Abstract
In this study we assessed the clinical significance of an epithelial-mesenchymal transition (EMT) gene signature and explored its association with the tumor microenvironment related to immunotherapy in patients with head and neck squamous cell carcinoma (HNSCC). Genes were selected when mRNA levels were positively or negatively correlated with at least one well-known EMT marker. We developed an EMT gene signature consisting of 82 genes. The patients were classified into epithelial or mesenchymal subgroups according to EMT signature. The clinical significance of the EMT signature was validated in three independent cohorts and its association with several immunotherapy-related signatures was investigated. The mesenchymal subgroup showed worse prognosis than the epithelial subgroup, and significantly elevated PD-1, PD-L1, and CTLA-4 levels, and increased interferon-gamma, cytolytic, T cell infiltration, overall immune infiltration, and immune signature scores. The relationship between PD-L1 expression and EMT status in HNSCC after treatment with TGF-β was validated in vitro. In conclusion, the EMT gene signature was associated with prognosis in HNSCC. Additionally, our results suggest that EMT is related to immune activity of the tumor microenvironment with elevated immune checkpoint molecules.
Subject terms: Cancer genomics, Prognostic markers, Head and neck cancer
Introduction
Epithelial-mesenchymal transition (EMT) refers to a process whereby the adhesive polarity of epithelial cancer cells dissipates and changes to mesenchymal cells. This occurs in conjunction with increased cell migration and invasiveness and is also known to play an important role in cytoskeletal remodeling and resistance to apoptosis1. Several studies have reported the association of EMT activation with cancer metastasis, resistance to anticancer drugs, and thus a poor prognosis2–4.
Head and neck squamous carcinoma (HNSCC) is the sixth most prevalent cancer worldwide, with mortality rates of 40–50% despite advances in radiation and surgical treatments5. Radiotherapy and cytotoxic chemotherapy for HNSCC are associated with substantial toxicity and morbidity. There is no biomarker that can predict response to treatments, such as radiotherapy, chemotherapy, and especially immunotherapy in patients with HNSCC. Immunotherapy has begun a new era in cancer treatment by using treatments such as checkpoint inhibitors that target the host immune system instead of tumors6. Immune checkpoint inhibitors have showed promising preliminary data and were approved for use by the FDA in patients with advanced HNSCC7–9. Few studies have reported the impact of EMT on the interactions between cancer and immune cells.
We sought to develop an EMT gene signature that can predict prognosis by systematically analyzing genomic data from several independent cohorts of patients with HNSCC. In addition, we analyzed the association between EMT gene signatures and several immunotherapy-related gene signatures with the aim of determining whether the activation status of EMT signatures corresponds to the tumor microenvironment related to immunotherapy.
Results
Discovery of EMT related gene signature in patients with HNSCC
We identified the genes where mRNA expression levels were significantly correlated with each EMT marker, namely E-cadherin (CDH1), vimentin (VIM), N-cadherin (CDH2), and fibronectin 1 (FN1). EMT related gene signature was developed and consisted of 82 genes correlated with the four EMT marker genes (Supplementary Table S1).
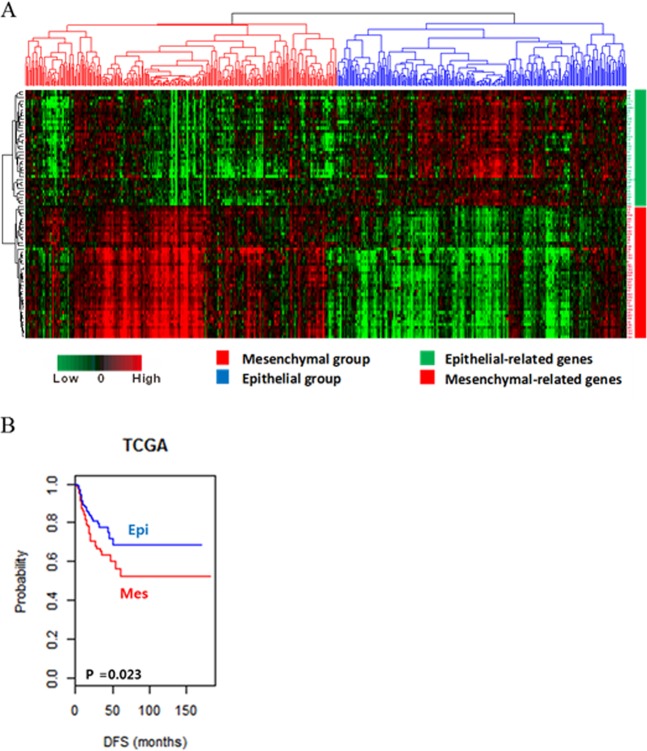
We identified two distinct subgroups of HNSCC - mesenchymal (Mes) and epithelial (Epi) subgroups – by hierarchical clustering of gene expression data of the training cohort (TCGA cohort). The genes of EMT signature were divided into distinct epithelial- and mesenchymal-related genes. The Mes subgroup had highly expressed mesenchymal-related genes, whilst the Epi subgroup had highly expressed epithelial-related genes (Fig. 1A). The Kaplan-Meier plots and the log-rank test showed that the disease-free survival (DFS) differed significantly between the two subgroups. The patients in the Mes subgroup showed worse DFS than those in the Epi subgroup (p = 0.02; Fig. 1B).
Figure 1.
Stratification of patients with HNSCC in the TCGA cohort with EMT signature. (A) Patients in the TCGA cohort were clustered by two-way hierarchical clustering using the Pearson correlation distance between genes (rows), Euclidean distance between EMT signature genes (columns), and Ward’s linkage rule. The TCGA cohort was divided into two distinct subgroups of HNSCC: the Mes group (red) and Epi group (blue). The 82 EMT signature genes were separated into distinct epithelial-related genes (green bar) and mesenchymal-related genes (red bar). (B) Kaplan-Meier plots of DFS of patients with HNSCC in the TCGA cohort.
EMT gene signature was associated with the prognosis of patients with HNSCC
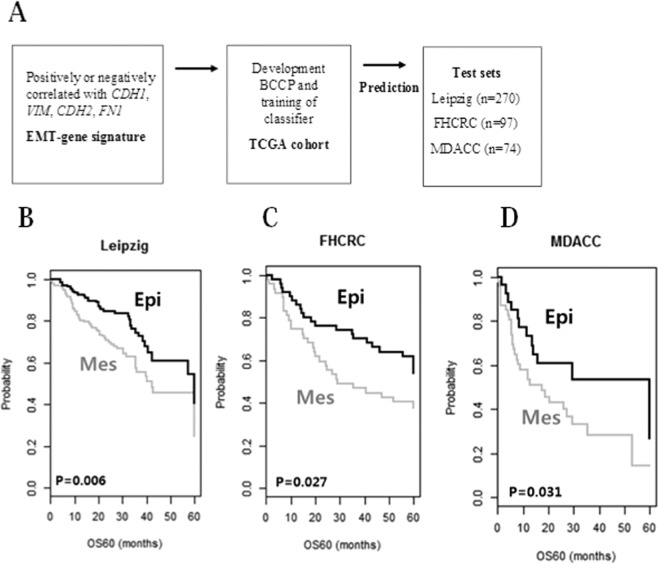
Three independent cohorts (Leipzig, FHCRC, and MDACC cohorts) were used to validate the robustness of the EMT gene signature. Validation was performed as outlined in the flow chart in Fig. 2A.
Figure 2.
Construction of the prediction model and evaluation of the predicted outcome. (A) Schematic overview of the strategy used for constructing prediction models and evaluating the predicted outcomes based on gene expression signatures. (B) Kaplan-Meier plots of OS of the Leipzig cohort. (C) Kaplan-Meier plots of OS of the FHCRC cohort. (D) Kaplan-Meier plots of OS of the MDACC cohort. Patients were stratified by EMT signatures. The differences between groups were statistically significant, as indicated by the log-rank test.
Consistent with the results from the training cohort, patients were classified patients into Epi and Mes subgroups based on the EMT signature. When patients in the Leipzig, FHCRC, and MDACC cohorts were stratified according to EMT gene signatures, significant prognostic differences between subgroups were identified in all independent validation data sets. Five-year overall survival (OS) probabilities for subgroups were significantly different in all cohorts [Leipzig (p = 0.006; Fig. 2B), FHCRC (p = 0.027; Fig. 2C), and MDACC (p = 0.031; Fig. 2D) cohorts]. These results demonstrated the robustness of the prognostic value of EMT signature.
Univariate and multivariate analyses were performed on data from the TCGA, Leipzig, and MDACC cohorts (n = 825). These analyses were based on available clinical data to verify that the prognostic effect of EMT signatures acts independently of other clinical variables. The variables include patient age, gender, smoking status, alcohol consumption, anatomic site, HPV status, primary tumor stage, lymph node metastasis, and TNM stage. Gender, age, smoking status, alcohol consumption, anatomic site, HPV status, lymph node metastasis, and the EMT gene signature were statistically significant variables associated with OS in the univariate analysis. Alternatively, HPV status, lymph node metastasis, TNM stage, and EMT gene signature were statistically significant in the multivariate analysis. (Table 1).
Table 1.
Univariate and multivariate Cox proportional hazard regression analysis of overall survival in the TCGA, Leipzig and MDACC cohorts (n = 825).
| Variables | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | |
| EMT signature (Epithelial) | 0.65 (0.52–0.83) | 0.00046* | 0.65 (0.50–0.84) | 0.0013* |
| Gender (male) | 0.76 (0.59–0.98) | 0.037* | 0.81 (0.60–1.11) | 0.197 |
| Age (>60 y) | 1.33 (1.05–1.67) | 0.015* | 1.27 (0.98–1.64) | 0.069 |
| Smoking (No) | 0.41 (0.19–0.90) | 0.028* | 0.53 (0.21–1.28) | 0.16 |
| Alcohol (No) | 0.56 (0.38–0.82) | 0.031* | 0.45 (0.26–0.79) | 0.056 |
| Anatomic site (Oropharynx) | 0.56 (0.32–0.95) | 0.034* | 0.74 (0.42–1.30) | 0.301 |
| HPV status (HPV-positive) | 0.41 (0.26–0.63) | 6.98e-05* | 0.53 (0.31–0.90) | 0.019* |
| Primary tumor (T3 & 4) | 1.34 (0.81–2.20) | 0.247 | 1.07 (0.51–2.24) | 0.851 |
| Regional lymph node (N+) | 1.75 (1.34–2.28) | 3.4e-05* | 1.71 (1.26–2.33) | 0.00054* |
| Stage (stage III & IV) | 4.14 (0.58–29.57) | 0.156 | 1.65 (1.00–2.71) | 0.048* |
HPV-positive and HPV-negative patients show distinct EMT gene signatures
The patients included in the TCGA and Leipzig cohorts were divided into HPV-positive and HPV-negative subtypes according to their HPV status. The survival analysis did not reveal statistically significant differences in 5-year OS between the Epi and Mes subgroups in HPV-positive patients (Log rank test p = 0.342; Fig. 3A). However, in HPV-negative patients, Mes subgroups had significantly worse 5-year OS than the Epi subgroup (p = 6.8e-06; Fig. 3B). These results suggest good predictability of the gene signature for HPV-negative patients.
Figure 3.
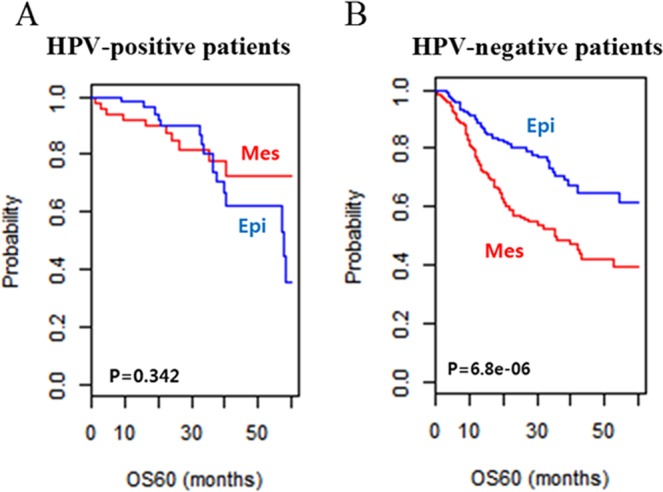
(A) Kaplan-Meier plots of OS of HPV-positive patients in the TCGA and Leipzig cohorts. (B) Kaplan-Meier plots of OS of HPV-negative patients in the TCGA and Leipzig cohorts. Patients were stratified based on EMT signatures. The differences between groups were statistically significant, as indicated by the log-rank test in HPV-negative patients.
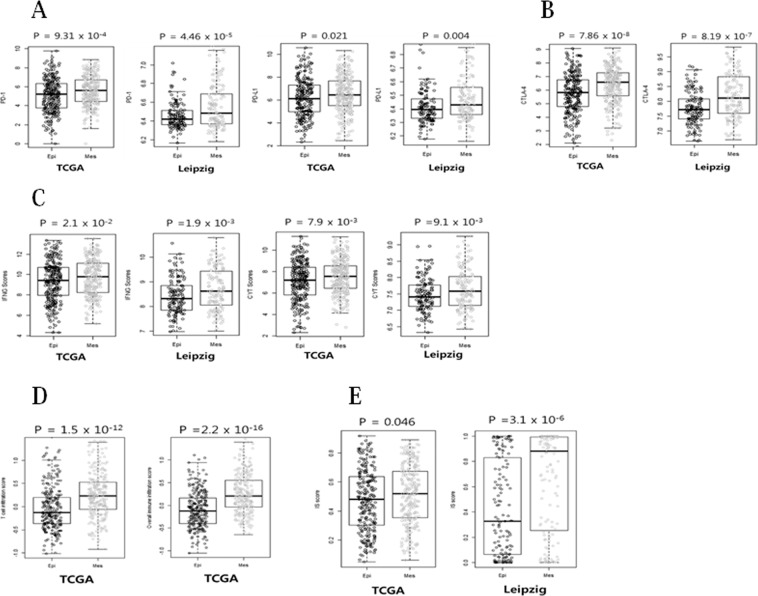
PD-1, PD-L1, and CTLA-4 levels were elevated in the Mes subgroup
In many cancers, the PD-1 (PD-L1/PD-L2) pathway is known to be involved in tumor evasion from immune activity10. As shown in Fig. 4A, PD-1 (PDCD1) and PD-L1 (CD274) levels were significantly elevated in the Mes subgroup as compared with those in the Epi subgroup in the TCGA and Leipzig cohorts (p = 9.3e-4, p = 4.5e-5, p = 0.021, and p = 0.004, respectively).
Figure 4.
Evidence of immune checkpoint molecules and immune scores in Mes compared with Epi subgroups of HNSCC. (A) PD-1 and PD-L1 levels were significantly elevated in Mes subgroup as compared with Epi subgroup in the TCGA and Leipzig cohorts. (B) CTLA-4 level was significantly higher in the Mes subgroup than that in the Epi subgroup in the TCGA and Leipzig cohorts. (C) The INFG and CYT scores were analyzed in the Mes subgroup in comparison with the Epi subgroup of HNSCC patients for gene expression data from the TCGA and Leipzig cohorts. (D) TIS and IIS were analyzed in the Mes subgroup in comparison with the Epi subgroup of HNSCC patients for gene expression data from the TCGA cohort. (E) The IS score was significantly elevated in Mes subgroup as compared with Epi subgroup in the TCGA and Leipzig cohorts.
Furthermore, CTLA-4 (CTLA4) expression was higher in the Mes subgroup than that in the Epi subgroup (Fig. 4B; p = 7.8e-8 and p = 8.2e-7, respectively). Notably, the level of immune checkpoint molecules, including CTLA-4 was significantly elevated in the TCGA and Leipzig cohorts.
INFG, CYT, TIS, IIS, and IS scores were elevated in the Mes subgroup
To assess the association of EMT signature with immune activity in the tumor microenvironment we analyzed the gene expression of several immunotherapy-related scores. Interferon gamma (INFG) and cytolytic activity (CYT) scores were significantly elevated in the Mes subgroup in the TCGA and Leipzig cohorts (p = 0.021, p = 0.0019, p = 0.0079, and p = 0.0091, respectively, Fig. 4C). The T cell infiltration score (TIS) and overall immune infiltration score (IIS) were also significantly elevated in the Mes subgroup in the TCGA cohort (p = 1.57e-12 and p = 2.2e-16, respectively). Furthermore, the immune signature (IS) score was higher in the Mes subgroup in the both cohorts (p = 0.046 and p = 3.1e-6, respectively; Fig. 4E).
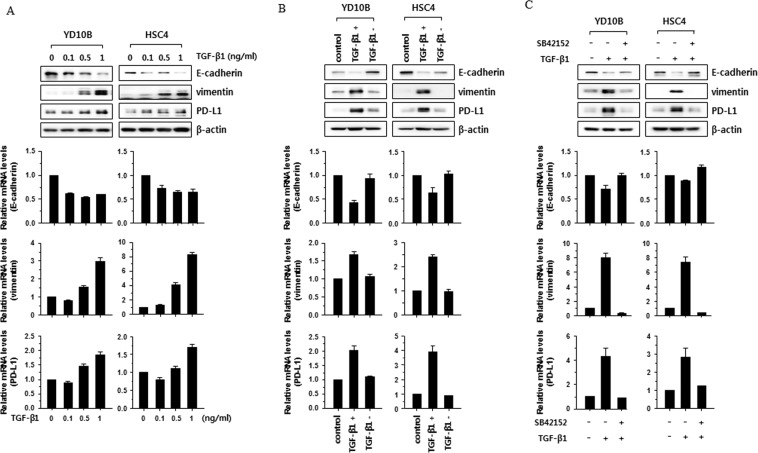
TGF-β-induced EMT enhances PD-L1 expression in HNSCC cell lines
The role of TGF-β as the primary inducer of EMT has been reported in several cancers11–13. Therefore, to investigate whether EMT status is associated with PD-L1 expression, we induced EMT status in YD-10B and HSC-4 HNSCC cells by treatment with TGF-β1 (0.1–1 ng/mL). To determine EMT status in YD-10B and HSC-4 HNSCC cells, expression levels of the epithelial cell marker (E-cadherin) and mesenchymal cell marker (vimentin) were analyzed using real-time PCR and western blotting. mRNA and protein levels of E-cadherin were decreased following treatment with TGF-β1 in a dose-dependent manner in YD-10B and HSC-4 cells, whereas mRNA and protein levels of vimentin were increased. Interesting, mRNA and protein levels of PD-L1 were increased (Fig. 5A). To determine whether PD-L1 expression was controlled by TGF-β-induced EMT, mRNA and protein levels of PD-L1 were analyzed in head and neck cancer cells under EMT and MET status using a reversion assay. mRNA and protein levels of PD-L1 were significantly altered under treatment with TGF-β1, as compared with control, and reverted to control levels after switching to culture medium without TGF-β1 (Fig. 5B). Furthermore, the TGF-β1-induced increase of PD-L1 expression was abolished by treatment with a TGF-β1 receptor kinase inhibitor (SB 431542) (Fig. 5C). These results suggest that PD-L1 expression is regulated by TGF-β-induced EMT status through the TGF-β signaling pathway.
Figure 5.
PD-L1 expression was enhanced by TGF-β-induced EMT. (A) YD-10B and HSC-4 cells were treated with the indicated concentrations of TGF-β1 (0.1, 0.5, and 1 ng/mL) for 2 days. (B) YD-10B and HSC-4 cells were incubated with medium containing 1 ng of TGF-β1 (TGF-β1 +) for 3 days, and then further incubated with medium without TGF-β1 (TFG-β1 –) for 5 days. (C) YD-10B and HSC-4 cells were treated with TGF-β1 (1 ng/mL) or TGF-β1 (1 ng/mL) plus SB 42152 (10 μM) for 2 days. (A–C) The levels of E-cadherin, vimentin, and PD-L1 were compared by western blotting (top) or real-time PCR (bottom). The real-time PCR data are presented as relative values normalized to those of the internal control (GAPDH). Full length uncropped western blotting images were shown in Supplementary Fig. S1.
Association between EMT signature and clinicopathologic characteristics of HNSCC
We sought to identify the association between EMT signature and clinical or pathological characteristics of HNSCC, and hence we performed subset analyses in the TCGA and Leipzig cohorts (Supplementary Table S2). In particular, the association between EMT subgroups and tumor sites, T stage, regional lymph node metastasis, TNM stage, HPV status, smoking status, and alcohol consumption was evaluated. We did not find any significantly different clinical or pathological characteristics of HNSCC with which to identify the Mes and Epi subgroups.
Relationship between EMT signature and somatic mutation
To identify somatic mutations that coincide with the activation of EMT signature in HNSCC, the somatic mutation data in the TCGA cohort (n = 493) were analyzed. Based on previous studies and results from the TCGA analysis, we selected six genes that are most likely to be associated with HNSCC14–18.
The somatic mutation frequencies of the six genes were compared between the Mes and Epi subgroups. The somatic mutation frequencies of EGFR and TP53 were significantly different between the two groups (the Mes subgroup presented more frequent mutations) (Supplementary Table S3). There was no difference in mutation rates between the Mes and Epi subgroups for the other four genes.
Biological process and pathway analysis
EMT signature gene analysis with DAVID bioinformatics resources 6.7 identified ten significant Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways based on enrichment of mutations (Supplementary Table S4): ECM-receptor interaction (p = 4.3e-11), focal adhesion (p = 2.7e-10), protein digestion and absorption (p = 5.3e-8), amoebiasis (p = 4.0e-6), platelet activation (p = 1.3e-5), axon guidance (p = 0.016), and arrhythmogenic right ventricular cardiomyopathy (p = 0.035). Moreover, several pathways known to be important in cancer and HNSCC pathogenesis were identified, including the PI3K-AKT signaling pathway (p = 7.2e-7), pathway in cancer (p = 0.022), and microRNAs in cancer (p = 0.03).
Discussion
In the present study, a robust EMT gene signature, clinically significant to patients with HNSCC, was developed and tested. The results revealed that Mes subgroup had poor OS in the training cohort. In the test cohorts, Mes subgroup showed worse OS and DFS. EMT signature showed independent prognostic effects in Cox proportional hazards analysis. Therefore, EMT gene signature may serve as a prognostic biomarker in HNSCC.
HPV is known to be involved in tumor formation through specific mechanisms, but it is unclear how this involvement affects EMT activation19. Jung et al. reported that HPV16 positivity correlates with the EMT transcription factor ZEB1 and that both E6 and E7 increase Slug, Twist, ZEB1, and ZEB2 expression20. Other studies have demonstrated that E6 or E7 inhibited E-cadherin at the cell surface21. In the present study, there was a significant difference in the OS of HPV-negative patients between the Mes and Epi subgroups, but not in the OS of HPV-positive patients. This suggests that EMT signature is another factor for predicting the prognosis of patients with HNSCC with respect to HPV status, which is line with a previous report showing that HPV positivity induces the expression of EMT-related genes20,21.
A number of authors reported that immune checkpoint inhibitors are effective in patients with elevated immune checkpoints, but biomarkers representing immune responses and immune checkpoint status are yet to be identified22–24. EMT is a biologic process related to decreased cell adhesion and increased invasiveness, and is known as a key program for metastasis and drug resistance in many cancers2,3,25,26. Studies have reported that EMT transcription factors (Snail or CCL2) can induce EMT process and are related to immunosuppressive cytokines activation and T-lymphocyte resistance in several cancers27–30. It has been reported that EMT can induce PD-L1 expression in non-small cell lung cancer31. However, the relationship between EMT and PD-L1 expression in HNSCC is not yet well known. We found that the levels of immune checkpoint genes, such as PD-1, PD-L1, and CTLA-4 were significantly elevated in HNSCC patients with “mesenchymal” phenotype. We also investigated whether EMT signature is closely associated with immune checkpoint genes in HNSCC cell lines. To study this, we induced EMT in HNSCC cell lines by treatment with TGF-β, and analyzed PD-L1 expression compared to cells under EMT and MET status. In this result, PD-L1 expression was enhanced by TGF-β-induced EMT, thereby indicating the relationship between EMT and immune checkpoint genes. Next, we sought to investigate whether EMT signature is associated with tumor microenvironment change using known immunotherapy-related gene signature. In the KEYNOTE-001 study, six gene signatures of INFG-related genes (IDO1, HLA-DRA, STAT1, CXCL9, IFNG and CXCL10) were previously developed in melanoma patients32. These six genes were significantly associated with the response to anti-PD-1 inhibitor in patients with melanoma. CYT was assessed by granzyme A (GZMA) and perforin (PRF1), and was dramatically associated with CD8 + T cell activation. Therefore, CYT could be used to predict clinical response to treatment with immune checkpoint inhibitors33. mRNA-based TIS was used for characterizing the computational immune cell decomposition for solid tumors. T cell infiltration in the tumor microenvironment is a key process in the balance of tumor and anti-tumor immune responses33,34. The IS score is known to be a transcriptional predictor of anti-CTLA-4 immune checkpoint inhibitor and was assessed in the genomic data from ~10,000 human tissues across 30 different cancer types to estimate the potential response to immunotherapy35. INFG, CYT, TIS, IIS, and IS scores were elevated in patients with “mesenchymal” phenotype as compared with those in patients with “epithelial” phenotype. Our findings demonstrated that EMT is associated with immune activation of the tumor microenvironment and elevation of multiple targetable immune checkpoint molecules, and may be used as a prognostic indicator in patients with HNSCC. EMT could induce cancer growth and metastasis by reprogramming immune activity in the tumor microenvironment. This suggests that patients in the Mes subgroup might have better response to immune checkpoint inhibitors, and targeting immune checkpoints may affect cancer metastasis and drug resistance associated with EMT. Therefore, our data suggest that EMT signature-based biomarkers may be valuable for identifying patients who can benefit from immune checkpoint blockade agents.
In conclusion, our results showed that the EMT signature was associated with the prognosis of patients with HNSCC, and in particular, patients in the Mes subgroup showed poor prognosis. In addition, there was an association between EMT and immune activity of the tumor microenvironment with elevated immune checkpoint molecules.
Methods
Patients and cohorts
For this study, clinical and gene expression data were collected from public databases representing four independent cohorts. Gene expression data of The Cancer Genome Atlas (TCGA cohort, n = 513) were downloaded from the UCSC Xena Browser (https://xena.ucsc.edu/)18. The data from the Institute for Medical Informatics, Statistics and Epidemiology (Leipzig cohort, GSE65858, n = 270)36, Fred Hutchinson Cancer Research Center (FHCRC cohort, GSE41613, n = 97)37, and MD Anderson Cancer Center (MDACC cohort, GSE42743, n = 74)37 were obtained from the National Center for Biotechnology Information Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) and used as the test sets. Gene expression data of TCGA and Leipzig were generated by Illumina HiSeq. 2000 and Illumina HumanHT-12 v4.0 Expression Beadchip, respectively. FHCRC and MDACC cohort data were generated by Affymetrix Human Genome U133 Plus 2.0 Array. Table 2 provides the details of the pathological and clinical characteristics of the patients in all cohorts.
Table 2.
Patient demographics and clinical characteristics of all cohorts.
| TCGA cohort (N = 513) |
Leipzig cohort (N = 270) |
FHCRC cohort (N = 97) |
MDACC cohort (N = 74) |
|
|---|---|---|---|---|
| Gender | ||||
| Male | 370 (73.7%) | 223 (82.6%) | 66 (68.0%) | 58 (78.3%) |
| Female | 132 (26.3%) | 47 (17.4%) | 31 (32.0%) | 16 (21.7%) |
| Age (mean ± SD) | 60.9 ± 11.9 | 60.1 ± 10.0 | NA | 58.1 ± 13.4 |
| Anatomic site | ||||
| Oral cavity | 301 (60.0%) | 83 (30.7%) | 86 (88.7%) | 71 (95.9%) |
| Oropharynx | 79 (15.7%) | 102 (37.8%) | 11 (11.3%) | 3 (4.1%) |
| Larynx | 113 (22.5%) | 48 (17.8%) | 0 | 0 |
| Hypopharynx | 9 (1.8%) | 33 (12.2%) | 0 | 0 |
| others | 0 | 4 (1.5%) | 0 | 0 |
| Primary tumor | ||||
| T1 | 33 (6.8%) | 35 (13.0%) | NA | 3 (4.1%) |
| T2 | 147 (30.2%) | 80 (29.6%) | NA | 27 (36.4%) |
| T3 | 129 (26.5%) | 58 (21.5%) | NA | 28 (37.8%) |
| T4 | 178 (36.6%) | 97 (35.9%) | NA | 16 (21.7%) |
| Regional lymph node | ||||
| N0 | 238 (49.5%) | 94 (34.8%) | NA | 29 (39.1%) |
| N1 | 79 (16.4%) | 32 (11.9%) | NA | 13 (17.5%) |
| N2 | 155 (32.2%) | 132 (48.9%) | NA | 32 (43.4%) |
| N3 | 9 (1.9%) | 12 (4.4%) | NA | 0 |
| Stage | ||||
| I | 20 (4.1%) | 18 (6.7%) | 30 (30.9%) | 3 (4.1%) |
| II | 96 (19.6%) | 37 (13.7%) | 11 (11.3%) | 16 (21.6%) |
| III | 101 (20.7%) | 37 (13.7%) | 15 (15.5%) | 15 (20.2%) |
| IV | 272 (55.6%) | 178 (65.9%) | 41 (42.3%) | 40 (54.1%) |
| HPV status | ||||
| Positive | 68 (19.9%) | 60 (23.4%) | 0 | NA |
| Negative | 274 (80.1%) | 196 (76.6%) | 97 (100%) | NA |
| Tobacco use | ||||
| Never | 114 (23.3%) | 48 (17.8%) | NA | 15 (20.3%) |
| Yes | 376 (76.7%) | 222 (82.2%) | NA | 59 (79.7%) |
| Alcohol use | ||||
| Never | 154 (42.1%) | 31 (11.5%) | NA | NA |
|
Yes EMT signature |
212 (57.9%) | 239 (88.5%) | NA | NA |
| Mesenchymal | 266 (51.8%) | 136 (50.3%) | 47 (48.4%) | 45 (60.8%) |
| Epithelial | 247 (48.2%) | 134 (49.7%) | 50 (51.6%) | 29 (39.2%) |
Abbreviations: TCGA, The Cancer Genome Atlas; FHCRC, Fred Hutchinson Cancer Research Center; MDACC, MD Anderson Cancer Center; HPV, Human papilloma virus; EMT, Epithelial-mesenchymal transition; NA, not available.
Development of EMT gene signature in HNSCC
The BRB-ArrayTools software program (http://brb.nci.nih.gov/BRB-ArrayTools/) was used for analysis of gene expression data38. The raw data were preprocessed using a robust multiarray averaging method for normalization39.
To find EMT-specific genes in HNSCC, gene expression data were analyzed from the TCGA cohort. Genes were selected when the mRNA expression levels were either positively or negatively correlated with at least one of the well-known EMT markers: E-cadherin (CDH1), vimentin (VIM), N-cadherin (CDH2), and/or fibronectin 1 (FN1). These markers were selected on the basis of their previously established roles as markers of EMT in lung cancer as well as other epithelial tumors40.
Using a gene feature and its correlated genes, hierarchical clustering analysis was performed with the centered correlation coefficient as the measure of similarity and a complete linkage clustering method. According to the patient clustering result, the patients were divided into two subgroups (mesenchymal and epithelial). Cluster analysis was performed with Cluster 3.041.
Construction of prediction models and validation in test cohorts
To test the ability of the gene expression signatures to predict the class of patients in an independent cohort, a previously developed model based on the Bayesian compound covariate predictor (BCCP) was adopted42. Gene expression data in the training set (TCGA cohort) were combined to form a series of classifiers according to the BCCP algorithm and the robustness of the classifier was estimated according to the misclassification rate determined during leave-one-out cross-validation of the training set. Validation was conducted in three independent patient groups (Leipzig, FHCRC, and MDACC cohorts).
Immunotherapy-related gene score
To determine the association of EMT signature with immunotherapy, we used a previously described and validated immunotherapy-related gene signature. Interferon gamma (INFG) score was calculated as the average of the normalized values of the INFG-related six genes (CXCL9, CXCL10, IDO1, IFNG, HLA-DRA, and STAT1)32. The cytolytic activity (CYT) score was calculated as the average of the expression level of two key cytolytic effectors (GZMA and PRF1)43. The T cell infiltration score (TIS) was defined as the mean of the standardized values for all T cell subsets, except for T gamma delta and T follicular helper cells: CD8 T, T helper, T, T central and effector memory, Th1, Th2, Th17, and Treg cells33. The overall immune infiltration score (IIS) was defined as the mean of the standardized values for macrophages, dendritic cell subsets (total, plasmacytoid, immature, and activated), B cells, cytotoxic cells, eosinophils, mast cells, neutrophils, natural killer (NK) cell subsets [total, CD56(bright), and CD56(dim)], and all T cell subsets, excluding T gamma delta and T follicular helper cells33. The immune signature (IS) score was obtained using 105 immune signature genes35. The IS score of TCGA HNSCC cohort was obtained from the data of a previous study35.
Pathway analysis
The list of EMT signature genes was submitted to the Database for Annotation, Visualization, and Integrated Discovery (DAVID) bioinformatics resources 6.7 to discover the gene ontology categories with significantly enriched gene numbers44. The default setting from the software was used to map the EMT signature genes to the reference set of direct and indirect relationships. Next, relevant inputs to the gene list, such as the molecular networks and biological functions were generated by the software’s algorithm. The significance of the gene annotation with a p-value less than 0.05 was determined with two-tailed Fisher’s exact test.
Antibodies, recombinant protein, and inhibitor
The following antibodies were used in this study: anti-E-cadherin, anti-N-cadherin, anti-vimentin, and anti-PD-L1 from Cell Signaling Technology (Danvers, MA, USA); and anti-β-actin from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Recombinant human transforming growth factor (TGF)-β1 was purchased from PeproTech (Rocky Hill, NJ, USA). SB 431542, a TGF-β inhibitor, was purchased from Tocris Bioscience (St. Louis, MO, USA).
Cell culture and treatment
Human HNSCC, YD-10B, and HSC-4 cells were used in this study. YD-10B was purchased from Korean cell line bank (Seoul, Korea). HSC-4 was purchased from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan). The cells were cultured in RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum. To induce EMT status in HNSCC, cells were incubated in the presence or absence of TGF-β1 (0.1–1 ng/mL) for 48 h. For reversion assay, cells were treated 1 ng/ml of TGF-β1 for 3 days, and then cultured in medium without TGF-β1 for 5 days.
Western blotting
Cells were lysed on ice for 30 min in a buffer containing 20 mM Tris-HCl (pH 7.4), 100 mM NaCl, 0.5% NP-40, 0.1 mM Na3VO4, 50 mM NaF, 30 mM Na4O7P2 · 10 H2O, and a protease inhibitor cocktail (GenDepot, Barker, TX, USA). Equal amounts of protein in the lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, electrotransferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA), and analyzed using specified antibodies with an ECL detection system (GE Healthcare, Chicago, IL, USA).
Real-time quantitative reverse transcriptase PCR
Total RNA was extracted from the indicated cell lines using an RNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. RNA was reverse-transcribed to cDNA with PrimeScript™ RT Master Mix (Takara, Shiga, Japan). Quantitative real-time PCR was performed using the TB Green™ Premix Ex Taq™ II (Takara). Primer sequences are shown in Table 3. Relative amounts of mRNA were calculated from the threshold cycle number using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an endogenous control. All experiments were performed in triplicate and the values were averaged.
Table 3.
Primer sequences used for qPCR experiments.
| Gene | Sequence | |
|---|---|---|
| E-cadherin | Forward | AAG AAG CTG GCT GAC ATG TAC GGA |
| (CDH1) | Reverse | CCA CCA GCA ACT GAT TTC TGC AT |
| Vimentin | Forward | AGA ACC TGC AGG AGG CAG AAG AAT |
| (VIM) | Reverse | TTC CAT TTC ACG CAT CTG GCG TT |
| PD-L1 | Forward | AGC CCT CAG CCT GAC ATG TC |
| (CD274) | Reverse | GGT GCC GAC TAC AAG CGA AT |
| GAPDH | Forward | TGC ACC ACC AAC TGC TTA GC |
| Reverse | GGC ATG GAC TGT GGT CAT GAG |
Statistical analysis
To test the prognostic significance, only gene expression data with available survival data were used. Prognostic significance was estimated by the Kaplan-Meier method and log-rank test between two predicted subgroups. Univariate and multivariate Cox proportional hazards regression analysis was performed to evaluate independent prognostic factors associated with survival. Pearson’s correlation was used for correlation analysis and Fisher’s exact test was used to assess the frequency difference of somatic mutation. All statistical analyses were performed using the R language program (http://www.r-project.org).
Supplementary information
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) Grant funded by the Korea Government (Ministry of Science and ICT) [grant number NRF-2018R1D1A1B07050154] and supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea [grant number HI18C1039].
Author contributions
Designed Study: A.R.J., C.H.J., J.K.N., Y.C.L., Y.G.E.; Collected data: A.R.J., C.H.J., J.K.N.; Performed analysis: A.R.J., C.H.J., Y.G.E.; Wrote the paper: A.R.J., C.H.J., Y.C.L., Y.G.E.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-60707-x.
References
- 1.Thiery JP. Epithelial–mesenchymal transitions in tumour progression. Nature Reviews Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 2.Prudkin L, et al. Epithelial-to-mesenchymal transition in the development and progression of adenocarcinoma and squamous cell carcinoma of the lung. Modern pathology. 2009;22:668–678. doi: 10.1038/modpathol.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zavadil, J., Haley, J., Kalluri, R., Muthuswamy, S. K. & Thompson, E. Epithelial-mesenchymal transition. Cancer Res.23, 9574–9577 (2008). [DOI] [PubMed]
- 4.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 5.Torre LA, et al. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 6.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Mehra, R. et al. (American Society of Clinical Oncology, 2016).
- 8.Ferris, R. L. et al. (American Society of Clinical Oncology, 2016).
- 9.Gillison, M. L. et al. In Cancer Research. (Amer Assoc Cancer Research 615 Chestnut St, 17th Floor, Philadelphia, PA 19106-4404 USA).
- 10.Blank C, Gajewski TF, Mackensen A. Interaction of PD-L1 on tumor cells with PD-1 on tumor-specific T cells as a mechanism of immune evasion: implications for tumor immunotherapy. Cancer Immunology, Immunotherapy. 2005;54:307–314. doi: 10.1007/s00262-004-0593-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horiguchi K, et al. TGF-beta drives epithelial-mesenchymal transition through deltaEF1-mediated downregulation of ESRP. Oncogene. 2012;31:3190–3201. doi: 10.1038/onc.2011.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vincent T, et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat. Cell. Biol. 2009;11:943–950. doi: 10.1038/ncb1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanmei L, et al. HMGB1 attenuates TGF-β-induced epithelial-mesenchymal transition of FaDu hypopharyngeal carcinoma cells through regulation of RAGE expression. Mol. Cell. Bochem. 2017;431:1–10. doi: 10.1007/s11010-017-2968-2. [DOI] [PubMed] [Google Scholar]
- 14.Lee JW, et al. Somatic mutations of EGFR gene in squamous cell carcinoma of the head and neck. Clinical Cancer Research. 2005;11:2879–2882. doi: 10.1158/1078-0432.CCR-04-2029. [DOI] [PubMed] [Google Scholar]
- 15.Oberholzer PA, et al. RAS mutations are associated with the development of cutaneous squamous cell tumors in patients treated with RAF inhibitors. Journal of Clinical Oncology. 2011;30:316–321. doi: 10.1200/JCO.2011.36.7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu W, et al. PIK3CA mutations in head and neck squamous cell carcinoma. Clinical Cancer Research. 2006;12:1441–1446. doi: 10.1158/1078-0432.CCR-05-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson YC, Wang E, Clayman GL. Genotypic analysis of tumor suppressor genes PTEN/MMAC1 and p53 in head and neck squamous cell carcinomas. The Laryngoscope. 1998;108:1553–1556. doi: 10.1097/00005537-199810000-00024. [DOI] [PubMed] [Google Scholar]
- 18.Network CGA. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillison, M. L. In Seminars in oncology. 744–754 (Elsevier). [DOI] [PubMed]
- 20.Jung Y-S, Kato I, Kim H-RC. A novel function of HPV16-E6/E7 in epithelial–mesenchymal transition. Biochemical and biophysical research communications. 2013;435:339–344. doi: 10.1016/j.bbrc.2013.04.060. [DOI] [PubMed] [Google Scholar]
- 21.Matthews K, et al. Depletion of Langerhans cells in human papillomavirus type 16-infected skin is associated with E6-mediated down regulation of E-cadherin. Journal of virology. 2003;77:8378–8385. doi: 10.1128/JVI.77.15.8378-8385.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herbst RS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizvi NA, et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji R-R, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunology, Immunotherapy. 2012;61:1019–1031. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomson S, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non–small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer research. 2005;65:9455–9462. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 26.Bell DW, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nature genetics. 2005;37:1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 27.Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer cell. 2009;15:195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 28.Kudo-Saito C, Shirako H, Ohike M, Tsukamoto N, Kawakami Y. CCL2 is critical for immunosuppression to promote cancer metastasis. Clinical & experimental metastasis. 2013;30:393–405. doi: 10.1007/s10585-012-9545-6. [DOI] [PubMed] [Google Scholar]
- 29.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 30.Lu H, Knutson KL, Gad E, Disis ML. The tumor antigen repertoire identified in tumor-bearing neu transgenic mice predicts human tumor antigens. Cancer research. 2006;66:9754–9761. doi: 10.1158/0008-5472.CAN-06-1083. [DOI] [PubMed] [Google Scholar]
- 31.Funaki SEA. Chemotherapy enhances programmed cell death 1/ligand 1 expression via TGF-β induced epithelial mesenchymal transition in non-small cell lung cancer. Oncol Rep. 2017;38:2277–2284. doi: 10.3892/or.2017.5894. [DOI] [PubMed] [Google Scholar]
- 32.Muro K, et al. Pembrolizumab for patients with PD-L1-positive advanced gastric cancer (KEYNOTE-012): a multicentre, open-label, phase 1b trial. The lancet oncology. 2016;17:717–726. doi: 10.1016/S1470-2045(16)00175-3. [DOI] [PubMed] [Google Scholar]
- 33.Senbabaoglu, Y. et al. The landscape of T cell infiltration in human cancer and its association with antigen presenting gene expression. bioRxiv, 025908 (2015).
- 34.Gajewski TF, Louahed J, Brichard VG. Gene signature in melanoma associated with clinical activity: a potential clue to unlock cancer immunotherapy. The Cancer Journal. 2010;16:399–403. doi: 10.1097/PPO.0b013e3181eacbd8. [DOI] [PubMed] [Google Scholar]
- 35.Ock C-Y, et al. Genomic landscape associated with potential response to anti-CTLA-4 treatment in cancers. Nature communications. 2017;8:1050. doi: 10.1038/s41467-017-01018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wichmann G, et al. The role of HPV RNA transcription, immune response‐related gene expression and disruptive TP53 mutations in diagnostic and prognostic profiling of head and neck cancer. International journal of cancer. 2015;137:2846–2857. doi: 10.1002/ijc.29649. [DOI] [PubMed] [Google Scholar]
- 37.Lohavanichbutr P, et al. A 13-gene signature prognostic of HPV-negative OSCC: discovery and external validation. Clinical Cancer Research. 2013;19:1197–1203. doi: 10.1158/1078-0432.CCR-12-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon R, et al. Analysis of gene expression data using BRB-array tools. Cancer informatics. 2007;3:11. doi: 10.1177/117693510700300022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolstad BM, Irizarry RA, Åstrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 40.Yauch RL, et al. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clinical Cancer Research. 2005;11:8686–8698. doi: 10.1158/1078-0432.CCR-05-1492. [DOI] [PubMed] [Google Scholar]
- 41.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim JH, et al. Genomic predictors for recurrence patterns of hepatocellular carcinoma: model derivation and validation. PLoS medicine. 2014;11:e1001770. doi: 10.1371/journal.pmed.1001770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.