Abstract
The allograft inflammatory factor (AIF) gene family consists of two identified paralogs – AIF1 and AIF1-like (AIF1L). The encoded proteins, AIF1 and AIF1L, are 80% similar in sequence and show conserved tertiary structure. While studies in human populations suggest links between AIF1 and metabolic diseases such as obesity and diabetes, such associations with AIF1L have not been reported. Drawing parallels based on structural similarity, we postulated that AIF1L might contribute to metabolic disorders, and studied it using mouse models. Here we report that AIF1L is expressed in major adipose depots and kidney but was not detectable in liver or skeletal muscle; in notable contrast to AIF1, AIF1L was also not found in spleen. Studies of AIF1L deficient mice showed no obvious postnatal developmental phenotype. In response to high fat diet (HFD) feeding for 6 or 18 weeks, WT and AIF1L deficient mice gained weight similarly, showed no differences in fat or lean mass accumulation, and displayed no changes in energy expenditure or systemic glucose handling. These findings indicate that AIF1L is not essential for the development of obesity or impaired glucose handling due to HFD, and advance understanding of this little-studied gene and its place in the AIF gene family.
Subject terms: Metabolism, Metabolic disorders, Obesity, Preclinical research
Introduction
The founding member of the AIF gene family, AIF1 (also called ionized binding adapter, or IBA1), has been described as an interferon (IFN)-γ responsive pro-inflammatory molecule1. Interestingly, AIF1 has also been linked to metabolic conditions in several human populations: serum AIF1 concentrations correlate with HgbA1c and waist circumference in a Japanese population2, sequence variants near the AIF1 locus are associated with adult obesity in Greeks3, and a recent report characterizes AIF1 as an adipokine secreted by macrophages in the white adipose tissue of obese women4. Furthermore, in vitro studies showed that conditioned media from the RAW 264.7 macrophage cell line overexpressing AIF1 induces pro-inflammatory genes (Tnf-α, IL-6), limits glucose uptake, and inhibits insulin signaling in differentiated 3T3L1 cells – a cellular model of white adipocytes5.
Despite strong structural similarity with AIF16, AIF1L (also called IBA2) – first identified by the Human Genome Project – has not been studied in a metabolic context. The limited literature describing AIF1L function links it to proliferation and migration of breast cancer cells7,8 and regulation of actomyosin contractility and filopodial extensions in podocytes9. Expression of Aif1l has been described in kidney, heart, lung, and brain7, but has not been reported in adipose tissues.
AIF1L has a predicted molecular weight of 17 kDa, with amino acid sequence identity of 63% and similarity of 80% when compared to AIF1. The two proteins belong to the EF hand-bearing superfamily of proteins10, share similar molecular structures, and have indistinguishable F-actin bundling activity in vitro6. Despite the similarities between AIF1 and AIF1L, it remains unclear whether these proteins have cooperative, complementary, or redundant roles in mammalian physiology or pathophysiology. Because of the homology of these members of the AIF family and reported AIF1 associations with metabolism and obesity, we hypothesized that AIF1L might play a role in diet-induced obesity (DIO) and associated glucose insensitivity.
Obesity is among the most important public health challenges of modern times. In the U.S. alone, approximately 2 in 3 adults are overweight or obese, 1 in 3 is obese, and 1 in 20 is morbidly obese. Although traditional views of obesity pathogenesis focused on lifestyle, it is now well established that interactions between diet, environment, and genetic factors play critical roles in the susceptibility and severity of several metabolic diseases, including obesity. In addition, obesity is either associated with or poses an independent risk for the development of several pathophysiological conditions such as cancer11, cardiovascular diseases12, and non-alcoholic-fatty liver disease13.
Obesity is a major risk factor for insulin resistance (IR), which is closely linked to Type 2 diabetes in human subjects. Obesity has also been associated with chronic inflammation in visceral adipose tissue14,15, which is in turn associated with the development of IR. However, emerging evidence and independent studies from different groups show that IR can develop after only 3 days16,17 or 4 weeks of HFD feeding18, whereas an increase in pro-inflammatory macrophages is not seen until 10 weeks of HFD feeding18. Early development of IR is initiated as an acute response to HFD consumption, while long term HFD-induced IR is dependent on adipose tissue inflammation characterized by macrophage infiltration and accumulation16. In addition, IR itself can lead to additional monocyte/macrophage infiltration18; creating a vicious cycle. However, detailed molecular mechanisms defining these events are yet to be identified and understood. Cells of macrophage lineage display remarkable phenotypic plasticity in response to external stimuli19. AIF1 has been primarily characterized as a macrophage cytoplasmic protein that plays a role in phagocytosis, migration, and survival20–23, and it also supports the expression of inflammatory factors including IL-6, IL-12, and IFN-gamma in vivo24. Whether AIF1L also promotes inflammatory signaling or cytokine production is not known.
This is the first study to investigate to report the functional role of AIF1L in the context of metabolism and obesity. Here, we note that AIF1L is detected in the major adipose depots of mice fed with either normal diet (ND) or high fat diet (HFD). We also report that mice lacking AIF1L and fed HFD exhibit no differences in food intake, weight gain, fat mass, or energy expenditure compared to control mice. In addition, we found that loss of AIF1L did not change systemic glucose or insulin sensitivity in 8–9-week-old mice fed ND or HFD-induced glucose insensitivity.
Results
AIF1L is expressed in metabolic tissues of adult mice at baseline and upon HFD feeding
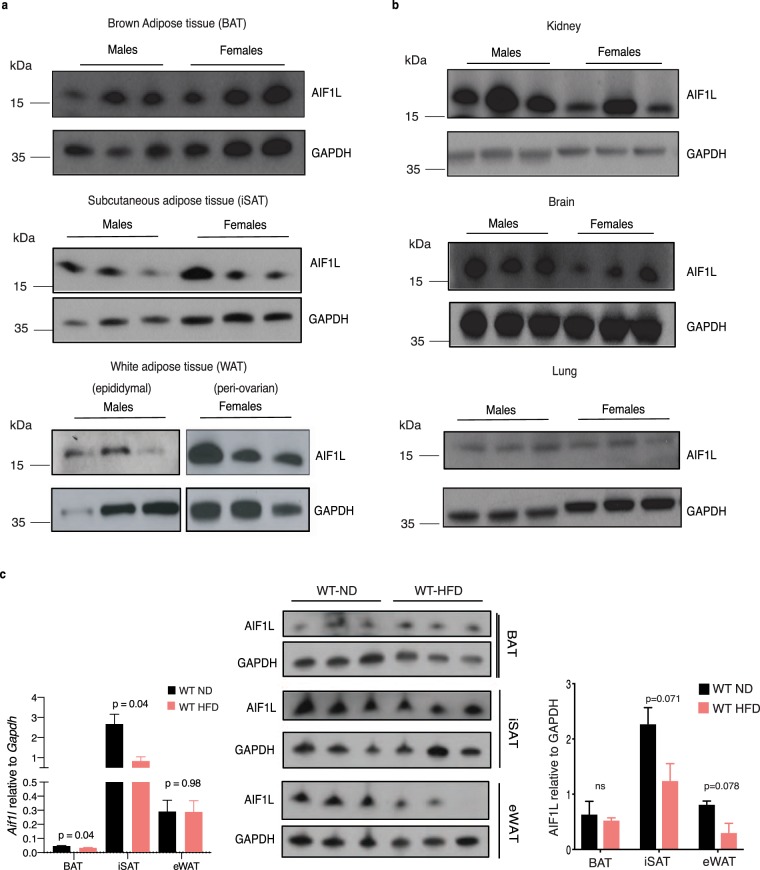
AIF1 is expressed predominantly in testis and immune-associated organs such as spleen25,26, whereas the tissue distribution of AIF1L has not been established. RT-PCR analysis showed Aif1l transcripts in mouse kidney, heart, brain, lung, spleen, and skeletal muscle7, but mRNA expression in adipose tissue has not been reported to date. To permit AIF1L protein detection, we first assessed the specificity of the anti-AIF1L antibody relative to AIF1, using AIF1L-deficient (described below) and AIF1-deficient tissues as controls. Immunoblotting with anti-AIF1L antibody showed an appropriate 17 kDa band in kidney, but not in spleen (where AIF1 is highly expressed), while an anti- AIF1 antibody gave the opposite pattern (Supplementary Fig. S1). This result indicates that this anti-AIF1L antibody can discriminate between AIF1 and AIF1L. In studies of ND-fed WT mice, we found AIF1L expression in all 3 adipose tissue depots – brown (BAT), inguinal subcutaneous (iSAT), and epididymal white adipose tissue (eWAT) (Fig. 1a). In addition to this signal in adipose depots, immunoblotting showed a strong 17 kDa band in kidney (Fig. 1b); this band was lacking in kidney lysates from AIF1L deficient mice(strategy described below and in methods section), confirming its identity as AIF1L (Supplementary Fig. S2). In addition, we observed moderate levels of AIF1L expression in brain, and low levels in lung (Fig. 1b), but no signal in liver or heart (Supplementary Fig. S2). Skeletal muscle from WT mice showed bands of ~14 and 17 kDa in immunoblotting; because similar bands were observed in skeletal muscle from AIF1L deficient mice, these most likely represent cross-reaction with an unknown protein, as further discussed in the figure legend, and we believe that expression of AIF1L in skeletal muscle is very low compared to other organs we tested and undetectable with the methodology used here. Possible cross-reactivity of the anti-AIF1L antibody with AIF1, which also migrates at 17 kDa, was excluded because the same antibody did not cross react with AIF1 in spleen, as noted above (Supplementary Fig. S1) further indicating that the anti-AIF1L antibody is cross-reacting with a protein of ~17 kDa expressed selectively in skeletal muscle that is neither AIF1L nor AIF1. The identity of this cross-reacting protein is unknown. While we did not find any significant differences in AIF1L expression between males and females in BAT, iSAT, and brain, we observed a trend towards increased expression in WAT from females compared to males (Supplementary Fig. S2). Considering the cellular heterogeneity of adipose tissues, we wanted to know if AIF1L is expressed in adipocytes, and used the well-known 3T3L1 cell line, an inducible in vitro model of white adipogenesis. We checked AIF1L expression at baseline, during differentiation, and after differentiation, and found that AIF1L is expressed in each of these stages. Interestingly, we could not detect AIF1L in the macrophage cell line, RAW 264.7, which is known to express AIF11 (Supplementary Fig. S1). Thus, AIF1 and AIF1L show distinct patterns of expression among different cell types.
Figure 1.
AIF1L is expressed in adipose depots, kidney, brain, and lung derived from adult male and female mice. (a) AIF1L protein expression in BAT, SAT and eWAT from 8-week-old adult male (n = 3) and female mice (n = 3). (b) AIF1L protein expression in kidney, brain, and lung. (c) Expression of Aif1l and AIF1L in adipose depots from age matched WT mice fed ND or HFD for 18 weeks, starting at 8 weeks of age (n = 3). Data are represented in mean ± SEM. ns – not significant.
We further assessed if the expression of AIF1L is regulated in adipose depots upon HFD feeding. Tissues harvested from WT mice fed with either ND or HFD for 18 weeks were tested for Aif1l levels using qRT-PCR. We found expression in all three major adipose depots with the lowest in BAT and highest in iSAT from ND-fed WT mice. Upon HFD- feeding, we still found Aif1l expression in these tissues but at a lower level in BAT and iSAT compared to ND-fed WT mice. However, although we found no significant differences in AIF1L protein levels between the diet groups, a trend toward decreased expression was observed in iSAT and eWAT of the HFD group (Fig. 1c).
AIF1L-deficient mice are born in predicted Mendelian ratios, grow normally, breed well, and reach adulthood
AIF1L-deficient mice have not been previously described. We utilized mice bearing a modified Aif1l allele developed by the Wellcome Trust Sanger institute (WTSI)/Knockout Mouse Project (KOMP); this knockout (KO)-first strategy inserts a splice acceptor-lacZ cassette into intron 2 of the Aif1l gene (Supplementary Fig. S3). Mice heterozygous for this allele (Aif1lWT/lacZ-flxneo-flxex3) were mated to obtain WT and homozygous Aif1l “KO-first” mice. Further crosses with E2a-Cre transgenic mice27 induced Cre-mediated recombination in early zygotes to yield a global Aif1l locus allele with the splice acceptor-lacZ cassette but lacking Aif1l exon 3 and the neomycin selection cassette (Supplementary Fig. S3) (Aif1llacZ/lacZ; referred as KO hereafter), in turn resulting in loss of AIF1L expression.
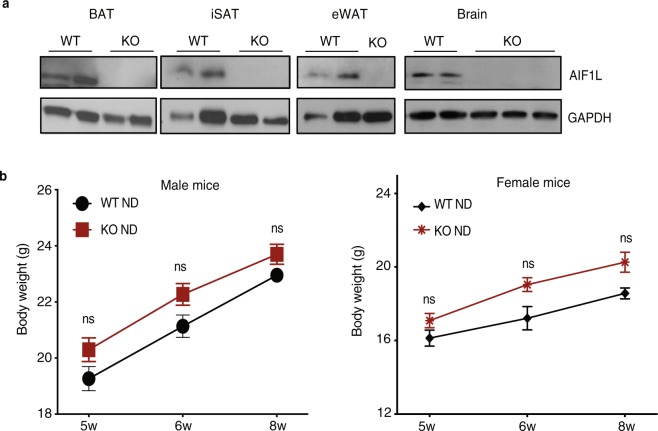
Genotyping by PCR analysis of genomic DNA from mutant mice confirmed the presence of lacZ, absence of the wild-type (WT) allele, and presence of the modified Aif1l allele (Supplementary Fig. S3). As predicted based on the gene targeting strategy, analysis by RT-PCR using primers from Aif1l exons 1 and 2 yielded the expected products in both WT and KO kidney, while analysis with primers spanning exons 1 and 5 detected transcripts in WT, but not KO, kidney (Supplementary Fig. S3). Furthermore, we observed distinct immunohistochemical staining for AIF1L in tubular epithelial cells and podocytes in the glomerulus of WT mice, and lack thereof in the KO mice (Supplementary Fig. S3). Protein lysates from adipose tissues and brain were tested by immunoblotting, which showed a predicted 17 kDa band in wild type animals and lack thereof in KO mice (Fig. 2a). In expanded breeding, we found that KO mice were born in Mendelian ratios (Table 1), reached adulthood, and did not show any obvious phenotypic differences when compared to WT controls after following for 52 weeks. Importantly, WT and KO mice of both sexes showed similar growth and weight gain patterns from 5 to 8 weeks of age when fed a ND (Fig. 2b).
Figure 2.
The modified Aif1l locus in mice results in loss of AIF1L expression and does not affect weight gain from 5 to 8 weeks of age. (a) Immunoblotting for AIF1L in adipose tissues from WT and KO mice (n = 2). (b) AIF1L-deficient male (WT n = 9, KO n = 7) and female (WT n = 3, KO n = 7) mice show similar weight and weight gain from 5 to 8 weeks of age when compared to WT mice on normal diet. Data are represented in mean ± SEM. ns – not significant.
Table 1.
Genotype frequency in the progeny of Aif1lWT/lacZ crossed with Aif1lWT/lacZ.
| Genotype | Expected # | Observed # | Expected % | Observed % |
|---|---|---|---|---|
| Wild type (Aif1lWT/WT) | 30 | 38 | 25 | 31.67 |
| Heterozygous (Aif1lWT/lacZ) | 60 | 54 | 50 | 45 |
| Homozygous (Aif1llacZ/lacZ) | 30 | 28 | 25 | 23.33 |
| Total | 120 | 120 | 100 | 100 |
No significant deviation observed from the expected mendelian ratios; Chi Square = 2.867 with 2 df, p = 0.2385.
To assess for possible compensatory regulation of AIF1 in AIF1L KO mice, we evaluated AIF1 expression levels in the key metabolic organs and spleen from adult mice (8–9 weeks old), using immunoblotting with lysates from bone marrow derived macrophages (BMDM) as controls. We found downregulation of AIF1 in BAT from AIF1L KO mice but a trend towards increased expression in eWAT (Supplementary Fig. S4). These contrasting patterns of AIF1L-dependent AIF1 regulation according to adipose tissue type are curious; the increase in AIF1 levels could reflect compensation in eWAT, but clearly a different mode of regulation is operating in BAT.
Loss of AIF1L does not affect body weight or body composition upon short-term high fat diet (HFD) feeding
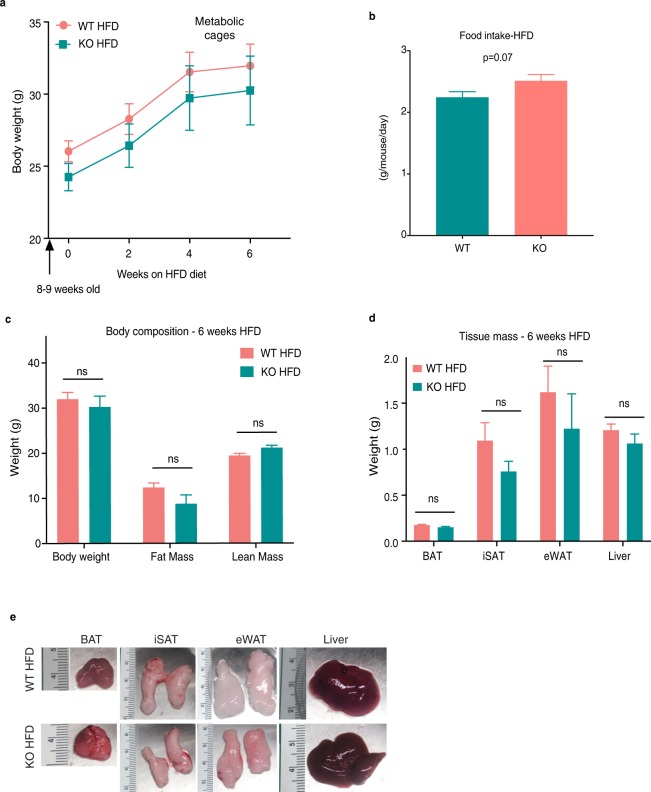
The expression of AIF1L in adipose tissues in both ND-and HFD-fed mice, together with the above-noted associations of its paralog AIF1 with metabolism and obesity, supported our rationale to further evaluate a potential role for AIF1L in obesity and glucose handling. After finding that mice lacking AIF1L showed no differences in growth or weight gain on ND through 8 weeks of age, we subjected adult WT and KO male mice to HFD and followed them for 6 weeks as a short-term HFD stress. We found no differences in weight gain curves (Fig. 3a), food intake (Fig. 3b) or body composition (Fig. 3c) between WT and AIF1L KO mice. Adipose depots (BAT, iSAT, eWAT) and liver were harvested from the KO animals at the end of the study (6 weeks HFD feeding) and did not show any differences in their mass (Fig. 3d) and macroscopic appearance (Fig. 3e), when compared to the WT mice. These results indicate that AIF1L is not essential for increased adiposity and weight gain upon short-term HFD feeding.
Figure 3.
Loss of AIF1L does not affect total weight gain or body composition during short term HFD feeding. (n = 3 for WT and n = 4 for KO). (a) Weight gain over 6 weeks of HFD feeding (Flattening of the curves at week 6 in these mouse cohorts followed a move to the Animal Physiology core facility for subsequent metabolic cage and MRI studies; other cohorts (e.g., Fig. 4a, Supplementary Fig. 5a) used in weight gain or glucose sensitivity studies did not show similar flattening of the weight curve.). (b) Food intake over 5 weeks of HFD feeding (n = 3 for WT and n = 4 for KO). (c) Body weight, fat, and lean mass measurements determined by MRI. (d) Brown (BAT), inguinal subcutaneous (iSAT), epididymal (eWAT) white adipose depot, and liver mass. (e) Macroscopic appearance of tissues - representative images. Data are represented as mean ± SEM. ns – not significant.
Loss of AIF1L does not affect age-dependent or HFD-induced body weight gain and body composition
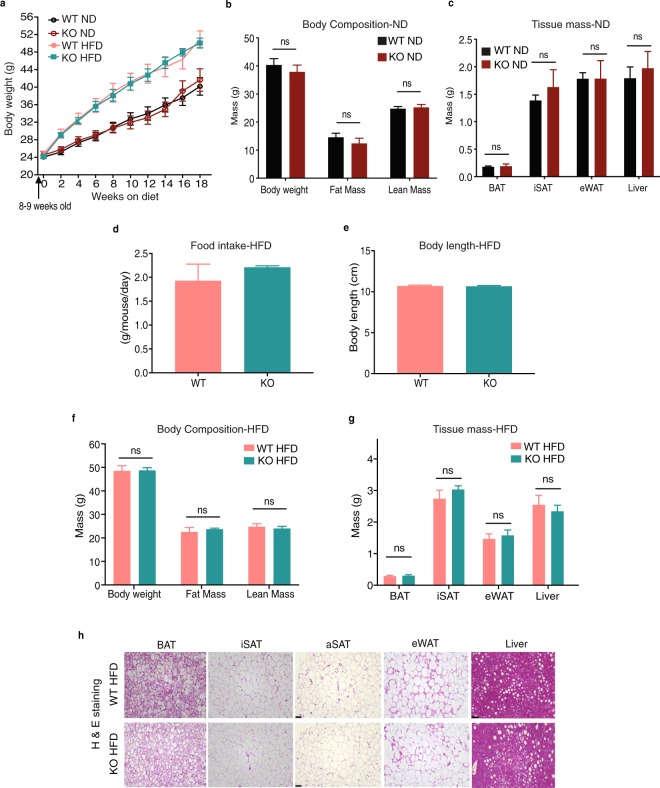
To evaluate the response to a longer metabolic stress, we either continued to follow the WT and KO mice on ND or switched to HFD feeding for 16–18 weeks, starting at 8–9 weeks of age. KO mice being followed on ND up to the age of 26 weeks displayed no change in weight gain curves (Fig. 4a) and body composition as measured by Echo-MRI compared to WT control mice (Fig. 4b). No differences were observed between the 2 groups in their adipose depot and liver mass (Fig. 4c) or the macroscopic appearance of these organs (Supplementary Fig. S5). These results show that AIF1L is not required for expected body weight increases on ND and age-dependent fat accumulation in adult mice.
Figure 4.
AIF1L does not affect total body weight or body composition during long term ND or HFD consumption. (a) Total body weight curves of WT (n = 6) and KO mice fed ND (n = 9) or HFD (n = 11 for WT, n = 8 for KO) starting at 8 weeks of age. (b–c) WT and KO mice on ND; age 26 weeks. (b) Body weight, fat, and lean mass measurements determined by MRI (n = 9 for WT, n = 8 for KO) (c) Brown (BAT), inguinal subcutaneous (iSAT), epididymal (eWAT) white adipose depot, and liver mass (n = 8 for WT, n = 3 for KO). (d–h) WT and KO mice fed with HFD for at least 18 weeks, starting at 8 weeks of age. (d) HFD consumption measured over 33 days (n = 6). (e) Body length (n = 3). (f) Body weight, fat mass and lean mass measured by MRI at the end of HFD feeding period for 18 weeks; age - 26 weeks. (n = 11 for WT, n = 8 for KO). (g) Brown (BAT), inguinal subcutaneous (iSAT), epididymal (eWAT) white adipose depot, and liver mass. (n = 11 for WT, n = 8 for KO). (h) H&E stained sections of adipose depot and liver sections - Representative images. (aSAT-anterior subcutaneous adipose depot). Data are represented as mean ± SEM. ns – not significant.
On HFD, both WT and KO mice gained more weight than with ND, with no genotype-dependent differences in absolute body weight or weight gain over the period of HFD feeding (Fig. 4a). WT and KO mice had similar food consumption, as measured over a period of 33 days (Fig. 4d). The body length of the two groups was similar, suggesting normal overall growth of the animals (Fig. 4e). We also performed micro-computed-tomography (μCT) scans, which showed no differences in fat accumulation in the visceral and subcutaneous adipose depots (Supplementary Fig. S5), and Echo MRI studies, which showed no differences in fat and lean masses between the two groups (Fig. 4f). Further analysis by harvesting and weighing the different adipose depots likewise revealed no differences in their masses and liver mass between WT and KO mice (Fig. 4g, Supplementary Fig. S5). Histological differences could still contribute to overall similar mass of the depots and liver. In that regard, we wanted to check if the observed similar fat accumulation corresponds to similar adipocyte morphology within the adipose tissue between the two groups. Haematoxylin and Eosin (H & E) staining of tissue sections (Fig. 4h) revealed no obvious morphological differences. We further confirmed this by analyzing the size distribution of adipocytes in iSAT and eWAT sections and found no differences between the WT and KO HFD-fed mice. Furthermore, quantification of crown-like structures in these sections revealed no differences between the two groups, suggesting comparable degrees of infiltration of macrophages to the visceral adipose site (Supplementary Fig. S5).
To further characterize the adipose tissue endocrine function, we checked gene expression of some of the major adipokines in iSAT and eWAT at baseline and from HFD-fed mice using quantitative PCR. These studies revealed genotype-dependent differences in adipokine expression: at baseline, leptin, adiponection, and resistin were lower in KO iSAT, consistent with lower serum levels of leptin found in KO animals, and this pattern of expression in adipose tissues was maintained with HFD (Supplementary Fig. S5). In eWAT, on the other hand, adipokine transcripts were not different at baseline, but with HFD, these levels increased, with the largest relative increase noted for leptin. Whether the depot specific mRNA differences observed upon long-term-HFD feeding are translated to differences in serum levels will require further testing.
To uncover any possible sexual dimorphism role for AIF1L, we performed HFD feeding studies in female mice as well and measured the same parameters as mentioned above for male mice and found no differences (Supplementary Fig. S6). Overall, despite these interesting effects on adipokine expression, these results combined suggest that AIF1L is not essential for the development of diet-induced obesity in male and female mice.
Loss of AIF1L has modest effects on metabolic profile and activity in HFD-fed mice
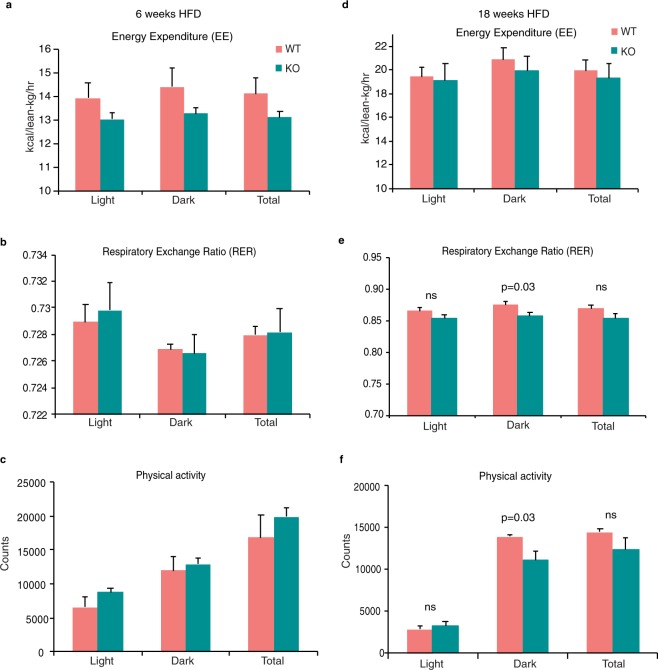
Since we did not observe differences between WT and AIF1L KO female mice upon HFD feeding, and that male mice have both higher sensitivity and lower variability in their susceptibility to metabolic syndrome28, we used male mice for the rest of the study. To determine if loss of AIF1L affects the overall metabolic profile of mice, we measured energy expenditure (EE), respiratory exchange ratio (RER), and physical activity of WT and KO mice after 6 or 18 weeks of HFD feeding. Measurement after 6 weeks of HFD feeding did not display any differences between WT and KO mice in their total EE, fuel source utilization, or physical activity either in the light or the dark cycle (Fig. 5a–c). Measurements over a 24 h period are shown in Supplementary Fig. S7.
Figure 5.
Loss of AIF1L does not alter metabolic profile with short- or long-term HFD feeding. (a–c) Metabolic cage studies performed on WT (n = 3) and KO (n = 4) mice fed HFD for 6 weeks (a) Energy expenditure (EE). Bar graph represents average EE values during day, night, and total. (b) Respiratory exchange ratio (RER; VCO2/VO2). Bar graph represents average RER values during day, night, and total. (c) Physical activity measured as beam breaks (Counts). Bar graph represents average beam break counts values during day, night, and total. (d–f) Metabolic cage studies performed on WT (n = 3) and KO (n = 3) mice fed HFD for 18 weeks. (d) Energy expenditure (EE). Bar graph represents average EE values during day, night, and total. (e) Respiratory exchange ratio (RER; VCO2/VO2). Bar graph represents average RER values during day, night, and total. (f) Physical activity measured as beam breaks (Counts). Bar graph represents average beam break counts values during day, night, and total. Data are represented in mean ± SEM. ns – not significant.
Even upon 18 weeks of HFD feeding, no significant differences were observed in EE between the 2 groups during the light or the dark cycle when monitored over a 24-hour period or average over 5 days of measurements (Fig. 5d, Supplementary Fig. S7). An RER of 0.7 indicates that fatty acid oxidation is the main source of energy, whereas an RER of 1.0 or greater indicates that carbohydrates are the primary fuel source. Although RER values were reduced in KO animals and statistically significant during the dark cycle, when light and dark cycles are collectively analyzed, they were not different (Fig. 5e). Similar observations were found for physical activity, showing different values between WT and KO mice during the dark cycle; with KO animals moving less; however, these differences disappeared when combined (Fig. 5f). Twenty four-hour period measurements for RER and physical activity are shown in Supplementary Fig. S7. These results indicate that loss of AIF1L did not change total energy expenditure after a short course of HFD (6 weeks), but modestly suppressed dark period RER and physical activity after an extended course of HFD (18 weeks).
Loss of AIF1L does not affect glucose handling at baseline or with HFD feeding
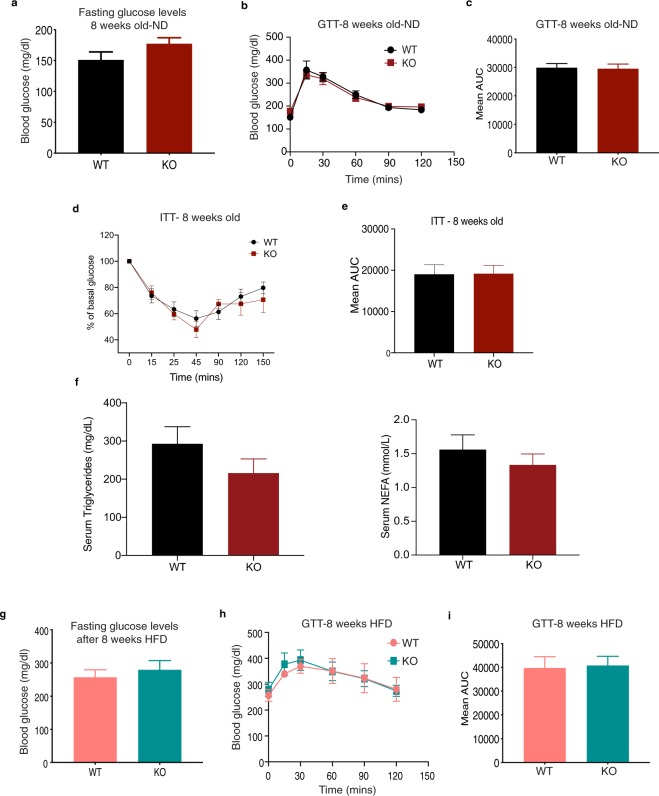
Although obesity (BMI >30) is a major risk factor for the onset of glucose insensitivity, impaired glucose tolerance can also develop independent of the obese state or degree of obesity29. To test if this occurs with loss of AIF1L in mice, we first performed glucose tolerance test (GTT) at baseline – 8 weeks of age, and before starting the HFD feeding. No significant differences were observed between the WT and KO mice in their fasting glucose levels (Fig. 6a), individual glucose measurements with GTT (Fig. 6b) and mean area under the curve (AUC) quantification (Fig. 6c). These results indicate that AIF1L is not required to maintain basal glucose sensitivity. Furthermore, we found that there were no differences between the groups in their glucose handling upon insulin challenge (Fig. 6d,e) indicating similar sensitivity to insulin stimulated glucose clearance. We also checked serum levels of triglycerides (TGs) and non-esterified fatty acids (NEFA) in these mice and found no differences between the two groups (Fig. 6f). These results combined suggest that AIF1L is not essential to maintain the overall metabolic health/key metabolic parameters in adult mice at baseline. We then checked if the HFD-induced stress could reveal a role for AIF1L in glucose handling. HFD feeding-induced glucose intolerance can manifest as early as 4 weeks after starting HFD consumption18. The same cohort of mice used for baseline GTT analysis were subjected to 8 weeks of HFD, after which significant increases in fasting blood glucose levels were observed in both groups compared to the ND-fed mice at age 8 weeks. However, the levels between the genotypes were similar (Fig. 6g). GTTs between the HFD-fed groups were not different either in their individual glucose measurements (Fig. 6h) or AUC calculations (Fig. 6i). These results show that both groups of mice are equally glucose intolerant. To differentiate the effect of age and HFD on glucose handling in mice receiving a HFD for 8 weeks, we performed GTT in 18-week-old male mice. Similar to our findings with mice raised on ND or fed a HFD for 8 weeks, we found no genotype-dependent differences between 18 week-old WT and AIF1L KO mice in terms of fasting blood glucose levels and AUC (Supplementary Fig. S8). Thus, moderate aging did not reveal an effect of AIF1L on glucose handling.
Figure 6.
Loss of AIF1L does not affect glucose handling at baseline, or after short or long term HFD feeding. (a–c) 8-week-old adult male mice on ND. (a) Blood glucose levels in WT (n = 4) and KO (n = 4) mice after overnight fasting. (b) GTT with 1 g/kg (according to body weight) of glucose. (c) Quantification of ipGTT as mean area under curve (AUC). (d–f) 8–12-week-old adult male mice on ND. (d) Drop in glucose levels over time in WT (n = 7) and KO (n = 6) mice upon 0.75U/kg (according to body weight) of insulin challenge. (e) Quantification of ipITT as mean area under curve (AUC). (f) Serum triglycerides and NEFA levels (n = 3). (g–i) 8 weeks of HFD feeding. (g) Blood glucose levels in WT (n = 8) and KO (n = 7) mice after overnight fasting. (h) ipGTT with 0.5 g/kg (according to lean mass) of glucose. (i) Quantification of ipGTT as mean area under curve (AUC). Data are represented as mean ± SEM. ns – not significant.
Discussion
In this study, we report that AIF1L expression in mice is detectable in the 3 major adipose depots – BAT, SAT, and eWAT – and also in kidney, lung, and brain. This corresponds to expression of AIF1L in human samples as reported in the Human Protein Atlas (https://www.proteinatlas.org/ENSG00000126878-AIF1L/tissue), thereby supporting the potential translational relevance of mouse models for understanding this little-studied gene. Using a genetic deletion in the mouse, we found that loss of AIF1L did not affect (1) embryonic development, (2) growth and maintenance of body weight from postnatal stage (5 weeks) to adult stage (8 weeks), (3) body composition or adiposity, (4) energy expenditure, or (5) glucose sensitivity. These findings indicate that AIF1L is not required for maintenance of chief metabolic parameters in mice. We demonstrate that loss of AIF1L does not affect age dependent weight gain and DIO up to age 26 weeks, nor does it modify basal glucose and insulin sensitivity or HFD-induced glucose insensitivity.
We observed a slight reduction in RER values for KO (0.85) mice during the dark period compared to WT mice (0.87), but these values are still within the range of the same substrate utilization as the WT mice. Interestingly, we found reduced physical activity levels during the dark period in AIF1L KO animals after long-term HFD feeding, despite similar body weight and energy expenditure. Given the robust expression of AIF1L in whole brain demonstrated here, and corresponding reports in human samples, these results suggest a possible role for AIF1L in the context of circadian biology bearing on activity. It should also be noted that this difference was only observed upon long term HFD feeding and not in the 6 week HFD-fed cohort; in view of the similar HFD-dependent body composition and metabolic functions among these groups, this difference in activity could reflect an AIF1L-dependent effect of aging rather than metabolism per se.
We also found reduced leptin mRNA expression in iSAT and reduced serum levels from 8–9-week-old AIF1L KO adult male mice when compared to the WT controls. Leptin is a critical regulator of appetite and energy expenditure30,31; while we did not observe any gross food intake differences in either sex when animals are group housed, the slightly lower physical activity observed during the dark phase in KO mice could be due to lower leptin levels in the KO animals. Because proper functioning of circadian rhythm itself regulates leptin transcription via Clock proteins32 and perturbations of such rhythms might be linked to the slight changes in dark-phase-specific physical activity in AIF1L KO animals, it is possible that these three observations are mechanistically linked, pointing to a need for further investigation of this potential novel regulator of leptin.
Loss of AIF1L did not affect glucose insensitivity in HFD-fed mice for 8 weeks. Although HFD-induced glucose intolerance could be seen as early as 4 weeks of HFD feeding17,33, effects of long-term (16–18 weeks) HFD feeding on glucose handling in these mice will require further testing. Considering that inflammation of the adipose organ is one of the key links between obesity and developing glucose intolerance15,34,35, we anticipate similarly impaired response between the control and AIF1L KO mice upon glucose challenge, based on the similar quantitation of crown like structures shown in Supplementary Fig. S5, one of the markers of inflammation.
Tremendous progress has been made in understanding adipose tissue function in the context of metabolic and its associated complications36–38. Despite this progress, how two major cell populations present in adipose tissue – adipocytes and macrophages – interact in an obese state is not fully understood, and this relationship is further complicated by sex-specific differences. Equally critical, the detailed molecular mechanisms by which macrophages in this inflammatory milieu endure and maintain their pro-inflammatory states have yet to be elucidated.
We hypothesized that the characterization of a novel candidate protein, AIF1L, in a mouse model of obesity could help to discover novel regulatory proteins acting in key pathways. Based on our study and published reports on AIF1, it appears that AIF1 and AIF1L proteins are both detectable in adipose tissues, but it is possible that they are expressed in different cell types of this heterogenous endocrine organ. We identified AIF1L expression in 3T3L1 cells under basal conditions and in differentiated white-adipocyte-like cells, but we could not detect it in mouse spleen or immortalized macrophages (Supplementary Fig. S1). In contrast, AIF1 is well characterized in macrophages. Our results also indicate that AIF1 expression in brown and visceral adipose depots at baseline is differentially affected by global loss of AIF1L. Further systematic studies will be required to determine if this trend continues after switching to a HFD and whether this regulation of expression has pathophysiological significance. These differences in expression among distinct cell types within adipose tissues and divergent regulation in different adipose depots also advocate for further investigation of these proteins in adipose tissue biology. The current study provides new information regarding the expression and functional significance of one member of the AIF family. The corresponding role of AIF1L’s paralog – AIF1 – in the same context has yet to be reported.
The downregulation of AIF1L expression observed in the intermediate stage of 3T3L1 cell differentiation suggests that AIF1L might limit adipogenesis and/or lipogenesis. However, our in vivo studies did not show any differences in the overall body weight during the adolescent period – starting at 5 w of age until 8 w of age – as shown in Fig. 2b. Taken together, these observations suggest that factors not present in the 3T3L1 system may compensate for the germline loss of AIF1L in our model and effectively obscure any adipogenesis-related phenotype. A prime candidate for such compensation is AIF1, the paralog of AIF1L; interestingly, AIF1 expression is differentially affected in BAT and eWAT depots by the loss of AIF1L, as shown in Supplementary Fig. S4.
Indeed, the existing literature that addresses AIF1L function is very limited. Recently, AIF1L was shown to enhance cell proliferation via cyclin D1 in cellular models of breast cancer7, and to promote migration when overexpressed in cell lines8. In human subjects with cancer, higher levels of AIF1L correspond to higher tumor grade and worse prognosis7,8. On the other hand, AIF1L has also been shown to have a role in regulating actomyosin contractility and filopodial extensions, in turn maintaining human podocyte morphology and integrity9.
At the protein level, AIF1L and AIF1 show strong conservation of amino acid identity as well as structure6. AIF1 has been extensively characterized as a macrophage cytoplasmic protein that functions in phagocytosis, migration, survival, and serves as a marker of activation in microglia39. This robust expression of AIF1 in macrophages has been linked to pathophysiological conditions such as rheumatoid arthritis (RA)40,41 in human subjects. Mouse models and in vitro studies display a significant role for AIF1 in autoimmune diseases such as experimental autoimmune encephalomyelitis (EAE), animal model of multiple sclerosis24,42, neuritis42,43, and type 1 diabetes in non-obese diabetic mice44. In addition, like AIF1L, AIF1 has also been reported to be upregulated in breast ductal tumor epithelia compared to normal epithelia45, and to promote cell proliferation45 and migration in breast cancer cell lines46. At the molecular level, the only known common function of these paralogs is their actin bundling activity6. These functional observations and disease associations for AIF1, along with the structural similarities of AIF1 and AIF1L led us to explore functions of the latter protein.
A possible limitation of our study is the use of mice with global loss of AIF1L throughout development, which could result in compensation by factors that mask a phenotype. Furthermore, our expression data for AIF1L and differences in leptin expression with AIF1L deficiency suggest a role in adipose tissue biology; spatiotemporally regulated loss of AIF1L could circumvent the problem of compensation and thereby reveal such a role. It is also difficult to fully exclude subtle phenotypes that might be revealed by either tissue specific deletion or by exposure to different metabolic stresses.
In conclusion, we have found that AIF1L is expressed in major metabolic tissues, but is not essential for survival, grossly normal development, or reproduction. In focused studies of AIF1L function in models of metabolic stress, including a HFD model of obesity, we found that loss of AIF1L did not affect maintenance of age-dependent body weight, HFD-induced weight gain, body composition, adipose depot size, or obesity-induced impairment of glucose handling. These findings show that while AIF1L is not required for the pathogenesis of diet induced obesity and glucose insensitivity, its expression pattern in adipose tissues and brain and regulation of AIF1 expression in AIF1L KO mice suggest that this protein family may yet have a regulatory role in this context, and use of temporally-controlled or tissue specific KO models may uncover these anticipated functions.
Materials and Methods
Mice
Mice bearing the Aif1L “KO-first” allele on a C57BL6/N background were obtained from Knockout mouse project repository (KOMP). They were backcrossed for 8 generations to the C57BL6/J strain before mating them with E2a-Cre transgenic mice (Jackson laboratories #003724) to generate AIF1L-deficient mice with the lacZ gene driven by the endogenous Aif1l promoter. All animals were housed in pathogen-free conditions, and procedures were in accordance with the rules and regulations of the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) and were approved by the Institutional Animal Care and Use Committee (IACUC) of the Albert Einstein College of Medicine. At least two of the cohorts used were littermates. Breeders of non- littermate cohorts came from the same founder animals. One of the three cohorts used in the weight gain studies came from breeding Aif1lWT/lacZ; E2a-CreTg with Aif1lWT/lacZ and rest of them came from either Aif1lWT/lacZ with Aif1lWT/lacZ or Aif1lWT/WT with Aif1lWT/WT or Aif1llacZ/lacZ with Aif1llacZ/lacZ.
Genotyping
Genotyping was done using genomic DNA extracted from tail tips using tail lysis buffer and proteinase K with overnight digestion at 56 °C. Proteinase K was deactivated at 92 °C for 20 minutes and samples were centrifuged to remove the debris. Supernatant was used to perform PCR reactions. Primers used for genotyping are mentioned in Table 2. To confirm if the recombination occurred in the mice bearing the E2a-Cre transgene, we designed primers spanning the intron 2-exon 3 junction.
Table 2.
Primers used.
| Primer name | Primer Sequence 5′ to 3′ | Position on modified AIF1L allele |
|---|---|---|
| For genotyping | ||
| Aif1l_39300_F | GCTCCTGCACTCTGCATCTC | 14699–14718 (From WTSI*) |
| Aif1l_39300_R | ACCTTCCCCACTCTGTGCTC | 22236–22255 (From WTSI) |
| CAS_R1_Term | TCGTGGTATCGTTATGCGCC | 15053–15072 (From WTSI) |
| LacZ_2_small_F | ATCACGACGCGCTGTATC | 18203–18220 |
| LacZ_2_small_R | ACATCGGGCAAATAATATCG | 18291–18310 |
| Exon3 forward | GATATCTCTATAGTCGCAGTAGGC | 22147–22161 |
| Exon3 reverse | CTTTGAAGGCTGCGAGCTTTTCC | 22621–22643 |
| For cDNA analysis | Primer Sequence 5′ to 3′ | |
| Exon1_F | ATGTCGGTCGCACTCAGCAA | |
| Exon5_R | CTGACACCCCCTGTCACCTC | |
| Exon2_R | CCGATTGATCTCCTCGAGCCTC | |
| Adiponectin_F | GGTCTTCTTGGTCCTAAGGGTGAG | |
| Adiponectin_R | GCGGCTTCTCCAGGCTCTC | |
| Resistin_F | GCTGCTCCTGTGGCTCTGC | |
| Resistin_R | GGCTGCTGTCCAGTCTATCCTTG | |
| Leptin_F | AGGGAGGAAAATGTGCTGGAGAC | |
| Leptin_R | GATACCGACTGCGTGTGTGAAATG | |
*WTSI- Wellcome trust Sanger Institute.
RT-PCR
Total RNA was isolated from frozen tissues using Trizol reagent (Invitrogen; 15596018) according to manufacturer’s instructions. The RNA concentration was calculated using NanoDrop technology (ThermoFisher Scientific; ND-1000) and reverse transcribed to cDNA using Superscript III first strand synthesis system (Invitrogen 18080-051). Primers that were used to perform PCR are mentioned in Table 2.
Recombination of the modified Aif1l locus with Cre recombinase disrupts splicing after exon 2, leaving the lacZ cassette, and deletes exon 3. Exon 3 encodes aa 32–53 of the predicted AIF1L protein sequence (NP_660126). This overlaps the start of the putative EF hand motif in AIF1L at aa 47–82, so loss of exon 3 after Cre-mediated recombination would likely significantly affect protein structure and function. In addition, in the rare event of a read-through transcript resulting from a skipped splice acceptor in the lacZ cassette, deletion of exon 3 is expected to cause a frame shift after exon 2 in all possible transcripts that include exons 4, 5, and 6, resulting in a premature stop codon. These transcripts would most likely be degraded by nonsense mediated decay (NMD). Finally, in theory it is possible that translation might arise from an alternative start site downstream of exon 3 and result in peptides or truncated proteins with an intact AIF1L C terminus. Such products should be detectable by the antibody we have used, but IHC staining in kidney sections from AIF1L KO mice do not show any signal with this antibody. To confirm the loss of Aif1l mRNA transcript, we designed 2 sets of primers (1) spanning exon1 and exon 2, and (2) spanning exon 1 and exon 5. With this, we expected to see an amplicon resulting from exon 1 and exon 2 in both WT and KO mice, as shown in the Supplementary Fig. 3. With Set 2 we confirmed the loss of full length Aif1l transcript, as demonstrated by the presence of the expected amplicon in WT mice and lack thereof in the KO mice.
Quantitative RT-PCR
RNA and cDNA were prepared as mentioned above. 10 μl reactions with 50 ng of cDNA were set up in triplicates using SYBR green (Applied biosystems, 4309155) and the assay was run in ViiA7 Real time PCR system (Applied biosystems). The data was analyzed with 2−ΔCt method in which ΔCt was calculated between the gene of interest and housekeeping gene, 18S rRNA or Gapdh, as indicated.
Immunoblotting
Frozen tissues were homogenized in RIPA lysis buffer (50 mM Tris-HCL pH 7.4, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 150 mM NaCl) with protease inhibitors (Complete, Roche) and phosphatase inhibitors (Roche; PhosSTOP Easypack 04906837001) for protein extraction and then centrifuged to defat and remove debris. Protein concentrations were measured by a BCA protein assay kit (Pierce). Equal amounts of protein (20–60 µg) were loaded, separated by 10–20% Tricine gel (Invitrogen; EC6625BOX) electrophoresis and blotted onto 0.2 μm pore size PVDF membranes (Immobilon-P, Millipore). After blocking in TBST (Tris pH 8.0, NaCl 150 mM, 0.1% Tween20) plus 5% (w/v) non-fat milk or BSA, blots were incubated at 4 °C overnight with primary antibodies. Signals were detected with horseradish peroxidase- conjugated secondary antibody and chemiluminescence (ECL blotting substrate, Pierce; 32106). Equivalent protein loading was tested with anti-GAPDH antibody (Santa Cruz, sc-25778, 1:5000 dilution). Primary antibodies used: anti-AIF1L (Atlas antibodies, HPA020522, 1:500 dilution), anti-AIF1 (Wako FUJIFILM, 016–20001, Abcam ab178847). Densitometric analysis was performed using ImageJ software.
DIO models
Three different cohorts of animals were used for the studies. For one of the cohorts, mice were monitored and weighed at 5, 6, and 8 weeks of age before starting the HFD studies. At 8 weeks of age, the mice were either continued on ND (PicoLab® Mouse Diet 20 5058) or switched to HFD (Research Diets, D12492i; 60% of kCal from fat) for 18 weeks. The mice were weighed before switching and thereafter every 2 weeks. At the end of the feeding period, mice with a final weight of either 2 SD below or above the mean were excluded (n = 1 for each genotype) from the analysis.
Food intake
A defined amount (2 g/day/mouse) of HFD food pellets were provided ad libitum to mice housed in groups of 3–5, and the amount remaining was measured on the third day. The amount consumed was averaged by the number of mice in the cage. This was done over a period of 33 days, with measurements every alternate day, starting at the first day of HFD feeding.
Body composition and imaging
Lean mass and fat mass were acquired using an EchoMRI-100 Body composition analyzer. µCT scan was performed with the following settings – 0.08 mm, 30 slices, and slow speed.
Indirect calorimetry analysis
Columbus Labs comprehensive lab animal monitoring systems (CLAMS) (Animal Physiology Core at Einstein) was used to collect multiparameter metabolic data, including energy expenditure (EE), physical activity as horizontal and vertical beam breaks, O2 consumption, and CO2 production to calculate respiratory exchange ratio (RER). All parameters were normalized to lean mass of each mouse. Data was continuously collected for 5 days after 48 h of acclimatization in the cages where mice were individually housed. These assays were done after either 5 weeks or 17 weeks of HFD feeding.
Glucose sensitivity
Fasting blood glucose levels and intraperitoneal GTT were measured in tail blood using a OneTouch Ultra 2 glucometer, after fasting the animals overnight. Details of the assay are mentioned in figure legends.
Insulin sensitivity
Adult WT and AIF1L KO mice (8–12 weeks old) were fasted for 10hrs before performing the assay. Fasting glucose levels were noted for both groups before challenging them with an ip injection of 0.75U/kg of insulin (Eli Lilly) according to body weight. Blood glucose levels were measured in tail blood at time points as labeled in the figure.
Serum analysis
Serum from 8–12 weeks old adult mice on normal diet was collected without fasting using standard protocol and stored at −80C until analysis. Triglycerides (TGs) and Non-esterified fatty acids (NEFA) were measured colorimetrically with Beckman Coulter AU480 using Beckman Coulter Triglycerides reagent (OSR60118) & Fuji Film HR Series NEFA-HR solutions. Leptin levels were measured by ELISA according to manufacturer’s instructions (EZML-82K Milipore).
Tissue preparation
Mice (8–9 weeks old) were anesthetized with an ip injection of ketamine and xylazine. Organs were isolated and a small piece from each was immediately frozen for RNA and protein isolation. Remaining parts were fixed in 10% formalin for 24 h and then transferred to 70% ethanol.
Histological analysis
Tissue samples fixed for 24 h were processed and embedded in paraffin at the Einstein Histopathology core facility. They were sectioned at 5 µm and a routine H&E staining was performed. Adipocyte size measurements were performed using Adiposoft plugin for ImageJ with manual supervision and frequency distribution of the cell size was graphed using GraphPad prism. Crown like structures in eWAT were calculated manually using at least 3 fields per mouse.
Statistics
Based on results of a pilot study for body weight gain curves on 60% HFD-fed WT mice, we determined the minimum number of mice per group that would be adequate to ensure 80% power to detect 75% change in mean weight gain, assuming a two-sided test and alpha of 0.05. For single time-point measurement, statistical analyses were performed using unpaired t tests with Welch’s correction for two groups to compare each group with WT mice unless otherwise specified. For repeated measurements, analysis of variance (ANOVA) was performed, with Sidak’s correction tests. AUC was measured for glucose and insulin tolerance tests. Chi square test was done to test statistical differences between observed and expected Mendelian ratios. Statistical analysis for frequency distribution curves were done with nonlinear regression fit gaussian model. All analysis was performed using GraphPad Prism 7 and data were expressed as mean ± SEM.
Supplementary information
Acknowledgements
We would like to thank the Einstein Diabetes Center Animal Physiology core facility for assistance with performing Echo-MRI, CT Scans, and metabolic cage studies. We would also like to thank the histopathology core facility and biomarker analytic research core (BARC) Einstein for help with H&E staining and serum analyte measurements respectively. These studies were supported by NIH grant R01HL128066 to N.E.S.S.
Author contributions
Conception and design N.E.S.S., D.P. Performance of experiments D.P., D.F.R-B., L.E.G., S.J., P.C., V.A. Analysis of data D.P., N.E.S.S. Figures and writing D.P., N.E.S.S. All authors reviewed the manuscript.
Data availability
Data presented here are available upon request, from the corresponding author.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-60433-4.
References
- 1.Sibinga NES, Feinberg MW, Yang H, Werner F, Jain MK. Macrophage-restricted and interferon γ-inducible expression of the allograft inflammatory factor-1 gene requires Pu.1. J. Biol. Chem. 2002;277:16202–16210. doi: 10.1074/jbc.M200935200. [DOI] [PubMed] [Google Scholar]
- 2.Fukui M, et al. The serum concentration of allograft inflammatory factor-1 is correlated with metabolic parameters in healthy subjects. Metabolism. 2012;61:1021–5. doi: 10.1016/j.metabol.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Rouskas K, et al. Common variants in FTO, MC4R, TMEM18, PRL, AIF1, and PCSK1 show evidence of association with adult obesity in the Greek population. Obesity (Silver Spring). 2012;20:389–95. doi: 10.1038/oby.2011.177. [DOI] [PubMed] [Google Scholar]
- 4.Lorente-Cebrián S, et al. Allograft inflammatory factor 1 (AIF-1) is a new human adipokine involved in adipose inflammation in obese women. BMC Endocr. Disord. 2013;13:54. doi: 10.1186/1472-6823-13-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ren J, Lin Y, Tang J, Yue H, Zhao Y. Allograft Inflammatory Factor-1 Mediates Macrophage-Induced Impairment of Insulin Signaling in Adipocytes. Cell. Physiol. Biochem. 2018;47:403–413. doi: 10.1159/000489952. [DOI] [PubMed] [Google Scholar]
- 6.Schulze JO, et al. Structural and functional characterization of human Iba proteins. FEBS J. 2008;275:4627–40. doi: 10.1111/j.1742-4658.2008.06605.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Wang S, Li L. EF hand protein IBA2 promotes cell proliferation in breast cancers via transcriptional control of cyclin D1. Cancer Res. 2016;76(15):4535–45. doi: 10.1158/0008-5472.CAN-15-2927. [DOI] [PubMed] [Google Scholar]
- 8.Liu P, Li W, Hu Y, Jiang Y. Absence of AIF1L contributes to cell migration and a poor prognosis of breast cancer. Onco. Targets. Ther. 2018;11:5485–5498. doi: 10.2147/OTT.S165874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yasuda-Yamahara, M. et al. AIF1L regulates actomyosin contractility and filopodial extensions in human podocytes. Plos One (2018). [DOI] [PMC free article] [PubMed]
- 10.Girard F, Venail J, Schwaller B, Celio MR. The EF-hand Ca2+-binding protein super-family: A genome-wide analysis of gene expression patterns in the adult mouse brain. Neuroscience. 2015;294:116–155. doi: 10.1016/j.neuroscience.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 11.Colditz GA, Lindsay L. Obesity and cancer: Evidence, impact, and future directions. Clin. Chem. 2018;64:154–162. doi: 10.1373/clinchem.2017.277376. [DOI] [PubMed] [Google Scholar]
- 12.Hubert HB, Feinleib M, McNamara PM, Castelli WP. Obesity as an independent risk factor for cardiovascular disease: a 26- year follow-up of participants in the Framingham Heart Study. Circulation. 1983;67:968–977. doi: 10.1161/01.CIR.67.5.968. [DOI] [PubMed] [Google Scholar]
- 13.Li L, et al. Obesity is an independent risk factor for non-alcoholic fatty liver disease: Evidence from a meta-analysis of 21 cohort studies. Obes. Rev. 2016;17:510–519. doi: 10.1111/obr.12407. [DOI] [PubMed] [Google Scholar]
- 14.Weisberg, S. P. et al. Obesity is associated with macrophage accumulation. 112 (2003). [DOI] [PMC free article] [PubMed]
- 15.Xu H, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invest. 2003;112:1821–1830. doi: 10.1172/JCI200319451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ham M, et al. Inflammation Is Necessary for Long-Term but Not Short-Term High-Fat Diet-Induced Insulin Resistance. Diabetes. 2011;60:2474–2483. doi: 10.2337/db11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams Lynda M., Campbell Fiona M., Drew Janice E., Koch Christiane, Hoggard Nigel, Rees William D., Kamolrat Torkamol, Thi Ngo Ha, Steffensen Inger-Lise, Gray Stuart R., Tups Alexander. The Development of Diet-Induced Obesity and Glucose Intolerance in C57Bl/6 Mice on a High-Fat Diet Consists of Distinct Phases. PLoS ONE. 2014;9(8):e106159. doi: 10.1371/journal.pone.0106159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimobayashi M, et al. Insulin resistance causes inflammation in adipose tissue. J. Clin. Invest. 2018;128:1538–1550. doi: 10.1172/JCI96139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mosser DM, Edwards JP. NIH Public Access. Genetics. 2009;8:958–969. [Google Scholar]
- 20.Kanazawa H. Macrophage/Microglia-specific Protein Iba1 Enhances Membrane Ruffling and Rac Activation via Phospholipase C-gamma -dependent Pathway. J. Biol. Chem. 2002;277:20026–20032. doi: 10.1074/jbc.M109218200. [DOI] [PubMed] [Google Scholar]
- 21.Yang ZF, et al. Allograft inflammatory factor-1 (AIF-1) is crucial for the survival and pro-inflammatory activity of macrophages. Int. Immunol. 2005;17:1391–1397. doi: 10.1093/intimm/dxh316. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki Y, Ohsawa K, Kanazawa H, Kohsaka S, Imai Y. Iba1 is an actin-cross-linking protein in macrophages/microglia. Biochem. Biophys. Res. Commun. 2001;286:292–297. doi: 10.1006/bbrc.2001.5388. [DOI] [PubMed] [Google Scholar]
- 23.Ohsawa K, Imai Y, Kanazawa H, Sasaki Y, Kohsaka S. Involvement of Iba1 in membrane ruffling and phagocytosis of macrophages/microglia. J. Cell Sci. 2000;113(Pt 1):3073–3084. doi: 10.1242/jcs.113.17.3073. [DOI] [PubMed] [Google Scholar]
- 24.Chinnasamy P, et al. Loss of Allograft Inflammatory Factor-1 Ameliorates Experimental Autoimmune Encephalomyelitis by Limiting Encephalitogenic CD4 T-Cell Expansion. Mol. Med. 2015;21:233–41. doi: 10.2119/molmed.2014.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Utans U, Arceci RJ, Yamashita Y, Russell ME. Cloning and characterization of allograft inflammatory factor-1: A novel macrophage factor identified in rat cardiac allografts with chronic rejection. J. Clin. Invest. 1995;95:2954–2962. doi: 10.1172/JCI118003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casimiro, I., Chinnasamy, P. & Sibinga, N. E. S. Genetic inactivation of the allograft inflammatory factor-1 locus. Genesis (2013). [DOI] [PMC free article] [PubMed]
- 27.Lakso M, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong J, Stubbins RE, Smith RR, Harvey AE, Nú̃ez NP. Differential susceptibility to obesity between male, female and ovariectomized female mice. Nutr. J. 2009;8:1–5. doi: 10.1186/1475-2891-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hospital H, Hospital K. Diabetologia. 1991;2:483–487. [Google Scholar]
- 30.Maffei M, et al. Leptin levels in human and rodent: Measurement of plasma leptin and ob RNA in obese and weight-reduced subjects. Nat Med. 1995;1:1155–1161. doi: 10.1038/nm1195-1155. [DOI] [PubMed] [Google Scholar]
- 31.Park HK, Ahima RS. Physiology of leptin: energy homeostasis, neuroendocrine function and metabolism. Metabolism. 2015;64:24–34. doi: 10.1016/j.metabol.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kettner NM, et al. Circadian dysfunction induces leptin resistance in mice. Cell Metab. 2015;22:448–459. doi: 10.1016/j.cmet.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta D, et al. Temporal characterization of β cell-adaptive and -maladaptive mechanisms during chronic high-fat feeding in C57BL/6NTac mice. J. Biol. Chem. 2017;292:12449–12459. doi: 10.1074/jbc.M117.781047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patsouris D, et al. Ablation of CD11c-Positive Cells Normalizes Insulin Sensitivity in Obese Insulin Resistant Animals. Cell Metab. 2008;8:301–309. doi: 10.1016/j.cmet.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saberi M, et al. Hematopoietic Cell-Specific Deletion of Toll-like Receptor 4 Ameliorates Hepatic and Adipose Tissue Insulin Resistance in High-Fat-Fed Mice. Cell Metab. 2009;10:419–429. doi: 10.1016/j.cmet.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gómez-Hernández A, Beneit N, Díaz-Castroverde S, Escribano Ó. Differential Role of Adipose Tissues in Obesity and Related Metabolic and Vascular Complications. Int. J. Endocrinol. 2016;2016:1–15. doi: 10.1155/2016/1216783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reilly SM, Saltiel AR. Adapting to obesity with adipose tissue inflammation. Nat. Rev. Endocrinol. 2017;13:633–643. doi: 10.1038/nrendo.2017.90. [DOI] [PubMed] [Google Scholar]
- 38.Kusminski CM, Bickel PE, Scherer PE. Targeting adipose tissue in the treatment of obesity-associated diabetes. Nat. Rev. Drug Discov. 2016;15:639–660. doi: 10.1038/nrd.2016.75. [DOI] [PubMed] [Google Scholar]
- 39.Ahmed Z, et al. Actin-binding proteins coronin-1a and IBA-1 are effective microglial markers for immunohistochemistry. J. Histochem. Cytochem. 2007;55:687–700. doi: 10.1369/jhc.6A7156.2007. [DOI] [PubMed] [Google Scholar]
- 40.Kimura M, et al. A critical role for allograft inflammatory factor-1 in the pathogenesis of rheumatoid arthritis. J. Immunol. 2007;178:3316–3322. doi: 10.4049/jimmunol.178.5.3316. [DOI] [PubMed] [Google Scholar]
- 41.Pawlik, A. et al. Association of allograft inflammatory factor-1 gene polymorphism with rheumatoid arthritis. Tissue Antigens (2008). [DOI] [PubMed]
- 42.Schluesener, H. J., Seid, K., Kretzschmar, J. & Meyermann, R. Allograft-inflammatory factor-1 in rat experimental autoimmune encephalomyelitis, neuritis, and uveitis: Expression by activated macrophages and microglial cells. Glia (1998). [DOI] [PubMed]
- 43.Pashenkov et al. Augmented expression of daintain/allograft inflammatory factor-1 is associated with clinical disease: Dynamics of daintain/allograft inflammatory factor-1 expression in spleen, peripheral nerves and sera during experimental autoimmune neuritis. Scand. J. Immunol. (2000). [DOI] [PubMed]
- 44.Zhao Y-Y, Huang X-Y, Chen Z-W. Daintain/AIF-1 (Allograft Inflammatory Factor-1) accelerates type 1 diabetes in NOD mice. Biochem. Biophys. Res. Commun. 2012;427:513–7. doi: 10.1016/j.bbrc.2012.09.087. [DOI] [PubMed] [Google Scholar]
- 45.Liu S, et al. Daintain/AIF-1 promotes breast cancer proliferation via activation of the NF-κB/cyclin D1 pathway and facilitates tumor growth. Cancer Sci. 2008;99:952–957. doi: 10.1111/j.1349-7006.2008.00787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, T. et al. Daintain/AIF-1 promotes breast cancer cell migration by up-regulated TNF-α via activate p38 MAPK signaling pathway. Breast Cancer Res. Treat. (2012). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data presented here are available upon request, from the corresponding author.