Abstract
We describe the use of MALDI-TOF mass spectrometry in the analysis of a suspected case of gamma heavy chain disease. The patient had an abnormal serum immunofixation result where a monoclonal gamma heavy chain band was present without a corresponding light chain. Analysis by MALDI-TOF mass spectrometry revealed large peaks in the spectrum following IgG-specific purification. The m/z values of the peaks were outside the expected range for normal heavy chains or light chains. Corresponding peaks were not present in mass spectra of the kappa- or lambda-specific purifications. MALDI-TOF MS confirmed the presence of a truncated heavy chain without associated light chains. This case report demonstrates the value of mass spectrometry in interpreting challenging cases such as the identification of heavy chain disease.
Keywords: MALDI-TOF mass spectrometry, heavy chain disease, monoclonal immunoglobulin
1. Introduction
Heavy chain disease (HCD) is a group of rare B-cell proliferative disorders characterized by the production of truncated monoclonal immunoglobulin heavy chains without associated light chains. There are three types of HCD based on the class of immunoglobulin heavy chain produced by the malignant lymphoplasma cell: alpha, gamma, and mu. All are extremely rare, with alpha HCD being the most common and mu HCD being the least common [1].
Diagnosis of HCD requires the identification of a truncated heavy chain without an associated light chain in serum or urine [1]. This is often accomplished through use of serum and urine immunofixation. However, recent case reports have illustrated the difficulty in correctly identifying HCD using this approach [2–4]. Light chain bands are sometimes not visible by immunofixation due to antigen excess, over-dilution of serum, different detection limits of the heavy- and light-chain antisera reagents, or masking of the light chain epitope. In addition, immunofixation interpretation can be challenging. Monoclonal heavy chains often appear as broad bands and can be mistaken for polyclonal immunoglobulins [3].
Recently, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) has been used to detect monoclonal immunoglobulins in serum and urine [5]. This method offers improved analytical sensitivity and specificity over current electrophoretic techniques and can analyze samples in a high-throughput, automated manner [5–7]. In addition, mass measurements provide structural information about the protein not available by immunofixation. Therefore, MALDI-TOF MS could greatly aid in the identification of HCD. Here, we describe a case of gamma HCD that was identified by serum immunofixation during routine clinical testing and was confirmed using MALDI-TOF MS.
2. Materials and Methods
2.1. Case description
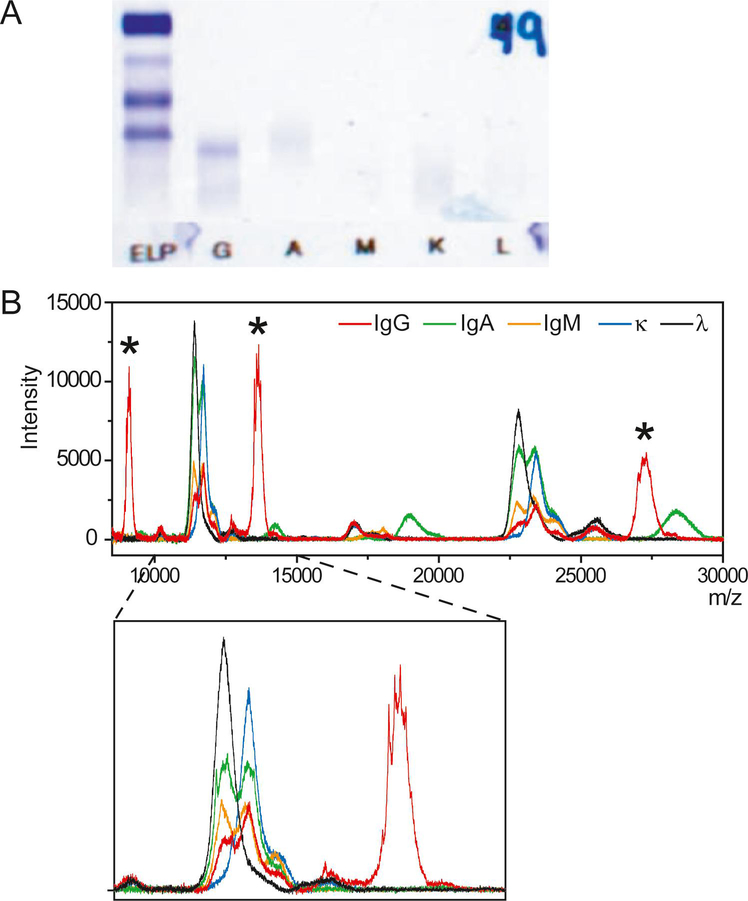
A 73-year old man presented to his primary care physician in late 2018 after several months of progressive malaise. Symptoms included fatigue, low grade fevers, cough, and body aches. The patient, a former smoker, had a history of low-grade lymphoma since 1998 with a splenectomy in 2013. In the workup of his malaise, chest X-ray was concerning for metastatic malignancy, and a PET scan showed extensive hypermetabolic lymphadenopathy involving almost all lymph nodes in the neck, chest, abdomen, pelvis and bilateral inguinal regions. A biopsy of the right axillary lymph node showed a diffuse atypical lymphoid infiltrate. Immunohistochemistry and flow cytometric analysis performed on the biopsy revealed an abnormal immunophenotype including abnormal expression of CD20 (bright), CD19 (bright), MUM1 and lambda light chain restriction without BCL2, BCL6 or MYC rearrangement by cytogenetic analysis. The diagnosis was diffuse large B-cell lymphoma (non-germinal center B-cell type). The abnormal B-cell population represented 17.3% of the white blood cells, though the cells from this biopsy had reduced viability. Serum immunofixation, which was performed on a Hydrasys 2 system using the Hydragel 9 IF kit (Sebia), revealed a monoclonal gamma heavy chain with no corresponding kappa or lambda light chain (Figure 1A).
Figure 1:
Immunofixation results (A) and MALDI-TOF mass spectra (B) of the patient sample. The different charge states of the gamma heavy chain are highlighted with an asterisk (*). The inset shows details of spectra from 10,000–15,000 m/z.
2.2. Sample purification
Prior to analysis by mass spectrometry, immunoglobulins were purified from serum using magnetic beads coated with polyclonal sheep antibodies specific for human IgG, IgA, or IgM heavy chains, or total kappa or total lambda light chains (The Binding Site). Serum was diluted 1:10 in phosphate buffered saline with 0.1% Tween (PBST) and divided into 5 separate aliquots. Magnetic beads were added to the aliquots and incubated for 15 minutes while shaking. Beads were washed 3 times with PBST and twice with deionized water. Captured immunoglobulins were eluted from beads using 5% acetic acid, 20 mM tris(2-carboxyethyl) phosphine (TCEP) in water.
Because gamma heavy chains are often truncated, three other purification methods were used. First, IgG was purified from the patient serum using Protein A spin columns (NAb Protein A Plus Spin Kit, Thermo Scientific). Briefly, the column was equilibrated with binding buffer (100 mM phosphate, 150 mM sodium chloride, pH 7.2) provided with the kit. Twohundred microliters of serum were added to the spin column and incubated at room temperature with mixing for 10 minutes. The column was centrifuged and the flow-through was discarded. The column was washed 3 times with 400 μL of binding buffer. Purified immunoglobulins were eluted in 3 fractions using 400 μL of elution buffer provided with the kit. Ten microliters of each fraction were mixed with 10 μL of 50 mM TCEP and analyzed by MALDI-TOF MS.
The sample was also purified using camelid-derived nanobody reagents that are specific to the constant 1 (CH1) domain of IgG (CaptureSelect IgG-CH1) or the Fc-region of IgG (CaptureSelect FcXL) (Thermo Fisher Scientific). The patient’s serum was diluted 1:10 with phosphate buffered saline (PBS). Nanobody beads were added to the diluted serum and incubated for 45 minutes with gentle shaking. Unbound material was discarded, and the beads were washed 3 times with PBS and 2 times with water. The immunoglobulins were eluted from the beads and the light chains were separated from heavy chains using 40 μL of 5% acetic acid, 50 mM TCEP.
2.3. Treatment with PNGase F
PNGase F (Promega) was used to confirm suspected glycosylation of the gamma heavy chain. The patient’s sample was diluted 1:10 with PBST. IgG-specific magnetic beads were added and were incubated with the diluted sample for 15 minutes while shaking. Beads were washed 3 times with PBST and 2 times with water to remove non-specific serum proteins. Thirty microliters of PBST and 2 μL (20 units) of PNGase F were added to the beads. The mixture was incubated overnight at 37°C. After incubation, the supernatant was removed and discarded. The beads were washed twice with PBST and twice with water to remove residual buffer and PNGase F. Captured immunoglobulins were eluted with 30 μL of 5% acetic acid, 20 mM TCEP in water. As a control, a second aliquot the patient sample was prepared the same way, except that PNGase F was not added to the mixture.
2.4. MALDI-TOF MS
Purified samples were mixed 1:9 (sample:matrix) with 11 mg/mL alpha-cyano-4-hydroxycinnamic acid in 50/50 (v/v) acetonitrile/water with 0.1% TFA. The sample/matrix mixture was spotted onto a polished stainless steel MALDI target plate. Mass spectra were acquired on a Microflex LT MALDI-TOF mass spectrometer (Bruker) using positive ion mode from 8,000–30,000 m/z. Data was also acquired at a higher mass range (8,000–60,000 m/z) for the IgG-specific purifications of the patient sample. Five hundred laser shots were taken per spot and summed. Spectra were reviewed in FlexAnalysis Software (Bruker) and visually inspected for the presence of monoclonal peaks.
3. Results
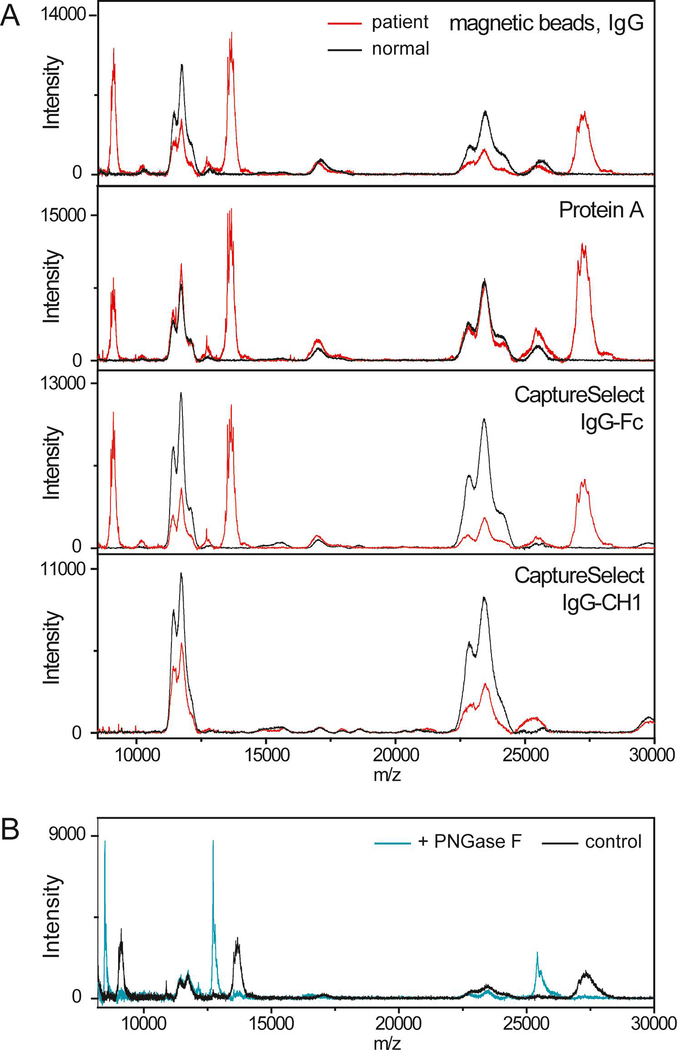
Analysis by MALDI-TOF MS demonstrated large peaks in the mass spectrum of the IgG-specific purification at ~27000, ~13500 and ~9000 m/z (Figures 1B). No abnormal peaks were observed at higher m/z values. These peaks have no corresponding signals in the mass spectra of the kappa- or lambda-specific purifications (Figure 1B). These peaks are not present in normal serum and are outside the expected mass range for normal IgG heavy chains (Figure 2A). These peaks were observed after purification with the IgG-specific magnetic beads, Protein A, and the CaptureSelect beads specific to the Fc region of IgG. The peaks were not observed after purification with the CaptureSelect beads specific for the CH1 domain of IgG (Figure 2A). Finally, single peaks at lower m/z values were observed after treatment with the deglycosylating enzyme, PNGase F (Figure 2B). This confirms that the protein is glycosylated (N-linked) and that the broad, multipeak signals resulted from glycosylation.
Figure 2:
(A) MALDI-TOF mass spectra of the patient sample (red) purified with IgG-specific magnetic beads, Protein A spin column, IgG-Fc-specific CaptureSelect reagents, and IgG-CH1-specific CaptureSelect reagents compared to that of a healthy volunteer (black). (B) MALDI-TOF mass spectra of the patient sample treated with PNGase F showing a shift toward lower m/z values compared to the sample without PNGase F (control).
4. Discussion
This case demonstrates the benefit of using MALDI-TOF MS to detect monoclonal immunoglobulins. Mass spectrometry provides direct detection of light and heavy chains and provides structural information about the proteins, both of which are particularly important for correctly identifying monoclonal heavy chains.
When interpreting mass spectra, the light chain charge states are primarily used to detect the presence of monoclonal proteins. The light chains produce larger signals by MALDI-TOF MS compared to heavy chains because they ionize more readily and are usually not glycosylated. When an intact monoclonal immunoglobulin is present, a monoclonal peak is visible in the light chain regions of both the heavy-chain-specific and light-chain-specific purification (Supplemental Figure 1). The peaks are produced from the same molecule (i.e. the monoclonal light chain) which has been purified from serum using two different reactions. A monoclonal peak is less apparent in the heavy chain regions of the spectra for several reasons. Heavy chains typically don’t ionize as well and produce lower signals by MALDI-TOF MS. Also, the presence of multiple glycoforms cause the signals to appear broad due to the lower resolution of the MALDI-TOF mass spectrometer.
Compared to the spectra observed with an intact monoclonal immunoglobulin, the spectra from our patient are unusual and indicate the presence of a truncated gamma heavy chain for several reasons. First, large signals in the spectrum of the IgG-specific purification are present with no corresponding signal from the light chain purifications. Second, treatment with PNGase F confirmed that the protein is glycosylated (Figure 2B). Finally, the m/z values of these broad peaks are outside the expected range for normal heavy chains and normal light chains, and the protein is likely missing at least part of the CH1 domain since the abnormal peaks were not observed following purification with the IgG-CH1-specific nanobody reagent. These observations are consistent with a gamma heavy chain; previous structural studies have shown that gamma heavy chains typically range in size from 27,000–49,000 Da and completely lack the CH1 domain [8]. The fact that this signal is relatively large suggests that the protein may ionize better than normal heavy chains (perhaps because of its smaller size) or may be concentrated. Deletion of the CH1 domain prevents degradation of the protein through heat shock protein 78. Therefore, heavy chains can be observed at high concentrations in serum, despite low disease burden in the marrow [9]. In this case, mass spectrometry helped identify a monoclonal gamma heavy chain associated with a lymphoplasma cell proliferative disorder not visible on the lymph node biopsy in a patient already diagnosed with diffuse large B-cell lymphoma.
Few other laboratory techniques are able to show both that the heavy chain is structurally abnormal and that there are no associated light chains. Serum and urine immunofixation are primarily used to identify HCD. However, due to analytical limitations, it is recommended that at least one additional method be used to confirm the presence of a monoclonal heavy chain [3]. In previous case reports, capillary electrophoresis combined with immunotyping, the heavy/light chain nephelometric immunoassay (Hevylite™, The Binding Site), or a modified immunofixation procedure have been used [3,10–12]. These techniques provide additional evidence that a free heavy chain without an associated light chain is present. However, they are indirect measures of the immunoglobulin and do not demonstrate that the heavy chain is truncated. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blotting can show that the heavy chain is truncated and has been used in the workup of HCD cases [13,14]. However, this is a highly manual technique and not commonly used in clinical laboratories.
MALDI-TOF MS has been used previously to analyze a potential case of HCD. In this previous case, MALDI-TOF MS detected a corresponding light chain confirming that an intact immunoglobulin, not an alpha heavy chain, was present. This finding was consistent with clinical history as the patient had no symptoms of alpha HCD [2]. This report used CaptureSelect nanobody reagents to purify immunoglobulins from serum. Readers should be aware that these reagents are available with various specificities to the heavy chain constant domains and that the IgG-CH1-specific beads may miss true cases of gamma heavy chain disease, as we observed in our patient (Figure 2A, bottom panel).
In conclusion, MALDI-TOF MS could greatly improve the recognition of HCD because it directly detects the light chains and heavy chains and provides structural information about the proteins. Although this technique currently has limited availability, MALDI-TOF mass spectrometers are relatively inexpensive, user-friendly and robust instruments. The development of automated sample preparation and data analysis software will help make this methodology more accessible to clinical laboratories in the future.
Supplementary Material
Acknowledgements
This work was supported by a grant from the Society of Memorial Sloan Kettering Cancer Center (to K.L.T.) and by the Memorial Sloan Kettering Core Grant (P30 CA008748) funded by the National Cancer Institute. K.L.T has received research support in the form of reagents and instrumentation from The Binding Site.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Wahner-Roedler DL, Kyle RA, Heavy chain diseases, Best Pract Res Clin Haematol. 18 (2005) 729–746. doi: 10.1016/j.beha.2005.01.029. [DOI] [PubMed] [Google Scholar]
- [2].Yu M, Bruns DE, Katzmann JA, Silverman LM, Murray DL, Restricted IgG-Kappa and free alpha-heavy-chain bands in an asymptomatic 62-year-old man, Clin. Chem 64 (2018) 265–268. doi: 10.1373/clinchem.2016.269050. [DOI] [PubMed] [Google Scholar]
- [3].Bosman MCJ, Schreurs RHP, Nieuwenhuizen L, Bakkeren DL, Jacobs JFM, Broad bands observed in serum electrophoresis should not be taken lightly, Clin. Chem 65 (2019) 618–621. doi: 10.1373/clinchem.2018.297176. [DOI] [PubMed] [Google Scholar]
- [4].Ramasamy I, Rudzki Z, Two cases of γ-heavy chain disease and a review of the literature, Case Rep Hematol. 2018 (2018). doi: 10.1155/2018/4832619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mills JR, Kohlhagen MC, Dasari S, Vanderboom PM, Kyle RA, Katzmann JA, Willrich MAV, Barnidge DR, Dispenzieri A, Murray DL, Comprehensive assessment of M-proteins using nanobody enrichment coupled to MALDI-TOF mass spectrometry, Clin. Chem 62 (2016) 1334–1344. doi: 10.1373/clinchem.2015.253740. [DOI] [PubMed] [Google Scholar]
- [6].Milani P, Murray DL, Barnidge DR, Kohlhagen MC, Mills JR, Merlini G, Dasari S, Dispenzieri A, The utility of MASS-FIX to detect and monitor monoclonal proteins in the clinic, Am. J. Hematol 92 (2017) 772–779. doi: 10.1002/ajh.24772. [DOI] [PubMed] [Google Scholar]
- [7].Moore LM, Cho S, Thoren KL, MALDI-TOF mass spectrometry distinguishes daratumumab from M-proteins, Clin. Chim. Acta 492 (2019) 91–94. doi: 10.1016/j.cca.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wahner-Roedler DL, Witzig TE, Loehrer LL, Kyle RA, γ-Heavy Chain Disease: Review of 23 Cases, Medicine. 82 (2003) 236–250. doi: 10.1097/01.md.0000085058.63483.7f. [DOI] [PubMed] [Google Scholar]
- [9].Munshi NC, Digumarthy S, Rahemtullah A, Case 13–2008, N. Engl. J. Med 358 (2008) 1838–1848. doi: 10.1056/NEJMcpc0800959. [DOI] [PubMed] [Google Scholar]
- [10].Gulli F, Napodano C, Pocino K, Cuccaro A, Hohaus S, Basile U, Heavy chain disease: our experience, Clin. Chem. Lab. Med 56 (2017) e10–e12. doi: 10.1515/cclm-2017-0254. [DOI] [PubMed] [Google Scholar]
- [11].Luraschi P, Infusino I, Zorzoli I, Merlini G, And CF, Franzini C, Heavy chain disease can be detected by capillary zone electrophoresis, Clin. Chem 51 (2005) 247–249. doi: 10.1373/clinchem.2004.043158. [DOI] [PubMed] [Google Scholar]
- [12].Kaleta E, Kyle R, Clark R, Katzmann J, Analysis of patients with γ-heavy chain disease by the heavy/light chain and free light chain assays, Clin. Chem. Lab. Med 52 (2014). doi: 10.1515/cclm-2013-0714. [DOI] [PubMed] [Google Scholar]
- [13].Johannis W, Blommer J, Klatt AR, Renno JH, Wielckens K, Gamma heavy chain disease in a patient with rheumatoid arthritis – a laboratory evaluation, Biochem Med (Zagreb). 22 (2012) 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Morita K, Kawamoto H, Takada H, Nakamura S, Ishii K, Okamoto Y, Unusual γ heavy chain disease protein in a patient with splenic marginal-zone lymphoma, Ann. Clin. Biochem 43 (2006) 161–164. doi: 10.1258/000456306776021490. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.