Abstract
Background
Fibroblast growth factor receptor (FGFR)1 and FGFR4 have been associated with tumorigenesis in a variety of tumour types. As a therapeutic approach, their inhibition has been attempted in different types of malignancies, including lung cancer, and was initially focused on FGFR1-amplified tumours, though with limited success.
Methods
In vitro and in vivo functional assessments of the oncogenic potential of downregulated/overexpressed genes in isogenic cell lines were performed, as well as inhibitor efficacy tests in vitro and in vivo in patient-derived xenografts (PDXs). mRNA was extracted from FFPE non-small cell lung cancer samples to determine the prognostic potential of the genes under study.
Findings
We provide in vitro and in vivo evidence showing that expression of the adhesion molecule N-cadherin is key for the oncogenic role of FGFR1/4 in non-small cell lung cancer. According to this, assessment of the expression of genes in different lung cancer patient cohorts showed that FGFR1 or FGFR4 expression alone showed no prognostic potential, and that only co-expression of FGFR1 and/or FGFR4 with N-cadherin inferred a poorer outcome. Treatment of high-FGFR1 and/or FGFR4-expressing lung cancer cell lines and patient-derived xenografts with selective FGFR inhibitors showed high efficacy, but only in models with high FGFR1/4 and N-cadherin expression.
Interpretation
Our data show that the determination of the expression of FGFR1 or FGFR4 alone is not sufficient to predict anti-FGFR therapy efficacy; complementary determination of N-cadherin expression may further optimise patient selection for this therapeutic strategy.
Keywords: FGFR1, FGFR4, N-cadherin, Predictive biomarker, FGFR inhibitors
Research in context.
Evidence before this study
FGFR1 and FGFR4 have been described as oncogenes in many types of cancer, including lung cancer. FGFR1 amplification was associated to FGFR inhibitor (FGFRi) sensitivity in preclinical studies. However, responses of selected patients to FGFR inhibition in clinical trials was poor, suggesting that FGFR amplification is not predictive of FGFRi efficacy.
Added value of this study
We provide in vitro, in vivo and clinical evidence for the context-dependant oncogenic role of both FGFR1 and FGFR4 in lung tumours, thereby expanding the body of knowledge addressing FGFR activity in lung cancer biology. Furthermore, we provide a potential predictive biomarker for high anti-FGFR therapy efficacy.
Implications of all the available evidence
These additional insights into the functions of FGFR will improve understanding of the behaviour of tumours overexpressing FGFR1/4, provide molecular criteria for the selection patients who could benefit from FGFR inhibition therapy, and thus pave the way for the design and improvement of targeted therapeutics for lung cancer patients.
Alt-text: Unlabelled box
1. Introduction
Lung cancer accounts for 27% of cancer-related deaths, representing the leading cause of cancer mortality [1] due to the late stage at which it is usually diagnosed and to the relative lack of effective systemic therapies [2]. The most prevalent lung cancer histology, non-small cell lung cancer (NSCLC), is a highly heterogeneous malignancy at the molecular level [3]. NSCLC is characterised by numerous genomic aberrations underlying the disease, some of which are druggable oncogenic drivers such as ALK translocations and EGFR mutations, whose targeting has improved patient outcomes and changed clinical practices [4], [5], [6]. However, there is a high percentage of NSCLC patients with tumours harbouring no targetable alteration who would benefit from the discovery of effective targets.
The fibroblast growth factor receptor (FGFR) family plays a role in the progression of a variety of human cancers [7], [8], [9], [10]. In lung cancer, FGFR1 amplification is detected in approximately 20% of squamous cell carcinoma cases [11,12]. FGFR1 amplification and expression have been identified as an indicator of sensitivity to FGFR inhibition in preclinical models of lung cancer [13], [14], [15], [16], [17], [18]; however, at the clinical level, FGFR inhibitors have shown limited responses in selected patients, thus highlighting the need for improved predictive biomarkers for these therapies [19]. FGFR4 expression has also been associated with poorer outcomes in several types of cancer [20], [21], [22], [23]. Regarding lung cancer, there is evidence that FGFR4 protein expression correlates with poor prognosis [24]. Despite these results suggesting an oncogenic role for the expression of both receptors in cancer, few studies have examined in depth the roles of FGFR1 and FGFR4 in lung tumorigenesis.
In the present study, we describe the molecular context-dependant role of FGFR1 and FGFR4 in lung cancer. We show that N-cadherin is essential for defining the role of both FGFRs in tumorigenesis, and we provide evidence that expression of N-cadherin is predictive of the potential efficacy of anti-FGFR therapy.
2. Methods
2.1. Cell lines
Characteristics of the cell lines used are shown in Supplementary Table S1. All cell lines except for H3122, which was kindly provided by Dr. Maina, were purchased from ATCC immediately prior to this work and were regularly tested for mycoplasma.
2.2. Transfections
All cell lines were transfected as described in [25]. TransIT-X2 Transfection Reagent (Mirus) was used to transfect the cell lines as indicated by the manufacturer. FGFR1 (RC202080) and FGFR4 (RG204230) cDNA clones were obtained from Origene in the pCMV6 plasmid (PS100001). Positive clones were isolated using G418 selection and were pooled in a monolayer. G418 was maintained in the medium to provide continuous selective pressure. For N-cadherin overexpression, N-cadherin cDNA in the pCCL-c-MNDU3c-PGK-EGFP plasmid (#38153) and the negative control PL-SIN-PGK-EGFP plasmid (#21316) were obtained from Addgene. Transfection-positive cells were selected by cell sorting based on EGFP reporter expression. For FGFR1 and FGFR4 silencing, shRNAs in the p-RS plasmid were purchased from Origene (TF320354 and TR320356, respectively). Puromycin was used to select positively transfected clones, which were then pooled in a monolayer. For N-cadherin silencing, shRNAs in the pB-RS plasmid were purchased from Origene (HC138304). Blasticidin was used to select positively transfected clones, which were then pooled in a monolayer. For shRNA silencing, two independent shRNAs were used to avoid off-target effects.
2.3. Immunoblotting
Western blots were performed as described in [26]. Protein was extracted from cell lines and PDX tumours using RIPA buffer (Sigma) supplemented with a protease inhibitor cocktail (cOmplete Mini EDTA-free, Roche) and a phosphatase inhibitor cocktail (PhosSTOP EASYpack, Roche). Protein extracts were quantified using the Bradford method and aliquoted. Electrophoresis was performed with miniProtean western blot accessories (BioRad), and proteins were transferred to a PVDF membrane (GE Healthcare) with the Trans-Blot Turbo semi-dry transfer system (BioRad). When necessary, different blots were performed in parallel with an internal reference sample and the images assembled. The following antibodies were used: FGFR1 (#9740, Cell signalling), FGFR4 (#8562, Cell signalling), pFGFR1 (06–1433, Millipore), pFGFR4 (MBS856043, MyBiosource), AKT (#9272, Cell signalling), pAKT (#9271, Cell signalling), p42/p44 (#9102, Cell signalling), p-p42/p44 (#9101, Cell signalling), STAT3 (#9139, Cell signalling), pSTAT3 (#9145, Cell signalling), α-tubulin (Sigma), EGFR (#4267, Cell signalling), pEGFR (#2234, Cell signalling), E-cadherin (#3195, Cell signalling) and N-cadherin (#3195, Cell signalling). All primary antibodies used had been previously validated in this specific assay and their specificities tested with adequate control samples. HRP-conjugated anti-rabbit and anti-mouse secondary antibodies were purchased from Cell signalling. Protein expression was assessed by chemiluminescence with a ChemiDoc Imaging System (BioRad). Western blot images with a high number of lanes were assembled from western blots run in parallel and with a common reference sample on both gels.
2.4. Foetal bovine serum (FBS) stimulation
Several cell lines were stimulated with FBS. The cells were cultured in FBS-free medium for five hours to induce basal phosphorylation levels. Cells were then stimulated with complete medium (10% FBS) for 15 min and protein extracts subsequently obtained as indicated above.
2.5. Surrogate assays
Clonogenicity, growth curves, and soft agar assays were performed as described in [23]. For clonogenic assays, cells were seeded in 10-cm cell culture plates at the appropriate clonogenic density (between 1000 and 10,000 cells/plate, depending on the cell line). Each condition was assayed in triplicate, and each assay was repeated at least three times. The medium containing selection antibiotics was renewed once a week during the assay. After a variable period of two or three weeks depending on the cell line, the plates were fixed with 0.5% glutaraldehyde for 20–30 min and cells then stained with a 1% crystal violet solution in water. After a washing step, the colonies were counted.
For growth curves, cells (~3500 per well) were seeded in 12-well culture plates in complete growth medium. Each point on the growth curve was assayed in triplicate, and at least three replicate growth curves were analysed for each experiment. For the first point on the curve (day 0), cells were fixed with a 0.5% glutaraldehyde solution 24 h after seeding. For the 0.5% FBS growth curves, cells were washed twice with PBS and then incubated with 0.5% FBS medium 24 h after seeding, before the day 0 plate was fixed. A plate was fixed every two days and cells stored in PBS at 4 °C until samples for the last point on the curve were fixed. Cells were then stained with a 1% crystal violet solution and washed. Crystal violet was dissolved in a 20% acetic acid/water solution, and the absorbence at 595 nm was measured in a VICTOR plate reader. The intensity of the absorbence was correlated to the number of cells, and values normalised to the day 0 absorbence. The normalised absorbence was then plotted versus time.
In the case of treatment growth curves, the medium was replaced with 0.5 µM or 1 µM BGJ398 or AZD4547-containing complete medium 24 h post-seeding, and the day 0 plate was fixed. A plate was then fixed every 24 h until a treatment period of 72 h was reached.
For soft agar assays, cells (100,000 per well) were seeded in 6-well plates in 0.35% agarose/growth medium on top of a base of 0.7% agarose/medium. Each condition was assayed in triplicate, and each assay was repeated at least three times. The day after plating, 3 mL of complete growth medium was added to each well, with the medium replaced twice per week until the end of the experiment. After sufficient colony growth, several images of each well were obtained with an Olympus camera (#U-CMAD3) fitted to an Olympus microscope (#IX2-SLP), and the size and number of colonies were quantified.
2.6. Immuno-colocalisation
Immunofluorescence assays were performed as described in [18] and [27]. Cells were fixed with 4% paraformaldehyde for 15 min, permeabilised with 0.1% Triton X-100 in PBS for 5 min, and then blocked with 1% BSA in 0.1% Triton X-100 in PBS for 1 h at room temperature. Cells were then incubated with mouse anti-N-cadherin (#MA1-159, Thermo Fisher) and rabbit anti-FGFR1 (#9740, CST) or anti-FGFR4 (#8562, CST) antibodies at a 1:100 dilution in 1% BSA and 0.1% Triton X-100 in PBS for 3 h at room temperature. The cells were then labelled with Alexa Fluor® 488 Goat Anti-Rabbit IgG (#R37116, Thermo Fisher) and Alexa Fluor® 555 Donkey Anti-Mouse IgG secondary antibodies (#A-31570, Thermo Fisher) at a dilution of 1:250 for 1 h at room temperature. Images of 15–20 cells per condition were obtained using a SP5-WLL confocal microscope. All primary antibodies used had been previously validated in this specific assay and their specificities tested.
2.7. Co-Immunoprecipitation
Co-immunoprecipitation was performed using the EZ View Red Protein G Affinity Gel (#E3403, Sigma). Protein extraction was performed using an extraction buffer prepared with 50 mM HEPES, 150 mM NaCl and 1% n-octylglucoside and supplemented with a protease inhibitor cocktail (cOmplete Mini EDTA-free, Roche) and a phosphatase inhibitor cocktail (PhosSTOP EASYpack, Roche). Protein extracts were quantified by the Bradford method using 2 mg aliquots of protein. The protein samples were precleared and incubated with antibody-conjugated beads. An N-cadherin (#3195, Cell signalling) antibody was used at a 1:100 dilution for the immunoprecipitation, and an equal amount of anti-IgG isotype control antibody (#3900, Cell signalling) was used as a negative control. Immunocomplexes were denatured by boiling in Laemmli buffer, and western blotting was performed to confirm the immunoprecipitation and assess the co-immunoprecipitation of FGFR1 and FGFR4.
2.8. Dovitinib sensitivity of cancer cell lines in a public database
Information concerning sensitivity to the FGFR inhibitor dovitinib, FGFR1, FGFR4 and N-cadherin mRNA expression, and FGFR1 amplification for a panel of NSCLC cell lines was obtained from the Cancer Cell Line encyclopaedia (CCLE) database (https://portals.broadinstitute.org/ccle/home).
2.9. In vivo experiments
In vivo experiments were performed as described in [18]. For cell line xenografts, cell lines were trypsinised, counted and diluted in PBS, following which the cell suspension was mixed with Matrigel (1:1) and 150 µL (containing 2×106 cells) of the mixture was injected into both flanks of female, 6-week-old athymic nude mice (nu+/nu+). Based on previous experiments in the laboratory and following review by the Animal Protection committee, a sufficient number of animals were included in each group to reach statistical significance. Tumours were measured weekly after implantation, and mice were sacrificed when the tumour volume exceeded 1000 mm3. Tumours were then harvested and stored.
We also used a collection of NSCLC PDX models established by our group at Institute of Biomedicine in Seville (IBIS). Early stage resected lung tumours from patients from HUVR (Hospital Universitario Virgen del Rocío, Sevilla, Spain) were obtained through the hospital biobank, inoculated subcutaneously and expanded in successive groups of nude mice. For this study, PDXs were selected depending on their histology, genetic background and FGFR1/4 and N-cadherin expression. Six models were used: TP43, TP60, TP91 (adenocarcinomas) and TP13, TP96 and TP114 (squamous cell carcinomas). Mutational profiles were determined by the OncoNIM Seq Lung Panel. The human tissue samples had been stored at the HUVR Biobank following written informed consent by all patients.
For the PDX treatments, previously amplified patient-derived tumours were cut into 100-mm3 pieces and subcutaneously inserted into one flank of female, 6-week-old athymic nude mice. Tumours were measured twice per week after implantation. As described above, a sufficient number of animals was included in each group to reach statistical significance. When tumour volumes had reached 150 mm3, mice were randomised into 2 groups of 3–5 mice each for the control and AZD4547 treatment groups, with tumour sizes for the two groups being of similar mean and standard deviation. Mice whose tumours presented no growth at this point were sacrificed. AZD4547 was administered by oral gavage 5 times per week at a concentration of 7.5 mg AZD4547/kg/day, in a 5-week-long treatment scheme unless the experiment had to be prematurely stopped due to excessive tumour growth. Mice were weighed once per week (to control for treatment toxicity) and then sacrificed at the end of treatment, following which tumour samples were collected and stored. These experiments were blinded, with one person treating the animals and a different person measuring the tumours and processing the data.
2.10. Ethics statement/Study approval
Regarding the collection, storage and use of human samples, written informed consent was provided by all donor patients. The project was approved by the Research Ethics Committee of the Virgen del Rocío University Hospital, Seville, Spain (Approval ID: 2012PI/241).
Procedures involving animals were approved by the Consejería de Agricultura of the Junta de Andalucia (Approval ref: SSA/SI/MD/pdm) and by Animal Protection of the Comunidad Autónoma de Madrid (Approval ID: PROEX 084/15 and PROEX134/16). All experiments were performed in compliance with animal use guidelines.
2.11. RNA extraction and analysis
RNA isolation and qPCR were performed as described in [18]. RNA was extracted from cell lines using TRIzol Reagent (Life Technologies) and RNA samples were reverse transcribed with the TaqMan Reverse Transcription Kit (Life Technologies). Gene expression was analysed using TaqMan probes from Life Technologies: Hs00917379_m1 FAM (FGFR1), Hs01106908_m1 FAM (FGFR4), Hs01023894_m1 FAM (E-cadherin), Hs00983056_m1 FAM (N-cadherin), Hs99999905_m1 FAM (GAPDH), and Hs99999907_m1 FAM (B2M). The last two probes targeted reference genes that were used to normalise the expression data: GAPDH for cell culture samples and B2M for patient samples.
2.12. Analyses of lung cancer patient databases
For the analysis of FGFR1, FGFR4, and N-cadherin expression, we used GSE19188, GSE18842, GSE19804, GSE33532, GSE10072, GSE31552, TCGA, GSE2109, GSE3141, GSE14018, GSE63074 and GSE43580 lung cancer] databases, which are available through the R2 genomics analysis and visualisation platform (http://r2.amc.nl/). Heatmaps were drawn using the Multiexperiment Viewer application software and plotting of the z-scores of gene expression data. All statistical analyses were performed using GraphPad Prism4, or directly from the R2 Genomics platform”.
2.13. Clinical data
The present study involved a discovery cohort of 109 subjects diagnosed with NSCLC (Supplementary Table E2) from the Virgen del Rocio University Hospital (Seville, Spain) who had undergone surgical resection. Tumour samples were sent to the pathology laboratory for diagnosis and were prepared for storage by formalin fixation and paraffin embedding. Inclusion criteria were: [1] confirmed NSCLC diagnosis; [2] access to patient clinical information, including age, gender, smoking status, the TNM Classification of Malignant Tumours (TNM) stage, diagnosis date, histologic subtype, date of relapse, date of the last revision and status at that time; [3] availability of tumour tissue obtained by surgical resection for immunohistochemistry. In addition, we obtained mRNA expression data from a publicly available cohort from the TCGA. These results are partly or fully based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. For the tumour marker prognostic study, the REMARK [28] reporting guidelines were followed.
2.14. Statistics
In vitro data are presented as the mean ± standard deviation to estimate variation within each group. Statistical analysis was performed with the SPSS statistical package (v19, IBM). The in vitro and in vivo experiments were analysed using an unpaired two-tailed non-parametric Mann-Whitney U test. p-values less than 0.05 were considered significant. For the prognosis analyses, the Kaplan-Meier method was used for survival analyses of the clinical data and cell line xenograft experiments. Overall survival (OS) was defined as the length of time from the date of starting treatment to the date of death or last follow up. Progression-free survival (PFS) was defined as the length of time from the date of treatment initiation to the date of progression/death or last follow up. A Log Rank test was used to analyse differences in survival amongst groups. The Cox proportional hazards model was used to obtain hazard ratio values.
3. Results
3.1. FGFR1 and FGFR4 expression can exert pro- or anti-tumorigenic effects in NSCLC cell lines
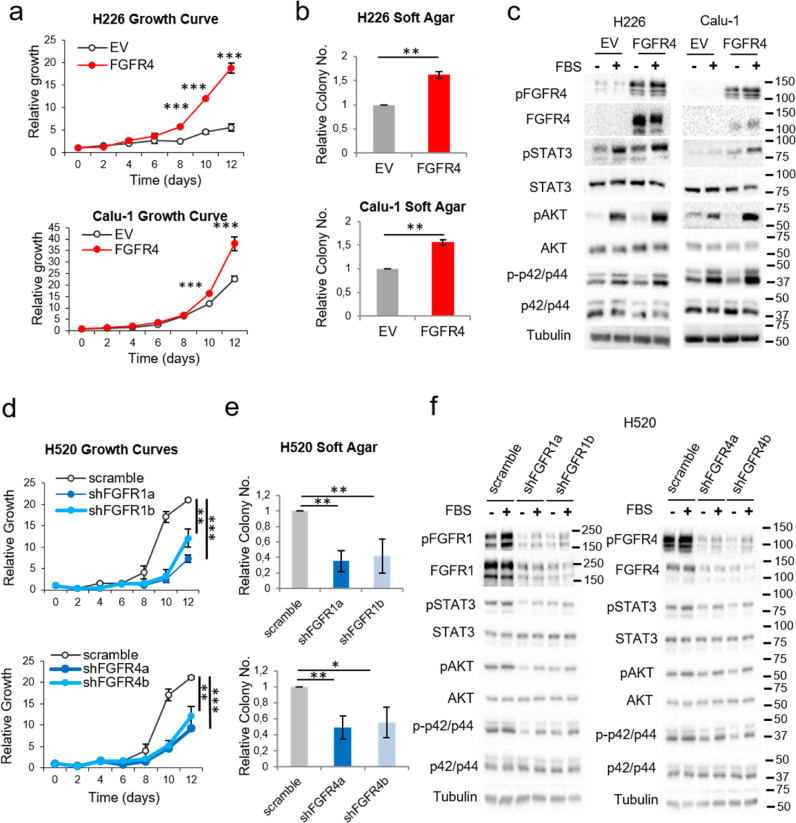
To study the role of FGFR1 and FGFR4, we downregulated/overexpressed either receptor in NSCLC cell lines (Supplementary Figure S1a-b and Supplementary Table S1). FGFR4 overexpression in two squamous cell carcinoma lines (H226 and Calu-1) which show different driver alterations, led to increased cell growth, clonogenicity and soft agar colony formation (Figs. 1a-b and Supplementary Figures S1c-d) compared to empty vector controls. As expected, FGFR4 overexpression led to its auto-activation in these cell lines. Furthermore, upon FBS stimulation, the STAT3, p42/p44 and AKT pathways were overactivated in the FGFR4-overexpressing cell lines (Fig. 1c). Since FGFR1 protein expression was high in all the squamous cell carcinoma lines examined, we did not attempt to overexpress this gene.
Fig. 1.
Effects of FGFR1 and FGFR4 on lung squamous carcinoma cell lines. See also Supplementary Figure S1. Growth curves in 10% FBS (a) and soft agar assays (b) of FGFR4-overexpressing lung squamous carcinoma cell lines. (c) Western blot analysis of the activation of FGFR-related signalling pathways in FGFR4-overexpressing lung squamous carcinoma cell lines compared to empty-vector-expressing cell lines after stimulation with FBS. Growth curves in 0.5% FBS (d) and soft agar assays (e) of FGFR1- and FGFR4-silenced H520 cells (lung squamous cell carcinoma). (f) Western blot analysis of the activation of FGFR-related signalling pathways in FGFR1- and FGFR4-silenced H520 cells. All experiments were reproduced a minimum of three times in the laboratory, and three technical replicates were obtained for each experiment. For growth curves and western blots, a representative figure/image is shown. On the growth curves, the means and standard deviations of the technical replicates are shown. In the soft agar assays, all values were normalised to the empty vector control, and the mean and standard deviation of all the normalised replicates are presented. Silencing of either gene was performed using two different shRNAs, referred to as “a” and “b”. p-values were obtained with the two-sided Mann-Whitney U test and are indicated by asterisks (* p<0.05; ** p<0.01; *** p<0.001). ADC = Adenocarcinoma, SCC = Squamous cell carcinoma, I = Immortalised, KRAS = KRAS-mutated, EGFR = EGFR-mutated, ALK = ALK translocation bearer, TN = “Triple negative” (referring to the absence of alterations in KRAS, EGFR and ALK), EV = empty vector control, FGFR1 = FGFR1-overexpressing, FGFR4 = FGFR4-overexpressing, scramble = scrambled shRNA control, shFGFR1 = FGFR1 shRNA, shFGFR4 = FGFR4 shRNA, FBS = foetal bovine serum. Western blot molecular weight references are indicated to the right of the images.
To assess the effect of FGFR1 on the oncogenic properties of the cells and to further study the role of FGFR4 in lung squamous cell carcinoma lines, either FGFR1 or FGFR4 were silenced in H520 cells (Supplementary Figure S1a). This led to reduced oncogenic properties (Figs. 1d-e and Supplementary Figures S1e-f), reduced phosphorylated FGFR1 and FGFR4, and reduced oncogenic signalling (Fig. 1f).
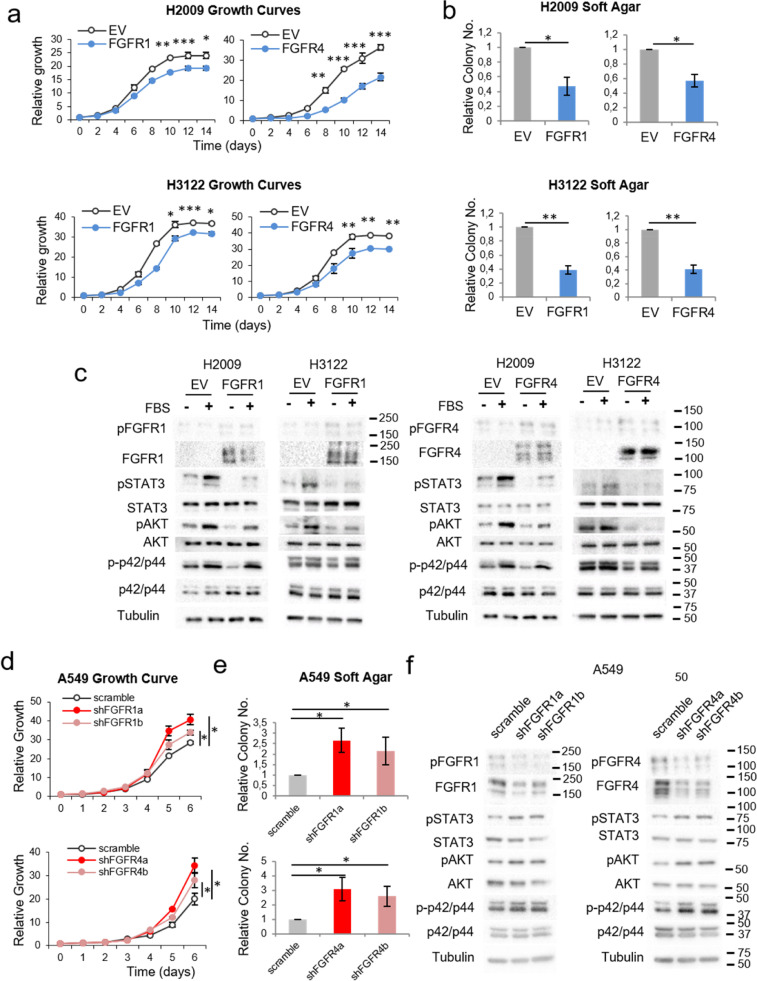
On the other hand, when we overexpressed FGFR1 or FGFR4 in non-squamous lung cell lines with different genetic backgrounds (H2009, H3122 and H460, Supplementary Table S1), this led to reduced oncogenicity (Figs. 2a-b and Supplementary Figures S2a-d). In these cell lines, activation of the STAT3 and AKT pathways was reduced after serum stimulation following FGFR1/4 overexpression. In contrast to the tested squamous cell carcinoma lines, no variations in FGFR1 or FGFR4 activation levels were observed after FGFR1 or FGFR4 overexpression (Fig. 2c and Supplementary Figure S2e).
Fig. 2.
Effects of FGFR1 and FGFR4 on non-squamous lung cell lines. See also Supplementary Figure S2. Growth curves in 10% FBS (a) and soft agar assays (b) of the FGFR1- or FGFR4-overexpressing lung adenocarcinoma cell lines H2009 and H3122. (c) Western blot analysis of the activation of FGFR-related signalling pathways in FGFR1-overexpressing H2009 and H3122 cells compared to empty-vector-expressing cells. Growth curves in 10% FBS (d) and soft agar assays (e) of FGFR1- and FGFR4-silenced A549 cells (adenocarcinoma cell line). (f) Western blot analysis of the activation of FGFR-related signalling pathways in FGFR1- and FGFR4-silenced A549 cells. All experiments were reproduced a minimum of three times in the laboratory, and three technical replicates were obtained for each experiment. For growth curves and western blots, a representative figure/image is shown. On the growth curves, the means and standard deviations of the technical replicates are shown. In the soft agar assays, all values were normalised to the empty vector control, and the mean and standard deviation of all the normalised replicates are presented. For western blots, cells were serum-starved for five hours prior to protein extraction. For the serum-stimulated conditions, serum-starved cells were incubated in serum-containing complete medium for 15 min before protein extraction. Silencing of either gene was performed using two different shRNAs, referred to as “a” and “b”. p-values were obtained with the two-sided Mann-Whitney U test and are indicated by asterisks (* p<0.05; ** p<0.01; *** p<0.001). EV = empty vector control, FGFR1 = FGFR1-overexpressing, FGFR4 = FGFR4-overexpressing, scramble = scrambled shRNA control, shFGFR1 = FGFR1 shRNA, shFGFR4 = FGFR4 shRNA, FBS = foetal bovine serum. Western blot molecular weight references are indicated to the right of the images.
Further to the above, silencing of either FGFR in non-squamous A549 cells led to increased oncogenicity (Figs. 2d-e and Supplementary Figure S2f). Despite relatively high levels of FGFR1 and FGFR4 protein expression in this cell line, activated FGFR1 and FGFR4 were barely detected. However, slightly increased activation of STAT3, AKT and p42/p44 signalling was observed after FGFR4 silencing in this cell line, while STAT3 signalling was upregulated, and a similar trend was observed for AKT and p42/p44 after FGFR1 silencing (Fig. 2f and Supplementary Figure S2g).
3.2. N-cadherin expression induces a pro-oncogenic role for FGFR1/4, independently of histology
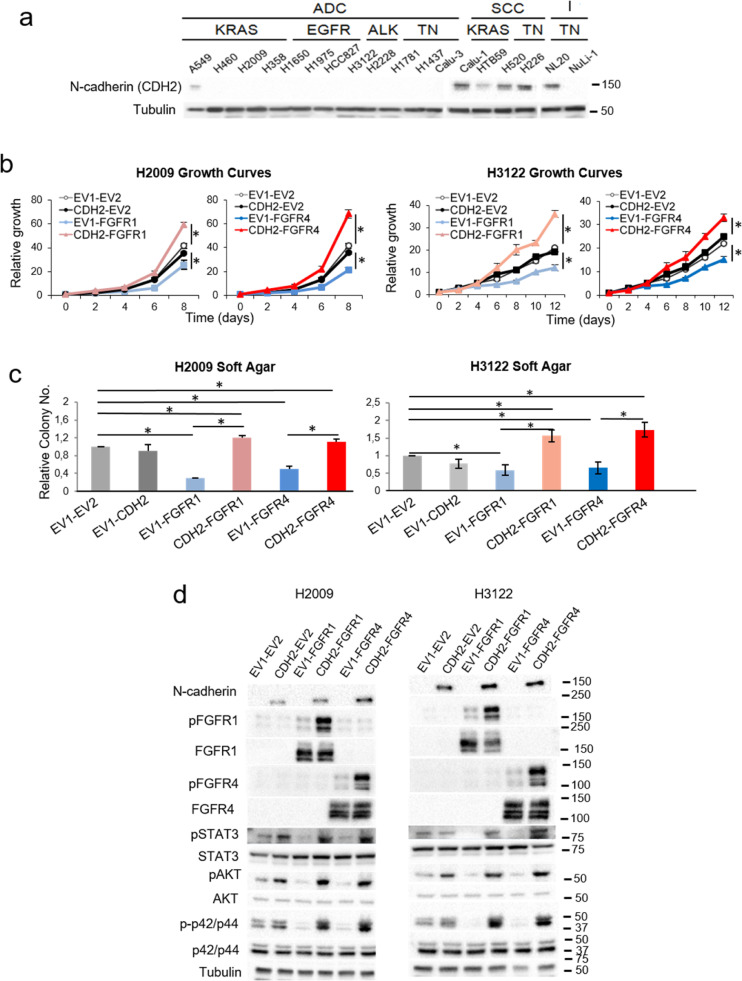
Evidence in the literature suggests that FGFRs may interact with N-cadherin, which results in increased FGFR signalling and pro-oncogenic effects, suggesting that this adhesion molecule may be triggering the activation and oncogenic potential of these receptors [29,30]. In line with these results, we found that N-cadherin expression was primarily detected in the squamous cell carcinoma lines where expression of either FGFR was pro-oncogenic (Fig. 3a).
Fig. 3.
Effects of N-cadherin on the pro-oncogenic role of FGFR1 and FGFR4. See also Supplementary Figures S3 and S4. (a) Western blots of N-cadherin and E-cadherin protein expression in our lung cell line panel. To assess the expression of these proteins in the 18 cell lines, different blots were performed in parallel with an internal reference sample and the assembled images are shown. 10% FBS growth curves (b) and soft agar assays (c) of H2009 and H3122 cells overexpressing N-cadherin and either FGFR1 or FGFR4. (d) Western blot analysis of the activation of FGFR-related signalling pathways in these cell lines. All experiments were reproduced a minimum of three times in the laboratory and three technical replicates were obtained for each experiment. For growth curves and western blots, a representative figure/image is shown. On the growth curves, the means and standard deviations of the technical replicates are shown. In the soft agar assays, all values were normalised to the empty vector control, and the mean and standard deviation of all the normalised replicates are presented. p-values were obtained with the two-sided Mann-Whitney U test and are indicated by asterisks (* p<0.05; ** p<0.01; *** p<0.001) non-SCC = non-Squamous, SCC = Squamous cell carcinoma, I = Immortalised, KRAS = KRAS-mutated, EGFR = EGFR-mutated, ALK = ALK translocation bearer, TN = “Triple negative” (referring to the absence of alterations in KRAS, EGFR and ALK), EV1 = empty vector 1, EV2 = empty vector 2, FGFR1 = FGFR1-overexpressing, FGFR4 = FGFR4-overexpressing, CDH2 = N-cadherin-overexpressing. Western blot molecular weight references are indicated to the right of the images.
As a proof of concept, we co-overexpressed N-cadherin and either FGFR in two lung non-squamous cell lines (H2009 and H3122) with no endogenous N-cadherin expression and studied the resultant effects on signalling pathways. Co-overexpression of either FGFR1 or FGFR4 with N-cadherin not only reversed the tumour suppressor effects of the overexpression of either FGFR alone, but also provoked increased oncogenic potential (Figs. 3b-c and Supplementary Figures S3a-b). An assessment of oncogenic signalling in these cell lines showed reduced pSTAT3, pAKT and p-p42/p44 levels in the absence of N-cadherin, whereas the opposite effects were observed when N-cadherin was co-overexpressed with the FGFRs. Furthermore, we found that FGFR1 or FGFR4 activation was increased after overexpression of either FGFR alone in the cell lines with induced N-cadherin expression (Fig. 3d).
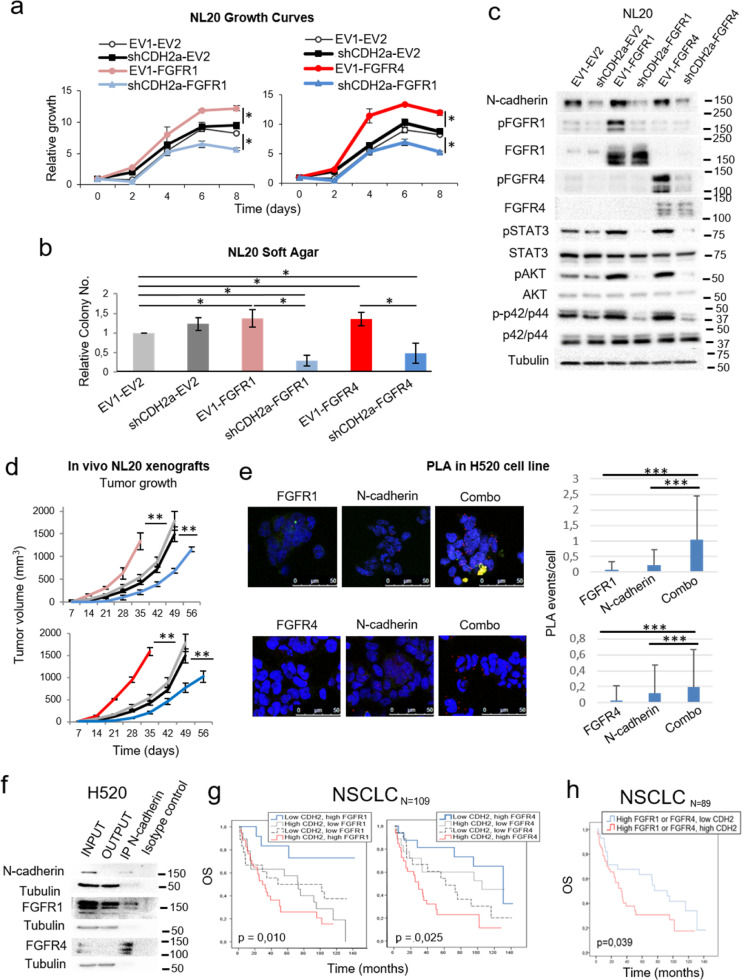
We also overexpressed either FGFR1 or FGFR4 and silenced N-cadherin expression in the immortalised lung epithelial cell line NL20, which exhibits high endogenous levels of N-cadherin (Fig. 3a). The overexpression of either FGFR increased the tumoral potential and oncogenic signalling of the cells under conditions of endogenous N-cadherin expression in vitro and in vivo in xenografts of these cells in immunodeprived mice. However, a reduction in N-cadherin protein levels reverted the tumour suppressor function of FGFR1 and FGFR4 (Fig. 4a-d and Supplementary Figure S3c-e). Supporting these data, analysis of four independent cohorts of human lung tumours with available mRNA data showed that FGFR1/4 expression positively correlated with N-cadherin mRNA levels (Supplementary Figure S4), suggesting that co-expression of these genes may represent an oncogenic advantage for these tumours.
Fig. 4.
Effects of N-cadherin on the pro-oncogenic role of FGFR1 and FGFR4 and the interaction of N-cadherin with FGFR1 and FGFR4. See also Supplementary Figure S4. (a) 0.5% FBS growth curves for FGFR1-overexpressing and N-cadherin-silenced (left) or FGFR4-overexpressing and N-cadherin-silenced (right) NL20 cells. (b) Soft agar assays of FGFR-overexpressing and N-cadherin-silenced NL20 cells. (c) Western blot analysis of the activation of FGFR-related signalling pathways in these cell lines. (d) Xenograft tumour volumes of the FGFR1, FGFR4 and N-cadherin interaction models in the immortalised NL20 cell line. (e) Proximity ligation assays (PLA) to assess the physical interaction of N-cadherin with FGFR1 (upper panel) or FGFR4 (lower panel). The interactions detected were quantified and normalised by cell number for each condition. As controls to distinguish signal from noise, interactions were quantified in the single antibody conditions (labelled as “N-cadherin”, “FGFR1” and “FGFR4”). To assess the interaction between the two proteins, both antibodies were used, either N-cadherin + FGFR1 (upper panel) or N-cadherin + FGFR4 (lower panel), in the condition labelled as “combo”. Representative images and quantifications are shown. (f) Co-immunoprecipitation of N-cadherin with FGFR1 and with FGFR4 in the H520 cell line. (g) Kaplan-Meier curves of overall survival (OS) for the entire NSCLC patient cohort (N = 109). Patients were grouped based on FGFR1 and N-cadherin expression levels or on FGFR4 and N-cadherin expression levels. (h) OS curve of patients in the cohort with high expression of FGFR1 and/or FGFR4 stratified by N-cadherin expression levels. In each analysis, for the FGFR1 and N-cadherin genes, the cut-off point was the median mRNA expression value for that variable. For FGFR4, the cut-off point was the first-quartile mRNA expression value in the TCGA adenocarcinoma cohort. The Kaplan-Meier method was used for survival analyses of the clinical data and cell line xenograft experiments, with a Cox proportional hazards model used to adjust for explanatory variables. A log Rank analysis was used to analyse differences in survival between groups. To obtain the hazard ratio values, the Cox proportional hazards model was used. All in vitro experiments were reproduced a minimum of three times in the laboratory, and three technical replicates were obtained for each experiment. For growth curves and western blots, a representative figure/image is shown. On the growth curves, the means and standard deviations of the technical replicates are shown. In the soft agar assays, all values were normalised to the empty vector control, and the mean and standard deviation of all the normalised replicates are presented. N-cadherin silencing was performed using two different shRNAs. Results generated with the alternative shRNA are shown in Supplementary Figure S3 c-e. p-values were obtained with the two-sided Mann-Whitney U test and are indicated by asterisks (* p<0.05; ** p<0.01; *** p<0.001). EV1 = empty vector 1, EV2 = empty vector 2, FGFR1 = FGFR1-overexpressing, FGFR4 = FGFR4-overexpressing, CDH2 = N-cadherin-overexpressing, scramble = scrambled shRNA control, shCDH2 = silenced with N-cadherin shRNA. Western blot molecular weight references are indicated to the right of the images.
3.3. N-Cadherin physically interacts with FGFR1 and FGFR4
N-Cadherin is known to physically interact with FGFR1 in breast cancer cell lines [30] and with FGFR4 in pancreatic cancer [31]. We conclude from the following pieces of evidence that this interaction also takes place in lung cell lines: the co-localisation and physical interaction of both FGFRs with N-cadherin was observed by proximity ligation assays (Fig. 4e) and co-immunoprecipitation experiments (Fig. 4f), respectively, in the H520 line which displays endogenous expression of these three genes (Supplementary Figure S1a and Fig. 3a).
3.4. FGFR1/4 and N-cadherin co-expression is a prognostic determinant in NSCLC
mRNA expression of FGFR1/4 and N-cadherin was determined in formalin-fixed, paraffin-embedded tumour samples from a cohort of 109 early-stage NSCLC patients (Supplementary Table S2) and correlated with patient outcome. Patients whose tumours exhibited high expression of FGFR1 and N-cadherin genes had the poorest prognosis, while the group with high FGFR1 and low N-cadherin expression had the best prognosis. Similar results were obtained for FGFR4 (Fig. 4g). Consistently, when both FGFR1 and FGFR4 expression were taken into account (n = 89), patients with high FGFR1/4 and low N-cadherin expression had increased overall survival (Hazard Ratio of 1.89 [1.02–3.49], p = 0.039) compared to patients with elevated expression of both genes (Fig. 4h).
3.5. Transcriptome analysis of human NSCLC tumours associates co-expression of FGFR1/4 and N-cadherin to a neuroendocrine transcriptional program
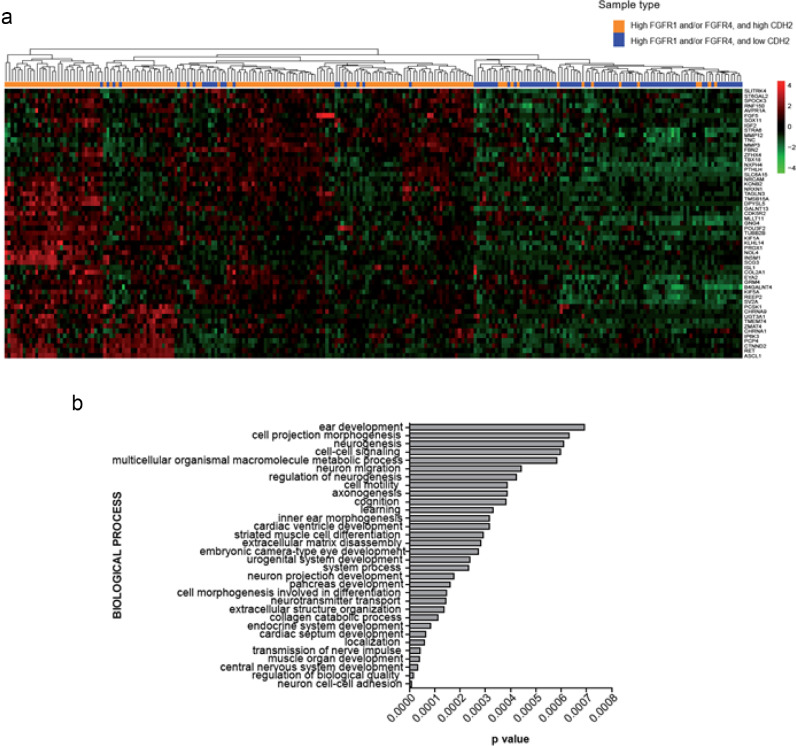
To explore the molecular mechanisms underlying the oncogenicity of FGFR1/4 in the context of high N-cadherin expression, we used the TCGA lung cancer (adenocarcinomas and squamous carcinomas) patient dataset for which gene expression data are available. We searched for those genes differentially expressed in FGFR1/4high- Cadherin-2(CDH2) high (n = 145) versus FGFR1/4high-CDH2low (n = 94) tumours. As an additional filter, we discarded those genes whose expression was dependant on a CDH2 high/low status regardless of FGFR1 and/or FGFR4 expression. This analysis unveiled a gene signature consisting of 55 upregulated genes (logFC>1, B>0) in tumours with high FGFR1/4 and N-cadherin expression (Fig. 5a and Supplementary Table S3). The gene signature was enriched in genes representing several processes related to nervous system functions (Fig. 5b). Furthermore, potential upstream transcription factors of the signature were identified by Ingenuity Pathway Analysis (IPA) using a conservative approach (p < 0.01 [Fisher's Exact test]) (Supplementary Table S4). These included master regulators of neuroendocrine differentiation, like ASCL1, NEUROD1 and POU2F3, and genes involved in lineage plasticity, such as SOX2, were identified as potential regulators of the FGFR1/4high-CDH2high gene signature. Notably, the FGFR1/4high-CDH2high gene signature obtained from patient data was significantly enriched in human lung cancer cell lines from the Cancer Cell Line encyclopaedia (CCLE) stratified by FGFR1/4 and N-cadherin expression levels (FGFR1/4high-CDH2high vs FGFR1/4high-CDH2low), suggesting that lung cancer cell lines are a suitable model where to test the functional implications of CDH2 in the context of FGFR1/4 expression (Supplementary Figure S4b-c).
Fig. 5.
RNAseq analysis of TCGA lung adenocarcinoma and squamous cell carcinoma datasets. (a) Differential gene expression analysis in FGFR1/4high-CDH2high (n = 145) versus FGFR1/4high-CDH2low (n = 94) patients. The gene dataset was filtered by discarding genes whose expression was dependant on CDH2 high/low status irrespective of FGFR1 and/or FGFR4 expression (FGFR1/4low-CDH2high patients, n = 21). Parameters were set up as logFC>1, B>0. (b) Query of defined gene expression signature against Gene Ontology. The results shown here are based in whole or in part upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
3.6. N-cadherin expression is predictive of FGFR inhibition efficacy in vitro
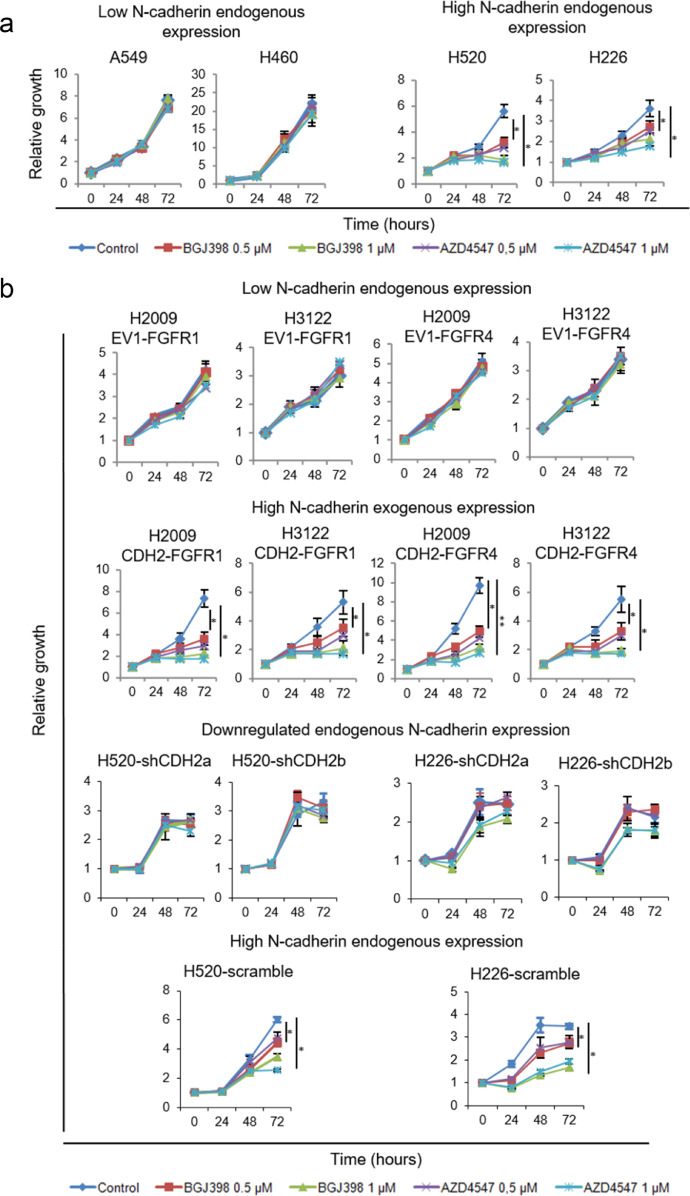
Our results suggested that in high FGFR1/FGFR4-expressing tumours, FGFR inhibition would be effective only when N-cadherin is highly expressed. To test this hypothesis, we assessed the effect of two selective FGFR inhibitors (BGJ398 and AZD4547) on a set of cell lines highly expressing these FGFRs, with variable endogenous (Fig. 6a) or exogenously downregulated/overexpressed (Fig. 6b) N-cadherin levels (Supplementary Figures S1a and Figs. 1–3). After treatment with FGFR inhibitors we observed reduced proliferation only in those cell lines where N-cadherin was highly co-expressed with FGFR1 and/or FGFR4 (Fig. 6).
Fig. 6.
Predictive potential of N-cadherin expression for anti-FGFR therapy in vitro. See also Supplementary Figure S5. Treatment for 72 h with AZD4547 or BGJ398 at a concentration of 0.5 or 1 µM was applied to cells with high endogenous expression of FGFR1 and/or FGFR4, with high or low endogenous expression of N-cadherin (a) and to cells either exogenously expressing FGFR1 or FGFR4, alone or in combination with N-cadherin, or to cells with high endogenous expression of the three genes with N-cadherin downregulation (b). All experiments were reproduced a minimum of three times in the laboratory. For growth curves, a representative figure is shown and the mean and standard deviation for the technical replicates are indicated. N-cadherin silencing was performed using two different shRNAs to avoid off-target effects. p-values were obtained with the two-sided Mann-Whitney U test and are indicated by asterisks (* p<0.05; ** p<0.01; *** p<0.001). EV1 = empty vector 1, EV2 = empty vector 2, FGFR1 = FGFR1-overexpressing, FGFR4 = FGFR4-overexpressing, CDH2 = N-cadherin-overexpressing, scramble = scrambled shRNA control, shCDH2 = silenced with N-cadherin shRNA.
As an external validation of our results, we analysed data from CCLE with an extensive panel of lung cancer cell lines treated with the FGFR1 inhibitor dovitinib (n = 92). This database includes data on cell line sensitivity to dovitinib, FGFR1 and N-cadherin mRNA expression, and FGFR1 amplification. We divided the cell lines into groups according to N-cadherin and FGFR1 mRNA expression levels by setting the median expression of either gene as the cut-off value to differentiate between low and high expressing cell lines. We found that sensitivity to dovitinib was only increased in those cell lines with high expression of both FGFR1 and N-cadherin genes (Supplementary Figure S5a). Of note, among nine cell lines with FGFR1 amplification, three out of three with high N-cadherin and FGFR1 levels were among the most sensitive to dovitinib, while five out of six cell lines with low N-cadherin and/or FGFR1 expression levels showed limited dovitinib sensitivity (Supplementary Figure S5b). When applied to our cell lines, we found FGFR inhibition sensitivity in FGFR-amplified H520 cells and in non-FGFR1-amplified H226 cells (Fig. 6b). The remaining cell lines tested did not harbour FGFR1 amplification. Further to this, as an additional external validation of our results, we analysed data from GDSC and performed experiments analogous to those whose results are presented in Figure S5a, with sensitivity data for the FGFR inhibitors (FGFRis) AZD4547 and PD173074 (Figures S5c-d). We found trends in common with our results, with cell lines with high FGFR1/FGFR4 and CDH2 expression showing the highest sensitivity to either inhibitor. Remarkably, in agreement with our results, in cell lines with high FGFR1 expression, those with high CDH2 expression showed a significantly higher sensitivity to AZD4547 (p = 0.004, Figure S5c). Taken together, these results suggest that FGFR1 and N-cadherin expression have a higher predictive potential than FGFR1 amplification for determining FGFR inhibition efficacy.
3.7. FGFR inhibition is highly effective in N-cadherin plus FGFR1 and/or FGFR4 co-expressing patient-derived xenografts (PDXs)
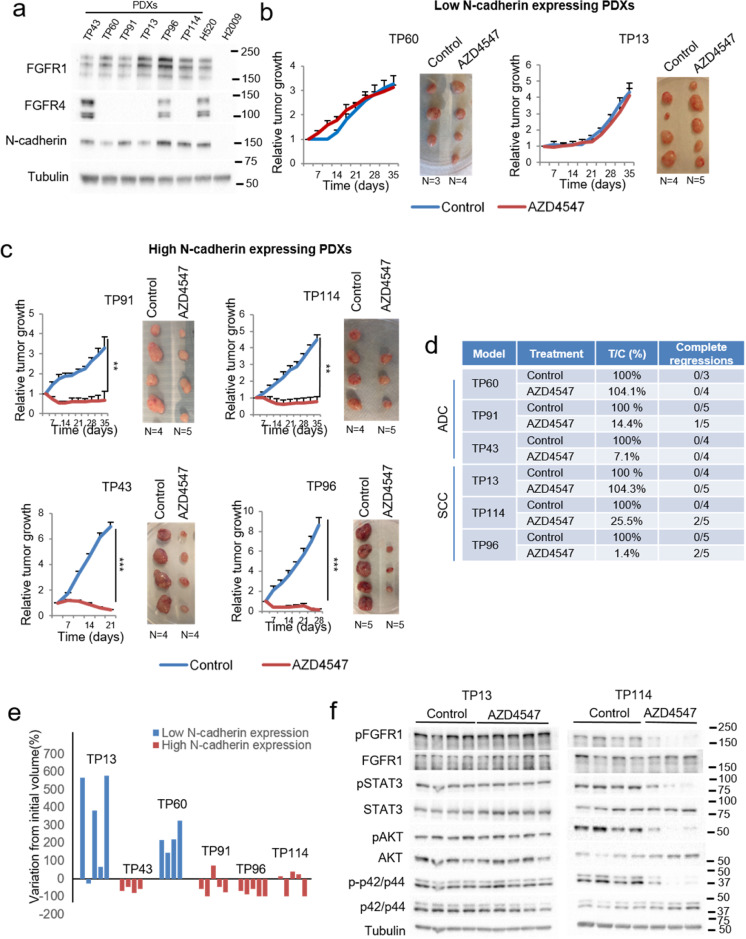
We selected several lung cancer PDX models with high FGFR1 expression and differential levels of N-cadherin: one adenocarcinoma and one squamous cell carcinoma PDX with high FGFR1 expression and low N-cadherin expression (TP60 and TP13, respectively), and one adenocarcinoma and one squamous cell carcinoma PDX with high expression of both genes (TP91 and TP114, respectively). In addition, we selected one adenocarcinoma and one squamous cell carcinoma PDX model with high expression of FGFR1, FGFR4 and N-cadherin (TP43 and TP96, respectively) (Fig. 7a and Supplementary Figure S6a). Of note, none of these models harboured FGFR1 amplification.
Fig. 7.
FGFR efficacy in N-cadherin, FGFR1 and/or FGFR4 co-expressing patient-derived xenografts (PDXs). See also Supplementary Figure S6. (a) Western blot showing FGFR1, FGFR4 and N-cadherin protein expression in five different lung PDXs. AZD4547 treatment of low (b) and high (c) N-cadherin-expressing PDXs. (d) Results of the PDX treatments in terms of tumour versus control volume (T/C), and complete regressions. T/C values are expressed as percentages. (e) Graph showing the median variation in tumour volume from the initial volume, for every model, calculated as the increase or decrease in volume and expressed as a percentage. (f) Western blot showing the effects of AZD4547 treatment on FGFR-related signalling pathways in one low-N-cadherin-expressing (TP13) and one high-N-cadherin-expressing (TP114) adenocarcinoma PDX. p-values were obtained with the two-sided Mann-Whitney U test and are indicated by asterisks (* p<0.05; ** p<0.01; *** p<0.001). Western blot molecular weight references are indicated to the right of the images.
We carried out in vivo experiments in which these PDXs were treated with AZD4547 inhibitor. No toxicity derived from the treatment was observed (Supplementary Figure S6b). This inhibitor had no effect on tumour growth in the PDX models with low N-cadherin expression (TP13 and TP60) (Fig. 7b). In contrast, in the models with higher N-cadherin expression (TP91 and TP114), AZD4547 treatment resulted in decreased tumour growth compared to the untreated tumours (tumour versus control volume (T/C)) of 14.4% and 25.5%, respectively). Indeed, AZD4547 treatment induced tumour shrinkage in these two models, and achieved 1/5 and 2/5 complete tumour regressions, respectively. Similar results were obtained in the TP43 model, with a median T/C value of 7.1% in the treated arm. In the TP96 model, the observed effects were more pronounced, including a dramatically reduced tumour volume compared to the untreated group (T/C of 1.4%), a dramatic reduction in tumour volume from the initial volume, and 2/5 complete tumour regressions (Fig. 7b-e). When we analysed the effects of AZD4547 treatment on oncogenic signalling pathways in these models, we found inhibition of the pathways specifically in those tumours that were responsive to therapy (Fig. 7f and Supplementary Figure S6c).
4. Discussion
We have shown here that FGFR1 and FGFR4 have molecular context-dependant roles in lung tumorigenesis, respectively suppressing or promoting oncogenic behaviour in vitro and in vivo depending on the absence or presence of N-cadherin expression. We have also provided evidence that the determination of FGFR1/4 levels alone is not predictive of a tumour's response to FGFR inhibitors. Complementary determination of N-cadherin levels may be necessary to select tumours against which anti-FGFR therapy may show efficacy, thus revealing the predictive potential of N-cadherin in this setting.
FGFR1 aberrations, particularly amplification, have been associated with NSCLC, especially in squamous cell carcinoma [11,12,32,33]. Recently, a potential oncogenic role for FGFR4 in NSCLC has also been reported [24,26,34]. However, we reported that both FGFRs exerted either pro-oncogenic or tumour suppressor effects depending on the cell line used. FGFR3 overexpression has also been reported to exert anti-oncogenic and pro-oncogenic effects, which were associated with epithelial-like and mesenchymal-like phenotypes, respectively, in pancreatic cell lines [29]. We hypothesised that the biological functions of FGFR1/4 could be influenced as well in the NSCLC context by an epithelial- or mesenchymal-like phenotype. In this way, we observed that N-cadherin, which is associated with mesenchymal phenotypes [35], is required for FGFR1/4 oncogenicity.
N-cadherin has been found to physically interact with FGFR1 and FGFR4 in breast and pancreatic tumours, respectively [31,36,37], and it has been reported that the N-cadherin/FGFR interaction promotes increased and sustained FGFR signalling, exerting oncogenic and cytoprotective effects [26,36,[38], [39], [40]]. In accordance with these data, we reported that N-cadherin co-localised and co-immunoprecipitated with FGFR1 and FGFR4 in lung cell lines, and found that FGFR1/4 activation and increased downstream signalling occurred only in the presence of N-cadherin, independently of histology. This suggests that, in the context of lung tumours, N-cadherin is needed for FGFR activation, as the activated form of these receptors is barely detected in the absence of this cadherin, even after forced overexpression of the FGFR, in contrast to what has been reported in other malignancies [41]. Taken together, these data indicate a molecular context-mediated role for FGFR1 and FGFR4 in lung tumours that is dependant on the expression of N-cadherin.
While this work strongly supports an oncogenic role for FGFR1/4 in lung tumours that is mediated by N-cadherin expression, it nevertheless, remains unclear how both FGFRs exert the tumour suppressive effects observed in the absence of this cadherin. We reported that FGFR1 or FGFR4 activation was hardly detected in cell lines with no expression of N-cadherin, which suggests that both FGFRs may exert their tumour suppressor activity through a mechanism independent of their activation. Recently, it was reported that FGFRs can translocate to the nucleus in the absence of activation and interact with nuclear proteins that regulate transcription [42,43]. This nuclear function of FGFRs may be responsible for the tumour suppressor effects observed in the lung cancer setting. However, further investigation is needed to describe these tumour suppressor effects in detail.
With a view to exploring the molecular mechanisms leading to FGFR1/4-N-cadherin oncogenic effects, we identified a gene expression signature specific to NSCLC tumours with high FGFR1/4 and N-cadherin expression, which was enriched in key transcription factors for lineage plasticity and neuroendocrine differentiation [44,45]. Among the regulatory kinases predicted to be present, the transmembrane tyrosine kinase RET was identified by IPA. RET is expressed in the central and peripheral nervous system and in neural crest-derived cells, and notably has been described as the target of one of the master regulators of neuroendocrine differentiation, ASCL1, in a subset of lung adenocarcinomas with neuroendocrine differentiation [28]. These observations suggest that both RET and ASCL1 could represent central regulatory nodes within the identified gene signature and, moreover, may provide a mechanistic explanation for the pro-tumorigenic role of FGFR1/4 and N-cadherin co-expression in lung cancer. In addition, we found that FGF5 expression was upregulated in tumours with high FGFR1/4 and N-cadherin expression. FGF5 shows preferential affinity for the FGFR1 IIIc splice variant, which is preferentially found in cells with a mesenchymal phenotype [46]. This result suggests a potential autocrine loop in lung cancer cells with high FGFR1 and N-cadherin (and thus, mesenchymal-like) co-expression. Therefore, autocrine FGFs may be involved in the FGFR1/FGFR4/CDH2 dependency, but were not interrogated in this study. Further research will be needed to address this potential tumorigenic mechanism.
Based on the above evidence, we hypothesised that N-cadherin expression may predict anti-FGFR therapy efficacy in FGFR1- and/or FGFR4-overexpressing tumours. We reported that FGFR inhibitors would only affect growth of those cell lines co-expressing FGFR1/4 and N-cadherin, and validated these results for FGFR1 in a publicly available panel of lung cancer cell lines treated with the FGFR1 inhibitor dovitinib. Although a similar trend could be observed in this database for FGFR4 expression, these analyses did not reach statistical significance, which may be due to the lower affinity of this inhibitor for the FGFR4 protein. On the other hand, when we tested the effects of selective FGFR inhibition on tumour growth in high FGFR1/4-expressing lung PDX models, only tumours with high N-cadherin expression responded to therapy, achieving complete tumour regression in some of the treated tumours. Classically, FGFR1 amplification has been associated with FGFR inhibition efficacy and has been established as an inclusion criterion for clinical trials with FGFR inhibitors [19,47,48]. However, so far the results of these trials have been very modest, with only 5–10% of FGFR1-amplified tumours showing partial responses [19,[48], [49], [50]], suggesting that this genomic aberration may not be a good predictive biomarker for anti-FGFR therapy. In the search for better predictive biomarkers for FGFR inhibition efficacy, it has been reported that FGFR1 mRNA and protein expression may be more predictive than FGFR1 amplification [13]. Also, different potential indicators of FGFR inhibition sensitivity (MYC overexpression and certain gene expression signatures), or resistance (NRAS amplification, DUSP6 deletion, MET upregulation) have been identified [51], [52], [53]. However, consistent in vitro functional assays and correlative in vivo sensitivity data have not been forthcoming thus far for any of the proposed biomarkers. Interestingly, within the PDX models tested, AZD4547 showed high efficacy in one of the models with the highest FGFR4 expression level (TP43) despite AZD4547 being highly active only to FGFR1-3. We hypothesise that these results may be due to the fact that this compound is also active against FGFR4, even if with a lower affinity compared to FGFR1-3 [54]. This result may have been possible due to the high FGFR4 expression in this model and to the relatively high AZD4547 doses used in these experiments. On the other hand, even if FGFR1/4 and N-cadherin co-expression at the protein level seem to predict high FGFRi efficacy, to determine this we relied on western blot, a technique that is not routinely used in the clinical context. Further work will be required to immunohistochemically determine the expression of these proteins and to establish cut-off expression levels predicting FGFRi efficacy.
It should also be noted that within the PDXs treated with a selective FGFR inhibitor, one KRAS-mutant lung adenocarcinoma PDX (TP91) showed high sensitivity, suggesting that FGFR1 inhibition may be an interesting therapeutic approach for selected KRAS-mutant tumours, which to date lack efficacious targeted therapies. In agreement with this result, the combined inhibition of MEK and FGFR1 reverted resistance to MEK inhibition and exhibited high efficacy in different in vivo KRAS-mutant lung cancer settings, including one PDX model [55]. We have shown that FGFR1/4 expression levels have low predictive potential as biomarkers of anti-FGFR therapy benefit, as these therapeutic strategies are not likely to be effective against tumours lacking N-cadherin expression. These findings could underlie the poor efficacy of FGFR inhibitors in clinical trials so far, with the exception of some isolated cases of high FGFR1 amplification or fusions [56].
Declaration of Competing Interest
Drs Quintanal-Villalonga, Molina-Pinelo, Carnero, Paz-Ares, and Ferrer jointly hold patent WO2019012174A1 and patent WO2019016422A1 (pending). Dr. Paz-Ares also reports personal fees from Roche, Lilly, MSD, BMS, AstraZeneca, Boehringer Ingelheim, Pfizer, Takeda, Novartis, Merck Serono, Amgen, Sanofi, Pharmamar, Clovis Oncology and Janssen outside the submitted work. JZ reports personal fees from Guardant Health. The remaining authors declare no conflict of interest.
Acknowledgments
Funding
This work was funded by the Community of Madrid, the ISCIII co-funded by FEDER from Regional Development European Funds (European Union), the Spanish Ministry of Economy and Competitiveness, the Mutua Madrileña Foundation, the Ministry of Health and Social Welfare of Junta de Andalucía, the AECC scientific foundation and the Spanish Ministry of Education, Culture and Sport.
Acknowledgments
The authors thank Diego Megias for their specialist assistance and support from CNIO Confocal Microscopy Unit, and Maria José Castro from the IBiS Flow Cytometry Core. The authors thank the donors and the Hospital Universitario Virgen del Rocío Biobank for the human specimens used in this study. L.P.A. was funded by the Community of Madrid, CAM, (B2017/BMD3884), ISCIII (PIE15/00076, PI17/00778 and DTS17/00089) and CIBERONC (CD16/12/00442), and co-funded by FEDER from Regional Development European Funds (European Union). A.C. was funded by grants from the Spanish Ministry of Economy and Competitiveness Plan Estatal de I+D+I 2018 co-funded by FEDER: RTI2018-097455-B-I00; CIBER de Cáncer (CB16/12/00275), co-funded by FEDER from Regional Development European Funds. S.M.P. is funded by the Mutua Madrileña Foundation (2014) Ministry of Health and Social Welfare of Junta de Andalucía (PI-0046-2012, Nicolas Monardes Program C-0040-2016), ISCIII (PI17/00033), and co-funded by FEDER from Regional Development European Funds (European Union). I.F. is funded by the AECC scientific foundation (AIO2015) and Consejería de Igualdad, Salud y Políticas Sociales de la Junta de Andalucía (PI-0029-2013) and ISCIII (PI16/01311) and co-funded by FEDER from Regional Development European Funds (European Union). A.Q. is funded by ISCIII (FI12/00429). L.O. is funded by the Spanish Ministry of Education, Culture and Sport (FPU13/02595). S.V. is supported by the Spanish Ministry of Economy and Competitiveness (MINECO, SAF2017-89944-R). Funders did not have any role in the study's design, data collection, data analysis, interpretation, or writing of the report.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102683.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Lu M., Tian H., Yue W., Li L., Li S., Qi L. TFIIB-related factor 2 over expression is a prognosis marker for early-stage non-small cell lung cancer correlated with tumor angiogenesis. PLoS ONE. 2014;9(2):e88032. doi: 10.1371/journal.pone.0088032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heist R.S., Engelman J.A. SnapShot: non-small cell lung cancer. Cancer Cell. 2012;21(3):448. doi: 10.1016/j.ccr.2012.03.007. e2. [DOI] [PubMed] [Google Scholar]
- 4.Casaluce F., Sgambato A., Maione P., Rossi A., Ferrara C., Napolitano A. ALK inhibitors: a new targeted therapy in the treatment of advanced NSCLC. Target Oncol. 2013;8(1):55–67. doi: 10.1007/s11523-012-0250-9. [DOI] [PubMed] [Google Scholar]
- 5.Jazieh A.R., Al Sudairy R., Abu-Shraie N., Al Suwairi W., Ferwana M., Murad M.H. Erlotinib in wild type epidermal growth factor receptor non-small cell lung cancer: a systematic review. Ann Thorac Med. 2013;8(4):204–208. doi: 10.4103/1817-1737.118503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quintanal-Villalonga A., Paz-Ares L., Ferrer I., Molina-Pinelo S. Tyrosine kinase receptor landscape in lung cancer: therapeutical implications. Dis Markers. 2016;2016 doi: 10.1155/2016/9214056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmad I., Iwata T., Leung H.Y. Mechanisms of FGFR-mediated carcinogenesis. Biochim Biophys Acta. 2012;1823(4):850–860. doi: 10.1016/j.bbamcr.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Nakanishi Y., Akiyama N., Tsukaguchi T., Fujii T., Satoh Y., Ishii N. Mechanism of oncogenic signal activation by the novel fusion kinase FGFR3-BAIAP2L1. Mol Cancer Ther. 2015;14(3):704–712. doi: 10.1158/1535-7163.MCT-14-0927-T. [DOI] [PubMed] [Google Scholar]
- 9.Capelletti M., Dodge M.E., Ercan D., Hammerman P.S., Park S.I., Kim J. Identification of recurrent FGFR3-TACC3 fusion oncogenes from lung adenocarcinoma. Clin Cancer Res. 2014;20(24):6551–6558. doi: 10.1158/1078-0432.CCR-14-1337. [DOI] [PubMed] [Google Scholar]
- 10.Tanizaki J., Ercan D., Capelletti M., Dodge M., Xu C., Bahcall M. Identification of oncogenic and drug-sensitizing mutations in the extracellular domain of FGFR2. Cancer Res. 2015;75(15):3139–3146. doi: 10.1158/0008-5472.CAN-14-3771. [DOI] [PubMed] [Google Scholar]
- 11.Perez-Moreno P., Brambilla E., Thomas R., Soria J.C. Squamous cell carcinoma of the lung: molecular subtypes and therapeutic opportunities. Clin Cancer Res. 2012;18(9):2443–2451. doi: 10.1158/1078-0432.CCR-11-2370. [DOI] [PubMed] [Google Scholar]
- 12.Seo A.N., Jin Y., Lee H.J., Sun P.L., Kim H., Jheon S. FGFR1 amplification is associated with poor prognosis and smoking in non-small-cell lung cancer. Virchows Arch. 2014;465(5):547–558. doi: 10.1007/s00428-014-1634-2. [DOI] [PubMed] [Google Scholar]
- 13.Wynes M.W., Hinz T.K., Gao D., Martini M., Marek L.A., Ware K.E. FGFR1 mRNA and protein expression, not gene copy number, predict FGFR TKI sensitivity across all lung cancer histologies. Clin Cancer Res. 2014;20(12):3299–3309. doi: 10.1158/1078-0432.CCR-13-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiss J., Sos M.L., Seidel D., Peifer M., Zander T., Heuckmann J.M. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2(62) doi: 10.1126/scitranslmed.3001451. 62ra93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotani H., Ebi H., Kitai H., Nanjo S., Kita K., Huynh T.G. Co-active receptor tyrosine kinases mitigate the effect of FGFR inhibitors in FGFR1-amplified lung cancers with low FGFR1 protein expression. Oncogene. 2016;35(27):3587–3597. doi: 10.1038/onc.2015.426. [DOI] [PubMed] [Google Scholar]
- 16.Singleton K.R., Hinz T.K., Kleczko E.K., Marek L.A., Kwak J., Harp T. Kinome RNAi screens reveal synergistic targeting of MTOR and FGFR1 pathways for treatment of lung cancer and HNSCC. Cancer Res. 2015;75(20):4398–4406. doi: 10.1158/0008-5472.CAN-15-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J., Zhang L., Su X., Li M., Xie L., Malchers F. Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clin Cancer Res. 2012;18(24):6658–6667. doi: 10.1158/1078-0432.CCR-12-2694. [DOI] [PubMed] [Google Scholar]
- 18.Quintanal-Villalonga A., Molina-Pinelo S., Cirauqui C., Ojeda-Marquez L., Marrugal A., Suarez R. FGFR1 cooperates with EGFR in lung cancer oncogenesis, and their combined inhibition shows improved efficacy. J Thorac Oncol. 2019 doi: 10.1016/j.jtho.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 19.Chae Y.K., Ranganath K., Hammerman P.S., Vaklavas C., Mohindra N., Kalyan A. Inhibition of the fibroblast growth factor receptor (FGFR) pathway: the current landscape and barriers to clinical application. Oncotarget. 2017;8(9):16052–16074. doi: 10.18632/oncotarget.14109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng S., Shao L., Yu W., Gavine P., Ittmann M. Targeting fibroblast growth factor receptor signaling inhibits prostate cancer progression. Clin Cancer Res. 2012;18(14):3880–3888. doi: 10.1158/1078-0432.CCR-11-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellor H.R. Targeted inhibition of the FGF19-FGFR4 pathway in hepatocellular carcinoma; translational safety considerations. Liver Int. 2014;34(6):e1–e9. doi: 10.1111/liv.12462. [DOI] [PubMed] [Google Scholar]
- 22.Penault-Llorca F., Bertucci F., Adelaide J., Parc P., Coulier F., Jacquemier J. Expression of FGF and FGF receptor genes in human breast cancer. Int J Cancer. 1995;61(2):170–176. doi: 10.1002/ijc.2910610205. [DOI] [PubMed] [Google Scholar]
- 23.Quintanal-Villalonga A, Molina-Pinelo S, Yagüe P, Marrugal Á, Ojeda-Márquez L, Suarez R, et al. FGFR4 increases EGFR oncogenic signaling in lung adenocarcinoma, and their combined inhibition is highly effective. Lung Cancer doi 10.1016/j.lungcan.2019.02.007. [DOI] [PubMed]
- 24.Huang H.P., Feng H., Qiao H.B., Ren Z.X., Zhu G.D. The prognostic significance of fibroblast growth factor receptor 4 in non-small-cell lung cancer. Onco Targets Ther. 2015;8:1157–1164. doi: 10.2147/OTT.S81659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrer I., Quintanal-Villalonga A., Molina-Pinelo S., Garcia-Heredia J.M., Perez M., Suarez R. MAP17 predicts sensitivity to platinum-based therapy, EGFR inhibitors and the proteasome inhibitor bortezomib in lung adenocarcinoma. J Exp Clin Cancer Res. 2018;37(1):195. doi: 10.1186/s13046-018-0871-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quintanal-Villalonga A., Ojeda-Marquez L., Marrugal A., Yague P., Ponce-Aix S., Salinas A. The FGFR4-388arg variant promotes lung cancer progression by N-Cadherin induction. Sci Rep. 2018;8(1):2394. doi: 10.1038/s41598-018-20570-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quintanal-Villalonga A., Molina-Pinelo S., Yague P., Marrugal A., Ojeda-Marquez L., Suarez R. FGFR4 increases EGFR oncogenic signaling in lung adenocarcinoma, and their combined inhibition is highly effective. Lung Cancer. 2019;131:112–121. doi: 10.1016/j.lungcan.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 28.Kosari F., Ida C.M., Aubry M.C., Yang L., Kovtun I.V., Klein J.L. ASCL1 and ret expression defines a clinically relevant subgroup of lung adenocarcinoma characterized by neuroendocrine differentiation. Oncogene. 2014;33(29):3776–3783. doi: 10.1038/onc.2013.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lafitte M., Moranvillier I., Garcia S., Peuchant E., Iovanna J., Rousseau B. FGFR3 has tumor suppressor properties in cells with epithelial phenotype. Mol Cancer. 2013;12:83. doi: 10.1186/1476-4598-12-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian X., Anzovino A., Kim S., Suyama K., Yao J., Hulit J. N-cadherin/FGFR promotes metastasis through epithelial-to-mesenchymal transition and stem/progenitor cell-like properties. Oncogene. 2014;33(26):3411–3421. doi: 10.1038/onc.2013.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavallaro U., Niedermeyer J., Fuxa M., Christofori G. N-CAM modulates tumour-cell adhesion to matrix by inducing FGF-receptor signalling. Nat Cell Biol. 2001;3(7):650–657. doi: 10.1038/35083041. [DOI] [PubMed] [Google Scholar]
- 32.Heist R.S., Mino-Kenudson M., Sequist L.V., Tammireddy S., Morrissey L., Christiani D.C. FGFR1 amplification in squamous cell carcinoma of the lung. J Thorac Oncol. 2012;7(12):1775–1780. doi: 10.1097/JTO.0b013e31826aed28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin A., Johnson A., Ross J.S., Miller V.A., Ali S.M., Schrock A.B. Detection of known and novel FGFR fusions in non-small cell lung cancer by comprehensive genomic profiling. J Thorac Oncol. 2018 doi: 10.1016/j.jtho.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Quintanal-Villalonga A., Carranza-Carranza A., Melendez R., Ferrer I., Molina-Pinelo S., Paz-Ares L. Prognostic role of the FGFR4-388Arg variant in lung squamous-cell carcinoma patients with lymph node involvement. Clin Lung Cancer. 2017;18(6):667–674. doi: 10.1016/j.cllc.2017.05.008. e1. [DOI] [PubMed] [Google Scholar]
- 35.Micalizzi D.S., Farabaugh S.M., Ford H.L. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia. 2010;15(2):117–134. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suyama K., Shapiro I., Guttman M., Hazan R.B. A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell. 2002;2(4):301–314. doi: 10.1016/s1535-6108(02)00150-2. [DOI] [PubMed] [Google Scholar]
- 37.Sanchez-Heras E., Howell F.V., Williams G., Doherty P. The fibroblast growth factor receptor acid box is essential for interactions with N-cadherin and all of the major isoforms of neural cell adhesion molecule. J Biol Chem. 2006;281(46):35208–35216. doi: 10.1074/jbc.M608655200. [DOI] [PubMed] [Google Scholar]
- 38.Hulit J., Suyama K., Chung S., Keren R., Agiostratidou G., Shan W. N-cadherin signaling potentiates mammary tumor metastasis via enhanced extracellular signal-regulated kinase activation. Cancer Res. 2007;67(7):3106–3116. doi: 10.1158/0008-5472.CAN-06-3401. [DOI] [PubMed] [Google Scholar]
- 39.Su Y., Li J., Witkiewicz A.K., Brennan D., Neill T., Talarico J. N-cadherin haploinsufficiency increases survival in a mouse model of pancreatic cancer. Oncogene. 2012;31(41):4484–4489. doi: 10.1038/onc.2011.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang C.X., Gu Y.X., Liu X.F., Tong S.Y., Ayanlaja A.A., Gao Y. Cross-link regulation of precursor N-cadherin and FGFR1 by GDNF increases U251MG cell viability. Oncol Rep. 2018;40(1):443–453. doi: 10.3892/or.2018.6405. [DOI] [PubMed] [Google Scholar]
- 41.Sarabipour S., Hristova K. Mechanism of FGF receptor dimerization and activation. Nat Commun. 2016;7:10262. doi: 10.1038/ncomms10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reilly J.F., Mizukoshi E., Maher P.A. Ligand dependent and independent internalization and nuclear translocation of fibroblast growth factor (FGF) receptor 1. DNA Cell Biol. 2004;23(9):538–548. doi: 10.1089/dna.2004.23.538. [DOI] [PubMed] [Google Scholar]
- 43.Bryant D.M., Stow J.L. Nuclear translocation of cell-surface receptors: lessons from fibroblast growth factor. Traffic. 2005;6(10):947–954. doi: 10.1111/j.1600-0854.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- 44.Mu P., Zhang Z., Benelli M., Karthaus W.R., Hoover E., Chen C.C. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355(6320):84–88. doi: 10.1126/science.aah4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Augustyn A., Borromeo M., Wang T., Fujimoto J., Shao C., Dospoy P.D. ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc Natl Acad Sci U S A. 2014;111(41):14788–14793. doi: 10.1073/pnas.1410419111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ornitz D.M., Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4(3):215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dutt A., Ramos A.H., Hammerman P.S., Mermel C., Cho J., Sharifnia T. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS ONE. 2011;6(6):e20351. doi: 10.1371/journal.pone.0020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim S.H., Sun J.M., Choi Y.L., Kim H.R., Ahn S., Lee J.Y. Efficacy and safety of dovitinib in pretreated patients with advanced squamous non-small cell lung cancer with FGFR1 amplification: a single-arm, phase 2 study. Cancer. 2016 doi: 10.1002/cncr.30135. [DOI] [PubMed] [Google Scholar]
- 49.Paik P.K., Shen R., Berger M.F., Ferry D., Soria J.C., Mathewson A. A phase ib open-label multicenter study of AZD4547 in patients with advanced squamous cell lung cancers. Clin Cancer Res. 2017;23(18):5366–5373. doi: 10.1158/1078-0432.CCR-17-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nogova L., Sequist L.V., Perez Garcia J.M., Andre F., Delord J.P., Hidalgo M. Evaluation of BGJ398, a fibroblast growth factor receptor 1-3 kinase inhibitor, in patients with advanced solid tumors harboring genetic alterations in fibroblast growth factor receptors: results of a global phase I, dose-escalation and dose-expansion study. J Clin Oncol. 2017;35(2):157–165. doi: 10.1200/JCO.2016.67.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malchers F., Dietlein F., Schottle J., Lu X., Nogova L., Albus K. Cell-autonomous and non-cell-autonomous mechanisms of transformation by amplified FGFR1 in lung cancer. Cancer Discov. 2014;4(2):246–257. doi: 10.1158/2159-8290.CD-13-0323. [DOI] [PubMed] [Google Scholar]
- 52.Malchers F., Ercanoglu M., Schutte D., Castiglione R., Tischler V., Michels S. Mechanisms of primary drug resistance in FGFR1-Amplified lung cancer. Clin Cancer Res. 2017;23(18):5527–5536. doi: 10.1158/1078-0432.CCR-17-0478. [DOI] [PubMed] [Google Scholar]
- 53.Kim H.R., Kang H.N., Shim H.S., Kim E.Y., Kim J., Kim D.J. Co-clinical trials demonstrate predictive biomarkers for dovitinib, an FGFR inhibitor, in lung squamous cell carcinoma. Ann Oncol. 2017;28(6):1250–1259. doi: 10.1093/annonc/mdx098. [DOI] [PubMed] [Google Scholar]
- 54.Fu W., Chen L., Wang Z., Kang Y., Wu C., Xia Q. Theoretical studies on FGFR isoform selectivity of FGFR1/FGFR4 inhibitors by molecular dynamics simulations and free energy calculations. Phys Chem Chem Phys. 2017;19(5):3649–3659. doi: 10.1039/c6cp07964d. [DOI] [PubMed] [Google Scholar]
- 55.Manchado E., Weissmueller S., Morris J.P.t., Chen C.C., Wullenkord R., Lujambio A. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature. 2016;534(7609):647–651. doi: 10.1038/nature18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lim S.H., Sun J.M., Choi Y.L., Kim H.R., Ahn S., Lee J.Y. Efficacy and safety of dovitinib in pretreated patients with advanced squamous non-small cell lung cancer with FGFR1 amplification: a single-arm, phase 2 study. Cancer. 2016;122(19):3024–3031. doi: 10.1002/cncr.30135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.