Abstract
Objective: Insufficient knowledge about the molecular pathology of diabetic foot ulcer (DFU) impedes the development of effective wound treatment. Circular RNAs (circRNAs) are a novel class of RNA recently discovered to be widely expressed and have important biological functions; however, their role in skin wound healing remains largely unexplored. In this study, we investigated the role of circRNAs in DFU.
Approach: CircRNA expression was profiled in normal wounds (NWs) and DFUs by microarray analysis, and hsa_circ_0084443 was identified as differentially expressed. The circularity and subcellular localization of hsa_circ_0084443 were characterized by northern blotting, real-time PCR, and fluorescence in situ hybridization. Cell migration, cell growth, and the transcriptome of human primary keratinocytes were analyzed after overexpression or RNA interference of hsa_circ_0084443.
Results: hsa_circ_0084443 is downregulated in NWs compared with intact skin, and its level is higher in DFUs than NWs. We confirmed its circularity and presence in the cytoplasm of human epidermal keratinocytes. We showed that hsa_circ_0084443 reduced motility while enhancing the growth of keratinocytes. Furthermore, we identified a gene network with the potential to mediate the biological effect of hsa_circ_0084443.
Innovation: CircRNAs have a functional role and a potential clinical significance in skin wound healing.
Conclusions: We identified hsa_circ_0084443, a circRNA downregulated during NW healing, as a negative regulator of keratinocyte migration. Higher levels of hsa_circ_0084443 were detected in DFU samples, suggesting that it plays a role in pathology. These findings pave the way to understanding the functional role of circRNAs in human skin wound healing.
Keywords: noncoding RNA, wound healing, keratinocyte, circular RNA, diabetic foot ulcer

Aoxue Wang, MD, PhD

Ning Xu Landén, MD, PhD
Introduction
Diabetes is a significant and escalating global burden, with 422 million patients worldwide.1 Up to 20% of them suffer from an impaired wound healing response and the most common form is diabetic foot ulcer (DFU).2 Owing to the complex nature of the diabetic wound environment, DFU is challenging to treat and up to 20% of patients with DFU receive lower extremity amputation.3 To develop more effective treatments, a better understanding of the underlying etiology of DFU is required. The diabetic wound environment is characterized by hyperglycemia, hypoxia, chronic inflammation, circulatory dysfunction, and impaired neuropeptide signaling.4 Epidermal keratinocytes in the wound-edge of DFU exhibit a phenotype of hyperproliferation with the inability to migrate, which prevents re-epithelialization.5 Understanding the molecular mechanisms underlying this impaired keratinocyte phenotype may help to reactivate the re-epithelialization process and to promote healing of DFU.
Circular RNA (circRNA) is a class of RNA with the 3′ and 5′ ends covalently linked, and it has recently been found to be widespread in eukaryotic organisms.6 CircRNA is formed by alternative splicing of pre-mRNA, in which an upstream splice acceptor is joined to a downstream splice donor in a process known as “back-splicing”.7 Although there still lacks consensus as to the molecular function of circRNAs, emerging data have revealed that they can shape gene expression by sponging microRNAs, regulating transcription and interfering with splicing.7 The crucial roles of circRNA-mediated regulation have been discovered in a variety of physiological and pathological processes, such as development, diabetes, neuropsychiatric disorders, and cancer.8–11 Recently, Yang et al. reported that ectopic application of a circRNA circ-Amotl1 accelerated wound healing of mouse skin, suggesting the therapeutic potential of circRNA in hard-to-heal wounds.12
To identify relevant circRNAs as therapeutic targets, we need to first understand their expression and function in human skin wound healing, which remains unexplored to date. In this study, we profiled circRNA expression in human DFU and normal acute wounds. We hypothesized that circRNAs that are dysregulated in DFUs may be involved in the pathophysiology of chronic wounds. We selected for further study a circular RNA named “hsa_circ_0084443”, which was shown in the microarray analysis to be significantly upregulated in the DFUs compared with normal wounds (NWs). Another rationale to focus on hsa_circ_0084443 is that it is derived from a parental gene, protein kinase DNA-activated catalytic polypeptide (PRKDC), which has important functions in DNA repair, aging, energy metabolism, and cancer.13–15 We suspected that hsa_circ_0084443 might be involved in these biological processes that are relevant to chronic wounds.
Clinical Problem Addressed
DFU is challenging to treat due to the complex nature of the diabetic wound environment. Further research on understanding the pathophysiology of DFU could help in the development of more effective treatments.
Materials and Methods
Tissue samples
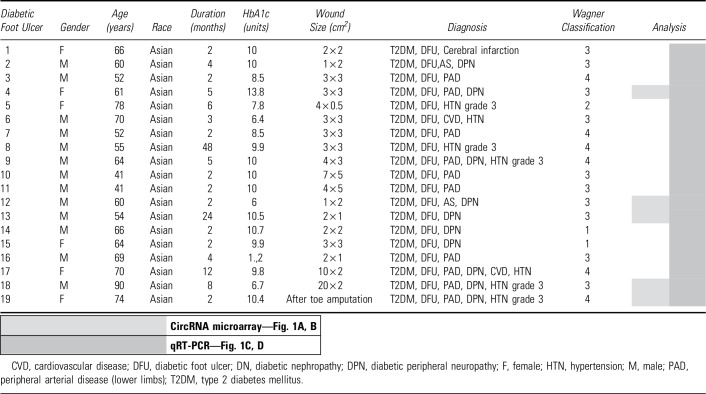
Healthy donors and DFU patients were enrolled at the Second Hospital of Dalian Medical University (SHDMU, Dalian, China). The healthy donor group consisted of 13 individuals with a mean age of 36 years (ranging from 25 to 63), of which 69% were women and 31% were men (Table 1). The exclusion criteria for healthy donors were diabetes, skin disease, unstable heart disease, infections, bleeding disorder, immune suppression, and any ongoing medical treatments. On the skin of each healthy donor, one excisional wound was created using a 3 mm punch and the excised skin from these surgical wounds was saved as intact skin control. The wound edges were collected using a 6 mm punch 7 days later (Fig. 1A). The DFU cohort consisted of 19 patients with a mean age of 63 years (ranging from 41 to 90), of which 32% were women and 68% were men (Table 2). Wound-edge samples were collected from patients with nonhealing DFU that, despite conventional therapy, persisted for >2 months (Fig. 1A).16 Patients were excluded with apparent soft tissue infection requiring systemic antibiotics, patients taking systemic antibiotics 24 h before biopsy, as well as immunocompromised patients. Biopsies were taken from the nonhealing edge of DFUs using a 4 mm punch. Local lidocaine injection was used for anesthesia while sampling. Written informed consent was obtained from all donors for the collection and use of clinical samples. This study was approved by the ethics committee of the SHDMU (Document number 2015-102 “Investigation of gene expression profile of human normal and chronic wounds”). The study was conducted according to the Declaration of Helsinki's principles.
Table 1.
Information of healthy donors
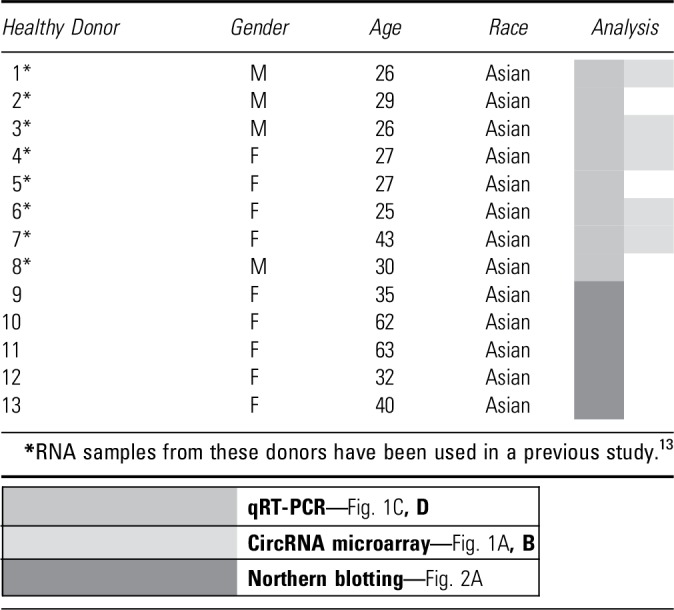 |
Figure 1.
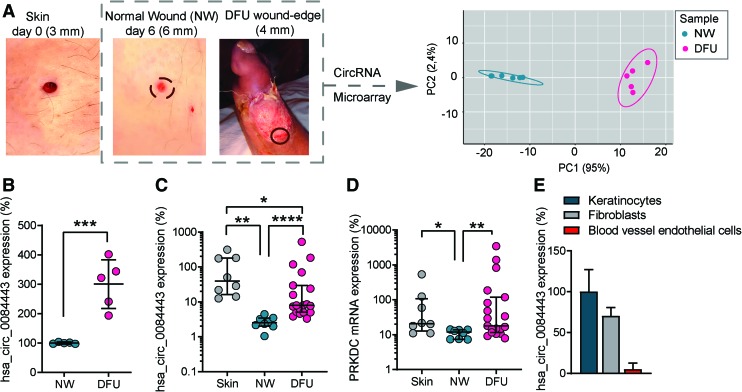
Circular RNA hsa_circ_0084443 is upregulated in human chronic diabetic wounds. (A) Biopsies (4 mm in diameter) were collected at wound edges of DFU (n = 5). A 3 mm-surgical wound was created on the skin of healthy volunteers (n = 5) and the wound edge was excised with a 6 mm biopsy punch 6 days later (NW). A principal component analysis was performed on the circRNA microarray data of DFU and NW. (B) The normalized expression levels of hsa_circ_0084443 were extracted from the microarray data. qRT-PCR analysis of hsa_circ_0084443 (C) and PRKDC mRNA (D) in skin (n = 8), NW (n = 8), and DFU (n = 19). (E) qRT-PCR analysis of hsa_circ_0084443 in human primary keratinocytes, fibroblasts and blood vessel endothelial cells. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 by unpaired two-tailed Student's t-test (B) and Wilcoxon matched pairs signed rank test or Mann–Whitney U test (C, D). Data are presented as mean ± SD. (B), median ± interquartile range (IQR) (C, D), and mean + SD. (E). DFU, diabetic foot ulcer; NW, normal wound; qRT-PCR, quantitative real-time polymerase chain reaction. Color images are available online.
Table 2.
List of patients with diabetic foot ulcer
RNA extraction and quantitative real-time polymerase chain reaction
Skin biopsies were homogenized using TissueLyser LT (Qiagen, Hilden, Germany) before RNA extraction. Total RNA was extracted from human tissue and cells using the miRNeasy Mini kit (Qiagen) or TRIzol reagent (ThermoFisher Scientific, Carlsbad, CA). Gene expression was determined by SybrGreen expression assays for qRT-PCR (ThermoFisher Scientific) and normalized based on the values of the housekeeping genes: B2M or β-actin or GAPDH. Accompanying information for all of the primers used in this study is listed in Table 3.
Table 3.
Sequence information of the primers used in this study
| Name | Sequence | Experiment | Company |
|---|---|---|---|
| hsa_circ_0084443 | for-CAGCCTTGTCAGAGAGATTC | SYBR™ qRT-PCR; sequencing; northern blot probe | Geneseed Biotech |
| rev-CAATTTGTAGAAACCACTGATGAG | |||
| for-cgGAATTCTGAAATATGCTATCTTACAGAGAGATTCTCCCTGAGAAACAAGC | Cloning | ||
| rev-cgGGATCCTCAAGAAAAAATATATTCACCTGACAAGGCTGAAGTCTTCAG | |||
| PRKDC | for-ACCAAAGTAGACTCACCAAATTGC | SYBR qRT-PCR | Eurofins |
| rev-TGTCCAGGTGTTCAGAAGTCTC | |||
| MALAT1 | for-GATCTAGCACAGACCCTTCAC | SYBR qRT-PCR | Eurofins |
| rev-CGACACCATCGTTACCTTGA | |||
| B2M | for-AAGTGGGATCGAGACATGTAAG | SYBR qRT-PCR | IDT |
| rev-GGAGACAGCACTCAAAGTAGAA | |||
| GAPDH | for-GGTGTGAACCATGAGAAGTATGA | SYBR qRT-PCR | IDT |
| rev-GAGTCCTTCCACGATACCAAAG | |||
| β-Actin | for-CATGTACGTTGCTATCCAGGC | SYBR qRT-PCR | Geneseed Biotech |
| rev-CTCCTTAATGTCACGCACGAT |
Circular RNA expression microarray
Circular RNA expression profiling in normal human wounds (n = 5) and DFUs (n = 5) was performed by using Arraystar Human Circular RNA Microarray V2.0 at KangChen Bio-tech (Shanghai, China). In brief, total RNAs were digested with RNase R (Epicentre, Madison, WI) to remove linear RNAs and enrich circular RNAs. Then, the enriched circular RNAs were amplified and transcribed into fluorescent cRNA utilizing a random priming method—Arraystar Super RNA Labeling Kit (Arraystar, Inc., Rockville, MD). The labeled cRNAs were hybridized onto the Arraystar Human circRNA Array v2 (8x15K; Arraystar). After having washed the slides, the arrays were scanned by the Agilent Scanner G2505C. Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images. Quantile normalization and subsequent data processing were performed using the R software limma package. CircRNAs showing at least twofold difference between NW and DFU and FDR value <0.001 were considered to be differentially expressed. The data discussed herein have been deposited in the NCBI's Gene Expression Omnibus database under accession number GSE114248 (token: qjcpiugkvvqbboj).
Cell culture and treatments
Human primary epidermal keratinocytes (Cascade Biologics, Portland, OR) were cultured in EpiLife medium supplemented with 10% Human Keratinocyte Growth Supplement (HKGS) and 1% penicillin/streptomycin at 37°C in 5% CO2 (ThermoFisher Scientific). To determine the stability of hsa_circ_0084443, we treated keratinocytes with 5 μg/mL Actinomycin D (Merck KGaA, Darmstadt, Germany) to block total cellular transcription. To knock down hsa_circ_0084443 expression, we transfected the third passage keratinocytes at 60–70% confluence in a 24-well plate with 20 nM siRNA targeting the diagnostic junction of hsa_circ_0084443 (si-hsa_circ0084443) or siRNA controls (Eurofins Genomics, Ebersberg, Germany) with Lipofectamine™ 2000 (ThermoFisher Scientific). To overexpress hsa_circ_0084443, we transfected keratinocytes with hsa_circ_0084443 overexpression plasmid or mock vector 0.05 or 0.1 μg using Fugene® HD Reagent (Promega Corporation, Fitchburg, WI). For the pathway inhibition experiments, transfected keratinocytes were treated with 1 μM Wortmannin (Merck KGaA), 500 nM PD153035 (Merck KGaA), or 10 μM U0126 (Merck KGaA), and dimethyl sulfoxide (Merck KGaA) was used as vehicle control.
Hsa_circ_0084443 expression was analyzed in different cell types that were maintained as follows: (1) Dermal fibroblasts were isolated from healthy skin and cultured as previously described.17 (2) Human umbilical vascular endothelial cells were isolated and cultured as previously described.18 (3) THP-1, a human monocytic cell line, was maintained in RPMI 1640 media supplemented with 2 mM l-glutamine, 100 U/mL penicillin/streptomycin, 25 mM HEPES, and 5% FBS (all from Gibco BRL, Gaithersburg, MD).
Northern blotting
Total RNA was extracted from keratinocytes and skin tissue using TRIzol. Half of the RNA was treated with RNase R at 37°C for 15 min after inactivation at 85°C for 3 min. RNA with or without RNase R treatment was separated by 1% formaldehyde denatured agarose gel and then transferred from gel to nylon membrane (Amersham, Buckinghamshire, England). RNA was immobilized on the membrane by baking it at 80°C for 2 h. Digoxigenin (Dig)-labeled DNA probes were prepared by PCR DIG Probe Synthesis Kit and DIG High Prime DNA Labeling and Detection Starter Kit II (Roche Applied Science, Penzberg, Germany) using specific primers for the diagnostic junction region of hsa_circ_0084443 (Table 4). The membrane was prehybridized by DIG Easy Hyb (3 mL/100 cm2) for 2 h at 50°C and hybridized by denatured DIG-labeled DNA probe (100 ng/mL) at 50°C overnight. Then, the membrane was washed twice with 2 × SSC, 0.1% SDS for 5 min at room temperature and twice with 0.1 × SSC, 0.1% SDS for 15 min at 68°C before being blocked by 1 × blocking buffer for 1 h with shaking at room temperature. Finally, the DIG-labeled DNA probe was detected by anti-Digoxigenin-AP, Fab fragments (Roche Applied Science).
Table 4.
Top 10 central nodes of hsa_circ_0084443-regulated gene network
| Gene ID | Betweenness | Closeness | Degree |
|---|---|---|---|
| PIK3CD | 6235.31 | 0.00316 | 16 |
| PXN | 3542.27 | 0.00296 | 12 |
| HIF1A | 2126.54 | 0.00267 | 7 |
| HSPA1A | 2777.4 | 0.00261 | 9 |
| BDNF | 2470.02 | 0.00261 | 7 |
| WASL | 2238.45 | 0.00239 | 7 |
| PTPN11 | 1735.89 | 0.0028 | 7 |
| RBBP4 | 296 | 0.0238 | 8 |
| S1PR1 | 966.66 | 0.00246 | 8 |
| CHRM4 | 181.85 | 0.00216 | 7 |
Cell fractionation
The cytoplasm and nucleus of keratinocytes were separated by using Nuclear Extract Kit (Active Motif, Carlsbad, CA) following the manufacturer's instructions. RNA was extracted from these fractions using TRIzol. qRT-PCR was performed to analyze the expression of hsa_circ_0084443, MALAT1, and GAPDH.
In situ hybridization
In situ hybridization was performed using a Digoxin-labeled DNA probe specific to the diagnostic junction region of hsa_circ_0084443: 5′-Digoxin-GGGAGAATCTCTCTGACAAGGCTGAAG.
TCT-Digoxin-3′. Keratinocytes cultured on cover glass were treated with 0.1 M HCl for 20 min, 0.5% TritonX-100 for 15 min, and then fixed with 4% paraformaldehyde for 5 min. After washing with PBS three times, the slides were immersed in 100% ethanol for 1 min and allowed to dry. After denaturation at 88°C for 5 min and equilibration at 4°C for 3 min, the DNA probe was added into hybridization buffer (40% formamide, 10% dextran sulfate, 1 × Denhardt's solution, 4 × SSC, 10 mM DDT, 1 mg/mL yeast transfer RNA, 1 mg/mL sheared salmon sperm DNA) and incubated with keratinocytes at 37°C overnight. The slide was washed with 2 × SSC, 0.1% NP-40 for 5 min for three times (once at room temperature and twice at 42°C), and blocked by 3% bovine serum albumin at 37°C for 30 min. The DIG-labeled DNA probe was detected by anti-Digoxigenin-FITC (Roche Applied Science). Nuclei were counterstained with 4,6-diamidino-2-phenylindole. The images were acquired on a Leica SP2 confocal microscope (Leica Microsystems, Wetzlar, Germany).
Plasmid construction
The hsa_circ_0084443 overexpression plasmid was constructed with the help of Guangzhou Geneseed Biotech Co. (Guangzhou, China). In brief, pLCDH-ciR vector, which includes front and back circular frames for the circularization of the transcripts, was used as the backbone plasmid. The front circular frame contains an endogenous flanking genomic sequence with the EcoRI restriction site and the back circular frame contains part of the inverted upstream sequence with the BamHI restriction site. The cDNA encoding hsa_circ_0084443 in HeLa cells was amplified using the primers listed in Table 4. The amplicon, which contained an EcoRI site, the splice acceptor AG, the hsa_circ_0084443 sequence, the splice donor GT, and a BamHI site, was then cloned into the backbone vector between the two frames. Vector construction was verified by Sanger sequencing. A mock vector containing only a nonsense sequence between the two circular frames was used as a control plasmid.
Analysis of cell motility
Cell motility was analyzed by using scratch assay and transwell migration assay. For the scratch assay, keratinocytes were transfected with si-hsa_circ_0084443 or scrambled siRNA, hsa_circ_0084443 overexpression plasmid or mock vector. Twenty-four hours later, cells at full confluence were treated with mitomycin C 5 μg/mL (Roche, Basel, Switzerland) to stop proliferation. A scratch was made with a 10-μL pipette tip and the cells were incubated with EpiLife medium without HKGS and photographed at the indicated timepoints. The wound areas were measured using Image J (National Institutes of Health, Bethesda, MD). Migration rate at each timepoint equals to (100%—the percentage of the initial wound area). For IncuCyte™ (Essen BioScience, Ann Arbor, MI) 96-well Real-Time Cell Migration Assay, keratinocytes were seeded in 96-well microtiter IncuCyte® ImageLock Plate (Essen BioScience) and transfected with si-hsa_circ_0084443 or scrambled siRNA. After 24 hours, when cells were at full confluence, a wound area was created using Essen® 96-pin WoundMaker™ (Essen BioScience). The assay plates were then incubated in the IncuCyte and equilibrated for at least 15 min before the first scan. The IncuCyte ZOOM software was set to scan the experiment every 2 h for 24 h. The data were analyzed by the integrated metrics in the IncuCyte ZOOM software package presented as migration rate.
Transwell migration assay was performed using the BD BioCoat Matrigel Chamber (BD Falcon, Erembodegen, Belgium). Keratinocytes were transfected with si-hsa_circ_0084443 or scrambled siRNA, hsa_circ_0084443 overexpression plasmid or mock vector for 24 h. 3 × 104 cells in HKGS-free medium were seeded into the upper chamber of the insert. Medium containing HKGS was added into the lower chamber. Twenty four hours later, the cells migrating through the membrane were stained with 0.1% crystal violet (Merck KGaA) and counted under a microscope.
Analysis of cell growth
For IncuCyte Live-cell imaging of cell growth, keratinocytes were transfected with si-hsa_circ_0084443 or scrambled siRNA, hsa_circ_0084443 overexpression plasmid or mock vector. After 24 h, the assay plates were incubated in the IncuCyte and equilibrated for at least 15 min before the first scan. The IncuCyte ZOOM software was set to scan the experiment every 2 h for 66 h. The cellular confluence was analyzed by the IncuCyte ZOOM software package and expressed as Phase Count Object (PCO). The results were presented as proliferation rate = (PCO timepoint x−PCO timepoint 0)*100/PCO timepoint 0.
CyQUANT® Cell Proliferation Assay was carried out to assess keratinocyte proliferation according to the manufacturer's instructions (Invitrogen, Carlsbad, CA).
For the colony formation assay, keratinocytes were transfected with si-hsa_circ_0084443 or hsa_circ_0084443 overexpression plasmids. After 24 h, transfected cells were reseeded at low density (6,000 cells/well in a six-well plate) and the cell growth was monitored daily. Cells were stained using 0.1% crystal violet 7 days after transfection. For quantification, crystal violet was dissolved with methanol and absorbance values were measured at 540 nm.
Gene expression microarray
Gene expression profiling of human primary keratinocytes transfected with either siRNA control (n = 3) or si-hsa_circ_0084443 (n = 3) for 24 h was performed using human Clariom™ S assay (ThermoFisher Scientific) at the microarray core facility at the Karolinska Institute. In brief, total RNA was extracted using the miRNeasy Mini Kit, and RNA quality and quantity were determined using Agilent 2200 Tapestation with RNA ScreenTape and Nanodrop 1000. One hundred fifty nanograms of total RNA was used to prepare cDNA following the GeneChip WT PLUS Reagent Kit labeling protocol. Standardized array processing procedures recommended by Affymetrix, including hybridization, fluidics processing, and scanning, were used. Expression data were analyzed using Transcriptome Analysis Console 4.0. Genes showing at least 1.5-fold difference from control and p-value <0.05 were considered to be differentially expressed. Heatmaps were generated with Multiple Experiment Viewer software. The data discussed herein have been deposited in the NCBI's Gene Expression Omnibus database under accession number GSE114236 (token: qbadiacchrchpmh).
Using the STRING App and Cytoscape (version 3.0.1), we generated a functional protein association network among the genes regulated by hsa_circ_0084443. Using CentiScape 2.2 App, three measures were made to characterize topological features of each node: (1) “Closeness” is defined as the inverse of the sum of node in distances to all other nodes, (2) “Betweenness” represents the number of edges running through a node, and (3) “Degree” defines the number of links for each node. The top 10 central nodes were chosen based on the highest value of all measures. The migration-related genes [which are defined as the genes included in the following GO groups: regulation of cell migration (GO: 0030334), regulation of locomotion (GO: 2000145), regulation of cell motility (GO: 0040012), positive regulation of cell migration (GO: 0030335), positive regulation of cell motility (GO: 2000147), and positive regulation of cytoskeleton organization (GO: 0051495)], and proliferation-related genes [negative regulation of cell proliferation (GO:0008285), negative regulation of epithelial cell proliferation (GO: 0050680), regulation of epithelial cell proliferation (GO: 0050678), positive regulation of cell proliferation (GO: 0008284), positive regulation of epithelial cell proliferation (GO: 0050679), epithelial cell proliferation (GO: 0050673), and cell proliferation (GO: 0008283)] were highlighted in this protein association network.
Statistics
Statistical significance was determined by two-tailed Student's t-test, Mann–Whitney U Test, or Wilcoxon Matched Pairs Signed Rank Test. Differences between groups were computed using two-way repeated-measures ANOVA. p-Value <0.05 was considered to be statistically significant.
Results
Dysregulation of circular RNA expression in human chronic diabetic wounds
To study circRNA expression in human skin wounds in vivo, we performed circRNA microarray analysis in human normal acute wounds (n = 5, Table 1) and DFUs (n = 5, Table 2). In total, 8,847 circRNAs passed the low intensity filtering criterion in all of the samples and were used for subsequent differential expression analysis. Based on the circular RNA expression, principal component analysis separated the DFUs from the NWs (Fig. 1A). This microarray analysis identified 115 upregulated and 111 downregulated circRNAs in the DFUs compared with the NWs [absolute fold-change (FC) ≥2, false discovery rate (FDR) <0.001, average raw intensity >100] (Supplementary Table S1).
Increased hsa_circ_0084443 expression in human chronic diabetic wounds
As a proof of a principle, “hsa_circ_0084443” was shown in microarray analysis to be significantly upregulated (FC = 2.9, FDR = 0.0006) in the DFUs compared with the NWs (Fig. 1B). By using quantitative real-time PCR (qRT-PCR) with primers designed to specifically amplify diagnostic junctions of circRNAs, which are absent in their cognate linear isoforms, we examined hsa_circ_0084443 expression in a bigger patient cohort.19 qRT-PCR analysis confirmed the microarray results, showing more hsa_circ_0084443 in the DFUs compared with the NWs (Fig. 1C). In addition, we found that hsa_circ_0084443 expression was downregulated in the NWs compared with the intact skin (Fig. 1C). In the same patient cohort, we showed that PRKDC exhibited a similar expression pattern as hsa_circ_0084443 (Fig. 1D). Furthermore, we characterized hsa_circ_0084443 expression in several cell types and found its highest level in keratinocytes, followed by fibroblasts, and least abundant in blood vessel endothelial cells (Fig. 1E).
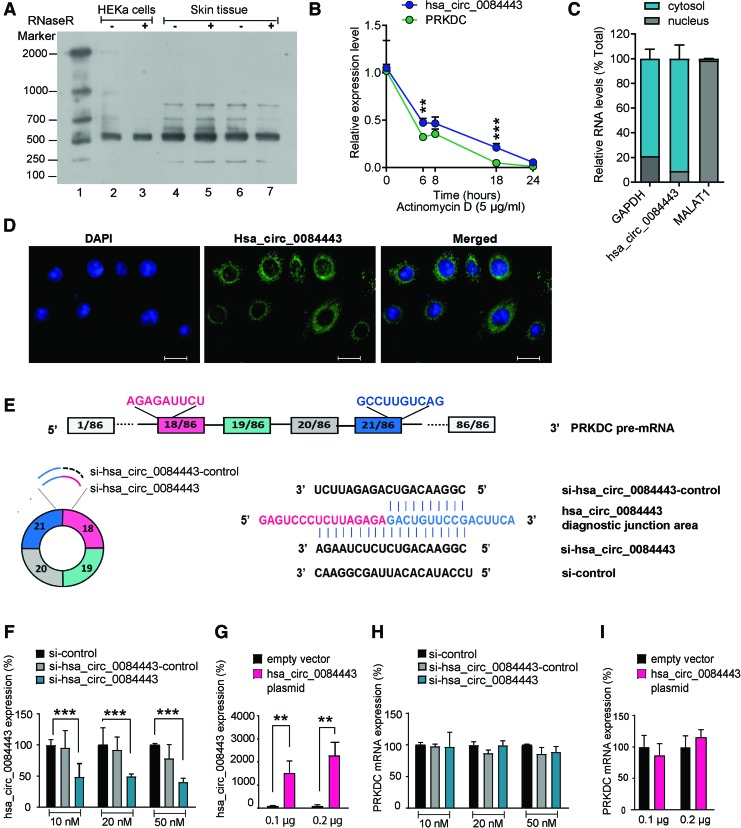
hsa_circ_0084443 is localized to the cytoplasm of human epidermal keratinocytes
As hsa_circ_0084443 is an annotated but uncharacterized circRNA, we next examined its circularity, stability, and subcellular localization. We performed northern blotting on total RNA extracted from human primary epidermal keratinocytes and skin biopsies (Fig. 2A). Using a denatured DNA probe spanning over the diagnostic junction of the circRNA, we detected the expression of hsa_circ_0084443 in all the samples with the expected size of 527 nt20 (Fig. 2A). Moreover, we observed that RNase R treatment did not change the intensity of hsa_circ_0084443 bands, confirming its circularity (Fig. 2A). To determine the stability of RNA molecules, we treated keratinocytes with Actinomycin D (5 μg/mL) to block total cellular transcription and then analyzed the relative amount of specific RNAs over time (Fig. 2B). qRT-PCR analysis revealed a half-life of 6.2 h for hsa_circ_0084443 and 4.6 h for PRKDC mRNA, suggesting that hsa_circ_0084443 is more stable than its cognate linear isoform PRKDC in keratinocytes (Fig. 2B). Next, we performed qRT-PCR on RNA extracted from either the nucleus or cytoplasm of keratinocytes. Enrichment of the nuclear lncRNA MALAT1 and GAPDH mRNA in the nuclear and cytoplasmic fractions, respectively, confirmed the purity of each fraction (Fig. 2C). We found that hsa_circ_0084443 was mainly present in the cytoplasm of keratinocytes (Fig. 2C), which was further confirmed and visualized by in situ hybridization analysis (Fig. 2D).
Figure 2.
hsa_circ_0084443 localizes in the cytoplasm of human epidermal keratinocytes and its aberrant expression does not change PRKDC levels. (A) Northern blotting analysis of hsa_circ_0084443 expression in human primary keratinocytes (HEKa) (lane 2, 3) and skin tissue (lane 4–7) with and without RNase R treatment. RNA molecular weight marker was used (lane 1). (B) qRT-PCR analysis of hsa_circ_0084443 and PRKDC mRNA in HEKa treated with Actinomycin D at the indicated timepoints (n = 3). (C) qRT-PCR analysis of hsa_circ_0084443, MALAT1, and GAPDH in nucleus or cytoplasm of HEKa (n = 3). (D) Fluorescence in situ hybridization to visualize hsa_circ_0084443 in HEKa. Scale bar 20 μm. (E) Illustration of hsa_circ_0084443 and pre-mRNA of PRKDC. Sequence information of si-hsa_circ_0084443, si-control, and si-hsa_circ_0084443-control. qRT-PCR of hsa_circ_0084443 (F) and PRKDC mRNA (H) in keratinocytes transfected with 10, 20, and 50 nM si-control, si-hsa_circ_0084443, or si-hsa_circ_0084443-control for 24 h (n = 3). QRT-PCR of hsa_circ_0084443 (G) and PRKDC mRNA (I) in keratinocytes transfected with hsa_circ_0084443 overexpression plasmid (0.1 or 0.2 μg) for 24 h (n = 3) **p < 0.01 and ***p < 0.001 by unpaired two-tailed Student's t test. Data are presented as mean + SD. Color images are available online.
Modulation of hsa_circ_0084443 expression does not affect PRKDC expression
To determine if hsa_circ_0084443 plays a functional role, we modulated its expression in human primary keratinocytes. To knock down hsa_circ_0084443 expression, we designed a siRNA targeting its diagnostic junction (si-hsa_circ_0084443). As controls, a scrambled siRNA sequence (si-control) and a siRNA partially (10 out of 21 nt) complementary to the junction sequence of hsa_circ_0084443 (si-hsa_circ_0084443-control) were used (Fig. 2E). To overexpress hsa_circ_0084443, we subcloned the DNA sequence corresponding to hsa_circ_0084443 and its endogenous flanking region, which also included complementary circular frames needed for circularization, into a plasmid expression cassette. qRT-PCR analysis showed that transfection of hsa_circ_0084443-specific plasmid or siRNA successfully changed its expression in keratinocytes (Fig. 2F, G), but did not affect the expression level of its parent gene PRKDC (Fig. 2H, I).
hsa_circ_0084443 reduces keratinocyte motility
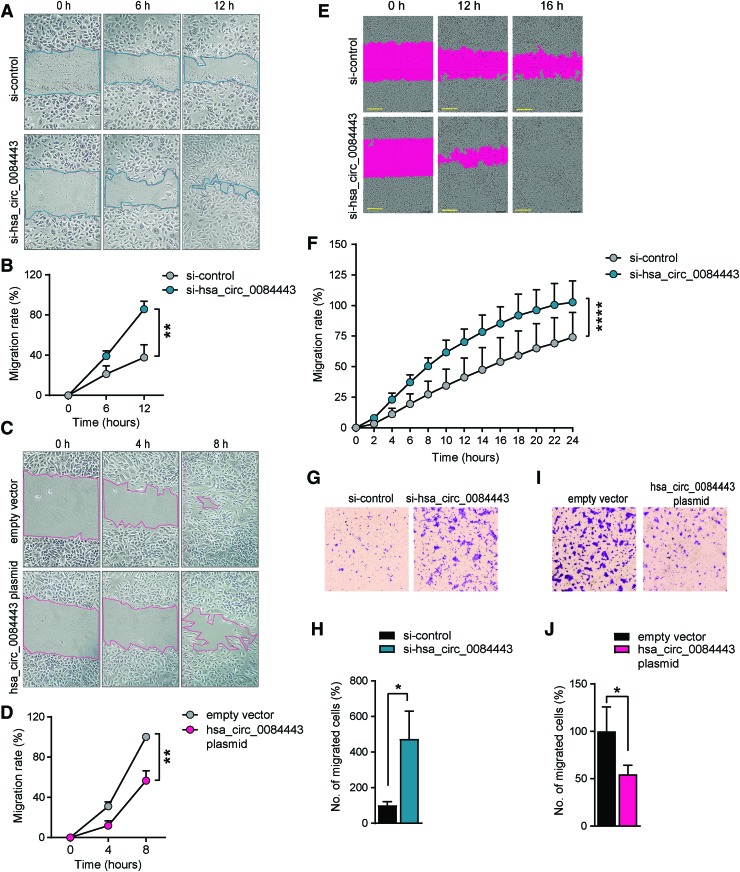
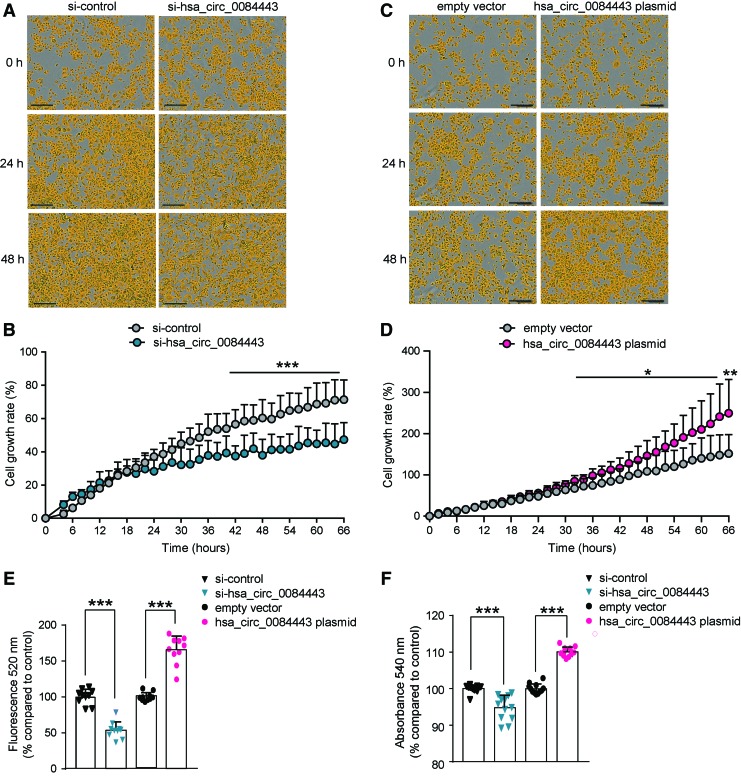
Proliferation and migration of epidermal keratinocytes are indispensable for wound re-epithelization.21 To study the effect of hsa_circ_0084443 on keratinocyte migration, we performed scratch wound assay on keratinocytes transfected with hsa_circ_0084443-specific plasmids or siRNAs. We showed that silencing hsa_circ_0084443 expression increased keratinocyte migration, whereas its overexpression exhibited the opposite effect (Fig. 3A–D). These results were confirmed by live-cell imaging using the IncuCyte System (Fig. 3E, F and Supplementary Movie S1). Consistent with these data, a haptotactic transwell migration assay showed that knocking down hsa_circ_0084443 enhanced the migration capacity of keratinocytes (Fig. 3G, H), whereas overexpression of hsa_circ_0084443 reduced their migration (Fig. 3I, J). Taken together, these findings point to hsa_circ_0084443 acting as a negative regulator of keratinocyte motility.
Figure 3.
hsa_circ_0084443 inhibits keratinocyte migration. Scratch wound assay of keratinocytes transfected with 20nM si-hsa_circ_0084443 and si-control (A, B) or 0.1 μg hsa_circ_0084443 plasmid and empty vector (C, D) for 24 h (n = 4). Photographs were taken at the indicated timepoints after the scratch (A, C). The healing rates were quantified (B, D). Live-cell imaging using the IncuCyte® system of migrating keratinocytes with hsa_circ_0084443 knockdown (n = 12). Representative photographs of migrating keratinocytes at the indicated timepoints. Scale bar = 300 μm (E). The migration rates were quantified by measuring the area of the scratched region (F). Representative photographs of the transwell migration assay for keratinocytes with hsa_circ_0084443 knockdown (n = 3) (G) or hsa_circ_0084443 overexpression (n = 3) (I). The number of cells passing through the membrane was counted (H, J). *p < 0.05, **p < 0.01, and ****p < 0.0001 by unpaired two-tailed Student's t test (B, D, H, J) and by two-way ANOVA (F). Data are presented as mean + SD. Color images are available online.
hsa_circ_0084443 promotes keratinocyte growth
To investigate the effect of hsa_circ_0084443 on cell proliferation, we monitored the growth of keratinocytes transfected with hsa_circ_0084443-specific siRNA (Fig. 4A, B) or overexpression plasmid (Fig. 4C, D) using time-lapse microscopy. We found that the keratinocytes with silenced hsa_circ_0084443 expression grew slower, whereas keratinocytes overexpressing hsa_circ_0084443 grew faster compared with the control cells. In line with this, the CyQuant proliferation assay, which measured cell number based on the amount of DNA, revealed that the cell number was lower after hsa_circ_0084443 knockdown, whereas more cells were observed upon hsa_circ_0084443 overexpression (Fig. 4E). In addition, we performed a colony formation assay and found that overexpression of hsa_circ_0084443 increased the long-term self-renewal of keratinocytes, whereas knocking it down had the opposite effect (Fig. 4F). Based on these data, we concluded that hsa_circ_0084443 played a pro-proliferative role in keratinocytes.
Figure 4.
hsa_circ_0084443 promotes keratinocyte growth. Growth of keratinocytes with hsa_circ_0084443 knockdown (A) or hsa_circ_0084443 overexpression (C) was monitored using the IncuCyte Live-Cell Imaging System (n = 5). Scale bar 300 μm. (B, D) The results were quantified and presented as cell growth rate. (E) Keratinocytes were transfected with si-hsa_circ_0084443 or hsa_circ_0084443 overexpression plasmid for 24 h. The number of viable cells was quantified with CyQUANT cell proliferation assay (n = 12). (F) Colony formation assay of keratinocytes with hsa_circ_0084443 knockdown or overexpression (n = 12). *p < 0.05 and ***p < 0.001 by two-way ANOVA (B, D) or by unpaired two-tailed Student's t-test (E, F). Data are presented as mean + SD. Color images are available online.
A gene network regulated by hsa_circ_0084443 in keratinocytes
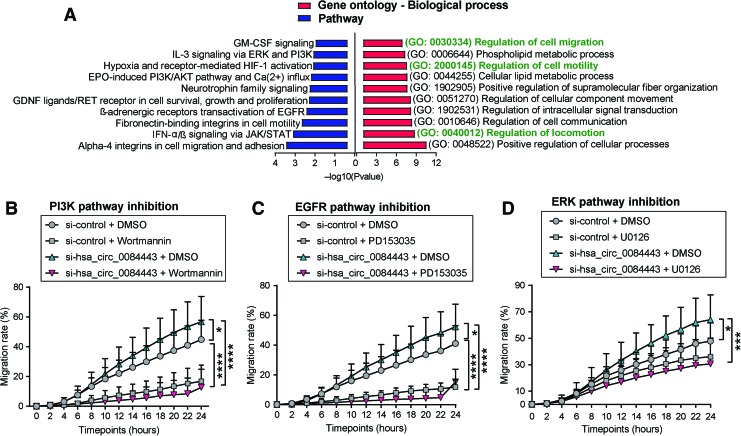
To determine the molecular basis underlying the biological function of hsa_circ_0084443, we performed a global transcriptomic analysis in human keratinocytes with hsa_circ_0084443 knockdown and identified 493 genes that were significantly regulated (absolute fold change ≥1.5, p < 0.05) by hsa_circ_0084443 (Supplementary Table S2). Gene ontology (GO) analysis revealed that among the top 10 enriched biological processes, 3 were related to cell migration: regulation of locomotion (GO: 0040012, p = 1,602E-09), regulation of cell motility (GO: 2000145, p = 2.357E-07), and regulation of cell migration (GO: 0030334, p = 1,111E-07) (Fig. 5A). Moreover, pathway enrichment analysis suggested that several signaling pathways important for skin wound healing, such as JAK/STAT, EGFR, PI3K, ERK, and HIF-1, may be regulated by hsa_circ_0084443 (Fig. 5A).
Figure 5.
PI3K, EGFR, and ERK signaling pathways are important for the antimigratory function of hsa_circ_0084443 in keratinocytes. Gene expression profile of keratinocytes with hsa_circ_0084443 knockdown (n = 3) was analyzed by microarray. (A) Gene ontology analysis of the genes regulated by hsa_circ_0084443 (absolute fold change ≥1.5, p < 0.05). Live-cell imaging using the IncuCyte system of migrating keratinocytes with hsa_circ_0084443 knockdown (n = 6) and treated with DMSO or Wortmannin (1 μM) (B), PD153035 (500 nM) (C) or U0126 (10 μM) (D). The migration rates were calculated by measuring the area of the scratched region (B–D). *p < 0.05, ***p < 0.001, and ****p < 0.001 by two-way ANOVA. Data are presented as mean + SD. Color images are available online.
To test if PI3K, EGFR, and ERK signaling pathways may mediate the biological effect of hsa_circ_0084443 in keratinocytes, we blocked these pathways using specific chemical inhibitors, which completely abolished the pro-migratory effect of hsa_circ_0084443 siRNAs (Fig. 5B–D). Of note, PI3K and EGFR pathways may play a more critical role in keratinocyte migration than ERK, as blockage of PI3K and EGFR pathways severely delayed cell migration (Fig. 5B–D). These results suggest that PI3K, EGFR, and ERK signaling pathways are important in mediating the antimigratory function of hsa_circ_0084443 in keratinocytes.
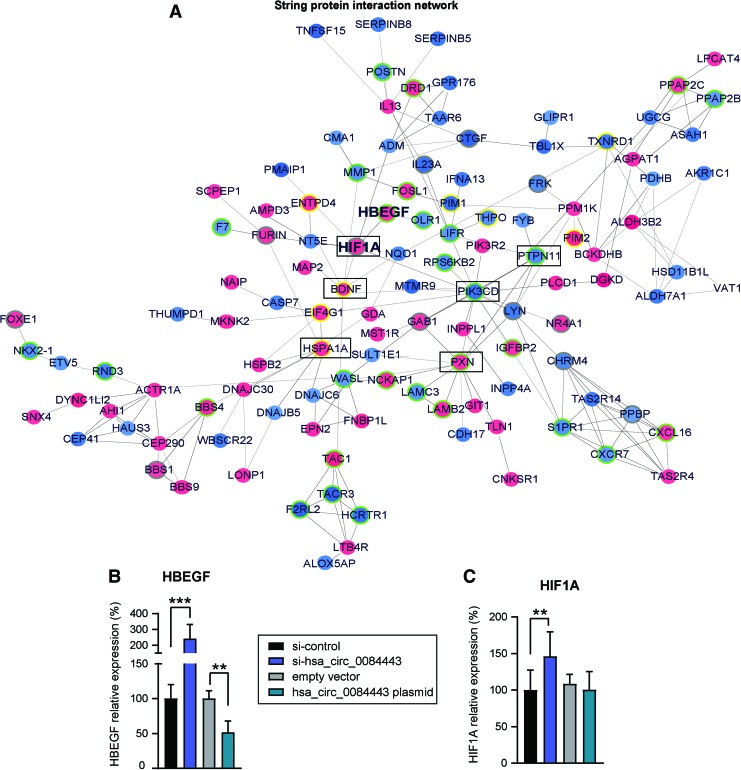
Furthermore, using the STRING database,22 we analyzed the functional protein association network among the protein-coding genes regulated by hsa_circ_0084443 (Fig. 6A). Of note, many genes in this network (e.g., HBEGF23,24 and HIF1A25) have been previously involved in regulating cell migration and proliferation. We confirmed that hsa_circ_0084443 regulated HBEGF and HIF1A expression by qRT-PCR analysis of keratinocytes by either silencing or overexpressing hsa_circ_0084443 (Fig. 6B, C). Based on topological features of the network, we also identified the centrally located nodes, which may contribute to a greater extent to the biological effects of hsa_circ_0084443 by possessing a central regulatory ability over other genes in the network (Table 4). Interestingly, the central hubs PXN, PTPN11, and PIK3CD have been previously implicated in cell migration,26–28 whereas HSPA1A and B can regulate cell proliferation,29,30 indicating their potential to mediate the biological function of hsa_circ_0084443 in keratinocytes.
Figure 6.
A gene network regulated by hsa_circ_0084443 in keratinocytes. (A) Functional protein association network was identified by String APP in Cytoscape software among the genes regulated by hsa_circ_0084443 shown in the microarray analysis. Genes up- or downregulated after transfection of si-hsa_circ_0084443 are in red or blue, respectively. Genes previously reported to have an impact on cell migration and proliferation are highlighted: yellow border = proliferation genes, green border = migration genes, grey border = migration and proliferation genes. Core nodes identified by centrality analysis with the CentiScaPe 2.2 App are marked with black rectangles. qRT-PCR of HBEGF mRNA (B) and HIF1A mRNA (C) in keratinocytes with hsa_circ_0084443 knockdown (n = 10) or overexpression (n = 4). **p < 0.01 and ***p < 0.001 by unpaired two-tailed Student's t-test. Data are presented as mean + SD. Color images are available online.
Discussion
In this study, we have determined the expression profiles of circRNAs in human normal skin wounds and DFUs, which provide the foundation for studying their function in skin wound healing. Among the circRNAs dysregulated in DFUs (Supplementary Table S1), a few have been previously reported. In brief, hsa_circ_0032704 (FC = 2,3, FDR = 0.0001) was found upregulated in cutaneous squamous cell carcinoma31; hsa_circ_0001946 (FC = −2,3, FDR = 0.0001), also known as Cdr1as or ciRS-7, was shown to sponge miR-7 and regulate brain function in vivo.32 However, the vast majority of these circRNAs have not been previously characterized. This study strongly suggests that hsa_circ_0084443 and other circRNAs play important functional roles in skin wound healing. Further studies are warranted to determine the biological functions of these DFU-related circRNAs and whether they may be involved in the pathogenesis of chronic nonhealing wounds. It is noteworthy that in the comparison of circRNA profiles, the DFU patients (mean age of 63 years) are older than the healthy donors (mean age of 29 years), and the men/women ratio is higher in the DFU group than the control group. Given that the gene expression pattern of human skin changes with aging33 and that sex hormones are known to modulate wound healing,34 the age and gender differences between the DFU and control groups are limitations of this study that need to be addressed in subsequent studies.
In this study, we showed that hsa_circ_0084443 was downregulated during NW healing, whereas its level was higher in DFUs than NWs. A functional study revealed that hsa_circ_0084443 reduced keratinocyte motility while enhancing their growth. Thus, the reduced hsa_circ_0084443 expression at the proliferative phase of wound healing may be important for wound re-epithelization, whereas its increased expression in DFU may be partially responsible for the impaired phenotype of wound-edge keratinocytes, which is characterized by deficient migratory ability and increased proliferation.35 Of note, the biological function of hsa_circ_0084443 is not mediated through its cognate linear isoform PRKDC, since neither overexpression nor knockdown of hsa_circ_0084443 expression affected the mRNA expression of PRKDC in keratinocytes.
To understand the mechanisms underlying the biological effect of hsa_circ_0084443, we performed global transcriptomic profiling of keratinocytes by knocking down hsa_circ_0084443, which revealed that several signaling pathways important for wound healing, such as EGFR, MAPK/ERK, Pl3K, and HIF-1, may be regulated by hsa_circ_0084443 in keratinocytes. EGFR signaling has been known to play a central role in regulating re-epithelialization, angiogenesis, cell proliferation, and survival.36 One of its ligands, HB-EGF, can increase keratinocyte migration while decreasing cell growth and it is indispensable for skin re-epithelialization.23,24,37 In this study, we found that HBEGF was upregulated after knocking down hsa_circ_0084443, and it was downregulated after overexpressing hsa_circ_0084443. This reinforces the observation that HB-EGF may mediate the antimigratory and pro-proliferative effects of hsa_circ_0084443 through EGFR signaling in keratinocytes. Interconnected with EGFR signaling, MAPK/ERK activation is crucial for wound healing by regulating cell adhesion and migration.38,39 We showed that by blocking MEK1/2, a MAPK/ERK kinase, the pro-migratory effect of hsa_circ_0084443 kockdown was abolished. Also, MAPK and PI3K cascades have been shown to activate HIF-1α.40 Alongside its role in maintaining oxygen homeostasis in tissue, HIF-1α is also a key player in controlling keratinocyte motility.25 Interestingly, we observed increased HIF1A levels in keratinocytes with enhanced motility after hsa_circ_0084443 depletion. Collectively, this gene network has a potential to mediate the biological effect of hsa_circ_0084443 on keratinocyte migration and proliferation.
In short, we have characterized the expression profiles of circRNAs in human normal skin wounds and in DFUs, which identified a list of circRNAs dysregulated in chronic nonhealing wounds. As a proof of principle to demonstrate the biological role of circRNAs in wound healing, we confirmed the involvement of an upregulated circRNA in DFU, hsa_circ_0084443, in human keratinocyte migration, and proliferation. Our findings strongly support the functional and potential clinical significance of circRNAs in skin wound healing. Further efforts are warranted to understand the exact roles of these DFU-related circRNAs and the underlying molecular mechanisms, which will reveal new and unanticipated biology, advancing our understanding of the pathophysiology of DFU.
Innovation
We identified a list of circRNAs dysregulated in human DFU, which paves the way to further study the functional role of circRNAs in human skin wound healing. Moreover, we found that hsa_circ_0084443 was upregulated in DFU and acts as a negative regulator of keratinocyte migration, suggesting a role in the pathology of the disease. Our findings strongly support the potential clinical significance of circRNAs in human skin wound healing. Further efforts to understand the exact functions of these DFU-related circRNAs will advance our understanding of the pathophysiology of DFU and offer the possibility for the development of more effective wound healing treatments.
Key Findings
We identified a circular RNA signature in DFU.
We found increased levels of hsa_circ_0084443 in DFU.
Hsa_circ_0084443 reduces motility and promotes the growth of keratinocytes, suggesting a pathological role in wound-edge keratinocytes of DFU.
Supplementary Material
Abbreviations and Acronyms
- B2M
beta-2-microglobulin
- CircRNA
circular ribonucleic acid
- DFU
diabetic foot ulcer
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- FC
fold change
- FDR
false discovery rate
- GO
gene ontology
- HB-EGF
heparin-binding EGF-like growth factor
- HIF1A
hypoxia inducible factor 1 alpha
- HKGS
human keratinocyte growth supplement
- HSPA1A
heat shock protein family A member 1A
- MAPK
mitogen-activated protein kinase
- mRNA
messenger ribonucleic acid
- NW
normal wound
- PCO
phase count object
- PI3K
phosphatidylinsitol-3-kinase
- PRKDC
protein kinase DNA-activated catalytic polypeptide
- PTPN11
protein tyrosine phosphatase, nonreceptor type 11
- PXN
paxillin
- qRT-PCR
quantitative real-time polymerase chain reaction
- SHDMU
Second Hospital of Dalian Medical University
- SiRNA
small interfering ribonucleic acid
Acknowledgements and Funding Sources
We express our gratitude to the patients and healthy donors participating in this study. We thank the Microarray core facility at Novum, BEA, which is supported by the board of research at the Karolinska Institute and by the research committee at the Karolinska University Hospital.
This study was supported by the Swedish Research Council (Vetenskapsrådet, Dnr. 2015-06246 and 2016-02051), Ragnar Söderbergs Foundation (M31/15), Hedlunds Foundation, Welander and Finsens Foundation (Hudfonden), Åke Wibergs Foundation, Jeanssons Foundation, Swedish Psoriasis Foundation, Ming Wai Lau Centre for Reparative Medicine, Tore Nilsons Foundation, Lars Hiertas Foundation and Karolinska Institutet. This study was also aided by the National Natural Science Foundation of China (No. 81611130075) and the Natural Fund of the Liaoning Provincial Science and Technology Department (No. 20180530070).
Author Disclosure and Ghostwriting
No competing financial interests exist. The content of this article was expressly written by the authors listed. No ghostwriters were used to write this article.
About the Authors
Aoxue Wang, MD, PhD, is a professor and physician in the department of dermatology, The Second Hospital of Dalian Medical University, College of Integrative Medicine, Dalian Medical University (DMU, Dalian, China).
Ning Xu Landén, PhD, is an associate professor and a principal investigator leading a research group working on regulatory RNAs and skin wound healing at Karolinska Institute (Stockholm, Sweden).
Maria A. Toma and Xi Li are PhD candidates, Dongqing Li, PhD, is an assistant professor, and Manika Vij, PhD, is a postdoctoral fellow in Dr. Xu Landén's Laboratory. Jingxin Ma, PhD, is an associate professor in the department of cell biology, DMU. Tongbin Chu, MD, is the director of department of wound regeneration, The Second Hospital of Dalian Medical University. Jing Wang is a master student working in Dr. Wang's research team.
Supplementary Material
References
- 1. World Health Organization. Global Report on Diabetes. Geneva, Switzerland: World Health Organization, 2016;978:88 [Google Scholar]
- 2. Guo S, DiPietro L. Factors affecting wound healing. J Dent Res 2010;89:219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Frykberg RG, Zgonis T, Armstrong DG, et al. Diabetic foot disorders. A clinical practice guideline (2006 revision). J Foot Ankle Surg 2006;45:S1–S66 [DOI] [PubMed] [Google Scholar]
- 4. Baltzis D, Eleftheriadou I, Veves A. Pathogenesis and treatment of impaired wound healing in diabetes mellitus: new insights. Adv Ther 2014;31:817–836 [DOI] [PubMed] [Google Scholar]
- 5. Catrina SB, Zheng X. Disturbed hypoxic responses as a pathogenic mechanism of diabetic foot ulcers. Diabetes Metab Res Rev 2016;32:179–185 [DOI] [PubMed] [Google Scholar]
- 6. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development 2016;143:1838–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Venø MT, Hansen TB, Venø ST, et al. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol 2015;16:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stoll L, Sobel J, Rodriguez-Trejo A, et al. Circular RNAs as novel regulators of β-cell functions in normal and disease conditions. Mol Metab 2018;9:69–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. You X, Vlatkovic I, Babic A, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 2015;18:603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kai D, Yannian L, Yitian C, et al. Circular RNA HIPK3 promotes gallbladder cancer cell growth by sponging microRNA-124. Biochem Biophys Res Commun 2018;503:863–869 [DOI] [PubMed] [Google Scholar]
- 12. Yang ZG, Awan FM, Du WW, et al. The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol Ther 2017;25:2062–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chung JH. The role of DNA-PK in aging and energy metabolism. FEBS J 2018;285:1959–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ferguson BJ, Mansur DS, Peters NE, et al. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife 2012;1:e00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goodwin JF, Knudsen KE. Beyond DNA repair: DNA-PK function in cancer. Cancer Discov 2014;4:1126–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li X, Li D, Wang A, et al. MicroRNA-132 with therapeutic potential in chronic wounds. J Invest Dermatol 2017;137:2630–2638 [DOI] [PubMed] [Google Scholar]
- 17. Li X, Li D, Wikstrom JD, et al. MicroRNA-132 promotes fibroblast migration via regulating RAS p21 protein activator 1 in skin wound healing. Sci Rep 2017;7:7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheung L, Fisher RM, Kuzmina N, et al. Psoriasis skin inflammation-induced microRNA-26b targets NCEH1 in underlying subcutaneous adipose tissue. J Invest Dermatol 2016;136:640–648 [DOI] [PubMed] [Google Scholar]
- 19. Panda AC, Gorospe M, Diseases A. Detection and analysis of circular RNAs by RT-PCR. Bio Protoc 2018;8:e2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA 2014;20:1666–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pastar I, Stojadinovic O, Yin NC, et al. Epithelialization in wound healing: a comprehensive review. Adv Wound Care 2014;3:445–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. von Mering C, Jensen LJ, Kuhn M, et al. STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res 2007;35:D358–D362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Poumay Y, De Rouvroit CL. HB-EGF, the growth factor that accelerates keratinocyte migration, but slows proliferation. J Invest Dermatol 2012;132:2129–2130 [DOI] [PubMed] [Google Scholar]
- 24. Shirakata Y. Heparin-binding EGF-like growth factor accelerates keratinocyte migration and skin wound healing. J Cell Sci 2005;118:2363–2370 [DOI] [PubMed] [Google Scholar]
- 25. Fitsialos G, Bourget I, Augier S, et al. HIF1 transcription factor regulates laminin-332 expression and keratinocyte migration. J Cell Sci 2008;121:2992–3001 [DOI] [PubMed] [Google Scholar]
- 26. Melchionna R, Bellavia G, Romani M, et al. C/EBPγ regulates wound repair and EGF receptor signaling. J Invest Dermatol 2012;132:1908–1917 [DOI] [PubMed] [Google Scholar]
- 27. Hartman ZR, Schaller MD, Agazie YM. The tyrosine phosphatase SHP2 regulates focal adhesion kinase to promote EGF-induced lamellipodia persistence and cell migration. Mol Cancer Res 2013;11:651–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Byzova TV, Goldman CK, Pampori N, et al. A mechanism for modulation of cellular responses to VEGF: activation of the integrins. Mol Cell 2000;6:851–860 [PubMed] [Google Scholar]
- 29. Wu FH, Yuan Y, Li D, et al. Extracellular HSPA1A promotes the growth of hepatocarcinoma by augmenting tumor cell proliferation and apoptosis-resistance. Cancer Lett 2012;317:157–164 [DOI] [PubMed] [Google Scholar]
- 30. Abbasi M, Gupta V, Chitranshi N, et al. Regulation of brain-derived neurotrophic factor and growth factor signaling pathways by tyrosine phosphatase Shp2 in the retina: a brief review. Front Cell Neurosci 2018;12:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sand M, Bechara FG, Gambichler T, et al. Circular RNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci 2016;83:210–218 [DOI] [PubMed] [Google Scholar]
- 32. Piwecka M, Glažar P, Hernandez-Miranda LR, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017;357:eaam8526. [DOI] [PubMed] [Google Scholar]
- 33. Li T, Yan X, Jiang M, et al. The comparison of microRNA profile of the dermis between the young and elderly. J Dermatol Sci 2016;82:75–83 [DOI] [PubMed] [Google Scholar]
- 34. Thomason HA, Williams H, Hardman MJ. Sex and sex hormones mediate wound healing. In: Klein, Sabra L, Roberts CW, eds. Sex and Gender Differences in Infection and Treatments for Infectious Diseases. Cham: Springer, 2015:31–48 [Google Scholar]
- 35. Stojadinovic O, Brem H, Vouthounis C, et al. Molecular pathogenesis of chronic wounds. Am J Pathol 2005;167;59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schneider MR, Werner S, Paus R, Wolf E. Beyond wavy hairs: the epidermal growth factor receptor and its ligands in skin biology and pathology. Am J Pathol 2008;173:14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stoll SW, Rittié L, Johnson JL, Elder JT. Heparin-binding EGF-like growth factor promotes epithelial-mesenchymal transition in human keratinocytes. J Invest Dermatol 2012;132:2148–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stoll SW, Kansra S, Elder JT. Metalloproteinases stimulate ErbB-dependent ERK signaling in human skin organ culture. J Biol Chem 2002;277:26839–26845 [DOI] [PubMed] [Google Scholar]
- 39. Escuin-Ordinas H, Li S, Xie MW, et al. Cutaneous wound healing through paradoxical MAPK activation by BRAF inhibitors. Nat Commun 2016;7:12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bárdos JI, Ashcroft M. Negative and positive regulation of HIF-1: a complex network. Biochim Biophys Acta 2005;1755:107–120 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.