Highlights
-
•
E. fetida microbiota is affected by pH and carbon content of rearing substrates.
-
•
This is the first report of the microbiota of E. fetida fed with brewers’ spent grains.
-
•
Brewers’ spent grains induced proliferation of bacterial taxa involved in cellulose degradation and nitrogen cycle.
-
•
Ammonia and nitrates were assimilated at high rates by the microbiota of E. fetida fed with brewers’ spent grains.
Keywords: Brewers’ spent grains, Phylogeny-based metabolic interference, Canonical correspondence analysis, LEfSe, Biolog
Abstract
Vermicomposting is a cost-effective biotechnology for the management of organic wastes that relies on the activity of earthworms and their associated microbiota. Here, the microbiotas of the earthworm Eisenia fetida fed with brewers’ spent grains (FBSG), cow manure (FCM) and a mix of brewers’ spent grains/cow manure (FMIX), were identified by high-throughput DNA sequencing (16S rRNA). Bacterial community variance was correlated with the pH and the organic carbon content of the rearing substrates. FBSG microbiota was enriched in Paenibacillaceae, Enterobacteriaceae, Chitinophagaceae and Comamonadaceae. In addition, FBSG microbiota had a predicted higher abundance of genes involved in cellulose degradation as well as in the nitrogen cycle and showed higher utilization of ammonia and nitrate. Results obtained will allow to optimize the vermicomposting of brewers’ spent grains and to evaluate the effect of vermicompost addition on nutrient dynamics in soil.
1. Introduction
Vermicomposting is an environmentally friendly process of bio-oxidation and stabilization of organic wastes such as paper residues [1], animal dungs [2], industrial wastes [3], municipal sewage sludges [4], etc. Nutrient recycling through vermicomposting involves joint action of earthworms and microorganisms [5]. Recently, one of the most suitable earthworm species for vermicomposting, Eisenia fetida, was shown to grow healthy on brewers’ spent grains (BSG) the most abundant waste of the brewing industry [6]. BSG are usually supplied to local farmers at low or null cost [7], however they are rich in cellulose, non-cellulosic polysaccharides, lignin as well as proteins [8]. Saba et al. [6] showed that the vermicompost obtained from BSG was rich in nitrogen, respected the biological and microbiological safety law parameters and was characterized by particularly high enzymatic activities. Considering that proper decomposition of organic wastes and nutrient availability of vermicompost depend mainly on the microbial communities in earthworms’ cast [9], the characterization of this microbiota appears necessary to optimize the process, to fully exploit BSG nutritional properties and to evaluate the effect of vermicompost addition on nutrient dynamics in soil. To this end, the microbiota in the casts of the earthworm Eisenia fetida fed with brewers’ spent grains (FBSG), cow manure (FCM) and a mix made of cow manure/brewers’ spent grains with a 50/50 (vol/vol) ratio (FMIX) was evaluated by high-throughput sequencing DNA approach.
2. Materials and methods
300 g of the red earthworm Eisenia fetida (Savigny, 1826) were inoculated into three experimental vermibeds (60 × 40 × 13 cm plastic containers) containing: (i) brewers’ spent grain (FBSG treatment); (ii) cow manure (FCM treatment) and (iii) a mix made of brewers’ spent grain and manure (FMIX treatment) with a 50/50 (vol/vol) ratio. After 90 days of acclimatization, mature earthworms were removed from FBSG, FCM and FMIX vermibeds and washed with sterile distilled water to remove adhesive residues and mucus. Earthworms were placed in sterile Petri dishes under sterile conditions. After 24 h of incubation at 25 °C, 0.5 g of fresh earthworms’ casts were collected from each Petri dish with a sterile spatula for subsequent sequencing. Overall, 6 cast specimens (two for each rearing substrate) were analyzed.
DNA from cast samples was extracted using the PowerSoil DNA Isolation kit (MoBio Laboratories Inc., Carlsbad, California) according to the manufacturer’s protocol. Microbiota composition was determined by sequencing of the PCR products of the 16S rRNA gene covering the V3-V4 region [10]. Barcoding and sequencing libraries for the Illumina MiSeq system with a paired-end 300 cycles protocol were generated, sequenced and their quality checked by BaseClear BV (Leiden, The Netherlands). Demultiplexed FASTQ files were analyzed with the Quantitative Insights into Microbial Ecology tool (QIIME 2 version 2019.1) [11]. Sequence quality filtering and OTU picking was carried out using the Deblur plugin implemented in QIIME2. Taxonomic classification of 16S OTUs was carried out using a pre-trained Naive Bayes classifier trained on the Greengenes 13_5_97 % OTUs full-length sequences. The significance of the differences at the phylum and at the family level were assessed using ANOVA. Benjamini-Hochberg FDR was used as multiple test correction method. LDA Effect Size (LEfSe) algorithm [12] was applied to identify differentially abundant features (OTUs) characterizing the differences between the three conditions tested (FBSG, FCM and FMIX).
The contribution of the chemical characteristics of the three rearing substrates to the variances of bacterial communities was assessed with variance partitioning analysis and canonical correspondence analysis (CCA) using the package vegan (https://CRAN.R-project.org/package=vegan) of the R environment (v.2.13.1; http://www.r-project.org/). The following chemical parameters in both the rearing substrates and vermicompost were considered: pH, total nitrogen, total extractable carbon, total organic carbon, CN ratio, humic-like substances. The values of these chemical parameters as well as the methods for their determination are reported in Saba et al. [6].
PICRUSt v.1.0.0 [13] software pipeline was used to predict the functional composition of bacterial enzymatic activity using 16S rDNA datasets (OTU table). Specific cellulose degradation and N cycling functionality was assessed by specifying KEGG orthologs for cellulose degradation (K01188, K01179), N fixation (K02588, K02586, K02591, K22896, K22897, K22898, K22899), assimilatory nitrate reduction (K00367, K10534, K00372, K00360, K00366, K17877), dissimilatory nitrate reduction (K00370, K00371, K00374, K02567, K02568, K00362, K00363, K03385, K15876), nitrification (K10944, K10945, K10946, K10535), denitrification (K00368, K15864, K04561, K02305, K00376), anammox (K00368, K20932, K20933, K20934, K20935) and ammonia assimilation (K00264, K00265, K00266, K01915, K01948).
Freshly obtained cast samples (0.20 g) were diluted into sterile water to a density of 84 % transmittance on a Biolog turbidimeter (Biolog, Hayward, CA, USA). The suspension was diluted 48-fold in IF-0 culture medium (Biolog, Hayward, CA, USA) added with sodium succinate (20 mM) as carbon source. 100 μL of the inoculating fluid were seeded into the Biolog PM3B plate wells containing ammonia (A01), sodium nitrite (A02), sodium nitrate (A03) and urea (A05) as nitrogen sources. Plates were incubated statically at 30 °C in an Omnilog Reader for 96 h. The quantitative color changes were recorded automatically every 15 min using a CCD camera, for each well. The kinetic responses were analyzed using Omnilog-PM software (Biolog, Inc., Hayward, CA, USA) as well as R (v.2.13.1; http://www.r-project.org/) and the package OPM (https://github.com/cran/opm) to calculate curve parameters via spline-fitting. The significance of the differences among average well color development (AWCD) values was determined by ANOVA followed by Tukey-HSD test (p < 0.05).
3. Results and discussion
Illumina (MiSeq) sequencing of 16S rRNA amplicons generated a data set ranging between 75,863 and 84,060 raw sequences per sample. Rarefaction analysis showed that the sequencing depth truly reflected the diversity of the microbial communities, as all the samples reached the sequencing plateau (data not shown). Sequence clustering produced significantly higher OTU counts in FMIX than in FCM (Table 1). Community evenness analyses indicated that the microbiota of FCM was dominated by significantly fewer taxa than that of FBSG and FMIX. Particularly, the Shannon index in the FBSG microbiota was in accordance with that reported during vermicomposting of wastes of different origins [14]. These results suggested that BSG used as feeding substrate for earthworms induced a high bacterial proliferation and diversification.
Table 1.
Operational taxonomic units (OTUs) and community evenness indices (Simpson and Shannon) of 16S rRNA sequences from the casts of earthworms fed with brewers’ spent grains (FBSG), cow manure (FCM) and brewer’s spent grains/cow manure (50/50 v/v) (FMIX).
| Treatment | |||
|---|---|---|---|
| OTUs | Simpson | Shannon | |
| FBSG | 453 ± 160ab | 0.910 ± 0.015a | 3.08 ± 0.21a |
| FCM | 485 ± 18.62b | 0.852 ± 0.025b | 2.65 ± 0.12b |
| FMIX | 661 ± 59.78a | 0.904 ± 0.03a | 3.41 ± 0.36a |
Data are mean and standard deviation. Different superscript letters in the same column indicate significant differences as determined by ANOVA followed by Tukey HSD test (p < 0.05).
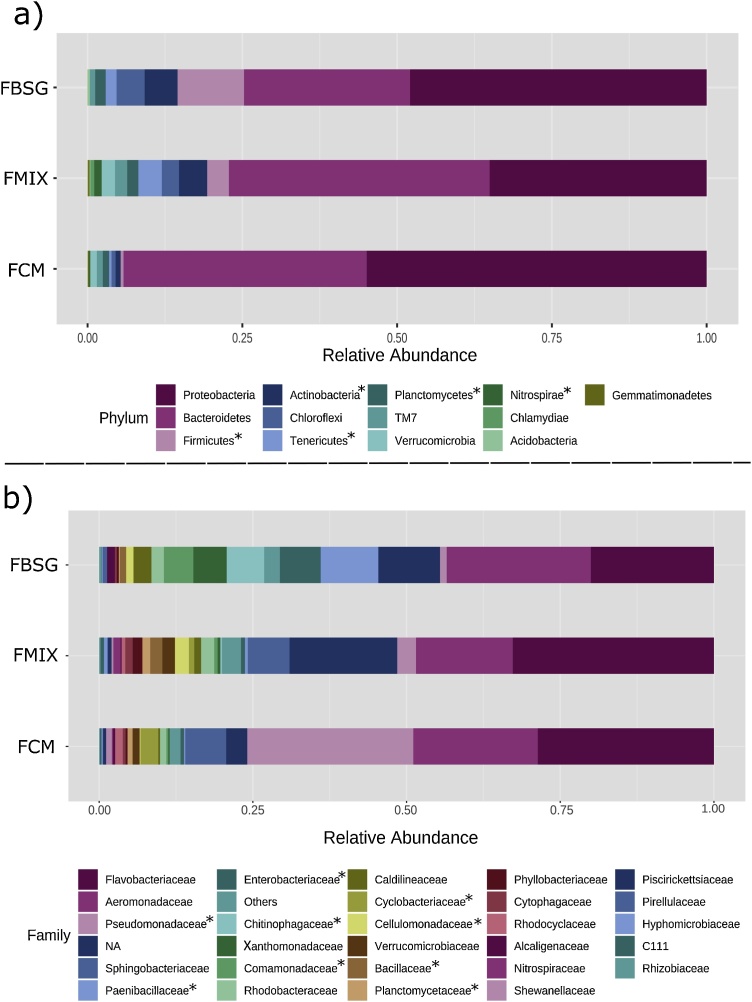
In accordance with previous reports [15], Proteobacteria and Bacteroidetes were the dominant phyla in E. fetida fed with the three rearing substrates (Fig. 1). Considering that earthworms’ microbiota is influenced by the diet [16,17], taxonomic differences were also identified. At the phylum level, Firmicutes and Actinobacteria were associated with the presence of BSG in the feeding substrate as their abundance was significantly higher in FBSG and FMIX than in FCM (Fig. 1a). A taxonomical profile similar to that of FBSG microbiota was reported by Yasir et al. [18] who found that vermicompost from paper sludge was dominated by Proteobacteria, Bacteroidetes, Firmicutes and Actinobacteria. Similarly, Huang et al. [19] reported that the bacterial community in vermicompost produced from vegetable wastes was dominated by Bacteroidetes and Actinobacteria. The low abundance of Firmicutes in FCM agrees with previous studies on earthworms’ cast from cow manure [20].
Fig. 1.
Relative abundance of bacterial OTUs at the phyla (a) and family (b) levels in the casts of E. fetida fed with brewer’s spent grains (FBSG), cow manure (FCM) and brewer’s spent grains/cow manure (50/50 v/v) (FMIX). Asterisks indicate significant different phyla (a) and families (b) as determined by ANOVA (adjusted p-value < 0.05).
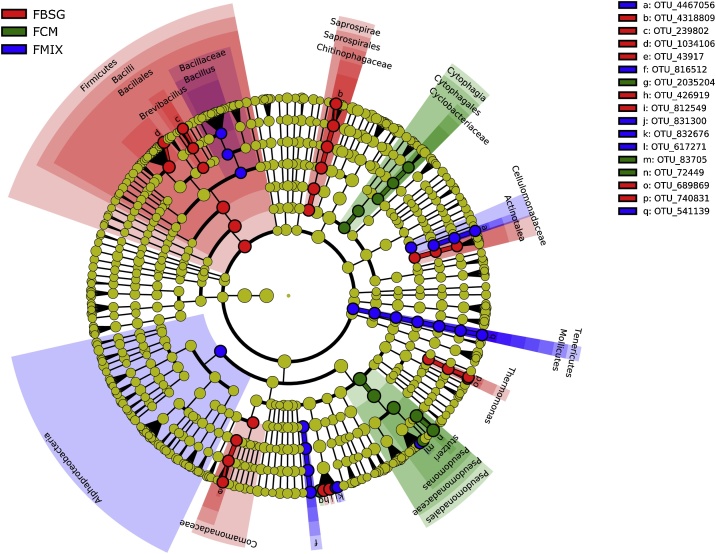
At the family level, FBSG microbiota was characterized by significantly higher abundance of Paenibacillaceae, Enterobacteriaceae, Chitinophagaceae and Comamonadaceae. FCM microbiota had higher abundance of Pseudomonadaceae while FMIX microbiota was enriched in Cellulomonadaceae and Planctomycetaceae (Fig. 1b). Finally, the LDA effect size (LEfSe) algorithm allowed to identify 16 OTUs as the genomic features characterizing the differences among the studied microbiotas (Fig. 2). Particularly, Paenibacillus, Bacillus and Thermomonas genus characterized FBSG microbiota, while Pseudomonas and Actinotalea were associated to the FCM and FMIX microbiota, respectively.
Fig. 2.
Cladogram of the taxa showing different abundance values in the casts of E. fetida as determined by LDA Effect Size (LEfSe) analysis. Earthworms were fed with brewer’s spent grains (FBSG), cow manure (FCM) and brewer’s spent grains/cow manure (50/50 v/v) (FMIX).
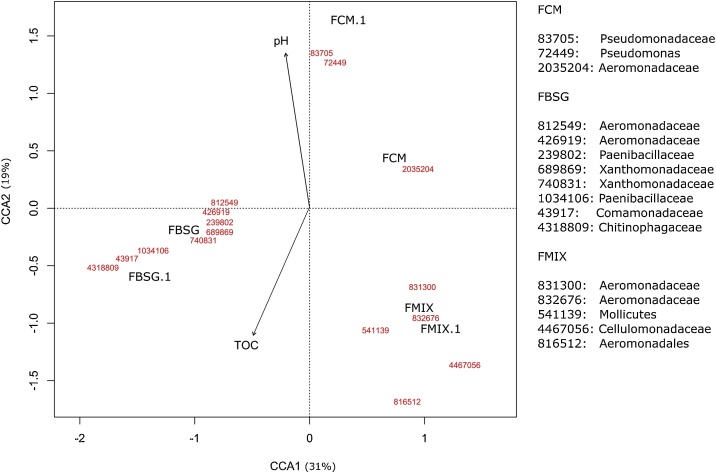
In order to identify the possible relationship between the microbial community structure and the chemical properties of the substrates, canonical correspondence analysis (CCA) was performed considering the above mentioned 16 significant OTUs and the chemical properties of the rearing substrates and the corresponding vermicomposts [6]. Based on Akaike’s Information Criterion (AIC), pH and total organic carbon (TOC) of the rearing substrates were identified as the significant variables and selected for the CCA biplot (Fig. 3).
Fig. 3.
Canonical correspondence analysis (CCA) of chemical characteristics of the substrates and significant OTUs in earthworms’ cast fed with brewer’s spent grains (FBSG and FBSG.1), cow manure (FCM and FCM.1) and brewer’s spent grains/cow manure (50/50 v/v) (FMIX and FMIX.1). Arrows indicate the direction and magnitude of measurable variables associated with bacterial community structures. TOC = total organic carbon.
The influence of pH on the structure of bacterial communities has been extensively documented. Considering that the three rearing substrates and vermicomposts were characterized by a significantly higher pH values in FCM than in the two other conditions [6], it is not surprising that the microbial community composition has been strongly influenced by this environmental factor. In particular, the higher abundance of Pseudomonadaceae in FCM could be explained by their sensitivity to pH values lower than 4.5 [21].
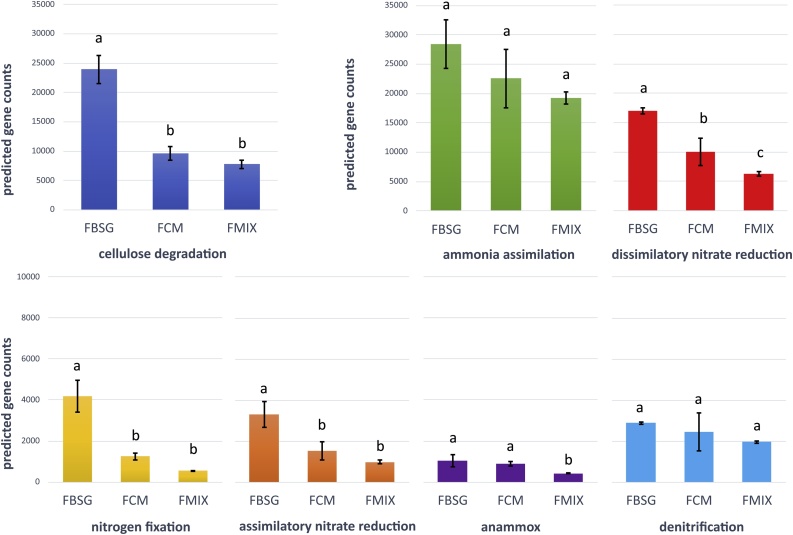
In addition to pH, bacterial community composition was also influenced by the TOC content of the substrates, which accounted for the 22.3 % (w/w) of cow manure and 34.8 % (w/w) of BSG [6]. Given that the lignocellulose fraction is the main component of TOC in BSG, it is conceivable that the FBSG microbiota was enriched in bacterial taxa involved in lignin, cellulose and hemicellulose degradation. Accordingly, members of Paenibacillaceae and Comamonadaceae are known producer of chitinase, cellulases, hemicellulases and lignin-modifying enzymes [[22], [23], [24], [25]], some active even at very low pH [26]. Enterobacter sp. isolated from agricultural wastes was shown to produce extracellular cellulolytic and hemicellulolytic enzymes [27]. Some members of Chitinophagaceae degrade chitin and cellulose and their abundance is positively correlated to high β‐glucosidase activity [28]. Accordingly to these observations, the previously determined β‐glucosidase activity [6], as well as the predicted abundance of genes involved in cellulose degradation (Fig. 4) were significantly higher in FBSG than in FCM.
Fig. 4.
PICRUSt predicted average gene counts in the microbiota of the cast of E. fetida fed with brewer’s spent grains (FBSG), cow manure (FCM) and brewer’s spent grains/cow manure (50/50 v/v) (FMIX). Data are means ± standard deviations. Different superscript letters in the same pathway indicate significant differences among treatments as determined by ANOVA followed by Tukey-HSD test (p < 0.05).
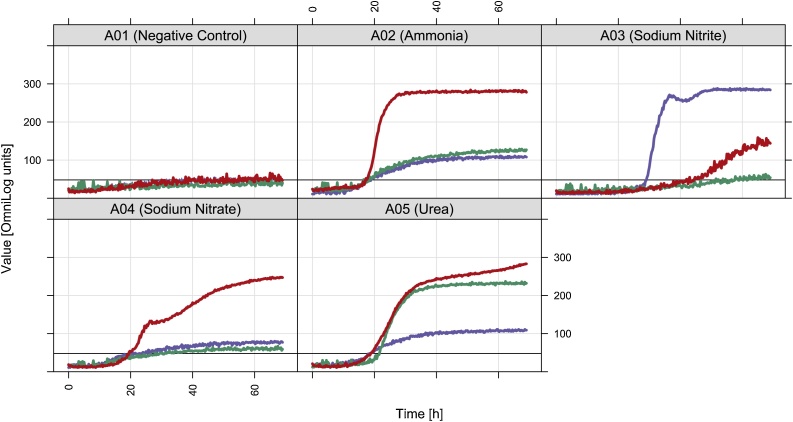
Considering that vermicompost from BSG is particularly rich in nitrogen, the abundance of genes involved in the N-cycling was evaluated by using PICRUSt software pipeline, which has been proven to be particularly accurate (∼98 %) in the prediction for nitrogen metabolism [13]. The predicted abundance of genes involved in different steps of the N-cycle was significantly higher in FBSG microbiota than in those of FCM and FMIX (Fig. 4). Thus, the FBSG microbiota have the potential to promote the fixation of molecular nitrogen (N2) and the assimilation into cell biomass of nitrogenous compounds, but also to enhance nitrate losses in form of N2 and gaseous nitrogen oxide products (e.g. N2O). Accordingly, phenotypic analyses showed that ammonia and nitrates were assimilated at higher rates by the FBSG microbiota (Fig. 5). Particularly, maximal AWCD values of FBSG microbiota in presence of ammonia (281 ± 11) and sodium nitrate (246 ± 36) were significantly higher than those measured from the FCM (126 ± 44 and 60 ± 16) and FMIX (113 ± 3 and 77 ± 1) microbiota. Considering that BSG bring nitrogen mainly in the form of ammonia while nitrates are poorly represented [29], it is plausible that ammonia assimilation could be the predominant step of the N-cycle in the vermicompost from BSG.
Fig. 5.
Metabolic outputs in wells A01, A02, A03, A04 and A05 of the PM3B plate (Biolog, Inc.). Data are average well color development (AWCD) values from FBSG (red), FCM (green) and FMIX (blue) microbiota. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
In conclusion, BSG used as feeding substrate strongly affected the microbiota in the cast of E. fetida. Particularly, the low pH and high organic carbon content of BSG were associated to higher abundance of bacterial taxa involved in cellulose degradation and showing high assimilation of ammonia and nitrates. The results obtained in this work will allow to optimize the vermicomposting of BSG and to identify the proper utilization of the resulting vermicompost as a soil conditioner.
Data availability
Data that support the findings of this study have been deposited at the European Nucleotide Archive (ENA) database and are publicly available under study accession number: PRJEB32972, or secondary accession number: ERP115720.
CRediT authorship contribution statement
Marilena Budroni: Funding acquisition, Investigation, Supervision, Writing - review & editing. Ilaria Mannazzu: Data curation, Methodology, Writing - review & editing. Severino Zara: Investigation, Writing - review & editing. Sara Saba: Validation, Writing - review & editing. Antonio Pais: Visualization, Writing - review & editing. Giacomo Zara: Investigation, Supervision, Data curation, Methodology, Formal analysis, Visualization, Writing - original draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by “Fondazione Banco di Sardegna, Italy” within the “Bando progetti di ricerca FBS – annualità 2015”. GZ gratefully acknowledges Sardinia Regional Government for the financial support of his research grant (Regional Operational Program of the European Social Fund (ROP ESF) 2014-2020 -C.U.P. J86C18000270002). The authors would like to thank the “Centro Servizi di Ateneo per la Ricerca (CeSAR)” of the University of Sassari, for the utilization of the Phenotype Microarray© platform.
References
- 1.Gajalakshmi S., Abbasi S.A. Vermiconversion of paper waste by earthworm born and grown in the waste-fed reactors compared to the pioneers raised to adulthood on cowdung feed. Bioresour. Technol. 2004;94:53–56. doi: 10.1016/j.biortech.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Garg V.K., Yadav A. Growth and reproduction of Eisenia foetida in various animal wastes during vermicomposting. Appl. Ecol. Environ. Res. 2005;3:51–59. [Google Scholar]
- 3.Garg P., Gupta A., Satya S. Vermicomposting of different types of waste using Eisenia foetida: a comparative study. Bioresour. Technol. 2006;97:391–395. doi: 10.1016/j.biortech.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Suthar S. Vermistabilization of municipal sewage sludge amended with sugarcane trash using epigeic Eisenia fetida (Oligochaeta) J. Hazard. Mater. 2009;163:199–206. doi: 10.1016/j.jhazmat.2008.06.106. [DOI] [PubMed] [Google Scholar]
- 5.Domínguez J., Aira M., Gómez-Brandón M. Vermicomposting: earthworms enhance the work of microbes. In: Insam H., Franke-Whittle I., Goberna M., editors. Microbes Work Wastes Resour. Springer; Berlin Heidelberg, Berlin, Heidelberg: 2010. pp. 93–114. [Google Scholar]
- 6.Saba S., Zara G., Bianco A., Garau M., Bononi M., Deroma M., Pais A., Budroni M. Comparative analysis of vermicompost quality produced from brewers’ spent grain and cow manure by the red earthworm Eisenia fetida. Bioresour. Technol. 2019;293:122019. doi: 10.1016/j.biortech.2019.122019. [DOI] [PubMed] [Google Scholar]
- 7.Lynch K.M., Steffen E.J., Arendt E.K. Brewers’ spent grain: a review with an emphasis on food and health. J. Inst. Brew. 2016;122:553–568. [Google Scholar]
- 8.Mussatto S.I., Dragone G., Roberto I.C. Brewers’ spent grain: generation, characteristics and potential applications. J. Cereal Sci. 2006;43:1–14. [Google Scholar]
- 9.Lazcano C., Domínguez J. Soil Nutr. Nova Science Publishers, Inc; 2011. The use of vermicompost in sustainable agriculture: impact on plant growth and soil fertility; pp. 230–254. Mohammad Miransari. [Google Scholar]
- 10.Bukin Y.S., Galachyants Y.P., Morozov I.V., Bukin S.V., Zakharenko A.S., Zemskaya T.I. The effect of 16S rRNA region choice on bacterial community metabarcoding results. Sci. Data. 2019;6 doi: 10.1038/sdata.2019.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langille M.G.I., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A., Clemente J.C., Burkepile D.E., Vega Thurber R.L., Knight R., Beiko R.G., Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y., Zhang Y., Zhang Q., Xu L., Li R., Luo X., Zhang X., Tong J. Earthworms modify microbial community structure and accelerate maize stover decomposition during vermicomposting. Environ. Sci. Pollut. Res. Int. 2015;22:17161–17170. doi: 10.1007/s11356-015-4955-z. [DOI] [PubMed] [Google Scholar]
- 15.Singh A., Singh D.P., Tiwari R., Kumar K., Singh R.V., Singh S., Prasanna R., Saxena A.K., Nain L. Taxonomic and functional annotation of gut bacterial communities of Eisenia foetida and Perionyx excavatus. Microbiol. Res. 2015;175:48–56. doi: 10.1016/j.micres.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Engel P., Moran N.A. The gut microbiota of insects - diversity in structure and function. FEMS Microbiol. Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 17.Knapp B.A., Podmirseg S.M., Seeber J., Meyer E., Insam H. Diet-related composition of the gut microbiota of Lumbricus rubellus as revealed by a molecular fingerprinting technique and cloning. Soil Biol. Biochem. 2009;41:2299–2307. [Google Scholar]
- 18.Yasir M., Aslam Z., Kim S.W., Lee S.-W., Jeon C.O., Chung Y.R. Bacterial community composition and chitinase gene diversity of vermicompost with antifungal activity. Bioresour. Technol. 2009;100:4396–4403. doi: 10.1016/j.biortech.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 19.Huang K., Li F., Wei Y., Chen X., Fu X. Changes of bacterial and fungal community compositions during vermicomposting of vegetable wastes by Eisenia foetida. Bioresour. Technol. 2013;150:235–241. doi: 10.1016/j.biortech.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Aira M., Bybee S., Pérez-Losada M., Domínguez J. Feeding on microbiomes: effects of detritivory on the taxonomic and phylogenetic bacterial composition of animal manures. FEMS Microbiol. Ecol. 2015;91 doi: 10.1093/femsec/fiv117. [DOI] [PubMed] [Google Scholar]
- 21.Tanner R.S., James S.A. Rapid bactericidal effect of low pH against Pseudomonas aeruginosa. J. Ind. Microbiol. 1992;10:229–232. [Google Scholar]
- 22.Grady E.N., MacDonald J., Liu L., Richman A., Yuan Z.-C. Current knowledge and perspectives of Paenibacillus: a review. Microb. Cell Factories. 2016;15:203. doi: 10.1186/s12934-016-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenberg E. The family chitinophagaceae. In: Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., Thompson F., editors. Prokaryotes Major Lineages Bact. Archaea. Springer; Berlin, Heidelberg: 2014. pp. 493–495. [Google Scholar]
- 24.Zheng Y., Chai L., Yang Z., Chen Y., Shi Y., Wang Y. Environmentally safe treatment of black liquor with Comamonas sp. B-9 under high-alkaline conditions. J. Basic Microbiol. 2014;54:152–161. doi: 10.1002/jobm.201200340. [DOI] [PubMed] [Google Scholar]
- 25.Mohammad B.T., Al Daghistani H.I., Jaouani A., Abdel-Latif S., Kennes C. Isolation and characterization of thermophilic Bacteria from Jordanian hot springs: Bacillus licheniformis and Thermomonas hydrothermalis isolates as potential producers of thermostable enzymes. Int. J. Microbiol. 2017 doi: 10.1155/2017/6943952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raddadi N., Cherif A., Daffonchio D., Fava F. Halo-alkalitolerant and thermostable cellulases with improved tolerance to ionic liquids and organic solvents from Paenibacillus tarimensis isolated from the Chott El Fejej, Sahara desert, Tunisia. Bioresour. Technol. 2013;150:121–128. doi: 10.1016/j.biortech.2013.09.089. [DOI] [PubMed] [Google Scholar]
- 27.Waghmare P.R., Patil S.M., Jadhav S.L., Jeon B.-H., Govindwar S.P. Utilization of agricultural waste biomass by cellulolytic isolate Enterobacter sp. SUK-Bio. Agric. Nat. Resour. 2018;52:399–406. [Google Scholar]
- 28.Bailey V.L., Fansler S.J., Stegen J.C., McCue L.A. Linking microbial community structure to β -glucosidic function in soil aggregates. ISME J. 2013;7:2044–2053. doi: 10.1038/ismej.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erdem N., Ok S.S. Effect of brewery sludge amendments on some chemical properties of acid soil in pot experiments. Bioresour. Technol. 2002;84:271–273. doi: 10.1016/s0960-8524(02)00046-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data that support the findings of this study have been deposited at the European Nucleotide Archive (ENA) database and are publicly available under study accession number: PRJEB32972, or secondary accession number: ERP115720.