Abstract
Human immunodeficiency virus type 1 (HIV-1) subtype C (C-HIV) is the most prevalent form of HIV-1 globally, accounting for approximately 50% of infections worldwide. C-HIV is the predominant and near-exclusive subtype in the low resource regions of India and Southern Africa. Given the vast diversity of HIV-1 subtypes, it is curious as to why C-HIV constitutes such a large proportion of global infections. This enriched prevalence may be due to phenotypic differences between C-HIV isolates and other viral strains that permit enhanced transmission efficiency or, pathogenicity, or might due to the socio-demographics of the regions where C-HIV is endemic. Here, we compare the mechanisms of C-HIV pathogenesis to less prominent HIV-1 subtypes, including viral genetic and phenotypic characteristics, and host genetic variability, to understand whether evolutionary factors drove C-HIV to predominance.
Keywords: HIV-1, Subtype C, Transmission, Pathogenesis
1. Background
HIV-1 remains a significant global health challenge, with approximately 38 million people infected worldwide [1]. HIV-1 demonstrates a broad diversity of subtypes and recombinant forms. Subtype C HIV-1 (C-HIV) is globally the most prevalent subtype, with 46% of global infections and is predominant in Southern Africa, India and Ethiopia. It is interesting that one subtype rose to global predominance while being the dominant subtype in only a few geographical locations. Perhaps this subtype has evolved to be more virulent than other strains? Such an advantage could involve a multitude of factors, including genetic differences between virus subtypes and within-host populations. However, the mechanisms that may permit such an advantage for C-HIV viruses to propagate within a given population are poorly understood. Alternatively, the high prevalence of C-HIV could also be a result of random introduction into high-risk populations during times of complex socio-political change, particularly in regions of Southern Africa [2]. This review discusses the mechanisms of pathogenesis, including replication efficiency, disease progression, and transmission efficiency to understand how C-HIV expanded faster than other subtypes, particularly within Sub-Saharan Africa.
2. HIV strains
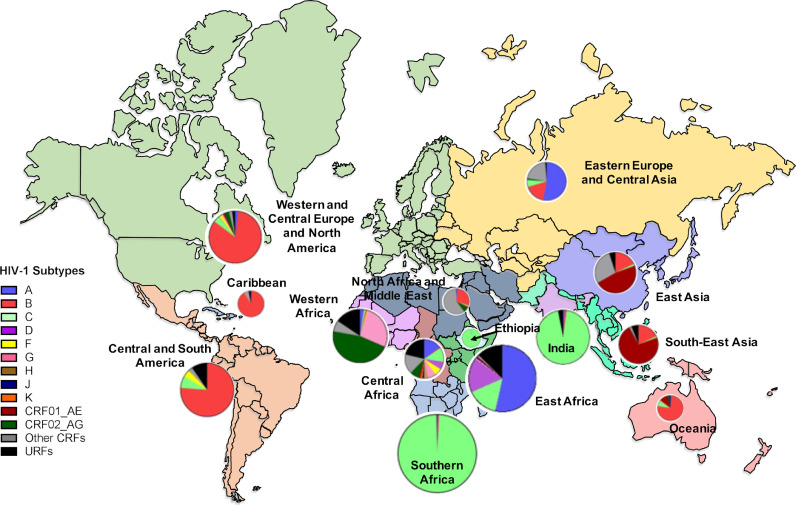
HIV-1 viruses are categorised into four groups: Main (M), Outlier (O), Non-M (N) and group P viruses [3], each originating from an independent cross-species transmission event between non-human primates (NHPs) and humans [3]. Group M viruses are further sub-categorised into ten distinct subtypes (A, B, C, D, F, G, H, I, J and K) and numerous circulating recombinant forms (CRFs) [4,5]. Inter-subtype sequence diversity ranges between 20 and 35% depending on the subtypes and genetic regions examined [6,7]. C-HIV demonstrates a disproportionately high prevalence, comprising four-fold more infections than any other subtype [8]. C-HIV is mostly confined to the low-income regions of India, Ethiopia and Southern Africa (Fig. 1), although the prevalence of C-HIV is increasing in Eastern Europe and Eastern Africa [8]. South Africa, in particular, bears the brunt of the HIV-1 epidemic, where approximately 20% of adults are living with HIV-11, and more than 98% of infections are caused by C-HIV [8]. Despite this, the majority of HIV-1 research has focused on subtype B (B-HIV), which constitutes only 12% of infections and is prevalent in Western and Central Europe, Latin and North America and Oceania (Fig. 1) [8].
Fig. 1.
World map illustrating the prevalence of HIV-1 group M subtypes within each region. Pie graphs show the percentage of each subtype that circulates within a region and the size of each pie represents the total number of infections in that region. Each region is colour coded. This map was adapted from subtype prevalence data from Hemelaar et al., 20198 and infection prevalence data from UNAIDS Data 2019 (https://www.unaids.org/en/resources/documents/2019/2019-UNAIDS-data).
3. Origin of subtype C HIV-1
Phylogenetic data places the origin of Group M HIV-1 in Kinshasa in the Democratic Republic of Congo (DRC) between 1909 and 1930 [9]. From here, group M strains have spread heterogeneously around the world [8]. Phylogenetic data suggests the origin of C-HIV was in Mbuji-Mayi, a diamond mining city in Southern DRC in the 1950s [10]. At the time, Mbuji-Mayi had an influx of immigrants from Lubumbashi and neighbouring countries primarily to work in the mines [10]. Such areas of economic growth were high-risk areas for HIV-1 transmission. From here, ancestral C-HIV strains were carried south to the city of Lubumbashi in the 1960s, which is proximal to the DRC-Zambia border. Ancestral C-HIV strains were introduced into Zimbabwe, Ethiopia, Kenya, Tanzania and Uganda by the 1970s, most likely through migration of mineworkers returning from the DRC [10], [11], [12]. Phylogenetic data suggest that multiple introductions of C-HIV into South Africa occurred from neighbouring countries Botswana, Malawi, Mozambique, Tanzania, Zambia and Zimbabwe between the 1980s and 1990s [13]. These introductions coincided with a period of complex socio-political change within South Africa (1985–2000) that lead to increased migration into and within South Africa, increased trade between neighbouring countries, and increased fertility rates, which facilitated the exponential growth of C-HIV infections during the 80 s and 90 s [13]. Overall, these studies suggest that the socio-demographic climate of Southern Africa between the 60 s and 90 s likely influenced the rapid expansion C-HIV in the region.
3.1. Does subtype C HIV-1 have a replication advantage?
Given that C-HIV is four-fold more prevalent than any other subtype, it is plausible that C-HIV viruses demonstrate improved replication fitness. Early studies assessed in vitro virus replication capacity by co-infection of susceptible cells with two viral isolates from diverse subtypes (termed a dual-competition assay) [14], [15], [16], [17]. These studies found that C-HIV viruses were outcompeted by other subtypes (A, B and D) in CD4+ T cells, macrophages and activated PBMC cultures, suggesting C-HIV viruses may have reduced replicative fitness. Overall, these dual-competition assays suggest that C-HIV viruses may demonstrate a reduced replication capacity compared to other group M viruses and may facilitate a slower disease progression of C-HIV infections, which in turn may increase the opportunity for new transmission events. Moreover, researchers have speculated that reduced replication kinetics may increase the half-life of productively infected cells and that these productively infected cells, if transmitted in genital fluid, would increase transmission risk compared to free virions [18].
3.2. Viral factors that may influence replication capacity
Viral factors intrinsically control virus replication capacity. Studies have found differential phenotypic properties between C-HIV isolates and other group M viruses in Env [16], Gag [18], the long-terminal repeat (LTR) region [19], [20], [21], reverse transcriptase (RT) [22], Protease [18,23] and Vif [24]. Marsozan et al. demonstrated that viruses that outcompete another in dual-competition assays mediate viral entry more efficiently [16], suggesting subtype C viruses may be less efficient at entering target cells than other group M isolates [14], [15], [16], [17]. Consistent with this, Venner et al. found C-HIV Envs from untreated, viremic individuals demonstrated reduced rates of viral fusion compared to other subtypes [17]. Additionally, the Ndung'u laboratory found that NL4.3 recombinant viruses containing clinical subtype B gag-protease sequences (n = 803) demonstrated significantly greater replication capacity in GXR cells (a CD4+ T cell line containing an HIV-1 LTR-driven green fluorescent protein reporter), compared to viruses containing C-HIV gag-protease sequences (n = 406) [18]. Given that Gag codes for the nucleocapsid, capsid and matrix structural proteins, it is possible that C-HIV viruses demonstrate reduced rates of viral assembly compared to other subtypes. Furthermore, a separate study found that viruses carrying reverse transcriptase (RT) from C-HIV isolates, as well as chimeric viruses containing either the C-HIV RT polymerase domain, connection domain, and/or RNase H domain, had decreased levels of cDNA accumulation, reduced integration and lower levels of viral replication compared to B-HIV RTs [22]. Overall, these studies suggest that several genetic components of C-HIV viruses may contribute to the lower in vitro replicative fitness compared to other HIV-1 subtypes.
Differential mechanisms of transcriptional regulation may occur between diverse subtypes, which influence replication kinetics downstream. Studies have found subtype-specific differences in the LTR, including sequence alterations in the negative regulatory element (NRE) [20], and differential activation in response to the cellular transcriptional activators Tat, Rel-p65 and NFAT [25]. C-HIV isolates also demonstrate an additional NF-κB site compared to subtypes A, B, D, F, G, and AE [19,20,26,27], which enhances LTR activity [20,25]. Furthermore, C-HIV LTRs have similar or increased activity under basal conditions or post-Tat trans-activation compared to other subtypes [19,20]. These studies suggest that C-HIV LTRs are either similar or more active than other subtypes. Overall, it may be that C-HIV viruses demonstrate similar levels of transcriptional activity while maintaining reduced replication fitness through dampened mechanisms of virulence (slower viral fusion, reduced rates of viral assembly, reduced RT efficiency).
3.3. Host genetic factors that may influence viral replication
Genetic variability within hosts can play a role in determining resistance or susceptibility to HIV-128. Families of genes that can influence disease susceptibility or progression include: (i) genes coding for human leukocyte antigens (HLAs); (ii) genes coding for coreceptors CCR5 or CXCR4 and their ligands, and (iii) other genes involved in the immune response to HIV-1 (reviewed by Lama and Planelles [28]).
The entry of HIV-1 into host cells requires engagement with the CD4 receptor as well as CCR5 or CXCR4 coreceptors. Viruses use CCR5 almost exclusively during the early stages of infection, and this usage is maintained throughout disease progression in 50% of PLWH [29], [30], [31], [32], [33], [34], [35]. Genetic differences in CCR5 variants between ethnicities may influence the efficiency of HIV-1 transmission between different populations. A 32-bp deletion within CCR5 (denoted as CCR5Δ32) produces a truncated protein that is not expressed on the cell surface and prevents viruses from engaging the host cell [36]. Individuals that are homozygous for this mutation are almost entirely resistant to infection, while some studies have shown that CCR5Δ32 heterozygotes may have a 2–4-year delay in disease progression to AIDS [37,38]. This mutation is present in 15% of European Caucasians (1% are homozygous) and is rarely detected in African and Asian populations [36,38]. Furthermore, a high prevalence of the CCR5 (−2459G>A) polymorphism that results in increased CCR5 expression was found to be present in 98% of adults in a study of 258 Papua New Guineans [39], a country where HIV-1 prevalence is high (> 0.8% of adults are living with HIV-1), and the majority of infections are caused by C-HIV (> 90%) [40]. Although further studies are required, these studies suggest that CCR5 alleles found within different populations and ethnicities may influence susceptibility to HIV-1 infection.
HLA is the most polymorphic gene locus in the human genome and encodes for surface proteins that present foreign antigens to elicit immune responses [41]. Humans encode two classes of HLA genes (HLA class I and II), of which class I HLAs have been associated with the prediction of disease progression in PLWH [41]. The HLA class I locus contains three genes, HLA-A, HLA-B and HLA-C. Given the primary function of HLAs is to present foreign antigens to initiate T cell responses, individuals who are heterozygous in the HLA locus recognise a broader repertoire of antigens than homozygotes [41]. Indeed, HLA class I homozygosity has been linked to rapid disease progression to AIDS in Caucasian individuals from the USA and the Netherlands, African Americans, and Rwandan women [42,43]. Several studies suggest that HLA alleles can differentially affect disease outcome in different HIV-1 subtypes. For instance, Kawashima et al. found that HLA-B*51:01 was protective against B-HIV during the early years of the epidemic [44], while being associated with disease progression during C-HIV infection [45]. Other examples include HLA-B*35:01 and HLA-B*07:02, which are associated with rapid disease progression in B-HIV infection but do not confer susceptibility to C-HIV infection [46,47]. Likewise, HLA-B*58:01 and HLA-B*58:02 are protective and disease-susceptible alleles respectively in C-HIV infection but have no known effects on B-HIV infection [42,45]. Overall, disease progression and susceptibility of different viral subtypes may be influenced by different HLA class I alleles. Future studies should focus on elucidating the frequencies of different HLA alleles that are known to be protective within populations that bear the brunt of the HIV-1 epidemic and how these may influence HIV-1 acquisition.
3.4. Does subtype C have a slower disease progression?
Differences in disease progression and transmission efficiency may have influenced the expansion of C-HIV infections within low-sociodemographic regions. Comparison of disease progression characteristics between subtypes remains challenging, mainly because C-HIV rarely cocirculates with other subtypes within the same geographical region (Fig 1). For this reason, comparing inter-subtype disease progression often involves surveying two different populations. For instance, Amornkul et al. tracked clinical characteristics of 491 untreated individuals from Kenya, Rwanda, South Africa, Uganda and Zambia between 2006 and 201148. The median duration of infection before enrolment was 54 days, with individuals followed for a median of 2.8 years. Viral subtype analysis found that C-HIV almost exclusively (> 97%) accounted for infections in South African and Zambian individuals, while subtypes A and D were prevalent in Kenya, Rwanda and Uganda. Following correction for age, HLA types, and sex, hazard ratio analysis found that individuals infected with subtypes C and D progressed to low CD4+ T cell counts (< 350 cells/μl) and AIDS endpoints (< 200 CD4+ T cells/µl, <14% CD4+ T cells or an AIDS-defining event) faster than subtype A infected individuals. One explanation for this, however, is that people living with (PLW) C-HIV also demonstrated higher baseline viral loads and lower baseline CD4+ T cell counts compared to those with subtype A. Furthermore, no significant difference was seen in CD4+ T cell decline over time in PLW C-HIV infected compared to those with subtype A HIV-1. A separate disease progression study of 246 untreated individuals from Brazil found that those infected with C-HIV had a faster progression to an AIDS endpoint (< 350 CD4+ T cells/µl or an AIDS-defining event) of 976 days compared to PLW B-HIV (1881 days), although this was not statistically significant due to low sample size for C-HIV infection [49]. Furthermore, plasma viral load and time individuals were infected was unknown in this study, which may have influenced disease progression to AIDS independent of subtype.
In contrast to these findings, Venner et al. longitudinally sampled 186 untreated women from Uganda (n = 112) and Zimbabwe (n = 174) for an average of five years (up to 9.5 years) [17]. Ugandan women were predominantly infected with subtype A (68%) and D strains (28%), while Zimbabwean women were infected with C-HIV exclusively. Here, no significant differences were seen between subtypes in plasma viral load at set point and during disease progression. Furthermore, the authors found that CD4+ T cell counts of PLW C-HIV declined 2.5-fold and 1.6-fold slower than those living with subtype D and A infections respectively. A large-scale analysis of 3364 seroconverters in the CASCADE collaboration found that PLW C-HIV had lower CD4+ T cell counts at seroconversion compared to subtype B [50]. Furthermore, PLW C-HIV demonstrated slower rates of CD4+ T cell loss compared to subtype B, although this did not reach statistical significance. Overall, it remains unclear whether C-HIV demonstrates differential rates of disease progression compared to other group M HIV-1 subtypes.
3.5. Does subtype C HIV-1 have a transmission advantage?
While C-HIV viruses seem to demonstrate reduced viral fitness in vitro, they may be transmitted with similar or higher efficiency compared to other HIV-1 strains. Limited large-scale studies have aimed to elucidate whether C-HIV has an improved transmission efficiency. Such epidemiological studies are logistically challenging, given that a large number of infected donor-recipient partners are required. One study of 317 untreated, HIV-1 infected pregnant Kenyan women found that vaginal secretions of those infected with C-HIV were 3- to 8-fold more likely to contain viral DNA compared to subtypes A and D viruses [51], suggesting virus subtype may influence the degree of shedding of infected cells in the genital mucosa. However, the authors found no difference in mother-to-child transmissions between subtypes [51]. One limitation of this study was that it did not control for the mother's duration of infection, which may have influenced viral shedding. A more recent study of 622 serodiscordant couples from Eastern and Southern Africa assessed the risk of transmission between subtypes A, C and D strains [52]. Virus subtyping was achieved by sequencing segments of gag and env, with the authors finding no difference in transmission risk between subtypes. Furthermore, the authors analysed viral RNA within the endocervical fluid and seminal plasma and found no difference in the level of viral RNA present in these sites between individuals infected with C-HIV and non-C-HIV. Overall these transmission studies between linked donor-recipient pairs have not revealed a difference in transmission efficiency between C-HIV and other subtypes.
To assess transmission efficiency using a molecular approach, investigators have primarily focused on the phenotypic properties of transmitted/founder (T/F) viruses. T/F viruses are not directly isolated and sequenced from PLWH during primary infection because the time to virus detection (Fiebig stages) is days-to-weeks following primary infection [53]. Instead, they are inferred by prediction of the most recent common ancestor (MRCA) of multiple viral sequences during early infection using mathematical modelling [30]. Numerous studies have assessed differences in infectivity and virus transmission efficiency between T/F viruses and isolates from later stages of infection [[31], [32], [33],54,55]. Two studies found that subtype B and C T/F infectious molecular clones (IMCs) demonstrated improved infectivity in TZM-bl cells compared to viruses isolated from chronic stages of disease in both subtypes [31,54], although these observations were not consistent in vitro CD4+ T cell infections [32,33,55].
When comparing infection kinetics of T/F viruses from different subtypes, Parrish et al. found that subtype B T/F IMCs demonstrated greater infectivity in TZM-bl cells than C-HIV T/F IMCs, suggesting subtype B viruses may demonstrate improved transmissibility compared to C-HIV [31]. Additionally, Chikere et al. previously used Affinofile cells, which allow for controlled induction of cell surface CD4 and CCR5 expression levels, to test the infectivity of viruses pseudotyped with acute-stage subtype A, B, C and D Envs [29]. This study found that pseudoviruses containing Envs from acute C-HIV infected donors demonstrated reduced infectivity compared to subtypes A, B and D in Affinofile cells expressing high levels of CD4 and low levels of CCR5, which reflect the receptor profile of CD4+ T cells. Overall, previous studies do not suggest that C-HIV T/F viruses have a transmission advantage against other group M subtypes in CD4+ T cells.
While T/F viruses appear to maintain similar infectivity in CD4+ T cells across subtypes, transmission efficiency to a new host may also be influenced by virus uptake by mononuclear phagocytes, such as macrophages or dendritic cells (DCs). Langerhans cells (LCs) are a subset of DCs that are found in the genital mucosal tissue and have been implicated as the first cells to encounter HIV-1 following transmission to a new host [56]. LCs and other DC subsets mediate infection of CD4+ T cells through uptake and release of virions [57,58], termed trans infection, or through de novo infection and release of new virions to infect CD4+ T cells, termed cis infection [59]. Indeed, several studies have assessed whether C-HIV viruses demonstrate differential uptake by DC subsets during transmission to a new host. Ball et al. demonstrated that C-HIV viruses demonstrated similar replication kinetics to B-HIV isolates in skin-derived LCs despite being outcompeted in activated PBMCs [15]. Parrish et al. found that T/F virus uptake was more efficient than chronic viruses regardless of subtype and that B-HIV T/F viruses were captured by monocyte-derived DCs and subsequently transferred to CD4+ T cells in co-culture experiments more efficiently than C-HIV T/F viruses [31]. Current findings suggest C-HIV T/F viruses are not transmitted more efficiently by DCs to CD4+ T cells, although the DC models used in these studies may not be entirely representative of those present in the mucosal tissue. Therefore, future studies should focus on the ability of C-HIV T/F viruses to mediate infection in lymphoid cells isolated from rectal and genital tissues [60,61]. Overall, the studies presented do not suggest that C-HIV carries a transmission advantage over other group M subtypes.
3.6. Coreceptor switching in subtype C HIV-1
The vast majority of T/F viruses are exclusively CCR5-using, suggesting that transmission to a new host selects for CCR5-using viruses [29], [30], [31], [32], [33], [34]. However, throughout a natural HIV-1 infection, coreceptor usage switches from CCR5 to CXCR4 in 50% of PLW B-HIV, which is associated with an accelerated disease progression to AIDS [35]. Why coreceptor switching occurs, particularly in only some individuals remains unclear. One explanation is that depletion of memory CD4+ T cells that coexpress CCR5 and CXCR4 may select for CXCR4-using strains, which would increase the range of target cells to increase naïve CD4+ T cell [62]. In contrast to B-HIV, studies of coreceptor switching in C-HIV have demonstrated a reduced prevalence of CXCR4-using variants in late disease stages and AIDS patients. Our laboratory longitudinally analysed coreceptor usage in untreated C-HIV infected subjects who progressed from chronic to late-stage disease and found 2/21 (9·5%) of individuals developed CXCR4-using viruses [63]. Similarly, in a large cross-sectional study of untreated C-HIV infected Botswanan women with CD4+ T cell counts below 200 cells/mL, 22/148 (15%) developed CXCR4-using viruses [64]. Coreceptor switching in B-HIV infected individuals is associated with the presence of positively charged residues at V3 positions 11 and 25 and an increase in V3 loop net charge [63]. Moreover, studies have demonstrated that in addition to V3 loop alterations, C-HIV viruses also require extensive mutations within the V1/V2, along with additional compensatory mutations within the V4 and V5 loops to mediate a coreceptor switch [63,[65], [66], [67]]. Thus, C-HIV viruses undergo coreceptor switching less frequently than other subtypes possibly due to the additional mutations required, which in turn may extend the window of opportunity for C-HIV transmission to new hosts through maintaining a CCR5-using state.
3.7. The proviral landscape of subtype C infection
In recent years, it has been well established that the majority of proviruses found in PLWH on suppressive ART contain defects (90%–95% for B-HIV) that block the production of infectious virus particles [68], [69], [70]. Such defects are a result of large internal, 5′ end or 3′ end deletions, sequence insertions or inversions, deletions of the packaging signal or major splice donor site, or hypermutation induced by APOBEC3G/F proteins [68], [69], [70]. The majority of these proviral deletions likely occur during minus-strand synthesis before the second strand transfer event of reverse transcription [71,72]. The degree of defective versus intact proviruses within PLWH (referred to as proviral landscape) is assessed through a single genome amplification and sequencing approach using two rounds of near full-length PCR at limited dilution [68,69]. In B-HIV studies, individuals with viremia during chronic infection have a proviral pool with 35% intact proviruses [69]. In contrast, individuals treated within 100 days of primary infection demonstrated a proviral pool containing 7% intact proviruses [69]. One difference in B-HIV proviral landscape between individuals treated in acute versus chronic infection is that those treated in acute infection demonstrated a higher proportion of hypermutations, which is likely a consequence of upregulation of APOBEC3F and APOBEC3G during acute infection [73]. Furthermore, a longitudinal study found that intact provirus numbers per million cells exponentially decayed over time in individuals treated during chronic infection [74].
C-HIV might maintain a higher proportion of intact proviruses than B-HIV, which may also improve the chance of transmission to a new host. To date, only one study has assessed the proviral landscape in the context of C-HIV infection. The Lichterfeld laboratory assessed the proviral landscape of two South African female participants enrolled in Fiebig stage II and two in stage V [75]. Fiebig II participants demonstrated lower frequencies of intact proviruses per million cells (0.8–2.2 versus 3.4–31.1) and higher percentages of intact proviruses in the total HIV-1 DNA pool (82%–100% versus 15%–35%) compared to participants identified in Fiebig stage V, suggesting that defective proviruses accumulate with more rounds of viral replication. Participants were followed for up one year, with one from each Fiebig group initiated ART at the first sampling time point. Over one-year, all participants showed reductions in intact proviruses per million cells and percentage of intact proviruses regardless of their treatment status. One limitation of this study was the low sample size and sampling depth within each time point. Overall, this study suggests the proviral landscape of C-HIV infections does not differ from that of B-HIV. However, it is difficult to compare the proviral landscape in acute infection with C-HIV to studies of individuals treated during chronic infection with B-HIV. Future studies should aim to determine the proviral landscape of C-HIV in individuals who are viremic during chronic infection, treated during chronic infection and how the proviral landscape changes longitudinally over many years of ART in PLW C-HIV.
4. Conclusion
C-HIV remains the most prevalent HIV-1 strain globally, causing 46% of infections. An interesting question that stems from this epidemiologic data is whether this enriched prevalence of C-HIV is a result of viral adaptation to enhance viral transmission, or the random introduction of this subtype into high-risk groups led to a more rapid transmission between individuals. It remains unclear whether C-HIV infection leads to a faster progression to AIDS compared to other isolates with contrasting results presented in large scale studies [17,48]. Early studies suggested that chronic C-HIV isolates were less fit for replication but maintained similar or enhanced transmission efficiencies compared to other subtypes [14], [15], [16], [17]. However, studies of T/F viruses do not suggest C-HIV is more fit for transmission than other subtypes [15,29,31]. An important future consideration is to assess transmission and replication capacity in lymphoid cells found within genital and rectal tissues. Furthermore, limited studies do not suggest any meaningful differences in the proviral landscape of C-HIV viruses compared to other subtypes. Future studies should determine the dynamics of the C-HIV proviral landscape in viremic and treated individuals. Current evidence suggests that the rapid expansion of C-HIV was not a result of viral evolutionary factors but could be the result of the introduction into high-risk populations during a time of complex socio-political changes.
4.1. Outstanding questions
Do genetic variabilities within geographical regions or different ethnicities influence transmission of C-HIV in endemic regions?
Do C-HIV viruses demonstrate an improved transmission efficiency in cell types that reflect transmission sites, such as residential memory CD4+ T cells, tissue resident macrophages, CD11c+ dendritic cells and Langerhans cells?
Is proviral landscape different between C-HIV infections and other subtypes in individuals that are viremic or virally suppressed and on ART?
4.2. Search strategy and selection criteria
This review includes data identified in searches of PubMed and references relevant articles using the search terms ‘subtype C HIV’, ‘subtype C transmission’, and ‘subtype C replication’. Furthermore, we include epidemiological data published online by The Joint United Nations Programme on HIV/AIDS (UNAIDS). Articles published between 1996 and 2019 were included.
Contributor Information
Paul R. Gorry, Email: paul.gorry@rmit.edu.au.
Jacqueline K. Flynn, Email: jacqueline.flynn@unimelb.edu.au, jacqueline.flynn@monash.edu.
References
- 1.UNAIDS. UNAIDS Data 2019. Geneva: UNAIDS 2019. (https://www.unaids.org/sites/default/files/media_asset/2019-UNAIDS-data_en.pdf. Accessed: 14th August 2019).
- 2.Wilkinson E., Engelbrecht S., de Oliveira T. History and origin of the HIV-1 subtype C epidemic in South Africa and the greater southern African region. Scientific Reports. 2015;5:16897. doi: 10.1038/srep16897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharp P.M., Hahn B.H. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1(1) doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buonaguro L., Tornesello M.L., Buonaguro F.M. Human Immunodeficiency Virus Type 1 Subtype Distribution in the Worldwide Epidemic: Pathogenetic and Therapeutic Implications. Journal of Virology. 2007;81(19):10209–10219. doi: 10.1128/JVI.00872-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jakobsen M.R., Ellett A., Churchill M.J., Gorry P.R. Viral tropism, fitness and pathogenicity of HIV-1 subtype C. Future Virology. 2010;5(2):219–231. [Google Scholar]
- 6.Hemelaar J., Gouws E., Ghys P.D., Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20(16):W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 7.Gaschen B., Taylor J., Yusim K., Foley B., Gao F., Lang D. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296(5577):2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 8.Hemelaar J., Elangovan R., Yun J., Dickson-Tetteh L., Fleminger I., Kirtley S. Global and regional molecular epidemiology of HIV-1, 1990-2015: a systematic review, global survey, and trend analysis. The Lancet Infectious Diseases. 2019;19(2):143–155. doi: 10.1016/S1473-3099(18)30647-9. [DOI] [PubMed] [Google Scholar]
- 9.Faria N.R., Rambaut A., Suchard M.A., Baele G., Bedford T., Ward M.J. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science. 2014;346(6205):56–61. doi: 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faria N.R., Vidal N., Lourenco J., Raghwani J., Sigaloff K.C.E., Tatem A.J. Distinct rates and patterns of spread of the major HIV-1 subtypes in Central and East Africa. PLoS Pathogens. 2019;15(12) doi: 10.1371/journal.ppat.1007976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalai S.C., de Oliveira T., Harkins G.W., Kassaye S.G., Lint J., Manasa J. Evolution and molecular epidemiology of subtype C HIV-1 in Zimbabwe. AIDS. 2009;23(18):2523–2532. doi: 10.1097/QAD.0b013e3283320ef3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tully D.C., Wood C. Chronology and evolution of the HIV-1 subtype C epidemic in Ethiopia. AIDS. 2010;24(10):1577–1582. doi: 10.1097/QAD.0b013e32833999e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkinson E., Rasmussen D., Ratmann O., Stadler T., Engelbrecht S., de Oliveira T. Origin, imports and exports of HIV-1 subtype C in South Africa: A historical perspective. Infection, Genetics and Evolution. 2016;46:200–208. doi: 10.1016/j.meegid.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Abraha A., Nankya I.L., Gibson R., Demers K., Tebit D.M., Johnston E. CCR5- and CXCR4-tropic subtype C human immunodeficiency virus type 1 isolates have a lower level of pathogenic fitness than other dominant group M subtypes: implications for the epidemic. Journal of Virology. 2009;83(11):5592–5605. doi: 10.1128/JVI.02051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ball S.C., Abraha A., Collins K.R., Marozsan A.J., Quinones-Mateu M.E., Penn-Nicholson A. Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. Journal of Virology. 2003;77(2):1021–1038. doi: 10.1128/JVI.77.2.1021-1038.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marozsan A.J., Moore D.M., Lobritz M.A., Fraundorf E., Abraha A., Reeves J.D. Differences in the fitness of two diverse wild-type human immunodeficiency virus type 1 isolates are related to the efficiency of cell binding and entry. Journal of Virology. 2005;79(11):7121–7134. doi: 10.1128/JVI.79.11.7121-7134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venner C.M., Nankya I., Kyeyune F., Demers K., Kwok C., Chen P.L. Infecting HIV-1 Subtype Predicts Disease Progression in Women of Sub-Saharan Africa. EBioMedicine. 2016;13:305–314. doi: 10.1016/j.ebiom.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiguoya M.W., Mann J.K., Chopera D., Gounder K., Lee G.Q., Hunt P.W. Subtype-Specific Differences in Gag-Protease-Driven Replication Capacity Are Consistent with Intersubtype Differences in HIV-1 Disease Progression. Journal of Virology. 2017;91(13) doi: 10.1128/JVI.00253-17. e00253-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeeninga R.E., Hoogenkamp M., Armand-Ugon M., de Baar M., Verhoef K., Berkhout B. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. Journal of Virology. 2000;74(8):3740–3751. doi: 10.1128/jvi.74.8.3740-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naghavi M.H., Schwartz S., Sonnerborg A., Vahlne A. Long terminal repeat promoter/enhancer activity of different subtypes of HIV type 1. AIDS Res Hum Retroviruses. 1999;15(14):1293–1303. doi: 10.1089/088922299310197. [DOI] [PubMed] [Google Scholar]
- 21.Hotter D., Bosso M., Jonsson K.L., Krapp C., Sturzel C.M., Das A. IFI16 Targets the Transcription Factor Sp1 to Suppress HIV-1 Transcription and Latency Reactivation. Cell Host & Microbe. 2019;25(6) doi: 10.1016/j.chom.2019.05.002. 858-72.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iordanskiy S., Waltke M., Feng Y., Wood C. Subtype-associated differences in HIV-1 reverse transcription affect the viral replication. Retrovirology. 2010;7:85. doi: 10.1186/1742-4690-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Velazquez-Campoy A., Todd M.J., Vega S., Freire E. Catalytic efficiency and vitality of HIV-1 proteases from African viral subtypes. Proc Natl Acad Sci U S A. 2001;98(11):6062–6067. doi: 10.1073/pnas.111152698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwabu Y., Kinomoto M., Tatsumi M., Fujita H., Shimura M., Tanaka Y. Differential anti-APOBEC3G activity of HIV-1 Vif proteins derived from different subtypes. Journal of Biological Chemistry. 2010;285(46):35350–35358. doi: 10.1074/jbc.M110.173286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montano M.A., Novitsky V.A., Blackard J.T., Cho N.L., Katzenstein D.A., Essex M. Divergent transcriptional regulation among expanding human immunodeficiency virus type 1 subtypes. Journal of Virology. 1997;71(11):8657–8665. doi: 10.1128/jvi.71.11.8657-8665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munkanta M., Handema R., Kasai H., Gondwe C., Deng X., Yamashita A. Predominance of three NF-kappaB binding sites in the long terminal repeat region of HIV Type 1 subtype C isolates from Zambia. AIDS Res Hum Retroviruses. 2005;21(10):901–906. doi: 10.1089/aid.2005.21.901. [DOI] [PubMed] [Google Scholar]
- 27.Bachu M., Mukthey A.B., Murali R.V., Cheedarla N., Mahadevan A., Shankar S.K. Sequence insertions in the HIV type 1 subtype C viral promoter predominantly generate an additional NF-kappaB binding site. AIDS Res Hum Retroviruses. 2012;28(10):1362–1368. doi: 10.1089/aid.2011.0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lama J., Planelles V. Host factors influencing susceptibility to HIV infection and AIDS progression. Retrovirology. 2007;4:52. doi: 10.1186/1742-4690-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chikere K., Webb N.E., Chou T., Born K., Sterjovski J., Gorry P.R. Distinct HIV-1 entry phenotypes are associated with transmission, subtype specificity, and resistance to broadly neutralizing antibodies. Retrovirology. 2014;11:48. doi: 10.1186/1742-4690-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keele B.F., Giorgi E.E., Salazar-Gonzalez J.F., Decker J.M., Pham K.T., Salazar M.G. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105(21):7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parrish N.F., Gao F., Li H., Giorgi E.E., Barbian H.J., Parrish E.H. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci U S A. 2013;110(17):6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parrish N.F., Wilen C.B., Banks L.B., Iyer S.S., Pfaff J.M., Salazar-Gonzalez J.F. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin alpha4beta7. PLoS Pathogens. 2012;8(5) doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilen C.B., Parrish N.F., Pfaff J.M., Decker J.M., Henning E.A., Haim H. Phenotypic and immunologic comparison of clade B transmitted/founder and chronic HIV-1 envelope glycoproteins. Journal of Virology. 2011;85(17):8514–8527. doi: 10.1128/JVI.00736-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ping L.H., Joseph S.B., Anderson J.A., Abrahams M.R., Salazar-Gonzalez J.F., Kincer L.P. Comparison of Viral Env Proteins from Acute and Chronic Infections with Subtype C Human Immunodeficiency Virus Type 1 Identifies Differences in Glycosylation and CCR5 Utilization and Suggests a New Strategy for Immunogen Design. Journal of Virology. 2013;87(13):7218–7233. doi: 10.1128/JVI.03577-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuitemaker H., van 't Wout A.B., Lusso P. Clinical significance of HIV-1 coreceptor usage. J Transl Med. 2011;9(Suppl 1):S5–SS. doi: 10.1186/1479-5876-9-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu R., Paxton W.A., Choe S., Ceradini D., Martin S.R., Horuk R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86(3):367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 37.Dean M., Carrington M., Winkler C., Huttley G.A., Smith M.W., Allikmets R. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273(5283):1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 38.Samson M., Libert F., Doranz B.J., Rucker J., Liesnard C., Farber C.M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382(6593):722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 39.Mehlotra R.K., Hall N.B., Bruse S.E., John B., Zikursh M.J.B., Stein C.M. CCR2, CCR5, and CXCL12 variation and HIV/AIDS in Papua New Guinea. Infection, Genetics and Evolution. 2015;36:165–173. doi: 10.1016/j.meegid.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan C.E., Gare J., Crowe S.M., Wilson K., Reeder J.C., Oelrichs R.B. The heterosexual HIV type 1 epidemic in Papua New Guinea is dominated by subtype C. AIDS Res Hum Retroviruses. 2007;23(7):941–944. doi: 10.1089/aid.2007.0043. [DOI] [PubMed] [Google Scholar]
- 41.Goulder P.J., Walker B.D. HIV and HLA class I: an evolving relationship. Immunity. 2012;37(3):426–440. doi: 10.1016/j.immuni.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang J., Costello C., Keet I.P., Rivers C., Leblanc S., Karita E. HLA class I homozygosity accelerates disease progression in human immunodeficiency virus type 1 infection. AIDS Res Hum Retroviruses. 1999;15(4):317–324. doi: 10.1089/088922299311277. [DOI] [PubMed] [Google Scholar]
- 43.Carrington M., Nelson G.W., Martin M.P., Kissner T., Vlahov D., Goedert J.J. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283(5408):1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 44.Kawashima Y., Pfafferott K., Frater J., Matthews P., Payne R., Addo M. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458(7238):641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carlson J.M., Listgarten J., Pfeifer N., Tan V., Kadie C., Walker B.D. Widespread impact of HLA restriction on immune control and escape pathways of HIV-1. Journal of Virology. 2012;86(9):5230–5243. doi: 10.1128/JVI.06728-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matthews P.C., Koyanagi M., Kloverpris H.N., Harndahl M., Stryhn A., Akahoshi T. Differential clade-specific HLA-B*3501 association with HIV-1 disease outcome is linked to immunogenicity of a single Gag epitope. Journal of Virology. 2012;86(23):12643–12654. doi: 10.1128/JVI.01381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kloverpris H.N., Adland E., Koyanagi M., Stryhn A., Harndahl M., Matthews P.C. HIV subtype influences HLA-B*07:02-associated HIV disease outcome. AIDS Res Hum Retroviruses. 2014;30(5):468–475. doi: 10.1089/aid.2013.0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amornkul P.N., Karita E., Kamali A., Rida W.N., Sanders E.J., Lakhi S. Disease progression by infecting HIV-1 subtype in a seroconverter cohort in sub-Saharan Africa. AIDS. 2013;27(17):2775–2786. doi: 10.1097/QAD.0000000000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leite T.C., Campos D.P., Coelho A.B., Teixeira S.L., Veloso V., Morgado M.G. Impact of HIV-1 Subtypes on AIDS Progression in a Brazilian Cohort. AIDS Res Hum Retroviruses. 2017;33(1):41–48. doi: 10.1089/AID.2016.0126. [DOI] [PubMed] [Google Scholar]
- 50.Touloumi G., Pantazis N., Pillay D., Paraskevis D., Chaix M.L., Bucher H.C. Impact of HIV-1 subtype on CD4 count at HIV seroconversion, rate of decline, and viral load set point in European seroconverter cohorts. Clinical Infectious Diseases. 2013;56(6):888–897. doi: 10.1093/cid/cis1000. [DOI] [PubMed] [Google Scholar]
- 51.John-Stewart G.C., Nduati R.W., Rousseau C.M., Mbori-Ngacha D.A., Richardson B.A., Rainwater S. Subtype C Is associated with increased vaginal shedding of HIV-1. Journal of Infectious Diseases. 2005;192(3):492–496. doi: 10.1086/431514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kahle E., Campbell M., Lingappa J., Donnel D., Celum C., Ondondo R. HIV-1 subtype C is not associated with higher risk of heterosexual HIV-1 transmission: a multinational study among HIV-1 serodiscordant couples. AIDS. 2014;28(2):235–243. doi: 10.1097/QAD.0000000000000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fiebig E.W., Wright D.J., Rawal B.D., Garrett P.E., Schumacher R.T., Peddada L. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17(13):1871–1879. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 54.Ashokkumar M., Aralaguppe S.G., Tripathy S.P., Hanna L.E., Neogi U. Unique Phenotypic Characteristics of Recently Transmitted HIV-1 Subtype C Envelope Glycoprotein gp120: Use of CXCR6 Coreceptor by Transmitted Founder Viruses. Journal of Virology. 2018;92(9) doi: 10.1128/JVI.00063-18. e00063-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deymier M.J., Ende Z., Fenton-May A.E., Dilernia D.A., Kilembe W., Allen S.A. Heterosexual Transmission of Subtype C HIV-1 Selects Consensus-Like Variants without Increased Replicative Capacity or Interferon-alpha Resistance. PLoS Pathogens. 2015;11(9) doi: 10.1371/journal.ppat.1005154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hladik F., Sakchalathorn P., Ballweber L., Lentz G., Fialkow M., Eschenbach D. Initial events in establishing vaginal entry and infection by human immunodeficiency virus type-1. Immunity. 2007;26(2):257–270. doi: 10.1016/j.immuni.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ballweber L., Robinson B., Kreger A., Fialkow M., Lentz G., McElrath M.J. Vaginal langerhans cells nonproductively transporting HIV-1 mediate infection of T cells. Journal of Virology. 2011;85(24):13443–13447. doi: 10.1128/JVI.05615-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ganor Y., Bomsel M. HIV-1 transmission in the male genital tract. Am J Reprod Immunol. 2011;65(3):284–291. doi: 10.1111/j.1600-0897.2010.00933.x. [DOI] [PubMed] [Google Scholar]
- 59.Nobile C., Petit C., Moris A., Skrabal K., Abastado J.P., Mammano F. Covert human immunodeficiency virus replication in dendritic cells and in DC-SIGN-expressing cells promotes long-term transmission to lymphocytes. Journal of Virology. 2005;79(9):5386–5399. doi: 10.1128/JVI.79.9.5386-5399.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bertram K.M., Botting R.A., Baharlou H., Rhodes J.W., Rana H., Graham J.D. Identification of HIV transmitting CD11c(+) human epidermal dendritic cells. Nature Communications. 2019;10(1):2759. doi: 10.1038/s41467-019-10697-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen R., Richter H.E., Smith P.D. Early HIV-1 target cells in human vaginal and ectocervical mucosa. Am J Reprod Immunol. 2011;65(3):261–267. doi: 10.1111/j.1600-0897.2010.00939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joseph S.B., Swanstrom R. The evolution of HIV-1 entry phenotypes as a guide to changing target cells. Journal of Leukocyte Biology. 2018;103(3):421–431. doi: 10.1002/JLB.2RI0517-200R. [DOI] [PubMed] [Google Scholar]
- 63.Jakobsen M.R., Cashin K., Roche M., Sterjovski J., Ellet A., Borm K. Longitudinal Analysis of CCR5 and CXCR4 Usage in a Cohort of Antiretroviral Therapy-Naive Subjects with Progressive HIV-1 Subtype C Infection. PloS One. 2013;8(6):e65950. doi: 10.1371/journal.pone.0065950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin N.H., Smeaton L.M., Giguel F., Novitsky V., Moyo S., Mitchell R.M. Prevalence and clinical associations of CXCR4-using HIV-1 among treatment-naive subtype C-infected women in Botswana. J Acquir Immune Defic Syndr. 2011;57(1):46–50. doi: 10.1097/QAI.0b013e318214fe27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coetzer M., Nedellec R., Cilliers T., Meyers T., Morris L., Mosier D.E. Extreme genetic divergence is required for coreceptor switching in HIV-1 subtype C. J Acquir Immune Defic Syndr. 2011;56(1):9–15. doi: 10.1097/QAI.0b013e3181f63906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cashin K., Gray L.R., Harvey K.L., Perez-Bercoff D., Lee G.Q., Sterjovski J. Reliable genotypic tropism tests for the major HIV-1 subtypes. Scientific Reports. 2015;5:8543. doi: 10.1038/srep08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cashin K., Jakobsen M.R., Sterjovski J., Roche M., Ellet A., Flynn J.K. Linkages between HIV-1 specificity for CCR5 or CXCR4 and in vitro usage of alternative coreceptors during progressive HIV-1 subtype C infection. Retrovirology. 2013;10:98. doi: 10.1186/1742-4690-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hiener B., Horsburgh B.A., Eden J.S., Barton K., Schlub T.E., Lee E. Identification of Genetically Intact HIV-1 Proviruses in Specific CD4(+) T Cells from Effectively Treated Participants. Cell Reports. 2017;21(3):813–822. doi: 10.1016/j.celrep.2017.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bruner K.M., Murray A.J., Pollack R.A., Soliman M.G., Laskey S.B., Capoferri A.A. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nature Medicine. 2016;22(9):1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee G.Q., Orlova-Fink N., Einkauf K., Chowdhury F.Z., Sun X., Harrington S. Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. Journal of Clinical Investigation. 2017;127(7):2689–2696. doi: 10.1172/JCI93289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu H., Jetzt A.E., Ron Y., Preston B.D., Dougherty J.P. The nature of human immunodeficiency virus type 1 strand transfers. Journal of Biological Chemistry. 1998;273(43):28384–28391. doi: 10.1074/jbc.273.43.28384. [DOI] [PubMed] [Google Scholar]
- 72.Jetzt A.E., Yu H., Klarmann G.J., Ron Y., Preston B.D., Dougherty J.P. High rate of recombination throughout the human immunodeficiency virus type 1 genome. Journal of Virology. 2000;74(3):1234–1240. doi: 10.1128/jvi.74.3.1234-1240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pillai S.K., Abdel-Mohsen M., Guatelli J., Skasko M., Monto A., Fujimoto K. Role of retroviral restriction factors in the interferon-alpha-mediated suppression of HIV-1 in vivo. Proc Natl Acad Sci U S A. 2012;109(8):3035–3040. doi: 10.1073/pnas.1111573109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pinzone M.R., VanBelzen D.J., Weissman S., Bertuccio M.P., Cannon L., Venanzi-Rullo E. Longitudinal HIV sequencing reveals reservoir expression leading to decay which is obscured by clonal expansion. Nature Communications. 2019;10(1):728. doi: 10.1038/s41467-019-08431-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee G.Q., Reddy K., Einkauf K.B., Gounder K., Chevalier J.M., Dong K.L. HIV-1 DNA sequence diversity and evolution during acute subtype C infection. Nature Communications. 2019;10(1):2737. doi: 10.1038/s41467-019-10659-2. [DOI] [PMC free article] [PubMed] [Google Scholar]