Abstract
Background
How the oncoprotein epidermal growth factor receptor (EGFR) evades proteolytic degradation and accumulates in non-small cell lung cancer (NSCLC) remains unclear, and ubiquitin pathway genes (UPGs) that are critical to NSCLC needs to be systematically identified.
Methods
A total of 696 UPGs (including E1, E2, E3, and deubiquitinases) were silenced by small interfering RNA (siRNA) library in NSCLC cells, the candidates were verified, and their significance was evaluated in patients with NSCLC. The effects of a candidate gene on EGFR were investigated in vitro and in vivo.
Findings
We report 31 candidates that are required for cell proliferation, with the E2 ubiquitin conjugase CDC34 as the most significant one. CDC34 is elevated in tumor tissues in 76 of 114 (66.7%) NSCLCs and inversely associated with prognosis, is higher in smoker patients than nonsmoker patients, and is induced by tobacco carcinogens in normal human lung epithelial cells. Forced expression of CDC34 promotes, whereas knockdown of CDC34 inhibits, NSCLC cell proliferation in vitro and in vivo. CDC34 competes with c-Cbl to bind Y1045 to inhibit polyubiquitination and degradation of EGFR. In EGFR-L858R and EGFR-T790M/Del (exon 19)-driven lung tumor growth in mouse models, knockdown of CDC34 significantly inhibits tumor formation.
Interpretation
These results demonstrate that an E2 enzyme is capable of competing with E3 ligase to stabilize substrates, and CDC34 represents an attractive therapeutic target for NSCLCs.
Funding
National Key Research and Development Program of China, National Natural Science Foundation of China, and the CAMS Innovation Fund for Medical Sciences.
Keywords: Lung cancer, Ubiquitin pathway genes, CDC34, EGFR, c-Cbl
Research in context.
Evidence before this study
The proteolysis of EGFR, an oncoprotein that is hyperactivated by somatic mutations or overexpression in more than a half of patients with NSCLC, is controlled by an E3 ligase c-Cbl and an E2 ubiquitin-conjugating enzyme Ubc4/5. How EGFR maintains a high level in NSCLCs remains obscure. In addition, the E2 conjugases facilitate proteasomal degradation by transferring the ubiquitin on their conserved cysteine residue to the ε-amino group of lysine residues on substrates. Whether the E2 conjugases have the function to protect substrates from catabolism needs to be determined.
Added value of this study
Our results showed that CDC34 was overexpressed and inversely associated with clinical outcome of the patients, and promoted NSCLC cell proliferation in vitro and in vivo. CDC34 stabilized EGFR by binding with it at Y1045 to dissociate c-Cbl and thus prevented subsequent ubiquitination and degradation. Silencing of CDC34 inhibited EGFR-L858R and EGFR-T790M/Del (exon 19)-driven lung cancer in mice.
Implications of all the available evidence
Our findings showed for the first time that CDC34 as an E2 emzyme is capable of protecting the protein substrate EGFR from ubiquitination and degradation by competing with E3 ligase c-Cbl. Inhibition of CDC34 induced EGFR catabolism and inhibited EGFR-L858R- and EGFR-T790M-driven lung cancer, providing a promising therapeutic target for this deadly disease.
Alt-text: Unlabelled box
1. Introduction
Hyperactivation of the epidermal growth factor receptor (EGFR) by gain-of-function mutations and overexpression has been found in more than a half of patients with non-small cell lung cancers (NSCLCs), and inhibition of constitutively activated EGFR significantly benefits lung adenocarcinoma patients with mutant EGFR [1,2]. The proteolysis of EGFR is controlled by an E3 ligase c-Cbl and E2 conjugase Ubc4/5 [3]. c-Cbl binds EGFR in the absence or presence of EGF [4]. c-Cbl mediates the ubiquitination, endosome fusion, and lysomal sorting of EGFR [5], [6], [7]. The conserved N-terminal of c-Cbl is sufficient to enhance EGFR ubiquitination [8], while the RING finger C-terminal flank controls EGFR fate downstream of receptor ubiquitination [9]. Somatic mutations and loss of heterozygosity (LOH) of c-Cbl had been reported in a proportion of NSCLCs [10], but how EGFR evades proteolytic degradation and thus accumulates in lung cancer cells remains to be elucidated.
The ubiquitin (Ub)-proteasome system is the principal pathway for diverse intracellular protein degradation, in which the E2 ubiquitin-conjugating enzymes play critical roles by transferring the Ub on their conserved cysteine residue to the ε-amino group of lysine residues on substrates [11]. The cell division cycle 34 (CDC34, or UBCH3, Ubc3, UBE2R1) is an E2 enzyme which contains a non-covalent Ub binding domain in its carboxyl terminus [12] and employs distinct sites to coordinate attachment of Ub to a substrate and assembly of polyubiquitin chains [13]. CDC34 functions in conjunction with the Skp1-Cullin 1-F-box (SCF) E3 Ub ligase to catalyze covalent attachment of polyubiquitin chains to substrates such as p27 [14], ATF5 [15], WEE1 [16], IκBα [17], Grr1 [18], c-Ski [19], Sic1 [20], and other substrate proteins. However, whether CDC34 could protect substrates from proteolytic degradation remains to be investigated.
To systematically identify ubiquitin pathway genes (UPGs) that are crucial to lung cancer cell proliferation and EGFR stabilization, we conducted a systematic silencing of the E1, E2, E3, and deubiquitinases in NSCLC cell lines. We reported that 31 UPGs were required for the survival of the two NSCLC lines, among them CDC34 was overexpressed and inversely associated with clinical outcome of NSCLC patients. Interestingly, CDC34 competitively bound EGFR and prevented it from proteolysis to promote lung cancer.
2. Materials and methods
2.1. Patient samples
The study was approved by the local research ethics committees of Sun Yat-Sen University Cancer Center and the Third Affiliated Hospital of Kunming Medical University. All lung cancer samples were collected with informed consent. The diagnosis of lung cancer was confirmed by at least two pathologists. Tissue samples were taken at the time of surgery and quickly frozen in liquid nitrogen. The tumor samples contained a tumor cellularity of greater than 60% and the matched control samples had no tumor content.
2.2. siRNA library
Four Human siGENOME SMARTpool siRNA Libraries were obtained from Thermo Scientific Dharmacon (Lafayette, CO, USA). These libraries included Human Deubiquitinating Enzymes (G-004,705-05), Human Ubiquitin Conjugation Subset 1 (G-005615-05), Human Ubiquitin Conjugation Subset 2 (G-005625-05), Human Ubiquitin Conjugation Subset 3 (G-005635-05), and siGENOME Controls Complete Kit (K-002800-C2-02). Transfections were performed by using DharmFECT transfection reagent (T-2001-01) according to the manufacturer's instructions.
2.3. Cell culture, cell cycle, and cell apoptosis
NSCLC lines A549, H226, H460, H1975, HCC827, human embryonic kidney line HEK293T (obtained from the American Tissue Culture Collection (ATCC; Manassas, VA, USA), and the human normal bronchial epithelial cell line 16HBE (Clonetics, Walkersville, MD) were cultured in Dulbecco Modified Eagle Medium (DMEM) or RPMI 1640 supplemented with 10% fetal bovine serum. Cell viability of A549 and H1975 in siRNA library screening was assayed by the CellTiter-Glo Reagent (Promega, Fitchburg, WI, USA) according to the manufacturer's instructions. The z-score [z=(x−m)/s], where x is the raw score to be standardized, m is the mean of the plate, and s is the standard deviation of the plate, was determined for each SMARTpool within the plate [21]. The z-scores from the three replicates for each SMARTpool were averaged and the SD determined.
To detect the cell cycle distribution, the cells were harvested and washed in PBS, fixed in 70% ethanol and kept in 4 °C overnight. The cells were centrifuged and washed with PBS containing 1% FBS, followed by the treatment with 1% RNaseA for 15 min at 37 °C, and then stained with 50 μg/ml of propidium iodide. The fluorescent intensity was measured by the flow cytometry (BD FACSVantage Diva, USA). Apoptosis in individual cells was detected using an in situ cell death detection kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions. Figures for exemplifying the gating strategy of flow cytometry are shown in Supplementary Fig. 8.
2.4. Antibodies and reagents
Antibodies used included mouse anti-β-Actin (#A5441, Sigma, St.Louis, MO, USA; 1:5000 for Western blot), mouse anti-Flag (#F1804, Sigma; 1:200 for immunoprecipitation, 1:5000 for Western blot), mouse anti-EGFR (#sc-373746, Santa Cruz Biotechnology; 1:100 for immunoprecipitation, 1:1000 for Western blot, 1:50 for immunofluorescence), rabbit anti-EGFR (#4267, Cell Signaling Technology, Beverly, MA, USA; 1:50 for immunohistochemistry (IHC) staining), mouse anti-CDC34 (#sc-28381, Santa Cruz Biotechnology; 1:100 for immunoprecipitation, 1:1000 for Western blot), rabbit anti-CDC34 (#A5457, ABclonal, Cambridge, MA, USA; 1:100 for immunofluorescence, 1:50 for IHC), goat anti-pEGFR-Y1173 (#sc-12351, Santa Cruz Biotechnology; 1:1000 for Western blot), rabbit anti-pAKT (#sc-7985-R, Santa Cruz Biotechnology; 1:1000 for Western blot), rabbit anti-AKT (#sc-8312, Santa Cruz Biotechnology; 1:1000 for Western blot), rabbit anti-p27 (#sc-528, Santa Cruz Biotechnology; 1:1000 for Western blot), rabbit anti-ERK (#sc-514302, Santa Cruz Biotechnology; 1:1000 for Western blot), rabbit anti-pERK (#4370, Cell Signaling Technology; 1:1000 for Western blot), mouse anti-pSTAT3-Y705 (#9138, Cell Signaling Technology; 1:1000 for Western blot), rabbit anti-pSTAT3- S727 (#9134, Cell Signaling Technology; 1:1000 for Western blot), mouse anti-STAT3 (#9139, Cell Signaling Technology; 1:1000 for Western blot), mouse anti-HA (#AE008, ABclonal; 1:2000 for Western blot), rabbit anti-GST (#A-5800, Invitrogen, Frederick, MD, USA; 1:5000 for Western blot), rabbit anti-Ki67 (#ab15580, Abcam, Cambridge, MA, USA; 1:400 for IHC), and rabbit anti-c-Cbl (#ab137375, Abcam; 1:1000 for Western blot). Reagents used included cycloheximide (CHX) (#94271, Amresco Inc., Solon, OH, USA), erlotinib (#HY-12008, MedChemExpress, USA), epoxomicin (#A2606, APExBIO, USA), Universal Tyrosine Kinase Assay Kit (#MK410, Clontech, Palo Alto, CA), MG132 (Sigma, #SML1135), chloroquine (Sigma, #PHR-1258), BaP (Sigma, #B1760), BAA (Sigma, #B2209), and DBA (Sigma, #91861).
2.5. siRNA, shRNA, plasmids and transfections
siRNA or shRNA were purchased from GenePharmaCo. Ltd (Shanghai, China) and the sequences are as follows: GCUCAGACCUCUUCUACGA (siCDC34-1#); GGACGAGGGCGAUCUAUAC (siCDC34-2#); GAUCGGGAGUACACAGACA (siCDC34-3#); UGAACGAGCCCAACACCUU (siCDC34-4#); GGCTCAGACCTCTTCTACGAC (human CDC34-shRNA-1#); GAGTGTGATCTCCCTCCTGAA (human CDC34-shRNA-2#); CTCTTCTACGACGACTACTAT (mouse CDC34-shRNA-1#); GAGTGTAATTTCGCTGCTGAA (mouse CDC34-shRNA-2#); GGCUGGUUAUGUCCUCAUU (siEGFR); GGAGACACAUUUCGGAUUA (siCBL). The sequence for CDC34res in CDC34 construct was 5′-GCTCAGATCTATTCTACGA3’ (CDC34res 1# for siCDC34 1#) and 5′-TGAACGAACCTAACACCTT-3′ (CDC34res 2# for siCDC34 2#), respectively.
FLAG-CDC34 vector was constructed based on pcDNA3.1 plasmid; HA-CDC34 vector was constructed based on pCS2 plasmid, and pCDH-CDC34 vector was constructed based on pCDH-GFP plasmid. All CDC34 mutants were subcloned from Flag or HA-tagged CDC34 vectors. FLAG-EGFR was cloned from the pCAG-3Flag-HA-EGFR vector and FLAG-EGFRintracellular domain (ICD) was subcloned from FLAG-EGFR vector. pFlag-CMV4-CBL vector was kindly provided by Dr. Jianhua Mao (Shanghai Institute of Hematology, Rui Jin Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, China). GST or His-tagged CDC34, EGFRICD, CBL were generated based on the backbone of pGEX-4T-1 and pET28a (kindly provided by Dr. Quan Chen, Institute of Zoology, Chinese Academy of Sciences, Beijing, China), respectively. The shCDC34 constructs were made with PLKO.1 backbone (kindly provided by Dr. Wanzhu Jin, Institute of Zoology, Chinese Academy of Sciences) using Age I and EcoR I sites. Cells were transfected with siRNA, shRNA or plasmids using the Lipofectamine 2000 or Lipofectamine 3000 (Invitrogen).
2.6. Lentivirus-mediated transfection
For lentiviral particle production, shCDC34 constructs in PLKO.1 or pCDH-CDC34 constructs in pCDH-GFP were co-transfected with psPAX2 and pMD2G into HEK293T cells. The culture medium was replaced with fresh medium after 6 h, and the supernatants were harvested 48 h and 72 h post transfection. A549-luciferase cells were infected with viral particles in the presence of 8 μg/mL polybrene to generate CDC34 stably knockdown or overexpression cells.
2.7. RT-PCR
The total RNA was isolated using the TRIZOL reagent (Invitrogen) and the phenol-chloroform extraction method according to the manufacturer's instruction. Total RNA (2 μg) was annealed with random primers at 65 °C for 5 min. The cDNA was synthesized using a 1st-STRAND cDNA Synthesis Kit (Fermentas, Pittsburgh, PA, USA). Quantitative real-time PCR was carried out using SYBR PremixExTaq (Takara Biotechnology, Dalian, China). The primers used for quantitative RT-PCR are as follows: human GAPDH, 5′-GAGTCAACGGATTTGGTCGT-3′ (forward) and 5′-GACAAGCTTCCCGTTCTCAG-3′ (reverse); mouse GAPDH 5′-AGTATGACTCCACTCACGGCAA-3′ (forward) and 5′-TCTCGCTCCTGGAAGATGGT-3′ (reverse); human CDC34, 5′- GACGAGGGCGATCTATACAACT-3′ (forward) and 5′- GAGTATGGGTAGTCGATGGGG-3′ (reverse); mouse CDC34, 5′- CCCCAACACCTACTATGAGGG-3′ (forward) and 5′- ACATCTTGGTGAGGAACCGGA-3′ (reverse); human EGFR, 5′-GGACTCTGGATCCCAGAAGGTG-3′ (forward) and 5′-GCTGGC CATCACGTAGGCTT-3′ (reverse); human CCND1, 5′- GCTGGAGCCCGTGAAAAAGA-3′ (forward) and 5′-CTCCGCCTCTGGCATTTTG-3′ (reverse). Each sample was analyzed in triplicate for three times.
2.8. Immunofluorescence microscopy
Cells grown on coverslip (24 mm × 24 mm) were fixed with 4% paraformaldehyde for 15 min, washed with 150 mM glycine in PBS, and permeabilized with 0.3% Triton X-100 in PBS for 20 min at room temperature. After blocking with 5% BSA, the cell smears were incubated with indicated primary antibodies overnight at 4 °C, washed, and FITC/PE-labeled secondary antibody in PBS was added to the cell smears. Images were taken by a laser scanning confocal microscopy (Zeiss, Oberkochen, Germany).
2.9. Immunohistochemistry analysis
IHC assay was performed with anti-CDC34, anti-Ki67 and anti-EGFR antibodies. Briefly, formalin-fixed, paraffin-embedded human or mouse lung cancer tissue specimens (5 µm) were deparaffinized through xylene and graded alcohol, and subjected to a heat-induced epitope retrieval step in citrate buffer solution. The sections were then blocked with 5% BSA for 30 min and incubated with indicated antibodies at 4 °C overnight, followed by incubation with secondary antibodies for 90 min at 37 °C. Detection was performed with 3, 3′-diaminobenzidine (DAB, Zhongshan Golden Bridge Biotechnology, Beijing, China) and counterstained with hematoxylin, dehydrated, cleared and mounted as in routine processing. The scoring of immunoreactivity was calculated as IRS (0–12)=RP (0–4) × SI (0–3), where RP is the percentage of staining-positive cells and SI is staining intensity.
2.10. Western blotting
Cells were lysed on ice for 30 min in RIPA buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 0.1% SDS, 1% deoxycholate, 1% Triton X-100, 1 mM EDTA, 5 mM NaF, 1 mM sodium vanadate, and protease inhibitors cocktail), and protein extracts were quantitated. Proteins (20 mg) were subjected to 8–15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), electrophoresed and transferred onto a nitrocellulose membrane. After blocking with 5% non-fat milk in Tris-buffered saline, the membrane was washed and incubated with the indicated primary and secondary antibodies and detected by Luminescent Image Analyzer LSA 4000 (GE, Fairfield, CO, USA).
2.11. CO-immunoprecipitation
Cells were treated either with or without 10 μM MG132 for 3–4 h before lysis. After wash with cold PBS for two times, cells were suspended in IP lysis buffer (40 mM Tris–HCl, pH 7.4, 137 mM NaCl, 1.5 mM MgCl2, 0.2% sodium deoxycholate, 1% Nonidet P-40, 2 mM EDTA, 1 mM PMSF, complete protease inhibitors cocktail) and cleared by centrifugation. Indicated antibody was added and incubated overnight with each cell lysate at 4 °C. Protein A/G PLUS-Agarose beads (Santa Cruz) were added after washing for 3 times with lysis buffer. After 2-h of incubation, beads were washed four times, 5 min per wash in IP wash buffer (40 mM Tris–HCl, pH 7.4, 137 mM NaCl, 1.5 mM MgCl2, 2 mM EDTA, 0.2% Nonidet P-40).
2.12. Protein purification, GST or His pull-down assay, and in vitro kinase assay
GST or His-tagged CDC34, EGFRICD, c-Cbl proteins were expressed in E. coli Rosetta (DE3). GST fusion proteins were purified on glutathione-Sepharose 4 Fast Flow beads (GE Health Science, Pittsburgh, PA, USA) and His fusion proteins were purified on HisPur Cobalt Resin (Thermo Scientific, Basingstoke, UK), respectively. The GST was removed with thrombin (Amresco). For the GST pull-down, 2 μg of GST-fusion protein was incubated with cell lysates or purified proteins for 2 h at 4 °C and then washed 5 times with 1 mL PBS buffer. The precipitate complex was boiled with sample buffer containing 1% SDS for 5 min at 95 °C and subjected to SDS-PAGE. The nitrocellulose membrane was stained with Ponceau S and followed by immunoblotting with indicated antibodies. In vitro EGFR kinase activity was determined using Universal Tyrosine Kinase Assay Kit (TaKaRa Biotechnology), following the manufacturer's instructions.
2.13. Animal studies
The animal studies were approved by the Institutional Review Board of Institute of Zoology, Chinese Academy of Sciences, and the methods were carried out in accordance with the approved guidelines. The mice were numbered, injected with indicated cells or virus particles, and randomized into groups. For xenograft tumor models, six-week-old SCID beige mice were maintained in the pathogen-free (SPF) conditions. A549-luciferase cells stably expressing the shCDC34 (1 × 106) or pCDH—CDC34 (2 × 105) were injected into the lateral tail veins of the mice, and the tumors were monitored by IVIS Spectrum Imaging System 30, 45, or 55 days after cell inoculation (Caliper Life Sciences, Hopkinton, MA, USA). For EGFRL858R or EGFRT790M/Del(exon19)-driven lung tumors, 6-week-old Tet-op-EGFRmutant/CCSP-rtTA FVB mice (kindly provided by Professor Liang Chen, Jinan University, Guangzhou, China) were intranasally administrated with shCDC34 or shNC lentiviral particles once a day for 3 days. One week after the first lentiviral administration, DOX was added to feed the mice and the lung tumors were analysed by micro-CT (PerkinElmer, Waltham, MA, USA) scanning, and tumor volume was quantitated by the Analyze 12.0 (PerkinElmer) Caliper microCT Analysis Tools according to the manufacturer's instruction. The A/J mice were exposed to cigarette smoke generated by DSI's Buxco Smoke Generator (Buxco, NC, USA) inside a perspex box, at a frequency of 5 cigarettes per day, 5 days per week for 2 months. Whole body cigarette smoke exposure per cigarette was 3 min followed by a 15-min period of fresh air [22]. Mice were anesthetized by mixture of oxygen/isoflurane inhalation and positioned with legs fully extended, and assayed according to manufacturers’ instruction. Survival of the mice was evaluated from the first day of DOX treatment until death or became moribund, at which time points the mice were sacrificed.
2.14. Statistical analysis
All experiments were repeated at least three times and the data were presented as the mean ± SD unless noted otherwise. Differences between data groups were evaluated for significance using Student's t-test of unpaired data or one-way analysis of variance. P values less than 0.05 indicate statistical significance.
2.15. Data availability
Cancer microarray data was downloaded from the Oncomine datasets Okayama Lung (can also be found at the Gene Expression Omnibus, GSE31210), Selamat Lung (the Gene Expression Omnibus, GSE32867), Stearman Lung (the Gene Expression Omnibus, GSE2415), Landi Lung (the Gene Expression Omnibus, GSE10072), and Su Lung (the Gene Expression Omnibus, GSE7670). The data of the reverse phase protein array of 235 lung adenocarcinomas were downloaded from the Cancer Genomics Hub (CGHub) (https://cghub.ucsc.edu/) with approval by the National Institutes of Health (NIH; approval number #24437-4). All the remaining data supporting the findings of this study are available within this paper and its supplementary information.
3. Results
3.1. A systematic silencing of UPGs in lung cancer cells
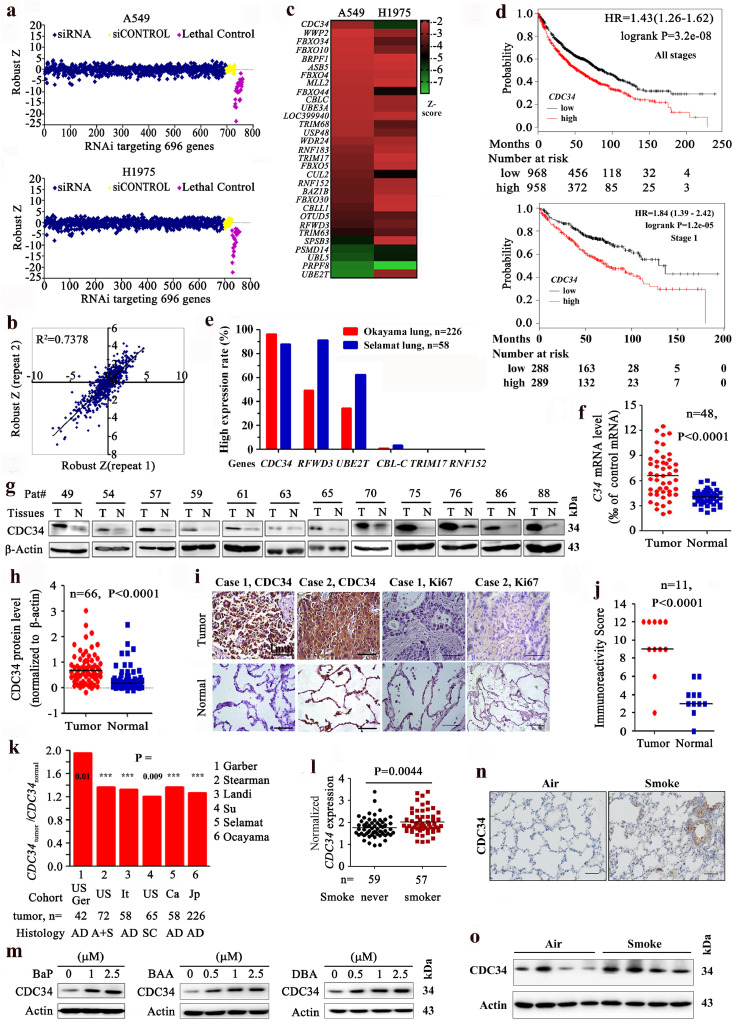
All the 696 UPGs found in human genome (Supplementary Table 1) were silenced by transfection of small interfering RNA (siRNA) of the Dharmacon human siGENOME SMARTpool library into the NSCLC lines A549 (with wild type EGFR) and H1975 (harboring L858R/T790M EGFR mutation). Cell viability was measured 72 h after transfection, and the Z-scores from duplicate experiments for each SMARTpool (Fig. 1a) were determined [21]. To validate the results, a secondary screen was performed in A549 and the results showed that the robust Z scores of most of the genes across repeats were strongly correlated (r = 0.86; Fig. 1b). A SMARTpool was considered a hit if the Z-score was ≤ −2 in both A549 and H1975 (Fig. 1c and Supplementary Table 1). Eighty hits (11.5%) were identified in A549 and 72 hits (10.3%) were uncovered in H1975 cells (Supplementary Fig. 1), with 31 hits discovered in both cells (Supplementary Fig. 1 and Fig. 1c). These hits include two E2 enzymes (UBE2T, CDC34), twenty E3 ligases (UBE3A, WWP2, TRIM68, and others), and nine others (e.g. USP48, PSMD14, OUTD5).
Fig. 1.
Identification of CDC34 as a crucial oncogene in NSCLCs by siGenome screening. (a) A549 and H1975 cells were treated with 50 nM siRNAs from the siGenome library (containing 696 UPGs) for 72 h and the cell viability was measured. The z score was calculated as described in materials and methods. (b) A second replicated screening was conducted in A549 to validate the results of the initial screening. (c) Heat map showing 31 candidates with Z score ≤ −2 in both A549 and H1975. (d) Overall survival of NSCLCs of all stages (upper) and stage 1 (lower) with high or low expression of CDC34. (e) The expression of 6 candidate genes in Okayama and Selamat cohorts (Tumor/Normal ≥ 1.5). (f–j) The expression of CDC34 was tested by qRT-PCR (f), Western blot (g, h), and IHC (i, j) assays of NSCLCs of our cohort. Molecular weight (kDa) of the protein bands was listed on the right side (g). Size bar, 1 mm. The densitometry analysis of the Western blot results (h) and the immunoreactivity score of CDC34 (j) was calculated. P values, Student's t-test. (k) CDC34 expression was detected by microarrays in tumor samples and normal lung tissues in Oncomine datasets. AD, adenocarcinoma; SC, squamous cell carcinoma; A + S, adenocarcinoma and squamous cell carcinoma; Ger, Germany; Ca, Canada; It, Italy; Jp, Japan; Tw, Taiwan, China. ***, P<0.001. (l) CDC34 in never smoker and smoker NSCLCs of the work of Selamat et al. [25] in Oncomine datasets. P value, Student's t-test. (m) 16HBE cells were treated with BaP, BAA, and DBA at indicated concentration for 24 h, and lyzed for Western blot assay. (n) IHC assays of CDC34 in the lungs of A/J mice exposed to tobacco smoke for two months. Scale bar, 250 µm. (o) Western blot analyses of lysates of lung tissues of A/J mice exposed to tobacco smoke for two months.
To identify genes that are critical to lung carcinogenesis, the association between the expression levels of the 31 genes and the clinical outcome of NSCLC patients was analyzed using the Online Survival Analysis Software [23] (http://kmplot.com/analysis/index.php?p=service&cancer=lung). We reported that the expression of six genes, CDC34 (Fig. 1d, upper panel), UBE2T, RNF152, TRIM17, CBLC, and RFWD3, was inversely associated with overall survival of the patients. The prognostic significance of CDC34 was further analyzed using stage-matched samples. For stage 1 patients (n = 577), those with higher expression of CDC34 had much shorter survival time than cases with lower level of CDC34 (Fig. 1d, lower panel). For stage 2 (Supplementary Fig. 2a) and 3 (Supplementary Fig. 2b) NSCLCs, patients with higher expression of CDC34 had slightly, but not statistically significantly, shorter overall survival than cases with lower CDC34. The significance of CDC34 in stage 4 NSCLCs was not analyzed due to small sample size of the patients (n = 4). The data [24,25] of the cancer microarray database Oncomine [26] (http://www.oncomine.org) showed that, of these 6 genes, CDC34 was most significantly upregulated in lung tumors compared to normal tissues (Fig. 1e). CDC34 was therefore chosen for further study.
3.2. Overexpression of CDC34 in NSCLCs
We tested the expression of CDC34 in 114 NSCLCs (Table 1) by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (Fig. 1f), Western blot (Fig. 1g and h) and IHC (Fig. 1i and j), and showed that in 76 (66.7%) of the patients the expression of CDC34 was significantly elevated in tumor samples than the counterpart normal lung tissues (Table 1). A two-sided Fisher exact test showed that the expression of CDC34 in smoker NSCLCs was significantly higher than in non-smoker patients (P = 0.027; Table 1). In works of Oncomine database [24, 25, [27], [28], [29]–30], CDC34 expression in tumor samples was higher than in their paired normal lung tissues (Fig. 1k), and smokers [25] had higher CDC34 expression in tumor tissues than nonsmokers (Fig. 1l). We then tested the effects of several tobacco carcinogens on normal human lung epithelial cell line 16HBE, and showed that benzo(a)pyrene (BaP), benzo[a]anthracene (BAA), and dibenzo(a,h)anthracene (DBA) induced upregulation of CDC34 in the cells (Fig. 1m). Upregulation of CDC34 was also seen in A/J mice exposed to tobacco smoke for two months (Fig. 1n and o).
Table 1.
Summary of baseline demographic characteristics of the 114 patients.
| Characteristics | Cases,n | CDC34 high#, n (%) | P values* |
|---|---|---|---|
| Total number | 114 | 76 (66.7) | |
| Age | |||
| <65 | 74 | 51 (68.9) | 0.264 |
| ≥65 | 28 | 16 (57.1) | |
| not determined | 12 | 9 (75) | |
| Gender | |||
| male | 71 | 50 (70.4) | 0.127 |
| female | 31 | 17 (54.8) | |
| not determined | 12 | 9 (75) | |
| Smoking | |||
| smoker | 59 | 44 (74.6) | 0.027 |
| non-smoker | 43 | 23 (53.5) | |
| not determined | 12 | 9 (75) | |
| Histology | |||
| adenocarcinoma | 61 | 36 (59) | 0.093 |
| squamous-cell carcinoma | 37 | 28 (75.7) | |
| not determined | 16 | 12 (75) | |
| TNM stage | |||
| I-II | 58 | 35 (60.3) | 0.219 |
| III-IV | 43 | 31 (72.1) | |
| not determined | 13 | 10 (76.9) |
The standard of CDC34 high is that CDC34 in tumors is at least 1.5 times higher than in counterpart normal lung tissues.
P values were calculated using a two-sided Fisher's exact test.
3.3. CDC34 is required for lung cancer cell proliferation
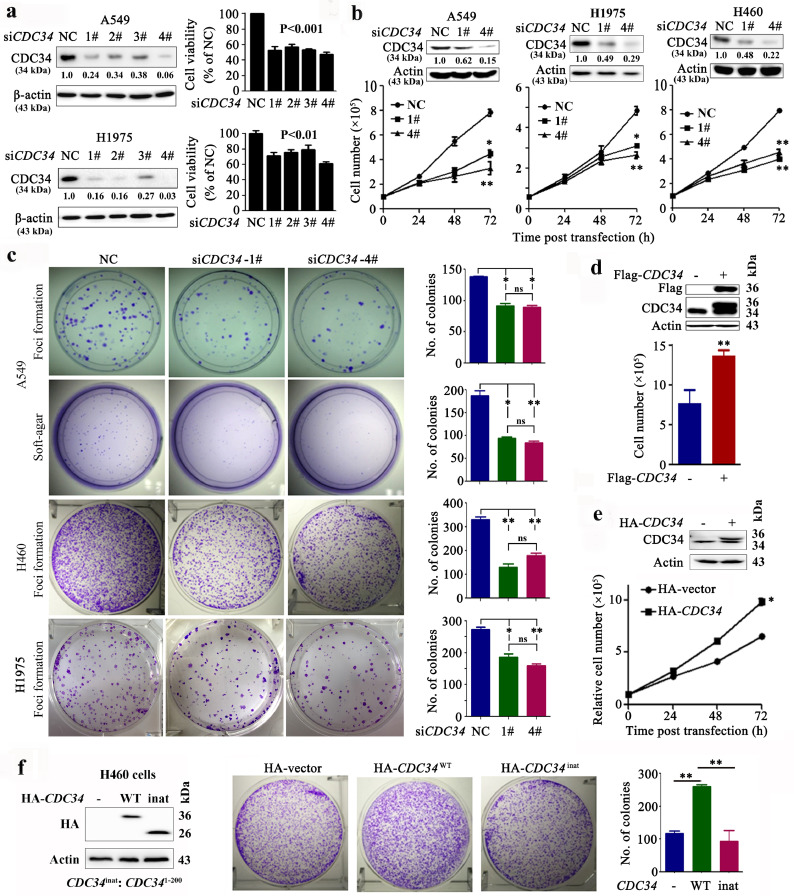
To evaluate the role of CDC34 in lung cancer, siRNA against CDC34 (siCDC34) was transfected into A549 and H1975 cells, which resulted in a reduction of CDC34 protein (Fig. 2a, left) and cell viability (Fig. 2a, right) of the cells, and even modest knockdown had significant effects. Silencing of CDC34 led to a significant inhibition of cell growth (Fig. 2b) and suppression of colony forming activity (Fig. 2c) of the NSCLC cells. On the contrary, exogenous expression of CDC34 (by transient transfection) significantly increased the proliferation (Fig. 2d), growth (Fig. 2e), and colony forming activity (Fig. 2f) of the cells.
Fig. 2.
CDC34 is required for proliferation of NSCLC cells in vitro and in vivo. (a) A549 and H1975 cells were transfected with siCDC34 (1# to 4#) and lyzed 72 h later for the detection of CDC34 expression by Western blot assays (left). Cell viability was assessed by the CellTiter-Glo Reagent (right). Numbers under the Western blot bands are the relative expression values to Actin determined by densitometry analysis. Error bars, sd. (b) A549, H1975, and H460 cells were transfected with siCDC34 (1# and 4#), and cell proliferation was assessed by trypan blue exclusion analysis. The relative expression of CDC34 was detected by Western blot, and the numbers under the Western blot bands are the relative expression values to Actin determined by densitometry analysis. Error bars, sd; P values, Student's t-test. (c) Foci formation and soft-agar assays of A549, H460, and H1975 cells transfected with siCDC34. Error bars, sd; P values, Student's t-test. (d–f) The H460 (d, f) or A549 (e) cells were transfected with Flag or HA-tagged CDC34 and cell proliferation was assessed by trypan blue exclusion analysis (d, e) or colony formation assays (f). The etopic expression of exogenous proteins was shown by Western blot bands. Inat: inactive; NC, negative control. Error bars, sd; P values, Student's t-test. *P< 0.05, **P< 0.01.
3.4. CDC34 positively regulates EGFR
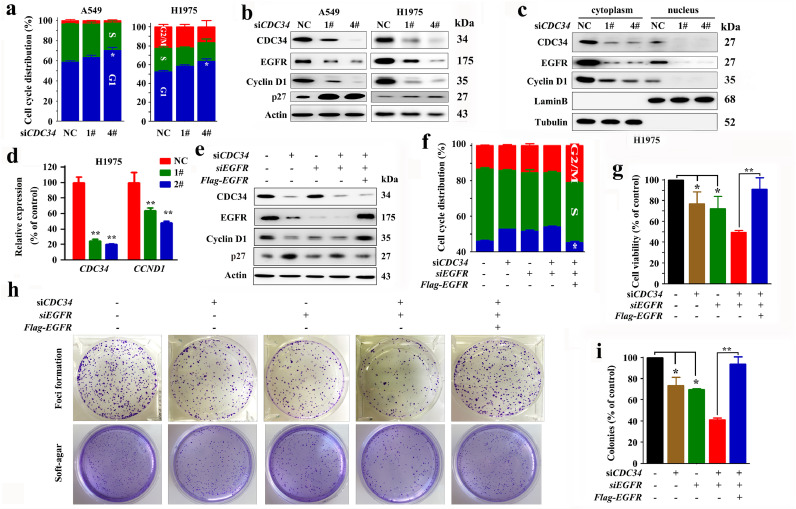
We aimed to identify the downstream target of CDC34 in lung cancer. By using the data of the reverse phase protein array of 235 lung adenocarcinomas from a previous work [31], we analyzed the potential association between CDC34 and some driver proteins of lung cancer such as EGFR, N-RAS, MEK1, and LKB1, as well as CDC34’s substrate protein p27 [32]. We reported that the expression of CDC34 was inversely associated with p27 expression (P = 0.013). Interestingly, CDC34 expression level was also correlated with EGFR (P = 0.0015, Fig. 3a), suggesting that CDC34 may have a role in determining EGFR expression. IHC assays of NSCLC samples showed that patients with higher CDC34 had higher EGFR in the tumor tissues (Fig. 3b Table 2). CDC34 expression at mRNA level in EGFR WT NSCLCs was slightly higher than in patients with EGFR mutations (Fig. 3c). However, some patients with mutant EGFR also showed high level CDC34 (Fig. 3c), suggesting that CDC34 may have a role in determining EGFR protein stability. We then showed that silencing of CDC34 in H460, H226 (Fig. 3d), and A549 (Supplementary Fig. 3a) cells resulted in downregulation of EGFR, in the absence and presence of EGF. In H1975 and HCC827 (with an E746-A750 in-frame deletion of EGFR) cells, siCDC34 also downregulated EGFR (Fig. 3d and Supplementary Fig. 3b). Moreover, knockdown of CDC34 resulted in inhibition of constitutively activated EGFR and EGF-induced phosphylated EGFR (pEGFR) (Fig. 3d, Supplementary Fig. 3a–c), and suppressed its kinase activity (Fig. 3e). Consistently, pERK1/2, pAKT and pSTAT3 were also dowregulated by siCDC34 (Fig. 3f, Supplementary Fig. 3c).
Fig. 3.
CDC34 positively regulates EGFR. (a) A scatter diagram of reverse phase protein array (RPPA) data showing a negative correlation between the mRNA level of CDC34 and protein level of p27, and a positive correlation between the mRNA level of CDC34 and protein level of EGFR. The data are from TCGA datasets. P values, Student's t-test. (b) IHC analysis of CDC34 and EGFR in NSCLCs of our setting. (c) The expression of CDC34 in EGFR WT and mutant NSCLCs in datasets and our setting. P values, Student's t-test. (d) Western blot analysis of EGFR/pEGFR in cells transfected with siCDC34 in H460, H226, and H1975 cells with or without EGF co-incubation. Numbers under the Western blot bands are the relative expression values to Actin determined by densitometry analysis. (e) H460 cells transfected with siCDC34 were lysed, the lysates were immunoprecipitated with an anti-EGFR antibody and assayed for tyrosine kinase activity. Error bars, sd. P values, Student's t-test. (f) Western blot analysis of EGFR/pEGFR and downstream molecules in EGF-stimulated, siCDC34-transfected H460 cells. (g) Immunoblotting of lysates of Flag-CDC34-expressing A549 (upper) or HA-CDC34-transfected H226 (lower) cells co-incubated with or without EGF. Numbers under the Western blot bands are the relative expression values to Actin determined by densitometry analysis. (h) H460 cells were transfected with Flag-CDC34, co-incubated with or without EGF, lysed, and the cytoplasm and nucleus proteins were harvested for immunoblotting using indicated antibodies. (i) A549 cells were transfected with Flag-CDC34, lysed 24 h later, and the lysates were immunoprecipitated with an anti-EGFR antibody and assayed for tyrosine kinase activity. Error bars,sd. P values, Student's t-test. (j – l) H460 cells were transfected with siCDC34 and/or CDC34res, which is mutated to resist siCDC34 targeting. Immunoblot assays were conducted using indicated antibodies (j), cell viability was analyzed by MTT assay (k), and colony forming activity was evaluated by foci formation and soft-agar assays (l). Error bars, sd; P values, Student's t-test. P values for indicated comparison: Group 1 vs group 2: < 0.05; group 1 vs group 3: < 0.01; group 2 vs group 4: < 0.05; group 3 vs group 5: < 0.05.
Table 2.
Correlation between CDC34 and EGFR in lung tumors of 24 paired NSCLCs.
| CDC34+ | CDC34− | Total | P value* | |
|---|---|---|---|---|
| EGFR+ | 12 | 0 | 12 | 0.023 |
| EGFR− | 5 | 7 | 12 | |
| Total | 17 | 7 | 24 |
P value was calculated using a two-sided Fisher's exact test.
On the contrary, exogenous expression of CDC34 in A549, H226, and H460 cells significantly increased protein levels of EGFR/pEGFR, in the absence and presence of EGF (Fig. 3g, Supplementary Fig. 3d). Furthermore, ectopic expression of CDC34 upregulated EGFR in cytoplasm and nucleus compartments of the H460 cells, in the absence and presence of EGF co-incubation (Fig. 3h). Forced expression of CDC34 in H460 cells also increased EGFR tyrosine kinase activity (Fig. 3i). Besides, the reduction of EGFR caused by silencing of CDC34 was rescued by overexpressed CDC34 whose CDS region was synonymously mutated in case of targeting by siRNA against CDC34 (CDC34res; Fig. 3j, supplementary Fig. 3e). CDC34res also rescued siCDC34-induced reduction in cell viability (Fig. 3k) and colony forming activity of the cells (Fig. 3l). These results indicate that CDC34 upregulates EGFR in an activation-independent manner.
3.5. Effects of CDC34 on lung cancer growth in vivo
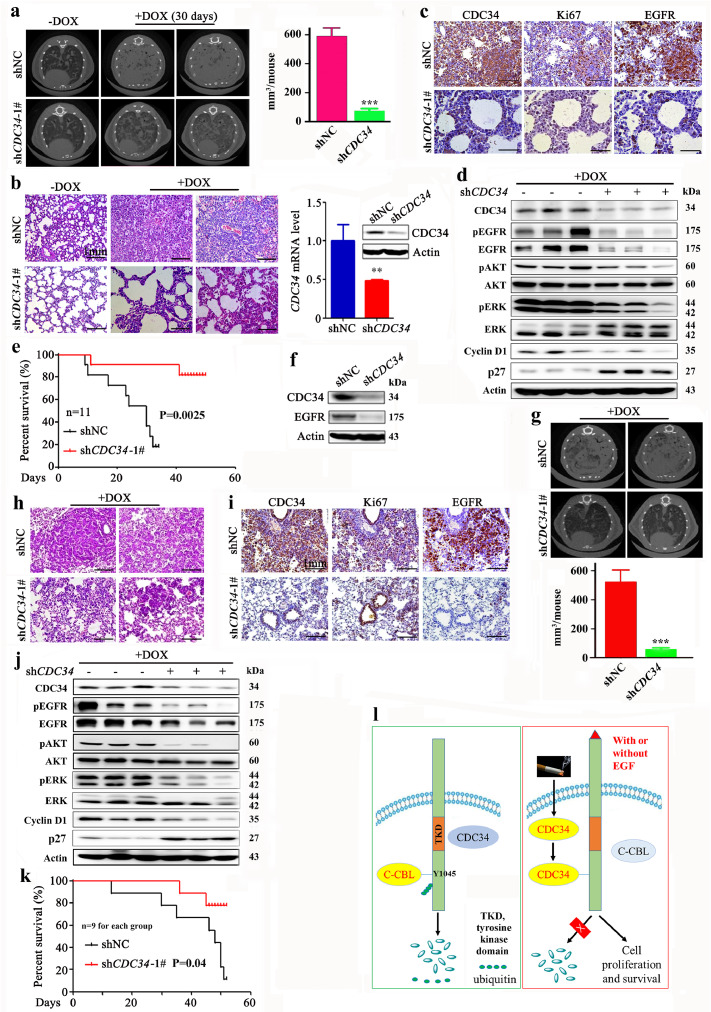
To study the effect of CDC34 on lung cancer cell proliferation in vivo, A549-luciferase cells transfected with shCDC34 (Supplementary Fig. 3f) were inoculated into SCID-beige mice via tail vein, and the results showed that the luciferase signal in CDC34 knockdown groups was significantly lower than in control group (Fig. 4a). Hematoxylin-eosin (HE) staing showed that the lungs from the control group were almost full of tumor cells, but lungs from the CDC34 knockdown mice had markedly less tumor cells (Fig. 4b). The expression levels of CDC34 and EGFR were tested in the tumor tissues by IHC (Fig. 4c) and Western blot assays (Fig. 4d), and the results showed that in the lungs of the mice inoculated with A549-luciferase-shCDC34 cells, the protein level of the two molecules was markedly lower than the control group. In addition, Ki67 staining in CDC34 knockdown cells was drastically decreased compared to tumor tissues of control group mice (Fig. 4c). Moreover, the overall survival of the mice harboring the CDC34-knockdown cells was significantly longer than the control group (Fig. 4e).
Fig. 4.
Effects of CDC34 on tumor growth in vivo. (a) SCID beige mice were injected with 1 × 106 A549-luciferase cells via tail vein, detected by the IVIS Spectrum system 30 days later (left), and the relative luciferase intensity in the mice was analyzed (right). Error bars, sd; P values, Student's t-test. (b) Hematoxylin-eosin (HE) staining of the lung sections from mice of each group. (c) IHC analysis of CDC34, EGFR and Ki67 in tumors from mice of each group. Scale bar, 1 mm. (d) The expression of CDC34 and EGFR in the tumor samples from the mice of each group. (e) Kaplan–Meier survival curve of the mice. P value, log-rank test. *, P < 0.05; **, P < 0.01. (f–j) pCDH—CDC34-expressing A549-luciferase cells (2 × 105) were injected into SCID-Beige mice, and detected for relative luciferase intensity by IVIS Spectrum at indicated time points (f, g). Error bars, sd; P values, Student's t-test. Kaplan–Meier survival curve of the mice was shown (h). P value, log-rank test. The mice were sacrified, the lung tissues were subjected to hematoxylin-eosin (HE) or IHC staining (i; scale bar, 0.5 mm), or lyzed for Western blot assays using indicated antibodies (j). *, P < 0.05; **, P < 0.01.
To further test the role of CDC34 in tumor growth in vivo, pCDH-CDC34 plasmid was transfected into A549-luciferase cells, which were then injected into SCID-beige mice via tail vein. To reflect the growth of the xenografted tumor, luciferase intensity of the mice was measured by IVIS Spectrum 30, 45, and 55 days after cell inoculation. We found that while A549-luciferase cells propogated gradually, CDC34 significantly increased tumor burden (Fig. 4f and g) and shortened life-span of the mice (Fig. 4h). HE and Ki67 staining of lung sections confirmed the tumor facilitating effect of CDC34 (Fig. 4i), while Western blot assays of tumor lysates indicated the upregulation of EGFR and Cyclin D1 in CDC34-expressing cells in vivo (Fig. 4j).
3.6. CDC34 interacts with EGFR
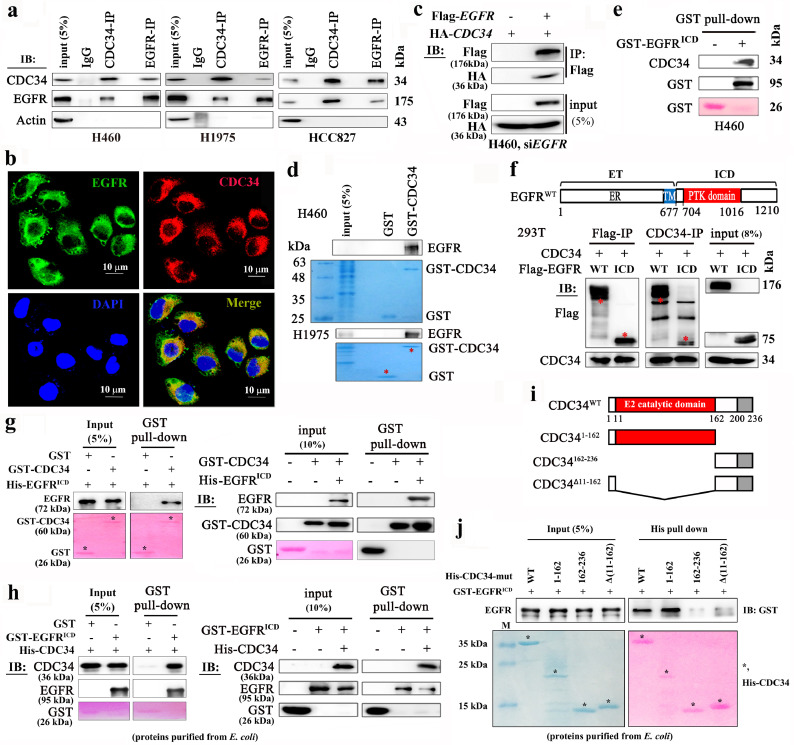
To elucidate the mechanism of CDC34 in regulating EGFR, we performed CO-immunoprecipitation (CO-IP) assay and found that EGFR was precipitated by a monoclonal anti-CDC34 antibody in H460, H1975, and HCC827 cells (Fig. 5a). Consistently, CDC34 was detected in a monoclonal anti-EGFR antibody-precipitated proteins (Fig. 5a). The immunofluorescence analysis showed that CDC34 colocalized with EGFR mainly in intracelluar region near the cell membrane (Fig. 5b). In H460 cells with EGFR-silenced by siEGFR, ectopic expressed Flag-EGFR interacted with HA-CDC34 (Fig. 5c). To confirm these findings, purified GST-CDC34 was co-incubated with lysates of H460 and H1975 cells, and precipitated EGFR was detected by immunoblot (Fig. 5d). On the other hand, the purified GST-EGFR (from bacteria E.Coli) precipitated CDC34 from H460 cell lysates (Fig. 5e). In H460 and 293T cells, CDC34 was capable of interacting with EGFR in the presence and absence of EGF stimulation (Supplementary Fig. 4a and b), suggesting that CDC34-EGFR interaction was independent of the phosphorylation status of EGFR.
Fig. 5.
CDC34 interacts directly with EGFR. (a) Co-IP and immunoblotting assays using indicated antibodies and cell lysates. (b) Immunofluorescence assays of H460 cells using antibodies against CDC34 (red) and EGFR (green), and 4′,6-diamidino-2-phenylindole (DAPI) to counterstain the nucleus (blue). (c) Co-IP and immunoblotting assays using indicated antibodies and lysates of H460 cells transfected with siEGFR, Flag-EGFR, and HA-CDC34. (d) Purified GST or GST-CDC34 was incubated with lysates of NSCLC cells in vitro to detect endogenous EGFR using indicated antibodies. (e) GST or GST-EGFRICD was incubated with lysates of H460 in vitro to detect endogenous CDC34 using indicated antibodies. ICD, intracellular domain. (f) The cells were transfected with indicated plasmids, lyzed, and Co-IP and immunoblot assays were performed using indicated antibodies. Schematic representation of EGFR protein shown at upper panel. ET, extracellular domain; (g) In vitro GST pull-down assays and immunoblot experiments using purified GST-CDC34 and His-EGFRICD proteins and indicated antibodies. GST- and His-tagged proteins were isolated from E. coli transfected with respective plasmids. (h) In vitro GST pull-down assays and immunoblot experiments using purified GST-EGFRICD and His-CDC34 proteins and indicated antibodies. GST- and His-tagged proteins were isolated from E. coli transfected with respective plasmids. (i) Schematic representation of CDC34 truncted mutants. (j) Indicated plasmids were transfected into E. Coli, proteins were purified, and His pull-down and immunoblotting assays were performed using indicated antibodies.
To determine the CDC34-binding region within EGFR, several deletion mutants were constructed, and the results showed that the intracellular domain (EGFRICD) could bind CDC34 in cells (Fig. 5f). In vitro assays using purified GST-CDC34 and His-EGFRICD (Fig. 5g) or GST-EGFRICD/His-CDC34 (Fig. 5h) harvested from bacteria E. Coli confirmed the direct binding of the two proteins, while GST itself did not interact with EGFR or CDC34. To unveil the region of CDC34 that binds to EGFR, deletion mutants were designed (Fig. 5i) and transfected into E. coli, and in vitro His-pull down and immunoblotting assays were performed. We found that while the C-terminal (162–236 amino acids) or the mutant lacking E2 catalytic domain could not bind, the full-length and the N-terminal of CDC34 (1–162 amino acids) bound EGFR (Fig. 5j). The active site residue Cys93 was not required for interaction with EGFR, because mutations in this amino acid (C93A) did not affect CDC34-EGFR interaction (Supplementary Fig. 4c).
3.7. CDC34 protects EGFR from proteolytic degradation
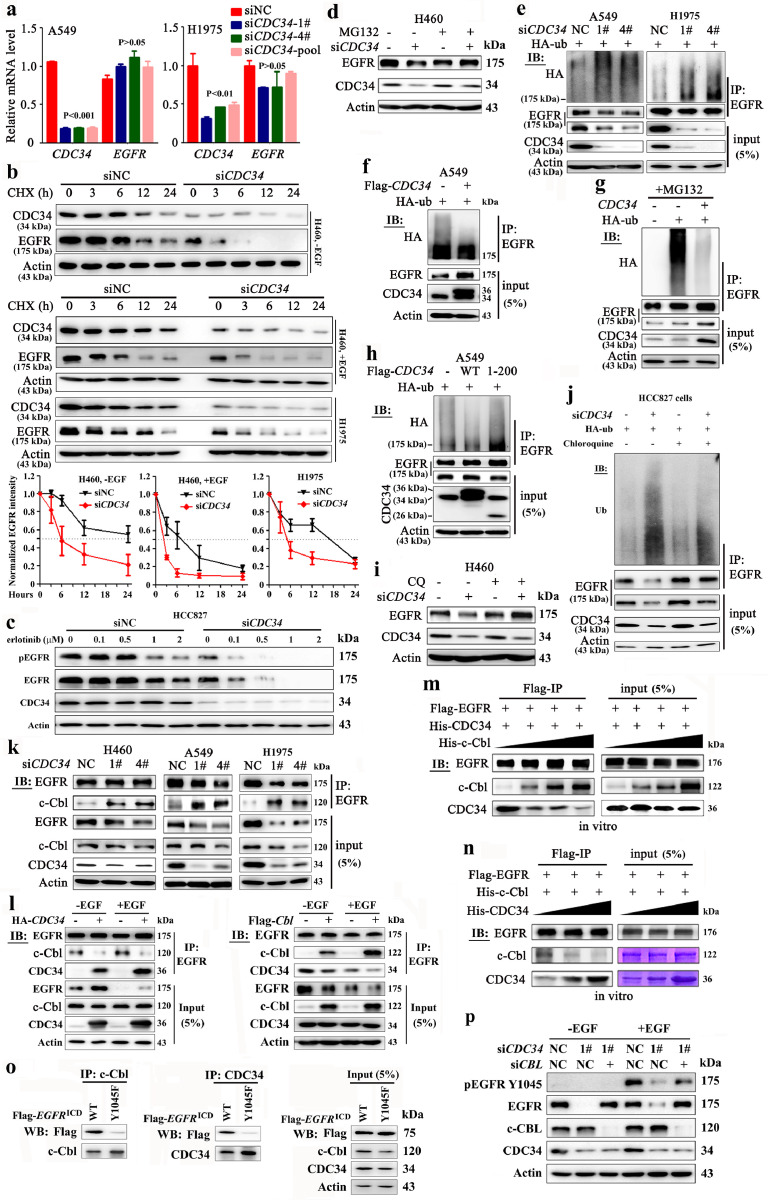
We noticed that knockdown of CDC34 in A549 and H1975 cells resulted in downregulation of EGFR at protein (Fig. 3d, f, Supplementary Fig. 3) but not mRNA level (Fig. 6a), suggestive of a role for CDC34 in stabilizing EGFR. The cycloheximide (CHX) chase assay was conducted in H460 cells in the absence and presence of EGF, and the half-life of EGFR was calculated [33]. We reported that knockdown of CDC34 led to downregulation of EGFR (Fig. 6b), and the half-life of EGFR in the absence of EGF was 3.9 and 2.6 h for siNC and siCDC34 treatment groups, respectively. In the presence of EGF, the half-life of EGFR was 2.9 and 1.5 h for siNC and siCDC34 treatment groups, respectively. Knockdown of CDC34 also reduced EGFR half-life in H1975 cells (Fig. 6b; 3.15 and 1.75 h for siNC and siCDC34 treatment groups, respectively). In HCC827 cells co-incubated with EGFR tyrosine kinase inhibitor erlotinib, siCDC34 drastically inhibited pEGFR and downregulated EGFR expression (Fig. 6c). In HCC827 cells without EGF co-incubation, silencing of CDC34 led to increased recruitment of c-Cbl by EGFR; upon erlotinib treatment, knockdown of CDC34 reduced EGFR/c-Cbl interaction (Supplementary Fig. 5a). We further showed that forced expression of CDC34 increased the stability of EGFR in H460 cells (Supplementary Fig. 5b).
Fig. 6.
CDC34 inhibits the ubiquitination of EGFR and protects it from proteolytic degradation. (a) qRT-PCR analysis of CDC34 and EGFR in A549 and H1975 cells 72 h after siCDC34 transfection. Error bars, sd; P values, Student's t-test. (b) Cycloheximide (CHX) chase assay to measure EGFR half-life in H460 (in the absence and presence of EGF) and H1975 cells transfected with siCDC34-1# (lower panel). The relative expression values of EGFR were the results determined by densitometry analysis normalized to Actin. Error bars, sd. (c) HCC827 cells were transfected with 50 nM siCDC34-1# for 24 h, treated with erlotinib for 24 h, and lyszed for Western blot assays using indicated antibodies. (d) Western blot analysis of the EGFR expression in H460 cells transfected with siCDC34-1# in the presence or absence of MG132. (e) A549 and H1975 cells were transfected with siCDC34 and HA-Ub, lysed, and the lysates were subjected to immunoprecipitation (IP) and immunoblot using indicated antibodies. (f) A549 cells were transfected with Flag-CDC34 and HA-Ub, and lysed for IP and immunoblot assays. (g) H460 cells were transfected with indicated constructs, treated with MG132, and lysed for IP and immunoblot assays. (h) The cells were transfected with CDC34 and HA-Ub, and lysed for IP and immunoblot assays using indicated antibodies. (i) Western blot analysis of EGFR in #1 siCDC34-transfected H460 cells in the presence or absence of chloroquine (CQ). (j) HCC827 cells were transfected with siCDC34 and HA-Ub for 24 h, treated with or without chloropuine for additional 24 h, lyzed, and subjected to IP and immunoblot using indicated antibodies. (k) H460, A549, and H1975 cells transfected with siCDC34 were lysed and subjected to Co-IP and immunoblotting using indicated antibodies. (l) H460 cells were transfected with CDC34 (left) or CBL (right) in the absence or presence of EGF for 48 h, and lysed for Co-IP and immunoblot using indicated antibodies. (m) In vitro Co-IP experiments using Flag-EGFR purified from 293T cells, His-CDC34 (from E. coli), and increasing amount of purified His-c-Cbl (from E. coli) proteins and indicated antibodies. (n) In vitro Co-IP experiments using Flag-EGFR purified from 293T cells, His-c-Cbl (from E. coli), and increasing amount of purified His-CDC34 (from E. coli) proteins and indicated antibodies. (o) 293T cells were transfected with the wild type or mutant (Y1045F) Flag-EGFRICD for 48 h, lyzed, and subjected to IP and immunobloting using indicated antibodies. (p) siCDC34-mediated loss of EGFR is rescued by knockdown of CBL. A549 cells were transfected with siCDC34 and/or siCBL for 72 h. Before lysis for immunoblot assays, the cells were serum-starved for 3 h and then stimulated by 50 ng/mL EGF for 15 min.
EGFR turnover is controlled by proteasome and lysosome pathways [5–7], whereas CDC34 undergoes proteasomal degradation [34]. We found that the proteasome/lysosome inhibitor MG132 inhibited siCDC34-induced down-regulation of CDC34 as well as EGFR (Fig. 6d). A more specific proteasome inhibitor epoxomicin also inhibited siCDC34-induced downregulation of EGFR (Supplementary Fig. 5c). Indeed, silencing of endogenous CDC34 increased the ubiquitination level of EGFR in A549 and H1975 cells (Fig. 6e), whereas exogenous expression of CDC34 reduced its ubiquitination in the absence (Fig. 6f) or presence of MG132 (Fig. 6g). Furthermore, compared with wild-type CDC34, a truncated mutant protein CDC341-200 failed to suppress EGFR ubiquitination in the absence (Fig. 6h) and presence of MG132 (Supplementary Fig. 5d), and failed to maintained EGFR levels in CDC34-silenced H460 cells (Supplementary Fig. 5e), suggesting that the intact CDC34 is required for CDC34 essential function [35]. The lysosome inhibitor chloroquine (CQ) also inhibited siCDC34-caused downregulation of EGFR at protein level (Fig. 6i), and suppressed EGFR ubiquitination in HCC827 cells transfected with siCDC34 (Fig. 6j).
c-Cbl was reported to mediate the degradation of EGFR [36]. By using the CO-IP experiment, we found that siCDC34 drastically enhanced c-Cbl-EGFR interaction in H460, A549, and H1975 cells (Fig. 6k). In contrast, the protein level of c-Cbl that binds EGFR was reduced upon CDC34 overexpression, in the absence and presence of EGF (Fig. 6l, left panel; supplementary Fig. 5f). When c-Cbl was overexpressed in H460 cells, EGFR-CDC34 binding was decreased, in the absence and presence of EGF (Fig. 6l, right panel). In in vitro experiments using His-CDC34 purified from E. coli and Flag-EGFR from 293T cells, we found that increased levels of c-Cbl supressed CDC34-EGFR interaction (Fig. 6m), while increased dosages of CDC34 diminished c-Cbl-EGFR binding (Fig. 6n). In vitro experiments using GST-EGFR and His-CDC34/His-c-Cbl from E. coli confirmed that c-Cbl competed with CDC34 to bind EGFR (Supplementary Fig. 5 g). c-Cbl binds to Y1045 and Y1045F mutation in EGFR abrogated binding between EGFR and c-Cbl tyrosine kinase binding (TKB) domain [37]. We found that Y1045F mutation in EGFR also impaired the interaction between EGFR and CDC34 (Fig. 6o), indicating that Y1045 is the binding site for both c-Cbl and CDC34. siCDC34-mediated loss of EGFR was rescued by knockdown of CBL (Fig. 6p), indicating that c-Cbl is critical for CDC34-mediated EGFR stabilization.
3.8. EGFR is critical to CDC34-induced cell proliferation
CDC34 is a key regulator of cell cycle. We tested the effects of CDC34 knockdown on NSCLC cells, and reported that siCDC34 treatment of H1975 and A549 cells blocked cell cycle progression at G1 phase (Fig. 7a) but did not induced significant apoptosis (Supplementary Fig. 6a). Lin et al. showed that nuclear localized EGFR functions as a transcription factor to activate genes such as Cyclin D1 that are required for highly proliferating activities [38]. EGFR also negatively regulates p27 [39]. We showed that silencing of CDC34 resulted in downregulation of Cyclin D1 and upregulation of p27 in the cells (Fig. 7b). Indeed, knowdown of CDC34 in H1975 (Fig. 7c) and HCC827 (Supplementary Fig. 6b) cells led to downregulation of EGFR and Cyclin D1 in both the cytoplasm and nucleus compartments. Transfection of siCDC34 into the cells reduced CDC34 and Cyclin D1 (CCND1) at mRNA level (Fig. 7d and Supplementary Fig. 6c). Silencing of CDC34 or EGFR by siRNA also induced downregulation of Cyclin D1 and upregulation of p27 (Fig. 7e), arrested cell cycle at G1 phase (Fig. 7f), inhibited cell viability (Fig. 7g), and suppressed colony forming activity (Fig. 7h and i) of H460 cells. Interesting, transfection of EGFR into siCDC34/siEGFR-treated H460 cells rescued the above inhibitory effects (Fig. 7e–i), indicating that downregulation of EGFR has a critical role in the anti-proliferative effects of CDC34 silencing.
Fig. 7.
CDC34 regulates Cyclin D1 and promotes lung cancer via EGFR oncoprotein. (a) The cells were transfected with siCDC34 and cell cycle distribution was determined by propidium iodide staining for DNA content followed by flow cytometric analysis. Error bars, sd; P values, Student's t-test. (b) The cells were transfected with siCDC34, lyzed 48 h later, and the expression of CDC34, EGFR, and Cyclin D1 were tested by Western blot. (c, d) H1975 cells were transfected with siCDC34, lysed 48 h later, and the cytoplasm and nucleus proteins were harvested for immunoblotting using indicated antibodies (c). The expression of CDC34 and CCND1 at mRNA level was tested by qRT-PCR (d). Error bars, sd; P values, Student's t-test. (e–i) H460 cells were transfected with siCDC34-1#, siEGFR, and Flag-EGFR, lyzed, and the lysates were subjected to immunoblot using indicated antibodies (e). The cell cycle distribution (f), cell viability (g), and colony forming activity (h, i) were tested. Error bars, sd; P values, Student's t-test.
3.9. Inhibition of CDC34 suppresses EGFR L858R-driven lung cancer
To evaluate the therapeutic potential of CDC34 inhibition, an EGFRL858R-driven lung cancer mouse model was established as described [40], and the lentiviral particles containing short hairpin RNA targeting CDC34 (shCDC34-1#) were generated and intranasally administrated into the lungs of the mice before they were treated with doxycycline (DOX) to induce lung cancer [40]. One month after the initiation of DOX treatment, the mice were detected by microscopic computed tomography (micro-CT). We found disseminated tumors in the lungs of control group mice, but only small tumors were seen in the CDC34 knockdown group mice (Fig. 8a and Supplementary Fig. 7a). Tumor volume of the mice was quantitated by the Analyze 12.0 (PerkinElmer) Caliper microCT Analysis Tools, and the results showed that shCDC34 treatment significantly reduced tumor volume of the mice (Fig. 8a, right panel). Histological examination of the lungs demonstrated that the shCDC34-treated mice developed lesions in alveoli, whereas the shNC group mice harbored disseminated adenocarcinomas (Fig. 8b). qRT-PCR, immunoblot, and IHC assays of the lung specimens revealed the downregulation of CDC34 at both mRNA and protein levels (Fig. 8b and c) and the decrease in EGFR as well as Ki67 (Fig. 8c) in shCDC34-treated mice. Western blot analysis further showed that DOX administration induced the expression of EGFR, and silencing of CDC34 downregulated EGFR, pEGFR, pAKT, pERK, and Cyclin D1, and upregulated p27 in lungs of the mice (Fig. 8d). In addition, the overall survival of the shCDC34 group mice was significantly prolonged as compared to shNC-treated mice (Fig. 8e).
Fig. 8.
Knockdown of CDC34 significantly suppresses L858R- and T790M-EGFR-driven lung cancer. (a) The EGFRL858R-transgenic mice were intranasally infected with virus particles containing shNC or shCDC34-1#, treated with DOX, scanned by micro-CT (left), and tumor volume was quantitated by microCT Analysis Tools (right). P values, Student's t-test. ***, P < 0.001. (b) Hematoxylin-eosin staining of the lung sections from mice of each group. The expression of CDC34 at both mRNA and protein levels in the lung tissues was tested by qRT-PCR (left panel) and Western blot (right panel), respectively, and the results were shown. Error bars, sd; P values, Student's t-test. **, P < 0.01. (c) IHC analysis of CDC34, Ki67, and EGFR in tumors from mice of each group. Scale bar, 1 mm. (d) The expression of indicated proteins in the tumor samples from the mice of each group was detected by Western blot. (e) Kaplan–Meier survival curve of the mice. P value, log-rank test. (f) The EGFRT790M/Del (exon19)-transgenic mice were intranasally infected with viral particles containing shNC or shCDC34-1#, treated with DOX, sacrified one month later, and the lung tissue lysates were subjected to immunoblot to detect the expression of CDC34 and EGFR. (g) The EGFRT790M/Del (exon19)-transgenic mice were intranasally infected with virus particles, treated with DOX, scanned by micro-CT (upper panel), and tumor volume was quantitated by microCT Analysis Tools (lower panel). P values, Student's t-test. ***, P < 0.001. (h) Hematoxylin-eosin staining of the lung sections from mice of each group. Scale bar, 1 mm. (i) IHC analysis of CDC34, Ki67, and EGFR in lung tumors from the mice. Scale bar, 1 mm. (j) The expression of indicated proteins in the tumor samples from the mice of each group. (k) Kaplan–Meier survival curve of the mice. P value, log-rank test. (l) Schematic representation of CDC34 in lung epithelial cells.
3.10. Knockdown of CDC34 inhibits EGFRT790M/Del (E746-A750) -driven lung cancer
The EGFR T790M mutation is associated with acquired resistance to erlotinib in NSCLCs. We tested the effects of shCDC34 on Tet-op-EGFRT790M/Del (E746-A750)/CCSP-rtTA transgenic mice [41]. Interestingly, we found that silencing of CDC34 (Fig. 8f) led to alleviated lung carcinogenesis in the mice, reflected by micro-CT (Fig. 8g, Supplementary Fig. 7b) and histological examination (Fig. 8h). Knockdown of CDC34 also reduced Ki67 (Fig. 8i) and downregulated the expression of EGFR, pEGFR, pAKT, pERK, and Cyclin D1, and upregulated p27 (Fig. 8j) in tumor tissues of the mice. The overall survival of the shCDC34 treatment group mice was also significantly prolonged compared to shNC-treated mice (Fig. 8k).
4. Discussion
In this study, we used a large-scale siRNA screening to identify UPGs that are critical to lung carcinogenesis, and unveiled 31 candidates that were required for proliferation of A549 and H1975 cells (Fig. 1, Supplementary Fig. 1). Among them, CDC34 represented the most significant one, which was elevated in 76 of 114 (66.7%) NSCLCs and was inversely associated with clinical outcome of the patients (Table 1, Fig. 1). CDC34 was overexpressed in acute lymphoblastic leukemia [42], multiple myeloma [43], breast cancer [44], hepatocellular carcinomas [45,46], and prostate cancer [47], indicating that this E2 conjugase palys a critical role in tumorigenesis.
Cigarette smoking is responsible for more than 1.44 million lung cancer deaths each year worldwide [48]. Tobacco smoke causes genomic mutations in tumors [49] and counterpart normal controls [50], [51], promotes cell proliferation, inhibits programmed cell death, facilitates angiogenesis, invasion and metastasis potentials, and enhances tumor promoting inflammation [52], [53], [54]. We found that CDC34 was overexpressed in 44 of 59 (74.6%) smoker NSCLCs and in 23 of 43 (53.5%) nonsmoker patients (P = 0.027; Table 1). In another cohort [25], CDC34 expression in smokers was significantly higher than in nonsmokers (Fig. 1). These results suggest that tobacco smoke may induce CDC34 expression to maintain hyperactivation of EGFR oncoprotein, thus contributing to lung carcinogenesis. This possibility was confirmed by the findings that tobacco smoke caused elevation of CDC34 in A/J mice, and tobacco carcinogens BaP, BAA, and DBA induced upregulation of CDC34 in the cells (Fig. 1). Therefore, tobacco may induce lung carcinogenesis in an unexpected way by perburbing the expression of genes like CDC34, IRX5 [55], and RFWD3 [56], which were identified by large-scale screening (Supplementary Table 1).
CDC34 functions as a K48 Ub chain-building enzyme and cognate E2 of SCF E3 ligases, and collaborates with HECT-, RING-, and RING-between-RING (RBR)-E3s to control proteolysis of substrates [57]. So far, no evidence suggests a role for CDC34 in EGFR turnover, which is tightly controlled by Ubc4/5/c-Cbl cascade [3]. We showed that knockdown of CDC34 resulted in downregulation of EGFR, whereas ectopic expression of CDC34 led to upregulation of EGFR and increase in its tyrosine kinase activity (Figs. 3 and 6). The downstream signaling molecules, pAKT and pERK, were accordingly affected (Fig. 3). Mechanistically, the E2 catalytic domain of CDC34 bound EGFR at its ICD region, and competed with c-Cbl to bind EGFR at Y1045 and inhibited the K48-linked polyubiquitination and subsequent degradation of the substrate (Fig. 8l). In cellular and animal models, silencing of CDC34 suppressed, while overexpression of CDC34 promoted, lung cancer cell proliferation (Figs. 2, 3, 4). The inhibitory effects of CDC34 on NSCLC cells was mediated by EGFR, since ectopic expression of EGFR rescued siCDC34-induced suppression of the cells (Fig. 7). These results demonstrate previously unreported functions of CDC34, and indicate that an E2 Ub conjugase can compete with E3 ligases to protect proteolysis of substrates.
CDC34 exerts oncogenic or tumor-promoting functions, and has been used as a therpeutic target for drug development [58]. Inhibition of CDC34 enhances anti-myeloma activity of proteasome inhibitor bortezomib [43] and contributes to the chemopreventive activity of Chinese herbs (anti-tumor B, ATB) in mouse model of lung cancer [59]. We showed that silencing of CDC34 inhibited cell proliferation and colony forming activity of NSCLC cells in vitro (Figs. 2 and 3), and suppressed tumor growth and prolonged lifespan of xenograft NSCLC mouse models in vivo (Fig. 4). In an EGFRL858R-driven mouse lung adenocarcinoma model, shCDC34 treatment significantly inhibited cancer progression and prolonged survival time of the mice (Fig. 8). These results indicate that CDC34 represents a rational drug target for NSCLC. Moreover, treatment of the patients with gefitinib and erlotinib will fail because of the development of EGFR T790M mutation [60], which affects the gatekeeper residue in the kinase catalytic domain and weakens the interaction of the inhibitors with the target [61]. Efforts have been made to overcome drug resistance [62,63]. Here, we showed that silencing of CDC34 dramatically suppressed tumor dissemination and cell proliferation in EGFRT790M/Del (exon 19)-driven lung cancer (Fig. 8). Knockdown of CDC34 downregulated the expression of EGFR, pEGFR, pAKT, and pERK in EGFRT790M/Del (exon 19) mice (Fig. 8). These data demonstrate that silencing of CDC34 can overcome T790M mutation-caused drug resistance, therefore represents an attrative therapeutic target in lung cancer. Preclinical and clinical studies of using CDC34 inhibitor to treat lung cancer with or without EGFR mutation warrant intensive investigation.
Declaration of Competing Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2016YFC0905501), the Key Project of the National Natural Science Foundation of China (81830093), the National Natural Science Funds for Distinguished Young Scholar (81425025), the National Natural Science Foundation of China (81672765 and 81802796), and the CAMS Innovation Fund for Medical Sciences (CIFMS; No. 2019-I2M-1-003). The study sponsor had no role in the design of the study; the data collection, analysis, or interpretation; the writing of the article; or the decision to submit for publication.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102689.
Appendix. Supplementary materials
References
- 1.Cataldo V.D., Gibbons D.L., Perez-Soler R., Quintas-Cardama A. Treatment of non-small-cell lung cancer with erlotinib or gefitinib. N Engl J Med. 2011;364(10):947–955. doi: 10.1056/NEJMct0807960. [DOI] [PubMed] [Google Scholar]
- 2.Paez J.G., Jänne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Umebayashi K., Stenmark H., Yoshimori T. Ubc4/5 and c-Cbl continue to ubiquitinate egf receptor after internalization to facilitate polyubiquitination and degradation. Mol Biol Cell. 2008;19(8):3454–3462. doi: 10.1091/mbc.E07-10-0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levkowitz G., Waterman H., Ettenberg S.A., Katz M., Tsygankov A.Y., Alroy I. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4(6):1029–1040. doi: 10.1016/s1097-2765(00)80231-2. PubMed PMID: 10635327. Epub 2000/01/15. [DOI] [PubMed] [Google Scholar]
- 5.Levkowitz G., Waterman H., Zamir E., Kam Z., Oved S., Langdon W.Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12(23):3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan L., Miura Y., Dimri M., Majumder B., Dodge I.L., Reddi A.L. Cbl-mediated ubiquitinylation is required for lysosomal sorting of epidermal growth factor receptor but is dispensable for endocytosis. J Biol Chem. 2003;278(31):28950–28960. doi: 10.1074/jbc.M304474200. [DOI] [PubMed] [Google Scholar]
- 7.Visser Smit G.D., Place T.L., Cole S.L., Clausen K.A., Vemuganti S., Zhang G. Cbl controls egfr fate by regulating early endosome fusion. Sci Signal. 2009;2(102):ra86. doi: 10.1126/scisignal.2000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lill N.L., Douillard P., Awwad R.A., Ota S., Lupher M.L., Miyake S. The evolutionarily conserved N-terminal region of cbl is sufficient to enhance down-regulation of the epidermal growth factor receptor. J Biol Chem. 2000;275(1):367–377. doi: 10.1074/jbc.275.1.367. [DOI] [PubMed] [Google Scholar]
- 9.Visser G.D., Lill N.L. The cbl ring finger C-terminal flank controls epidermal growth factor receptor fate downstream of receptor ubiquitination. Exp Cell Res. 2005;311(2):281–293. doi: 10.1016/j.yexcr.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 10.Tan Y.-H.C., Krishnaswamy S., Nandi S., Kanteti R., Vora S., Onel K. CBL is frequently altered in lung cancers: its relationship to mutations in met and egfr tyrosine kinases. PLoS One. 2010;5(1):e8972. doi: 10.1371/journal.pone.0008972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6(1):79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 12.Choi Y.-.S., Wu K., Jeong K., Lee D., Jeon Y.H., Choi B.-.S. The human cdc34 carboxyl terminus contains a non-covalent ubiquitin binding activity that contributes to SCF-dependent ubiquitination. J Biol Chem. 2010;285(23):17754–17762. doi: 10.1074/jbc.M109.090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gazdoiu S., Yamoah K., Wu K., Pan Z.-.Q. Human cdc34 employs distinct sites to coordinate attachment of ubiquitin to a substrate and assembly of polyubiquitin chains. Mol Cell Biol. 2007;27(20):7041–7052. doi: 10.1128/MCB.00812-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butz N., Ruetz S., Natt F., Hall J., Weiler J., Mestan J. The human ubiquitin-conjugating enzyme cdc34 controls cellular proliferation through regulation of p27Kip1 protein levels. Exp Cell Res. 2005;303(2):482–493. doi: 10.1016/j.yexcr.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Wei Y., Jiang J., Liu D., Zhou J., Chen X., Zhang S. Cdc34-mediated degradation of ATF5 is blocked by cisplatin. J Biol Chem. 2008;283(27):18773–18781. doi: 10.1074/jbc.M707879200. [DOI] [PubMed] [Google Scholar]
- 16.Schwob E., Böhm T., Mendenhall M.D., Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to s transition in S. cerevisiae. Cell. 1994;79(2):233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 17.Strack P., Caligiuri M., Pelletier M., Boisclair M., Theodoras A., Beer-Romero P. SCFβ-TRCP and phosphorylation dependent ubiquitination of iκbα catalyzed by ubc3 and ubc4. Oncogene. 2000;19:3529. doi: 10.1038/sj.onc.1203647. [DOI] [PubMed] [Google Scholar]
- 18.Skowyra D., Koepp D.M., Kamura T., Conrad M.N., Conaway R.C., Conaway J.W. Reconstitution of G1 cyclin ubiquitination with complexes containing scf grr1 and rbx1. Science. 1999;284(5414):662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 19.Macdonald M., Wan Y., Wang W., Roberts E., Cheung T.H., Erickson R. Control of cell cycle-dependent degradation of c-Ski proto-oncoprotein by cdc34. Oncogene. 2004;23(33):5643–5653. doi: 10.1038/sj.onc.1207733. [DOI] [PubMed] [Google Scholar]
- 20.Verma R., Feldman R.M., Deshaies R.J. SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol Biol Cell. 1997;8(8):1427–1437. doi: 10.1091/mbc.8.8.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malo N., Hanley J.A., Cerquozzi S., Pelletier J., Nadon R. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;24:167. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 22.Wang G.-.Z., Zhang L., Zhao X.-.C., Gao S.-.H., Qu L.-.W., Yu H. The aryl hydrocarbon receptor mediates tobacco-induced PD-L1 expression and is associated with response to immunotherapy. Nat Commun. 2019;10(1):1125. doi: 10.1038/s41467-019-08887-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gyorffy B., Surowiak P., Budczies J., Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okayama H., Kohno T., Ishii Y., Shimada Y., Shiraishi K., Iwakawa R. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72(1):100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 25.Selamat S.A., Chung B.S., Girard L., Zhang W., Zhang Y., Campan M. Genome-scale analysis of dna methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22(7):1197–1211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhodes D.R., Kalyana-Sundaram S., Mahavisno V., Varambally R., Yu J., Briggs B.B. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garber M.E., Troyanskaya O.G., Schluens K., Petersen S., Thaesler Z., Pacyna-Gengelbach M. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98(24):13784–13789. doi: 10.1073/pnas.241500798. PubMed PMID: PMC61119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landi M.T., Dracheva T., Rotunno M., Figueroa J.D., Liu H., Dasgupta A. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3(2):e1651. doi: 10.1371/journal.pone.0001651. PubMed PMID: PMC2249927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su L.-.J., Chang C.-.W., Wu Y.-.C., Chen K.-.C., Lin C.-.J., Liang S.-.C. Selection of DDX5 as a novel internal control for q-rt-pcr from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. PubMed PMID: PMC1894975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stearman R.S., Dwyer-Nield L., Zerbe L., Blaine S.A., Chan Z., Paul A. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 2005;167(6):1763–1775. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Network TCGAR Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pagano M., Tam S., Theodoras A., Beer-Romero P., Del Sal G., Chau V. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269(5224):682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 33.Belle A., Tanay A., Bitincka L., Shamir R., O'Shea E.K. Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci USA. 2006;103(35):13004–13009. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagglund R., Van Sant C., Lopez P., Roizman B. Herpes simplex virus 1-infected cell protein 0 contains two E3 ubiquitin ligase sites specific for different E2 ubiquitin-conjugating enzymes. Proc Natl Acad Sci USA. 2002;99(2):631–636. doi: 10.1073/pnas.022531599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolman C.J., Toth J., Gonda D.K. Identification of a portable determinant of cell cycle function within the carboxyl-terminal domain of the yeast CDC34 (UBC3) ubiquitin conjugating (E2) enzyme. EMBO J. 1992;11(8):3081–3090. doi: 10.1002/j.1460-2075.1992.tb05380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubin C., Gur G., Yarden Y. Negative regulation of receptor tyrosine kinases: unexpected links to c-Cbl and receptor ubiquitylation. Cell Res. 2005;15:66. doi: 10.1038/sj.cr.7290268. [DOI] [PubMed] [Google Scholar]
- 37.Takeshita K., Tezuka T., Isozaki Y., Yamashita E., Suzuki M., Kim M. Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of cbl-c. J Biochem. 2012;152(5):487–495. doi: 10.1093/jb/mvs085. [DOI] [PubMed] [Google Scholar]
- 38.Lin S.-.Y., Makino K., Xia W., Matin A., Wen Y., Kwong K.Y. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 39.Lenferink A.E.G., Simpson J.F., Shawver L.K., Coffey R.J., Forbes J.T., Arteaga C.L. Blockade of the epidermal growth factor receptor tyrosine kinase suppresses tumorigenesis in MMTV/Neu + MMTV/TGF-α bigenic mice. Proc Natl Acad Sci USA. 2000;97(17):9609–9614. doi: 10.1073/pnas.160564197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Politi K., Zakowski M.F., Fan P.-.D., Schonfeld E.A., Pao W., Varmus H.E. Lung adenocarcinomas induced in mice by mutant EGF receptors foundin human lung cancers respondto a tyrosine kinase inhibitor orto down-regulation of the receptors. Genes Dev. 2006;20(11):1496–1510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li D., Shimamura T., Ji H., Chen L., Haringsma H.J., McNamara K. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant egfr respond to HKI-272 and rapamycin combination therapy. Cancer Cell. 2007;12(1):81–93. doi: 10.1016/j.ccr.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 42.Eliseeva E., Pati D., Diccinanni M.B., Yu A.L., Mohsin S.K., Margolin J.F. Expression and localization of the CDC34 ubiquitin-conjugating enzyme in pediatric acute lymphoblastic leukemia. Cell Growth Differ. 2001;12(8):427–433. [PubMed] [Google Scholar]
- 43.Chauhan D., Li G., Hideshima T., Podar K., Shringarpure R., Mitsiades C. Blockade of ubiquitin-conjugating enzyme CDC34 enhances anti-myeloma activity of bortezomib/proteasome inhibitor PS-341. Oncogene. 2004;23(20):3597–3602. doi: 10.1038/sj.onc.1207458. [DOI] [PubMed] [Google Scholar]
- 44.Price G.R., Armes J.E., Ramus S.J., Provenzano E., Kumar B., Cowie T.F. Phenotype-directed analysis of genotype in early-onset, familial breast cancers. Pathology. 2006;38(6):520–527. doi: 10.1080/00313020601024052. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka K., Kondoh N., Shuda M., Matsubara O., Imazeki N., Ryo A. Enhanced expression of mRNAs of antisecretory factor-1, gp96, DAD1 and CDC34 in human hepatocellular carcinomas. Biochim Biophys Acta. 2001;1536(1):1–12. doi: 10.1016/s0925-4439(01)00026-6. [DOI] [PubMed] [Google Scholar]
- 46.Takagi K., Takayama T., Midorikawa Y., Hasegawa H., Ochiai T., Moriguchi M. Cell division cycle 34 is highly expressed in hepatitis c virus-positive hepatocellular carcinoma with favorable phenotypes. Biomed Rep. 2017;7(1):41–46. doi: 10.3892/br.2017.912. PubMed PMID: PMC5492648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng Y., Abdallah A., Lu J.-P., Wang T., Chen Y.-H., Terrian D.M. δ-Catenin promotes prostate cancer cell growth and progression by altering cell cycle and survival gene profiles. Mol. Cancer. 2009;8(1):19. doi: 10.1186/1476-4598-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. PubMed PMID: 30207593. [DOI] [PubMed] [Google Scholar]
- 49.Alexandrov L.B., Ju Y.S., Haase K., Van Loo P., Martincorena I., Nik-Zainal S. Mutational signatures associated with tobacco smoking in human cancer. Science. 2016;354(6312):618–622. doi: 10.1126/science.aag0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang D., Qu L., Zhou B., Wang G., Zhou G. Genomic variations in the counterpart normal controls of lung squamous cell carcinomas. Front Med. 2018;12(3):280–288. doi: 10.1007/s11684-017-0580-1. PubMed PMID: 29185122. [DOI] [PubMed] [Google Scholar]
- 51.Qu L.W., Zhou B., Wang G.Z., Chen Y., Zhou G.B. Genomic variations in paired normal controls for lung adenocarcinomas. Oncotarget. 2017;8(61):104113–104122. doi: 10.18632/oncotarget.22020. PubMed PMID: 29262625. PMCID: PMC5732791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sobus S.L., Warren G.W. The biologic effects of cigarette smoke on cancer cells. Cancer. 2014;120(23):3617–3626. doi: 10.1002/cncr.28904. [DOI] [PubMed] [Google Scholar]
- 53.Zhou B., Wang G.-.Z., Wen Z.-.S., Zhou Y.-.C., Huang Y.-.C., Chen Y. Somatic mutations and splicing variants of focal adhesion kinase in non–small cell lung cancer. J Natl Cancer Inst. 2018;110(2):195–204. doi: 10.1093/jnci/djx157. [DOI] [PubMed] [Google Scholar]
- 54.Zhou G. Tobacco, air pollution, environmental carcinogenesis, and thoughts on conquering strategies of lung cancer. Cancer Biol Med. 2019;16:700–713. doi: 10.20892/j.issn.2095-3941.2019.0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang D.-L., Qu L., Ma L., Zhou Y.-C., Wang G.-Z., Zhao X.-C. Genome-wide identification of transcription factors that are critical to non-small cell lung cancer. Cancer Lett. 2018;434:132–143. doi: 10.1016/j.canlet.2018.07.020. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y., Zhao X., Zhou Y., Wang M., Zhou G. Identification of an E3 ligase-encoding gene, RFWD3, in non-small cell lung cancer. Front Med. 2019 doi: 10.1007/s11684-019-0708-6. [DOI] [PubMed] [Google Scholar]
- 57.Stewart M.D., Ritterhoff T., Klevit R.E., Brzovic P.S. E2 enzymes: more than just middle men. Cell Res. 2016;26:423. doi: 10.1038/cr.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ceccarelli D.F., Tang X., Pelletier B., Orlicky S., Xie W., Plantevin V. An allosteric inhibitor of the human cdc34 ubiquitin-conjugating enzyme. Cell. 2011;145(7):1075–1087. doi: 10.1016/j.cell.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Z., Wang Y., Yao R., Li J., Yan Y., Regina M.L. Cancer chemopreventive activity of a mixture of Chinese herbs (antitumor B) in mouse lung tumor models. Oncogene. 2004;23(21):3841–3850. doi: 10.1038/sj.onc.1207496. [DOI] [PubMed] [Google Scholar]
- 60.Kobayashi S., Boggon T.J., Dayaram T., Janne P.A., Kocher O., Meyerson M. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352(8):786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 61.Pao W., Miller V.A., Politi K.A., Riely G.J., Somwar R., Zakowski M.F. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the egfr kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jia Y., Yun C.H., Park E., Ercan D., Manuia M., Juarez J. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature. 2016;534(7605):129–132. doi: 10.1038/nature17960. PubMed PMID: 27251290. PMCID: PMC4929832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou W., Ercan D., Chen L., Yun C.H., Li D., Capelletti M. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462(7276):1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cancer microarray data was downloaded from the Oncomine datasets Okayama Lung (can also be found at the Gene Expression Omnibus, GSE31210), Selamat Lung (the Gene Expression Omnibus, GSE32867), Stearman Lung (the Gene Expression Omnibus, GSE2415), Landi Lung (the Gene Expression Omnibus, GSE10072), and Su Lung (the Gene Expression Omnibus, GSE7670). The data of the reverse phase protein array of 235 lung adenocarcinomas were downloaded from the Cancer Genomics Hub (CGHub) (https://cghub.ucsc.edu/) with approval by the National Institutes of Health (NIH; approval number #24437-4). All the remaining data supporting the findings of this study are available within this paper and its supplementary information.