Abstract
This study was conducted to evaluate the applicability of crAssphage, pepper mild mottle virus (PMMoV), and tobacco mosaic virus (TMV) as indicators of the reduction of human enteric viruses during wastewater treatment. Thirty-nine samples were collected from three steps at a wastewater treatment plant (raw sewage, secondary-treated sewage, and final effluent) monthly for a 13-month period. In addition to the three indicator viruses, eight human enteric viruses [human adenoviruses, JC and BK polyomaviruses, Aichi virus 1 (AiV-1), enteroviruses, and noroviruses of genogroups I, II, and IV] were tested by quantitative PCR. Indicator viruses were consistently detected in the tested samples, except for a few final effluents for crAssphage and TMV. The mean concentrations of crAssphage were significantly higher than those of most tested viruses. The concentrations of crAssphage in raw sewage were positively correlated with the concentrations of all tested human enteric viruses (p <0.05), suggesting the applicability of crAssphage as a suitable indicator to estimate the concentrations of human enteric viruses in raw sewage. The reduction ratios of AiV-1 (1.8 ± 0.7 log10) were the lowest among the tested viruses, followed by TMV (2.0 ± 0.3 log10) and PMMoV (2.0 ± 0.4 log10). Our findings suggested that the use of not only AiV-1 and PMMoV but also TMV as indicators of reductions in viral levels can be applicable during wastewater treatment.
Subject terms: PCR-based techniques, Water microbiology, Pathogens
Introduction
Wastewater treatment plants (WWTPs) are currently facing numerous issues regarding insufficient treatment of human enteric viruses during the treatment process1. Although a number of treatment procedures to remove physical, chemical, and microbiological waste from sewage are performed, these processess still need to be improved to reduce the viral content of water2. The advanced multiple-barrier concept has been proposed for wastewater reclamation and reuse; however, it is insufficient to achieve complete reduction of human enteric viruses3,4. In fact, advanced wastewater treatment has been developed to achieve reduction of viruses by facilitating chemical clarification, ultrafiltration, reverse osmosis, and advanced oxidation at different levels of regulatory control5. However, despite implementing these modifications, the efficiency with which treatment can reduce viral levels in wastewater is still unsatisfactory. Moreover, no authorized regulatory standard for the reduction of viruses in wastewater treatment has yet been established6.
Human enteric viruses are the leading cause of waterborne illnesses and usually transmitted via the fecal–oral route5–7. Infected individuals discharge millions of viral particles that ultimately enter sewage systems7. In this context, it is important to study the efficiency of virus reduction at WWTPs to ascertain whether all viruses are removed from effluent samples or not. Fecal indicator bacteria, such as total coliforms and Escherichia coli, are not reliable indicators of the presence and removal of human enteric viruses during wastewater treatment8,9. Human enteric viruses are resistant to adverse conditions, such as wide ranges of pH, low temperature, and chlorination7, making their removal difficult even using complex treatment technology including activated sludge process, oxidation ponds, activated carbon treatment, filtration and lime coagulation, and chlorination10.
Numerous studies have been carried out to establish a single indicator that can predict the presence of human enteric viruses in aquatic environments7,11,12. Previous studies successfully demonstrated that several types of viruses, such as human adenoviruses (HuAdVs), polyomaviruses, enteroviruses (EVs), Aichi virus 1 (AiV-1), and pepper mild mottle virus (PMMoV), can be used as potential indicators to illustrate the adequate reduction of viruses in wastewater13–15. Studies showed that PMMoV and tobacco mosaic virus (TMV) are the most abundant and frequently detected viruses in sewage15–17. They are distributed globally and more abundant in environmental samples than human entric viruses, without any pronounced seasonal variation18,19. It may thus be beneficial to use PMMoV to evaluate the performance of drinking water and wastewater treatment processes15,20, although no studies have been conducted to date to evaluate the efficiency of reduction of TMV during wastewater treatment. Given that AiV-1, a member of genus Kobuvirus in the family Picornaviridae21, is more abundant and reduced less than other viruses during the wastewater treatment process, it could be used as an indicator of virus reduction15,22. Similarly, based on studies conducted in Japan, among four individual genogroups (I–IV), F-specific RNA coliphages of genogroup I have been proposed as a possible indicator of reduction23,24. Selecting an appropriate viral indicator is still a major challenge due to various factors, such as human population dynamics, seasonal variations, and the types of treatment processes used8. Moreover, it is almost impossible to analyze all pathogenic viruses in wastewater at the same time due to lack of cost and time and laboring25.
CrAssphage, a bacteriophage infecting the human gut bacterium Bacteroides intestinalis26, was first discovered from human fecal microbiomes27. Human metagenomics has also demonstrated its abundance in the human gut28. Subsequently, quantitative PCR (qPCR) assays were developed and applied to quantify crAssphage in stool29–31 and environmental water samples11,29,32–35. Moreover, the sequences of crAssphage variants detected in Europe were found to differ from those in the USA11. Both assays revealed the abundance of crAssphage of human origin; however, lower quantities (~3 log10 lower) were also noted in samples of animal origin11,29,33,36, raising questions about the specificity of the assay. Although the appropriateness of crAssphage as a human fecal marker has been tested in many studies11,29,32–35,37,38, to the best of our knowledge, no studies evaluating crAssphage as an indicator of virus reduction during water treatment processes have yet been reported. Farkas et al.37 conducted one-year monitoring of crAssphage in influent and effluent of WWTPs in UK, reporting that crAssphage concentrations were 105–109 copies/L in influent and 107–108 copies/L in effluent. However, since the purpose of that study was also to evaluate the applicalibity of crAssphage as a human-derived wastewater contamination marker, no discussion is provided regarding the suitability of crAssphage as an indicator of virus reductions during wastewater treatment.
Based on this background, this study aimed to evaluate crAssphage along with PMMoV and TMV for their applicability as indicators of the reduction of human enteric viruses during wastewater treatment. In addition, the applicability of crAssphage to estimate the concentrations of human enteric viruses in wastewater samples was evaluated. To our knowledge, this is the first study to demonstrate the applicability of crAssphage and TMV as an indicator of virus reductions during wastewater treatment.
Results
Detection of viruses in three steps of WWTP
Seven of the eight human enteric viruses were detected in the tested samples at different frequencies, while noroviruses of genogroup IV (NoVs-GIV) were not detected in any of the examined samples, as shown in Table 1. Thirteen (100%) raw sewage, 11 (85%) secondary-treated sewage, and 12 (92%) final effluent samples were positive for at least one of the eight human enteric viruses tested. NoVs of genogroup I (NoVs-GI) and BK polyomaviruses (BKPyVs), which were present in 12 (92%) samples, were the most prevalent human enteric viruses in raw sewage. Six enteric viruses were detected in the secondary-treated sewage, where NoVs of genogroup II (NoVs-GII) (46%) were the most frequently detected enteric viruses. The concentrations of NoVs-GII were significantly higher than those of other human enteric viruses, such as EVs, NoVs-GI, and BKPyVs [analysis of variance (ANOVA); p < 0.05]. For the final effluent samples, NoVs-GI (7/13; 54%) were the most prevalent human enteric viruses.
Table 1.
Detection of human enteric viruses and indicator viruses in wastewater samples.
| Virus analyzed | No. of tested samples | Raw sewage | Secondary-treated sewage | Final effluent | ||||
|---|---|---|---|---|---|---|---|---|
| No. of positive samples (%) | Concentration (mean ± standard deviation) (log10 copies/L) | No. of positive samples (%) | Concentration (mean ± standard deviation) (log10 copies/L) | No. of positive samples (%) | Concentration (mean ± standard deviation) (log10 copies/L) | |||
| Human enteric virus | AiV-1 | 13 | 9 (69) | 5.0 ± 0.4 | 4 (31) | 3.7 ± 0.3 | 5 (38) | 3.8 ± 0.5 |
| EVs | 13 | 11 (85) | 4.8 ± 0.6 | 2 (15) | 3.2 ± 0.2 | 1 (8) | 3.4 | |
| NoVs-GI | 13 | 12 (92) | 5.7 ± 0.5 | 5 (38) | 4.2 ± 0.3 | 7 (54) | 4.1 ± 0.4 | |
| NoVs-GII | 13 | 10 (77) | 7.5 ± 0.8 | 6 (46) | 6.1 ± 0.5 | 5 (38) | 6.0 ± 0.2 | |
| NoVs-GIV | 13 | 0 (0) | Not applicable | 0 (0) | Not applicable | 0 (0) | Not applicable | |
| HuAdVs | 13 | 11 (85) | 6.7 ± 0.5 | 4 (31) | 5.1 ± 0.3 | 5 (38) | 5.2 ± 0.2 | |
| JCPyVs | 13 | 9 (69) | 7.1 ± 0.7 | 0 (0) | Not applicable | 3 (23) | 6.0 ± 1.0 | |
| BKPyVs | 13 | 12 (92) | 7.3 ± 0.6 | 3 (23) | 5.4 ± 0.3 | 1 (8) | 5.4 | |
| Indicator virus | CrAssphage | 13 | 13 (100) | 10.3 ± 1.3 | 13 (100) | 7.7 ± 1.3 | 11 (85) | 7.4 ± 1.1 |
| PMMoV | 13 | 13 (100) | 7.1 ± 0.5 | 13 (100) | 5.4 ± 0.6 | 13 (100) | 5.1 ± 0.7 | |
| TMV | 13 | 13 (100) | 5.9 ± 0.5 | 13 (100) | 4.3 ± 0.5 | 12 (92) | 4.0 ± 0.5 | |
Three indicator viruses (PMMoV, TMV, and crAssphage) were consistently detected in all wastewater samples tested, except for a few final effluents for TMV (38/39; 97%) and crAssphage (37/39; 95%). The mean concentrations of crAssphage in raw sewage, secondary-treated sewage, and final effluent were significantly higher than those of other tested viruses, except for BKPyVs and JC polyomaviruses (JCPyVs) in final effluent samples (ANOVA; p < 0.05).
Reduction ratios of viruses during wastewater treatment process
Figure 1 shows the annual log10 reduction ratios of tested viruses (except for NoVs-GIV, which were not detected in any of the tested samples) during the whole wastewater treatment process. Among the seven human enteric viruses tested, BKPyVs showed the highest mean reduction ratio of 3.1 ± 0.8 log10 (n = 12), followed by NoVs-GII (2.5 ± 1.5 log10, n = 10), JCPyVs (2.5 ± 1.0 log10, n = 9), EVs (2.4 ± 0.5 log10, n = 11), HuAdVs (2.1 ± 0.7 log10, n = 11), NoVs-GI (2.0 ± 0.4 log10, n = 12), and AiV-1 (1.8 ± 0.7 log10, n = 9) as the lowest. For indicator viruses, PMMoV (2.0 ± 0.4 log10, n = 13) and TMV (2.0 ± 0.3 log10, n = 13) showed comparable reduction ratios.
Figure 1.
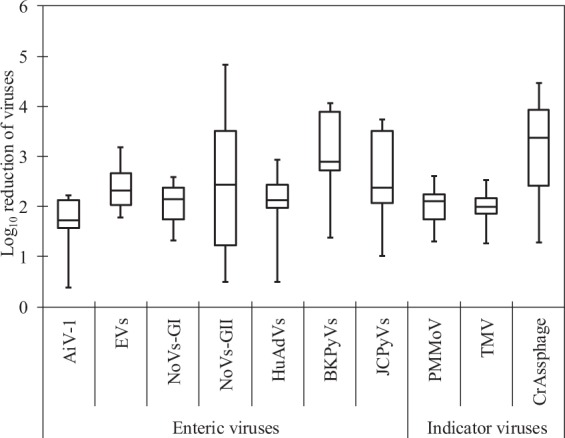
Annual reduction ratios of human enteric viruses and indicator viruses during the wastewater treatment process. Lines within the boxes represent median values, the upper and lower lines of the boxes represent 25th and 75th percentiles, respectively, and the bars outside the boxes represent minimum and maximum values.
On the other hand, a higher reduction ratio was obtained for crAssphage (3.3 ± 1.0 log10, n = 13), which was the highest among all of the viruses tested in this study. Compared with this study, Farkas et al.37 reported lower reduction of crAssphage (1–2 log10) at two WWTPs tested (activated sludge and filter beds treatment), which suggests that the reduction efficiency of crAssphage during wastewater treatment can vary greatly depending on treatment processes. Further studies are needed to clarify the mechanism of reduction of crAssphage.
The mean reduction ratios of the tested viruses during primary–secondary treatment varied greatly from 1.7 ± 0.4 log10 (n = 13; for TMV) to 2.9 ± 0.8 log10 (n = 12; for BKPyVs), whereas those during chlorination varied from no reduction to maximum reduction of 0.7 ± 0.7 log10 (n = 13; for crAssphage).
The virus concentration values obtained from raw sewage were analyzed for any possible seasonality during the 13-month study period. For this purpose, the months were categorized into four seasons [i.e., summer (June–August), fall (September–November), winter (December–February), and spring (March–May)]. The concentrations of human enteric viruses and indicator viruses were relatively stable throughout the study period (Tukey–Kramer test; p > 0.05). High concentrations of NoVs-GII were observed in March, April, and December (8.4–8.6 log10 copies/L), but the results were not statistically significant (Tukey–Kramer test; p > 0.05). Total concentrations of human enteric viruses tested (7.7 ± 0.7 log10 copies/L) were also equally dissiminated in raw sewage, showing no apparent seasonal variation (Tukey–Kramer test; p > 0.05).
Relationships between human enteric viruses and indicator viruses
To determine whether the concentrations of the indicator viruses were correlated with the concentrations of human enteric viruses tested, the data were analyzed for raw sewage. As shown in Table 2, a significant positive correlation was observed between the concentrations of human enteric viruses and those of each indicator virus in raw sewage, except between NoVs-GI and PMMoV and/or TMV.
Table 2.
Relationship between the concentrations of human enteric viruses and of indicator viruses in raw sewage.
| Virus analyzed | r-value | ||
|---|---|---|---|
| PMMoV | TMV | CrAssphage | |
| AiV-1 | 0.66* | 0.72* | 0.88* |
| EVs | 0.73* | 0.75* | 0.70* |
| NoVs-GI | 0.30 | 0.46 | 0.70* |
| NoVs-GII | 0.81* | 0.85* | 0.91* |
| HuAdVs | 0.85* | 0.72* | 0.71* |
| JCPyVs | 0.78* | 0.87* | 0.89* |
| BKPyVs | 0.86* | 0.88* | 0.91* |
*Statistically significant (p < 0.05).
Discussion
Only a few studies have been conducted for the quantitation and reduction of crAssphage during wastewater treatment throughout the year33,37. CrAssphage, a recently identified human fecal marker, was detected in all wastewater samples tested, at significantly higher concentrations than human enteric viruses and other indicator viruses. Previous studies also reported that crAssphage was highly abundant in various environmental samples11,29,33,34,36,39. Despite its abundance with high concentrations in water samples, several studies reported cross-reactions with feces from different animals11,29,34,38, raising questions about its suitability as a human fecal marker.
The concentrations of EVs and AiV-1 in wastewater samples were lower than those of other human enteric viruses, in agreement with the results of previous studies conducted in New Zealand and the USA40,41. The ratio of positivity of AiV-1 (69%) in wastewater was comparable to those reported in previous studies42,43, which exhibited no seasonal variation43. AiV-1 has been detected from stools of gastroenteritis patients and wastewater in Japan and elsewhere42–45, suggesting its widespread presence in the environment. A consistently high prevalence of NoVs-GI and GII was noted from the wastewater samples tested, and their positivity ratios were comparable with those reported in previous studies40,41,46–48. This variation in the data may depend on the epidemiology of viruses or diseases in specific locations. However, unlike most previous studies49–51, no remarkable seasonal variation was found in the concentrations of NoVs in wastewater, although slightly higher concentrations were observed in March, April, and December. A previous study reported the observation of two peaks (October–December and March–May) in concentrations of NoVs-GII52, indicating that the seasonal trend can vary depending on the region. Concentrations of NoVs in the final effluent samples were found comparatively higher than those reported by Kitajima et al.53 and similar to those reported by Dias et al.54. The observation of higher concentrations and undefined seasonal variation of tested viruses in our study might be because of enrollment of the limited number of samples during the study period. Due to their high prevalence in sewage, HuAdVs and JCPyVs have been proposed as an indicator of fecal contamination. We found great variations in the concentrations and reduction values for these viruses compared with those of other tested indicator viruses (PMMoV, TMV, and crAssphage). The data were in agreement with previous studies conducted in Egypt55 and USA53. Molecular methods are rapid and reliable techniques for the detection and quantitation of viruses in environmental samples, but the presence of inhibitory substances in the sample itself could cause this variation during analysis55.
This study focused on a possibility of using crAssphage, along with PMMoV and TMV, as indicators of the reduction of human enteric viruses during wastewater treatment. For this purpose, the efficiency of reduction of the levels of these indicator viruses was determined and compared with that of human enteric viruses. In addition to PMMoV (2.0 ± 0.4 log10) and TMV (2.0 ± 0.3 log10), the reduction ratios of AiV-1 (1.8 ± 0.7 log10) were also found to be lower than those of other human enteric viruses tested. The variation in the reduction efficacy of viruses during operational conditions in the secondary treatment and chlorination depends on various factors, such as retention time, water temperature, and flow volume56. For example, secondary biological treatment processes typically achieve less than 2-log10 reduction of viruses57, while modified membrane bioreactor secondary treatment could achieve greater reductions (3.0 to >6.7 log10) than conventional secondary treatment (1.5–4.2 log10)58. One of the most important findings of this study is the demonstratation of the applicability of these viruses as indicators of human enteric virus reduction during the wastewater treatment process due to their lower reduction efficiencies. Previous studies proposed PMMoV and AiV-1 as viral indicators during the wastewater treatment process15,21, which agreed with the results of this study. Furthermore, this is the first study that quantified the occurrence of TMV at a WWTP, indicating the usefulness of this virus as an indicator.
Since this study was performed using only qPCR, another interesting issue is whether the viruses detected after chlorination remained infectious. Cell culture-based techniques are a gold standard to measure the infectivity of viruses, but suitable cultivation facilities are still not available for some viruses. Therefore, new and effective methods should be developed to evaluate the reduction of infectivity of non-cultivatable viruses3. Previous studies focused on the unsuccessful reduction of viruses by chlorination at WWTPs15,41,49,50.
In this study, the reduction ratio of crAssphage (3.3 ± 1.0 log10) was the highest among all tested viruses. Regardless of its remarkably high concentrations in wastewater and significant association with human enteric viruses, crAssphage cannot be proposed as an indicator of virus reduction during the wastewater treatment process due to its high reduction ratio. Similarly, BKPyVs, JCPyVs, and NoVs-GII were reduced more efficiently than other human enteric viruses, in agreement with the results of previous studies15,22. In some cases, human enteric viruses were not detected in secondary-treated sewage but were detected in final effluent, which was also reported in a previous study conducted in the USA15. Results of “no detection” are commonly obtained during the quantification of viruses at WWTPs3,57. This variation may occur due to the lack of homogeneity of viral particles or obstruction of the analysis by organic matter or suspended solids present in wastewater57,58. Moreover, the efficiency of virus removal also depends on the adsorptive affinities and the adsorbents such as sand, pure clays, bacterial cells, naturally occurring suspended colloids, estuarine silts, and sediments59,60.
In summary, this study successfully quantified all tested viruses in wastewater samples of a WWTP in the USA during a 13-month period. Three indicator viruses were consistently abundant year-round in the wastewater samples. Despite the abundance with high concentrations, crAssphage was judged to be inappropriate as an indicator of virus reduction because of its higher reductions than human enteric viruses. However, crAssphage was suggested to be a suitable indicator to estimate the concentrations of human enteric viruses in raw sewage. More studies need to be conducted to obtain a better understanding of the occurrence and reduction of crAssphage during the wastewater treatment process for the effective management of risks associated with wastewater in various geographical settings. Not only the previously proposed AiV-1 and PMMoV but also TMV were judged as appropriate indicators of the reduction of human enteric viruses. Further studies are needed to determine the reductions of crAssphage and TMV at WWTPs with different treatment systems.
Materials and Methods
Collection of wastewater samples
The WWTP in New Orleans, USA, treats 57,000,000 gallons of wastewater daily. A conventional treatment system combined with equipment for the activated sludge process is installed at this WWTP and disinfection was performed by chlorination. The final effluent is discharged from this WWTP into the Mississippi River, which is later filtered and used as a freshwater source for populations further down the river. Biochemical oxygen demand (BOD) values were 50–134 mg/L in the influent samples and <5 mg/L in the final effluent samples, pH of the final effluent was ~7.3. Raw sewage (before primary sedimentation), secondary-treated sewage (after sedimentation), and final effluent (after chlorination) were collected as grab samples monthly for a 13-month period between March 2017 and March 2018. Water samples (100 mL for raw sewage and 1,000 mL for secondary-treated sewage and final effluent) were collected in sterile bottles, transported to the laboratory in a cooler on ice, and processed within 6 h of sample collection.
Virus concentration
Water samples were concentrated using an electronegative filter method, as previously described61 with slight modifications. Briefly, 2.5 M MgCl2 was added to these samples (100 mL of raw sewage and 1 L each of secondary-treated sewage and final effluent) to obtain a final concentration of 25 mM. Samples were subsequently passed through a mixed cellulose ester membrane filter (90-mm diameter and 0.45-µm pore size; Merck Millipore, Billerica, USA) attached to a glass filter holder (Advantec, Tokyo, Japan). Magnesium ions were removed by passing 200 mL of 0.5 mM H2SO4 through the filter, and the viruses were eluted with 10 mL of 1.0 mM NaOH. The eluate was recovered in a tube containing 50 µL of 100 mM H2SO4 and 100 µL of 100× Tris-EDTA buffer for neutralization. Further centrifugation was performed using a Centriprep YM-50 (Merck Millipore) to obtain a final volume of approximately 650 µL.
DNA/RNA extraction and reverse transcription (RT)
Viral DNA and RNA were extracted using a ZR Viral DNA/RNA Kit (Zymo Research, Irvine, USA) to obtain a final volume of 100 µL, in accordance with the manufacturer’s protocol. RT was performed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, USA).
qPCR for viral genomes
In this study, HuAdVs, JCPyVs, BKPyVs, AiV-1, EVs, and NoVs-GI, GII, and GIV were tested as human enteric viruses, whereas crAssphage, PMMoV, and TMV were tested as indicator viruses. Briefly, 2.5 µL of the viral DNA or cDNA was mixed with 22.5 µL of a qPCR mixture containing 12.5 µL of Probe qPCR Mix (Takara Bio, Kusatsu, Japan), 0.4 µM each of forward and reverse primers, and 0.2 µM of a TaqMan (MGB) probe (Thermo Fisher Scientific, Waltham, USA). Subsequently, PCR tubes containing the mixtures were placed in a Thermal Cycler Dice Real Time System TP800 (Takara Bio) and incubated at 95 °C for 30 s, followed by 45 cycles of 95 °C for 5 s and 60 °C for 30 s for crAssphage29, HuAdVs62, BKPyVs, and JCPyVs63. Similarly, TaqMan (MGB)-based qPCR was performed for seven RNA viruses [AiV-164, EVs61,65, NoVs-GI, GII66, GIV67, PMMoV16,68, and TMV69] with the following thermal conditions: 95 °C for 30 s, followed by 45 cycles of 95 °C for 5 s and 58 °C for 30 s (for AiV-1 and NoVs-GI and GII), 60 °C for 30 s (NoVs-GIV), or 60 °C for 60 s (for PMMoV, TMV, and EVs).
Six 10-fold dilutions of the artificially synthesized plasmid DNA were used to generate a standard curve, whereas molecular-grade water was used to prepare negative controls. Water samples, standard samples, and negative controls were tested in duplicate. The samples were considered negative if the cycle threshold value was greater than 40. One-tenth of the limit-of-detection value (2.0–2.4 log10 copies/L for secondary-treated sewage and final effluent) was given to a virus-negative sample. For calculation of the reduction ratios during primary–secondary treatment (from raw sewage to secondary-treated sewage), chlorination (from secondary-treated sewage to final effluent), and the whole treatment process (from raw sewage to final effluent), the results from months when a target virus was detected in the pretreated sample were used.
Process control
As previously recommended70, coliphage MS2 (ATCC 15597-B1) was used as a molecular process control to evaluate the efficiency during extraction, RT, and qPCR. In brief, 1 µL of coliphage MS2 was inoculated to 100 µL of the virus concentrate, followed by DNA/RNA extraction and RT, as shown in the Section 2.3. Subsequently, a qPCR assay specific for F-specific RNA coliphages of genogroup I71, where MS2 belongs, was performed to quantify MS2-cDNA.
In addition, qPCR-control DNA was used to evaluate the level of inhibition during qPCR amplification. Artificially synthesized plasmid DNA containing sequences amplified by qPCR assays for chicken parvovirus72 was used for RNA viruses as a process control, as described previously73,74.
The efficiency was calculated from the ratio of the copy number of cDNA or plasmid DNA in the sample qPCR tube to that in the non-inhibition control tube. For MS2 molecular process control, the lowest extraction-RT-qPCR efficiency of 6.4% was observed for the secondary-treated sewage sample which had been collected in June 2017, and more than half of the tested samples (22/39, 56%) yielded the recovery of >100%. Similarly, the qPCR amplification efficiency ranged from 55% to 140%, with a mean of 86% (n = 39). These results indicate that there was no significant loss and/or inhibition during any of the detection procedures, such as DNA/RNA extraction, RT, and qPCR. However, there might have been any loss during virus concentration, which was not tested in this study.
Statistical analysis
Data on the concentrations of all types of viuses was compared using ANOVA. Pearson’s correlation coefficients (r) were calculated to identify the relationships between total concentrations of human enteric viruses and concentrations of indicator viruses using bivariate correlation with Pearson’s coefficients. The Tukey–Kramer multiple comparison procedure was used to determine possible significant variation in the concentrations of indicator viruses and human enteric viruses among the seasons. A p value of <0.05 was considered significant.
Acknowledgements
This study was partially supported by d the Japan Society for the Promotion of Science through Grant-in-Aid for Scientific Research (B) (grant number JP17H03332). The authors thank all participants for their cooperation during the study, escpecially for Dr. Jia Xue and Mr. Collin Potter (Tulane University, USA), and Dr. Bikash Malla, Dr. Rajani Ghaju Shrestha, Mr. Ocean Thakali, Ms. Niva Sthapit, Mr. Khadga Bahadur Shrestha, and Mr. Bijay Man Shakya (University of Yamanashi, Japan).
Author contributions
S.P.S. collected water samples. S.T. and S.P.S. analyzed the samples and wrote the draft of the manuscript. S.T. and E.H. conducted the data analysis. E.H. revised the manuscript. All authors reviewed and approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Osuolale O, Okoh A. Human enteric bacteria and viruses in five wastewater treatment plants in the Eastern Cape, South Africa. J. Infect. Public. Heal. 2017;10:541–547. doi: 10.1016/j.jiph.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Ramírez-Castillo FY, et al. Waterborne pathogens: Detection methods and challenges. Pathogens. 2015;4:307–334. doi: 10.3390/pathogens4020307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sano D, Ueki Y, Watanabe T, Omura T. Memberance separation of indigenous noroviruses from sewage sludge and treated wastewater. Water Sci. Technol. 2006;54:77–82. doi: 10.2166/wst.2006.451. [DOI] [PubMed] [Google Scholar]
- 4.Amarasiri M, Kitajima M, Nguyen TH, Okabe S, Sano D. Bacteriophage removal efficiency as a validation and operational monitoring tool for virus reduction in wastewater reclamation: review. Water Res. 2017;121:258–269. doi: 10.1016/j.watres.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 5.Gerba CP, Betancourt WQ, Kitajima M, Rock CM. Reducing uncertainty in estimating virus reduction by advanced water treatment processes. Water Res. 2018;133:282–288. doi: 10.1016/j.watres.2018.01.044. [DOI] [PubMed] [Google Scholar]
- 6.Qiu Y, et al. Assessment of human virus removal during municipalwastewater treatment in Edmonton, Canada. J. Appl. Microbiol. 2015;119:1729–1739. doi: 10.1111/jam.12971. [DOI] [PubMed] [Google Scholar]
- 7.Fong TT, Lipp EK. Enteric viruses of humans and animals in aquatic environments: Health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 2005;69:357–371. doi: 10.1128/MMBR.69.2.357-371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerba, C. P., Kitajima, M. & Iker, B. C. Viral presence in waste water and sewage and control methods. In Viruses in Food and Water; Risks, Surveillance and Control (pp. 293–315). Cook, N., Ed.; Woodhead Publishing Ltd. Cambridge, UK (2013).
- 9.Fletcher SM, Stark D, Harkness J, Ellis J. Enteric protozoa in the developed world: A public health perspective. Clin. Microbiol. Rev. 2012;25:420–449. doi: 10.1128/CMR.05038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okoh AI, Sibanda T, Siyabulela S. Inadequately Treated Wastewater as a Source of Human Enteric Viruses in the Environment. Int. J. Env. Res. Public. Health. 2010;7:2620–2637. doi: 10.3390/ijerph7062620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García-Aljaro C, Ballesté E, Muniesa M, Jofre J. Determination of crAssphage in water samples and applicability for tracking human faecal pollution. Microb. Biotechnol. 2017;10:1775–1780. doi: 10.1111/1751-7915.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korajkic A, McMinn BR, Harwood V. Relationships between microbial indicators and pathogens in recreational water settings. Int. J. Env. Res. Public. Health. 2018;15:2842. doi: 10.3390/ijerph15122842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albinana-Gimenez N, et al. Analysis of adenoviruses and polyomaviruses quantified by qPCR as indicators of water quality in source and drinking-water treatment plants. Water Res. 2009;43:2011–2019. doi: 10.1016/j.watres.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 14.Hamza IA, Jurzik L, Uberla K, Wilhelm M. Evaluation of pepper mild mottle virus, human picobirnavirus and Torque teno virus as indicators of fecal contamination in river water. Water Res. 2011;45:1358–1368. doi: 10.1016/j.watres.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 15.Kitajima M, Iker BC, Pepper IL, Gerba CP. Relative abundance and treatment reduction of viruses during wastewater treatment processes identification of potential viral indicators. Sci. Total. Environ. 2014;488:290–296. doi: 10.1016/j.scitotenv.2014.04.087. [DOI] [PubMed] [Google Scholar]
- 16.Haramoto E, et al. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl. Env. Microbiol. 2013;79:7413–7418. doi: 10.1128/AEM.02354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shrestha S, Shrestha S, Shindo J, Sherchand JB, Haramoto E. Virological quality of irrigation water sources and pepper mild mottle virus and tobacco mosaic virus as index of pathogenic virus contamination level. Food Env. Virol. 2018;10:107–120. doi: 10.1007/s12560-017-9324-2. [DOI] [PubMed] [Google Scholar]
- 18.Kitajima M, Rachmadi AT, Iker BC, Haramoto E, Gerba CP. Temporal variations in genotype distribution of human sapoviruses and Aichi virus 1 in wastewater in southern Arizona, United States. J. Appl. Microbiol. 2018;124:1324–1332. doi: 10.1111/jam.13712. [DOI] [PubMed] [Google Scholar]
- 19.Tandukar Sarmila, Sherchand Jeevan, Bhandari Dinesh, Sherchan Samendra, Malla Bikash, Ghaju Shrestha Rajani, Haramoto Eiji. Presence of Human Enteric Viruses, Protozoa, and Indicators of Pathogens in the Bagmati River, Nepal. Pathogens. 2018;7(2):38. doi: 10.3390/pathogens7020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato R, Asami T, Utagawa E, Furumai H, Katayama H. Pepper mild mottle virus as a process indicator at drinking water treatment plants employing coagulation-sedimentation, rapid sand filtration, ozonation, and biological activated carbon treatments in Japan. Water Res. 2018;132:61–70. doi: 10.1016/j.watres.2017.12.068. [DOI] [PubMed] [Google Scholar]
- 21.Reuter G, Boros A, Pankovics P. Review Kobuviruses - a comprehensive review. Rev. Med. Virol. 2011;21:32–41. doi: 10.1002/rmv.677. [DOI] [PubMed] [Google Scholar]
- 22.Schmitz BW, Kitajima M, Campillo ME, Gerba CP, Pepper IL. Virus reduction during advanced bardenpho and conventional wastewater treatment processes. Environ. Sci. Technol. 2016;50:9524–9532. doi: 10.1021/acs.est.6b01384. [DOI] [PubMed] [Google Scholar]
- 23.Hata A, Kitajima M, Katayama H. Occurrence and reduction of human viruses, F-specific RNA coliphage genogroups and microbial indicators at a full-scale wastewater treatment plant in Japan. J. Appl. Microbiol. 2013;114:545–554. doi: 10.1111/jam.12051. [DOI] [PubMed] [Google Scholar]
- 24.Haramoto E, Fujino S, Otagiri M. Distinct behaviors of infectious F-specific RNA coliphage genogroups at a wastewater treatment plant. Sci. Total. Environ. 2015;520:32–38. doi: 10.1016/j.scitotenv.2015.03.034. [DOI] [PubMed] [Google Scholar]
- 25.Abdel-Raoufa N, Al-Homaidanb AA, Ibraheem IBM. Microalgae and wastewater treatment. Saudi J. Biol. Sci. 2012;19:257–275. doi: 10.1016/j.sjbs.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shkoporov AN, et al. CrAsshage represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun. 2018;9:4781. doi: 10.1038/s41467-018-07225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutilh BE, et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 2014;5:1–11. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stachler E, Bibby K. Metagenomic evaluation of the highly abundant human gut bacteriophage crAssphage for source tracking of human fecal pollution. Env. Sci. Technol. Lett. 2014;1:405–409. doi: 10.1021/ez500266s. [DOI] [Google Scholar]
- 29.Stachler E, et al. Quantitative crAssphage PCR assays for human fecal pollution measurement. Env. Sci. Technol. 2017;51:9146–9154. doi: 10.1021/acs.est.7b02703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cinek O, et al. Quantitative CrAssphage real-time PCR assay derived from data of multiple geographically distant populations. J. Med. Virol. 2018;90:767–771. doi: 10.1002/jmv.25012. [DOI] [PubMed] [Google Scholar]
- 31.Liang Y, Jin X, Huang Y, Chen S. Development and application of a real-time polymerase chain reaction assay for detection of a novel gut bacteriophage (crAssphage) J. Med. Virol. 2018;90:464–468. doi: 10.1002/jmv.24974. [DOI] [PubMed] [Google Scholar]
- 32.Stachler E, Akyon B, de Carvalho NA, Ference C, Bibby K. Correlation of crAssphage qPCR markers with culturable and molecular indicators of human fecal pollution in an impacted urban watershed. Env. Sci. Technol. 2018;52:7505–7512. doi: 10.1021/acs.est.8b00638. [DOI] [PubMed] [Google Scholar]
- 33.Ahmed W, et al. Evaluation of the novel crAssphage marker for sewage pollution tracking in storm drain outfalls in Tampa, Florida. Water Res. 2018;131:142–150. doi: 10.1016/j.watres.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 34.Ahmed W, Payyappat S, Cassidy M, Besley C, Power K. Novel crAssphage marker genes ascertain sewage pollution in a recreational lake receiving urban stormwater runoff. Water Res. 2018;145:769–778. doi: 10.1016/j.watres.2018.08.049. [DOI] [PubMed] [Google Scholar]
- 35.Kongprajug A, Mongkolsuk S, Sirikanchana K. CrAssphage as a potential human sewage marker for microbial source tracking in Southeast Asia. Env. Sci. Technol. Lett. 2019;6:159–164. doi: 10.1021/acs.estlett.9b00041. [DOI] [Google Scholar]
- 36.Balleste E, et al. Dynamics of crAssphage as a human source tracking marker inpotentially faecally polluted environments. Water Res. 2019;155:233–244. doi: 10.1016/j.watres.2019.02.042. [DOI] [PubMed] [Google Scholar]
- 37.Farkas K, et al. Critical Evaluation of CrAssphage as a Molecular Marker for HumanDerived Wastewater Contamination in the Aquatic Environment. Food Env. Virol. 2019;11:113–119. doi: 10.1007/s12560-019-09369-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malla B, Gahju RS, Tandukar S, Sherchand JB, Haramoto E. Performance evaluation of human-specifc viral markers and application of pepper mild mottle virus and crAssphage to environmental water samples as fecal pollution markers in the Kathmandu Valley, Nepal. Food Env. Virol. 2019;11:274–287. doi: 10.1007/s12560-019-09389-x. [DOI] [PubMed] [Google Scholar]
- 39.Stachler E, Crank K, Bibby K. Co-occurrence of crAssphage with antibiotic resistance genes in an impacted urban watershed. Env. Sci. Technol. Lett. 2019;6:216–221. doi: 10.1021/acs.estlett.9b00130. [DOI] [Google Scholar]
- 40.Hewitt J, Greening GE, Leonard M, Lewis GD. Evaluation of human adenovirus and human polyomavirus as indicators of human sewage contamination in the aquatic environment. Water Res. 2013;47:6750–6761. doi: 10.1016/j.watres.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 41.Montazeri N, et al. Pathogenic enteric viruses and microbial indicators during secondary treatment of municipal wastewater. Appl. Env. Microbiol. 2015;81:6436–6445. doi: 10.1128/AEM.01218-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alcala A, et al. Molecular detection and characterization of Aichi Viruses in sewage-polluted waters of Venezuela. Appl. Env. Microbiol. 2010;76:4113–4115. doi: 10.1128/AEM.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kitajima M, Haramoto E, Phanuwan C, Katayama H. Prevalence and genetic diversity of Aichi viruses in wastewater and river water in Japan. Appl. Env. Microbiol. 2011;77:2184–2187. doi: 10.1128/AEM.02328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sdiri-Loulizi K, et al. Aichi virus IgG seroprevalence in Tunisia Parallels genomic detection and clinical presentation in children with gastroenteritis. Clin. Vaccine Immunol. 2010;17:1111–1116. doi: 10.1128/CVI.00059-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kitajima M, Sassi HP, Torrey JR. Pepper mild mottle virus as a water quality indicator. npj Clean. Water. 2018;1:19. doi: 10.1038/s41545-018-0019-5. [DOI] [Google Scholar]
- 46.Skraber S, et al. Concentration and diversity of noroviruses detected in Luxembourg wastewaters in 2008–2009. Appl. Env. Microbiol. 2011;77:5566–5568. doi: 10.1128/AEM.00632-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maunula L, et al. Presence of human noro- and adenoviruses in river and treated wastewater, a longitudinal study and method comparison. J. Water Health. 2012;10:87–99. doi: 10.2166/wh.2011.095. [DOI] [PubMed] [Google Scholar]
- 48.Pouillot R, et al. Meta-analysis of the reduction of norovirus and male-specific coliphage concentrations in wastewater treatment plants. Appl. Env. Microbiol. 2015;81:4669–4681. doi: 10.1128/AEM.00509-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haramoto E, et al. Seasonal profiles of human noroviruses and indicator bacteria in wastewater treatment plant in Tokyo, Japan. Water Sci. Technol. 2006;54:301–308. doi: 10.2166/wst.2006.888. [DOI] [PubMed] [Google Scholar]
- 50.Katayama H, et al. One-year monthly quantitative survey of noroviruses, enteroviruses, and adenoviruses in wastewater collected from six plants in Japan. Water Res. 2008;42:1441–1448. doi: 10.1016/j.watres.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 51.Kitajima M, Haramoto E, Phanuwan C, Katayama H. Molecular detection and genotyping of human noroviruses in influent and effluent water at a wastewater treatment plant in Japan. J. Appl. Microbiol. 2012;112:605–613. doi: 10.1111/j.1365-2672.2012.05231.x. [DOI] [PubMed] [Google Scholar]
- 52.Farkas K, et al. Seasonal and spatial dynamics of enteric viruses in wastewater and inriverine and estuarine receiving. Env. Sci. Technol. 2018;634:1174–1183. doi: 10.1016/j.scitotenv.2018.04.038. [DOI] [PubMed] [Google Scholar]
- 53.Kitajima M, Iker BC, Pepper IL, Gerba CP. Relative abundance and 388 treatment reduction of viruses during wastewater treatment processes — Identification of potential viral indicators. Sci. Total. Environ. 2014;488–489:290–296. doi: 10.1016/j.scitotenv.2014.04.087. [DOI] [PubMed] [Google Scholar]
- 54.Dias E, Ebdon J, Taylor H. The application of bacteriophages as novel indicators of viralpathogens in wastewater treatment systems. Water Res. 2018;129:172–179. doi: 10.1016/j.watres.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 55.Hamza H, Rizk NM, Gad MA, Hamza IA. Pepper mild mottle virus in wastewater in Egypt: a potential indicator of wastewater pollution and the efciency of the treatment process. Arch. Virol. 2019;164:2707–2713. doi: 10.1007/s00705-019-04383-x. [DOI] [PubMed] [Google Scholar]
- 56.Grandclement C, et al. From the conventional biological wastewater treatment to hybrid processes, the evaluation of organic micropollutant removal: A review. Water Res. 2017;111:297–317. doi: 10.1016/j.watres.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 57.Worley-Morse T, Mann M, Khunjar W, Olabode L, Gonzalez R. Evaluating the fate of bacterial indicators, viral indicators, and viruses in water resource recovery facilities. Water Env. Res. 2019;91:830–842. doi: 10.1002/wer.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Francy D, et al. Comparative effectiveness of membrane bioreactors,conventional secondary treatment, and chlorine and UV disinfection to remove microorganisms from municipalwastewaters. Water Res. 2012;16:4167–4178. doi: 10.1016/j.watres.2012.04.044. [DOI] [PubMed] [Google Scholar]
- 59.Varela MF, et al. Sapovirus in wastewater treatment plants in Tunisia: Prevalence, removal, and genetic characterization. Appl. Env. Microbiol. 2018;84:e02093–17. doi: 10.1128/AEM.02093-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.da Silva AK, et al. Norovirus removal and particle association in a waste stabilization pond. Env. Sci. Technol. 2008;42:9151–9157. doi: 10.1021/es802787v. [DOI] [PubMed] [Google Scholar]
- 61.Katayama H, Shimasaki A, Ohgaki S. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Env. Microbiol. 2002;68:1033–1039. doi: 10.1128/aem.68.3.1033-1039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heim A, Ebnet C, Harste G, Pring-Åkerblom P. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 2003;70:228–239. doi: 10.1002/jmv.10382. [DOI] [PubMed] [Google Scholar]
- 63.Pal A, Sirota L, Maudru T, Peden K, Lewis AM. Realtime, quantitative PCR assays for the detection of virus-specific DNA in samples with mixed populations of polyomaviruses. J. Virol. Methods. 2006;135:32–42. doi: 10.1016/j.jviromet.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 64.Kitajima M, et al. Development of a reverse transcription-quantitative PCR system for detection and genotyping of Aichi viruses in clinical and environmental samples. Appl. Env. Microbiol. 2013;79:3952–3958. doi: 10.1128/AEM.00820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shieh YS, Wait D, Tai L, Sobsey MD. Methods to remove inhibitors in sewage and other fecal wastes for enterovirus detection by the polymerase chain reaction. J. Virol. Methods. 1995;54:51–66. doi: 10.1016/0166-0934(95)00025-p. [DOI] [PubMed] [Google Scholar]
- 66.Kageyama T, et al. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 2003;41:1548–1557. doi: 10.1128/jcm.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trujillo AA, et al. Use of TaqMan real‐time reverse transcription‐PCR for rapid detection, quantification, and typing of norovirus. J. Clin. Microbiol. 2006;44:1405–1412. doi: 10.1128/JCM.44.4.1405-1412.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang T, et al. RNA viral community in human feces: Prevalence of plant pathogenic viruses. PLoS Biol. 2005;4:108–118. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Balique F, et al. Tobacco mosaic virus in the lungs of mice following intra-tracheal inoculation. PLoS One. 2013;8:1–12. doi: 10.1371/journal.pone.0054993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haramoto E, et al. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 71.Friedman SD, Cooper EM, Calci KR, Genthner FJ. Design and assessment of a real time reverse transcription-PCR method to genotype single-stranded RNA male-specific coliphages (Family Leviviridae) J. Virol. Methods. 2011;173:196–202. doi: 10.1016/j.jviromet.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 72.Carratalà A, et al. A novel tool for specific detection and quantification of chicken/turkey parvoviruses to trace poultry fecal contamination in the environment. Appl. Env. Microbiol. 2012;78:7496–7499. doi: 10.1128/AEM.01283-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haramoto E, Katayama H, Asami M, Akiba M. Development of a novel method for simultaneous concentration of viruses and protozoa from a single water sample. J. Virol. Methods. 2012;182:62–69. doi: 10.1016/j.jviromet.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 74.Tandukar S, et al. Co-infection by waterborne enteric viruses in children with gastroenteritis in Nepal. Healthcare. 2019;7:9. doi: 10.3390/healthcare7010009. [DOI] [PMC free article] [PubMed] [Google Scholar]


