Abstract
Background
Endothelial cells (ECs) function as an instructive platform to support haematopoietic stem cell (HSC) homeostasis. Our recent studies found that impaired bone marrow (BM) ECs are responsible for the defective haematopoiesis in patients with poor graft function (PGF), which is characterised by pancytopenia post-allotransplant. Although activated autophagy was reported to benefit ECs, whether EC autophagy plays a critical role in supporting HSCs and its effect on PGF patients post-allotransplant remain unclear.
Methods
To evaluate whether the autophagy status of ECs modulates their ability to support haematopoiesis, human umbilical vein endothelial cells (HUVECs) and primary BM ECs derived from healthy donors were subjected to knockdown or overexpression of Beclin-1 (an autophagy-related protein). Moreover, BM ECs derived from PGF patients were studied.
Findings
Beclin-1 knockdown significantly reduced the haematopoiesis-supporting ability of ECs by suppressing autophagy, which could be restored by activating autophagy via Beclin-1 upregulation. Moreover, autophagy positively regulated haematopoiesis-related genes in HUVECs. Subsequently, a prospective case-control study demonstrated that defective autophagy reduced Beclin-1 expression and the colony-forming unit (CFU) plating efficiency in BM ECs from PGF patients compared to matched patients with good graft function. Rapamycin, an autophagy activator, quantitatively and functionally improved BM ECs from PGF patients in vitro and enhanced their ability to support HSCs by activating the Beclin-1 pathway.
Interpretation
Our results suggest that the autophagy status of ECs modulates their ability to support haematopoiesis by regulating the Beclin-1 pathway. Defective autophagy in BM ECs may be involved in the pathogenesis of PGF post-allotransplant. Rapamycin provides a promising therapeutic approach for PGF patients.
Funding
Please see funding sources.
Keywords: Autophagy, Endothelial cells, Haematopoietic stem cells, Poor graft function, Allotransplant
Research in context.
Evidence before this study
We conducted a review of the published work relating to autophagy in endothelial cells (ECs) as it relates to the regulation their haematopoiesis-supporting ability before initiating this study on January 1, 2017. We repeated this review of the literature before submission of our paper on October 19, 2019. We conducted a systematic search on PubMed using the MeSH terms (“endothelial cells” [Title/Abstract] AND “autophagy” [Title/Abstract] AND “haematopoiesis” [Title/Abstract]). We did not restrict the search by date, language, or article type. We also screened the reference lists of articles identified for relevant titles. ECs, which have been identified as preferential elements of the bone marrow (BM) microenvironment, possess the capacity to support and regulate haematopoiesis in animal studies. The impaired BM ECs were recently reported to be responsible for the defective haematopoiesis in patients with poor graft function (PGF), which is characterised by pancytopenia post-allotransplant. Autophagy, an essential homeostatic process responsible for nutrient deprivation, can be activated as a cytoprotective mechanism. Although activated autophagy was reported to benefit ECs, whether EC autophagy plays a critical role in supporting HSCs and its effect on PGF patients post-allotransplant remain to be elucidated.
Added value of this study
To the best of our knowledge, this is the first study to demonstrate that Beclin-1 knockdown significantly reduced the haematopoiesis-supporting ability of ECs by suppressing autophagy, and this ability could be restored by activating autophagy through Beclin-1 upregulation. Subsequently, a prospective case-control study illustrated that defective autophagy in BM ECs may be involved in the pathogenesis of PGF post-allotransplant. The impaired haematopoiesis-supporting ability of ECs could be improved by activating autophagy in PGF patients, thus providing a promising therapeutic approach.
Implications of all the available evidence
The current study demonstrated for the first time that the autophagy status of ECs modulates their ability to support haematopoiesis via regulation of the Beclin-1 pathway. Defective autophagy in BM ECs may be involved in the pathogenesis of PGF post-allotransplant. Rapamycin could improve the impaired HSC-supporting ability of ECs by activating autophagy, thus providing a promising therapeutic approach for PGF patients post-allotransplant in the future.
Alt-text: Unlabelled box
1. Introduction
Endothelial cells (ECs), which line the interior of blood vessels, are not only passive channels for nutrient transfer but also scaffolds that provide essential paracrine signals for tissue maintenance and regeneration [1], [2], [3]. Emerging evidence from mouse studies indicates that ECs are key elements that support haematopoiesis in the bone marrow (BM) microenvironment [4], [5], [6], [7] by providing critical signals, such as colony stimulating factor-1 (CSF-1) [8], that regulate haematopoietic stem cells (HSCs). Moreover, EC infusion promoted haematopoietic recovery in mice [9]. Poor graft function (PGF), which is characterised by pancytopenia after allogeneic HSC transplantation (allo-HSCT), is an appropriate model of human BM failure for studying haematopoiesis [10], [11], [12], [13], [14], [15], [16], [17]. Our series of reports have shown that impaired BM ECs are responsible for defective haematopoiesis post-allotransplant [14,15,[17], [18], [19], [20]], whereas prophylactic strategies to improve BM ECs promoted haematopoietic reconstitution in this context [18], further validating the vital role of BM ECs in accelerating the haematopoietic recovery of engrafted donor HSCs post-allotransplant. These findings suggest that BM ECs play an essential role in regulating HSC homeostasis. However, the underlying mechanism by which ECs regulate HSCs remains unclear.
Autophagy is a cellular pathway involved in protein and organelle degradation and is primarily a protective cellular response [21]. Cytoprotective autophagy in ECs exerts an antithrombotic effect by regulating von Willebrand factor secretion [22]. Paradoxically, overactivated autophagy in ECs exacerbates vascular dysfunction by decreasing nitric oxide production [23]. Beclin-1, an autophagy-related protein, plays a critical role in the formation of autophagosomes [24], [25], [26]. In human umbilical vein endothelial cells (HUVECs), the activation of autophagy prevents cell death upon exposure to oxidative stress by activating Beclin-1 [27]. Moreover, rapamycin, a pharmacological activator of autophagy, was shown to attenuate cardiac hypertrophy by promoting autophagy via modulation of Beclin-1 expression [24]. Accumulating evidence has demonstrated the essential roles of ECs in supporting HSCs and of autophagy in regulating the antithrombotic function of ECs, raising the question of whether EC autophagy plays a critical role in supporting HSCs. Moreover, it is unknown whether the effect of autophagy on the HSC-supporting ability of ECs can be validated in a human pancytopenia model.
Therefore, the current study was performed to investigate whether the autophagy status in ECs regulates their ability to support haematopoiesis. Moreover, we evaluated the effect of EC autophagy on HSC support in PGF patients, and the results may illuminate a promising therapeutic target for PGF patients post-allotransplant.
2. Materials and methods
2.1. Cultivation of HUVECs
HUVECs were obtained from ScienCell Research Laboratories (Carlsbad, CA, USA) and cultured in EC medium supplemented with EC growth supplement, 5% foetal bovine serum, and penicillin/streptomycin solution (ScienCell, Carlsbad, CA, USA) at 37 °C in an incubator containing 5% CO2. HUVECs were treated with rapamycin (an autophagy activator, 5 μM) or hydroxychloroquine (HCQ, an autophagy inhibitor, 10 μM) for 6 h.
Adherent cells were washed with PBS and gently detached with trypsin containing 0.25% EDTA. The number of live cells per well was counted with trypan blue (Solarbio, Beijing, China) by independent investigators blinded to the group allocation.
2.2. Transfection of HUVECs or primary BM ECs derived from healthy donors
For Beclin-1 knockdown, small interfering RNA (siRNA) targeting Beclin-1 (sBECN) was synthesised (target sequence: 5′-GGAGCCATTTATTGAAACTTT-3′). HUVECs or BM ECs derived from healthy donors were plated at 4 × 105 cells/60-mm dish 18 h before transfection, which was performed using the Lipofectamine 3000 protocol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. For Beclin-1 overexpression, pcDNA3.1-BECN1 were transfected into HUVECs or BM ECs derived from healthy donors using Lipofectamine 3000. Forty-eight hours after transfection, the cells were harvested.
2.3. Functional analysis of HUVECs
The levels of apoptosis and reactive oxygen species (ROS) were analysed by flow cytometry as previously reported [14,16,19,28,29]. For intracellular protein detection, HUVECs were fixed, permeabilised, and incubated with antibodies against microtubule-associated protein 1A light chain 3 (LC3), p62 and Beclin-1. Intracellular protein levels were analysed using an LSRFortessa and the associated software (Becton Dickinson Biosciences, San Jose, CA, USA), and the results are expressed as the mean fluorescence intensity (MFI).
Cell counting and migration assays were performed with HUVECs as previously reported [14,16,19,28,29]. For DiI-acetylated low density lipoprotein (DiI-AcLDL) uptake and fluorescein isothiocyanate-labelled Ulex Europaeus Agglutinin-I (UEA-I)-binding assays, HUVECs were incubated with DiI-AcLDL (Life Technologies, Gaithersburg, MD, USA) and FITC-UEA-I (Sigma, St. Louis, MO, USA). To determine the number of double-positive-stained ECs per well, three random high-power fields under a fluorescence microscope (Olympus, Tokyo, Japan) were analysed.
2.4. Monodansylcadaverine (MDC) staining assay
HUVECs were cultured in 24-well plates. After three washes with PBS, the cells were stained with MDC at 37 °C for 40 min, washed again, and inspected immediately. MDC-stained autophagic vacuoles were evaluated using fluorescence microscopy (excitation wavelength: 390 nm, emission wavelength: 460 nm).
2.5. Electron micrograph
HUVECs were fixed with 2% paraformaldehyde and 2% glutaraldehyde in 50 mM phosphate buffer (pH 7.4) for 10 min at 4 °C. The buffer was then replaced with 2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4), and the samples were incubated at 4 °C overnight. Sample preparation and imaging were conducted at Tokai Electron Microscopy Inc. (Nagoya, Japan).
2.6. RNA sequencing (RNA-seq) and real-time quantitative polymerase chain reaction (qRT-PCR)
To elucidate the regulatory mechanism underlying the impaired haematopoiesis-supporting ability of HUVECs via Beclin-1 inhibition, RNA-seq analyses were performed in HUVECs with siRNA-mediated Beclin-1 knockdown as described previously [30]. Differential gene expression between the Beclin-1-silenced (H-sBECN) group and the control group was analysed by the DESeq2 package in R (1.16.1). Gene ontology (GO) enrichment analysis of differentially expressed genes was performed with the clusterProfiler package in R.
To confirm the RNA-seq results, the relative mRNA levels of Beclin-1, JAG1, CSF-1, CSF-2, CSF-3, ETS1, IL-7, DLL1, THPO, CXCL-12, VEGFR2 and E-selectin in the H-sBECN and control groups were analysed with SYBR Green-based qRT-PCR. The levels of the aforementioned genes were evaluated after normalisation to Actin mRNA levels (detailed in Supplementary methods).
2.7. Western blot analysis
Cell lysis and protein concentration determination were performed according to a previously described technique [14,28]. The membranes containing the separated proteins were blocked and then incubated overnight at 4 °C with antibodies against LC3 (1:3000, Proteintech, Wuhan, China), Beclin-1 (1:1000, Cell Signalling Technology, Danvers, MA), p62 (1:3000, Proteintech, Wuhan, China) and GAPDH (1:3000, Cell Signalling Technology). Next, the membranes were washed and incubated with secondary antibodies (1:5000, Santa Cruz Biotechnology, Santa Cruz, California, USA) at room temperature for 60 min. The protein bands were observed on X-ray films after incubation with ECL reagents (Millipore, Bedford, MA). Different sample quantities were used to establish a working range of sample loading. Band intensities were compared with those of GAPDH by using ImageJ software.
2.8. Coculture of BM CD34+ cells with HUVECs for colony-forming unit (CFU) assays
BM mononuclear cells (BMMNCs) were isolated by density gradient centrifugation. BM CD34+ cells were isolated from BMMNCs from healthy donors with a CD34 MicroBead Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) and cocultured as nonadherent cells with adherent HUVECs for 7 days in StemSpan™ SFEM (Stem Cell Technologies, Vancouver, BC, Canada) [14,28].
CFU assays were performed using MethoCult™ H4434 Classic (Stem Cell Technologies, Vancouver, BC, Canada) as described previously. Cocultured CD34+ cells (2 × 103) were plated in 24-well plates and cultured for 14 days. Colony-forming unit erythroid (CFU-E), burst-forming unit erythroid (BFU-E), colony-forming unit-granulocyte/macrophage (CFU-GM), and colony-forming unit-granulocyte, erythroid, macrophage and megakaryocyte (CFU-GEMM) values were obtained by viewing the cultures under an inverted light microscope. Cultures were assayed in duplicate, and the results are expressed as the mean ± SEM.
2.9. Subjects
A prospective case-control study was conducted to evaluate autophagy levels in BM ECs from patients with PGF and those from matched patients with good graft function (GGF). Transplant recipients were identified from among individuals who received an allotransplant from March 13, 2017, to July 10, 2018, at Peking University Institute of Hematology. A total of 40 patients who developed PGF were eligible. For each case, one matched transplant recipient with GGF was selected from the same cohort after matching for age, pretransplant disease state and posttransplant interval (“risk-set sampling”) [31]. None of the clinical characteristics, such as transplanted CD34+ cell dose, history of graft versus host disease (GvHD) or cytomegalovirus (CMV) infection, or anti-CMV therapy with ganciclovir, were significantly different between the PGF and GGF patients (Table S1).
BM samples from healthy donors (N = 23, 13 males and 10 females; age range, 21–50 years; median age, 35 years) served as normal controls. The study was approved by the Ethics Committee of Peking University People's Hospital, and written informed consent was obtained from all subjects in accordance with the Declaration of Helsinki.
3. Definition of PGF and GGF
As previously reported, PGF [10], [11], [12], [13], [14], [15], [16], [17], [18] is defined as hypo- or aplastic BM with at least 2 of the following criteria: (1) absolute neutrophil count (ANC) ≤0.5 × 109/L; (2) platelets ≤20 × 109/L; and (3) hemoglobin concentration ≤70 g/L for at least 3 consecutive days after day +28 after HSCT or requiring platelet and/or RBC transfusion and/or G-CSF support in the presence of complete donor chimerism. GGF [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20] was characterised by ANC >0.5 × 109/L for 3 consecutive days, platelet count >20 × 109/L for 7 consecutive days, and haemoglobin level >70 g/L without transfusion support beyond day +28 after HSCT. Patients with evidence of haematologic relapse after allo-HSCT were excluded.
3.1. Transplantation protocols
Donor selection, HLA typing, graft harvesting, conditioning therapy and GvHD prophylaxis were performed as previously reported [32], [33], [34]. The subjects were screened for CMV infection by serology. qRT-PCR was performed twice per week to detect CMV reactivation in blood samples, and CMV infection was treated with ganciclovir or foscarnet as previously described [14]. After allo-HSCT, rhG-CSF was administered as previously reported [14].
3.2. Isolation, cultivation, characterisation and function analysis of primary BM ECs
The isolation, cultivation and characterisation of primary BM ECs from PGF patients, GGF patients and healthy donors were performed following our previously published protocols [14,19,28,29]. BMMNCs (1 × 106/well) were seeded in 24-well culture plates precoated with fibronectin (Sigma) and cultured in EGM-2-MV SingleQuots (Lonza, Walkersville, MD, USA) [35] supplemented with 10% foetal bovine serum (Gibco, MA, USA) at 37 °C in a humidified incubator with 5% CO2. After 7 days of cultivation, BM ECs were treated with rapamycin (5 μM) [24,36,37] or HCQ (10 mΜ) [38] for 6 h. The cultured BM ECs were characterised by staining with monoclonal mouse antibodies targeting human CD34, CD133, and vascular endothelial growth factor receptor 2 (VEGFR2, CD309) (Becton Dickinson) and running the labelled cells through a BD LSRFortessa cell analyser (Becton Dickinson) [14,19,28,29,39,40]. Aliquots of isotype-matched antibodies served as negative controls. Data were analysed with BD LSRFortessa software (Becton Dickinson).
BM ECs were evaluated with migration assays and DiI-AcLDL and FITC-UEA-I double-staining assays as previously reported [14,19,28,29]. The levels of apoptosis, ROS and intracellular proteins in BM ECs were detected by flow cytometry according to previously published protocols [14,19,28,29]. The MDC staining assay, western blot analysis and the coculture of BM CD34+ cells with BM ECs were performed using the same protocol in HUVECs as described above.
3.3. Statistical analyses
Statistical analyses were performed using one-way analysis of variance (ANOVA) for comparisons among groups. Wilcoxon matched-pairs signed rank test for paired data was used to identify drug effects [41], [42], [43]. Subject variables were compared using a chi-squared test for categorical variables and a Mann-Whitney U test for continuous variables. Analyses were performed with GraphPad Prism 8.0 (GraphPad Software, La Jolla, CA, USA), and P-values <0.05 were considered significant.
4. Results
4.1. Inhibiting autophagy by knocking down Beclin-1 impaired the haematopoiesis-supporting ability of HUVECs
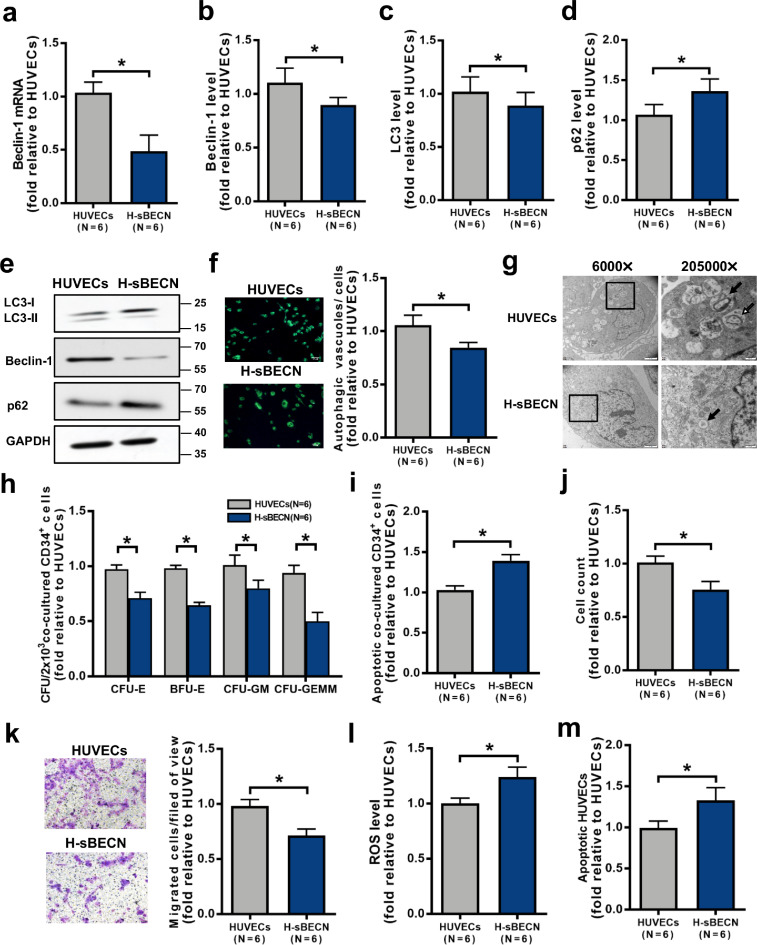
To investigate whether genetic regulation of autophagy modulates the haematopoiesis-supporting ability of ECs, Beclin-1 was knocked down in HUVECs by siRNA. Beclin-1 knockdown in HUVECs decreased Beclin-1 mRNA expression (Fig. 1a; 0.49 ± 0.15-fold; P = 0.03) and the intracellular levels of Beclin-1 protein (Fig. 1b; 0.89 ± 0.07-fold; P = 0.03; Fig. 1e and Fig. S1b; 0.33 ± 0.09-fold; P = 0.02) and LC3-II (Fig. 1c; 0.89 ± 0.12-fold; P = 0.03; Fig. 1e and Fig. S1a; 0.55 ± 0.008-fold; P = 0.0003) but increased the levels of p62 (Fig. 1d; 1.36 ± 0.15-fold; P = 0.03; Fig. 1e and Fig. S1c; 2.54 ± 0.14-fold; P = 0.009) compared to the control. Moreover, Beclin-1 knockdown in HUVECs decreased the numbers of autophagosomes and autolysosomes (Fig. 1f; 0.85 ± 0.05-fold; P = 0.03; Fig. 1g) compared to the control. Importantly, Beclin-1 knockdown decreased the ability of HUVECs to support CD34+ cells from healthy donors (Fig. 1h), as determined by the CFU-E (0.71 ± 0.05-fold; P = 0.03), BFU-E (0.65 ± 0.03-fold; P = 0.03), CFU-GM (0.80 ± 0.07-fold; P = 0.03) and CFU-GEMM (0.50 ± 0.08-fold; P = 0.03) values, and increased the apoptosis rate of CD34+ cells from healthy donors (Fig. 1i; 1.39 ± 0.07-fold; P = 0.03). Beclin-1 knockdown decreased the HUVEC cell count (Fig. 1j; 0.76 ± 0.08-fold; P = 0.03) and migration abilities (Fig. 1k; 0.71 ± 0.06-fold; P = 0.03) but increased the levels of ROS (Fig. 1l; 1.24 ± 0.09-fold; P = 0.03) and apoptosis (Fig. 1m; 1.33 ± 0.16-fold; P = 0.03). These results suggest that downregulating autophagy by Beclin-1 knockdown can quantitatively and functionally impair HUVECs, with a noted decrease in their haematopoiesis-supporting ability.
Fig. 1.
Inhibiting autophagy via Beclin-1 knockdown impaired the haematopoiesis-supporting ability of HUVECs. (a) Beclin-1 mRNA levels were analysed by qRT‐PCR after Beclin-1 knockdown in HUVECs. Intracellular (b) Beclin-1, (c) LC3 and (d) p62 levels in the Beclin-1 knockdown group (H-sBECN) and the control group were analysed by flow cytometry. (e) Representative western blots of LC3, Beclin-1 and p62. (f) Representative images (left panel) and quantification (right panel) of intracellular autophagosomes and autophagic vacuoles in the H-sBECN and control groups (original magnification, 10 × , scale bars represent 50 μm). The numbers of intracellular autophagosomes and autophagic vacuoles (green) per cell were counted in three random high-power fields and averaged. (g) Transmission electron microscope image of HUVECs in the H-sBECN and control groups. Images were obtained with a JEOL 1200 EX transmission electron microscope. Double-membrane autophagosomes (open arrows) and single-membrane autolysosomes (filled arrows) containing degraded material clustered at perinuclear sites (magnification: left 6000 × , scale bars represent 2 μm; right 20,500 × , scale bars represent 200 nm). (h) The CFU plating efficiency and (i) apoptosis rate of BM CD34+ cells from healthy donors were analysed after 7 days of coculture with HUVECs. (j) HUVEC count after Beclin-1 knockdown. (k) Representative images (left panel) and quantification (right panel) of the transwell migration assays in the H-sBECN and control groups (original magnification, 10 × , scale bars represent 50 μm). The number of migrated HUVECs per field of view was compared between the H-sBECN and control groups. In addition, (l) intercellular ROS levels (MFI, mean±SEM) and (m) the apoptosis rate were analysed by flow cytometry. Wilcoxon's test for paired data was used to identify drug effects. All P-values <0.05 were considered significant and are provided in the figure. *P<0.05, **P<0.005, ***P<0.0005.
4.2. The impaired haematopoiesis-supporting ability of ECs was restored by activating autophagy via Beclin-1 upregulation
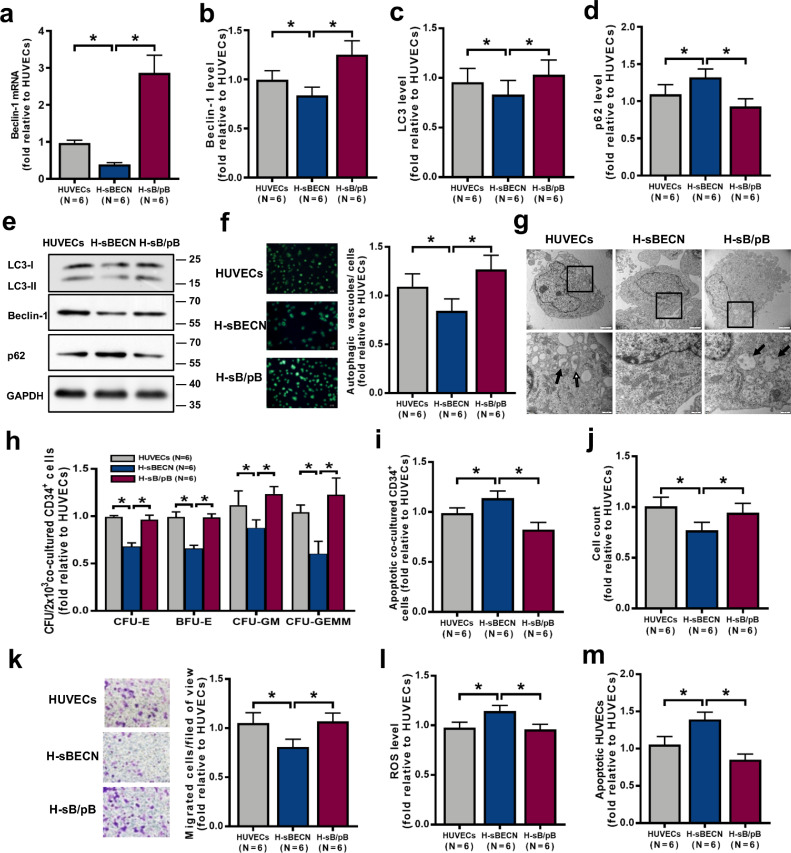
To further investigate whether reconstituted expression of Beclin-1 improves EC function by rescuing the deficient autophagy induced by Beclin-1 knockdown, Beclin-1 was reconstituted expressed in HUVECs, which resulted in increased Beclin-1 mRNA expression (Fig. 2a; 2.86 ± 0.48-fold vs. 0.39 ± 0.05-fold; P = 0.03) and intracellular protein levels (Fig. 2b; 1.25 ± 0.14-fold vs. 0.84 ± 0.09-fold; P = 0.03; Fig. 2e and Fig. S1e; 1.15 ± 0.09-fold vs. 0.29 ± 0.05-fold; P = 0.007) and increased LC3-II expression (Fig. 2c; 1.03 ± 0.15-fold vs. 0.83 ± 0.14-fold; P = 0.03; Fig. 2e and Fig. S1d; 0.87 ± 0.009-fold vs. 0.42 ± 0.04-fold; P = 0.01) but decreased p62 levels (Fig. 2d; 0.93 ± 0.10-fold vs. 1.32 ± 0.11-fold; P = 0.03; Fig. 2e and Fig. S1f; 1.21 ± 0.04-fold vs. 3.11 ± 0.20-fold; P = 0.01) compared to the levels of the corresponding genes/proteins in cells with genetic knockdown of Beclin-1 (H-sBECN). Compared to H-sBECN cells, reconstituted expression of Beclin-1 increased the numbers of autophagosomes and autolysosomes (Fig. 2f; 1.27 ± 0.15-fold vs. 0.84 ± 0.12-fold; P = 0.03; Fig. 2g). Importantly, reconstituted expression of Beclin-1 decreased the apoptosis rate of CD34+ cells (Fig. 2i; 0.82 ± 0.07-fold vs. 1.14 ± 0.07-fold; P = 0.03) from healthy donors and enhanced the ability of HUVECs to support these CD34+ cells (Fig. 2h), as determined by the CFU-E (0.96 ± 0.03-fold vs. 0.70 ± 0.04-fold; P = 0.03), BFU-E (0.99 ± 0.04-fold vs. 0.66 ± 0.03-fold; P = 0.03), CFU-GM (1.24 ± 0.08-fold vs. 0.88 ± 0.08-fold; P = 0.03) and CFU-GEMM (1.23 ± 0.18-fold vs. 0.60 ± 0.13-fold; P = 0.03) values. Reconstituted expression of Beclin-1 increased the cell number (Fig. 2j; 0.94 ± 0.09-fold vs. 0.77 ± 0.08-fold; P = 0.03) and migration (Fig. 2k; 1.07 ± 0.09-fold vs. 0.81 ± 0.08-fold; P = 0.03) and decreased the levels of ROS (Fig. 2l; 0.96 ± 0.05-fold vs. 1.14 ± 0.06-fold; P = 0.03) and apoptosis (Fig. 2m; 0.85 ± 0.08-fold vs. 1.39 ± 0.10-fold; P = 0.03). Similar results were found in primary BM ECs derived from healthy donors (Fig. S2). These results suggest that autophagy mediated by Beclin-1 can improve EC function, especially their haematopoiesis-supporting ability.
Fig. 2.
The impaired haematopoiesis-supporting ability of HUVECs was restored by activating autophagy via Beclin-1 upregulation. (a) Beclin-1 mRNA levels were analysed by qRT-PCR after Beclin-1 knockdown (H-sBECN) and overexpression (H-sB/pB) in HUVECs. The intracellular levels of (b) Beclin-1, (c) LC3 and (d) p62 in the H-sBECN, H-sB/pB and control groups were analysed by flow cytometry. (e) Representative western blots of LC3, Beclin-1 and p62. (f) Representative images (left panel) and quantification (right panel) of intracellular autophagosomes and autophagic vacuoles in the H-sBECN, H-sB/pB and control groups (original magnification, 10 × , scale bars represent 50 μm). The numbers of intracellular autophagosomes and autophagic vacuoles (green) per cell were determined in three random high-power fields and averaged. (g) Transmission electron microscope image of HUVECs in the H-sBECN, H-sB/pB and control groups. Double-membrane autophagosomes (open arrows) and single-membrane autolysosomes (filled arrows) containing degraded material clustered at perinuclear sites (magnification: above 6000 × , scale bars represent 2 μm; below 20,500 × , scale bars represent 200 nm). (h) The CFU plating efficiency and (i) apoptosis rate of BM CD34+ cells from healthy donors were analysed after 7 days of coculture with HUVECs. (j) HUVEC count and (k) representative images (left panel) and quantification (right panel) of the transwell migration assays for the H-sBECN, H-sB/pB and control groups (original magnification, 10 × , scale bars represent 50 μm). The numbers of migrated HUVECs per field of view were compared among the H-sBECN, H-sB/pB and control groups. (l) Intercellular ROS levels (MFI, mean±SEM) and (m) the apoptosis rate were analysed by flow cytometry. Wilcoxon's test for paired data was used to identify drug effects. All P-values <0.05 were considered significant and are provided in the figure. *P<0.05, **P<0.005, ***P<0.0005.
4.3. Autophagy positively regulated haematopoiesis-related genes in HUVECs
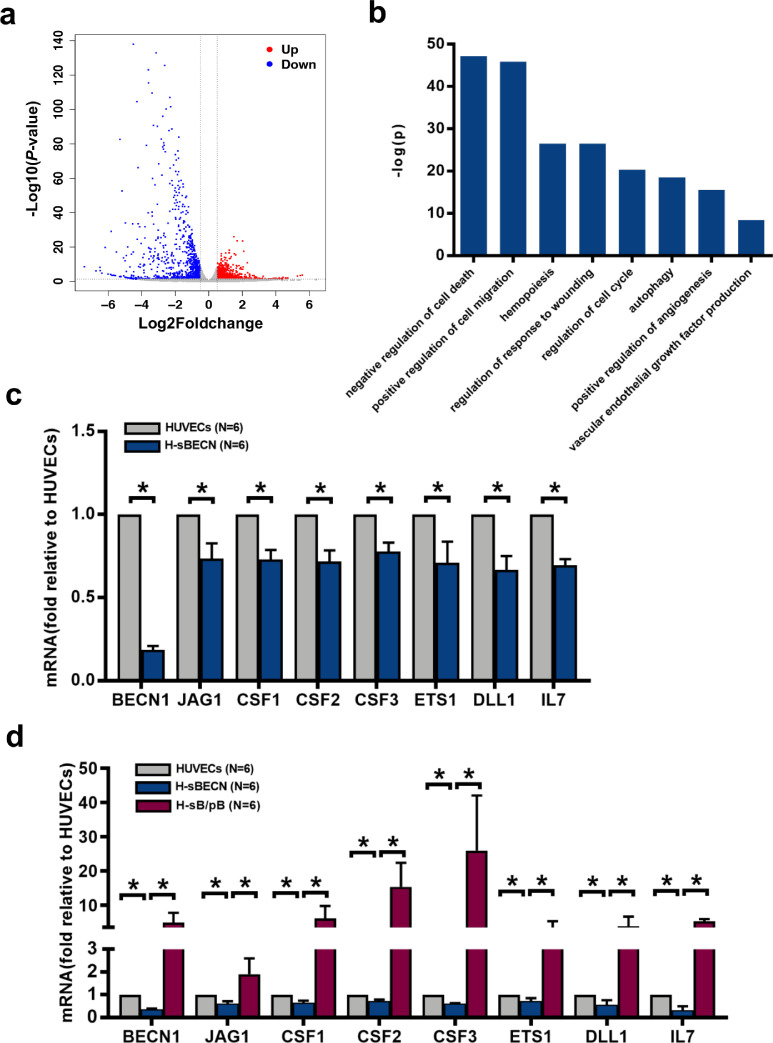
RNA-seq analyses were performed in HUVECs with siRNA-mediated knockdown of Beclin-1. Among the 27,652 genes in the volcano plot, 2579 were significantly differentially expressed between the control group (HUVECs) and the H-sBECN group. In the H-sBECN group, 639 genes were upregulated, and 789 genes were downregulated (Fig. 3a). GO term enrichment analysis demonstrated that the downregulated genes in the H-sBECN group were enriched in haematopoiesis-associated biological processes (Fig. 3b). The relative mRNA expression levels of Beclin-1 and haematopoiesis-regulating genes, including CSF-1, CSF-2, CSF-3, JAG1, ETS1, IL-7 and DLL1, were further analysed using qRT-PCR in the H-sBECN, control, and Beclin-1 reconstitution (H-sB/pB) groups. Beclin-1 mRNA levels were significantly different between the H-sBECN and control groups (Fig. 3c, 0.19 ± 0.02-fold, P = 0.03; Fig. 3d, 0.37 ± 0.02-fold, P = 0.03). Consistent with the RNA-seq results, CSF-1, CSF-2, CSF-3, JAG1, ETS1, IL-7 and DLL1 mRNA levels were significantly lower in the H-sBECN group than in the control group (Fig. 3c). Beclin-1 mRNA levels were significantly upregulated in the H-sB/pB group (Fig. 3d, 5.0 ± 2.80-fold, P = 0.03) compared to the H-sBECN group. In accordance with these results, the CSF-1, CSF-2, CSF-3, JAG1, ETS1, IL-7 and DLL1 mRNA levels were significantly higher in the H-sB/pB group than in the H-sBECN group (Fig. 3d). However, the mRNA levels of THPO, CXCL-12, VEGFR2 and E-selectin in HUVECs were not modulated by their autophagy status (Fig. S3). These data suggest that Beclin-1-dependent autophagy might be one of the underlying mechanisms by which ECs support HSCs.
Fig. 3.
Haematopoiesis-regulating genes, which were downregulated by Beclin-1 knockdown, were upregulated by Beclin-1 overexpression in HUVECs. (a) Volcano plots of the differentially expressed genes between the H-sBECN and control groups. Red dots represent upregulated genes with log2[H-sBECN/control]>0.5 and P<0.05. Blue dots represent downregulated genes with log2[H-sBECN/control]<−0.5 and P<0.05. Grey dots represent genes with no significant difference between the H-sBECN and control groups. (b) Bar charts show the biological processes of the enriched downregulated genes in the H-sBECN group. The bar indicates the -log10 of the P-value. (c) The relative mRNA levels of Beclin-1, JAG1, CSF-1, CSF-2, CSF-3, ETS1, IL-7 and DLL1 in the H-sBECN and control groups were determined using qRT‐PCR. (d) The relative mRNA levels of Beclin-1, JAG1, CSF-1, CSF-2, CSF-3, ETS1, IL-7 and DLL1 in the H-sBECN, H-sB/pB and control groups were determined using qRT‐PCR.
4.4. Upregulating autophagy with rapamycin improved the hematopoiesis-supporting ability of HUVECs
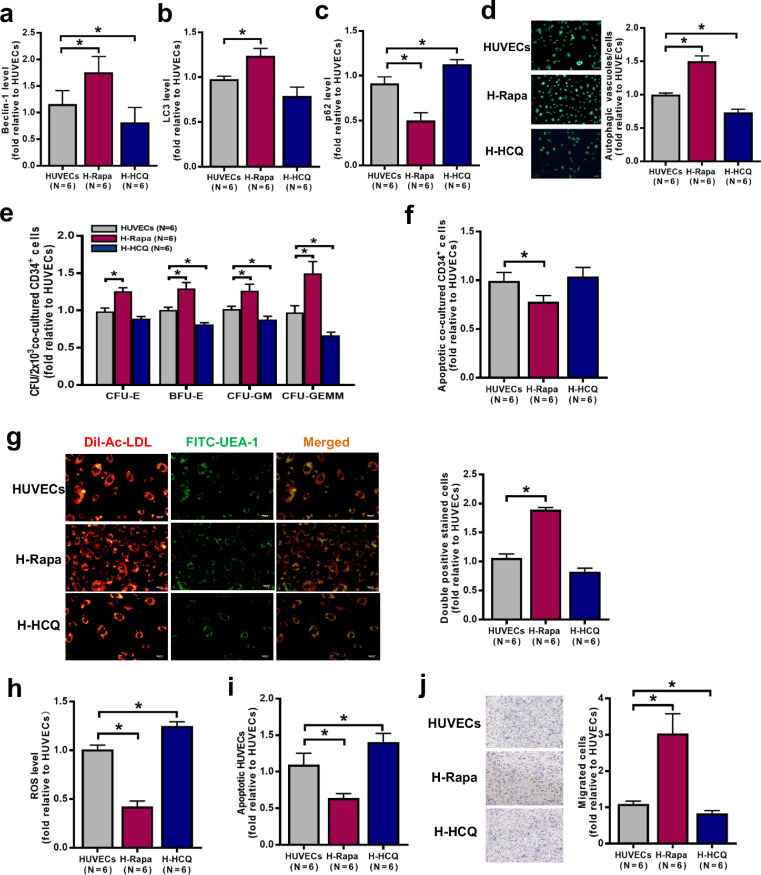
To investigate whether a pharmacological activator of autophagy (rapamycin) improves HUVEC function, the expression of autophagy-related proteins and the quantity and function of HUVECs were analysed. Compared with the control (no treatment), rapamycin increased LC3 levels (Fig. 4b; 1.24 ± 0.08-fold; P = 0.03) but decreased p62 levels (Fig. 4c; 0.50 ± 0.08-fold; P = 0.03) by upregulating Beclin-1 expression (Fig. 4a; 1.76 ± 0.30-fold; P = 0.03). To further confirm the increase in autophagy upon rapamycin treatment, MDC staining was performed to analyse autophagosomes and autolysosomes. Increased numbers of autophagosomes and autolysosomes were present in HUVECs after rapamycin treatment compared with the control treatment (Fig. 4d; 1.50 ± 0.08-fold; P = 0.03).
Fig. 4.
The effect of pharmacologic regulation of HUVEC autophagy on cocultured BM CD34+ cells. Intracellular levels of (a) Beclin-1, (b) LC3 and (c) p62 in HUVECs were analysed by flow cytometry after 6 h of treatment with rapamycin (autophagy activator, 5 μM) or HCQ (autophagy inhibitor, 10 μM). (d) The effects of the different treatments on autophagic vacuoles in HUVECs were determined: representative images (left panel) and quantification per well (right panel) (original magnification, 10 × , scale bars represent 50 μm). (e) The CFU plating efficiency and (f) the apoptosis rate of BM CD34+ cells from healthy donors were analysed after 7 days of coculture with HUVECs. (g) Representative images (left panel) of HUVECs. Typical HUVECs were characterised as double-positive for DiI-AcLDL (red) and FITC-UEA-I (green) (original magnification, 10 × , scale bars represent 50 μm). Quantification (right panel) of HUVECs double-positive for DiI-AcLDL (red) and FITC-UEA-I (green) after 6 h of treatment with rapamycin or HCQ (original magnification, 10 × , scale bars represent 50 μm). The (h) intercellular ROS levels (MFI, mean±SEM) and (i) the apoptosis rate of HUVECs were analysed by flow cytometry. (j) Representative images (left panel) and quantification (right panel) of HUVEC migration assays after 6 h of treatment with rapamycin or HCQ (original magnification, 10 × , scale bars represent 50 μm). The number of migrated BM ECs per field of view was counted in three random high-power fields and averaged. Wilcoxon's test for paired data was used to identify drug effects. All P-values <0.05 were considered significant and are provided in the figure. *P<0.05, **P<0.005, ***P<0.0005.
To explore whether pharmacological activation of autophagy affects the ability of HUVECs to support HSCs, BM CD34+ cells from healthy donors were cocultured with HUVECs after rapamycin treatment. As shown in Fig. 4E, compared with the control, rapamycin improved the ability of HUVECs to support CD34+ cells from healthy donors, as determined by increases in the CFU-E (1.26 ± 0.05-fold; P = 0.03), BFU-E (1.30 ± 0.08-fold; P = 0.03), CFU-GM (1.26 ± 0.09-fold; P = 0.03) and CFU-GEMM (1.50 ± 0.16-fold; P = 0.03) values. Rapamycin-treated HUVECs showed a decreased apoptosis rate (Fig. 4f; 0.8 ± 0.11-fold; P = 0.03) of CD34+ cells from healthy donors. The in vitro upregulation of autophagy significantly increased the number of cells with DiI-AcLDL and FITC-lectin-UEA-I double-stained cells (Fig. 4g; 1.89 ± 0.04-fold; P = 0.03) and the migration capacity (Fig. 4j; 3.04 ± 0.54-fold; P = 0.03) and decreased the levels of ROS (Fig. 4h; 0.42 ± 0.06-fold; P = 0.03) and apoptosis (Fig. 4i; 0.64 ± 0.06-fold; P = 0.03) compared with respective values of the control group. These data suggest that upregulated autophagy in HUVECs can enhance their haematopoiesis-supporting ability.
4.5. Inhibiting autophagy with hydroxychloroquine reduced the haematopoiesis-supporting ability of HUVECs
To investigate whether a pharmacological inhibitor of autophagy (HCQ) impairs HUVEC function, the expression of autophagy-related proteins and the quantity and function of HUVECs were analysed. Compared with control treatment, HCQ decreased LC3 levels (Fig. 4b; 0.79 ± 0.10-fold; P = 0.16) and increased p62 levels (Fig. 4c; 1.13 ± 0.05-fold; P = 0.03) by downregulating Beclin-1 expression (Fig. 4a; 0.81 ± 0.28-fold; P = 0.03). Moreover, HCQ decreased the numbers of autophagosomes and autolysosomes (Fig. 4d, 0.73 ± 0.05-fold; P = 0.03).
BM CD34+ cells from healthy donors were cocultured with HUVECs after HCQ treatment to explore whether pharmacological inhibition of autophagy affects the ability of HUVECs to support HSCs. Compared to the control treatment, HCQ decreased the ability of HUVECs to support CD34+ cells from healthy donors (Fig. 4e), as determined by the CFU-E (0.89 ± 0.03-fold; P = 0.06), BFU-E (0.81 ± 0.02-fold; P = 0.03), CFU-GM (0.88 ± 0.05-fold; P = 0.03) and CFU-GEMM (0.67 ± 0.04-fold; P = 0.03) values. Coculture with HCQ-treated HUVECs increased the apoptosis rate of CD34+ cells from healthy donors (Fig. 4f; 1.04 ± 0.09-fold; P = 0.22). Furthermore, compared with the control treatment, HCQ decreased the number of DiI-AcLDL and FITC-lectin-UEA-I double-stained cells (Fig. 4g; 0.83 ± 0.06-fold; P = 0.06) and migration abilities (Fig. 4j; 0.84 ± 0.07-fold; P = 0.03) and increased ROS levels (Fig. 4h; 1.25 ± 0.04-fold; P = 0.03) and apoptosis (Fig. 4i; 1.41 ± 0.12-fold; P = 0.03). These results suggest that inhibiting autophagy with HCQ can quantitatively and functionally impair HUVECs, with a specific reduction in their haematopoiesis-supporting ability by regulating the Beclin-1 pathway.
4.6. Defective autophagy and Beclin-1 pathway downregulation in BM ECs from PGF patients
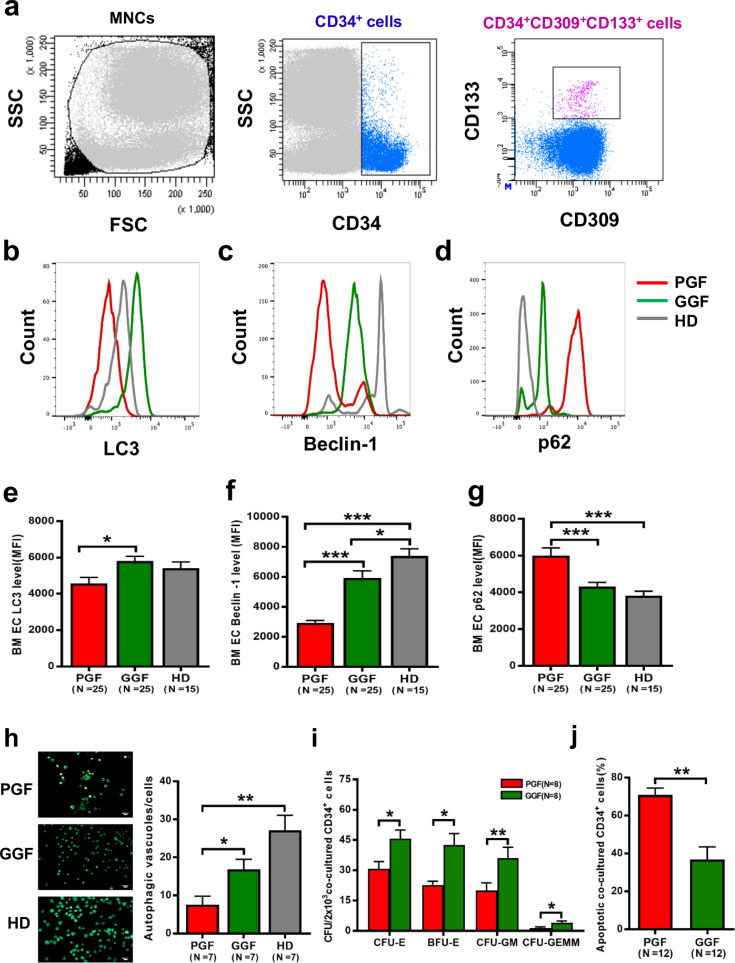
Subsequently, a prospective case-control study was conducted to investigate the functional role of EC autophagy in patients with PGF, a human model of BM failure after allo-HSCT. White blood cell (WBC) count, ANC, platelet count and hemoglobin levels were significantly lower in PGF patients than in GGF patients, but there were no differences in other demographic and clinical characteristics (Table S1).
The representative EC phenotype was characterised as positive expression of CD34, CD133 and CD309 as measured by flow cytometry (Fig. 5a). Defective autophagy in BM ECs, characterised by decreased LC3 levels (Fig. 5b and e; 4579 ± 321.5 vs. 5802 ± 256.9; P = 0.006), Beclin-1 downregulation (Fig. 5c and f; 2928 ± 166.1 vs. 5924 ± 477.3; P<0.0001), increased p62 levels (Fig. 5d and g; 6012 ± 410.6 vs. 4324 ± 217.0; P = 0.0003), and fewer intracellular autophagosomes and autophagic vacuoles (Fig. 5h; 7.6 ± 2.3 vs. 16.9 ± 2.7; P = 0.02) was observed in PGF patients compared with the corresponding levels in GGF patients. These results suggest that autophagy and Beclin-1 are significantly decreased in BM ECs from PGF patients compared to those from GGF patients.
Fig. 5.
Defective autophagy in BM ECs from PGF patients. (a) The EC phenotype was characterised by the positive expression of CD34, CD133 and CD309, and representative intracellular levels of (b) LC3, (c) Beclin-1 and (d) p62 in precultured BM ECs were analysed by flow cytometry. The intracellular levels of (e) LC3, (f) Beclin-1 and (g) p62 in precultured BM ECs from PGF patients, GGF patients and healthy donors were analysed by flow cytometry (MFI, mean±SEM). (h) Representative images (left panel) and quantification (right panel) of intracellular autophagosomes and autophagic vacuoles in cultivated BM ECs from PGF patients, GGF patients and healthy donors (original magnification, 10 × , scale bars represent 50 μm). (i) The CFU plating efficiency and (j) apoptosis rates of BM CD34+ cells after 7 days of coculture with BM ECs from PGF patients and GGF patients. Continuous variables were compared using the Mann–Whitney U test. All P-values<0.05 were considered significant and are provided in the figure. *P<0.05, **P<0.005, ***P<0.0005.
4.7. Reduced haematopoiesis-supporting ability of BM ECs from PGF patients
To explore whether impaired BM ECs affect HSC function in vitro, BM CD34+ cells from healthy donors were cocultured with BM ECs from PGF patients and GGF patients. As shown in Fig. 5i and j, the coculture of BM CD34+ cells with BM ECs from PGF patients led to significantly decreased CFU plating efficiency, as evidenced by lower CFU-E (30.75 ± 3.57 vs. 45.63 ± 4.33; P = 0.01), BFU-E (22.75 ± 1.87 vs. 42.50 ± 5.64; P = 0.01), CFU-GM (19.88 ± 3.92 vs. 36.0 ± 5.42; P = 0.004) and CFU-GEMM values (1.38 ± 0.6 vs. 4.0 ± 0.93; P = 0.03), in PGF patients compared to that in GGF patients. Coculture of BM CD34+ cells with BM ECs from PGF patients evoked a significantly higher apoptosis rate (70.98% ± 3.51% vs. 36.88% ± 6.59%; P = 0.001) than did coculture with BM ECs from GGF patients.
4.8. Rapamycin-induced EC autophagy enhanced HSC-supporting ability in PGF patients
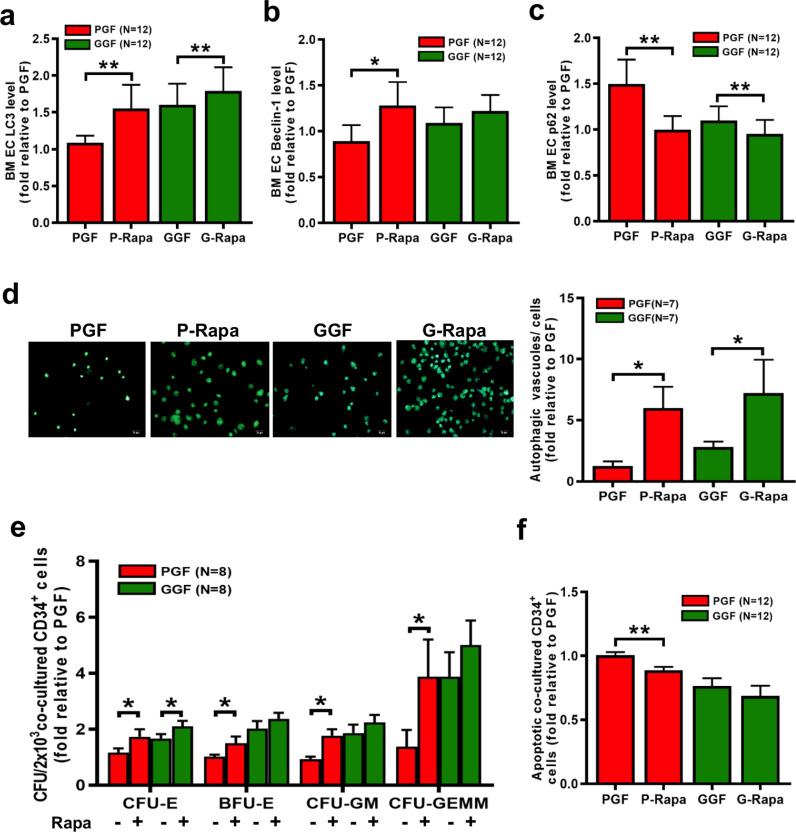
To investigate whether rapamycin enhances autophagy in BM ECs from PGF patients, the levels of LC3, Beclin-1 and p62 expression after rapamycin treatment were examined by flow cytometry and western blot. Compared to the control treatment, rapamycin increased the levels of LC3 (Fig. 6a; 1.55 ± 0.32-fold; P = 0.0005) and Beclin-1 (Fig. 6b; 1.28 ± 0.26-fold; P = 0.005) but decreased the levels of p62 (Fig. 6c; 1.0 ± 0.15-fold; P = 0.0005) in BM ECs from PGF patients. In addition, autophagosomes and autolysosomes were more abundant (Fig. 6d; 6.0 ± 1.75-fold; P = 0.02) in BM ECs from PGF patients after rapamycin treatment. Taken together, these results suggest that rapamycin can induce autophagy in BM ECs from PGF patients.
Fig. 6.
Rapamycin-induced autophagy of ECs from PGF patients enhanced their HSC-supporting ability. The intracellular levels of (a) LC3, (b) Beclin-1 and (c) p62 in BM ECs from PGF patients and GGF patients were analysed after 6 h of treatment with rapamycin. (d) Representative images (left panel) and quantification (right panel) of autophagic vacuoles in BM ECs after 6 h of treatment with rapamycin (original magnification, 10 × , scale bars represent 50 μm). Three random high-power fields were analysed, and the results were averaged per condition. (e) The CFU plating efficiency and (f) apoptosis rate of BM CD34+ cells after 7 days of coculture with BM ECs from PGF and GGF patients after 6 h of treatment with rapamycin. Wilcoxon's test for paired data was used to identify drug effects. All P-values<0.05 were considered significant and are provided in the figure. *P<0.05, **P<0.005, ***P<0.0005.
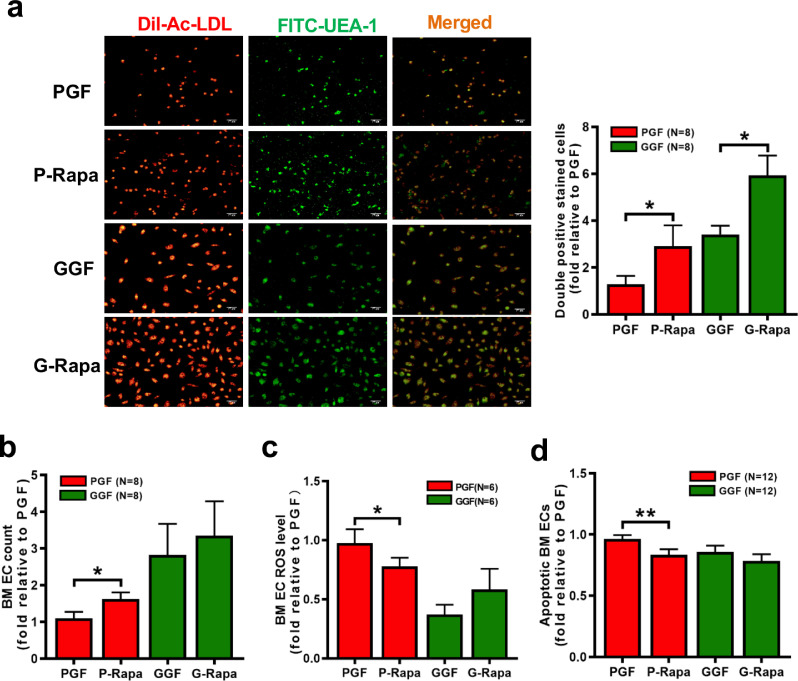
As shown in Fig. 6e, rapamycin improved the ability of BM ECs from PGF patients to support CD34+ cells from healthy donors, as determined by the CFU-E (1.73 ± 0.26-fold; P = 0.008), BFU-E (1.50 ± 0.24-fold; P = 0.008), CFU-GM (1.48 ± 0.23-fold; P = 0.008) and CFU-GEMM (3.63 ± 1.35-fold; P = 0.008) values. Rapamycin-treated BM ECs from PGF patients decreased the apoptosis rate of CD34+ cells from healthy donors (Fig. 6f; 0.89 ± 0.03-fold; P = 0.0005). Rapamycin-mediated upregulation of autophagy in BM ECs from PGF patients in vitro increased BM EC number (Fig. 7b; 1.62 ± 0.19-fold; P = 0.009) and the extent of DiI-AcLDL and FITC-lectin-UEA-I double-staining (Fig. 7a; 2.90 ± 0.89-fold; P = 0.005) and reduced the ROS levels (Fig. 7c; 0.78 ± 0.07-fold; P = 0.03) and apoptosis (Fig. 7d; 0.84 ± 0.05-fold; P = 0.0005) compared with the corresponding values in the control treatment group. These results suggest that activating autophagy can improve the function of BM ECs from PGF patients, with a particular increase in their HSC-supporting ability.
Fig. 7.
Rapamycin-induced autophagy quantitatively and functionally improved BM ECs from PGF patients. (a) Representative images (left panel) of BM ECs. Typical ECs from PGF and GGF patients were characterised by double-positive staining (merge, yellow) with DiI-AcLDL (red) and FITC-UEA-I (green) at day 7 of culture (original magnification, 10 × , scale bars represent 50 μm). Quantification (right panel) of double-positive BM ECs (merge, yellow) stained with DiI-AcLDL (red) and FITC-UEA-I (green) after 6 h of treatment with rapamycin (original magnification, 10 × , scale bars represent 50 μm). The (b) cell count, (c) intercellular ROS levels and (d) apoptosis rate of BM ECs from PGF and GGF patients were analysed after 6 h of treatment with rapamycin. Wilcoxon's test for paired data was used to identify drug effects. All P-values <0.05 were considered significant and are provided in the figure. *P<0.05, **P<0.005, ***P<0.0005.
5. Discussion
The current study is the first to demonstrate that autophagy in ECs modulates their ability to support haematopoiesis. Activating autophagy in ECs enhanced their haematopoiesis-supporting ability by upregulating the Beclin-1 pathway. Moreover, defective autophagy in BM ECs may be involved in the pathogenesis of PGF after allo-HSCT, which could be rescued by rapamycin through improvement of the impaired HSC-supporting ability of BM ECs by activating autophagy. Our data may provide new insights into the underlying mechanism by which ECs support HSCs.
Autophagy in ECs contributes to inflammation and the recruitment and regulation of innate and adaptive immune cells [44,45]. In haemolytic anaemia, upregulating autophagy protected ECs from a heme-induced oxidative state by repairing dysfunctional mitochondria [46]. Rezabakhsh et al. reported that upregulating autophagy protected HUVECs from damage induced by high glucose levels [47]. Villalba et al. showed that upregulating autophagy maintained the vasomotor function of the endothelium by regulating NO signalling [48]. In the current study, upregulating autophagy quantitatively and functionally improved HUVECs and primary BM ECs derived from healthy donors, with a particular increase in their haematopoiesis-supporting ability. Conversely, downregulating autophagy in HUVECs resulted in opposite outcomes. We provide further experimental evidence in patients with PGF, which is characterised by pancytopenia after allo-HSCT, to validate the vital role of autophagy in EC function, especially in their ability to support HSCs. Based on our previous work [14,15,17,18] and the current study, we speculate that insufficient induction of autophagy in BM ECs may hamper the haematopoietic reconstitution of successfully engrafted donor HSCs, ultimately leading to PGF after allo-HSCT. By contrast, improvement of autophagy in BM ECs may be a promising therapeutic approach to enhance haematopoietic activity of HSCs in PGF patients. However, the pathogenesis of PGF is complicated, and further studies with key functional data obtained from primary BM ECs of PGF patients are needed to clarify the functional roles of defective EC autophagy in PGF after allo-HSCT.
Recently, rapamycin has been suggested as an adjunct to cyclosporine and mycophenolate mofetil as immunosuppressive agents in patients receiving a non-myeloablative HSCT [49], [50], [51], [52]. Consistently, we found that rapamycin, an autophagy activator, improved the impaired HSC-supporting ability of ECs from PGF patients by activating autophagy in vitro. However, it should be noted that the anti-leukemic effects of rapamycin and their underlying mechanism in patients after allo-HSCT remain controversial [53], [54], [55], [56], [57], [58]. Considering the potential adverse effects of rapamycin, further clinical studies are needed to validate our preliminary findings in the future.
Beclin-1 plays a role in the formation of autophagosomes [24,25] and regulates autophagy and apoptosis, two processes crucial for tissue homeostasis [59,60]. The Beclin-1 gene has been implicated in numerous cases of prostate, breast, and ovarian cancers and non-Hodgkin lymphoma [61], [62], [63], [64], [65], [66], whereas its role in ECs is rather limited. Nicotra et al. reported a significant correlation of Beclin-1 with autophagy in non-Hodgkin lymphoma cells [61]. Wang et al. revealed that Beclin-1 knockdown inhibited autophagy and survival in ox-LDL-treated HUVECs [67]. Similarly, the current data indicate that Beclin-1 knockdown disrupted the ability of ECs to support HSCs by inhibiting autophagy. Conversely, upregulating Beclin-1 ameliorated this effect on ECs. RNA-seq data further revealed that classical haematopoiesis-regulating genes such as CSF-1, CSF-2, CSF-3, TLR3 and JAG1 were significantly downregulated [8,[68], [69], [70], [71], [72]], whereas no significant different levels of THPO, CXCL-12, VEGFR2 or E-selectin were detected in Beclin-1-silenced HUVECs [6,73,74]. Therefore, our data provide further evidence that autophagy plays a critical role in regulating EC function, especially for supporting haematopoiesis.
Considering the crucial role of ECs in supporting haematopoiesis in BM microenvironment [1,[3], [4], [5], [6], [7], [8]] and the current study, it is conceivable that higher autophagy in ECs regulated by the Beclin-1 pathway might be one of the underlying mechanisms by which ECs enhance haematopoietic activity of HSCs. However, we are aware that the underlying molecular mechanism of autophagy-related effect of ECs in the homeostasis of HSCs, such as the upstream and downstream proteins related to Beclin-1 pathway, need to be further explored.
In summary, the current study demonstrated that autophagy in ECs modulated their ability to support haematopoiesis by regulating the Beclin-1 pathway, and defective autophagy in BM ECs may be responsible for the pathogenesis of PGF. Although further validation is required, our data suggest that improving autophagy in BM ECs may be a potential therapeutic approach in PGF patients after allo-HSCT.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
Funding sources
This work was supported by the National Key Research and Development Program (2017YFA0104500), National Natural Science Foundation of China (81870139 and 81530046), the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (81621001), Clinical Medicine Plus X - Young Scholars Project of Peking University (PKU2019LCXQ016), the Science and Technology Project of Guangdong Province of China (2016B030230003) and Peking University Medicine Fund of Fostering Young Scholars’ Scientific & Technological Innovation (BMU2020PY007). The funding agencies had no role in study design, data collection, data analysis, interpretation, and writing of the manuscript.
Acknowledgements
The authors thank all of the core facilities at Peking University Institute of Hematology for patient care and sample collection. American Journal Experts (www.journalexperts.com) provided editorial assistance to the authors during the preparation of the manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102677.
Contributor Information
Yuan Kong, Email: successky@163.com.
Xiao-Jun Huang, Email: xjhrm@medmail.com.cn.
Appendix. Supplementary materials
References
- 1.Butler J.M., Kobayashi H., Rafii S. Instructive role of the vascular niche in promoting tumour growth and tissue repair by angiocrine factors. Nat Rev Cancer. 2010;10(2):138–146. doi: 10.1038/nrc2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu W.H., Shi Y.X., Ma Z.L. Proper autophagy is indispensable for angiogenesis during chick embryo development. Cell Cycle. 2016;15(13):1742–1754. doi: 10.1080/15384101.2016.1184803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sasine J.P., Yeo K.T., Chute J.P. Concise review: paracrine functions of vascular niche cells in regulating hematopoietic stem cell fate. Stem Cells Transl Med. 2017;6(2):482–489. doi: 10.5966/sctm.2016-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding L., Saunders T.L., Enikolopov G. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481(7382):457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendelson A., Frenette P.S. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med. 2014;20(8):833–846. doi: 10.1038/nm.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison S.J., Scadden D.T. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505(7483):327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scadden D.T. The stem-cell niche as an entity of action. Nature. 2006;441(7097):1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 8.He H., Xu J., Warren C.M. Endothelial cells provide an instructive niche for the differentiation and functional polarization of M2-like macrophages. Blood. 2012;120(15):3152–3162. doi: 10.1182/blood-2012-04-422758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salter A.B., Meadows S.K., Muramoto G.G. Endothelial progenitor cell infusion induces hematopoietic stem cell reconstitution in vivo. Blood. 2009;113(9):2104–2107. doi: 10.1182/blood-2008-06-162941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong Y. Poor graft function after allogeneic hematopoietic stem cell transplantation-an old complication with new insights. Semin Hematol. 2019;56(3):215–220. doi: 10.1053/j.seminhematol.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Zhao H.Y., Lyu Z.S., Duan C.W. An unbalanced monocyte macrophage polarization in the bone marrow microenvironment of patients with poor graft function after allogeneic haematopoietic stem cell transplantation. Br J Haematol. 2018;182(5):679–692. doi: 10.1111/bjh.15452. [DOI] [PubMed] [Google Scholar]
- 12.Song Y., Zhao H.Y., Lyu Z.S. Dysfunctional bone marrow mesenchymal stem cells in patients with poor graft function after allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transpl. 2018;24(10):1981–1989. doi: 10.1016/j.bbmt.2018.06.021. [DOI] [PubMed] [Google Scholar]
- 13.Kong Y., Wang Y.T., Cao X.N. Aberrant t cell responses in the bone marrow microenvironment of patients with poor graft function after allogeneic hematopoietic stem cell transplantation. J Transl Med. 2017;15(1):57. doi: 10.1186/s12967-017-1159-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi M.M., Kong Y., Song Y. Atorvastatin enhances endothelial cell function in posttransplant poor graft function. Blood. 2016;128(25):2988–2999. doi: 10.1182/blood-2016-03-702803. [DOI] [PubMed] [Google Scholar]
- 15.Kong Y., Wang Y.T., Hu Y. The bone marrow microenvironment is similarly impaired in allogeneic hematopoietic stem cell transplantation patients with early and late poor graft function. Bone Marrow Transpl. 2016;51(2):249–255. doi: 10.1038/bmt.2015.229. [DOI] [PubMed] [Google Scholar]
- 16.Kong Y., Song Y., Hu Y. Increased reactive oxygen species and exhaustion of quiescent CD34-positive bone marrow cells may contribute to poor graft function after allotransplants. Oncotarget. 2016;7(21):30892–30906. doi: 10.18632/oncotarget.8810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong Y., Chang Y.J., Wang Y.Z. Association of an impaired bone marrow microenvironment with secondary poor graft function after allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transpl. 2013;19(10):1465–1473. doi: 10.1016/j.bbmt.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 18.Kong Y., Wang Y., Zhang Y Y. Prophylactic oral NAC reduced poor hematopoietic reconstitution by improving endothelial cells after haploidentical transplantation. Blood Adv. 2019;3(8):1303–1317. doi: 10.1182/bloodadvances.2018029454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong Y., Shi M.M., Zhang Y.Y. N-acetyl-l-cysteine improves bone marrow endothelial progenitor cells in prolonged isolated thrombocytopenia patients post allogeneic hematopoietic stem cell transplantation. Am J Hematol. 2018;93(7):931–942. doi: 10.1002/ajh.25056. [DOI] [PubMed] [Google Scholar]
- 20.Kong Y., Hu Y., Zhang X.H. Association between an impaired bone marrow vascular microenvironment and prolonged isolated thrombocytopenia after allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transpl. 2014;20(8):1190–1197. doi: 10.1016/j.bbmt.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 21.Mizushima N., Levine B., Cuervo A.M. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torisu T., Torisu K., Lee I.H. Autophagy regulates endothelial cell processing, maturation and secretion of von Willebrand factor. Nat Med. 2013;19(10):1281–1287. doi: 10.1038/nm.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pestana C.R., Oishi J.C., Salistre-Araujo H.S. Inhibition of autophagy by chloroquine stimulates nitric oxide production and protects endothelial function during serum deprivation. Cell Physiol Biochem. 2015;37(3):1168–1177. doi: 10.1159/000430240. [DOI] [PubMed] [Google Scholar]
- 24.Gu J., Hu W., Song Z.P. Rapamycin inhibits cardiac hypertrophy by promoting autophagy via the MEK/ERK/Beclin-1 pathway. Front Physiol. 2016;7:104. doi: 10.3389/fphys.2016.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kihara A., Kabeya Y., Ohsumi Y. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2(4):330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie Y., You S.J., Zhang Y.L. Protective role of autophagy in AGE-induced early injury of human vascular endothelial cells. Mol Med Rep. 2011;4(3):459–464. doi: 10.3892/mmr.2011.460. [DOI] [PubMed] [Google Scholar]
- 28.Kong Y., Cao X.N., Zhang X.H. Atorvastatin enhances bone marrow endothelial cell function in corticosteroid-resistant immune thrombocytopenia patients. Blood. 2018;131(11):1219–1233. doi: 10.1182/blood-2017-09-807248. [DOI] [PubMed] [Google Scholar]
- 29.Cao X.N., Kong Y., Song Y. Impairment of bone marrow endothelial progenitor cells in acute graft-versus-host disease patients after allotransplant. Br J Haematol. 2018;182(6):870–886. doi: 10.1111/bjh.15456. [DOI] [PubMed] [Google Scholar]
- 30.Kong Y., Wu Y.L., Song Y. Ruxolitinib/nilotinib cotreatment inhibits leukemia-propagating cells in Philadelphia chromosome-positive ALL. J Transl Med. 2017;15(1):184. doi: 10.1186/s12967-017-1286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wacholder S., Silverman D.T., Mclaughlin J.K. Selection of controls in case-control studies. III. Design options. Am J Epidemiol. 1992;135(9):1042–1050. doi: 10.1093/oxfordjournals.aje.a116398. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y., Chang Y.J., Xu L.P. Who is the best donor for a related HLA haplotype-mismatched transplant? Blood. 2014;124(6):843–850. doi: 10.1182/blood-2014-03-563130. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., Chen H., Chen J. The consensus on the monitoring, treatment, and prevention of leukemia relapse after allogeneic hematopoietic stem cell transplantation in China. Cancer Lett. 2018;438:63–75. doi: 10.1016/j.canlet.2018.08.030. [DOI] [PubMed] [Google Scholar]
- 34.Xu L., Chen H., Chen J. The consensus on indications, conditioning regimen, and donor selection of allogeneic hematopoietic cell transplantation for hematological diseases in China-recommendations from the Chinese Society of Hematology. J Hematol Oncol. 2018;11(1):33. doi: 10.1186/s13045-018-0564-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingram D.A., Lien I.Z., Mead L.E. In vitro hyperglycemia or a diabetic intrauterine environment reduces neonatal endothelial colony-forming cell numbers and function. Diabetes. 2008;57(3):724–731. doi: 10.2337/db07-1507. [DOI] [PubMed] [Google Scholar]
- 36.Cordani M., Sanchez-Alvarez M., Strippoli R. Sestrins at the interface of ROS control and autophagy regulation in health and disease. Oxid Med Cell Longev. 2019;2019 doi: 10.1155/2019/1283075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang Y., Jacobi A., Vater C. Icariin promotes angiogenic differentiation and prevents oxidative stress-induced autophagy in endothelial progenitor cells. Stem Cells. 2015;33(6):1863–1877. doi: 10.1002/stem.2005. [DOI] [PubMed] [Google Scholar]
- 38.Sun C., Li C., Li X. Scutellarin induces apoptosis and autophagy in NSCLC cells through ERK1/2 and AKT signaling pathways in vitro and in vivo. J Cancer. 2018;9(18):3247–3256. doi: 10.7150/jca.25921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peichev M., Naiyer A.J., Pereira D. Expression of VEGFR-2 and AC133 by circulating human CD34(+) cells identifies a population of functional endothelial precursors. Blood. 2000;95(3):952–958. [PubMed] [Google Scholar]
- 40.Gehling U.M., Ergun S., Schumacher U. In vitro differentiation of endothelial cells from AC133-positive progenitor cells. Blood. 2000;95(10):3106–3112. [PubMed] [Google Scholar]
- 41.Berenstein R., Blau O., Nogai A. Multiple myeloma cells alter the senescence phenotype of bone marrow mesenchymal stromal cells under participation of the DLK1-DIO3 genomic region. BMC Cancer. 2015;15:68. doi: 10.1186/s12885-015-1078-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Son D., Chung M.H. In vitro synergism between chloroquine and antibiotics against orientia tsutsugamushi. Infect Chemother. 2014;46(3):182–188. doi: 10.3947/ic.2014.46.3.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosner B., Glynn R.J., Lee M.L. The wilcoxon signed rank test for paired comparisons of clustered data. Biometrics. 2006;62(1):185–192. doi: 10.1111/j.1541-0420.2005.00389.x. [DOI] [PubMed] [Google Scholar]
- 44.Sachdev U., Lotze M.T. Perpetual change: autophagy, the endothelium, and response to vascular injury. J Leukoc Biol. 2017;102(2):221–235. doi: 10.1189/jlb.3RU1116-484RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duraes F.V., Niven J., Dubrot J. Macroautophagy in endogenous processing of self- and pathogen-derived antigens for mhc class ii presentation. Front Immunol. 2015;6:459. doi: 10.3389/fimmu.2015.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Higdon A.N., Benavides G.A., Chacko B.K. Hemin causes mitochondrial dysfunction in endothelial cells through promoting lipid peroxidation: the protective role of autophagy. Am J Physiol Heart Circ Physiol. 2012;302(7):H1394–H1409. doi: 10.1152/ajpheart.00584.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rezabakhsh A., Fathi F., Bagheri H.S. Silibinin protects human endothelial cells from high glucose-induced injury by enhancing autophagic response. J Cell Biochem. 2018;119(10):8084–8094. doi: 10.1002/jcb.26735. [DOI] [PubMed] [Google Scholar]
- 48.Villalba N., Sonkusare S.K., Longden T.A. Traumatic brain injury disrupts cerebrovascular tone through endothelial inducible nitric oxide synthase expression and nitric oxide gain of function. J Am Heart Assoc. 2014;3(6) doi: 10.1161/JAHA.114.001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allison T.L. Immunosuppressive therapy in transplantation. Nurs Clin N Am. 2016;51(1):107–120. doi: 10.1016/j.cnur.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 50.Ceberio I., Dai K., Devlin S.M. Safety of voriconazole and sirolimus coadministration after allogeneic hematopoietic SCT. Bone Marrow Transpl. 2015;50(3):438–443. doi: 10.1038/bmt.2014.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsieh M.M., Fitzhugh C.D., Weitzel R.P. Nonmyeloablative HLA-matched sibling allogeneic hematopoietic stem cell transplantation for severe sickle cell phenotype. JAMA. 2014;312(1):48–56. doi: 10.1001/jama.2014.7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pinana J.L., Perez-Pitarch A., Garcia-Cadenas I. A time-to-event model for acute kidney injury after reduced-intensity conditioning stem cell transplantation using a Tacrolimus- and Sirolimus-based graft-versus-host disease prophylaxis. Biol Blood Marrow Transpl. 2017;23(7):1177–1185. doi: 10.1016/j.bbmt.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 53.Boehm A., Mayerhofer M., Herndlhofer S. Evaluation of in vivo antineoplastic effects of rapamycin in patients with chemotherapy-refractory AML. Eur J Intern Med. 2009;20(8):775–778. doi: 10.1016/j.ejim.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 54.Chiarini F., Fala F., Tazzari P.L. Dual inhibition of class IA phosphatidylinositol 3-kinase and mammalian target of rapamycin as a new therapeutic option for T-cell acute lymphoblastic leukemia. Cancer Res. 2009;69(8):3520–3528. doi: 10.1158/0008-5472.CAN-08-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiarini F., Grimaldi C., Ricci F. Activity of the novel dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor NVP-BEZ235 against T-cell acute lymphoblastic leukemia. Cancer Res. 2010;70(20):8097–8107. doi: 10.1158/0008-5472.CAN-10-1814. [DOI] [PubMed] [Google Scholar]
- 56.Mayerhofer M., Valent P., Sperr W.R. BCR/ABL induces expression of vascular endothelial growth factor and its transcriptional activator, hypoxia inducible factor-1alpha, through a pathway involving phosphoinositide 3-kinase and the mammalian target of rapamycin. Blood. 2002;100(10):3767–3775. doi: 10.1182/blood-2002-01-0109. [DOI] [PubMed] [Google Scholar]
- 57.Recher C., Beyne-Rauzy O., Demur C. Antileukemic activity of rapamycin in acute myeloid leukemia. Blood. 2005;105(6):2527–2534. doi: 10.1182/blood-2004-06-2494. [DOI] [PubMed] [Google Scholar]
- 58.Recher C., Dos Santos C., Demur C. mTOR, a new therapeutic target in acute myeloid leukemia. Cell Cycle. 2005;4(11):1540–1549. doi: 10.4161/cc.4.11.2159. [DOI] [PubMed] [Google Scholar]
- 59.Itakura E., Kishi C., Inoue K. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19(12):5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong Y., Wang Q.J., Li X. Distinct regulation of autophagic activity by Atg14l and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11(4):468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nicotra G., Mercalli F., Peracchio C. Autophagy-active beclin-1 correlates with favourable clinical outcome in non-Hodgkin lymphomas. Mod Pathol. 2010;23(7):937–950. doi: 10.1038/modpathol.2010.80. [DOI] [PubMed] [Google Scholar]
- 62.Gao X., Zacharek A., Salkowski A. Loss of heterozygosity of the BRCA1 and other loci on chromosome 17q in human prostate cancer. Cancer Res. 1995;55(5):1002–1005. [PubMed] [Google Scholar]
- 63.Saito H., Inazawa J., Saito S. Detailed deletion mapping of chromosome 17q in ovarian and breast cancers: 2-cM region on 17q21.3 often and commonly deleted in tumors. Cancer Res. 1993;53(14):3382–3385. [PubMed] [Google Scholar]
- 64.Futreal P.A., Soderkvist P., Marks J.R. Detection of frequent allelic loss on proximal chromosome 17q in sporadic breast carcinoma using microsatellite length polymorphisms. Cancer Res. 1992;52(9):2624–2627. [PubMed] [Google Scholar]
- 65.Eccles D.M., Russell S.E., Haites N.E. Early loss of heterozygosity on 17q in ovarian cancer. The abe ovarian cancer genetics group. Oncogene. 1992;7(10):2069–2072. [PubMed] [Google Scholar]
- 66.Russell S.E., Hickey G.I., Lowry W.S. Allele loss from chromosome 17 in ovarian cancer. Oncogene. 1990;5(10):1581–1583. [PubMed] [Google Scholar]
- 67.Wang K., Yang C., Shi J. Ox-LDL-induced lncRNA MALAT1 promotes autophagy in human umbilical vein endothelial cells by sponging miR-216a-5p and regulating Beclin-1 expression. Eur J Pharmacol. 2019;858 doi: 10.1016/j.ejphar.2019.04.019. [DOI] [PubMed] [Google Scholar]
- 68.Zengin E., Chalajour F., Gehling U.M. Vascular wall resident progenitor cells: a source for postnatal vasculogenesis. Development. 2006;133(8):1543–1551. doi: 10.1242/dev.02315. [DOI] [PubMed] [Google Scholar]
- 69.Uenishi G.I., Jung H.S., Kumar A. NOTCH signaling specifies arterial-type definitive hemogenic endothelium from human pluripotent stem cells. Nat Commun. 2018;9(1):1828. doi: 10.1038/s41467-018-04134-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bowers E., Slaughter A., Frenette P.S. Granulocyte-derived TNFalpha promotes vascular and hematopoietic regeneration in the bone marrow. Nat Med. 2018;24(1):95–102. doi: 10.1038/nm.4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin D.K., Shido K., Kopp H.G. Cytokine-mediated deployment of SDF-1 induces revascularization through recruitment of CXCR4+ hemangiocytes. Nat Med. 2006;12(5):557–567. doi: 10.1038/nm1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clarke R.L., Robitaille A.M., Moon R.T. A quantitative proteomic analysis of hemogenic endothelium reveals differential regulation of hematopoiesis by SOX17. Stem Cell Rep. 2015;5(2):291–304. doi: 10.1016/j.stemcr.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moore M.A. Waking up HSCs: a new role for E-selectin. Nat Med. 2012;18(11):1613–1614. doi: 10.1038/nm.2992. [DOI] [PubMed] [Google Scholar]
- 74.Crane G.M., Jeffery E., Morrison S.J. Adult haematopoietic stem cell niches. Nat Rev Immunol. 2017;17(9):573–590. doi: 10.1038/nri.2017.53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.