Abstract
Adeno-associated viral vectors have been successfully used in laboratory and clinical settings for efficient gene delivery. In these vectors, 96% of the adeno-associated virus (AAV) genome is replaced with a gene cassette of interest, leaving only the 145 bp inverted terminal repeat (ITR) sequences. These cis-elements, primarily from AAV serotype 2, are required for genome rescue, replication, packaging, and vector persistence. Previous work from our lab and others have demonstrated that the AAV ITR2 sequence has inherent transcriptional activity, which may confound intended transgene expression in therapeutic applications. Currently, AAV capsids are extensively study for vector contribution; however, a comprehensive analysis of ITR promoter activity of various AAV serotypes has not been described to date. Here, the transcriptional activity of AAV ITRs from different serotypes (1–4, 6, and 7) was compared in numerous cell lines and a mouse model. Under the conditions used here, all ITRs tested were capable of promoting transgene expression both in vitro and in vivo. However, we observed three classes of AAV ITR expression in vitro. Class I ITRs (AAV2 and 3) generated the highest level, whereas class II (AAV 4) had intermediate levels, and class III (AAV1 and 6) had the lowest levels. These expression levels were consistent across multiple cell lines. Only ITR7 demonstrated cell-type dependent transcriptional activity. In vivo, all classes had promoter activity. Next-generation sequencing revealed multiple transcriptional start sites that originated from the ITR sequence, with most arising from within the Rep binding element. The collective results demonstrate that the serotype ITR sequence may have multiple levels of influence on transgene expression cassettes independent of promoter selection.
Keywords: AAV, ITR, promoter
Introduction
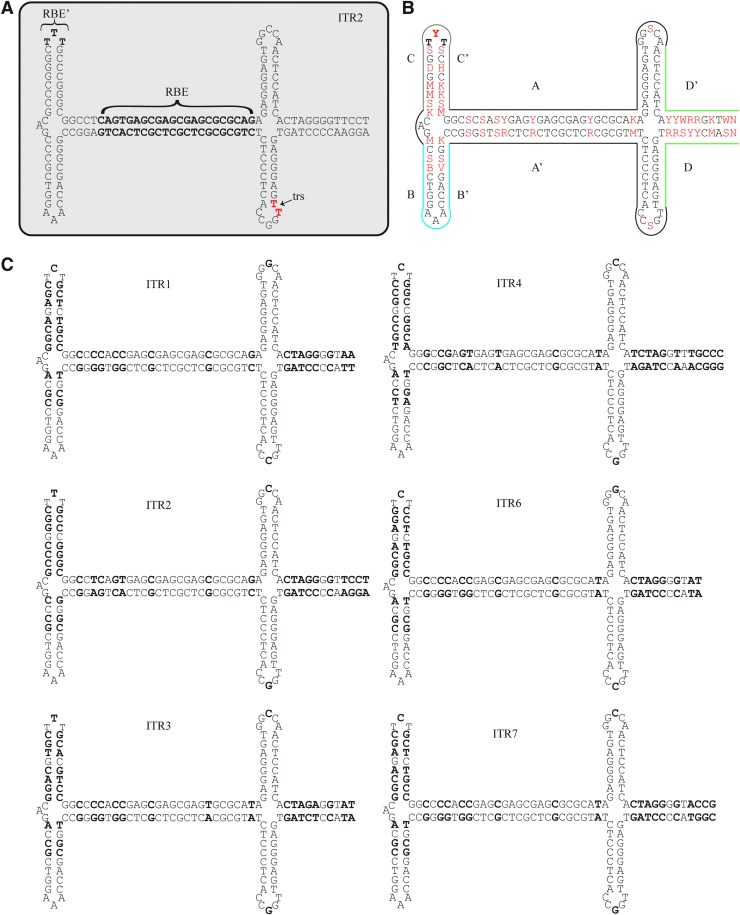
The wild-type viral genome of the canonical adeno-associated virus (AAV) serotype 2 is a ssDNA genome of ∼4,700 nucleotides (nts) and contains multiple genes with overlapping reading frames. The ends of the genome are flanked by 145 nt inverted terminal repeat (ITR) sequences that are predicted to 45-fold back on themselves to form hairpin structures (Fig. 1A). The Cap gene produces the capsid viral proteins 1, 2, and 3 and also contains the reading frame for assembly-activating protein (AAP), which helps in assembly of the capsid.1 The AAV2 Rep gene produces four proteins named for their approximate weights: Rep78, Rep68, Rep50, and Rep42. The small Reps, 50 and 42, can act as motor proteins to package nascent genomes into preformed capsids.2,3 The large Reps, 78 and 68, have endonuclease and ATP-dependent helicase functions that are necessary for genomic replication.4 These large Reps can initiate genome replication by binding to the Rep binding element (RBE) in the A region of the ITR (Fig. 1A–C).5–7 This initial binding helps to unwind the DNA strands and form a nicking stem that is cleaved by Rep at the dinucleotide TT terminal resolution site (trs).4,8 In addition, the large Rep proteins also make contact with the RBE’ region at the tip of the C-loop (Fig. 1A).5 The ITR plays a fundamental role in the life cycle of AAV by containing the replication of origin, packaging signals, and the ability to confer persistence to AAV genomes after infection. For AAV serotypes 1–4 and 6–7, the predicted structure of the ITR is alike but there are sequence differences throughout, notably in the number of GAGC repeats in the RBE, the TTT or TCT at the RBE’, the nucleotides in the hairpin loops, and the nucleotides in the D-region that do not participate in the formation of the nicking stem (Fig. 1B, C). Even with these differences, the AAV2 Reps are capable of replicating and cross-packaging genomes from serotypes 1, 3, 4, and 6 into numerous, non-AAV2 capsids.9,10
Figure 1.
Sequence and structures of AAV ITRs from serotypes 1–4, 6, and 7. (A) ITR2 with the RBE and RBE’ in bold. The terminal resolution nicking site TT dinucleotide is in red. (B) Consensus ITR sequence. Locations of nucleotide differences between ITR sequences 1–4, 6–7 are highlighted in red. The red nucleotides are in IUPAC code, where Y is C or T, R is A or G, S is G or C, W is A or T, K is G or T, M is A or C, B is G or T or C, V is G or C or A, and N is any nucleotide. Colored outlines denote the A (black), B (blue), C (gray), and D (green) regions in the ITR. (C) The sequence and structures of ITRs 1–4, 6–7. Bold letters denote non-conserved nucleotides between the ITR sequences. AAV, adeno-associated virus; RBE, Rep binding element; trs, terminal resolution site.
Currently, recombinant AAV (rAAV) vector production platforms rely on an AAV2 Rep—AAV2 ITR replication and packaging system.11 In rAAV, the internal genes of AAV are removed, leaving only the ITRs to flank the therapeutic cassette. Thus, in a clinical setting, patients receiving gene therapy are exposed not only to the capsid proteins but also to the native viral AAV2 ITR sequences. The impact of these sequences in cells has been historically understudied, but it is known that the AAV ITR interacts with a number of host proteins and can stimulate anti-viral and DNA damage response pathways.12–16 In addition, the ITR sequence from AAV2 is a promoter that is capable of driving transgene expression. This was first described by Flotte et al. in 1993 during work to find a small promoter suitable for cystic fibrosis gene therapy.17 Later work by Rubenstein et al. demonstrated that both CFTR mRNA and protein from ITR-promoted CFTR vectors were detectable in the lungs of injected rabbits.18 Our lab became interested in the promoter ability of the ITR2 after high levels of background expression were observed from an inducible AAV-based reporter system.19 Subsequent work identified a 37-nt region in the A/D junction as important for ITR promoter activity.20
Given that ITR sequences vary by AAV genotype, characterizing the promoter activity of non-AAV2 ITRs may shed light on the mechanism or sequence requirements needed for transgene expression. Here, various cell lines were infected with AAV vectors containing the ITR sequences from AAV serotypes 1, 2, 3, 4, 6, and 7 and their ability to promote luciferase expression was measured in vitro. In addition, the transcription start sites (TSS) for ITRs 1–4, 6, or 7 were determined by using amplification of luciferase specific complementary DNA (cDNA). Finally, floxed-luciferase mice were injected with ITR-cre recombinase vectors to assess ITR promoter ability in a mouse model to determine whether non-ITR2 sequences could also elicit transgene expression in vivo.
Methods
Plasmid construction
The ITR sequences from AAV serotypes 1–4 and 6–7 sequences were ordered from Genscript with unique restriction enzyme sites flanking the sequences for downstream cloning. These ITRs were ordered with one ITR per plasmid to prevent potential intermolecular recombination during synthesis and propagation. These plasmids were electroporated into SURE Electroporation-Competent Cells (200227; Agilent). Colony plasmids were screened for intact ITR sequences by using restriction enzymes specific to the ITR genotype and then cloned into a pUC19 backbone with a 20 nt stuffer sequence chosen randomly from lambda phage DNA (each plasmid contained the same stuffer sequence), followed by the reading frame for luciferase or cre recombinase, an SV40 early polyA signal, and 2,172 nts of lambda phage DNA stuffer sequence to bring the total length of the AAV vector genome to 4,395 or 3,778 bases, respectively (Supplementary Fig. S1). After plasmid construction, the ITR sequences were verified by using the illustra TempliPhi Sequence Resolver Kit (28903529; GE Life Sciences) followed by Sanger sequencing. After sequence confirmation, every subsequent plasmid prep was digested with multiple ITR-specific restriction enzymes to ensure the presence and stability of the ITR sequence. 5′ ITR sequences for ITR1, 2, 3, 4, and 7 were obtained from GenBank: ITR1: NC_002077.1, nts 1-143, ITR2: NC_001401.2, nts 1-145, ITR3: JB292182.1, nts 1-143, ITR4: NC_001829.1, nts 1-146, ITR7: NC_006260.1, nts 1-145. ITR6 was obtained from Grimm et al. 2006.10 3′ ITR sequences were the reverse complement of the 5′ sequence.
Cell lines
HEK293, HeLa, and Huh7 cells were maintained at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium with 10% bovine calf serum and 1% penicillin–streptomycin.
Virus production
rAAV vectors were produced by using the triple transfection method as previously described.11,21 Briefly, 15 cm plates of HEK293 cells at ∼80% confluency were transfected with ITR-containing luciferase or cre recombinase vector plasmids (Supplementary Fig. S1), an AAV helper plasmid containing AAV2 Rep and AAV1, 2, or 9 Cap genes,9 and the Ad helper plasmid pXX6–80. Two days post-transfection, the cells were collected, lysed, and subjected to a CsCl gradient ultracentrifugation. Fractions corresponding to the highest concentration of virus were taken and dialyzed in phosphate buffered saline (Slide-A-Lyzer Dialyses Cassettes MWCO 30,000, #66003; Thermo Fisher). Virus titer was determined in triplicate by quantitative polymerase chain reaction (qPCR) using transgene or stuffer-sequence specific primers and a viral standard containing the same transgene or stuffer sequence. A new batch of virus was made for every triplicate experiment, viruses were only used in the same experiment if they had been produced and titered together (i.e., for Fig. 2E–G, 5 batches of virus were made three times to test the AAV1–4, and 6 ITRs were used in triplicate for a total of 15 batches of virus).
Figure 2.
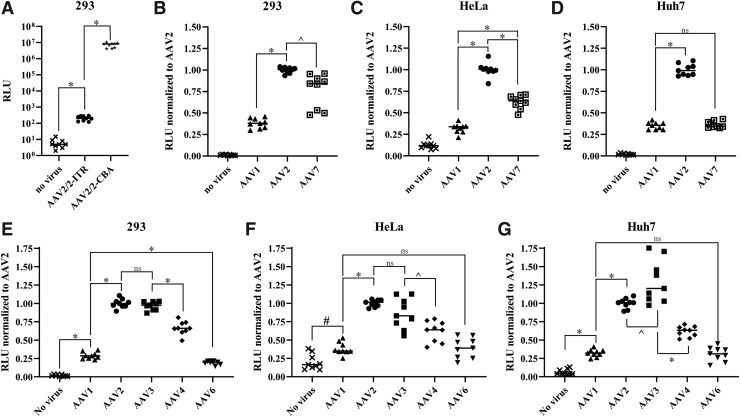
Luciferase activity from cell lines infected with AAV(1–4, 6–7)/2—ITR-luciferase vectors. (A) HEK293 cells were infected with AAV2/2-ITR-luciferase or AAV2/2-CBA-luciferase at 1E5 vg/cell. Two days post-infection, the cells were lysed and luciferase activity was measured by using luciferin substrate. Values shown are raw RLUs. (B–D) Indicated cell lines were infected with AAV1/2-ITR-luciferase, AAV2/2-ITR-luciferase, or AAV7/2-ITR-luciferase at 2E5 vg/cell. Two days post-infection, the cells were lysed and luciferase activity was measured by using luciferin substrate. RLU values were normalized to total cellular protein added to the luciferase assay, as measured by BCA; then, all values were normalized to ITR2. Each AAVN/2-ITR-luciferase was made in triplicate batches and titered together; then, cells were infected in triplicate. (E–G) Indicated cell lines were infected with AAV1/2-ITR-luciferase, AAV2/2-ITR-luciferase, AAV3/2-ITR-luciferase, AAV4/2-ITR-luciferase, or AAV6/2-ITR-luciferase at 2E5 vg/cell. Two days post-infection, the cells were lysed and luciferase activity was measured by using luciferin substrate. RLU values were normalized to total cellular protein added to the luciferase assay, as measured by BCA; then, all values were normalized to ITR2. Each AAVN/2-ITR-luciferase was made in triplicate batches and titered together; then, cells were infected in triplicate. p-Values are indicated as *<0.0001, #<0.001, and ^<0.01. ns, nonsignificant; RLU, relative light units.
Figure 5.
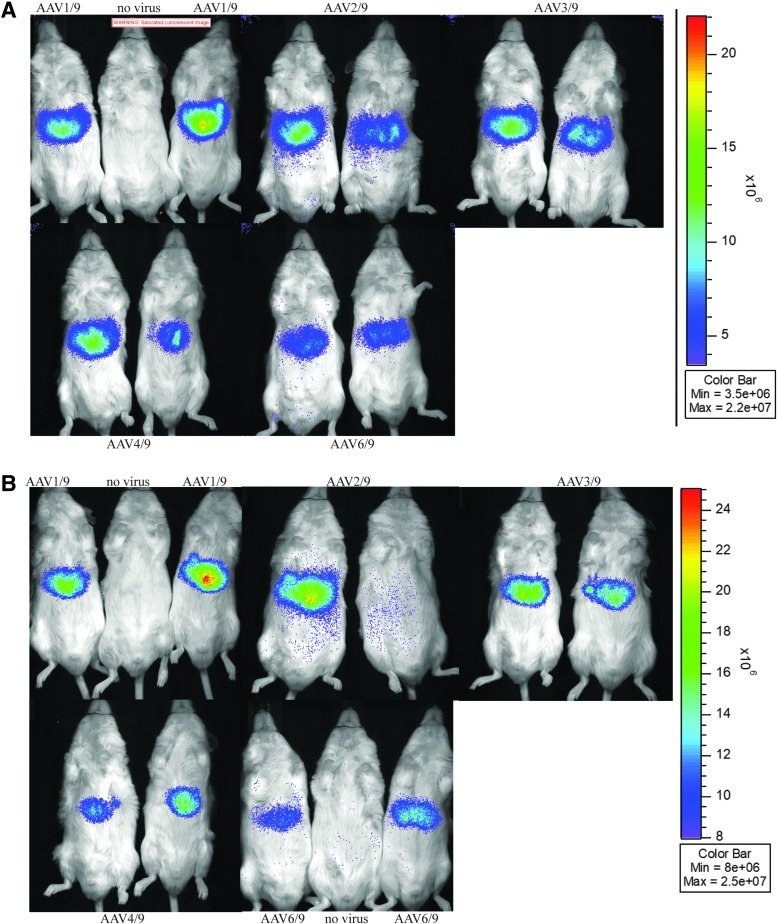
Luciferase activity from mice injected with AAV(1–4, 6)/9-ITR-cre recombinase. Four- to 6 week-old male FVB.129S6(B6)-Gt(ROSA)26Sortm1(Luc)Kael/J mice were injected with 100 μL of 1E9 vg of AAV(1–4, 6)/9-ITR-cre recombinase. At 3 weeks (A) and at 9 weeks (B) post-AAV injection, mice were given 100 μL of luciferase substrate i.p. and photons were recorded.
In vitro infection and luciferase assays
For comparison of AAV2/2-ITR-luciferase and AAV2/2-CBA-luciferase, 3.2E5 HEK293 cells were plated per well in 12-well plates and were infected the next day with 1E5 vg/cell. Two days post-infection, cells were lysed in 200 μL of Passive Lysis Buffer (PLB) for 20 min at room temperature. Cell lysate from AAV2/2-CBA-luciferase was diluted 1:50 in PLB. Twenty-five microliters of cell lysate was combined with 100 μL of luciferin (Luciferase Assay System, E1500; Promega) in a 96-well opaque white assay plate, and luminescence was measured with the Perkin Elmer Victor3 plate reader. Relative light units (RLUs) values from AAV2/2-CBA-luciferase were multiplied by their dilution factor. The nomenclature used here to denote the ITR and capsid serotype is AAV(ITR genotype)/capsid serotype.
For comparison of ITR-luciferase vectors, HEK293, HeLa, and Huh7 cells were plated individually into 6-well plates. The next day, the cells were infected with 2E5 vg/cell of AAV(N)/2-ITR-luciferse where N is the indicated ITR genotype. Two days later, the medium was removed and the cells were lysed in 350 μL of PLB for 20 min at room temperature. The lysate was transferred to 1.5 mL tubes and spun at 13,000 g for 10 min at 4°C. Twenty-five microliters of lysate was used in a BCA (Pierce™ BCA Protein Assay Kit, 23225) assay to determine total cellular protein concentration. One hundred microliters of cell lysate was combined with 100 μL of luciferin (Luciferase Assay System, E1500; Promega) in a 96-well opaque white assay plate, and luminescence was measured with the Perkin Elmer Victor3 plate reader. RLUs were normalized to total protein added.
Identification of TSS
HEK293 cells were infected with AAV(1–4, 6, or 7)/2-ITR-luciferase at 2E5 vg/cell and cultured for 3 days before RNA harvest using a Qiagen RNeasy kit. Following the manufacturer's instructions from the 5′/3′ rapid amplification of cDNA ends (RACE) second-generation kit (3353621001; Roche), ∼750 ng of RNA was reverse transcribed by using a primer located within the luciferase coding sequence (5′-GTGACGAACGTGTACATCGAC-3′). The synthesized cDNA was then purified by using the QIAquick PCR purification kit (28104; QIAGEN), and a polyA tail was added by terminal transferase as per the manufacturer's instructions. PCR was then conducted by using the supplied forward primer, 5′-GACCACGCGTATCGATGTCGACTTTTTTTTTTTTTTTTV-3′, and a nested reverse primer in the luciferase coding sequence, 5′-CTTAGAACCGGTCGAACACCACGGTAGGCT-3′. The resulting PCR product was purified and used as a template for an additional PCR reaction with the kit supplied forward primer 5′-GACCACGCGTATCGATGTCGAC-3′, and another nested reverse primer within luciferase sequence 5′-TTAGTTGGATCCGGTTCCATCTTCCAGCGG-3′. The product was purified and normalized to 20 ng/ul EB buffer; and 25 μL was sent for EZ amplicon sequencing by using next-generation sequencing (NGS) by Genewiz. Resulting NGS data were analyzed by the UNC Lineberger Bioinformatics Core using STAR v2.7.0a22 to align reads to the reference genomes. The bam files were processed in R to tabulate the frequency of the alignment start site. Sequences with multiple mismatches (>3) in the first 10 bases of alignment were filtered, as we could not infer whether the alignment should start before or after the mismatches. Read pairs with an insert size greater than expected (1,000 bp) were also removed.
Animal study
Animal experiments performed in this study were conducted with FVB.129S6(B6)-Gt(ROSA)26Sortm1(Luc)Kael/J mice23 (Stock No: 005125; Jackson Laboratories). The mice were maintained in accordance to National Institutes of Health guidelines, as approved by the UNC Institutional Animal Care and Use Committee (IACUC; Protocol number 19.023-1). Male mice were housed individually due to fighting. Each mouse was injected via tail vein with 100 μL of 1E9 viral genomes. Luciferase expression was imaged by using the IVIS Kinetic (Caliper Lifesciences, Waltham, MA) following a 100 μL i.p. injection of D-luciferin substrate (XenoLight D-Luciferin, 122799; Perkin Elmer, Waltham, MA). Bioluminescent images were analyzed by using Living Image (PerkinElmer). Acquisition was performed by using Living Image software version 2.20 using photon values.
Statistics
All statistical calculations were performed by using statistical software (GraphPad Prism 8.2). Data are presented as individual points with the group mean. Data for single comparisons were evaluated by using an unpaired two-tailed t test. Differences between different groups were considered statistically significant when p-values were less than 0.05.
Results
ITR serotype sequences have variable ability to promote luciferase expression in vitro.
To determine the promoter activity of ITRs from various AAV genotypes, luciferase reporter vector plasmids were constructed by using an AAV ITR sequence as the promoter (Supplementary Fig. S1). In vector plasmids, ITR sequences were assessed with multiple restriction enzymes to confirm the presence of the ITRs and their genotype identity. Initially, the activity of ITR2 was compared with the “strong” CBA promoter. AAV2-ITR-luciferase and AAV2-CBA-luciferase were packaged into AAV2 capsids and used to infect HEK293 cells at 1E5 vg/cell. Two days post-infection, luciferase activity was measured and found to be more than 4 logs higher from the CBA-promoted luciferase compared with ITR2-promoted luciferase, which is similar to previous findings24 (Fig. 2A). This demonstrated that ITR2 promoter activity could be successfully measured by using a luciferase reporter system.
The use of ITR7 in an AAV2 Rep packaging system has yet to be reported in the literature. To test the feasibility of using ITR7 in combination with AAV2 Rep and an AAV2 capsid, ITR7 containing vectors were transfected into HEK293 cells with an adenoviral helper and pXR2. Resulting virus was titered by qPCR. ITR7 vectors had similar titers to ITR1 and ITR2 vectors made at the same time (Table 1). HEK293, HeLa, and Huh7 cells were infected with AAV1/2, AAV2/2, and AAV7/2-ITR-luciferase at 2E5 vg/cell. Luciferase activity was measured 2 days post-infection. RLUs were normalized to total amount of cellular protein added to the luciferase assay as determined by a BCA assay and then further normalized to ITR2 RLU values (Fig. 2B–D). Interestingly, RLUs from ITR1-promoted luciferase were consistently lower than those of ITR2 across all three cell lines (p < 0.0001). In HEK293 cells, ITR1 had an average of 29% activity compared with ITR2 (Fig. 2B). This activity was slightly higher in HeLa cells at 35% (Fig. 2C) and in Huh7s at 32% (Fig. 2D). In contrast, ITR7 displayed different promoter activity across the cell lines. In Huh7 cells, ITR7 and ITR1 had the same expression level, 33% and 31% respectively, compared with ITR2 (Fig. 2D), but ITR7 had higher expression than ITR1 in HeLa cells at 62% (p < 0.0001) (Fig. 2C). In HEK293 cells, ITR7-promoted luciferase activity rose to an average of 83%, with one preparation of virus having reduced activity compared with the other two preparations of virus (Fig. 2B), but this overall activity was still lower than that of ITR2 (p = 0.0029).
Table 1.
Titers from vector preps using pXX6-80 and pXR2 helper plasmids in vg/μL
| Batch 1 | Batch 2 | Batch 3 | |||
|---|---|---|---|---|---|
| AAV1 | 8.88E+08 | AAV2 | 1.39E+09 | AAV2 | 2.92E+09 |
| AAV2 | 8.66E+08 | AAV1 | 1.11E+09 | AAV1 | 2.77E+09 |
| AAV7 | 7.28E+08 | AAV7 | 1.05E+09 | AAV7 | 1.87E+09 |
| Batch 1 | Batch 2 | Batch 3 | |||
|---|---|---|---|---|---|
| AAV2 |
3.04E+08 |
AAV2 |
8.72E+08 |
AAV2 |
1.01E+09 |
| AAV4 |
2.42E+08 |
AAV1 |
7.65E+08 |
AAV4 |
9.23E+08 |
| AAV3 |
2.11E+08 |
AAV4 |
6.89E+08 |
AAV1 |
8.15E+08 |
| AAV1 |
1.97E+08 |
AAV3 |
3.79E+08 |
AAV3 |
5.83E+08 |
| AAV6 | 1.55E+08 | AAV6 | 2.23E+08 | AAV6 | 4.18E+08 |
AAV, adeno-associated virus.
To determine whether ITRs 3, 4, and 6 also displayed promoter activity in these cell lines, ITR-luciferase plasmids were used to create AAV(1–4, 6)/2-ITR-luciferase vectors where AAVN/N is AAV(ITR genotype)/capsid serotype. All ITRs were able to be replicated and packaged by AAV2 Rep, and batch titers were within 4-fold of each other.10 Of note, under these replication, packaging, and purification conditions, titers from ITR2-containing vector plasmids were usually the highest, whereas ITR6 or ITR7 were always the lowest (Table 1). The lower yield with ITR6 has also been previously reported.10
HEK293, HeLa, and Huh7 cells were infected with AAV(1–4, 6, or 7)/2-ITR-luciferase vectors at 2E5 vg/ cell. Luciferase activity was measured as described earlier. ITRs 3, 4, and 6 also resulted in luciferase activity in the cell lines tested, but to varying degrees (Fig. 2E–G). In all three cells lines, ITR2 and ITR3 resulted in the highest luciferase activity. Only in Huh7 cells was there a significant difference between ITR2 and ITR3 (p = 0.0082), with ITR3 averaging 30% more activity than ITR2 (Fig. 2G). Across all cell types, ITR1 and ITR6 had the lowest activity and were not statistically different than each other, except in HEK293 cells in which ITR1 was 10% lower than ITR6 (p < 0.0001) with a mean of 19% compared with 29% (Fig. 2E). ITR4 consistently had 62–66% luciferase activity compared with ITR2 (Fig. 2E–G) and was significantly lower than ITR3 as well in all three cell lines (p < 0.01). Thus, the observed activity from the ITRs fell into three classes: Class I ITRs with the highest relative activity: ITR2 and ITR3, Class II with an intermediate level of activity: ITR4, and Class III with the lowest activity: ITR1 and ITR6. Of all the ITRs tested, only ITR7 showed cell-specific activity (Fig. 2B–D).
The differing levels of luciferase activity from ITR sequences 1–4, 6, and 7 implied that the ITR sequence itself was a significant determinate of luciferase activity, but alternatively, the high luciferase activity from the ITR2-containing vectors could be due to a capsid-specific interaction since this ITR was paired with its cognate capsid. To test whether ITR sequences packaged into their corresponding capsid influenced luciferase production, ITR1 and ITR2 luciferase vectors were packaged into AAV1 capsids and used to infect HEK293 cells at 2E5 vg/cell. Although the overall activity was reduced compared with AAV1/2 and AAV2/2, luciferase activity from AAV1/1-ITR-luciferase vectors was still lower than AAV2/1 vectors. When normalized to AAV2, the activity was equivalent, regardless of the capsid used (Fig. 3). Hence, ITR1 is still a Class III ITR, even when paired with its cognate capsid.
Figure 3.
Luciferase activity from HEK293 cells infected with AAV1/1 and AAV2/1—ITR-luciferase vectors. HEK293 cells were infected in triplicate with three biological replicates of AAV1/1, AAV2/1, AAV1/2, or AAV2/2-ITR-luciferase vectors at 2E5 vg/cell. Two days post-infection, the cells were lysed and luciferase activity was measured by using luciferin substrate. RLU values were normalized to total cellular protein added to the luciferase assay, as measured by BCA; then, all values were normalized to ITR2 values within each capsid group (i.e., AAV1/1 and 2/1 were normalized to AAV2/1. AAV1/2 and 2/2 were normalized to 2/2). There is no statistical difference between AAV1/1 and AAV1/2.
The ITR sequences contain multiple TSS
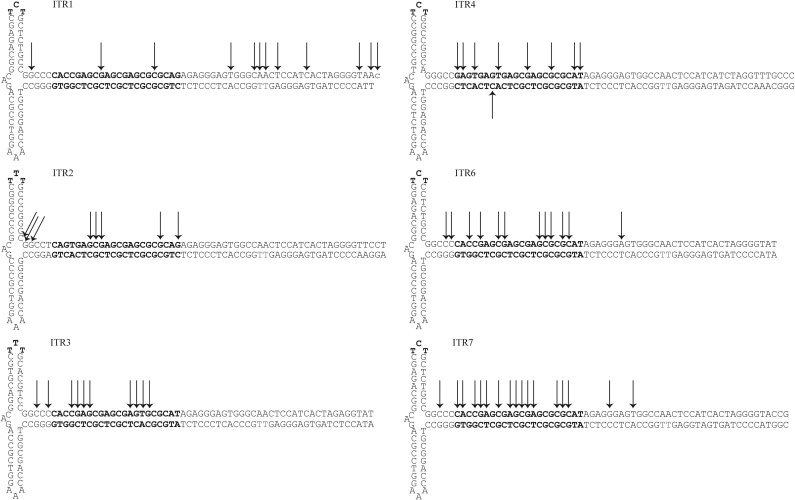
The ITR sequences of all the genotypes tested are high in CG content (64–70%) and lack a traditional TATA-box consensuses sequence. To determine whether the luciferase transcripts were originating from a single, focused TSS or multiple, dispersed TSSs, 5′ RACE was employed to find the originating nucleotide position(s). HEK293 cells were infected with AAV(1–4, 6, or 7)/2-ITR-luciferase at 2E5 vg/cell. Three days post-infection, total RNA was isolated and reverse transcribed by using a 5′ RACE kit. Luciferase-specific cDNA was amplified and analyzed by NGS. For all ITRs, multiple TSSs were found within each sequence and tended to cluster at the RBE, although ITR1 had more widespread start sites than the other ITRs (Fig. 4). For each ITR, 3–4 nts represented the majority of reads, but these hot spots were different for each ITR (Fig. 4). These results indicate that the transcripts for ITR-promoted luciferase can originate from multiple start sites within the ITR.
Figure 4.
TSS from ITR1–4, 6, or 7 promoted luciferase mRNA. HEK293 cells were infected with AAV(1–4, 6, or 7)/2-ITR-luciferase at 2E5 vg/cell, and total RNA was harvested 3 days post-infection. RNA was reverse transcribed by using luciferase specific primers. Final cDNA products were analyzed by next-generation sequencing. Black arrows indicate a TSS with 1% or higher of read sequences. The RBE’ and RBE are indicated in bold. The trs is denoted in red. TSS, transcription start sites.
Cre-recombinase driven by ITRs sequences is capable of activating luciferase production in vivo
To see how our in vitro findings translated to an in vivo model, the ITR promoter ability was tested in a floxed luciferase reporter mouse strain. The FVB.129S6(B6)-Gt(ROSA)26Sortm1(Luc)Kael/J mouse line contains a luciferase open reading frame inserted into the ROSA26 locus.23 Luciferase expression is prevented by a loxP-stop-LoxP sequence, which can be removed by cre recombinase. Four- to 6 week-old male mice were injected with 100 μL of 1E9 vg of AAV(1–4, 6)/9-ITR-cre recombinase via the tail vein (n = 2). AAV9 was chosen for its ability to highly transduce most mouse tissues.25 We reasoned that this capsid would be the best to identify tissue-specific differences, if any, between the ITR promoters. As a positive control, two mice were injected with AAV2/9-CMV-Cre vectors. Mice were imaged at 3, 5, 7, and 9 weeks post-injection. By 3 weeks post-injection, luciferase activity could be observed in the abdominal area of all mice (Fig. 5A). As expected, the positive control mice that had been injected with CMV-promoted cre recombinase had more recombined cells expressing luciferase and under the same imaging setting, these mice entirely saturated the camera (not shown). By 4 weeks post-injection, luciferase signal from one of the mice injected with AAV2/9-ITR-cre recombinase could no longer be detected. During the 9-week time course, luciferase signal remained steady in the remaining mice (Fig. 5B). We suspect that the loss of expression from the mouse injected with AAV2/9-ITR-cre recombinase was likely due to capsid antigen reactive CD8+ T cells, but we did not specifically investigate this.
Discussion
In this study, we show for the first time that the ITR sequences for AAVs 1–4, 6, and 7 have varying ability to promote transgene expression in vitro and that these sequences contain multiple TSS. In addition, we utilized a sensitive reporter mouse strain to demonstrate that at clinically relevant doses, the ITRs 1–4 and 6 have the ability to promote enough cre recombinase protein in vivo to have biological effects at the cellular level.
ITR serotype sequence influences promoter activity in vitro
The ITR promoter activity for ITRs 1–4 and 6 was consistent enough that they could be broken into three Classes: I, II, and III, with I being the highest activity. Across each cell line, ITR2 and ITR3 (Class I) consistently had the highest values for luciferase activity, implying that these sequences also have more promoter activity than the other ITRs tested, whereas ITR1 and ITR6 (Class III) had the lowest (Fig. 2). Similar expression values for ITR1 and ITR6 would be expected given the high degree of similarity between the two sequences, which differ from each other only by the last nucleotide in the D-region and 4 nts outside of the D region (Fig. 1C). A sequence analysis between Class I and Class III sequences revealed several points of variance that could explain the different activities (Fig. 1C). Specifically, ITR2 and ITR3 contained a TTT sequence in the RBE’, whereas ITR1 and ITR6 contained TCT.
There was also a consistent difference in the B-loop (positions 45:59 and 46:60) and C-loop (68:80 and 70:78), and a C to G change in the tips of the nicking stem loops at positions 3 and 122. ITR7 is also similar to the ITR1 sequence, but it had a different promoter activity profile (Fig. 2B–D). Since there are only a few nucleotides that differ between ITR1 and ITR7, a mutational analysis may be able to find the specific sequence(s) involved in the differential expression of ITR7-promoted luciferase in various cell types. The T:A pair in ITR7 at position 110:15 is the same pair seen at ITR3, 4, and 6 (Fig. 1C), so it is unlikely to be involved in the varying levels of luciferase activity we observed across the cells types tested. Similar to ITR2, 3, and 4, ITR7 also has a G near the nicking site at position 3 and a C at position 122, so these nucleotides may influence promoter strength. Another variable region of interest that could be influencing luciferase expression among the ITRs is the last 11 nt of the D region where only a CTAG motif is conserved, but there is no readily discernable pattern between the different classes of ITRs. Still, this region could be of interest since several host proteins have been shown to interact with the D region of ITR2.12,13 The question of which sequences have effects on transgene production may be addressed with position-specific ITR mutants, but given that complex DNA secondary structure may play diverse roles in transcription,14,26 this question may be difficult to unravel fully. The mechanism behind these expression differences is still under investigation. Under the conditions used here, ITR1 was still a Class III, even when packaged into an AAV1 capsid. This argues against an ITR-capsid interaction as having a strong influence on promoter activity. Previous work by Ling et al. found that using an entirely cognate system for AAV3 resulted in higher titer and greater transduction than using an AAV2 Rep to package an AAV2 ITR into an AAV3 capsid.27 In our study here, we exclusively used an AAV2 Rep, so it may still be that having the cognate Rep for these ITR sequences could influence various aspects of replication, packaging, and transducing units of rAAV.
Start sites for ITR-promoted luciferase transcripts.
Previous work done by Haberman et al. identified the A region of ITR2 as important for ITR2-promoted green fluorescent protein transgene expression.20 Here, we also found that the A region was a hotspot for transcriptional activity, but by using NGS we were able to identify multiple starts throughout the ITR sequences, primarily focused within a 40 bp region that included the RBE. This brought us to ask: What are the mechanisms by which the ITR is acting as a promoter? Clearly lacking a traditional TATA-box within the defined ITR sequence, but enriched in cytosine and guanine, these sequences bear striking similarity to the transcriptionally active CpG islands (CGIs) found in vertebrate genomes. It is now appreciated that CGIs are the most common promoter type in the vertebrate genome, occurring at 60–70% of annotated genes.28,29 CGIs are commonly defined as sequences with a C + G ratio of greater than 50% and observed-to-expected CpG dinucleotides at 60% or higher.30 The AAV ITR sequences fit this definition in both C + G content and CpG frequency (Table 2).
Table 2.
C + G content and observed-to-expected ratio of CpGs in AAV 5′ inverted terminal repeat sequences
| AAV genotype | C + G content (%) | Observed-to-expected CpG ratio (%) |
|---|---|---|
| AAV1 | 68.5 | 83.4 |
| AAV2 | 70.3 | 94.8 |
| AAV3 | 64.3 | 94.7 |
| AAV4 | 64.5 | 66.4 |
| AAV6 | 67.1 | 86.9 |
| AAV7 | 68.3 | 88.8 |
C + G content was calculated as: (C+G)/N, where C is the number of cytosines, G is the number of guanines, and N is the number of nucleotides in the 5′ ITR sequence of the indicated AAV genotype. Observed-to-expected CpG ratio was calculated by using the formula by Gardiner-Garden and Frommer29: [(CpGs)/(C × G)] × N, where CpG is the number of observed CpGs, C is the number of cytosines, G is the number of guanines, and N is the number of nucleotides in the sequence.
ITR, inverted terminal repeat.
In addition, CGIs are often origins of replication31–33 and are generally associated with multiple TSSs dispersed over a 50–100 bp region.34,35 This is in contrast to promoters with a single, focused TSS that are more commonly associated with specifically positioned core promoter elements, including the TATA-box, INR, TCT, and XCPE motifs.36 The data from our studies support a hypothesis that the ITR sequences from the AAV genotypes examined are functioning as CGI type promoters in the context of transgene promotion. That said, another interpretation of this data could be that these TSS are actually arising from distinct and variable episomal sequences.37 Since the resulting sequence that arises from the recombination of the two ITR ends after infection is variable, it may be that each episome has a different start site.
In the context of wild-type AAV, the p5 promoter has been well mapped out and contains a TATA box,38 so in this setting, these sequences could be acting in coordination with the TATA-containing promoter. Stutika et al. were able to map small RNAs from the wild-type AAV2 genome in the presence and absence of adenovirus and it is intriguing to note that in both scenarios, multiple small RNAs were present in the ITR regions with a hotspot within the RBE.39 In the absence of a helper virus when Rep78 is acting to auto-suppress transcription from the p5 promoter,40 the sequence reads from the ITR region were actually higher than those from p5,39 which raises the question of whether this promoter activity might serve a role in the life cycle of wild type AAV minimally during latency. Indeed, the impact of integration on this promoter activity is unknown, but it would be an interesting avenue for more research.
ITR1–4, 6 are capable of promoting cre recombinase in a mouse model
The in vivo data demonstrate that ITRs 1–4, 6 were able to promote high enough levels of cre recombinase to induce recombination and luciferase production. It is still an open question whether the difference in promoter activity observed in vitro is similar in vivo. It could be that in vivo, there are no differences and all ITRs promote transgene production in roughly equal amounts. Regardless, it is clear that in this strain of mice, all ITR sequences 1–4, 6 are active promoters and this may have important implications for gene therapy applications. These mice were injected with 1E11 vg, which is an approximate equivalent to 5E12 vg/kg and thus a clinically relevant dose. In the context of a strong ubiquitous promoter, these ITR sequences would likely have no effect on overall transgene production, as previously shown,10 but there are scenarios in which more targeted or sensitive applications could be affected, such as when using AAV-delivered cre recombinase or CRISPR.
More importantly, the bidirectional activity of the ITR2 promoter may be inducing the double-strand RNA (dsRNA) response pathway.21 Shao et al. found that AAV transduction stimulated MDA5, a dsRNA response protein that recognizes dsRNA products more than 2,000 nt long, at 8 days post-infection. It was proposed that the promoter activity of the ITR when in an episome confirmation may be driving minus strand RNA production, which could bind to positive strand RNA and accumulate in transduced cells.21 In this scenario, a promoter with less activity would be desirable to help blunt this arm of the innate immune response. Unfortunately, it may be an impossible task to completely eliminate promoter activity since CGI, TSS, and origins of replication are often associated together.31 Eliminating all the CGIs in the ITR would necessitate changing the RBE sequence, which has five CpGs, such that Rep could no longer efficiently bind it.5 Faust et al. were able to eliminate the CGI in the hairpins arms and still produce vector, so some CpG depletion is certainly viable but interpreting this effect on transcription alone could be complicated by innate immune pathways such as TLR9.41,42 Other strategies such as adding insulating sequences that flank the ITRs to prevent transcription read through may prove fruitful.
In summary, the data presented here show that the ITRs sequences from AAV serotypes 1–4, 6, and 7 have inherent promoter activity and this promoter activity is not at equal strength among the ITRs. Specifically, ITR2 and ITR3 sequences resulted in higher luciferase expression across multiple cell types when compared with ITRs 1, 4, and 6. ITR7 was the only ITR to display cell-specific differences in luciferase expression. The TSS were mapped to multiple locations within each ITR sequence, of which the bulk originated from a 40 bp region that contained the RBE. In vivo, all the ITRs tested had the ability to promote cre recombinase at high enough levels to induce cre-mediated recombination by 3 weeks post-injection. These data may help inform vector design strategies when sensitive or cell-specific therapies are needed.43
Supplementary Material
Acknowledgments
The authors would like to thank the UNC core services that helped with this work, specifically, Joel Parker and Wedia Gong at the Lineburger Cancer Center Bioinformatics Core for their NGS analysis. We are grateful to the veterinary and animal care staff as well as the Small Animal Imaging Facility at the UNC Biomedical Imaging Research Center for providing the luciferase imaging facility. They would also like to thank members of the UNC Gene Therapy Center Liujiang Song, Charles Askew, and Xintao Zheng for their technical advice and expertise as well as all of the authors' undergraduate staff members.
Author Disclosure
M.L.H. is an inventor on technology not assessed herein that has been licensed to Asklepios BioPharmaceutical and Tamid Bio. M.L.H. has also received royalties from Asklepios BioPharmaceutical related to a patent (9447433). M.L.H. and C.L. are co-founders of Bedrock Therapeutics. R.J.S. is the founder of and a shareholder at Asklepios BioPharmaceutical and Bamboo Therapeutics, Inc. He holds a patent (9475845) that has been licensed by the University of North Carolina at Chapel Hill to Asklepios BioPharmaceutical, for which he receives royalties. He has consulted for Baxter and has received payment for speaking. L.F.E. is currently an employee at Shape Therapeutics.
Author Contributions
L.F.E., M.L.H., and R.J.S. conceptualized and designed the experiments. L.F.E. conducted the experiments with assistance from V.M.L., L.M.C., and A.L.D. Manuscript writing was done by L.F.E. with review and editing from M.L.H. and R.J.S.
Funding Information
The imaging core is supported in part by an NCI cancer core grant, P30-CA016086-40. This work was supported by National Institutes of Health grants RO1AI072176-06A and RO1AR064369-01A (MLH). L.F.E. was funded by the Pfizer—NC Biotech Distinguished Postdoctoral Fellowship in Gene Therapy (GTF) Program (Agreement 2018-GTF-6907).
Supplementary Material
References
- 1. Sonntag F, Schmidt K, Kleinschmidt JA. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc Natl Acad Sci U S A 2010;107:10220–10225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. King JA, Dubielzig R, Grimm D, et al. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J 2001;20:3282–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Myers MW, Carter BJ. Assembly of adeno-associated virus. Virology 1908;102: 71–82 [DOI] [PubMed] [Google Scholar]
- 4. Im DS, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell 1990;61:447–457 [DOI] [PubMed] [Google Scholar]
- 5. Ryan JH, Zolotukhin S, Muzyczka N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J Virol 1996;70:1542–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McCarty DM, Pereira DJ, Zolotukhin I, et al. Identification of linear DNA sequences that specifically bind the adeno-associated virus rep protein. J Virol 1994;68:4988–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bishop BM, Santin AD, Quirk JG, et al. L. Role of terminal repeat GAGC trimer, the major Rep78 binding site, in adeno-associated virus DNA replication. FEBS Lett 1996;397:97–100 [DOI] [PubMed] [Google Scholar]
- 8. Brister JR, Muzyczka N. Mechanism of Rep-mediated adeno-associated virus origin nicking. J Virol 2000;74:7762–7771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rabinowitz JE, Rolling F, Li C, et al. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J Virol 2002;76:791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grimm D, Pandey K, Nakai H, et al. Liver transduction with recombinant adeno-associated virus is primarily restricted by capsid serotype not vector genotype. J Virol 2006;80:426–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xiao X, Li J, Samulski RJ. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol 1998;72:2224–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Julien L, Chassagne J, Peccate C, et al. RFX1 and RFX3 transcription factors interact with the D sequence of adeno-associated virus inverted terminal repeat and regulate AAV transduction. Sci Rep 2018;8:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qing K, Li W, Zhong L, et al. Adeno-associated virus type 2-mediated gene transfer: role of cellular FKBP52 protein in transgene expression. J Virol 2001;75:8968–8976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Satkunanathan S, Thorpe R, Zhao Y. The function of DNA binding protein nucleophosmin in AAV replication. Virology 2017;510:46–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raj K, Ogston P, Beard P. Virus-mediated killing of cells that lack p53 activity. Nature 2001;412:914–917 [DOI] [PubMed] [Google Scholar]
- 16. Hirsch ML, Fagan BM, Dumitru R, et al. Viral single-strand DNA induces p53-dependent apoptosis in human embryonic stem cells. PLoS One 2011;6:e27520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Flotte TR, Solow R, Owens RA, et al. Gene expression from adeno-associated virus vectors in airway epithelial cells. Am J Respir Cell Mol Biol 1992;7:349–356 [DOI] [PubMed] [Google Scholar]
- 18. Rubenstein RC, McVeigh U, Flotte TR, et al. CFTR gene transduction in neonatal rabbits using an adeno-associated virus (AAV) vector. Gene Ther 1997:4;384–392 [DOI] [PubMed] [Google Scholar]
- 19. Haberman RP, McCown TJ, Samulski RJ. Inducible long-term gene expression in brain with adeno-associated virus gene transfer. Gene Ther 1998;5:1604–1611 [DOI] [PubMed] [Google Scholar]
- 20. Haberman RP, McCown TJ, Samulski RJ. Novel transcriptional regulatory signals in the adeno-associated virus terminal repeat A/D junction element. J Virol 2000;74:8732–8739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shao W, Earley LF, Chai Z, et al. Double-stranded RNA innate immune response activation from long-term adeno-associated virus vector transduction. JCI Insight 2018;3:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Safran M, Kim WY, Kung AL, et al. Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre-mediated recombination. Mol Imaging 2003;2:297–302 [DOI] [PubMed] [Google Scholar]
- 24. Wang D, Fischer H, Zhang L, et al. Efficient CFTR expression from AAV vectors packaged with promoters—the second generation. Gene Ther 1999;6:667–675 [DOI] [PubMed] [Google Scholar]
- 25. Inagaki K, Fuess S, Storm TA, et al. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol Ther 2006;14:45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet 2012;13:770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ling C, Yin Z, Li J, et al. Strategies to generate high-titer, high-potency recombinant AAV3 serotype vectors. Mol Ther Methods Clin Dev 2016;3:16029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A 2006;103:1412–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu J, He F, Hu S, et al. On the nature of human housekeeping genes. Trends Genet 2008;24:481–484 [DOI] [PubMed] [Google Scholar]
- 30. Gardiner-Garden M, Frommer M. CpG Islands in vertebrate genomes. J Mol Biol 1987;196:261–282 [DOI] [PubMed] [Google Scholar]
- 31. Delgado S, Gómez M, Bird A, et al. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J 1998;17:2426–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sequeira-Mendes J, Díaz-Uriarte R, Apedaile A, et al. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet 2009;5:e1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Antequera F, Bird A. CpG islands as genomic footprints of promoters that are associated with replication origins. Curr Biol 1999;9:R661–R667 [DOI] [PubMed] [Google Scholar]
- 34. Carninci P, Sandelin A, Lenhard B, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 2006;38:626–635 [DOI] [PubMed] [Google Scholar]
- 35. Juven-Gershon T, Hsu JY, Theisen JW, et al. The RNA polymerase II core promoter—the gateway to transcription. Curr Opin Cell Biol 2008;20:253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Danino YM, Even D, Ideses D, et al. The core promoter: at the heart of gene expression. Biochim Biophys Acta 2015;1849:1116–1131 [DOI] [PubMed] [Google Scholar]
- 37. Duan D, Yan Z, Yue Y, et al. Structural analysis of adeno-associated virus transduction circular intermediates. Virology 1999;261:8–14 [DOI] [PubMed] [Google Scholar]
- 38. Green MR, Roeder RG. Definition of a novel promoter for the major adenovirus-associated virus mRNA. Cell 1980;22:231–242 [DOI] [PubMed] [Google Scholar]
- 39. Stutika C, Mietzsch M, Gogol-Döring A, et al. Comprehensive small RNA-Seq of adeno-associated virus (AAV)-infected human cells detects patterns of novel, non-coding AAV RNAs in the absence of cellular miRNA regulation. PLoS One 2016:11;e0161454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Beaton A, Palumbo P, Berns KI. Expression from the adeno-associated virus p5 and p19 promoters is negatively regulated in trans by the rep protein. J Virol 1989;63:4450–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhu J, Huang X, Yang Y. The TLR9-MyD88 pathway is critical for adaptive immune responses to adeno-associated virus gene therapy vectors in mice. J Clin Invest 2009;119:2388–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Faust SM, Bell P, Cutler BJ, et al. CpG-depleted adeno-associated virus vectors evade immune detection. J Clin Invest 2013;123:2994–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wilmott P, Lisowski L, Alexander IE, et al. user's guide to the inverted terminal repeats (ITR) of adeno-associated virus. Hum Gene Ther Methods 2019;30:206–213 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.