Abstract
Background
Tuberculosis (TB) continues to be a critical global health problem, which killed millions of lives each year. Certain circulating cell subsets are thought to differentially modulate the host immune response towards Mycobacterium tuberculosis (Mtb) infection, but the nature and function of these subsets is unclear.
Methods
Peripheral blood mononuclear cells (PBMC) were isolated from healthy controls (HC), latent tuberculosis infection (LTBI) and active tuberculosis (TB) and then subjected to single-cell RNA sequencing (scRNA-seq) using 10 × Genomics platform. Unsupervised clustering of the cells based on the gene expression profiles using the Seurat package and passed to tSNE for clustering visualization. Flow cytometry was used to validate the subsets identified by scRNA-Seq.
Findings
Cluster analysis based on differential gene expression revealed both known and novel markers for all main PBMC cell types and delineated 29 cell subsets. By comparing the scRNA-seq datasets from HC, LTBI and TB, we found that infection changes the frequency of immune-cell subsets in TB. Specifically, we observed gradual depletion of a natural killer (NK) cell subset (CD3-CD7+GZMB+) from HC, to LTBI and TB. We further verified that the depletion of CD3-CD7+GZMB+ subset in TB and found an increase in this subset frequency after anti-TB treatment. Finally, we confirmed that changes in this subset frequency can distinguish patients with TB from LTBI and HC.
Interpretation
We propose that the frequency of CD3-CD7+GZMB+ in peripheral blood could be used as a novel biomarker for distinguishing TB from LTBI and HC.
Fund
The study was supported by Natural Science Foundation of China (81770013, 81525016, 81772145, 81871255 and 91942315), National Science and Technology Major Project (2017ZX10201301), Science and Technology Project of Shenzhen (JCYJ20170412101048337) and Guangdong Provincial Key Laboratory of Regional Immunity and Diseases (2019B030301009). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Keyword: Tuberculosis, Single-cell RNA sequencing, CD3-CD7+GZMB+, NK cells
Research in context.
Evidence before this study
Tuberculosis continues to be a critical global health problem, which killed millions of lives each year. A major challenge for TB control is the lack of biomarkers to distinguish active TB from LTBI. While many efforts have been made to identify the blood transcript profiles that define TB, the proportion of individual immune-cell subsets in the blood can vary considerably depending on the severity of disease or treatment exposure. Sizeable fluctuations in subset-specific genes, particularly those that characterize a minority cell subset, may also be overlooked when whole blood is examined. Certain circulating cell subsets are thought to differentially modulate the host immune response towards Mycobacterium tuberculosis (Mtb) infection, but the nature and function of these subsets is unclear.
Added value of this study
We performed scRNA-seq on PBMCs from HC, LTBI and TB in an unbiased, surface-marker-free approach, to delineate the transcriptomic profile of individual immune-cell subsets. Our data represent the first scRNA-seq-based description of PBMC immune-cell subsets in LTBI and TB We found 29 subsets in PMBC and identified a large number of additional markers to these subsets, which could be used as references for further study to investigate the role of immune-cell subsets of PBMC in TB pathogenesis as well as protective immunity against TB. In addition, we observed a NK cell subsets, CD3-CD7+GZMB+ gradually depletion from HC, to LTBI and TB. The levels of CD3-CD7+GZMB+ is sensitive and specific to discriminate active TB from LTBI and HC.
Implications of all the available evidence
Our scRNA-seq analyses confirm many important observations made previously, and highlight key ongoing research areas and challenges. Circulating cell subsets in HC, LTBI and TB provides a useful framework to examine the role of cell subsets in TB disease progression. A cytotoxic NK cell subsets, CD3-CD7+GZMB+ could be used as a novel biomarker for distinguishing TB from LTBI and HC. Further dissecting the mechanism behind dysregulation of TB-associated cell subsets such as CD3-CD7+GZMB+ NK cell, might open new avenues for therapeutic intervention against TB.
Alt-text: Unlabelled box
1. Introduction
Tuberculosis (TB) is a chronic disease caused by Mycobacterium tuberculosis (Mtb) infection. TB represents a major global health problem, claiming ∼1.7 million lives annually [1]. As such, the World Health Organization ranks this disease as the leading cause of death from a single infectious agent. Nevertheless, the majority of those infected with Mtb generate an effective immune response to eliminate or control the infection; these patients remain clinically asymptomatic in a state known as ‘latent TB infection’ (LTBI). A relatively small proportion (5–10%) of patients with LTBI develop active TB.
A major challenge for TB control is the lack of biomarkers to distinguish active TB from LTBI, estimate disease severity, monitor drug treatment efficacy, predict disease outcome (recovery or relapse) and evaluate vaccine-induced protection[2,3]. In considering high proportions of active TB with negative bacteriology, an ideal TB biomarker should be sputum-free, reliable, high sensitivity and specificity. Blood is the most readily accessible sample in humans with high immune cell content. Better understanding the impact of Mtb infection on the host defense, through which identify any underlying differences in the immune-cell and gene expression profile in blood between healthy and diseased conditions, is one approach to reveal TB biomarker. Indeed, transcriptomic profiling of blood has provided an unbiased analysis and comprehensive overview of the host factors that are perturbed in TB [4], [5], [6], [7], [8], [9], [10]. However, bulk RNA-sequencing (RNA-seq) or microarray cannot reflect the proportion of individual immune-cell subsets in the blood that vary considerably depending on the status and severity of TB disease [11,12]. Consequently, sizeable fluctuations in subset-specific genes, particularly those that characterize a minority cell subset, may also be overlooked.
Compared to bulk RNA-seq which only provides the average expression signal for millions of cells, single-cell RNA sequencing (scRNA-seq) now allows for simultaneous analysis of >10,000 single-cell transcriptomes and thus characterization of novel cell subsets [13], [14], [15], [16], [17]. Consequently, scRNA-seq can reliably identify even closely-related cell populations [17], and reveal changes that render each individual cell type unique, and elucidate the heterogeneity in gene expression patterns in peripheral blood cell populations in health and disease [7,15]. In this study, we performed scRNA-seq on PBMCs from healthy controls (HC), LTBI and TB in an unbiased, surface-marker-free approach, to delineate the transcriptomic profile of individual immune-cell subsets.
2. Methods
2.1. Ethics statement
This study was approved by the Institutional Review Board of the Shenzhen University School of Medicine, China. and informed written consent was obtained from each participant. All experiments and samplings were carried out in accordance with ethical and biosafety protocols approved by the Institutional guidelines.
2.2. Subjects and clinical sample collection
Whole blood samples were collected from HC, LTBI and TB admitted to the Shenzhen Third People's Hospital (China), Shenzhen University General Hospital (China), Yuebei Second People's Hospital (China) and Shenzhen Baoan Hospital (China) between Aug 2018 and Oct 2019. The first cohort included HC (n = 2), LTBI (n = 2) and TB (n = 3) was used for the 10 × genomics scRNA-seq. The second cohort included HC (n = 81) and TB (n = 50) and the third cohort included HC (n = 39), LTBI (n = 27) and TB (n = 37) were used for flow cytometry analysis (Table S1 in the Supplementary Appendix). 11 TB patients in the second cohort were follow-up to 3 months after anti-TB treatment. At the time of enrolment, all TB patients had no record of prior TB disease of anti-TB chemotherapy. We also recruited 73 TB patients who received anti-TB treatment from 1–10 months (Table S1 in the Supplementary Appendix). Diagnosis of active TB was based on clinical symptoms, chest radiography and microscopy for acid fast bacilli (AFB), sputum Mtb culture and response to anti-TB chemotherapy. A previously established Mtb-specific IFN-γ ELISPOT assay (IGRA) was used to differentiate individuals with LTBI from uninfected HC [18].
2.3. 10 × genomics single-cell sample processing and cDNA library preparation
PBMCs were isolated from whole blood as previously described [19]. Cell viability was assessed by trypan blue staining and the samples (cell viability >90%) were prepared using a 10 × Genomics Single Cell 3′ v2 Reagent Kit according to the manufacturer's instructions. Single-cell libraries were prepared as previously described [20] and sequenced on an Illumina HiSeq X Ten system (Illumina).
2.4. Single-cell data preprocessing, gene expression quantification and cell-type determination
For each sample, the cleaned data filtered for the low quality reads and unrelated sequences were imported to CellRanger (version 2.2.0) and aligned to human reference genome (hg19, GRCh37). Cells were sorted according to the barcodes and the unique molecular identifiers (UMIs) were counted per gene for each cell. In total, 7711–10,928 (averagely 9767) cells were captured for individual libraries, and 835–1292 (averagely 1046) genes were detected with UMIs per cell (Table S2 in the Supplementary Appendix). To remove the possible doublets, we removed the top 7% cells with the highest number of UMIs and the cells with >3500 total expressed genes, followed by another round of doublet check and filtering with Scrublet [21]. The cells with <500 total expressed genes or with >7% mitochondria-expressed genes were also removed. With this procedure, averagely 8946 (7064–9871) cells were retained for each sample (Table S2 in the Supplementary Appendix). Gene expression matrices for the remaining cells were normalized to the total cellular and mitochondrial read counts using a linear regression model, implemented with the Seurat Regress Out function of the Seurat R package (version 2.3.4) [21]. Differentially expressed genes were selected based on a normalized expression value between 0.0125 and 8.0, and a quantile-normalized variance >0.4. Principle component analysis (PCA) was performed and tSNE were used for dimensionality reduction [22]. Cells were represented in a two-dimensional tSNE plane, and clusters were identified and annotated according to the marker gene composition. A marker gene was defined as being >0.25 log-fold higher than the mean expression value in the other sub-clusters, and with a detectable expression in > 25% of all cells from the corresponding sub-cluster. Raw sequencing reads are available at the NCBI Short Read Archive (SRA) under the accession numbers SRR11038989-SRR11038995.
2.5. Gene set variation analysis (GSVA)
Pathway analyses were performed on the 50 hallmark pathways annotated in the molecular signature database [23], which was exported using the GSEA Base package (version 1.40.1). The GSVA package (version 1.26.0) was applied with default settings to assign pathway activity estimates to individual cells [24]. A previous strategy was adopted to assess differential activities of pathways between sets of cells, with modifications [16]. Briefly, the activity scores for each cell were compared using a generalized linear model (GLM). The outputs of these GLMs were visualised in heat maps.
2.6. Flow cytometry and intracellular cytokine staining
Fresh whole blood samples (200 μl each) from HC, LTBI and TB were obtained for flow cytometry. Erythrocytes were removed with lysing solution (BD Biosciences), and the samples were first stained with the surface markers using mAbs against CD3, CD7, CD56, CD14, CD19 (BD Biosciences) and ghost dye (Tonbobio) for 30 min at 4 °C following permeabilization for 30 min at room temperature and then stained with mAbs against GZMB for 30 min at room temperature (BD Biosciences). All antibodies were validated by the manufacturer for flow cytometry application. The cells were re-suspended in 200 μl 2% paraformaldehyde and then acquired using FACSDiva software (BD Biosciences) and analysed using FlowJo software version 10.
2.7. Statistical analysis
A one-way ANOVA Newman–Keuls multiple comparison test was used to compare the differences among multiple groups. An unpaired t-test was used to analyze the differences between two groups. All statistical analyses were performed in GraphPad Prism (version 5.0). Two-tailed statistical tests were conducted and a P <0.05 was considered statistically significant.
3. Results
3.1. scRNA-seq reveals new cell type markers in PMBC
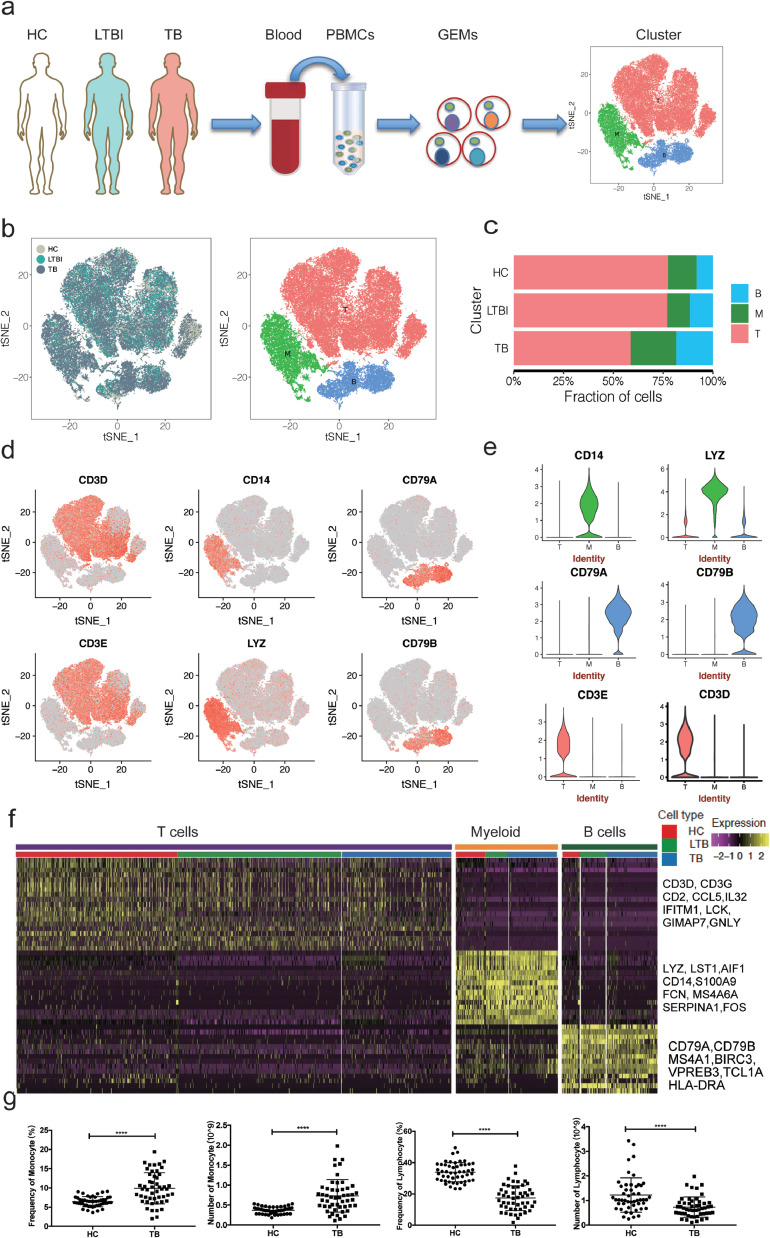
We first isolated and sequenced a total of 68,369 cells from PBMC suspensions derived from seven individuals, including two HC (20,711 cells), two LTBI (17,185 cells), and three TB (30,473 cells). After removing ∼8.5% cells that could represent doublets, empty droplets and low-quality ones (refer to Materials and Methods), we took forward 62,628 cells for further analysis (Fig. 1a, Fig. S1 and Table S2 in the Supplementary Appendix). Unsupervised clustering using the Seurat package [25] identified three distinct cell clusters across all three groups (Fig. 1b and). Cluster 1 (∼69.5% of all cells) comprised T cells (Fig. 1c and Table S3 in the Supplementary Appendix) expressing CD3D, CD3E, IL32 and CD2 (Fig. 1d–f and Fig. S3 in the Supplementary Appendix). Cluster 2 (∼17.3% of all cells) comprised myeloid cells expressing Lyz, S100A9, S100A8, S100A12 and CD14 (Fig. 1c–e and Fig. S3 in the Supplementary Appendix). Cluster 3 (∼13.2% of all cells) comprised B cells expressing CD79A, CD79B and MS4A1 (Fig. 1c–e and Fig. S3 in the Supplementary Appendix).
Fig. 1.
Single-cell transcriptional profiling of PBMCs from HC, LTBI and TB. (a) Schematic of experimental workflow for defining and comparing PBMC between all three donor groups. (b) tSNE of single cell profile with each cell color-coded for sample type and associated cell type. (c) The fraction of cells for three cell type in HC, LTBI and TB. (d)Expression of marker genes for the cell types defined above each panel. (e) The expression of known cell type discriminating genes in T, B and Myeloid from all donors.(f) Differential expression analysis was performed comparing cells from HC, LTBI and TB within T, B and Myeloid cells. Heatmaps are shown representing the up- and down-regulated genes in T, B and Myeloid cells. (g) Blood routine analysis of the numbers and frequencies of monocytes and lymphocyte from HC and TB. Unpaired t-test were used and the data represent the means ± SEM. ***P < 0. 001, ***P < 0. 0001.
We also identified a large number of additional markers: FCN1, CST4, SERPINA1, MNDA, LST1 and MS4A6A for myeloid cells; BANK1, VPREB3, FCER2 and ADAM28 for B cells; and IFITM1, GIMAP7, CD247 and LCK for T cells. (Fig. S3 and Table S4 in the Supplementary Appendix). These markers might be valuable PBMC cell-specific markers for future analyses.
3.2. Myeloid and B cells are enriched and T cells are depleted in TB
It has been reported that the frequencies of lymphocytes (T cells, B cells and NK cells) vary are in the range of 70–90%, monocytes from 10 to 20%, while dendritic cells are rare, accounting for only 1–2% in human [26]. Consistently, our scRNA-seq indicated that the isolated PBMCs from the HC consisted largely of T cells (∼78%), followed by myeloid (∼14%) and B cells (∼8%) (Fig. 1c and Table S3 in the Supplementary Appendix); these frequencies were largely similar in LTBI (Fig. 1c and Table S3 in the Supplementary Appendix). Conversely, we found higher frequencies of B cells (∼18%) and myeloid (∼23%) and a lower frequency of T cells (∼59%) in TB compared to HC or LTBI (Fig. 1c and Table S3 in the Supplementary Appendix). These described patterns in TB are consistent with previous reports [27], [28], [29]. In addition, the decrease of lymphocytes in TB was further confirmed by routine blood analysis in an independent cohort (Fig. 1g).
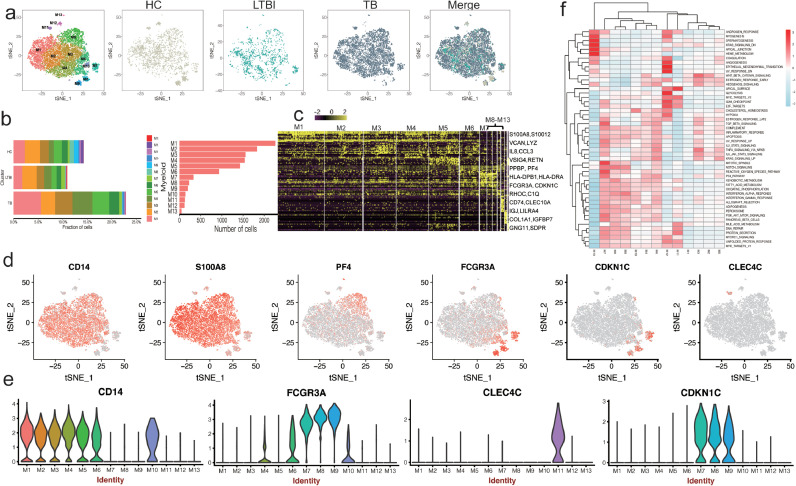
3.3. PBMC scRNA-seq identifies thirteen myeloid subsets
TB induces an accumulation of myeloid cells that express high levels of the inflammatory markers S100A8, S100A9 and S100A12 [30,31]. We therefore looked more closely at our Cluster 2 myeloid cells to determine whether the subset frequency changed between the three donor groups (HC, LTBI and TB). First we aimed to detect the three monocyte subsets defined as classical (CD14+CD16–), non-classical (CD14lowCD16+) and intermediate (CD14+CD16+) monocytes [32], scRNA-seq analysis confirmed the presence of these three distinct monocyte populations in all donor groups (Fig. 2a,b and Fig. S4 in the Supplementary Appendix). We were also able to subdivide the classical CD14+CD16- cells into six subsets (M1-M5) based on distinct marker expression (Fig. 2c,d and Table S5 in the Supplementary Appendix). Specifically, M1, M2 and M4 represented inflammatory monocytes, expressing high levels of S100A9, S100A12, RETN and S100A8 [33] (Fig. 2d,e and Fig. S5a in the Supplementary Appendix).M3 contained inflammatory monocyte/macrophage showing enrichment in IL1B, CCL3 and IL8 [34]. M5 cells enriched for PPBP (CXCL7) and PF4 (CXCL4) (Fig. 2d,e and Fig. S5a in the Supplementary Appendix), indicative of monocyte migration and autocrine, receptor-desensitizing chemokine ligand release [35,36]. Intermediate monocytes (M6) express high level of HLA-DPB1, HLA-DPA1 and CCL3 (Table S5 in the Supplementary Appendix). Non-classical monocytes (M7-M9) shared CDKN1C, RHOC and LYPD2 marker expression without S100A12 (Fig. 2d,e, Fig. S5a and Table S5 in the Supplementary Appendix); these cells were previously reported to belong to CD16+ (FCGR3A) monocytes [37]. We found a DC specific marker CLEC10A [37] distributed in M6 but enriched in M10 (Fig. S5a and Table S5 in the Supplementary Appendix), indicative of a DC-like subset. In addition, we also found a pDC subset M11, which specific expressed high level of pDC markers, including LILRA4, MZB1, ITM2C and CLEC4C [37], [38], [39] (Fig. 2d,e, Fig. S5a and Table S5 in the Supplementary Appendix). M12 specific enriched for mesenchymal cell-like makers, including COL1A1, IGFBP7, COL1A2 and SERPINH1 [40] (Fig. S5a and Table S5 in the Supplementary Appendix). We also find a small group of megakaryocyte-like cells (M13; PPBP, PF4 and GNG11) (Fig. 2a–d and Table S5 in the Supplementary Appendix) [41]. In conclusion, our scRNA-seq revealed thirteen subsets in myeloid cells with distinct markers, although the functions of these subsets remained to be elucidated. These findings provided a point of reference for examining the role of myeloid subsets in the circulating immune cells.
Fig. 2.
Myeloid clusters in PBMC from HC, LTBI and TB. (a) tSNE of single cell profile with each cell color coded for myeloid subsets (left to right): the associated cell type, the corresponding status (HC, LTBI and TB) and merged all status. (b)The fraction of cells for myeloid subsets in HC, LTBI and TB. (c) Differential expression analysis was performed comparing myeloid subsets from HC, LTBI and TB. Heatmaps are shown representing the up- and down-regulated genes in Myeloid subsets. (d) Expression of marker genes for the myeloid subsets. (e) Differential expression analysis was performed comparing cells from HC, LTBI and TB within Myeloid cell subsets. (f) Differences in pathway activities scored per cell by GSVA between the different myeloid subsets.
3.4. Four subsets of monocytes are enriched in TB
The frequency of intermediate monocyte CD14+CD16+ typically increases in inflammatory diseases and cancers [29,[42], [43], [44]]. In TB, these CD16+ circulating monocytes are more prone to produce TNF-α and undergo cell death in response to Mtb infection [29]. Consistently, we found that M6 (CD14+CD16+) monocytes were enriched in patients with TB (2%) compared to HC (1.19%) and LTBI (0.9%) in our cohort (Fig. 2b and Table S3 in the Supplementary Appendix). In addition, M1 (∼5.6%), M4 (∼4.0%) and M5 (∼3.4%)subsets were also more abundant in patients with TB than in HC (∼2.3%,∼1.0% and ∼1.7%, respectively) and LTBI (∼1.8%, ∼1.5% and 1.2%, respectively) (Fig. 2b and Table S3 in the Supplementary Appendix). Pathway analysis showed that M1 and M5 showed down-regulated Notch signaling, Myc-Targets, suggesting that these cells are exhausted. M4 and M6 subsets had up-regulated hedgehod signaling, with strong inflammatory response(Fig. 2f), suggestive of immune activity. Together, our data confirm and extend the abundant of CD14+ cell in TB and further reveal that three subsets of CD14+ cells (M1, M4 and M6) were specific enriched in TB. Future studies are required to reveal their functions in TB.
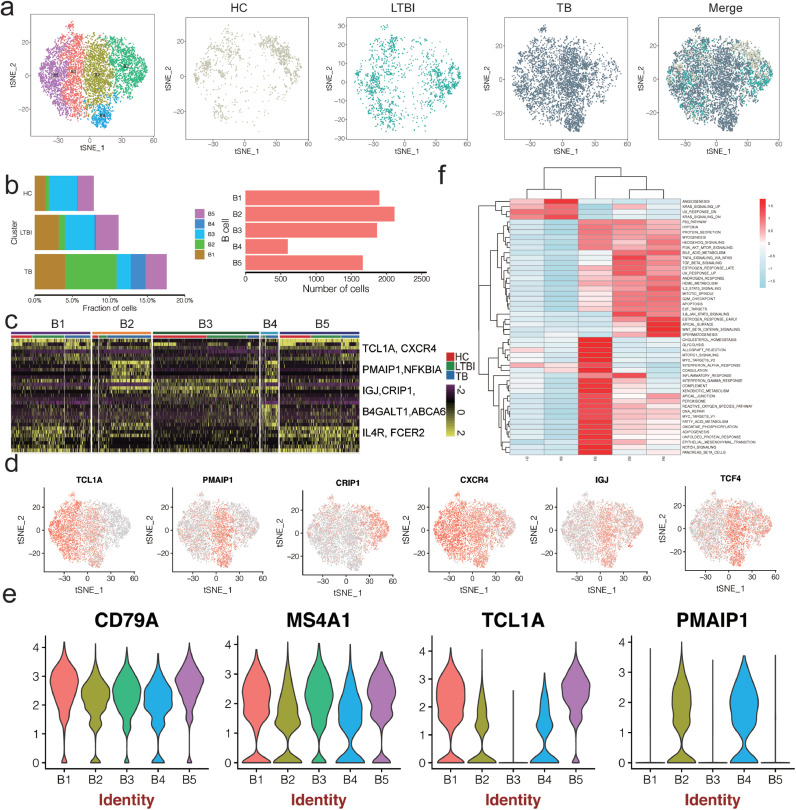
3.5. PBMC scRNA-seq identifies five B-cell subsets
The role of B cells in combating Mtb infection is unclear, with only a few studies explaining their function in TB [45]. Furthermore, information on B-cell phenotype and function is limited due to difficulties in manipulating B cells. Our scRNA-seq analysis identified six distinct B-cell subsets, each representing different stages of B-cell development (Fig. 3a–c and Fig. S6 in the Supplementary Appendix): B1 and B5 expressed high TCL1A, CD79A, CD79B, MS4A1 levels and lacked CD27 and CD138 expression. This expression pattern is indicative of follicular B cells [46,47] (Fig. 3c–e and Table S6 in the Supplementary Appendix). B2 and B4 was specifically enriched in PMAIP1(NOXA), ABCA6 and TCF4 (Fig. 3d and e, Fig. S5B and Table S6 in the Supplementary Appendix), which is a critical mediator of B-cell development and apoptosis in activated B cells [48,49]. B3 expressed high MS4A1, CD79A, CRIP1 and IGJ levels and low TCL1A, PMAIP1 levels, indicative of mature B cells (Fig. 3d and e, Table S6 and Fig. S5B in the Supplementary Appendix) [47].
Fig. 3.
B cell clusters in PBMC from HC, LTBI and TB. (a) tSNE of single cell profile with each cell color coded for B cell subsets (left to right): the associated cell type, the corresponding status (HC, LTBI and TB) and merged all status. (b)The fraction of cells for B cell subsets in HC, LTBI and TB. (c) Differential expression analysis was performed comparing B cell subsets from HC, LTBI and TB. Heatmaps are shown representing the up- and down-regulated genes in Myeloid subsets. (d) Expression of marker genes for the B cell subsets. (e) Differential expression analysis was performed comparing cells from HC, LTBI and TB within B cell subsets. (f) Differences in pathway activities scored per cell by GSVA between the different B clusters.
Studies enumerating B-cells in patients with TB disease have yielded conflicting results [27,50,51]. In our scRNA-seq data, all five B-cell subsets were present in all seven donors, albeit in variable proportions. Of these subsets, B2 and B4 were enriched in patients with TB (Fig. 3b and Table S3 in the Supplementary Appendix). We thus characterised the functions of B2 and B4 cells by pathway analysis (Fig. 3e). GSEA analysis showed that B2 and B4 were similar in terms of exhibiting high rates of TNFα-, TGFβ-, PI3K-AKT- signaling, but differed in terms of WNTβ catenin signaling and inflammatory response, indicating immune function differences (Fig. 3e). Together, these data reveals five B cell subsets with distinct markers in PBMC and exhausted B cells (B1) are abundant in TB. Our data will be useful in studying the roles of B-cell subsets in TB disease.
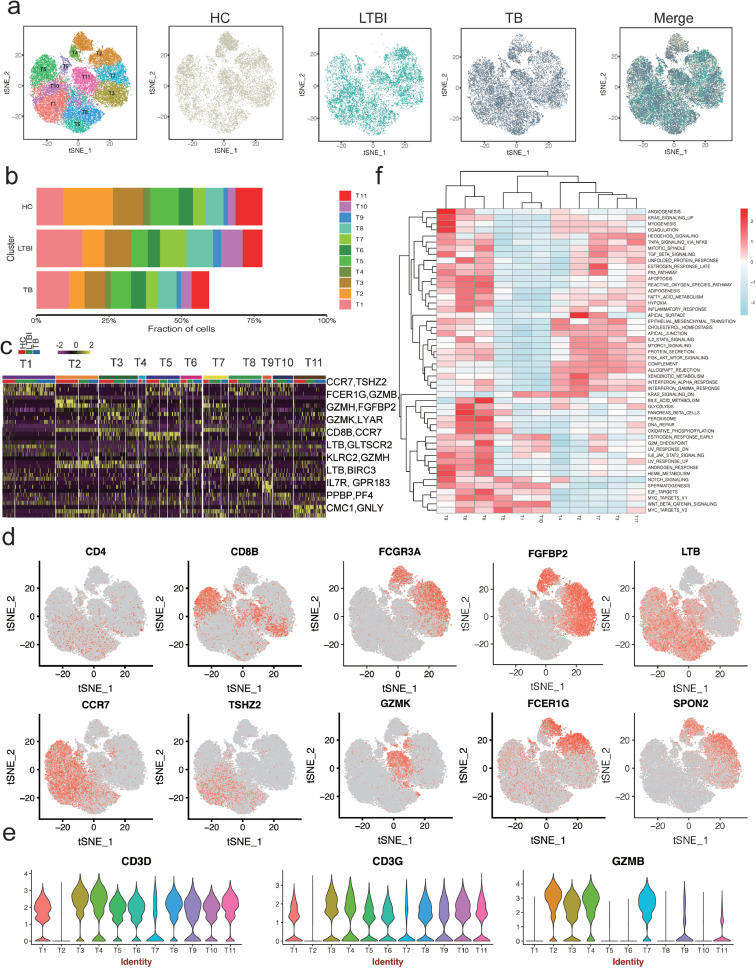
3.6. scRNA-seq identifies nine CD4 and CD8 subsets and reveals an altered T-cell subsets distribution in TB
T cells have a critical role in controlling Mtb infection in patients with TB [52,53]. Our scRNAseq analysis detected 43,537 T cells in all three donor groups that could be sub-clustered into 11 subsets (Fig. 4a–d and Fig. S7). We thus attempted to identify marker genes for each of these subsets and to assign them to known T-cell types based on the expression of established markers reported in the CellMarker database [54]. Among 11 subsets, there are 9 subsets expression high level of CD3 (Fig. 4e). We next identified four distinct CD4+ T-cell subsets: T1, T6, T8 and T10 (Fig. 4a,d). T1 expressed high CCR7 levels, indicative of naïve-like CD4 T cells [55]. Both T6 and T8 expressed high levels of the activated CD4 T-cell marker LTβ (TNFSF3) [56] and other functional markers such as AQP3, GPR183 and LDHB (Table S7 in the Supplementary Appendix), which regulate T-cell trafficking and migration [57,58]. GSEA revealed that T9 exhibits a high rate of myogenesis, angiogenesis, coagulation and KRAS signaling, suggesting these cells are strongly activated (Fig. 4f). T6 and T8 displayed up-regulated pathways associated with peroxisome, E2F- and MYC- targets and glycolysis (Fig. 4f), indicating that these cells are exhausted. The frequencies of T1, T6 and T8 were not significantly differences between the three donor groups (Table S3 in the Supplementary Appendix).
Fig. 4.
T cell clusters in PBMC from HC, LTBI and TB. (a) tSNE of single cell profile with each cell color coded for T cell subsets (left to right): the associated cell type, the corresponding status (HC, LTBI and TB) and merged all status, (b) The fraction of cells for T cell subsets in HC, LTBI and TB. (c) Differential expression analysis was performed comparing T cell subsets from HC, LTBI and TB. Heatmaps are shown representing the up- and down-regulated genes in T subsets. (d) Expression of marker genes for the T cell subsets. (e) Differential expression analysis was performed comparing cells from HC, LTBI and TB within B cell subsets. (f) Differences in pathway activities scored per cell by GSVA between the different T clusters.
We also identified five diverse CD8 T-cell subsets, including naïve CD8 T cells (T5; CCR7) [59], cytotoxic CD8 effector T cells (T3 and T4; GZMH, NKG7 and FGFBP2), transitional CD8 effector T cells (T11; GZMK, KLRB1 and LYAR4) [60] and a small group of megakaryocyte-like cells (T9; PPBP, PF4 and GNG11) (Fig. 4a–d, Fig. S5C and Table S7 in the Supplementary Appendix) [41]. The proportions of CD8 T-cell subsets were slightly increased in HC, compared to LTBI and TB (Fig. 4b and Table S3 in the Supplementary Appendix).
3.7. A cytotoxic NK cell subset is depleted in TB
Sub-cluster analysis also revealed two distinct clusters of NK cells: T2 and T7 (Fig. 4a–e). T2 and T7 expressed lower level of CD3 (Fig. 4e) indicating that they are most likely NK cells. T2 was expressed high level of FCER1G, GZMB, GNLY, SPON2, PRF1, CD7, MYOM2 and KLRF1 expression (Fig. S5C and Table S7 in the Supplementary Appendix), indicative of high cytotoxic activities. T7 was enriched in GZMH, FGFBP2, GNLY and selectively expressed KLRC2 (NKG2) suggesting the memory-like NK cells (Fig. 4e, Table S7 and Fig. S5C in the Supplementary Appendix) [61]. We found that some pathways were differentially regulated between the NK-like cell subsets. T2 and T7 showed high allograft rejection activities and strong IFN-γ and IFN-α responses, indicative of cell activation (Fig. 4f). Although T2 and T7 showed a high level of similarity based on the pathway analyses, the proportions of these subsets varied between HC and patients with LTBI and TB. Specifically, T2 gradually decreased from LTBI to TB, compared to HC; by contrast, T7 was most frequent in LTBI (Fig. 4b). Gene Ontology enrichment analysis of the upregulated expression genes in T2 of TB revealed that regulation of cell death pathway were highly enriched in T2 from TB, compared to HC and LTBI (Table S8 in the Supplementary Appendix), which may be responsible for the depletion of T2 in TB. Taken together, our scRNA-seq identified two NK-like subsets which are variable proportions among three donor groups and a cytotoxic NK subset (CD3-CD7+GZMB+) with high level of SPON2 and MYOM2 is depletion in TB.
3.8. CD3-CD7+GZMB+ subset efficiently differentiates TB from HC and LTBI
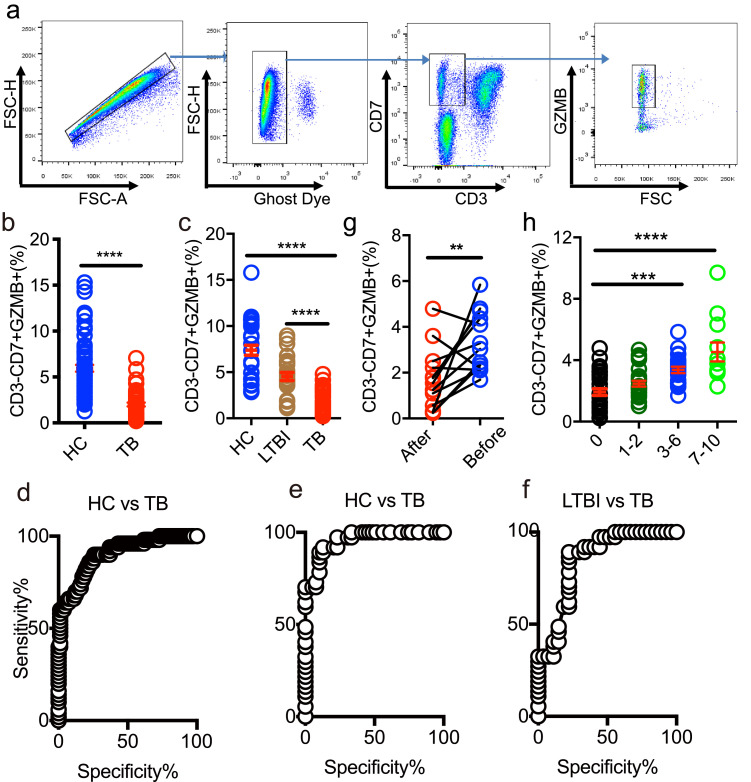
To better understand the nature of the T2 NK-cell subset, we validated T2 by flow cytometry. To do so, we used the markers CD7, CD3 and GZMB that identified from scRNA-seq data to gate the cells. We first identified that CD3-CD7+GZMB+ cells were all CD56+ and CD14-CD19-, supporting that T2 comprises NK cells (Fig. 5a and Fig. S8 in the Supplementary Appendix). Next, we compared the frequency of this subset among HC, LTBI and TB in two independent cohorts. Results showed that the frequency of CD3-CD7+GZMB+ cells was significantly decreased in patients with TB, compared to HC and LTBI (Fig. 5b,c); this finding is consistent with our original sc-RNAseq results (Fig. 4b). To further evaluate the diagnostic value of this subset for TB biomarker, we performed receiver-operating characteristic (ROC) curve analysis in these two cohorts. ROC curve analysis of TB vs HC or TB vs LTBI indicated that CD3-CD7+GZMB+ subset could serve as valuable biomarker, with the area under curve being 0.93 (Fig. 5d, TB vs HC in second cohort), 0.96 (Fig. 5e, TB vs HC in third cohort), 0.85 (Fig. 5f, TB vs LTBI in third cohort). We further analyzed the frequency of this subset after anti-TB treatment. As shown in Fig. 5g and h, the level of CD3-CD7+GZMB+ increased 3 months after initiation of treatment. Thus, by scRNA-seq and flow cytometry, we confirmed CD3-CD7+GZMB+ NK cell is useful for differentiating TB from LTBI and HC.
Fig. 5.
Flow cytometry analysis of CD3-CD7+GZMB+ subsets in HC, LTBI and TB. (a) Gating strategy of CD3-CD7+GZMB+ by flow cytometry. (b) The frequency of CD3-CD7+GZMB+ in HC and TB in the second cohort (HC=81, TB=50). (c) The frequency of CD3-CD7+GZMB+ in HC, LTBI and TB in the third cohort (HC=39, LTBI=27, and TB=37). (d–f) ROC curve for CD3-CD7+GZMB+ to separate TB from HC (d, e) or LTBI (f). AUC=0.93 (d), AUC=0.96 (e), AUC=0.85 (f). AUC, area under curve. (g,h)The frequency of CD3-CD7+GZMB+ is increased after anti-TB treatment in TB patients. (g) follow-up TB patients (n = 11) and (h) TB patients received anti-TB treatment from 1 to 10 months (n = 73). A one-way ANOVA Newman–Keuls multiple comparison test was used to compare the differences among multiple groups. An unpaired t-test was used to analyze the differences between two groups. An paired t-test was used to analyze the differences in the follow-up patients. The data represent the means ± SEM. **P < 0. 001, ***P < 0. 001, ****P < 0. 0001.
4. Discussion
In this study, we aimed to understand the impact of Mtb infection on circulating immune-cell subsets during TB development through scRNA-seq of PBMC from HC,LTBI and TB. We isolated >60,000 and performed scRNAseq followed by pathway analysis to annotate the nature and frequencies of PBMC subsets between donor groups. At this resolution, we distinguished three major cell types (T cells, B cells and myeloid cells) and subsequently sub-clustered these into 29 subsets based on quantitative gene expression. Although some of the markers were already known, we identified a large number of additional markers, including IFITM1, GIMAP7, CD247 and LCK for T cells, BANK1, VPREB3, FCER2 and ADAM28 for B cells and FCN1, CST4, SERPINA1, MNDA, LST1 and MS4A6A for myeloid cells. These markers might be further explored as PBMC cell-specific markers and use as references for further study to investigate the role of immune-cell subsets in TB pathogenesis as well as protective immunity against TB.
Comparing scRNA-seq datasets between HC and LTBI and TB revealed that disease gives rise to changes in subsets of known cell types, especially cell subsets that seem to be specific for disease, such as M1, M4 and B2 specific enriched and T2 depleted in TB. The cellular composition of PBMCs changes during pathological stress [62], [63], [64]. Consistently, our routine blood analysis and scRNA-seq confirmed that circulating immune cells frequencies change among HC, LTBI and TB. Our PBMC cell atlas not only provides additional insights into the cell subsets of PMBC in humans, but also provides insights into cell subsets related to different outcome of Mtb infection.
NK cells can kill Mtb-infected cells directly or through antibody-dependant cellular cytotoxicity (ADCC) [65]. However, NK cells were shown to have a minimal role in host protection in a mouse model of Mtb infection [66]. Although NK cell depletion has been noted during HIV infection [67], many studies have shown that the frequency of NK cells does not change in TB compared to HC or TB plus HIV co-infected patients [64,68]. Others cell frequency between pre- and post- TB treatment [62]. In line with previous reports, we also found that total NK cell was not significantly change between TB and HC. However, no prior studies have analysed NK cell subsets in TB. Here, we identified qualitative and quantitative changes that occur in circulating NK subsets among HC, and LTBI and TB. Interestingly, in our scRNA-seq, we found two NK subsets (T2 and T7) differentially represented between the three groups. T2 was gradually depletion from HC to LTBI to TB. By contrast, T7 was most frequent in LTBI, compared to HC and TB. These two subsets were specifically enriched for distinct markers, indicative of functional differences. Specifically, T7 selectively expressed NKG2, an activating NK cell receptor [69] that is critical for the pulmonary clearance of bacteria [70,71]. This subset was increased in LTBI, compared to HC and patients with TB, suggesting that this subset might be involved in controlling Mtb infection in LTBI [68]. The depletion of T2 in TB were further verified with two independent cohorts by flow cytometry. T2 was specifically enriched for GZMB and SPON2. SPON2 is a secreted ECM protein [72] that binds to integrin receptors and bacterial lipopolysaccharide [73]; this mechanism is essential for initiating an immune response and represents a unique pattern recognition molecule for microbial pathogens [74]. SPON2 also functions as an integrin ligand for inflammatory cell recruitment and T-cell priming [75,76]. This subset may be involved in host immunity to Mtb infection. A probable scenario of T2 depletion in human in TB is that this subset is easier to die in active TB, as we found that the upregulated genes in T2 from TB are enrichment in cell death pathway, compared to HC and LTBI.
While TB biomarker research is a highly active field, the impact of the data thus far has been limited. Except for sputum culture, the only WHO-endorsed tests for active TB detection are based on DNA detection in sputum, which is not perfect in TB diagnosis as sputum is not available with some of TB patients. Here, we found that the change in peripheral CD3-CD7+GZMB+ levels is sensitive and specific to discriminate active TB from LTBI and HC. As blood is the most readily accessible sample in humans, we propose that CD3-CD7+GZMB+ could be used as a novel biomarker for distinguishing TB from LTBI and HC.
There are also limitations to our study that highlight the need for further work to optimize and expand single-cell RNA-Seq datasets for TB. First, our scRNA-Seq dataset includes a relatively small samples and might create gender bias due to sex of samples. Even in this small cohort, however, we were able to identify many of know and novel markers in PBMC subsets, and validate a subset (CD3-CD7+GZMB+) by flow cytometry with two independent cohorts. Second, due to the antibody availability, we could not further confirmed the changing of some subsets with the markers from scRNA-seq in HC, LTBI and TB, such as B and myeloid cell subsets. Third, the present study data do not have the resolution to characterize the less-frequent immune-cell populations. We thus suggest that these clusters are likely multiple cell populations. For example, mucosal associated invariant T (MAIT) cells likely fall within our myeloid and T-cell subsets, as we found the MAIT cell marker CD161 (KLRB1) [77] distributed in T-cell subsets. Further characterization of these subsets will require including more cells and additional single-cell analysis tools, such as surface protein labeling (10 × Genomics Single Cell 3′ v3 Reagent Kit). These approaches will improve our ability to detect and identify these important cell populations in future analyses.
In describing key molecular differences among HC, LTBI and TB, our analyses confirm many important observations made previously, and highlight key ongoing research areas and challenges. For example, how specific circulating cell populations change and contribute to the development of TB disease is an ongoing topic of discussion. To the best of our knowledge, our data represent the first scRNA-seq-based description of PBMC immune-cell subsets in LTBI and TB. We consider that our characterization of circulating cell subsets in HC, LTBI and TB provides a useful framework to examine the role of cell subsets in TB disease progression. Further dissecting the mechanism behind dysregulation of TB-associated cell subsets such as CD3-CD7+GZMB+ NK cell, might open new avenues for therapeutic intervention against TB.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgments
The authors gratefully acknowledge all the study participants and study staff for the help and cooperation during the study, and Dr. Jessica Tamanini (Shenzhen University and ET editing) for editing the manuscript prior to submission.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102686.
Appendix. Supplementary materials
Fig. S1. Consistency of cell capture and identification in PBMC scRNA-seq. Number of unique molecular identifiers (nUMI) and genes identified, and fraction of reads mapping to mitochondrial or ribosomal genes across recovered cell types and nUMI and genes identified, and fraction of reads mapping to mitochondrial or ribosomal genes across all donors.
Fig. S2. tSNE projection of PBMC from seven donors. Upper panel (left to right): all donor merged PBMC single cells, the corresponding status (HC, LTBI and TB). Lower panel (left to right): the associated cell type, the corresponding status in associated cell type (HC, LTBI and TB)
Fig. S3. Expression of known and new marker genes for the myeloid, T and B cell. (a-b) Differential expression analysis was performed comparing cells from HC, LTBI and TB in PBMC. (a) tSNE; (b) Violin plot.
Fig. S4 tSNE projection of myeloid subsets from seven donors. Upper panel (left to right): all donor merged myeloid single cells, the corresponding status (HC, LTBI and TB). Lower panel (left to right): the associated cell type, the corresponding status in associated cell type (HC, LTBI and TB)
Fig. S5. Expression of known and new marker genes for the myeloid, T and B subsets. (a) Myeloid subsets; (b) B cell subsets; (c) T cell subsets
Fig. S6. tSNE projection of B cell subsets from seven donors. Upper panel (left to right): all donor merged B single cells, the corresponding status (HC, LTBI and TB). Lower panel (left to right): the associated cell type, the corresponding status in associated cell type (HC, LTBI and TB)
Fig. S7. tSNE projection of T subsets from seven donors. Upper panel (left to right): all donor merged T single cells, the corresponding status (HC, LTBI and TB). Lower panel (left to right): the associated cell type, the corresponding status in associated cell type (HC, LTBI and TB)
Fig. S8. Flow cytometry analysis of CD3-CD7+GZMB+ subsets
Table S1 Demographic characteristics of study populations
Table S2. Characteristics of scRNA-seq of the seven PBMC samples included in this study.
Table S3. The cell numbers and frequency of all subsets in PBMC from HC, LTBI and TB.
Table S4. Marker genes of PBMC major cell types identified by scRNA-seq
Table S5. Marker genes of myeloid subsets identified by scRNA-seq
Table S6. Marker genes of B cell subsets identified by scRNA-seq
Table S7. Marker genes of T cell subsets identified by scRNA-seq
Table S8. Gene oncology enrichment analysis of upregulated genes in T2 from TB
References
- 1.Floyd K., Glaziou P., Zumla A., Raviglione M. The global tuberculosis epidemic and progress in care, prevention, and research: an overview in year 3 of the end tb era. Lancet Respir Med. 2018;6(4):299–314. doi: 10.1016/S2213-2600(18)30057-2. [DOI] [PubMed] [Google Scholar]
- 2.Kaufmann S.H., Parida S.K. Tuberculosis in africa: learning from pathogenesis for biomarker identification. Cell Host Microbe. 2008;4(3):219–228. doi: 10.1016/j.chom.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Wallis R.S., Kim P., Cole S., Hanna D., Andrade B.B., Maeurer M. Tuberculosis biomarkers discovery: developments, needs, and challenges. Lancet Infect Dis. 2013;13(4):362–372. doi: 10.1016/S1473-3099(13)70034-3. [DOI] [PubMed] [Google Scholar]
- 4.Singhania A., Wilkinson R.J., Rodrigue M., Haldar P., O'Garra A. The value of transcriptomics in advancing knowledge of the immune response and diagnosis in tuberculosis. Nat Immunol. 2018;19(11):1159–1168. doi: 10.1038/s41590-018-0225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry M.P., Graham C.M., McNab F.W., Xu Z., Bloch S.A., Oni T. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466(7309):973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cliff J.M., Lee J.S., Constantinou N., Cho J.E., Clark T.G., Ronacher K. Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. J Infect Dis. 2013;207(1):18–29. doi: 10.1093/infdis/jis499. [DOI] [PubMed] [Google Scholar]
- 7.Roe J.K., Thomas N., Gil E., Best K., Tsaliki E., Morris-Jones S. Blood transcriptomic diagnosis of pulmonary and extrapulmonary tuberculosis. JCI Insight. 2016;1(16):e87238. doi: 10.1172/jci.insight.87238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singhania A., Verma R., Graham C.M., Lee J., Tran T., Richardson M. A modular transcriptional signature identifies phenotypic heterogeneity of human tuberculosis infection. Nat Commun. 2018;9(1):2308. doi: 10.1038/s41467-018-04579-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zak D.E., Penn-Nicholson A., Scriba T.J., Thompson E., Suliman S., Amon L.M. A blood rna signature for tuberculosis disease risk: a prospective cohort study. Lancet. 2016;387(10035):2312–2322. doi: 10.1016/S0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai Y., Yang Q., Tang Y., Zhang M., Liu H., Zhang G. Increased complement C1q level marks active disease in human tuberculosis. PLoS One. 2014;9(3):e92340. doi: 10.1371/journal.pone.0092340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veenstra H., Baumann R., Carroll N.M., Lukey P.T., Kidd M., Beyers N. Changes in leucocyte and lymphocyte subsets during tuberculosis treatment; prominence of CD3dimCD56+ natural killer t cells in fast treatment responders. Clin Exp Immunol. 2006;145(2):252–260. doi: 10.1111/j.1365-2249.2006.03144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thum M.Y., Bhaskaran S., Abdalla H.I., Ford B., Sumar N., Shehata H. An increase in the absolute count of CD56dimCD16+CD69+ nk cells in the peripheral blood is associated with a poorer ivf treatment and pregnancy outcome. Hum Reprod. 2004;19(10):2395–2400. doi: 10.1093/humrep/deh378. [DOI] [PubMed] [Google Scholar]
- 13.Reyfman P.A., Walter J.M., Joshi N., Anekalla K.R., McQuattie-Pimentel A.C., Chiu S. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med. 2019;199(12):1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zilionis R., Engblom C., Pfirschke C., Savova V., Zemmour D., Saatcioglu H.D. Single-Cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity. 2019;50(5):1317–1334. doi: 10.1016/j.immuni.2019.03.009. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gladka M.M., Molenaar B., de Ruiter H., van der Elst S., Tsui H., Versteeg D. Single-Cell sequencing of the healthy and diseased heart reveals cytoskeleton-associated protein 4 as a new modulator of fibroblasts activation. Circulation. 2018;138(2):166–180. doi: 10.1161/CIRCULATIONAHA.117.030742. [DOI] [PubMed] [Google Scholar]
- 16.Lambrechts D., Wauters E., Boeckx B., Aibar S., Nittner D., Burton O. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018;24(8):1277–1289. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 17.Puram S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S. Single-Cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171(7):1611–1624. doi: 10.1016/j.cell.2017.10.044. e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X., Yang Q., Zhang M., Graner M., Zhu X., Larmonier N. Diagnosis of active tuberculosis in china using an in-house gamma interferon enzyme-linked immunospot assay. Clin Vaccine Immunol. 2009;16(6):879–884. doi: 10.1128/CVI.00044-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X., Zhou B., Li M., Deng Q., Wu X., Le X. CD4(+)CD25(+)FoxP3(+) regulatory t cells suppress mycobacterium tuberculosis immunity in patients with active disease. Clin Immunol. 2007;123(1):50–59. doi: 10.1016/j.clim.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Xie T., Wang Y., Deng N., Huang G., Taghavifar F., Geng Y. Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep. 2018;22(13):3625–3640. doi: 10.1016/j.celrep.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolock S.L., Lopez R., Klein A.M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019;8(4):281–291. doi: 10.1016/j.cels.2018.11.005. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleiveland C.R. Peripheral blood mononuclear cells. In: Verhoeckx K, Cotter P, Lopez-Exposito I, Kleiveland C, Lea T, Mackie A, editors. The impact of food bioactives on health: in vitro and ex vivo models. 2015. pp. 161–167. editors. Cham (CH) [PubMed] [Google Scholar]
- 27.Wu Y.E., Zhang S.W., Peng W.G., Li K.S., Li K., Jiang J.K. Changes in lymphocyte subsets in the peripheral blood of patients with active pulmonary tuberculosis. J Int Med Res. 2009;37(6):1742–1749. doi: 10.1177/147323000903700610. [DOI] [PubMed] [Google Scholar]
- 28.Deveci F., Akbulut H.H., Celik I., Muz M.H., Ilhan F. Lymphocyte subpopulations in pulmonary tuberculosis patients. Mediators Inflamm. 2006;2006(2):89070. doi: 10.1155/MI/2006/89070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castano D., Garcia L.F., Rojas M. Increased frequency and cell death of CD16+ monocytes with mycobacterium tuberculosis infection. Tuberculosis. 2011;91(5):348–360. doi: 10.1016/j.tube.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Maertzdorf J., McEwen G., Weiner J., 3rd, Tian S., Lader E., Schriek U. Concise gene signature for point-of-care classification of tuberculosis. EMBO Mol Med. 2016;8(2):86–95. doi: 10.15252/emmm.201505790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaforou M., Wright V.J., Oni T., French N., Anderson S.T., Bangani N. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood rna expression signatures: a case-control study. PLoS Med. 2013;10(10) doi: 10.1371/journal.pmed.1001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziegler-Heitbrock L. Blood monocytes and their subsets: established features and open questions. Front Immunol. 2015;6:423. doi: 10.3389/fimmu.2015.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeiner P.S., Preusse C., Blank A.E., Zachskorn C., Baumgarten P., Caspary L. MIF receptor CD74 is restricted to microglia/macrophages, associated with a M1-Polarized immune milieu and prolonged patient survival in gliomas. Brain Pathol. 2015;25(4):491–504. doi: 10.1111/bpa.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cochain C., Vafadarnejad E., Arampatzi P., Pelisek J., Winkels H., Ley K. Single-cell RNA-seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ Res. 2018;122(12):1661–1674. doi: 10.1161/CIRCRESAHA.117.312509. [DOI] [PubMed] [Google Scholar]
- 35.Schwartzkopff F., Petersen F., Grimm T.A., Brandt E. CXC chemokine ligand 4 (CXCL4) down-regulates cc chemokine receptor expression on human monocytes. Innate Immun. 2012;18(1):124–139. doi: 10.1177/1753425910388833. [DOI] [PubMed] [Google Scholar]
- 36.Elmesmari A., Fraser A.R., Wood C., Gilchrist D., Vaughan D., Stewart L. MicroRNA-155 regulates monocyte chemokine and chemokine receptor expression in rheumatoid arthritis. Rheumatology. 2016;55(11):2056–2065. doi: 10.1093/rheumatology/kew272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villani A.C., Satija R., Reynolds G., Sarkizova S., Shekhar K., Fletcher J. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356(6335) doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao W., Bover L. Signaling and ligand interaction of ILT7: receptor-mediated regulatory mechanisms for plasmacytoid dendritic cells. Immunol Rev. 2010;234(1):163–176. doi: 10.1111/j.0105-2896.2009.00867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murray L., Xi Y., Upham J.W. CLEC4C gene expression can be used to quantify circulating plasmacytoid dendritic cells. J Immunol Methods. 2019;464:126–130. doi: 10.1016/j.jim.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 40.Winterhoff B.J., Maile M., Mitra A.K., Sebe A., Bazzaro M., Geller M.A. Single cell sequencing reveals heterogeneity within ovarian cancer epithelium and cancer associated stromal cells. Gynecol Oncol. 2017;144(3):598–606. doi: 10.1016/j.ygyno.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinha D., Kumar A., Kumar H., Bandyopadhyay S., Sengupta D. dropClust: efficient clustering of ultra-large scRNA-seq data. Nucleic Acids Res. 2018;46(6):e36. doi: 10.1093/nar/gky007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eljaszewicz A., Kleina K., Grubczak K., Radzikowska U., Zembko P., Kaczmarczyk P. Elevated numbers of circulating very small embryonic-like stem cells (VSELs) and intermediate CD14++CD16+ monocytes in iga nephropathy. Stem Cell Rev. 2018;14(5):686–693. doi: 10.1007/s12015-018-9840-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moniuszko M., Bodzenta-Lukaszyk A., Kowal K., Lenczewska D., Dabrowska M. Enhanced frequencies of CD14++CD16+, but not CD14+CD16+, peripheral blood monocytes in severe asthmatic patients. Clin Immunol. 2009;130(3):338–346. doi: 10.1016/j.clim.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 44.Saleh M.N., Goldman S.J., LoBuglio A.F., Beall A.C., Sabio H., McCord M.C. CD16+ monocytes in patients with cancer: spontaneous elevation and pharmacologic induction by recombinant human macrophage colony-stimulating factor. Blood. 1995;85(10):2910–2917. [PubMed] [Google Scholar]
- 45.du Plessis W.J., Walzl G., Loxton A.G. B cells as multi-functional players during mycobacterium tuberculosis infection and disease. Tuberculosis. 2016;97:118–125. doi: 10.1016/j.tube.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 46.Moyron-Quiroz J.E., Partida-Sanchez S., Donis-Hernandez R., Sandoval-Montes C., Santos-Argumedo L. Expression and function of CD22, a B-cell restricted molecule. Scand J Immunol. 2002;55(4):343–351. doi: 10.1046/j.1365-3083.2002.01063.x. [DOI] [PubMed] [Google Scholar]
- 47.Baker D., Marta M., Pryce G., Giovannoni G., Schmierer K. Memory b cells are major targets for effective immunotherapy in relapsing multiple sclerosis. EBioMedicine. 2017;16:41–50. doi: 10.1016/j.ebiom.2017.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wensveen F.M., Derks I.A., van Gisbergen K.P., de Bruin A.M., Meijers J.C., Yigittop H. BH3-only protein noxa regulates apoptosis in activated b cells and controls high-affinity antibody formation. Blood. 2012;119(6):1440–1449. doi: 10.1182/blood-2011-09-378877. [DOI] [PubMed] [Google Scholar]
- 49.Bretz J., Garcia J., Huang X., Kang L., Zhang Y., Toellner K.M. Noxa mediates p18INK4c cell-cycle control of homeostasis in b cells and plasma cell precursors. Blood. 2011;117(7):2179–2188. doi: 10.1182/blood-2010-06-288027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barcelos W., Martins-Filho O.A., Guimaraes T.M., Oliveira M.H., Spindola-de-Miranda S., Carvalho B.N. Peripheral blood mononuclear cells immunophenotyping in pulmonary tuberculosis patients before and after treatment. Microbiol Immunol. 2006;50(8):597–605. doi: 10.1111/j.1348-0421.2006.tb03834.x. [DOI] [PubMed] [Google Scholar]
- 51.Hernandez J., Velazquez C., Valenzuela O., Robles-Zepeda R., Ruiz-Bustos E., Navarro M. Low number of peripheral blood b lymphocytes in patients with pulmonary tuberculosis. Immunol Investig. 2010;39(3):197–205. doi: 10.3109/08820130903586346. [DOI] [PubMed] [Google Scholar]
- 52.Winslow G.M., Cooper A., Reiley W., Chatterjee M., Woodland D.L. Early T-cell responses in tuberculosis immunity. Immunol Rev. 2008;225:284–299. doi: 10.1111/j.1600-065X.2008.00693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lalvani A., Millington K.A. T cells and tuberculosis: beyond interferon-gamma. J Infect Dis. 2008;197(7):941–943. doi: 10.1086/529049. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X., Lan Y., Xu J., Quan F., Zhao E., Deng C. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47(D1):D721–D7D8. doi: 10.1093/nar/gky900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gattorno M., Prigione I., Morandi F., Gregorio A., Chiesa S., Ferlito F. Phenotypic and functional characterisation of CCR7+ and CCR7- CD4+ memory t cells homing to the joints in juvenile idiopathic arthritis. Arthritis Res Ther. 2005;7(2):R256–R267. doi: 10.1186/ar1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aggarwal B.B., Gupta S.C., Kim J.H. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey. Blood. 2012;119(3):651–665. doi: 10.1182/blood-2011-04-325225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emgard J., Kammoun H., Garcia-Cassani B., Chesne J., Parigi S.M., Jacob J.M. Oxysterol sensing through the receptor GPR183 promotes the lymphoid-tissue-inducing function of innate lymphoid cells and colonic inflammation. Immunity. 2018;48(1):120–132. doi: 10.1016/j.immuni.2017.11.020. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hara-Chikuma M., Chikuma S., Sugiyama Y., Kabashima K., Verkman A.S., Inoue S. Chemokine-dependent t cell migration requires aquaporin-3-mediated hydrogen peroxide uptake. J Exp Med. 2012;209(10):1743–1752. doi: 10.1084/jem.20112398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forster R., Davalos-Misslitz A.C., Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol. 2008;8(5):362–371. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- 60.Li H., van der Leun A.M., Yofe I., Lubling Y., Gelbard-Solodkin D., van Akkooi A.C.J. Dysfunctional CD8 t cells form a proliferative, dynamically regulated compartment within human melanoma. Cell. 2019;176(4):775–789. doi: 10.1016/j.cell.2018.11.043. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ram D.R., Manickam C., Hueber B., Itell H.L., Permar S.R., Varner V. Tracking KLRC2 (NKG2C)+ memory-like nk cells in SIV+ and rhCMV+ rhesus macaques. PLoS Pathog. 2018;14(5) doi: 10.1371/journal.ppat.1007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garand M., Goodier M., Owolabi O., Donkor S., Kampmann B., Sutherland J.S. Functional and phenotypic changes of natural killer cells in whole blood during mycobacterium tuberculosis infection and disease. Front Immunol. 2018;9:257. doi: 10.3389/fimmu.2018.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sauzullo I., Mengoni F., Mascia C., Rossi R., Lichtner M., Vullo V. Treatment of latent tuberculosis infection induces changes in multifunctional mycobacterium tuberculosis-specific CD4+ t cells. Med Microbiol Immunol. 2016;205(1):37–45. doi: 10.1007/s00430-015-0424-z. [DOI] [PubMed] [Google Scholar]
- 64.Ardain A., Domingo-Gonzalez R., Das S., Kazer S.W., Howard N.C., Singh A. Group 3 innate lymphoid cells mediate early protective immunity against tuberculosis. Nature. 2019;570(7762):528–532. doi: 10.1038/s41586-019-1276-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Esin S., Batoni G. Natural killer cells: a coherent model for their functional role in mycobacterium tuberculosis infection. J Innate Immun. 2015;7(1):11–24. doi: 10.1159/000363321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Junqueira-Kipnis A.P., Kipnis A., Jamieson A., Juarrero M.G., Diefenbach A., Raulet D.H. NK cells respond to pulmonary infection with mycobacterium tuberculosis, but play a minimal role in protection. J Immunol. 2003;171(11):6039–6045. doi: 10.4049/jimmunol.171.11.6039. [DOI] [PubMed] [Google Scholar]
- 67.Mansour I., Doinel C., Rouger P. CD16+ nk cells decrease in all stages of hiv infection through a selective depletion of the CD16+CD8+CD3- subset. AIDS Res Hum Retroviruses. 1990;6(12):1451–1457. doi: 10.1089/aid.1990.6.1451. [DOI] [PubMed] [Google Scholar]
- 68.Roy Chowdhury R., Vallania F., Yang Q., Lopez Angel C.J., Darboe F., Penn-Nicholson A. A multi-cohort study of the immune factors associated with m. tuberculosis infection outcomes. Nature. 2018;560(7720):644–648. doi: 10.1038/s41586-018-0439-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jamieson A.M., Diefenbach A., McMahon C.W., Xiong N., Carlyle J.R., Raulet D.H. The role of the nkg2d immunoreceptor in immune cell activation and natural killing. Immunity. 2002;17(1):19–29. doi: 10.1016/s1074-7613(02)00333-3. [DOI] [PubMed] [Google Scholar]
- 70.Wesselkamper S.C., Eppert B.L., Motz G.T., Lau G.W., Hassett D.J., Borchers M.T. NKG2D is critical for nk cell activation in host defense against pseudomonas aeruginosa respiratory infection. J Immunol. 2008;181(8):5481–5489. doi: 10.4049/jimmunol.181.8.5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Borchers M.T., Harris N.L., Wesselkamper S.C., Zhang S., Chen Y., Young L. The NKG2D-activating receptor mediates pulmonary clearance of pseudomonas aeruginosa. Infect Immun. 2006;74(5):2578–2586. doi: 10.1128/IAI.74.5.2578-2586.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Higashijima S., Nose A., Eguchi G., Hotta Y., Okamoto H. Mindin/F-spondin family: novel ecm proteins expressed in the zebrafish embryonic axis. Dev Biol. 1997;192(2):211–227. doi: 10.1006/dbio.1997.8760. [DOI] [PubMed] [Google Scholar]
- 73.Li Y., Cao C., Jia W., Yu L., Mo M., Wang Q. Structure of the F-spondin domain of mindin, an integrin ligand and pattern recognition molecule. EMBO J. 2009;28(3):286–297. doi: 10.1038/emboj.2008.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He Y.W., Li H., Zhang J., Hsu C.L., Lin E., Zhang N. The extracellular matrix protein mindin is a pattern-recognition molecule for microbial pathogens. Nat Immunol. 2004;5(1):88–97. doi: 10.1038/ni1021. [DOI] [PubMed] [Google Scholar]
- 75.Jia W., Li H., He Y.W. The extracellular matrix protein mindin serves as an integrin ligand and is critical for inflammatory cell recruitment. Blood. 2005;106(12):3854–3859. doi: 10.1182/blood-2005-04-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H., Oliver T., Jia W., He Y.W. Efficient dendritic cell priming of t lymphocytes depends on the extracellular matrix protein mindin. EMBO J. 2006;25(17):4097–4107. doi: 10.1038/sj.emboj.7601289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Le Bourhis L., Mburu Y.K., Lantz O. MAIT cells, surveyors of a new class of antigen: development and functions. Curr Opin Immunol. 2013;25(2):174–180. doi: 10.1016/j.coi.2013.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Consistency of cell capture and identification in PBMC scRNA-seq. Number of unique molecular identifiers (nUMI) and genes identified, and fraction of reads mapping to mitochondrial or ribosomal genes across recovered cell types and nUMI and genes identified, and fraction of reads mapping to mitochondrial or ribosomal genes across all donors.
Fig. S2. tSNE projection of PBMC from seven donors. Upper panel (left to right): all donor merged PBMC single cells, the corresponding status (HC, LTBI and TB). Lower panel (left to right): the associated cell type, the corresponding status in associated cell type (HC, LTBI and TB)
Fig. S3. Expression of known and new marker genes for the myeloid, T and B cell. (a-b) Differential expression analysis was performed comparing cells from HC, LTBI and TB in PBMC. (a) tSNE; (b) Violin plot.
Fig. S4 tSNE projection of myeloid subsets from seven donors. Upper panel (left to right): all donor merged myeloid single cells, the corresponding status (HC, LTBI and TB). Lower panel (left to right): the associated cell type, the corresponding status in associated cell type (HC, LTBI and TB)
Fig. S5. Expression of known and new marker genes for the myeloid, T and B subsets. (a) Myeloid subsets; (b) B cell subsets; (c) T cell subsets
Fig. S6. tSNE projection of B cell subsets from seven donors. Upper panel (left to right): all donor merged B single cells, the corresponding status (HC, LTBI and TB). Lower panel (left to right): the associated cell type, the corresponding status in associated cell type (HC, LTBI and TB)
Fig. S7. tSNE projection of T subsets from seven donors. Upper panel (left to right): all donor merged T single cells, the corresponding status (HC, LTBI and TB). Lower panel (left to right): the associated cell type, the corresponding status in associated cell type (HC, LTBI and TB)
Fig. S8. Flow cytometry analysis of CD3-CD7+GZMB+ subsets
Table S1 Demographic characteristics of study populations
Table S2. Characteristics of scRNA-seq of the seven PBMC samples included in this study.
Table S3. The cell numbers and frequency of all subsets in PBMC from HC, LTBI and TB.
Table S4. Marker genes of PBMC major cell types identified by scRNA-seq
Table S5. Marker genes of myeloid subsets identified by scRNA-seq
Table S6. Marker genes of B cell subsets identified by scRNA-seq
Table S7. Marker genes of T cell subsets identified by scRNA-seq
Table S8. Gene oncology enrichment analysis of upregulated genes in T2 from TB