6, Supplementary Material online).
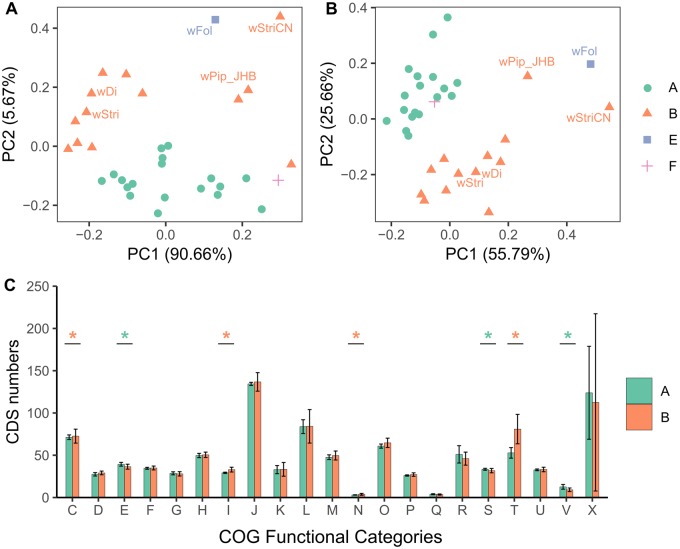
PCA based on the proportion of genes in each of the COG categories showed that supergroups A and B clustered separately by PC2. The factor that contributing to the PC1 (90.66% of total variance) and PC2 (5.67% of total variance) was the mobilome category which includes mainly prophages and transposons, and signal transduction mechanisms. The signal transduction category mainly consists of ANKs (fig. 3A). PCA without the mobilome category did not change the clustering of supergroups A and B (fig. 3B). wCle clustered with supergroup A Wolbachia, and wFol (the Wolbachia infecting springtail Folsomia candida) clustered on the side of supergroup B Wolbachia (fig. 3A), although they are phylogenetically distinct from each other (fig. 1). wStriCN’s closest neighbor in the PCA figure was wFol, which may be because of the high number of mobilome genes and ANKs in both genomes (supplementary table S6, Supplementary Material online).
Fig. 3.
—Comparison of COG categories from different Wolbachia supergroups. (A) PCA of Wolbachia genomes based on the proportion of annotated genes in each COG category. (B) PCA of Wolbachia genomes excluding X category genes. (C) Bar chart comparing COG categories between Wolbachia supergroups A and B. Abbreviations of COG categories are C, energy production and conversion; D, cell cycle control, cell division, and chromosome partitioning; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; J, translation, ribosomal structure, and biogenesis; K, transcription; L, replication, recombination, and repair; M, cell wall/membrane/envelope biogenesis; N, cell motility; O, posttranslational modification, protein turnover, and chaperones; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport, and catabolism; R, general function prediction only; S, function unknown; T, signal transduction mechanisms; U, intracellular trafficking, secretion, and vesicular transport; V, defense mechanisms; X, mobilome, prophages, and transposons.
Additionally, the number of genes belonging to clusters in COG categories of amino acid transport and metabolism (E), defense mechanisms (V), and function unknown (S) were significantly more in supergroup A Wolbachia than that of supergroup B (Wilcoxon rank-sum test, P value < 0.05). The genes in COG categories related to energy production (C), lipid transport and metabolism (L), cell mobility (N), and signal transduction (T) were much more in supergroup B Wolbachia than supergroup A (Wilcoxon rank-sum test, P value < 0.05) (fig. 3C). Above data indicate distinct evolutionary pattern of Wolbachia in the two supergroups.
Ortholog analysis of Prokka-predicted proteins from the 33 Wolbachia genomes yielded 367 single-copy orthologs, from which 346 genes were assigned to COG database. Category analysis showed that half of the annotated genes were related to five COG categories: biological processes of translation, ribosomal structure and biogenesis (J), energy production and conversion (C), posttranslational modification (O), protein turnover, chaperones, replication, recombination and repair (L), and nucleotide transport and metabolism (F) (supplementary fig. S5, Supplementary Material online). These pathways are usually conserved and are usually involved in basic biological processes of an organism.
Secreted and Transmembrane Proteins in wStriCN
SignalP and TMHMM predicted that the wStriCN genome has 33 proteins harboring signal peptide and 326 proteins with transmembrane helices. The secreted proteins were mainly proteins involved in forming the outer membrane, basic metabolite transportation, energy production, and secreted systems. The genome also contains proteins that may be associated with bacterial infection, such as the Invasion-associated locus B (IalB) (wStriCN_00836), a peptidoglycan hydrolase Rare lipoprotein A (RlpA) (wStriCN_00335), a Peptidoglycan deacetylase (PgdA) (wStriCN_00552), and the Outer membrane protein OmpA (wStriCN_00807). The proteins with predicted transmembrane-domains included mostly ANKs (37), energy production and conversion related proteins (34), proteins of secretion systems (20), inorganic ion transport related proteins (18), and phage tail proteins (12).
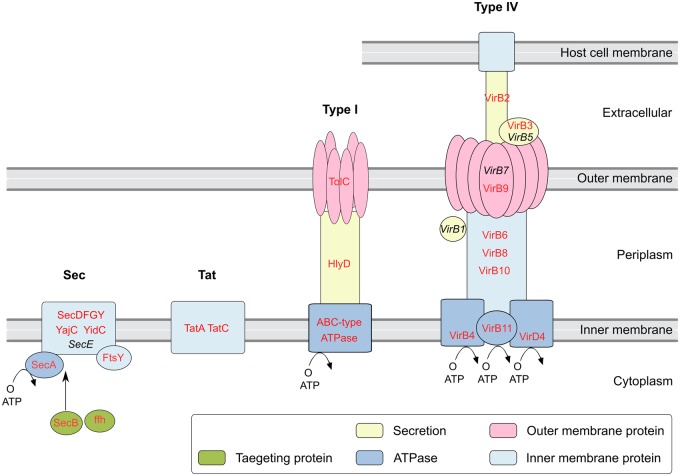
wStriCN has various secretion pathways including the Type I secretion system (T1SS), Type II secretion system (T2SS), Type IV secretion system (T4SS), Type VI secretion system (T6SS), Sec secretion system, and Tat secretion system. Several proteins related to T1SS, the ABC-type protease ATPase (wStriCN_00721), outer membrane protein TolC (wStriCN_00600), and the fusion protein HlyD (wStriCN_01572) were detected in wStriCN (fig. 4). T1SS bypasses the periplasm and allows the secretion of proteins of diverse sizes (Delepelaire 2004). For instance, T1SS has been reported to secrete some ANKs in Wolbachia’s relative Rickettsia (Kaur et al. 2012). wStriCN contains one GspD/PulD gene (wStriCN_00409), which may create a pore in the outer membrane of the bacterial cell through which proteins can be secreted in the Type II secretion system (Nivaskumar and Francetic 2014). The GspD/PulD gene was possibly obtained by horizontal transfer as it is the only T2SS gene in wStriCN genome.
Fig. 4.
—Schematic view of secretion systems identified in the wStriCN genome. The figure is based on KEGG pathway map 03070. Proteins detected in wStriCN are colored red and those missing from wStriCN are shown in italics and black. The GspD from T2SS and PAAR proteins from T6SS were excluded, as they were too few to assemble the secretion systems.
The wStriCN genome has a T4SS with 17 genes organized in 2 operons and 6 individual genes (fig. 4). One operon contains 4 gene copies of virB6, one virB4, and one virB3 (wStriCN_00590 - wStriCN_00595). The other operon contains virB8, virB9, virB10, virB11, and virD4 (wStriCN_01296 - wStriCN_01300). Three duplicated genes: virB4 (wStriCN_00628), virB9 (wStriCN_01017), and virB8 (wStriCN_01520) are scattered in other parts of the genome. The gene organization of the T4SS is the same as it is in most other Wolbachia strains (Pichon et al. 2009). In addition, virB2 has three homologs (wStriCN_00829, wStriCN_01226, and wStriCN_01313) in wStriCN.
We also identified eight genes harboring Zn-binding PAAR domain (supplementary table S8, Supplementary Material online), which are reported to be essential for T6SS-mediated secretion. Six genes forming the Sec secretion pathway (secA, secB, secD, secF, secY, and secG) and one tatC (wStriCN_01495) and two copies of tatA (wStriCN_00457 and wStriCN_01583) related to Tat translocases in the twin arginine translocation (Tat) pathway were identified as well (fig. 4). The Sec and Tat pathways are most commonly used in bacterial secretion systems to transport proteins across the cytoplasmic membrane. The Sec pathway primarily translocates proteins in their unfolded state, whereas the Tat pathway primarily secretes folded proteins (Green and Mecsas 2016).
CI Genes in wStriCN
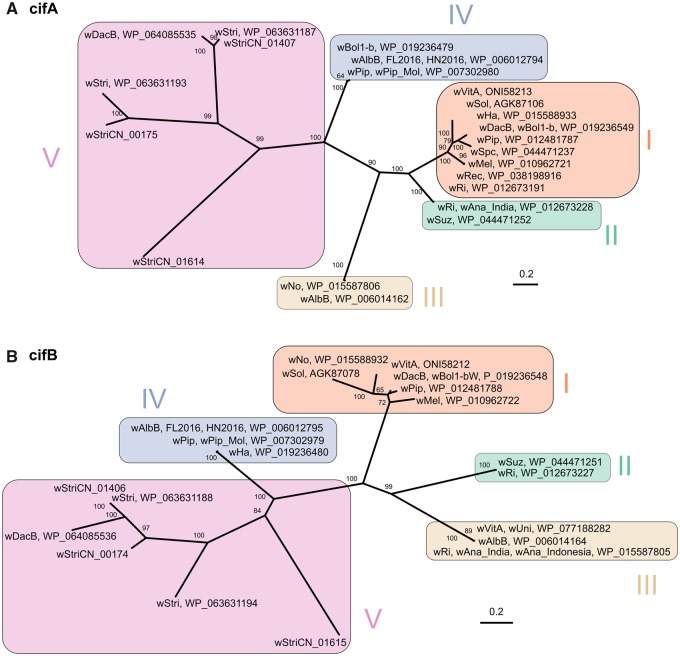
wStriCN has three tandem pairs of cifA–cifB genes (fig. 5). Phylogenetic analysis of CifA and CifB protein sequences showed they were all distinct from the four previously identified “Types” (Lindsey et al. 2018) and were assigned as Type V. The pair of wStriCN_01406 and wStriCN_01407 had similar homologs in wStri and wDacB. The pair of wStriCN_00174 and wStriCN_00175 were only closely related in wStri. The third pair, wStriCN_01614 and wStriCN_01615, were phylogenetically monophyletic (fig. 5). Altogether, CifA and CifB are abundant in wStriCN and have been diverged for a while.
Fig. 5.
—Phylogeny of CifA (A) and CifB (B) proteins. The names of Wolbachia strains and the corresponding NCBI accession numbers of Cif proteins are shown. The tree was constructed using a JTT + F + G4 substitution model for ML analysis. Bootstrap values are indicated at the respective node (only values >50% are shown). The scale bar represents the average number of substitutions per site.
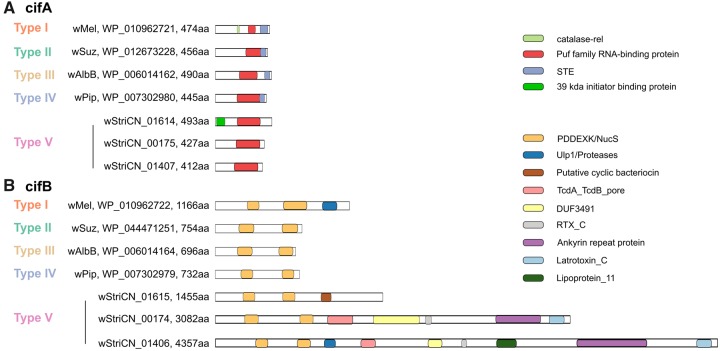
The CifA proteins in wStriCN have two modules: a 39-kDa initiator inhibitor binding domain, which is a new module and locates only in wStriCN_01614, and a Puf family RNA-binding domain that locates in all three CifA homologs and other types of known CifA (fig. 6A). The STE-like transcription factor domain that was present in all other four types was not found in wStriCN CifA proteins. All wStriCN CifB proteins were predicted to have two PDDEXK/endonuclease NucS domains (fig. 6A), which are the same as other types of CifB. In addition, more modules were predicted from wStriCN CifB proteins. One putative cyclic bacteriocin was predicted downstream of the PDDEXK domains in wStriCN_01615. Both wStriCN_00174 and wStriCN_01406 were much longer than any CifB proteins across Types I-IV and contained the following domains: a TcdA_TcdB_pore domain, a DUF3491 domain of unknown function, a RTX_C domain, an ANK, and a latrotoxin_C domain, respectively (fig. 6B). The Ulp1/Proteases domain, which has been experimentally proved to cause CI and exists only in Type I CifB proteins (Beckmann et al. 2017; Lindsey et al. 2018), was also detected in wStriCN_01406. As the longest CifB, wStriCN_01406 also encodes one lipoprotein domain, which was similar to Lepidopteran low molecular weight (30 kDa) lipoprotein. It is noteworthy that none of the three cifA–cifB gene-pairs are located in predicted WO regions based on the prediction results of PHASTER.
Fig. 6.
—Representative predicted structures of CifA (A) and CifB (B) proteins. Representative structures are shown for each type, with the Wolbachia strain name, accession number, and length of the protein indicated at the N-terminus. The structures of all three sets of Cif proteins from wStriCN are also shown.
Biotin and Riboflavin Synthesis Pathways in wStriCN
Like most other Wolbachia strains, wStriCN contained complete pathways for only essential energy-related metabolisms like the tricarboxylic acid cycle, biosynthesis of fatty acid and lipoid acid, and oxidative phosphorylation, which indicate that wStriCN has very limited capabilities of biosynthesizing biological macromolecules and metabolic intermediates. As a result, wStriCN has to reply on host cells to get nutrients for survival. On the other hand, the wStriCN genome has genes for synthesizing the vitamin B members, biotin and riboflavin, which suggests wStriCN could contribute essential nutrients that are poor in plant sap to host planthoppers.
Discussion
The characteristics of the wStriCN genome revealed in this study, such as small genome size (compared with other free-living bacteria), low GC content, and incomplete metabolic abilities, indicate that wStriCN falls in the range of facultative symbionts (Moran et al. 2008; McCutcheon and Moran 2012; Lo et al. 2016). Previous phylogenetic analyses of Wolbachia have assigned wStriCN into the supergroup B (Zhang et al. 2013). Our phylogenic trees of both ribosomal proteins and single-copy ortholog proteins are consistent with this classification and show that wStriCN is closely related to wDi. Our genome-wide ANI analysis confirms the close relationship between wStriCN and wDi. Therefore, wStriCN and wDi may have evolved from the same Wolbachia ancestor. Although genomes of wStriCN and wDi are highly collinear and have similar GC content, the genome size of wStriCN is almost 50% larger than that of wDi and is the largest of all reported supergroup B strains to date. Because the size of a genome is positively correlated with the number of CDSs in bacteria (Lo et al. 2016), wStriCN is hypothesized to have more biological functions than other Wolbachia strains.
Some arthropod Wolbachia strains carry a large number of mobile elements, such as IS and WO (Kent and Bordenstein 2010; Bouchon et al. 2011), which may explain the expansion of Wolbachia genomes. Our results show that Wolbachia strains have a diverse range of IS genes, ranging from 17 to 217. The distribution pattern of IS genes also varies significantly among different strains. The incomplete draft genome of wStriCN has 78 IS genes, more than the medium number in known complete Wolbachia genomes. Furthermore, IS genes of wStriCN belong to 10 IS families, which is the second highest number in all analyzed 33 Wolbachia genomes. The draft genome of wBol1 has 22 IS genes scattered in 11 IS families, which is the highest record in Wolbachia so far (Duplouy et al. 2013). IS are simple small transposase-encoding transposable elements that are frequently detected in prokaryotic genomes (Siguier et al. 2006). They can move from one position on a chromosome to a different position on the same or a different one and have important and spectacular effects in shaping and reshuffling bacterial genomes. Although the accumulation of nucleotide substitutions and deletions over time degraded the vast majority of Wolbachia IS copies (>70) (Bouchon et al. 2011), the high diversity of IS genes in the wStriCN genome showed frequent events of horizontal gene transfer between wStriCN and other organisms. The expansion of wStriCN genome may be a result of multiple exchanges of DNA fragments mediated by IS elements.
Bacteriophages can carry out lateral gene transfer and shuttle large portions of DNA into recipient genomes (Bordenstein and Wernegreen 2004). Nearly, all sequenced Wolbachia harbor prophage WO, except for those acting as obligate mutualistic symbionts (Gavotte et al. 2007; Bordenstein and Bordenstein 2016). The WO in wStriCN, with a size of 233.9 kb, are around twice the size of WO in wAlbB (Sinha et al. 2019). Large fragments of WO were hypothesized to be responsible for particular phenotypes because they can transfer new functions between phylogenetically distinct strains (Kent and Bordenstein 2010).
Many WO genomes encode putative effectors and toxins, like SpvB, VrlC, and Patatin, that potentially interact with host cells (Kent and Bordenstein 2010). Three copies of Patatin-like phospholipase (PLP) were scattered in the prophage regions of wStriCN. PLPs are utilized to facilitate many pathogens’ infection and dissemination (Sitkiewicz et al. 2007). Pathogens and symbionts had significantly higher numbers of PLP-containing genes in their genomes than free‐living bacteria (Banerji and Flieger 2004), which implies that they interact with host cells. For instance, the PLP of intracellular pathogen Legionella pneumophila can cleave fatty acids from membrane lipids (Zhu et al. 2013). The release of fatty acids destabilizes the membrane and causes the release of cytochrome C from the mitochondria in the host cells. Cytochrome C activates caspase 3, which in turn activates programed cell death pathways in the host (Zhu et al. 2013). Further studies are needed to determine whether wStriCN induces host immune responses in L. striatellus.
ANKs can modulate the transcription of host genes by interacting with specific regions of the host chromatin (Al-Khodor et al. 2010). The wStriCN genome encodes 113 ANKs, the largest number to date in all reported Wolbachia strains. This agrees with previous statistical analysis that showed the number of ANKs is significantly higher in supergroup B than in supergroup A (Lindsey et al. 2016). In addition, ANKs are significantly enriched in the phage regions, which suggests that WO have an important role. The wStriCN prophage regions contain 8 genes encoding proteins harboring Zn-binding PAAR, which is a key component of T6SS that can deliver lethal effectors upon direct contact with a target cell (Alteri and Mobley 2016). Bacteria that possess a T6SS have a specific advantage to discriminate, recognize, and kill potential competitors in a population of mixed bacteria.
Apart from T6SS, the wStriCN genome encodes secreted proteins and secretion systems that may be key in interacting with host cells. T4SS is prevalent in multiple Wolbachia strains (Pichon et al. 2009). It is required for bacterial infection, proliferation, and persistence within hosts. Some pathogenic bacteria use T4SSs to translocate virulence factors into the host cell or to mediate horizontal gene transfer (Grohmann et al. 2018). T1SS bypasses the periplasm and allows the secretion of proteins of diverse sizes as well (Delepelaire 2004). Many ANK-containing effectors of Wolbachia relatives, such as Rickettsia and Anaplasma spp., were found to be translocated by T4SS (Klasson et al. 2008; Al-Khodor et al. 2010; Sinha et al. 2019) and T1SS (Kaur et al. 2012).
Wolbachia is well known for inducing CI in many arthropods. Recent genetic studies have shown that the CifA and CifB proteins are mainly responsible for CI in Drosophila flies and Culex mosquitos (Beckmann et al. 2017; LePage et al. 2017). In addition, CI strength appears to be positively correlated with the number of copies of cifA and cifB genes in a strain (LePage et al. 2017). For instance, strains with only one copy, such as wMel, have a comparatively weak CI phenotype, whereas those with two or three copies, such as wRi and wHa, cause strong CI (LePage et al. 2017). The facts that wStriCN causes strong CI in L. striatellus (Noda et al. 2001; Bing et al. 2019) and its genome harbored three sets of cifA–cifB gene-pairs strongly support this correlation.
wStriCN CifA proteins are phylogenetically and structurally different from other known CifA proteins. The STE domain, which is conserved among all four types of CifA, was missing in all three wStriCN CifA proteins. wStriCN CifA proteins also do not have the antioxidant catalase-rel domain that may function in response to reactive oxygen. A genetic analysis suggests that Type I CifA protein alone was able to rescue CI in Drosophilamelanogaster (Shropshire et al. 2018). An evolutionary analysis showed that purifying selection is much stronger on the catalase-rel domain than on the Puf family RNA-binding domain and STE domain (Shropshire et al. 2018). So far the catalase-rel domain has been found only in Type I CifA proteins (Lindsey et al. 2018). Although the critical domain that is responsible for the rescue is unclear yet, we speculate that the Puf family RNA-binding domain is important as it is the only domain that exists across all CifA orthologs.
All three wStriCN CifB proteins harbor two PDDEXK modules. The Ulp1 ubiquitin proteases module of CifB, which was predicted to be unique and completely conserved in the Type I groups (Lindsey et al. 2018), was also identified in one wStriCN CifB (wStriCN_01406). A Wolbachia deubiquitylating enzyme (DUB, or ubiquitin proteases) and a PDDEXK (or PD-(D/E)XK) nuclease domain (DUF1703) were predicted to induce embryonic death in CI (Beckmann et al. 2017). All strains that are able to induce or rescue CI have two or more recovered modules, though they do not necessarily have the Ulp1 ubiquitin proteases module (Beckmann et al. 2017; Lindsey et al. 2018). However, whether or not these Cif proteins contribute to CI in L. striatellus needs experimental confirmation.
The CifB proteins in wStriCN contain many toxin-related domains. wStriCN_01615 encodes one putative cyclic bacteriocin domain, which was reported to have antimicrobial activity (Sawa et al. 2009). Both wStriCN_00174 and wStriCN_01406 encode a TcdA/TcdB pore-forming domain, an RTX C-terminal domain, and a black widow latrotoxin C-terminal domain. The TcdA and TcdB are known as toxins that mediate the pathogenicity of Clostridium difficile. They primarily disrupt the cytoskeletal structure and the tight junctions of target cells causing cell rounding and ultimately cell death (Di Bella et al. 2016). The RTX toxin superfamily is a group of cytolysins and cytotoxins produced by bacteria. During transport, the C-terminal repeats of the RTX toxin were recognized by the T1SS and transferred first through the channel (Linhartová et al. 2010). Although latrotoxins are the main toxin generally considered to be exclusively found in spiders, homologs of the latrotoxin C-terminal domain have been reported in bacteria such as Wolbachia, and Rickettsiella grylli (Zhang et al. 2012), which implies that latrotoxin genes were horizontally transferred from spiders to their bacterial endosymbiont. The latrotoxin C-terminal domain was shown as the most prevalent eukaryoticlike domain in WO (Bordenstein and Bordenstein 2016), indicating that phage might be facilitating its transfer. However, the wStriCN Cif genes are distributed out of prophage regions, implying that their evolution is complicated. Further studies are needed to determine whether those toxin-coding regions are active and play roles in interacting with other prokaryotic competitors or eukaryotic hosts.
Wolbachia can significantly enhance the fecundity of L. striatellus. Wolbachia-infected L. striatellus females laid 30% more eggs than uninfected females (Guo et al. 2018). Our genomic analysis showed that wStriCN contained complete pathways for biological synthesis of biotin and riboflavin, whose concentrations are relatively low in plant sap (Douglas 2017). In contrast to essential amino acids, vitamin Bs were thought to be unimportant nutrition factors for plant sap feeders. However, recent studies in aphids and red cotton bugs showed vitamin Bs contribute to host survival and development as well (Salem et al. 2014; Meseguer et al. 2017). Besides, our recent experimental study stressed the importance of wStriCN-provided biotin and riboflavin in L. striatellus (Ju et al. 2019).
Conclusions
The genome of wStriCN displays hallmarks of an insect symbiont, including a low GC content and reduced genome size compared with free-living bacteria. On the other hand, wStriCN has one of the largest Wolbachia genomes with a large number of mobile elements, which have considerable effects on Wolbachia genome evolution and gene content. Although the Cif proteins in wStriCN are phylogenetically and structurally distinct from all known Cif types, they contain the catalytic domains that correlate with the phenotype of CI and may explain the strong CI phenotype wStriCN induces in L. striatellus. The genome retains pathways for synthesizing biotin and riboflavin, which helps to explain how wStriCN might benefit its host, which feeds on low-nutrient plant sap. Altogether, the wStriCN genome is a resource that will provide further insight into the phylogeny of Wolbachia and enable further biochemical, molecular, and genetic analyses of wStriCN and related symbionts. The genome will also provide clues to the interactions between Wolbachia and its host that may lead to advances in pest and disease control.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Dré Kampfraath from Vrije Universiteit Amsterdam for the help on illustrating phage structure.
This work was supported by the National Natural Science Foundation of China (Grant No. 31672035 and 31871976 to X.-Y.H.) and the startup grant from Nanjing Agricultural University (Grant No. 804015 and 80900224 to X.-L.B.).
Data deposition: The draft genome sequences of wStriCN data have been deposited at NCBI GenBank database under the accession MUIX00000000.
Literature Cited
- Al-Khodor S, Price CT, Kalia A, Abu Kwaik Y.. 2010. Functional diversity of ankyrin repeats in microbial proteins. Trends Microbiol. 18(3):132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alteri CJ, Mobley HLT.. 2016. The versatile type VI secretion system. Microbiol Spectrum 4(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt D, et al. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44(W1):W16–W21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustinos AA, et al. 2011. Detection and characterization of Wolbachia infections in natural populations of aphids: is the hidden diversity fully unraveled? PLoS One 6(12):e28695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, Bordenstein S, Wernegreen JJ, Werren JH.. 2006. Widespread recombination throughout Wolbachia genomes. Mol Biol Evol. 23(2):437–449. [DOI] [PubMed] [Google Scholar]
- Baldo L, Dunning Hotopp JC, et al. 2006. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 72(11):7098–7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji S, Flieger A.. 2004. Patatin-like proteins: a new family of lipolytic enzymes present in bacteria? Microbiology 150(3):522–525. [DOI] [PubMed] [Google Scholar]
- Beckmann JF, Ronau JA, Hochstrasser M.. 2017. A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat Microbiol. 2(5):17007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, et al. 2018. GenBank. Nucleic Acids Res. 46(D1):D41–D47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bing X-L, Zhao D-S, Hong X-Y.. 2019. Bacterial reproductive manipulators in rice planthoppers. Arch Insect Biochem Physiol. 101(2):e21548. [DOI] [PubMed] [Google Scholar]
- Bing XL, et al. 2014. Diversity and evolution of the Wolbachia endosymbionts of Bemisia (Hemiptera: Aleyrodidae) whiteflies. Ecol Evol. 4(13):2714–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleidorn C, Gerth M.. 2017. A critical re-evaluation of multilocus sequence typing (MLST) efforts in Wolbachia. FEMS Microbiol Ecol. 94:fix163. [DOI] [PubMed] [Google Scholar]
- Bordenstein SR, Bordenstein SR.. 2016. Eukaryotic association module in phage WO genomes from Wolbachia. Nat Commun. 7(1):13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein SR, Wernegreen JJ.. 2004. Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol Biol Evol. 21(10):1981–1991. [DOI] [PubMed] [Google Scholar]
- Bouchon D, Leroy E, Cerveau N, Leclercq S, Cordaux R.. 2011. Short- and long-term evolutionary dynamics of bacterial insertion sequences: insights from Wolbachia endosymbionts. Genome Biol Evol. 3:1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10(1):421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. [DOI] [PubMed] [Google Scholar]
- Comandatore F, et al. 2013. Phylogenomics and analysis of shared genes suggest a single transition to mutualism in Wolbachia of nematodes. Genome Biol Evol. 5(9):1668–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delepelaire P. 2004. Type I secretion in gram-negative bacteria. Biochim Biophys Acta Mol Cell Res. 1694(1–3):149–161. [DOI] [PubMed] [Google Scholar]
- Di Bella S, Ascenzi P, Siarakas S, Petrosillo N, di Masi A.. 2016. Clostridium difficile toxins A and B: insights into pathogenic properties and extraintestinal effects. Toxins 8(5):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE. 2017. The B vitamin nutrition of insects: the contributions of diet, microbiome and horizontally acquired genes. Curr Opin Insect Sci. 23:65–69. [DOI] [PubMed] [Google Scholar]
- Duplouy A, et al. 2013. Draft genome sequence of the male-killing Wolbachia strain wBol1 reveals recent horizontal gene transfers from diverse sources. BMC Genomics. 14(1):20–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16(1):157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Makarova KS, Wolf YI, Koonin EV.. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43(D1):D261–D269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavotte L, et al. 2007. A survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol Biol Evol. 24(2):427–435. [DOI] [PubMed] [Google Scholar]
- Glowska E, Dragun-Damian A, Dabert M, Gerth M.. 2015. New Wolbachia supergroups detected in quill mites (Acari: Syringophilidae). Infect Genet Evol. 30:140–146. [DOI] [PubMed] [Google Scholar]
- Green ER, Mecsas J.. 2016. Bacterial secretion systems: an overview. Microbiol Spectrum 4(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann E, Christie PJ, Waksman G, Backert S.. 2018. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol Microbiol. 107(4):455–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, et al. 2018. Wolbachia-induced apoptosis associated with increased fecundity in Laodelphax striatellus (Hemiptera: Delphacidae). Insect Mol Biol. 27(6):796–807. [DOI] [PubMed] [Google Scholar]
- Haegeman A, et al. 2009. An endosymbiotic bacterium in a plant-parasitic nematode: member of a new Wolbachia supergroup. Int J Parasitol. 39(9):1045–1054. [DOI] [PubMed] [Google Scholar]
- Hoshizaki S, Shimada T.. 1995. PCR-based detection of Wolbachia, cytoplasmic incompatibility microorganisms, infected in natural populations of Laodelphax striatellus (Homoptera: Delphacidae) in central Japan: has the distribution of Wolbachia spread recently? Insect Mol Biol. 4(4):237–243. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, et al. 2016. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44(D1):D286–D293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins FM, Hurst GDD.. 2011. Rapid insect evolution by symbiont transfer. Science 332(6026):185–186. [DOI] [PubMed] [Google Scholar]
- Ju J-F, et al. 2019. Wolbachia supplement biotin and riboflavin to enhance reproduction in planthoppers. ISME J. Advance Access published November 25, 2019, doi: 10.1038/s41396-019-0559-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampfraath AA, et al. 2019. Genome expansion of an obligate parthenogenesis-associated Wolbachia poses an exception to the symbiont reduction model. BMC Genomics. 20(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S.. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur SJ, et al. 2012. TolC-dependent secretion of an ankyrin repeat-containing protein of Rickettsia typhi. J Bacteriol. 194(18):4920–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent BN, Bordenstein SR.. 2010. Phage WO of Wolbachia: lambda of the endosymbiont world. Trends Microbiol. 18(4):173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L, et al. 2008. Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol Biol Evol. 25(9):1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L, et al. 2009. The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc Natl Acad Sci U S A. 106(14):5725–5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Kurtz S, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol . 5(2):R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefoulon E, et al. 2016. Breakdown of coevolution between symbiotic bacteria Wolbachia and their filarial hosts. PeerJ 4:e1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LePage DP, et al. 2017. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 543(7644):243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, et al. 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20(2):265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey ARI, Werren JH, Richards S, Stouthamer R.. 2016. Comparative genomics of a parthenogenesis-inducing Wolbachia symbiont. G3 (Bethesda) 6:2113–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey ARI, et al. 2018. Evolutionary genetics of cytoplasmic incompatibility genes cifA and cifB in prophage WO of Wolbachia. Genome Biol Evol. 10(2):434–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhartová I, et al. 2010. RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol Rev. 34(6):1076–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo N, et al. 2007. Taxonomic status of the intracellular bacterium Wolbachia pipientis. Int J Syst Evol Microbiol. 57(3):654–657. [DOI] [PubMed] [Google Scholar]
- Lo W-S, Huang Y-Y, Kuo C-H.. 2016. Winding paths to simplicity: genome evolution in facultative insect symbionts. FEMS Microbiol Rev. 40(6):855–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loetscher A, et al. 2016. OrthoDB v9.1: cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res. 45:D744–D749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. 2018. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res. 46:D851–D860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA.. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10(1):13–26. [DOI] [PubMed] [Google Scholar]
- Meseguer AS, et al. 2017. Buchnera has changed flatmate but the repeated replacement of co-obligate symbionts is not associated with the ecological expansions of their aphid hosts. Mol Ecol. 26(8):2363–2378. [DOI] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A.. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 42(1):165–190. [DOI] [PubMed] [Google Scholar]
- Moriyama M, Nikoh N, Hosokawa T, Fukatsu T.. 2015. Riboflavin provisioning underlies Wolbachia’s fitness contribution to its insect host. mBio 6(6):e01732–01715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nault LR. 1994. Transmission biology, vector specificity and evolution of planthopper-transmitted plant viruses In: Denno RF, Perfect TJ, editors. Planthoppers: their ecology and management. Boston (MA: ): Springer; p. 429–448. [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh N, et al. 2014. Evolutionary origin of insect–Wolbachia nutritional mutualism. Proc Natl Acad Sci U S A. 111(28):10257–10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivaskumar M, Francetic O.. 2014. Type II secretion system: a magic beanstalk or a protein escalator. Biochim Biophys Acta Mol Cell Res. 1843(8):1568–1577. [DOI] [PubMed] [Google Scholar]
- Noda H. 1984a. Cytoplasmic incompatibility in a rice planthopper. J Hered. 75(5):345–348. [Google Scholar]
- Noda H. 1984b. Cytoplasmic incompatibility in allopatric field populations of the small brown planthopper, Laodelphax striatellus, in Japan. Entomol Exp Appl. 35(3):263–267. [Google Scholar]
- Noda H, Koizumi Y, Zhang Q, Deng K.. 2001. Infection density of Wolbachia and incompatibility level in two planthopper species, Laodelphax striatellus and Sogatella furcifera. Insect Biochem Mol Biol. 31(6–7):727–737. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H.. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 8(10):785–786. [DOI] [PubMed] [Google Scholar]
- Pichon S, et al. 2009. Conservation of the type IV secretion system throughout Wolbachia evolution. Biochem Biophys Res Commun. 385(4):557–562. [DOI] [PubMed] [Google Scholar]
- R Core Team. (2018). R: a language and environment for statistical computing [Internet]. Vienna (Austria: ): R Foundation for Statistical Computing; Available from: http://www.R-project.org/ [Google Scholar]
- Ros VID, Fleming VM, Feil EJ, Breeuwer JAJ.. 2009. How diverse is the genus Wolbachia? Multiple-gene sequencing reveals a putatively new Wolbachia supergroup recovered from spider mites (Acari: Tetranychidae). Appl Environ Microbiol. 75(4):1036–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem H, et al. 2014. Vitamin supplementation by gut symbionts ensures metabolic homeostasis in an insect host. Proc R Soc B 281(1796):20141838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa N, et al. 2009. Identification and characterization of lactocyclicin Q, a novel cyclic bacteriocin produced by Lactococcus sp. strain QU 12. Appl Environ Microbiol. 75(6):1552–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MJ, et al. 2018. Wolbachia wStri blocks Zika virus growth at two independent stages of viral replication. mBio 9(3):e00738–00718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14):2068–2069.24642063 [Google Scholar]
- Shen W, Le S, Li Y, Hu F.. 2016. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One 11(10):e0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shropshire JD, On J, Layton EM, Zhou H, Bordenstein SR.. 2018. One prophage WO gene rescues cytoplasmic incompatibility in Drosophila melanogaster. Proc Natl Acad Sci U S A. 115(19):4987–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M.. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34(90001):D32–D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A, Sun L, Li Z, Carlow CKS.. 2019. Complete genome sequence of the Wolbachia wAlbB endosymbiont of Aedes albopictus. Genome Biol Evol. 11(3):706–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitkiewicz I, Stockbauer KE, Musser JM.. 2007. Secreted bacterial phospholipase A2 enzymes: better living through phospholipolysis. Trends Microbiol. 15(2):63–69. [DOI] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN.. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, et al. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 35(3):543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Baldo L, Clark ME.. 2008. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol . 6(10):741–751. [DOI] [PubMed] [Google Scholar]
- Wu M, et al. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2(3):e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S-H, Ha S-M, Lim J, Kwon S, Chun J.. 2017. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110(10):1281–1286. [DOI] [PubMed] [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y.. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 8(1):28–36. [Google Scholar]
- Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L.. 2012. Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 7(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K-J, Han X, Hong X-Y.. 2013. Various infection status and molecular evidence for horizontal transmission and recombination of Wolbachia and Cardinium among rice planthoppers and related species. Insect Sci. 20(3):329–344. [DOI] [PubMed] [Google Scholar]
- Zhu W, Hammad LA, Hsu F, Mao Y, Luo Z-Q.. 2013. Induction of caspase 3 activation by multiple Legionella pneumophila Dot/Icm substrates. Cell Microbiol. 15:1783–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zug R, Hammerstein P.. 2012. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One 7(6):e38544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zug R, Hammerstein P.. 2015. Bad guys turned nice? A critical assessment of Wolbachia mutualisms in arthropod hosts. Biol Rev. 90(1):89–111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.