Abstract
Gastrointestinal parasites can induce low productivity in livestock by causing acute or chronic enteritis. Veterinarians make great efforts to design rational and effective hygienic protocols for both the prevention and treatment of diarrhea. Although prevalences can vary depending on the examined areas or the ages of the hosts, and the methods used for detections, it is helpful to accumulate data across many areas to evaluate parasitic distribution. A coprological survey in cattle was conducted in Tangerang, Banten Province of Indonesia, in order to determine the prevalence of the parasites, including those of diarrhea-associated diseases. Furthermore, the risk of transmission of Giardia intestinalis and Cryptosporidium spp. to human was genetically analyzed. Gastrointestinal parasites were detected in 87 of 109 cattle samples, including 85 carrying Eimeria spp., 36 carrying Fasciola gigantica, 35 carrying Strongyloides spp., 33 carrying Paramphistomum spp., and 15 carrying Capillaria spp. Giardia intestinalis and Cryptosporidium spp., parasites with zoonotic potential, were detected in 9 and 1 cattle samples, respectively. Molecular analyses identified the G. intestinalis isolate as a member of Assemblage E, which has been recently detected in humans in another country. These results may be helpful in understanding the hygienic risk affecting the livestock productivity and zoonotic potential of cattle in Indonesia.
Keywords: Assemblage E; Cryptosporidium; Giardia intestinalis; Indonesia, Tangerang
Introduction
Gastrointestinal parasites, including Protozoa, Nematoda, and Trematoda, can negatively affect productivity in livestock (Corwin 1997; Grisi et al. 2014). These parasites cause acute or chronic enteritis and lead to watery or bloody (and sometimes lethal) diarrhea, especially in younger animals like calves and chicks. Thus, parasitic infections often result in economic losses due to reduced weight gain of the hosts or the need to administer therapeutic treatments (Cho and Yoon 2014). The infected hosts shed environmentally robust stages of the parasites in feces, including oocysts or cysts for the Protozoa and ova or encysted larvae for the Nematoda and Trematoda. These stages are resistant to commonly used disinfectants and can survive for several months or more, potentially being transmitted to other hosts that in turn serve as potential sources of further infection. Therefore, veterinarians and farmers make great efforts to determine the prevalence of such infections, and to design rational and effective hygienic protocols for both the prevention and treatment of diarrhea (Smith 2015).
To date, four surveys of gastrointestinal parasites have (to our knowledge) been conducted in cattle in some areas of Indonesia, including West Java, Central Java, and East Java (Estuningsih et al. 2009; Ananta et al. 2014; Ekawasti et al. 2019; Hastutiek et al. 2019). Regarding Protozoa, Eimeria spp. were the most frequently detected, and the prevalences were reported to range from 22.4 to 53.4%; in contrast, Cryptosporidium spp. were rarely found (0.5% and 0.6%) (Ananta et al. 2014; Ekawasti et al. 2019; Hastutiek et al. 2019). However, very limited information is available for other protozoan parasites, including Giardia intestinalis, and nematodes and trematodes in Indonesia. Because prevalences can vary depending on a number of parameters, such as the examined areas, the ages of the hosts, and the methods used for detections, it is helpful to accumulate data across many areas to evaluate parasitic distribution.
In Indonesia, saturated salt flotation methods like the Whitlock and McMaster method are commonly used due to their simplicity and rapidity. In the present study, we examined the protozoan parasites using the sugar flotation method, which is more sensitive (Ekawasti et al. 2019), and conducted immunofluorescence assays (IFAs) to confirm the presence of Cryptosporidium spp. and G. intestinalis. Here, intestinal parasites in cattle were investigated by the flotation method for Protozoa and Nematoda and by the sedimentation method for Trematoda, to understand the prevalence and hygiene status in cattle toward the improvement of production. Additionally, among these parasites, Cryptosporidium spp. and G. intestinalis are known to infect humans and a wide range of animals. To assess the potential zoonotic risk of transmission from animals to humans, molecular analyses such as PCR and sequencing are needed. Because genetic identification of detected parasites typically have not been performed previously in Indonesia, further molecular analysis of the isolates of Giardia and Cryptosporidium spp. was attempted to determine the species/genotype and potential risk to humans.
Materials and methods
Study area and samples
During February 2017, we collected 109 fecal samples from adult beef cattle in 11 villages of the Tangerang district of the Banten Province, located on the island of Java, Indonesia. The average annual temperature is 25 °C. No animals showed clinical symptoms when fecal samples were collected. All samples were taken from the rectums of cattle, placed in individual plastic bags, and stored at 4 °C until laboratory examination.
Fecal examinations
Fecal samples were examined by the sugar flotation centrifugation method, which was modified based on a previous report (Matsubayashi et al. 2005; Fujino et al. 2006), and by a sedimentation method (Charlier et al. 2008). For the flotation method, 1 g of fecal sample was diluted in 9 mL of distilled water and centrifuged at 800 × g for 5 min. The supernatant was discarded and 10 mL of sugar solution with a specific gravity of 1.2 [e.g., 100 g of sugar (Gulaku Indonesia, Lampung, Indonesia) was added into the 120 mL of distilled water] was added to the sediment, followed by centrifugation at 800 × g for 5 min. The floated material was transferred to a glass slide. We examined the entire smear by light microscopy under 200× or 400× magnification. For detection of trematode eggs, 3 grams of fecal samples were used for the sedimentation method, as reported previously (Charlier et al. 2008). Briefly, weighed feces were mixed in 250 mL of water in a measuring cup and filtered through a tea sieve. Filtrates were allowed to stand for at least 10 min to allow the eggs to settle; and the supernatant then was discarded. This step was repeated twice. Finally, the collected sediments were stained with 5% methylene blue and observed by microscopy.
To confirm the presence of Giardia cysts or Cryptosporidium oocysts, the parasites were purified using the remaining feces (approximately 5–10 g). Briefly, feces were diluted in distilled water and filtered through a steel mesh. After centrifugation at 800 × g for 5 min, the above sugar solution was added to the sediments, and the mixture was overlaid with distilled water; the gradient then was centrifuged at 1200 × g for 10 min. The oocysts or cysts that floated on the surface of the sugar solution were recovered using a Pasteur pipette and were washed three times with distilled water. Finally, the purified oocysts were resuspended with 1–2 mL of phosphate-buffered saline and stored at 4 °C. The IFA was conducted using 10–20 μL of purified samples with a commercial Cryptosporidium/Giardia detection kit (EasyStain™; BTF, Australia) according to the manufacturer’s instructions.
Molecular identification of Giardia and Cryptosporidium spp.
For DNA extraction, 400 µL of Giardia cysts or Cryptosporidium oocysts purified as described above were used. The cysts/oocysts were subjected to five cycles of freeze–thaw to release the genomic DNA, and the resulting suspensions were centrifuged at 5400 × g for 3 min. An aliquot (200 µL) of each of the resulting supernatants then was processed using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
For identification of G. intestinalis, diagnostic fragments were amplified using primer pairs G7 and G759, which are specific for the Giardia β-giardin gene (yielding a fragment of approximately 760 bp) (Cacciò et al. 2002) and GDH1 and GDH4, which are specific for the glutamate dehydrogenase gene (GDH) (yielding a fragment of approximately 770 bp) (Homan et al. 1998). For identification of Cryptosporidium spp., nested PCR with primer pairs F1 and R1 and F2 and R2 (targeting the Cryptosporidium 18S ribosomal RNA (18S rRNA) gene; yielding a fragment of approximately 770 bp) were performed as previously described (Xiao et al. 1999; Nagano et al. 2007). Additionally, PCR targeting for Cryptosporidium oocyst wall protein (COWP) genes (Spano et al. 1997) was conducted. All of the PCR-negative samples were amplified using Illustra GenomiPhi DNA Amplification Kit (GE Healthcare Life Sciences, Tokyo, Japan), and the PCRs were performed again using amplified DNAs.
PCR products were subjected to electrophoretic separation on an agarose gel, stained with ethidium bromide, and visualized on a UV transilluminator. PCR products from positive samples were sequenced in both directions using the amplifying primers and an ABI 3130 automated sequencer (Applied Biosystems Inc., CA, USA). The obtained sequences were aligned with the representative nucleotide sequences deposited in GenBank (https://www.ncbi.nlm.nih.gov/) using ClustalW with initial fixed parameter values (DNA Data Bank of Japan (DDBJ), http://clustalw.ddbj.njg.ac.jp/top-j.html). Homology searches using the obtained partial gene sequences were performed using the FASTA program (http://www.ddbj.nig.ac.jp/search/fasta-j.html) (Yui et al. 2014).
Results
Fecal samples from 30 farms in Tangerang, Banten province, Indonesia, were screened for the presence of gastrointestinal parasites. Among the 109 examined samples, 87 were positive (Table 1). The most prevalent parasites were Eimeria spp. (78.0%), followed by Fasciola gigantica (33.0%), Strongyloides spp. (32.1%), and Paramphistomum spp. (30.3%). Although the species of Eimeria could not be identified based on the morphological examinations, oval-shaped oocysts at a size of approximately 25 μm (resembling those of E. bovis) and spherical oocysts at a size of approximately 18 μm (resembling those of like E. zuernii) were detected. The cysts of G. intestinalis and oocysts of Cryptosporidium spp. (morphologically resembling C. parvum) were found in 9 and 1 samples, respectively, by IFA.
Table 1.
Prevalence of gastrointestinal parasite infection in cattle in the Tangerang district of the Banten province, Indonesia
| Sub district | Names of villages | Nos. of farms | Nos. of cattle | Positive nos. of parasites | Protozoan | Nematoda | Trematoda | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Eimeria spp. | Gi*1 | Cp*2 | Strongyloides spp. | Capillaria spp. | Trichuris spp. | Fasciola gigantica | Paramphistomum spp. | |||||
| Panongan | Serdang Kulon | 6 | 12 | 12 | 12 | 1 | 0 | 9 | 7 | 0 | 10 | 7 |
| Mekar Jaya | 2 | 4 | 4 | 4 | 0 | 0 | 4 | 3 | 1 | 0 | 1 | |
| Ranca Iyuh | 1 | 7 | 7 | 7 | 0 | 0 | 4 | 0 | 1 | 2 | 2 | |
| Ranca Kelapa | 1 | 3 | 3 | 3 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | |
| Ciakar | 1 | 3 | 3 | 3 | 1 | 0 | 0 | 0 | 0 | 3 | 3 | |
| Tigaraksa | Cileles | 10 | 38 | 33 | 28 | 4 | 1 | 18 | 4 | 1 | 12 | 14 |
| Margasari | 2 | 8 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Bantar Panjang | 1 | 4 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | |
| Kadu Agung | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Solear | Cikareo | 4 | 18 | 16 | 14 | 2*3 | 0 | 9 | 1 | 0 | 7 | 4 |
| Cisauk | Dandang | 1 | 9 | 8 | 8 | 0 | 0 | 6 | 1 | 0 | 0 | 0 |
| Total (%) | 30 | 109 | 93 (85.3) | 83 (76.1) | 9 (8.3) | 1 (0.9) | 52 (47.7) | 16 (14.7) | 3 (2.8) | 36 (33.0) | 31 (28.4) | |
*1Gi; Giardia intestinalis, *2Cp; Cryptosporidium spp., *3One of samples was successfully identified as Assemblage E
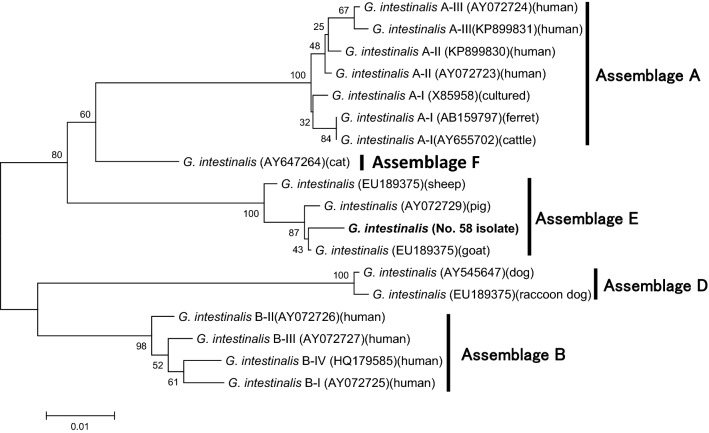
By PCR analyses, only a product specific for the Giardia β-giardin gene was obtained. Notably, this amplicon was obtained in only one (isolate No. 58) of the nine samples that had tested positive for G. intestinalis by IFA. The sequence of the isolates (Accession No. LC484286) possessed 2-bp or 4-bp differences from those of previous data [Assemblage E from pig in Czech Republic (Accession No. AY072729) or Assemblage E from sheep (Accession No. DQ116624)], respectively (Cacciò et al. 2002; Di Giovanni et al. 2006). The phylogenetic analyses revealed that the isolate was sorted with the cluster of Assemblage E (Fig. 1). The products of the other samples G. intestinalis samples and one Cryptosporidium spp. sample were not successfully amplified by PCR.
Fig. 1.
Phylogenetic tree of Giardia intestinalis inferred by the neighbor-joining method using partial Giardia β-giardin gene sequences. Accession numbers and source hosts are shown in parentheses. The scale bar represents substitutions per nucleotide, and bootstrap values are indicated (N = 1000)
Discussion
Gastrointestinal parasites in cattle reared in the Tangerang district of Banten province, Indonesia, were investigated coprologically. Eimeria infections were highly common in the examined areas, representing the most abundant class of parasites detected. Although a molecular analysis of these Eimeria isolates could not be conducted, the oocysts were similar in size and morphology to those of E. bovis and E. zuernii, known pathogenic species. Recently, it has been reported that E. bovis is highly prevalent in other areas of Indonesia (Ekawasti et al. 2019). Therefore, the Eimeria spp. identified in the present study might represent potentially lethal agents capable of causing bloody diarrhea, especially in younger calves. The second-most-prevalent class of parasites, present in approximately 30% of the examined cattle, were Strongyloides spp., Fasciola gigantica, and/or Paramphistomum spp. None of these cattle exhibited clinical symptoms when the feces were collected. Thus, these animals may have been lightly infected with the parasites or may have possessed acquired immunity against the parasites. Furthermore, the parasites may not have been spreading to other animals because the farms investigated were relatively small and managed by the family units, harboring < 10 cattle each (data not shown).
Giardia intestinalis and Cryptosporidium were detected in 9 and 1 cattle, respectively. We were able to PCR amplify a corresponding organism-specific product from only one of the samples that were positive by IFA. This failure to obtain PCR products may indicate that that the number of the cysts and oocysts was small, that the genomic DNAs were recovered at concentrations too low to permit PCR, or that the extracted DNAs contain some PCR inhibitors. However, the successful PCR amplification did permit the first (to our knowledge) identification of an isolate from Assemblage E of G. intestinalis isolate from cattle in Indonesia. G. intestinalis is a species complex consisting of eight assemblages (A–H), with assemblages A and B the dominant assemblages in humans (Ryan and Zahedi, 2019). Notably, while Assemblage E is known to infect hoofed livestock, isolates of this assemblage recently have been shown to have zoonotic potential (Abdel-Moein and Saeed 2016). Although there have been some reports of G. intestinalis infections in humans and animals in Indonesia, only Assemblage B previously has been detected in this country, specifically in orangutans (Mynářová et al. 2016). Additionally, Cryptosporidium spp. has been detected in humans from several areas of Indonesia, including Jakarta (Katsumata et al. 2000; Kurniawan et al. 2009 and 2013), which is close to the Tangerang district examined in the present study. Therefore, further genetic epidemiological surveys across wider geographic regions are needed to assess the zoonotic risk of these parasites.
Conclusion
To our knowledge, only four papers have reported surveys of the parasites in cattle in Indonesia, specifically in West Java, Central Java, East Java, and Yogyakarta as described in Introduction. Although we cannot easily compare the prevalences among these various reports, all of these results together are expected to facilitate our understanding of the potential risks affecting livestock production and human health in Indonesia.
Acknowledgements
The authors gratefully acknowledge Mrs. Rika Sekiguchi and Mrs. Noriko Asama for helping with the fecal and molecular examinations. This study was supported, in part, by a Grant-in-Aid from the Japan Society for the Promotion of Science (JSPS) (Grant No. 16H05803).
Author’s contribution
SDH, WAH, ME, EF, DDA, and MM collected the fecal samples, examined the parasites, and summarized the results and information of the farm. WAH and UBN organized the survey at the examined areas. ST, KM, SK, and MM performed molecular analyses of the parasites. SDH, TM, and MM mainly provided the manuscript. SDH, TM, WAH and MM equally contributed in the manuscript writing. All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The research work was conducted under the Indonesian Research Center for Veterinary Science, Bogor, Indonesia. The handling of animals in the study was approved by the ethics committee of Indonesia Agency for Agricultural Research and Development, Ministry of Agriculture, Indonesia (No. Balitbangtan/BB Litvet/AT_HL/01.01/2017).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
April Hari Wardhana, Email: wardhana24id@yahoo.com.
Makoto Matsubayashi, Email: matsubayashi@vet.osakafu-u.ac.jp.
References
- Abdel-Moein KA, Saeed H. The zoonotic potential of Giardia intestinalis assemblage E in rural settings. Parasitol Res. 2016;115:3197–3202. doi: 10.1007/s00436-016-5081-7. [DOI] [PubMed] [Google Scholar]
- Ananta SM, Suharno Hidayat A, Matsubayashi M. Survey on gastrointestinal parasites and detection of Cryptosporidium spp. on cattle in West Java, Indonesia. Asian Pac J Trop Med. 2014;7:197–201. doi: 10.1016/S1995-7645(14)60020-1. [DOI] [PubMed] [Google Scholar]
- Cacciò SM, De Giacomo M, Pozio E. Sequence analysis of the beta-giardin gene and development of a polymerase chain reaction-restriction fragment length polymorphism assay to genotype Giardia duodenalis cysts from human faecal samples. Int J Parasitol. 2002;32:1023–1030. doi: 10.1016/S0020-7519(02)00068-1. [DOI] [PubMed] [Google Scholar]
- Charlier J, De Meulemeester L, Claerebout E, Williams D, Vercruysse J. Qualitative and quantitative evaluation of coprological and serological techniques for the diagnosis of fasciolosis in cattle. Vet Parasitol. 2008;153:44–51. doi: 10.1016/j.vetpar.2008.01.035. [DOI] [PubMed] [Google Scholar]
- Cho YI, Yoon KJ. An overview of calf diarrhea - infectious etiology, diagnosis, and intervention. J Vet Sci. 2014;15:1–17. doi: 10.4142/jvs.2014.15.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corwin RM. Economics of gastrointestinal parasitism of cattle. Vet Parasitol. 1997;72:451–457. doi: 10.1016/S0304-4017(97)00110-6. [DOI] [PubMed] [Google Scholar]
- Di Giovanni GD, Betancourt WQ, Hernandez J, Assadian NW, Flores Margez JP, Lopez EJ. Investigation of potential zooanthroponotic transmission of cryptosporidiosis and giardiasis through agricultural use of reclaimed wastewater. Int J Environ Health Res. 2006;16:405–418. doi: 10.1080/09603120601095100. [DOI] [PubMed] [Google Scholar]
- Ekawasti F, Nurcahyo W, Wardhana AH, Shibahara T, Tokoro M, Sasai K, Matsubayashi M. Molecular characterization of highly pathogenic Eimeria species among beef cattle on Java Island, Indonesia. Parasitol Int. 2019 doi: 10.1016/j.parint.2019.101927. [DOI] [PubMed] [Google Scholar]
- Estuningsih E, Spithill T, Raadsma H, Law R, Adiwinata G, Meeusen E, Piedrafita D. Development and application of a fecal antigen diagnostic sandwich ELISA for estimating prevalence of Fasciola gigantica in cattle in central Java, Indonesia. J Parasitol. 2009;95:450–455. doi: 10.1645/GE-1672.1. [DOI] [PubMed] [Google Scholar]
- Fujino T, Matsuo T, Okada M, Matsui T. Detection of a small number of Cryptosporidium parvum oocysts by sugar flotation and sugar centrifugation methods. J Vet Med Sci. 2006;68:1191–1193. doi: 10.1292/jvms.68.1191. [DOI] [PubMed] [Google Scholar]
- Grisi L, Leite RC, Martins JR, Barros AT, Andreotti R, Cançado PH, León AA, Pereira JB, Villela HS. Reassessment of the potential economic impact of cattle parasites in Brazil. Rev Bras Parasitol Vet. 2014;23:150–156. doi: 10.1590/S1984-29612014042. [DOI] [PubMed] [Google Scholar]
- Hastutiek P, Yuniarti WM, Djaeri M, Lastuti NDR, Suprihati E, Suwanti LT. Prevalence and diversity of gastrointestinal protozoa in Madura cattle at Bangkalan Regency, East Java, Indonesia. Vet World. 2019;12:198–204. doi: 10.14202/vetworld.2019.198-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan WL, Gilsing M, Bentala H, Limper L, van Knapen F. Characterization of Giardia duodenalis by polymerase-chain-reaction fingerprinting. Parasitol Res. 1998;84:707–714. doi: 10.1007/s004360050474. [DOI] [PubMed] [Google Scholar]
- Katsumata T, Hosea D, Ranuh IG, Uga S, Yanagi T, Kohno S. Short report: possible Cryptosporidium muris infection in humans. Am J Trop Med Hyg. 2000;62:70–72. doi: 10.4269/ajtmh.2000.62.70. [DOI] [PubMed] [Google Scholar]
- Kurniawan A, Karyadi T, Dwintasari SW, Sari IP, Yunihastuti E, Djauzi S, Smith HV. Intestinal parasitic infections in HIV/AIDS patients presenting with diarrhoea in Jakarta, Indonesia. Trans R Soc Trop Med Hyg. 2009;103:892–898. doi: 10.1016/j.trstmh.2009.02.017. [DOI] [PubMed] [Google Scholar]
- Kurniawan A, Dwintasari SW, Connelly L, Nichols RA, Yunihastuti E, Karyadi T, Djauzi S. Cryptosporidium species from human immunodeficiency-infected patients with chronic diarrhea in Jakarta, Indonesia. Ann Epidemiol. 2013;23:720–723. doi: 10.1016/j.annepidem.2013.07.019. [DOI] [PubMed] [Google Scholar]
- Matsubayashi M, Takami K, Kimata I, Nakanishi T, Tani H, Sasai K, Baba E. Survey of Cryptosporidium spp. and Giardia spp. infections in various animals at a zoo in Japan. J Zoo Wildl Med. 2005;36:331–335. doi: 10.1638/04-032.1. [DOI] [PubMed] [Google Scholar]
- Mynářová A, Foitová I, Kváč M, Květoňová D, Rost M, Morrogh-Bernard H, Nurcahyo W, Nguyen C, Supriyadi S, Sak B. Prevalence of Cryptosporidium spp., Enterocytozoon bieneusi, Encephalitozoon spp. and Giardia intestinalis in Wild, Semi-Wild and Captive Orangutans (Pongo abelii and Pongo pygmaeus) on Sumatra and Borneo, Indonesia. PLoS ONE. 2016;11:e0152771. doi: 10.1371/journal.pone.0152771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano S, Matsubayashi M, Naushima KT, Kimata T, Iseki I, Hajiri M, Tani T, Sasai H, Baba KE. Detection of a mixed infection of a novel Cryptosporidium andersoni and its subgenotype in Japanese cattle. Vet Parasitol. 2007;149:213–218. doi: 10.1016/j.vetpar.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Ryan U, Zahedi A. Molecular epidemiology of giardiasis from a veterinary perspective. Adv Parasitol. 2019;106:209–254. doi: 10.1016/bs.apar.2019.07.002. [DOI] [PubMed] [Google Scholar]
- Smith G. Antimicrobial decision making for enteric diseases of cattle. Vet Clin North Am Food Anim Pract. 2015;31:47–60. doi: 10.1016/j.cvfa.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spano F, Putignani L, McLauchlin J, Casemore DP, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- Xiao L, Morgan UM, Limor J, Escalante A, Arrowood M, Shulaw W, Thompson RC, Fayer R, Lal AA. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl Environ Microbiol. 1999;65:3386–3391. doi: 10.1128/AEM.65.8.3386-3391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui T, Nakajima T, Yamamoto N, Kon M, Abe N, Matsubayashi M, Shibahara T. Age-related detection and molecular characterization of Cryptosporidium suis and Cryptosporidium scrofarum in pre- and post-weaned piglets and adult pigs in Japan. Parasitol Res. 2014;113:359–365. doi: 10.1007/s00436-013-3662-2. [DOI] [PubMed] [Google Scholar]