Abstract
Cycas panzhihuaensis L. Zhou & S. Y. Yang (Cycadaceae) is an endangered gymnosperm species endemic to the dry-hot valley of the Jinsha River basin in southwest China. Although the wild C. panzhihuaensis population from Panzhihua Cycad Natural Reserve is well protected and its genetic diversity has been well assessed, the genetic characteristics of populations outside the nature reserve, which face larger risks of extinction, remain unknown. Furthermore, the population genetics and historical dynamics of this endemic and endangered species have not been examined across its entire range. In this study, to analyze the genetic diversity, phylogeographical structure and demographic history of C. panzhihuaensis from all its seven known locations, we sequenced and compared molecular data from chloroplastic DNA (psbA-trnH, psbM-trnD, and trnS-trnG), single-copy nuclear genes (PHYP, AC5, HSP70, and AAT) from 61 individuals, as well as 11 nuclear microsatellite loci (SSR) from 102 individuals. We found relatively high genetic diversity within populations and high genetic differentiation among populations of C. panzhihuaensis, which is consistent with the patterns of other Asian inland cycads. Although no significant phylogeographical structure was detected, we found that small and unprotected populations possess higher genetic diversity and more unique haplotypes, which revises our understanding of diversity within this species and deserves due attention. Analysis of demographic dynamics suggest that human activity might be the key threat to C. panzhihuaensis. Based on the genetic characterization of C. panzhihuaensis, we propose several practical guidelines for the conservation of this species, especially for the populations with small sizes.
Keywords: Cycas panzhihuaensis, Conservation, Genetic diversity, Phylogeography, Chloroplast and nuclear DNA, Microsatellite
1. Introduction
Cycads represent an ancient lineage originating around 250 million years ago (Ma), although recent studies date the crown age of most extant cycad species to around 12 Ma, much younger than proposed by the ‘Miocene radiations’ hypothesis (Nagalingum et al., 2011, Salas-Leiva et al., 2013). Regardless, even though Cycadales have undergone rapid diversification, most species remain endangered. As the most threatened group in gymnosperms, 88% of cycads (IUCN, 2017) are threatened with extinction, which is mainly due to human activities including habitat disturbance, over-collecting for ornamental and medical purposes (Mankga and Yessoufou, 2017).
Cycas panzhihuaensis L. Zhou & S. Y. Yang, the only species from Cycas sect. Panzhihuaenses (D.Yue Wang) K.D.Hill, is sister to two coastal species in China and Japan from C. sect. Asiorientales Schuster, Cycas revoluta and Cycas taitungensis, and seems to be the earliest diverged species from Cycas (Liu et al., 2018). There seems to be great discrepancy in the genetic patterns between these two coastal species (C. revoluta and C. taitungensis) and other species distributed in the southwest China due to the differences in historical topography and the evolutionary process of species. The two coastal species were considered to have experienced population expansion in Quaternary glaciation, characterized by high gene flow, shared alleles among populations, and no significant genetic structure among populations (Huang et al., 2001, Wu et al., 2007). Correspondingly, population retreating was generally detected in southwest China Cycas species, with large genetic differences and significant genetic structure among populations (Zheng et al., 2017). However, few studies have examined the genetic characteristics of populations of C. panzhihuaensisis, which is the inland species most closely related to coastal Cycas species.
In addition, the habitat of C. panzhihuaensis is xerothermic valley along the Jinsha River in southwest China, a basal zone adjacent to the Hengduan Mountains Region. This northernmost dry hot valley is a fragile ecosystem inhabited by a high number of endemic and endangered plant species (Jin, 1999, Yang et al., 2016), suggesting that the valley was a refugium in the quaternary glacial period (Zhao and Gong, 2015). Given the fragmented habitat and limited gene flow among populations that result from the precipitous terrains and deep gorges, the endemic flora of this region occupies generally low levels of genetic diversity within populations and high genetic differentiation among populations, with a correlation between lineages and geographic distribution (Zhao and Gong, 2015), e.g., in the cases of Munronia delavayi (Jia et al., 2014), Aristolochia delavayi (Yang et al., 2014) and Psammosilene tunicoides (Zhang et al., 2011a). Comparing the population genetic diversity, genetic structure, and population dynamics of C. panzhihuaensis with other endemic species from the dry hot valley would help elucidate the role of geological, climatic, and evolutionary factors in shaping the population history of Cycas species.
The population of C. panzhihuaensis is vulnerable and has suffered serious declining in previous 40 years for illegal trading since it was described in 1980s (Zhou et al., 1981), after the cycads became popular in horticulture in China since 1970s. Combined with frequently deforestation, grazing, agricultural reclamation and mining, half of primitive habitat was decreased (45.22% estimated by He and Li, 1999, 20%–50% by Hill et al., 2003). He and Li (1999) found that only five of the 13 populations remained by 1999. Through a comprehensive investigation along the Jinsha River basin, C. panzhihuaensis was mainly found to restrict in the Sichuan Panzhihua Cycad Natural Reserve, which the census population size (Nc) is large to 240,000, with other isolated populations only accounting for about 1% individuals (Hao, 2011).
Several studies have been conducted on C. panzhihuaensis previously including geographical distribution (He and Li, 1999), habits (Zhou et al., 2009), morphological characteristics (Liu et al., 1990; Wang et al., 2011), breeding (Liu et al., 2016, Tang and Su, 2004, Wang et al., 1997; Yu, 2015) and community characteristics (Hao, 2011, Wang, 2016) as well as the whole chloroplast (cp) genome sequencing and characterization (Han et al., 2016). The genetic diversity of this species has also been reported in a series of studies. Relatively high level of genetic variation (percentage of polymorphic loci %P = 90.38%) was found in population Panzhihua Natural Reserve using 13 inter-simple sequence repeat (ISSR) markers (Wang et al., 2010). Subpopulation structure in this reserve was explored and two evolutionary significant units were defined via amplified fragment length polymorphism (AFLP) (Yang et al., 2015). However, the above studies only included one wild population, as most wild populations are facing their risk of extinction. Zhang (2012) performed a genetic diversity analysis of this species by sampling seven populations (Zhang, 2012, unpublished). However, this study was conducted basing on two chloroplastic DNA markers with limited variation sites, which made their conclusions less comprehensive and convincing. Considering the declining population size and extinction risk of C. panzhihuaensis, a more comprehensive revisiting and survey on the population genetics of this species by applying more informative molecular data is urgently required.
In this study, we characterized the population genetic diversity, genetic structure, and population dynamics of all populations of C. panzhihuaensis using three sets of molecular markers. Our findings allow us to compare genetic characteristics of C. panzhihuaensis with other Cycas species or taxa that live in similar habitats, determine how populations in small size affect conservation guidelines, and understand how different sets of molecular data affect determination of key parameters when examining population genetic characteristics in Cycas.
2. Materials and methods
2.1. Sampling
All individuals in this study were collected from seven locations located in either Yunnan or Sichuan province, China. Five of these populations are wild, whereas two populations, JPD and GH, are cultivated. Cultivated plants were sampled because the nearby habitats were completely destroyed. From each population, 10 and 20 individuals were randomly selected for nucleotide sequencing and microsatellite data analysis, respectively. All individuals were used in populations with less than 10–20 individuals. In total, we sampled 102 adult individuals for microsatellite analysis and 61 adult individuals for chloroplast and nuclear DNA sequencing. Details of populations and sample sizes (N) are shown in Table S1.
2.2. DNA extraction and PCR amplification
Total DNA was extracted from leaves dried in silica and stored at −20 °C using a modified CTAB method (Doyle, 1991). For all analyses of C. panzhihuaensis populations, we screened each population using three chloroplast DNA (cpDNA) intergenic spacers, psbA-trnH, psbM-trnD, and trnS-trnG (Hamilton, 1999; Shaw et al., 2005); four low-copy nuclear genes (nDNA), PHYP (phytochrome P gene), AC5 (Cycas revoluta actin 5), HSP70 (heat shock protein 70 kDa gene), and AAT (aspartate aminotransferase) (unpublished); and 11 nuclear SSR markers with high polymorphism (Li et al., 2009, Wang et al., 2008, Yang et al., 2008, Zhang et al., 2009, Zhang et al., 2010). Previous studies have used these molecular markers to analyze both the phylogeny and population genetics of Cycadaceae (Feng et al., 2017, Gong and Gong, 2016, Liu et al., 2018, Sangin et al., 2010, Zheng et al., 2016). Previous studies have used these molecular markers to analyze both the phylogeny and population genetics of Cycadaceae (Feng et al., 2017, Gong and Gong, 2016, Liu et al., 2018, Sangin et al., 2010, Zheng et al., 2016). Primer information is listed in Table S2 and Table S3.
For nDNA, the PCR reaction mix contained 3.0 μL 10 × PCR buffer, 2.25 μL MgCl2, 2.25 μL dNTP, 2.25 μL DMSO/BSA, 0.6 μL of each primer, 0.6 μL Taq DNA polymerase (5 U/μL; TsingKe Biological Technology, Kunming, China), 10–50 ng DNA and replenished double-distilled water to a total volume of 30 μL. PCR sequence amplifications for nDNA were an initial 5 min denaturation at 94 °C; 35 cycles of 5 min denaturation at 95 °C, 1 min annealing at 50–55 °C, and 2 min extension at 72 °C; with a final 10 min extension at 72 °C. For cpDNA, the PCR reaction mix contained 3.0 μL 10 × PCR buffer, 1.5 μL MgCl2, 1.5 μL dNTP, 1.5 μL DMSO, 0.45 μL of each primer, 0.45 μL Taq DNA polymerase (5 U/μL), 10 ng DNA and 19.5 μL double-distilled water to a total volume of 30 μL. PCR sequence amplifications for cpDNA were initiated with a 5-min denaturation at 80 °C; 30 cycles of 5 min denaturation at 95 °C, 1 min annealing at 50–55 °C, and 1.5 min extension at 65 °C; with a final 1.5 min extension at 65 °C. The PCR products were purified and sequenced in both directions by TsingKe Biological Technology, Kunming, China. For microsatellite markers, primers were labeled fluorescently with FAM, capillary electrophoresis was performed with ABI 3730xl DNA analyser, and SSR fragments were analysed using Genemapper 4.0 at Shanghai Majorbio Bio-pharm Technology Company Ltd. GenBank accession numbers of the DNA sequences used in this study are available in Table S2.
2.3. Data analysis
2.3.1. Nucleotide sequence
Nucleotide sequences were visually adjusted and generated by SeqMan (Swindell and Plasterer, 1997) in DNASTAR biological information package, and aligned using clustal W (Thompson et al., 1994) with subsequent manual adjustment in BioEdit v7.0.4.1 (Hall, 1999). Before the combination of sequence data, the incongruence length difference test (Barker and Lutzoni, 2002) was conducted by PAUP v4.0 (Swofford, 2002) with a heuristic search and 100 replicates to verify the congruency of different markers. After the test, the three cpDNA fragments (P = 1.00), as well as three nuclear gene sequences AC5, HSP70, and AAT (P = 0.86), were combined by PAUP v4.0. The combined nDNA matrix was named N3 for subsequent analysis and PHYP was analyzed separately as this nuclear gene was incongruent with other nuclear genes (P < 0.05). To estimate the genetic diversity and differentiation level of C. panzhihuaensis, the nDNA sequences were phased and the nucleotide diversity (π) and haplotype diversity (Hd) (Nei, 1987) of nucleotide sequences were calculated using DnaSP v5.0 (Rozas, 2009). The total genetic diversity (HT), genetic diversity within populations (HS), and the coefficient of genetic differentiation for non-ordered alleles (GST, NST) were calculated using Permut v1.0 (Pons and Peti, 1996). The genetic variations within and among populations were evaluated by AMOVA in Arlequin v3.11 (Schneider et al., 2000). The FST acquired from AMOVA analysis was used to calculate the gene flow (Nm) via the formula Nm = (1−FST)/4FST (Wright, 1931). The correlation of geographic distance and genetic distance among populations of C. panzhihuaensis was detected by Mantel tests in GenAlEx package v6.5 (Peakall and Smouse, 2012), which evaluated the isolation by a distance (IBD) model.
Maximum Parsimony (MP) 50% majority trees and Bayesian trees were constructed to infer the relationship of haplotypes by using Cycas guizhouensis Lan & R.F. Zou as an outgroup. The optimal substitution model of each dataset was tested by IQ-TREE Modelfinder (Kalyaanamoorthy et al., 2017). For the MP trees, the aligned dataset was input to PAUP v4.0 and a multi-tree heuristic search was conducted with 1000 replicates of bootstrap analysis. Ten trees were retained at each step and tree-bisection-reconnection (TBR) branch swapping was engaged in the following analyses. Bayesian inference was implemented in MrBayes v3.2.1 (Fredrik et al., 2012), Markov chain Monte Carlo (MCMC) was run for 106 generations and trees were sampled every 100 generations, with the first 2500 trees discarded. The unrooted phylogenetic relationship network plots among haplotypes were generated by NETWORK v 5.0.0.0 (Forster, 2015) and artificially adjusted, with indel(s) treated as single mutation events.
The Bayesian method implemented in BEAST v1.84 (Drummond et al., 2012) was used to estimate the coalescence time across haplotypes (the most recent common ancestor, TMRCA) of C. panzhihuaensis, with a strict clock model and a Constant Size Process tree model prior as suggested for cycads (Condamine et al., 2015). C. guizhouensis was also used as a reference outgroup to estimate the divergence between these two species, as haplotypes of C. revoluta could not be separated from those of C. panzhihuaensis because of still scarce data and the recent radiation of this genus. The best-fit model in BEAST (HKY, Hasegawa-Kishino-Yano model applied to cpDNA, PHYP and N3) was also defined by IQ-TREE Modelfinder (Kalyaanamoorthy et al., 2017) according to the Bayesian Information Criterion (BIC). Because there is no substitution rate of closer taxa that can be applied to Cycas, and no exact divergence time could be applied to the split of C. panzhihuaensis and its outgroup, we adopted the average nucleotide synonymous substitution rate estimated as 6.7 × 10−10 per site per year mutation for nDNA to estimate the haplotype divergence, which we derived from the average rates of Cycadales (De La Torre et al., 2017). The three chloroplastic sequences used in this study (psbA-trnHpsbM-trnD, and trnS-trnG) were also available for Cycas angulata and Cycas bifida from GeneBank. We estimated the nucleotide substitution rates of these three cp markers as 2.14 × 10−10 using the same formula as in De La Torre et al. (2017) based on the estimated divergence time of these two species (12Ma) in Salas-Leiva et al. (2013). Three independent MCMC runs were performed and combined with each run for 108 generations and sampled every 104 generations in BEAST. We used Tracer, v1.7 (Rambaut et al., 2018) to check if the parameters converged and the effective sample size (ESS) exceeded 200, which indicates that there is acceptable mixing and sufficient sampling. The BEAST tree was generated after the first 25% of trees were discarded as burn-in.
To infer the past population fluctuation events of C. panzhihuaensis, we used DnaSP v5.0 to calculate Tajima's D and Fu's Fs neutrality tests and to generate mismatch distributions plots, and calculated the sum of squared deviation (SSD) and Harpending's raggedness (r) of mismatch distributions by Arlequin v3.11 (Schneider et al., 2000). Meanwhile, the Bayesian Skyline Plot (BSP) analyses based on all individuals from different datasets (cpDNA, PHYP and N3) were performed in BEAST v1.84 to generate the historical demographical plot of C. panzhihuaensis. Posterior distributions of priors were obtained by MCMC analyses with a total of 108 generations and sampled every 104 generations under a strict clock prior and HKY nucleotide substitution model. The effective sample sizes (ESS>200) and convergence to the stationary distribution were also examined in Tracer, v1.6.
2.3.2. Microsatellite data analysis
The microsatellite data was firstly used to evaluate the genetic diversity and differentiation by GenAlEx v6.3. The AMOVA analysis and calculations of gene flow were the same as the nucleotide sequence. Additionally, the Bayesian clustering method was also conducted in STRUCTURE v2.2 (Pritchard et al., 2000) to group the genetic component of all seven populations by microsatellite data. Twenty separate runs with group number K from one to ten were performed. Each run was estimated as 105 MCMC steps and a 105 iterations burn-in. The best fit number of genetic clusters was deduced by ΔK using STRUCTURE HARVESTER (Earl and vonHoldt, 2012). Principal coordinate analysis (PCoA) based on all individuals was also generated by MVSP v3.12 (Kovach, 1998) and compared with the STRUCTURE grouping analyses. The BOTTLENECK v1.2.02 (Piry et al., 1999) software was used to reveal possible severe and recent bottleneck events at the population level based on the heterozygosity excess test. The Sign tests and Wilcoxon sign rank tests were performed under the stepwise mutation model (SMM) and two-phased model (TPM). In addition, we used the Garza-Williamson index (GWI; M-ratio) implemented in Arlequin v3.1 to detect any early shrink in population size of C. panzhihuaensis. GWI is the ratio of allelic number to range in allele size and the loss of alleles of different lengths will lead to the asynchronous changes of the two values. As a reference, GWI below 0.68 index suggests a relative early reduction in population size when more than seven loci are analyzed (Schneider et al., 2000). In addition, to estimate effective population sizes of different C. panzhihuaensis populations (Ne), we used LDNeInterface v1.3.1 (Waples and Do, 2008), which implemented the linkage disequilibrium (LD) method at a comparison-wise 0.01 level of lowest allele frequency for a 95% confidence interval.
3. Results
3.1. DNA sequences characterization
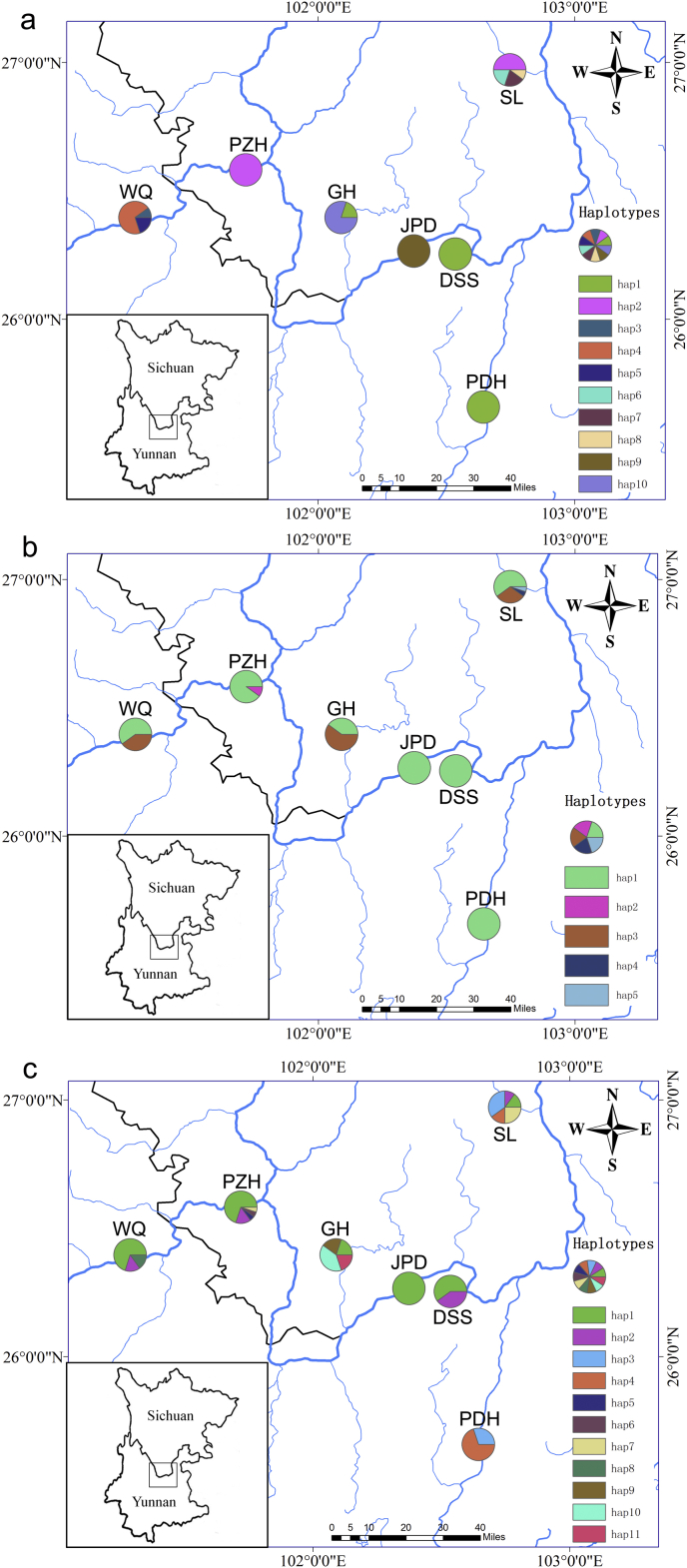
The combined cpDNA alignment of psbA-trnH, psbM-trnD and trnS-trnG was 2741 bp, and 137 bp of variable sites were detected. Among these variable sites, four substitutions and three inserts/indels from combined cpDNAs generated 10 haplotypes in seven populations of C. panzhihuaensis. The cpDNA haplotype Hap1 (cp_H1) is the most widely shared haplotype, which was detected in three populations (PDH, DSS and GH). Haplotype cp_H2 was shared by two populations (PZH and SL). The remaining haplotypes were specific to single populations (Table S1, Fig. 1a).
Fig. 1.
Geographical distributions of haplotypes identified in seven populations of C. panzhihuaensis (a) cpDNA; (b) nDNA PHYP; (c) N3. Blue lines represent Jinsha River and its tributaries. Frequencies of haplotypes in each population are indicated by the proportions of pie diagrams. DSS, Dasongshu town; PDH, Puduhe zones, Natural Reserve of Jiaozishan; PZH, Natural Reserve of Panzhihua; WQ, Wenquan town; SL, Songling town; JPD, Jiaopingdu town; GH, Guanhe town. Maps were drawn by ArcGIS v10.2 (http://desktop.arcgis.com).
PHYP sequence was aligned with a length of 1007 bp containing 42 polymorphic sites, and five haplotypes were derived from the seven populations. The most common haplotype, P_H1, occurred in all populations. P_H3 was shared by three unprotected populations (WQ, SL and GH); P_H2 was private to one population (PZH); and P_H4 and P_H5 were only found in one population (SL) (Fig. 1b). The nuclear genes AC5, HSP70 and AAT were aligned as 862 bp, 716 bp, and 571 bp respectively. When combined, the matrix was 2149 bp in length, containing eight polymorphic sites in total. For the combined three nuclear genes, we found 11 haplotypes from seven populations. Among them, N3_H1 and N3_H2 were the most widely distributed haplotypes, which were detected in six and four populations respectively. In contrast, N3_H5 and N3_H6 were only found in PZH, N3_H8 only occurred in WQ, and N3_H9, N3_H10 and N3_H11 were only detected in GH (Fig. 1c).
3.2. Genetic diversity and differentiation
We found higher total nucleotide diversity (π) and higher haplotype diversity (Hd) of C. panzhihuaensis in combined cpDNA (π = 0.00250, Hd = 0.8033) than nDNA (PHYP: π = 0.00040, Hd = 0.3302; N3: π = 0.00056, Hd = 0.7350) (Table 1). Populations WQ, SL and GH displayed haplotype variation at the cpDNA level (Table S1). For nuclear DNA, populations PZH, WQ, SL and GH showed haplotype variation from PHYP, and six populations except JPD showed haplotype variation, which was revealed by N3. Results of cpDNA, nDNA and microsatellites were consistent, with three small and unprotected populations (WQ, SL and GH) displaying higher within-population haplotype differentiation, and population SL possessing the most private haplotypes in all datasets. Population SL also had the highest genetic diversity detected by SSR (Table 2).
Table 1.
Parameters of genetic diversity and genetic differentiation for the combined cpDNA, nDNA PHYP, and three combined nDNA markers (N3) in seven populations of C. panzhihuaensis.
| Marker | Hd | π × 103 | HS | HT | GST | NST | U test |
|---|---|---|---|---|---|---|---|
| cpDNA | 0.8033 | 2.59 | 0.235 (0.1168) | 0.881 (0.0926) | 0.733 (0.1314) | 0.782 (0.1298) | non-significant |
| PHYP | 0.3302 | 0.4 | 0.257 (0.1025) | 0.366 (0.1133) | 0.296 (0.0480) | 0.296 (0.0652) | non-significant |
| N3 | 0.7350 | 0.56 | 0.506 (0.1016) | 0.769 (0.1078) | 0.342 (0.0919) | 0.420 (0.1202) | non-significant |
Hd: Haplotype (gene) diversity; π: Nucleotide diversity; HS: Sequence diversity within population; HT: Total sequence diversity; GST, NST: The coefficient of genetic differentiation for non-ordered alleles.
Table 2.
Parameters of genetic diversity and genetic differentiation for 11 microsatellite markers in seven populations of C. panzhihuaensis.
| Pop | N | %P |
Na |
Ne |
I |
Ho |
He |
F |
Ne | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | Mean | SE | Mean | SE | Mean | SE | Mean | SE | ||||
| DSS | 20 | 63.64% | 1.909 | 0.251 | 1.605 | 0.179 | 0.461 | 0.116 | 0.286 | 0.095 | 0.298 | 0.075 | 0.061 | 0.160 | 0.9 |
| PDH | 20 | 72.73% | 2.545 | 0.455 | 1.589 | 0.204 | 0.515 | 0.137 | 0.118 | 0.028 | 0.282 | 0.073 | 0.456 | 0.100 | 3.3 |
| PZH | 20 | 72.73% | 3.273 | 0.662 | 1.861 | 0.409 | 0.606 | 0.175 | 0.182 | 0.060 | 0.306 | 0.082 | 0.440 | 0.126 | 17.6 |
| WQ | 20 | 81.82% | 2.818 | 0.536 | 1.953 | 0.379 | 0.630 | 0.162 | 0.236 | 0.064 | 0.353 | 0.081 | 0.347 | 0.117 | 15.4 |
| SL | 11 | 90.91% | 3.273 | 0.524 | 2.373 | 0.374 | 0.825 | 0.179 | 0.107 | 0.042 | 0.445 | 0.090 | 0.643 | 0.126 | 1.8 |
| JPD | 6 | 72.73% | 2.091 | 0.315 | 1.835 | 0.265 | 0.552 | 0.139 | 0.227 | 0.079 | 0.347 | 0.081 | 0.246 | 0.180 | – |
| GH | 5 | 54.55% | 1.909 | 0.315 | 1.541 | 0.178 | 0.428 | 0.134 | 0.164 | 0.065 | 0.262 | 0.079 | 0.302 | 0.165 | – |
| Total | 102 | 72.73% | 2.545 | 0.178 | 1.822 | 0.113 | 0.574 | 0.056 | 0.189 | 0.025 | 0.328 | 0.030 | 0.374 | 0.054 | 39.0 |
N: sample size, Na: number of alleles, Ne: number of effective alleles, I: Shannon's index, Ho: observed heterozygosity, He: expected heterozygosity, F: fixation index, %P: percentage of polymorphic loci. Ne of population JPD and GH could not be detected because of the insufficient sample size.
The total genetic diversity of C. panzhihuaensis inferred by chloroplast and nuclear DNA data was higher than average intrapopulation diversity (cpDNA: HT = 0.881, HS = 0.235; PHYP: HT = 0.366, HS = 0.257; N3: HT = 0.769, HS = 0.506) (Table 1), which was consistent with AMOVA analysis (Table 3). For cpDNA, nearly 87% of variations existed among populations, with only 13% occurring within populations. For nuclear DNA, in contrast, the variations among-population and within-population were 32.73% vs 67.27% (PHYP) and 40.83% vs 67.27% (N3), respectively, showing higher levels of intra-population variation than inter-population variation of nDNA. The estimated gene flow of each dataset was less than 1, and FST for each dataset was more than 0.25, suggesting high levels of differentiation among populations (Table 3).
Table 3.
Results of analysis of molecular variance (AMOVA) of combined cpDNA, nuclear gene PHYP, combined nDNA sequences (N3) sequences and microsatellite markers of C. panzhihuaensis.
| Markers | Source of variation | d.f. | Sum of squares | Variance components | Percentage of variation (%) | FST | Nm |
|---|---|---|---|---|---|---|---|
| cpDNA | Among populations | 6 | 157.166 | 2.980 | 86.60 | 0.866 | 0.039 |
| Within populations | 54 | 24.900 | 0.461 | 13.40 | |||
| PHYP | Among populations | 6 | 211.481 | 1.824 | 32.73 | 0.327 | 0.514 |
| Within populations | 115 | 431.150 | 3.749 | 67.27 | |||
| N3 | Among populations | 6 | 29.848 | 0.266 | 40.83 | 0.408 | 0.362 |
| Within populations | 115 | 44.300 | 0.385 | 59.17 | |||
| microsatellite | Among populations | 6 | 165.258 | 0.912 | 33.06 | 0.331 | 0.506 |
| Within populations | 197 | 363.830 | 1.847 | 66.94 |
d.f.: degrees of freedom; FST: fixation index; Nm: gene flow = (1-FST)/4*FST.
3.3. Genetic structure of populations
No significant difference was detected between each pair of GST and NST of C. panzhihuaensis (Table 1), implying that this species has no phylogeographical pattern. Furthermore, Mantel tests based on all DNA datasets detected no significant correlation between genetic distance and geographical distance (P > 0.05) (Fig. S1), which indicates a non-significant effect of the isolation by distance (IBD) model.
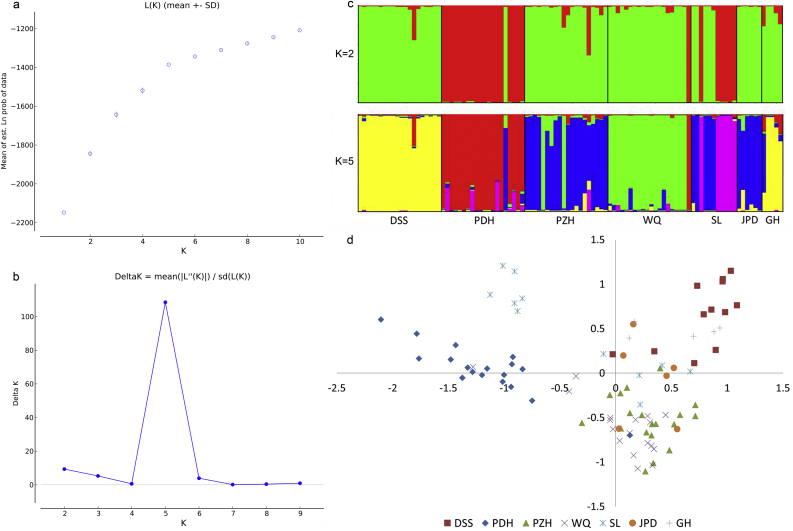
Based on the ΔK method in STRUCTURE analysis, the optimal and sub-optimal K values were five and two respectively (Fig. 2a, b). For the other K value options, inconsistent clustering results were encountered. Under the K = 2 scenario (Fig. 2c), population PDH, as well as half of the SL population component, were divided from other populations. For the result K = 5, all 102 individuals from seven populations were generally grouped into five clusters, corresponding to DSS, PDH, PZH, WQ and part of SL. Individuals from JPD and part of SL were closer to PZH, and GH shared similar components with DSS. PCoA analysis, however, showed that all individuals are roughly divided into four subgroups: DSS with GH, PDH, half of SL with PZH, plus JPD, plus WQ, and the other part of SL (Fig. 2d). The WQ population was judged as unique by STRUCTURE analysis.
Fig. 2.
STRUCTURE analyses and Principal Coordinate Analyses of 102 individuals from seven populations of C. panzhihuaensis based on phenotypes of 11 microsatellite markers. Determining the optimal K value using mean likelihood L(K) (a) and delta K values (b) generated by STRUCTURE HARVESTER, with 20 replications, respectively; (c) results of the STRUCTURE analyses (K = 2 and k = 5); (d) Principal coordinate analysis.
3.4. Haplotype phylogeny and network analysis
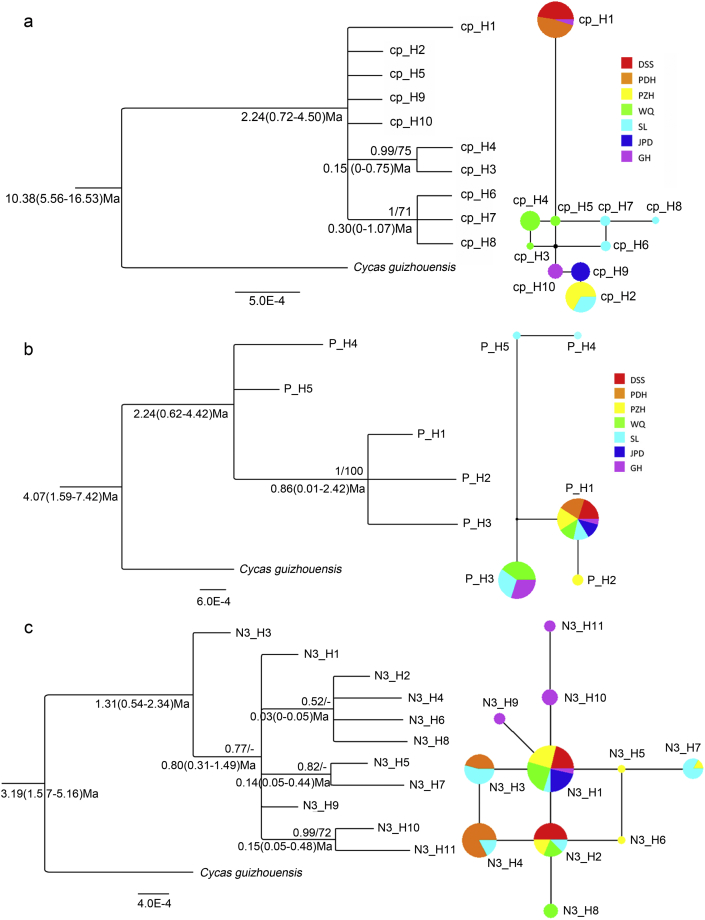
The network inferred from cpDNA showed a relatively closer relationship between private haplotypes within each population (cp_H3-5 from WQ and cp_H6-8 from SL), and all private haplotypes shared distant relationships with more widely shared Hap_1 (Fig. 3a). For PHYP, a missing haplotype occurred between the widely shared P_H1, P_H3 and private P_H4-5, which belong to SL (Fig. 3b). For the combined N3, the most widely shared haplotype N3_H1 and N3_H2 were located at the center of the network, connecting the other private haplotypes in a ring-and-star manner by one step (Fig. 3c).
Fig. 3.
Phylogenetic relationship and median-joining network of haplotypes. (Left) Phylogenetic relationship of haplotypes based on Bayesian Inference (BI) and Maximum Parsimony (MP) with Cycas guizhouensis used as outgroup. Numbers above clades are the posterior probability (PP) and bootstrap values (BP) respectively with PP < 0.50 and BP < 50 not displayed. Numbers below clades indicate the divergence time as well as their 95% highest posterior density (HPD). (Right) Median-joining network of haplotypes inferred from different datasets: (a) cpDNA; (b) PHYP; (c) N3. Color and size of pies correspond to haplotypes and proportion of individuals respectively.
Consistent topologies were generated by Maximum Parsimony and Bayesian inference based on the haplotypes. The results showed most haplotypes from each dataset formed separate comb-like cladograms with poor support values due to insufficient informative sites (Fig. 3). Taking this into consideration, the estimated divergence times were somewhat unconvincing, even though Bayesian analysis using either cpDNA, PHYP or N3 data indicated that the divergence times between C. panzhihuaensis and the outgroup C. guizhouensis were 10.38 Ma, 4.07 Ma and 3.19 Ma respectively. Inferences of the divergence times in the crown node of haplotypes were 2.24 Ma, 2.24 Ma and 1.31 Ma from corresponding datasets, and the estimated times among different haplotypes on the internal divergence node were 0.03–0.86 Ma (Fig. 3). This indicates the divergence among haplotypes occurred in the Early Pleistocene.
3.5. Neutrality test, mismatch analysis and the Bayesian Skyline Plot
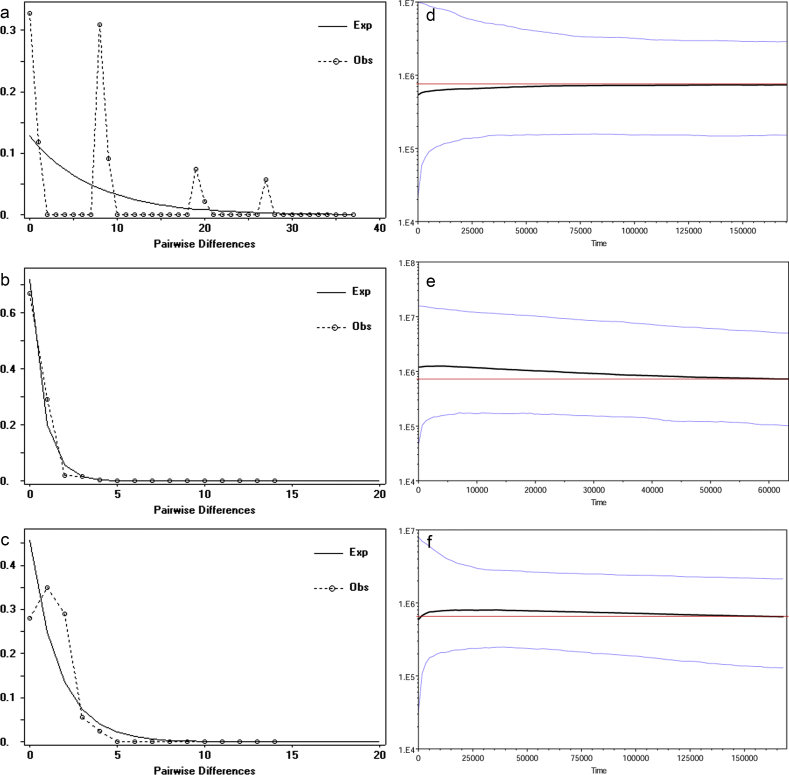
The Tajima's D and Fu's Fs test based on cpDNA displayed positive values, suggesting a historical contraction of C. panzhihuaensis population, although only the Fu's Fs value was significant. The mismatch distribution under stable populations model displayed multimodal curves for cpDNA, indicating the demographic equilibrium of populations (Fig. 4a–c). For nDNA, non-significant negative Tajima's D and Fu's Fs (Table 4), and unimodal mismatch distributions, were detected from both PHYP and N3, suggesting a history of population expansion. Non-significant SSD and raggedness index showed that the unimodal model could not be rejected, except the SSD value of PHYP.
Fig. 4.
Mismatch distribution plots and Bayesian Skyline Plots of C. panzhihuaensis based on different datasets. (a–c) Mismatch distribution plots based on different datasets: (a) cpDNA; (b) PHYP; (c) N3. Multimodal graphs indicate demographic equilibrium and unimodal graphs often indicate population expansions. (d–f) The plots reflect the effective population size fluctuation over time. (d) cpDNA; (e) PHYP; (f) N3. Black lines: median estimation; area between blue lines: 95% confidence interval; Red lines: horizontal lines for reference.
Table 4.
Parameters of neutrality tests and mismatch analysis based on cpDNA and nuclear genes of C. panzhihuaensis.
| Marker | Tajima's D | Fu and Li's D | Fu and Li's F | Fu's Fs | SSD | R |
|---|---|---|---|---|---|---|
| cpDNA | 0.493,325 | 1.82,738 | 1.59,999 | 13.648 | 0.06186 | 0.16,882 |
| PHYP | −1.19,639 | −0.06795 | −0.52,146 | −2.021 | 0.20,020# | 0.34,830 |
| N3 | −0.19,000 | 1.17,849 | 0.84,329 | −2.791 | 0.00920 | 0.11,837 |
SSD: the sum-of-squared deviations; R: raggedness index; #: P < 0.05 significant difference.
The Bayesian Skyline Plot of different datasets showed a coherent relatively stable (cpDNA) or a slightly increasing trend (nuclear genes) of population size in history, which is consistent with the results of the neutral test and mismatch analysis. In addition, a slight, recent decline about 5000 years ago was detected (Fig. 4). The Wilcoxon and Sign tests revealed significant differences for populations DSS, JP and SL, and the shifted mode was confirmed by the mode-shift test for DSS and JPD (Table 5), suggesting these populations had experienced severe bottleneck events, which may have contributed to the slight decline in the BSP result. Furthermore, the Garza-Williamson indices of every population were below 0.68 (Table 5), indicating a relatively early reduction in population size.
Table 5.
Bottleneck analysis and Garza-Williamson index based on microsatellite loci from seven populations of C. panzhihuaensis.
| Population | TPM |
SMM |
Mode shift | Garza-Williamson index | ||
|---|---|---|---|---|---|---|
| Sign test | Wilcoxon test | Sign test | Wilcoxon test | |||
| DSS | 0.01328* | 0.00073** | 0.01422* | 0.02100* | shifted mode | 0.45,034 |
| PDH | 0.08215* | 0.17,480 | 0.43,846 | 0.51,953 | L-shaped distribution | 0.41,138 |
| PZH | 0.34,274 | 0.57,715 | 0.33,621 | 0.70,020 | L-shaped distribution | 0.43,259 |
| WQ | 0.22,812 | 0.06738* | 0.47,564 | 0.36,523 | L-shaped distribution | 0.39,331 |
| SL | 0.02812* | 0.00098** | 0.02932* | 0.00684** | L-shaped distribution | 0.45,275 |
| JPD | 0.02284* | 0.00342** | 0.03018* | 0.00684** | shifted mode | 0.39,471 |
| GH | – | – | – | – | – | 0.34,077 |
| Mean | – | – | – | – | – | 0.41,083 |
*: P < 0.05, significant difference; **: P < 0.01, highly significant difference. Indices of population GH could not be detected by Bottleneck analysis because of its insufficient sample size.
4. Discussion
4.1. C. panzhihuaensis populations have high levels of genetic diversity but no significant genetic structure
The total genetic diversity of C. panzhihuaensis populations estimated by cpDNA was 0.881, which is higher than the mean value of total genetic diversity (HT = 0.67) for 170 plant species (Petit et al., 2005). This indicates that C. panzhihuaensis has a high level of genetic diversity. We found that, based on cpDNA analyses, the genetic diversity and fixation index of C. panzhihuaensis (π = 0.00259; Hd = 0.8033; FST = 0.866) was similar to those of previously reported Asian inland Cycas species (π = 0.0009–0.0038; Hd = 0.492–0.940; FST = 0.6982–9980, Zheng et al., 2017). Our finding is in accordance with previous studies on C. panzhihuaensis (π = 0.0038; Hd = 0.571; FST = 0.7903, Zhang, 2012, unpublished). This may because Asian Cycas are long-lived dioecious gymnosperms.
Earlier studies reported that C. panzhihuaensis populations have higher genetic diversity than other inland Cycas species, suggesting that C. panzhihuaensis populations possess more ancestral polymorphisms (Yang et al., 2015). However, our results contradict these findings. The total percentage of polymorphic loci and expected heterozygosity that we calculated for C. panzhihuaensis using SSRs (%P = 72.73%, He = 0.328) were similar to other Cycas species (%P = 66.67–94.12, aver. 87.19, He = 0.2470–0.5430, aver. 0.4416, Zheng et al., 2017). Apparent discrepancies between our values for polymorphic loci and expected heterozygosity for C. panzhihuaensis and those of earlier studies may be due to differences in study design. In our study, all known populations of C. panzhihuaensis were screened for SSRs, cpDNA, and nDNA markers. Earlier studies that reported higher genetic diversity for C. panzhihuaensis used only one population and relied on AFLP and ISSR markers (%P = 94.12%, He = 0.335, Yang et al., 2015; and %P = 90.38%, Wang et al., 2010).
Previous studies have shown that genetic variation is strongly influenced by large geographical distances between sampled populations (Reisch and Bernhardt-Römermann, 2014). The inland Asian Cycas species, which have similar habitat, morphological traits, life history, and reproductive characteristics, share similar genetic traits, e.g., lower levels of genetic variation and significant genetic differentiation among populations (Zheng et al., 2017). In contrast, although C. panzhihuaensis seems to have diverged much earlier than other Cycas species and occupied larger Nc and narrower distribution area, we found no significant differentiation between this species and other inland Asian Cycas species. Furthermore, C. panzhihuaensis genetic variation differs from that of two coastal species (C. revoluta and C. taitungensis) that have been shown to be sister to C. panzhihuaensis (Liu et al., 2018); these two coastal species display much higher genetic diversity (π = 0.0127–0.0581; Hd = 0.959–0.998; FST = 0.056–0.864. Chiang et al., 2009, Huang et al., 2001).
Southwest China is a known hotspot of biodiversity where the terrain complexity has produced unique and diverse climate types (Myers et al., 2000). The Jinsha River dry hot valley provided refugia for various surviving species in the Quaternary glaciations (Gong et al., 2011, Zhang et al., 2011a, Zhang et al., 2011b), including the ancestors of C. panzhihuaensis. Unlike previous studies of inland Cycas species (Liu et al., 2015, Yang et al., 2017, Zheng et al., 2016) or the sympatric species distributed in the dry and hot valley of Jinsha River, e.g., M. delavayi (Jia et al., 2014) and Nouelia insignis (Gong et al., 2011), our study of C. panzhihuaensis detected no significant population genetic structure, which is consistent with previous studies on this species (Wang et al., 2010, Yang et al., 2015, Zhang, 2012). Furthermore, the phylogenetic relationships and median-joining networks of C. panzhihuaensis haplotypes showed neither a center of significant genetic diversity nor migration routes between populations. The distribution pattern of C. panzhihuaensis, which differs from other species, has yet to be explored in depth.
Our estimates of gene flow inferred from DNA datasets were lower than the critical value 1 (Table 3), indicating that intraspecific gene flow between C. panzhihuaensis populations is low. As chloroplast genomes of Cycas are maternally inherited, whereas nuclear genomes are biparentally inherited, thus reflecting seed flow and pollen flow, our finding that the chloroplast FST was higher than the nuclear DNA and SSR FST (Table 3) indicates that in C. panzhihuaensis pollen flow is much greater than seed flow. This also explains AMOVA results why cpDNA analyses indicated that there was more genetic variation between populations and less genetic variation within populations, whereas nuclear DNA and microsatellites indicated the opposite.
On the other hand, the dispersal of C. panzhihuaensis seeds, which are sinkable and large (~2.5 cm), mainly relies on gravity. Up to 98.5% of these gravity-dispersed seeds tumble less than 10 m away from mother plants (Hall and Walter, 2011), which increases the chances of inbreeding within populations. In rare cases, C. panzhihuaensis seeds are dispersed long distances; this long-distance dispersal has been attributed to the squirrel-like northern tree shrew, Tupaia belangeri (Yu et al., 2007). Our field survey of C. panzhihuaensis found few seedlings in most populations; hence, we reason that pollination and seed dispersal barriers may be one of the reasons that C. panzhihuaensis has relatively low natural fruiting and germination rates in the wild.
4.2. Demographic history of C. panzhihuaensis
Historical demographic dynamics inferred from genetic data provides an indication of how do habitat loss, landscape changes, and climate changes affect population viability (Selwood et al., 2015). According to our results, most of the private haplotypes were found in WQ, SL, GH, which are small populations located on the boundary of the distribution range. This may be due to limited seed flow among populations of C. panzhihuaensis, which has resulted in multiple evolutionary lineages in different isolated locations. As we did not detect significant demographic contractions in entire species using Bayesian skyline plots or mismatch distribution estimates (Fig. 4), AMOVA analyses showed that gene flow between C. panzhihuaensis populations was low, suggesting that the ability of C. panzhihuaensis to migrate is weak. Taken together, these findings suggest at some point in their history these three small populations underwent contraction and experienced rapid adaptive evolution due to the geographical and climatic complexity of their distribution range. According to this hypothesis, small populations should be regarded as essential sources of genetic diversity. Peripheral populations, which tend to be less affected by gene flow and more likely to preserve unique genetic lineages, may be more likely to adapt to the colonized habitat and expand the distribution boundaries of species due to their exposure to different selection pressures (Araújo and Williams, 2001, Kark et al., 1999). Furthermore, these genetic variations will exist as potential candidates for new adaptive traits.
In terms of population divergence, despite the unclear dating results from BEAST, our results indicate that C. panzhihuaensis has experienced haplotype differentiation from ca. 0.03 Ma to 2.24 Ma in the Early Pleistocene, which is consistent with the conclusions of Yang et al. (2015), who estimated that C. panzhihuaensis populations grew from ca. 0.08 Ma - 1.6 Ma. Our results are also consistent with the haplotype divergence time estimation of other inland Asian Cycas species, e.g., Cycas chenii, 0.030 Ma - 1.175 Ma (Yang et al., 2017); C. guizhouensis, 0.048 Ma - 1.334 Ma (Feng et al., 2016); Cycas diannanensis, 0.075 Ma - 2.525 Ma (Liu et al., 2015). Affected by climate oscillation, small populations of these Cycas species were formed and isolated during the Quaternary ice age (Zheng et al., 2017). Furthermore, instead of retaining distinct ancestral haplotypes, the populations of C. panzhihuaensis from isolated mountains or valleys diverged significantly from each other and maintained their private haplotypes. This genetic heterogeneity from isolated regions may also be caused by the frequently altered alpine landforms of inland Asia (Zhao and Gong, 2015).
Given that no significant correlations were found between genetic and geographical distance of C. panzhihuaensis populations, we deduced the ancestor lineages of C. panzhihuaensis might have formed an interconnected population in the Jinsha River basin before or during the Early Pleistocene, which was subsequently isolated to form several fragmented populations, each of which has possibly experienced a loss of ancestral polymorphisms, resulting in the random and unordered genetic structure pattern of extant populations. The revealed Garza-Williamson index below 0.68 can be one of the consequences of population declines. As a result, populations SL, DSS and JPD appear to have undergone serious bottleneck events, especially the latter two.
Although the results of the neutral test and mismatch analyses were not significant, the demographic dynamics these analyses revealed when we used cpDNA suggest that C. panzhihuaensis populations have undergone recent bottleneck events, whereas our analyses based on nDNA showed evidence of possible population expansions (Table 4; Fig. 4).
This population growth scenario of C. panzhihuaensis is supported by previous research (Yang et al., 2015). Similarly, the results of the Bayesian skyline plot (BSP), which are consistent with the results of the neutral test and the mismatch distribution based on nuclear genes, also showed a slight increase in the C. panzhihuaensis population at some point in its history; in contrast, when chloroplast DNA was used in this same method, our results showed a stable population size over time (Fig. 4). The contradiction of results between cpDNA and nDNA results may be due to differences in their genomic backgrounds, each of which have been exposed to different selective pressures by repeated climatic oscillations of the Pleistocene (Ren et al., 2017, Zhang et al., 2011a).
Furthermore, the Garza-Williamson indices indicate a relative early reduction in population size. The results of the mode-shift test and the significant difference between TPM and SMM indicate that populations SL, DSS and JPD have experienced severe bottleneck events (Table 5). Results of the Bayesian Skyline Plot imply that population decline events occurred in the past five thousand years (Fig. 4). Therefore, Holocene climate fluctuations in the dry-hot valley of the Jinsha River seem to have affected the population size of C. panzhihuaensis, especially the northeastern populations (SL, DSS and JPD).
4.3. Implications for conservation
To successfully conserve endangered species, it is often insufficient to raise the abundance of offspring. Maximum genetic diversity must be maintained as it partially drives evolutionary potential (Etterson, 2004, Jump and Penuelas, 2005, Li et al., 2012). Our estimated value of the effective population size (Ne) for population PZH is only 17.6 (Table 2), despite its considerable Nc. The value is lower than but close to the previous study (Mode: 29.369, Median: 30.750, Yang et al., 2015). We estimate that the total Ne for C. panzhihuaensis is 39, which indicates that the current conservation efforts are far from effective. Generally, if a species has a total Ne lower than 50, inbreeding depression cannot be prevented over five generations in wild plants (Frankham et al., 2014). To completely avoid inbreeding (Ne > 100), conservation efforts should protect as much genetic diversity as possible.
Genetic diversity is not often correlated with population size in cycads, and the longevity cycads as well as their overlapping generations allows rare alleles to remain within the populations for longer periods of time (James et al., 2018). This is consistent with our finding that the genetic diversity of large populations was not as high as expected in C. panzhihuaensis, whereas small and unprotected populations possessed unexpectedly higher genetic diversity. The high Nc of population PZH suggests that the PZH population is the center of the C. panzhihuaensis refugium. However, although the current and past distribution areas of populations WQ and GH are far smaller than that of PZH, the haplotype diversity is higher in these populations than in PZH (Table S1).
Small populations of C. panzhihuaensis have been suffering from severe exploitation and habitat loss (He and Li, 1999). For example, before large-scale collection, population SL had an area of 20 hm2 that contained 2000 adult individuals in 1979. Currently, there are fewer than 20 individuals remaining (Wang, 2016). PDH and PZH are the only two populations containing more than 100 adult individuals, but both populations are still threatened by deforestation from mining and grazing. Excluding the PZH population, the habitat of the existing six isolated populations is fragmented and most of their restricted individuals are young, which leaves populations vulnerable to extirpation by demographic stochasticity, as well as environmental or reproductive disturbance (Segarra-Moragues et al., 2005). Only populations PDH and PZH are effectively protected at present; therefore, the unprotected haplotypes of cpDNA, nDNA PHYP and N3 accounted for 90%, 80% and 63.6% of the total haplotypes, respectively (Table S1, Fig. 3), and the threatened effective population size accounted for 53.58% of the total effective population size. Given the current threat of extinction to these small populations, much more attention should be paid their protection.
Although genetic diversity of populations is directly related to population size, some studies have suggested that the reduction of genetic diversity in small populations may be slower than expected. As it was suggested, large population declines do not immediately result in great losses of genetic diversity and population instability (Schou et al., 2017). For example, this may be because soil seed banks serve as a genetic resource (Münzbergová et al., 2018). A meta-analysis indicated that the adaptability of small groups is not necessarily lower, and small groups are not necessarily subject to more significant selection pressure (Wood et al., 2016). On the other hand, the proportion of genetic diversity within species may be unevenly concentrated in small populations through models (Rauch and Bar-Yam, 2004). This undoubtedly gives us hope for the future of small populations and raises the value of their conservation.
The criteria for selecting priority protected populations must include the uniqueness and diversity of the population, particularly in its allelic composition (Petit et al., 2008). For C. panzhihuaensis, given the distinct private haplotypes of SL, PZH, WQ and GH, and the estimated genetic clustering obtained by STRUCTURE analyses (Fig. 2c), five evolutionary significant units (ESUs) combining JPD and PZH, DSS and GH separately were defined and should be protected separately as independent management units (MUs) (Griffith et al., 2015). Results of PCoA almost matched this relation as the clustering discrepancy of the WQ population may be due to the limitations of two-dimensional principal coordinate analysis. In situ conservation should be implemented as it can effectively reduce the high risk of genetic erosion to small populations. Because the dry-hot valley of the Jinsha River is a fragile ecosystem that has experienced serious artificial disturbance, and the entire range of dispersed populations of C. panzhihuaensis are difficult protect, ex-situ conservation is likely to be the most effective approach to preserving genetic diversity. It is imperative and urgent to collect seeds from these populations for nearby reserves or botanical gardens while ensuring the sustainability of in situ conservation (Griffith et al., 2017). Interbreeding between JPD and PZH, as well as DSS and GH, may considerably improve the genetic variation of these MUs(Kaulfuss and Reisch, 2017). The smallest populations of C. panzhihuaensis (SL, WQ, GH, JPD, and DSS) should be protected. Specifically, suitable habitat should be evaluated; then these populations should be reconstructed and recovered at appropriate localities. This will restore the viability and evolutionary potential of these populations and help to avoid the adverse effects of small populations.
5. Conclusions
Our study on the population genetics of C. panzhihuaensis revealed a divergence period and pattern of genetic diversity similar to other Asian inland cycad species, but a non-significant phylogeographic structure, which may be attributed to rapid in-situ evolution driven by limited seed flow. These findings suggest that population of C. panzhihuaensis may have been widely distributed in the dry hot valley of the Jinsha River before the Pleistocene; however, subsequent population expansions and contractions during the glacial–interglacial cycles of the Early Pleistocene reduced ancestral genetic polymorphisms and formed randomly isolated populations at present. Furthermore, it cannot be neglected that the sum of Ne in small populations with 1% of total individuals is equal to a large population with 99% of total individuals, suggesting the necessity and urgency to conserve those populations with small size, which deserve due attention in future studies. For conservation guidelines, we defined five ESUs for C. panzhihuaensis in this study and three of them are small and unprotected populations which require special and imperative protection efforts.
Author contributions
Conceived and designed the experiments: Xun Gong, Yunheng Ji. Performed the experiments: Siyue Xiao. Analyzed the data: Siyue Xiao. Collected samples: Yunheng Ji. Wrote the paper: Siyue Xiao, Jian Liu.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported by the National Key R & D Program of China (2017YF0505200). The authors thank Fangming Zhang for her assistance with plant sampling, and Xiuyan Feng, Rui Yang and Yujuan Zhao for their help and discussion of data analyses.
Editor: Yingxiong Qiu
Footnotes
Peer review under responsibility of Editorial Office of Plant Diversity.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2019.10.001.
Contributor Information
Siyue Xiao, Email: xiaosiyue@mail.kib.ac.cn.
Yunheng Ji, Email: jiyh@mail.kib.ac.cn.
Jian Liu, Email: liujian@mail.kib.ac.cn.
Xun Gong, Email: gongxun@mail.kib.ac.cn.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- Araújo M.B., Williams P.H. The bias of complementarity hotspots toward marginal populations. Conserv. Biol. 2001;15:1710–1720. [Google Scholar]
- Barker F.K., Lutzoni F.M. The utility of the incongruence length difference test. Syst. Biol. 2002;51:625–637. doi: 10.1080/10635150290102302. [DOI] [PubMed] [Google Scholar]
- Chiang Y.C., Hung K.H., Moore S.J., Ge X.J., Huang S., Hsu T.W., Schaal B.A., Chiang T. Paraphyly of organelle DNAs in Cycas Sect. Asiorientales due to ancient ancestral polymorphisms. BMC Evol. Biol. 2009;9:161. doi: 10.1186/1471-2148-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condamine F.L., Nagalingum N.S., Marshall C.R., Morlon H. Origin and diversification of living cycads: a cautionary tale on the impact of the branching process prior in Bayesian molecular dating. BMC Evol. Biol. 2015;15:65. doi: 10.1186/s12862-015-0347-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Torre A.R., Li Z., Van de Peer Y., Ingvarsson P.K. Contrasting rates of molecular evolution and patterns of selection among gymnosperms and flowering plants. Mol. Biol. Evol. 2017;34:1363–1377. doi: 10.1093/molbev/msx069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. DNA protocols for plants. In: Hewitt G.M., Johnston A.W.B., Young J.P.W., editors. Molecular Techniques in Taxonomy. Springer; Berlin, Heidelberg: 1991. pp. 283–293. [Google Scholar]
- Drummond A.J., Suchard M.A., Xie D., Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl D.A., vonHoldt B.M. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012;4:359–361. [Google Scholar]
- Etterson J.R. Evolutionary potential of Chamaecrista fasciculata in relation to climate change. II. Genetic architecture of three populations reciprocally planted along an environmental gradient in the great plains. Evolution. 2004;58:1459–1471. doi: 10.1111/j.0014-3820.2004.tb01727.x. [DOI] [PubMed] [Google Scholar]
- Feng X.Y., Zheng Y., Gong X. Middle-Upper Pleistocene climate changes shaped the divergence and demography of Cycas guizhouensis (Cycadaceae): evidence from DNA sequences and microsatellite markers. Sci. Rep. 2016;6:27368. doi: 10.1038/srep27368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X., Liu J., Chiang Y.C., Xun G. Investigating the genetic diversity, population differentiation and population dynamics of Cycas segmentifida (Cycadaceae) endemic to southwest China by multiple molecular markers. Front. Plant Sci. 2017;8:839. doi: 10.3389/fpls.2017.00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster Michael. Network; 2015. Network 5.0.0.0 User Guide; pp. 1–55.http://fluxus-engineering.com/Network5000_user_guide.pdf Downloaded on [Google Scholar]
- Frankham R., Bradshaw C.J.A., Brook B.W. Genetics in conservation management: revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol. Conserv. 2014;170:56–63. [Google Scholar]
- Fredrik R., Maxim T., Paul V.D.M., Ayres D.L., Aaron D., Sebastian H., Bret L., Liang L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y.Q., Gong X. Pollen-mediated gene flow promotes low nuclear genetic differentiation among populations of Cycas debaoensis (Cycadaceae) Tree Genet. Genomes. 2016;12:93. [Google Scholar]
- Gong X., Luan S.S., Hung K.H., Hwang C.C., Lin C.J., Chiang Y.C., Chiang T.Y. Population structure of Nouelia insignis (Asteraceae), an endangered species in southwestern China, based on chloroplast DNA sequences: recent demographic shrinking. J. Plant Res. 2011;124:221–230. doi: 10.1007/s10265-010-0363-0. [DOI] [PubMed] [Google Scholar]
- Griffith M.P., Calonje M., Meerow A.W., Tut F., Kramer A.T., Hird A., Magellan T.M., Husby C.E. Can a botanic garden Cycad collection capture the genetic diversity in a wild population? Int. J. Plant Sci. 2015;176:1–10. [Google Scholar]
- Griffith M.P., Calonje M., Meerow A.W., Francisco-Ortega J., Knowles L., Aguilar R., Tut F., Sánchez V., Meyer A., Noblick L.R., Magellan T.M. Will the same ex situ protocols give similar results for closely related species? Biodivers. Conserv. 2017;26:2951–2966. [Google Scholar]
- Hall T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hall J.A., Walter G.H. Does pollen aerodynamics correlate with pollination vector? Pollen settling velocity as a test for wind versus insect pollination among cycads (Gymnospermae: Cycadaceae: Zamiaceae) Biol. J. Linn. Soc. 2011;104:75–92. [Google Scholar]
- Hamilton M.B. Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Mol. Ecol. 1999;8:521–523. [PubMed] [Google Scholar]
- Han J., Wang M.C., Qiu Q., Guo R.Y. Characterization of the complete chloroplast genome of Cycas panzhihuaensis. Conserv. Genet. Resour. 2016;9:21–23. [Google Scholar]
- Hao Y.Q. Florogeographical analysis of spermatophytes in Cycas panzhihuaensis assemblage. J. Sichuan Forest Sci. Technol. 2011;32:29–33. (In Chinese with English abstract) [Google Scholar]
- He Y.H., Li C.L. The ecological geographic distribution, spatial pattern and collecting history of Cycas panzhihuaensis populations. Acta Phytoecol. Sin. 1999;23:23–30. (In Chinese with English abstract) [Google Scholar]
- Hill K., Chen C., Loc P. Chapter 5: regional overview: Asia. In: Donaldson J., editor. Cycads: Status Survey and Conservation Action Plan. IUCN/SSC Cycad Specialist Group., IUCN; Gland, Switzerland and Cambridge: 2003. pp. 25–30. [Google Scholar]
- Huang S., Chiang Y.C., Schaal B.A., Chou C.H., Chiang T.Y. Organelle DNA phylogeography of Cycas taitungensis, a relict species in Taiwan. Mol. Ecol. 2001;10:2669–2681. doi: 10.1046/j.0962-1083.2001.01395.x. [DOI] [PubMed] [Google Scholar]
- IUCN . 2017. The IUCN Red List of Threatened Species.http://www.iucnredlist.org Version 2017-3. [Google Scholar]
- James H.E., Forster P.I., Lamont R.W., Shapcott A. Conservation genetics and demographic analysis of the endangered cycad species Cycas megacarpa and the impacts of past habitat fragmentation. Aust. J. Bot. 2018;66:173–189. [Google Scholar]
- Jia J., Wu H., Wang J.F., Xun G. Genetic diversity and structure of Munronia delavayi Franch. (Meliaceae), an endemic species in the dry-hot valley of Jinsha River, south-western China. Genet. Resour. Crop Evol. 2014;61:1381–1395. [Google Scholar]
- Jin Z. The floristic study on seed plants in the dry-hot valleys in Yunnan and Sichuan. Guihaia. 1999;19:1–14. (In Chinese with English abstract) [Google Scholar]
- Jump A.S., Penuelas J. Running to stand still: adaptation and the response of plants to rapid climate change. Ecol. Lett. 2005;8:1010–1020. doi: 10.1111/j.1461-0248.2005.00796.x. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kark S., Alkon P.U., Safriel U., Randi E. Conservation priorities for chukar partridge in Israel based on genetic diversity across an ecological gradient. Conserv. Biol. 1999;13:542–552. [Google Scholar]
- Kaulfuss F., Reisch C. Reintroduction of the endangered and endemic plant species Cochlearia bavarica-Implications from conservation genetics. Ecol. Evol. 2017;7:11100–11112. doi: 10.1002/ece3.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach W. Kovach Comput Serv; Wales, UK 137: 1998. MVSP-A Multivariate Statistical Package for Windows, Ver. 3.1.https://mvsp-a-multivariate-statistical-package.apponic.com/Downloaded on [Google Scholar]
- Li L., Wang Z.F., Jian S.G., Zhu P., Zhang M., Ye W.H., Ren H. Isolation and characterization of microsatellite loci in endangered Cycas changjiangensis (Cycadaceae) Conserv. Genet. 2009;10:793–795. [Google Scholar]
- Li Y.Y., Tsang E.P.K., Cui M.Y., Chen X.Y. Too early to call it success: an evaluation of the natural regeneration of the endangered Metasequoia glyptostroboides. Biol. Conserv. 2012;150:1–4. [Google Scholar]
- Liu G.F., Deng T.X., Zhou L., Yang S.Y. Karyotype analysis of Cycas panzhihuaensis L. Zhou & S.Y.Yang. Chin. J. Plant Ecol. 1990;14:23–30. (In Chinese with English abstract) [Google Scholar]
- Liu J., Zhou W., Gong X. Species delimitation, genetic diversity and population historical dynamics of Cycas diannanensis (Cycadaceae) occurring sympatrically in the Red River region of China. Front. Plant Sci. 2015;6:696. doi: 10.3389/fpls.2015.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Lin W.Y., Li X.Y. Chinese nature reserve continues experimental use of fire to benefit the threatened Cycas panzhihuaensis. Oryx. 2016;50:582. [Google Scholar]
- Liu J., Zhang S.Z., Nagalingum N.S., Chiang Y.C., Lindstrom A.J., Gong X. Phylogeny of the gymnosperm genus Cycas L. (Cycadaceae) as inferred from plastid and nuclear loci based on a large-scale sampling: evolutionary relationships and taxonomical implications. Mol. Phylogenetics Evol. 2018;127:87–97. doi: 10.1016/j.ympev.2018.05.019. [DOI] [PubMed] [Google Scholar]
- Mankga L.T., Yessoufou K. Factors driving the global decline of cycad diversity. AoB Plants. 2017;9:plx022. doi: 10.1093/aobpla/plx022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münzbergová Z., Šurinová M., Husáková I., Brabec J. Strong fluctuations in aboveground population size do not limit genetic diversity in populations of an endangered biennial species. Oecologia. 2018;187:863–872. doi: 10.1007/s00442-018-4152-0. [DOI] [PubMed] [Google Scholar]
- Myers N., Mittermeier R.A., Mittermeier C.G., da Fonseca G.A., Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Nagalingum N.S., Marshall C.R., Quental T.B., Rai H.S., Little D.P., Mathews S. Recent synchronous radiation of a living fossil. Science. 2011;334:796. doi: 10.1126/science.1209926. [DOI] [PubMed] [Google Scholar]
- Petit R., Duminil J., Fineschi S., Hampe A., Salvini D., Giovanni Giuseppe V. Invited review: comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 2005;14:689–701. doi: 10.1111/j.1365-294X.2004.02410.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press; New York, USA: 1987. [Google Scholar]
- Peakall R., Smouse P.E. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit R., Abdelhamid E.M., Pons O. Identifying populations for conservation on the basis of genetic markers. Conserv. Biol. 2008;12:844–855. [Google Scholar]
- Piry S., Luikart G., Cornuet J. Computer note. BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. Hereditas. 1999;90:502–503. [Google Scholar]
- Pons, Peti R.J. Measwring and testing genetic differentiation with ordered versus unordered alleles. Genetics. 1996;144:1237–1245. doi: 10.1093/genetics/144.3.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J.K., Stephens M., Rosenberg N.A., Donnelly P. Association mapping in structured populations. Am. J. Hum. Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Systematic Biology. 2018;syy032 doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch E.M., Bar-Yam Y. Theory predicts the uneven distribution of genetic diversity within species. Nature. 2004;431:449–452. doi: 10.1038/nature02745. [DOI] [PubMed] [Google Scholar]
- Reisch C., Bernhardt-Römermann M. The impact of study design and life history traits on genetic variation of plants determined with AFLPs. Plant Ecol. 2014;215:1493–1511. [Google Scholar]
- Ren G., Mateo R.G., Liu J., Suchan T., Alvarez N., Guisan A., Conti E., Salamin N. Genetic consequences of Quaternary climatic oscillations in the Himalayas: Primula tibetica as a case study based on restriction site-associated DNA sequencing. New Phytol. 2017;213:1500–1512. doi: 10.1111/nph.14221. [DOI] [PubMed] [Google Scholar]
- Rozas J. DNA sequence polymorphism analysis using DnaSP. Methods Mol. Biol. 2009;537:337–350. doi: 10.1007/978-1-59745-251-9_17. [DOI] [PubMed] [Google Scholar]
- Salas-Leiva D.E., Meerow A.W., Calonje M., Griffith M.P., Francisco-Ortega J., Nakamura K., Stevenson D.W., Lewis C.E., Namoff S. Phylogeny of the cycads based on multiple single-copy nuclear genes: congruence of concatenated parsimony, likelihood and species tree inference methods. Ann. Bot. 2013;112:1263–1278. doi: 10.1093/aob/mct192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangin P., Lindstrom A., Kokubugata G., Chaiprasongsuk M., Mingmuang M. Phylogenetic relationships within Cycadaceae inferred from non-coding regions of chloroplast DNA. Kasetsart J. (Nat. Sci.) 2010;44:544–557. [Google Scholar]
- Schneider S., Roessli D., Excoffier L. ARLEQUIN: a software for population genetics data analysis. Evol. Bioinform. Online. 2000;1 [PMC free article] [PubMed] [Google Scholar]
- Schou M.F., Loeschcke V., Bechsgaard J., Schlotterer C., Kristensen T.N. Unexpected high genetic diversity in small populations suggests maintenance by associative overdominance. Mol. Ecol. 2017;26:6510–6523. doi: 10.1111/mec.14262. [DOI] [PubMed] [Google Scholar]
- Segarra-Moragues J., Palop-Esteban M., Gonzalez-Candelas F., Catalan P. On the verge of extinction: genetics of the critically endangered Iberian plant species, Borderea chouardii (Dioscoreaceae) and implications for conservation management. Mol. Ecol. 2005;14:969–982. doi: 10.1111/j.1365-294X.2005.02482.x. [DOI] [PubMed] [Google Scholar]
- Selwood K.E., McGeoch M.A., Mac Nally R. The effects of climate change and land-use change on demographic rates and population viability. Biol. Rev. Camb. Philos. Soc. 2015;90:837–853. doi: 10.1111/brv.12136. [DOI] [PubMed] [Google Scholar]
- Shaw J., Lickey E., Beck J.T. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- Swindell S.R., Plasterer T.N. SEQMAN. In: Swindell S.R., editor. Sequence Data Analysis Guidebook. Springer New York; Totowa, NJ: 1997. pp. 75–89. [Google Scholar]
- Swofford D.L. Sinauer Associates, Inc; Sunderland, Massachusetts: 2002. PAUP*: Phylogenetic Analysis Using Parsimony (And Other Methods), Version 4.0b10. [Google Scholar]
- Tang Y.L., Su Z.X. The research actuality and prospect of a rare and endangered plant named Cycas panzhihuaensis. J. Shaanxi Normal Univ. (Nat. Sci. Ed.) 2004;18:87–92. (In Chinese) [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.C. Characteristics of Cycas panzhihuaensis community in Pudu valley of Yunnan. For. Inventory Plan. 2016;41:32–39. (In Chinese with English abstract) [Google Scholar]
- Wang Q., Li C.L., Yang S.Y., Huang R., Chen F. Pollination biology of Cycas panzhihuaensis L. Zhou et S. Y. Yang. Acta Bot. Sin. 1997;39:156–163. (In Chinese with English abstract) [Google Scholar]
- Wang Z.F., Ye W.H., Cao H.L., Li Z.C., Peng S.L. Identification and characterization of EST-SSRs and cpSSRs in endangered Cycas hainanensis. Conserv. Genet. 2008;9:1079–1081. [Google Scholar]
- Wang J., Wang Y.Y., Zhou H., Gong Z.D., Shi M.M., Huang R.R., Na H.Y. ISSR analysis on genetic diversity of Cycas panzhihuaensisi L. Zhou et S. Y. Yang. J. Sichuan Univ. (Nat. Sci. Ed.) 2010;47:366–370. (In Chinese with English abstract) [Google Scholar]
- Wang Y.Y., Zhou H., Su B., Huang R., Wu J., Na H.Y. Study on seed coat structure and trachary elements in xylems of Cycas panzhihuaensis L. Zhou et S. Y. Yang. J Sichuan Univ (Nat Sci Ed) 2011;48:247–252. (In Chinese with English abstract) [Google Scholar]
- Waples R.S., Do C. ldne: a program for estimating effective population size from data on linkage disequilibrium. Mol. Ecol. Resour. 2008;8:753–756. doi: 10.1111/j.1755-0998.2007.02061.x. [DOI] [PubMed] [Google Scholar]
- Wood J., Yates M., Fraser D. Are heritability and selection related to population size in nature? Meta-analysis and conservation implications. Evol. Appl. 2016;9:640–657. doi: 10.1111/eva.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Evolution in mendelian populations. Genetics. 1931;16:97. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.S., Wang Y.N., Liu S.M., Chaw S.M. Chloroplast genome (cpDNA) of Cycas taitungensis and 56 cp protein-coding genes of Gnetum parvifolium: insights into cpDNA evolution and phylogeny of extant seed plants. Mol. Biol. Evol. 2007;24:1366–1379. doi: 10.1093/molbev/msm059. [DOI] [PubMed] [Google Scholar]
- Yang Y., Li Y., Li L.F., Ge X.J., Gong X. Isolation and characterization of microsatellite markers for Cycas debaoensis Y. C. Zhong et C. J. Chen (Cycadaceae) Mol. Ecol. Resour. 2008;8:913–915. doi: 10.1111/j.1755-0998.2008.02114.x. [DOI] [PubMed] [Google Scholar]
- Yang Z.Y., Yi T.S., Zeng L.Q., Xun G. The population genetic structure and diversification of Aristolochia delavayi (Aristolochiaceae), an endangered species of the dry hot valleys of the Jinsha River, Southwestern China. Botany. 2014;92:579–587. [Google Scholar]
- Yang Y.Q., Huang B.H., Yu Z.X., Liao P.C. Inferences of demographic history and fine-scale landscape genetics in Cycas panzhihuaensis and implications for its conservation. Tree Genet. Genom. 2015;11:78. [Google Scholar]
- Yang J., Zhang Z., Shen Z., Ou X., Geng Y., Yang M. Review of research on the vegetation and environment of dry-hot valleys in Yunnan. Biodivers. Sci. 2016;24:462–474. (In Chinese with English abstract) [Google Scholar]
- Yang R., Feng X., Gong X. Genetic structure and demographic history of Cycas chenii (Cycadaceae), an endangered species with extremely small populations. Plant Diver. 2017;39:44–51. doi: 10.1016/j.pld.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W.B. ATBC Asia Pacific Chapter Meeting; 2015. Floral and Reproductive Ecology of Cycas panzhihuaensis. [Google Scholar]
- Yu Z.X., Yang Y.Q., Liu J., Gong L.L., Zhan C.L. Preliminary experimenton propagation of Cycas panzhihuaensis. J. Southwest Forestry Coll. 2007;27:36–41. (In Chinese) [Google Scholar]
- Zhang F.M. Kunming Institute of Botany, Chinese Academy of Sciences; 2012. Conservation Genetics of Cycas Panzhihuaensis. [Google Scholar]
- Zhang M., Wang Z.F., Jian S.G., Ye W.H., Cao H.L., Zhu P., Li L. Isolation and characterization of microsatellite markers for Cycas hainanensis C. J. Chen (Cycadaceae) Conserv. Genet. 2009;10:1175–1176. [Google Scholar]
- Zhang F.M., Su T., Yang Y., Zhai Y.H., Ji Y.H., Chen S. Development of seven novel EST-SSR markers from Cycas panzhihuaensis (Cycadaceae) Am. J. Bot. 2010;97:159–161. doi: 10.3732/ajb.1000377. [DOI] [PubMed] [Google Scholar]
- Zhang Q.Q., Zhao Y.J., Gong X. Genetic variation and phylogeography of Psammosilene tunicoides (Caryophyllaceae), a narrowly distributed and endemic species in south-western China. Aust. J. Bot. 2011;59:450–459. [Google Scholar]
- Zhang T.C., Comes H.P., Sun H. Chloroplast phylogeography of Terminalia franchetii (Combretaceae) from the eastern Sino-Himalayan region and its correlation with historical river capture events. Mol. Phylogenet. Evol. 2011;60:1–12. doi: 10.1016/j.ympev.2011.04.009. [DOI] [PubMed] [Google Scholar]
- Zhao Y.J., Gong X. Diversity and conservation of plant species in dry valleys, southwest China. Biodivers. Conserv. 2015;24:2611–2623. [Google Scholar]
- Zheng Y., Liu J., Gong X. Tectonic and climatic impacts on the biota within the Red River Fault, evidence from phylogeography of Cycas dolichophylla (Cycadaceae) Sci. Rep. 2016;6:33540. doi: 10.1038/srep33540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Liu J., Feng X.Y., Gong X. The distribution, diversity, and conservation status of Cycas in China. Ecol. Evol. 2017;7:3212–3224. doi: 10.1002/ece3.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Yang S.Y., Fu L.G., Cheng S.Z. Two new species of Cycas from Sichuan. Acta Phytotaxon. Sin. 1981;19:335–338. (In Chinese) [Google Scholar]
- Zhou H., Wang J., Huang R., Na H.Y. Callus induction and browning inhibition in Cycas panzhihuaensis L. Zhou et S. Y. Yang. J. Sichuan Univ. (Nat. Sci. Ed.) 2009;46:849–852. (In Chinese with English abstract) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.