Abstract
Background
As a reader of histone H3K4me3, BPTF associated protein of 18 kDa (BAP18) is involved in modulation of androgen receptor action in prostate cancer. However, the function of BAP18 on oral squamous cell carcinoma (OSCC) and its molecular mechanism remains to be elusive.
Methods
OSCC-derived cell lines carrying silenced BAP18 were established by Lentiviral infection. Quantitative PCR (qPCR), western blot, and ChIP assay were performed to detect gene transcription regulation and the possible mechanism. Colony formation, cell growth curve and xenograft tumor experiments were performed to examine cell growth and proliferation.
Findings
Our study demonstrated that BAP18 was highly expressed in OSCC samples compared with that in benign. BAP18 depletion obviously influenced the expression of a series of genes, including cell cycle-related genes. We thus provided the evidence to demonstrate that BAP18 depletion significantly decreases CCND1 and CCND2 (CCND1/2) transcription. In addition, BAP18 is recruited to the promoter regions of CCND1/2, thereby facilitating the recruitment of the core subunits of MLL1 complex to the same regions, to increase histone H3K4me3 levels. Furthermore, BAP18 depletion delayed G1-S phase transition and inhibited cell growth in OSCC-derived cell lines.
Interpretation
This study suggests that BAP18 is involved in modulation of CCND1/2 transcription and promotes OSCC progression. BAP18 could be a potential target for OSCC treatment and diagnosis.
Fund
This work was funded by National Natural Science Foundation of China (31871286, 81872015, 31701102, 81702800, 81902889), Foundation for Special Professor of Liaoning Province, and Supported project for young technological innovation-talents in Shenyang (No. RC170541).
Keywords: BAP18, CCND1/2, Transcription regulation, Histone methylation, Oral squamous cell carcinoma
Research in context.
Evidence before this study
Oral squamous cell carcinoma (OSCC) displays a poor prognosis with above half of patients surviving less than 5 years. Rapid tumor growth and tumor recurrence remains the big challenges for OSCC. Thus, finding the biomarker for tumor progression would be helpful to understand tumor development and find the new therapeutic target for OSCC. BPTF associated protein of 18 kDa (BAP18) as a reader of histone H3K4me3 is a subunit of MLL1 complex involved in maintenance of active transcription. BAP18 plays a vital role in prostate cancer progression. However, the biological function and molecular mechanism of BAP18 in OSCC is largely unknown.
Added value of this study
This study has demonstrated that BAP18 is highly expressed in OSCC tissues. The depletion of BAP18 significantly decreases CCND1/2 transcription. Moreover, BAP18 facilitates the recruitment of core subunits of MLL1/WDR5 complex to the promoter region of CCND1/2, thereby increasing histone H3K4me3 levels. BAP18 depletion inhibits the cell growth in OSCC-derived cells. These data suggest that BAP18 possibly contributes to rapid tumor growth and BAP18 could act as a potential biomarker and therapeutic target in OSCC.
Implications of all the available evidence
Our results implicate the biological function of BAP18 in OSCC, and its molecular mechanism underlying the regulation function of BAP18 on CCND1/2 transcription. Thus, our study may provide a new therapeutic target for OSCC.
Alt-text: Unlabelled box
1. Introduction
Oral squamous cell carcinoma (OSCC) displays a significant health threat and poor prognosis with above half of patients surviving less than 5 years [1], [2], [3], [4], [5]. There are difficulties for obtaining acceptable results for advanced OSCC (stages III and IV), though early OSCC considered stages I/II can be alleviated by surgery or radiotherapy [6,7]. Some approaches just like targeted therapy, immunotherapy, and radioactive seed implantations seem not to be fully adequate in clinic due to the tumor malignant proliferation [8]. Patients who are not candidates for salvage surgery or re-irradiation usually receive chemotherapy, but even with the most recent combinations of drugs the prognosis remains poor and cure is rare [8], [9], [10], [11]. Moreover, OSCC has an easy-characterized progression from teratogenesis through dysplasia to carcinoma with a multi-step process including the accretion of diverse genetic and epigenetic in oncogenes, inducing dysregulation of multiple signaling pathways, which disturbed the cell cycle and the balance between cell proliferation and cell death [12]. Rapid tumor growth and tumor recurrence remains the big challenges for OSCC. Thus, finding the biomarker for tumor progression would be helpful to understand tumor development and find the new therapeutic target for OSCC.
Cell cycle progression is mainly dominated by the interplay of cyclin-dependent kinases and cyclin family members, which are characterized by a dramatic periodicity in protein abundance throughout the cell cycle [13]. Three types of cyclin family members are reported as Cyclin-Ds, including CyclinD1, D2 and D3, which are involved in controlling cell cycle phase transition and cell mitotic growth [13], [14], [15]. Cyclin-Ds are encoded by diverse genes (CCNDs) with significant amino acid similarity [16,17]. The gene transcription of CCNDs can be respectively induced by transcription factor. GATA3 cooperates with PARP1 to induce CCND1 gene transcription in breast cancer cells [18]. Once induced, Cyclin-Ds associate with partner CDKs to cooperatively drive cells from G1 phase to S phase. Cyclin-Ds dysregulation have been shown to contribute to tumorigenesis, tumor malignant proliferation and poorer outcomes in number of mammalian cancers, including breast cancer, ovarian cancer, leukemia and so on [19], [20], [21], [22], [23], [24], [25], [26]. Thus, Cyclin-Ds play crucial roles in regulating cell cycle process. However, the molecular mechanism for upstream modulation of CCNDs gene transcription in OSCC remains to be elusive.
BPTF associated protein of 18 kDa (BAP18) as a reader of histone H3K4me3 is a subunit of MLL1/WDR5 complex involved in active transcription. Carrying a SANT domain, BAP18 is considered that it may possess a key role in chromatin remodeling as well as histone modification [27], [28], [29]. In our previous study, BAP18 was identified as a coactivator of androgen receptor (AR) and promoted prostate cancer progression [30]. However, the biological function of BAP18 and the molecular mechanism underlying the regulation function of BAP18 on gene transcription in OSCC is largely unknown.
In this study, our results have demonstrated that BAP18 is highly expressed in OSCC samples, compared with that in non-cancerous oral epithelial tissues by western blotting and immunohistochemistry (IHC) experiments. Furthermore, BAP18 depletion influenced the transcription of a series of genes, including cell cycle-related genes. We provided the evidence to show that knockdown of BAP18 significantly decreased the transcription of genes, such as CCND1, CCND2, ATF4 etc. Meanwhile, BAP18 depletion abrogated the protein expression of CyclinD1 and CyclinD2. Importantly, chromatin immunoprecipitation (ChIP) assays results showed that BAP18 depletion decreased the recruitment of the core subunits of MLL1/WDR5 complex to promoter regions of CCND1 or CCND2, thereby reducing histone H3K4me3 level. Our results further demonstrated that knockdown of BAP18 decreased the cell growth/proliferation in OSCC-derived cells. Moreover, the promotion of cell growth by BAP18 was reduced by CCND1/2 depletion. Taken together, our data suggested the biological function of BAP18 on OSCC progression is related to Cyclin D1/2, and thus implicated that BAP18 could act as a potential biomarker and therapeutic target in OSCC.
2. Materials and methods
2.1. Cell culture
Cal-27 cells and SCC9 cells were all grown in Roswell Park Memorial Institute medium 1640 (RPMI-1640) with 10% FBS (CLARK) and penicillin/streptomycin. Both two cells were cultured with 5% CO2 at 37 °C.
2.2. Plasmids and antibodies
BAP18 expression plasmid was constructed with pcDNA3-FLAG carrier plasmid and BAP18 cDNA as previously described [30].
The antibodies were used in our study as followed: anti-BAP18 (Bethyl #A304-207A-1), anti-Cyclin D1 (Cell signaling #DCS6), anti-Cyclin D2 (ABclonal #A1773), anti-GAPDH (Kangchen #KC5G4), anti-WDR5 (Bethyl #A302-429A-2), anti-ASH2 (Bethyl #A300-107A-2), anti-DPY30 (Abcam #ab187690), anti-H3K4me3/H3Ac/H3K9Ac/H4Ac/H4K16Ac (Millipore), anti-Rabbit/Mouse (ABclonal), anti-IgG (Santa #sc-2025).
2.3. RNA isolation, quantitative real-time PCR (qPCR)
Total RNA was isolated by Trizol (TAKARA) and cDNAs were reversed by PrimeSripte™RT-PCR Kit (TAKARA). q-PCR analysis was performed with the SYBR Premix Ex Taq kit (TAKARA) and runon LightCycler® 96 (Roche). Gene expressions were normalized to β-Actin. All results were from an individual experiment that was representative of at least three independent experiments. All primers for qPCR were listed in Supplementary Table S1.
2.4. siRNA and lentivirus
siRNA sequences against BAP18, CCND1 and CCND2 used in this work were shown in Supplementary Table S2. All siRNAs were transfected by jet-PRIME transfection reagent (Polyplus).
Lentiviral production of BAP18 targeted the same sequence as siBAP18#1 and the lentivirus were purchased from Shanghai GeneChem Company. Lentivirus system were transfected into cell following manufacturer's instructions.
2.5. Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed with the previously described protocol [31]. Two kinds of OSCC cells were transfected with siRNAs and cultured for at least 48 h until 90% cell confluency. The purified DNA was analyzed by q-PCR. Results were shown as percentage of input chromatin, and every experiment was represented from at least three independent experiments. The primers for q-PCR were listed in the Supplementary Table S3.
2.6. FACS analysis, colony formation and cell growth curve
The cells were grown in six-well plates for 24 h. Then, the cells were dissociated with trypsin, resuspended in PBS, and then fixed in ice-cold 70% ethanol. Next, the cells were incubated in propidium iodide/RNAse solution at 37 °C for 1 h. The cell-cycle analysis was performed by a FACS flow cytometer.
Two kinds of OSCC cell were planted into 35 mm dishes for 400 cells per dish for colony formation and 2000 cells for cell growth. In colony formation, cells were cultured for at least 14 days. After individual colony formation appearance, the dishes were washed with ice PBS and immobilized with 4% paraformaldehyde for 30 min. Pictures were taken after dyed with R250 for 120 min.
Cell growth curve were drawn by everyday’ cell accounts.
2.7. Xenograft tumor
About 5.0 million OSCC cells stably knockdown BAP18 (shBAP18) and its negative control (shCtrl) were appended in 50 μl medium and 50 μl Metrigel (BD Biosciences) mixture before injections. Tumor size were measured by electronic caliper and the tumor volume formal was as followed: V = (π/6) × (L × W)3/2, V for volume (mm3), L for biggest diameter (mm), W for smallest diameter (mm). Animal works were approved and supervised by the Animal Ethics Committee of China Medical University.
2.8. Tissue samples and immunohistochemistry (IHC)
All OSCC samples and non-cancerous tissues were obtained from the hospital of stomatology of China Medical University, and all patients were treated with curative intent. Tumor samples from OSCC patients were obtained from surgeries without primary treatments, including radiotherapy and chemotherapy or adjuvant therapies. Non-cancerous oral epithelium tissues (non-cancerous tissues) from patients were got from general outpatient surgeries. The non-cancerous tissues examined in this study included focal epithelial hyperplasia tissues and gingival epithelial tissues. Clinical OSCC Sections were prepared from the hospital of stomatology of China Medical University, and the IHC experiments were performed as previously described [30]. The collection of tissue samples and all investigations were approved by the Human Research Ethics Committee of China Medical University. Patient informed consents from all samples were already obtained.
The immunohistochemical staining results were assigned with the German immunoreactive scoring system using immunoreactive scores (IRSs). Briefly, the intensity was scored as follows: 0, negative; 1, weak; 2, moderate and 3, strong. The proportion of positive-staining cells was scored as follows: 0, less than 5%; 1, 5% to 25%; 2, 26% to 50%; 3, 51% to 75%; 4, greater than 75%. The final score, which ranged from 0 to 12, was determined by summing the intensity score multiplied by the corresponding proportion score.
2.9. Statistics
All statistical analysis was performed with Prism Graphpad program (https://www.graphpad.com/). Students’ t-tests using two-tails were adopted for determination of relevance of two groups. For clinical data analysis, Chi-square test was adopted. Statistically significant were considered as P-values of *p<0.05; **p<0.01; and ***p<0.001.
3. Results
3.1. BAP18 is highly expressed in clinical OSCC samples
Our previous results have demonstrated that BPTF associated protein of 18 kDa (BAP18) as a reader of histone H3K4me3 displays oncogenic function in prostate cancer growth [30]. However, the biological function and molecular mechanism underlying the modulation of BAP18 in gene transcription in OSCC characterized as rapid growth and malignant recurrence still need to be elusive. To evaluate BAP18 expression in OSCC, we first turned to western blotting analysis using the fresh clinical OSCC tissues. The results showed that BAP18 was significantly higher expressed in OSCC tissues, compared with that in non-cancerous oral epithelial tissues (non-cancerous tissues, NC) (Fig. 1a and b). To further confirm the expression level of BAP18 in OSCC samples, IHC assays were performed in amount of OSCC samples from surgery patients. The results have demonstrated thatBAP18 was strongly stained in OSCC tissues, whereas weakly stained in non-cancerous tissues. In OSCC samples, higher expression of BAP18 was shown in stage III/IV, and lower expression in stage I/II), indicating that BAP18 expression is positively correlated with clinical stages (Fig. 1c and d). Moreover, univariate analysis demonstrated that high expression ofBAP18 was significantly correlated with primary tumor size and clinical stages, but no correlation was found between BAP18 expression and gender, age, tumor lymphatic metastasis, distal metastasis, or location. Multivariate analysis of primary tumor size and clinical stages suggested that clinical stage is an independent predictor of BAP18 expression (Table 1). Ki67 expression has been considered to assess the ability of tumor proliferation. To determine the correlation between the expression level of BAP18 and tumor proliferation, we thus turned to examine the expression of Ki67 and BAP18 in paraffin sections from the same OSCC samples. The results showed that BAP18 expression was positively correlated with that of Ki67 (Fig. 1e and f). In addition, we further analyzed the correlation between BAP18 expression and disease-free survival using head and neck squamous cell carcinomas (HNCC) cohort data in the cancer genome atlas (TCGA), although no evidences in OSCC patients in TCGA. The results showed that high expression of BAP18 in HNCC patients moderately contributed to poor prognosis in short-term survival (0–100 months), but no significant correlation with the prognosis from 100–200 months (Fig. 1g). Taken together, these results suggest that BAP18 is highly expressed in OSCC tissues. BAP18 might act as a potential biomarker for early OSCC diagnosis.
Fig. 1.
BAP18 is highly expressed in clinical OSCC samples. (a) Expression of BAP18 in unpaired fresh tissues of oral non-cancerous (NC) tissues and OSCC tissues. (b) Grayscale values of western blotting in (a), Student t-test was used for statistical significance between two groups. (c) BAP18 expression in non-cancerous (NC) tissues and OSCC samples with different clinical stages. Magnification: 4* and 20*; Scale bars: 500 μm and 100 μm. (d) Statistical significance among non-cancerous tissues, early stages tissues (I and II) and terminal stages tissues (III and IV). Average score of three independent view calculation represents the sample final score. Mann-Whitney U test were used, ***p<0.001. (e) Ki67 and BAP18 expression in OSCC samples. Magnification: 10*; Scale bars: 100 μm. (f) Scatter diagram of two proteins (BAP18 and Ki67) expression in the same sample, and the straight line was generated by Prism Graphpad7.0. (g) Disease free survival curves was generated from GEPIA showing BAP18 contribution to HNCC patients living from TCGA cohorts (http://gepia.cancer-pku.cn/index.html).
Table 1.
Expression of BAP18 in OSCC samples with clinical characteristics.
| Variables | Groups | Cases (n = 90) | BAP18 low expression (n = 26) | BAP18 high expression (n = 64) | p-value |
|---|---|---|---|---|---|
| Univariate analysis | |||||
| Age | ≤55 | 26 | 6 | 20 | |
| >55 | 64 | 20 | 44 | 0.6039 | |
| Gender | Male | 66 | 17 | 49 | |
| Female | 24 | 9 | 15 | 0.4100 | |
| T | T1/T2 | 42 | 18 | 24 | |
| T3+ | 48 | 8 | 40 | 0.003** | |
| N | N0 | 26 | 6 | 20 | |
| N0+ | 64 | 20 | 44 | 0.6039 | |
| M | M0 | 63 | 18 | 45 | |
| M0+ | 27 | 8 | 19 | 0.9192 | |
| Clinical stage | I/II | 24 | 17 | 7 | |
| III+ | 66 | 9 | 57 | <0.001*** | |
| Differentiation stage | Well/Moderately | 86 | 20 | 66 | |
| Poor | 4 | 1 | 3 | 0.9999 | |
| Location | Tongue | 25 | 4 | 21 | |
| other | 65 | 22 | 43 | 0.1575 | |
| Multivariate analysis | |||||
| T | T1/T2 | 42 | 18 | 24 | |
| T3+ | 48 | 8 | 40 | 0.276 | |
| Clinical stage | I/II | 24 | 17 | 7 | |
| III+ | 66 | 9 | 57 | <0.001*** | |
Abbreviations: T= tumor; N=lymph Node; M=metastasis.
Notes: In univariate analysis, Chi-square test were used, ***p<0.001, **p<0.01, *p<0.05; and in multivariate analysis, Hosmer and Lemeshow test were used, ***p<0.001.
3.2. BAP18 depletion inhibits the transcription of a series of genes in OSCC-derived cell lines
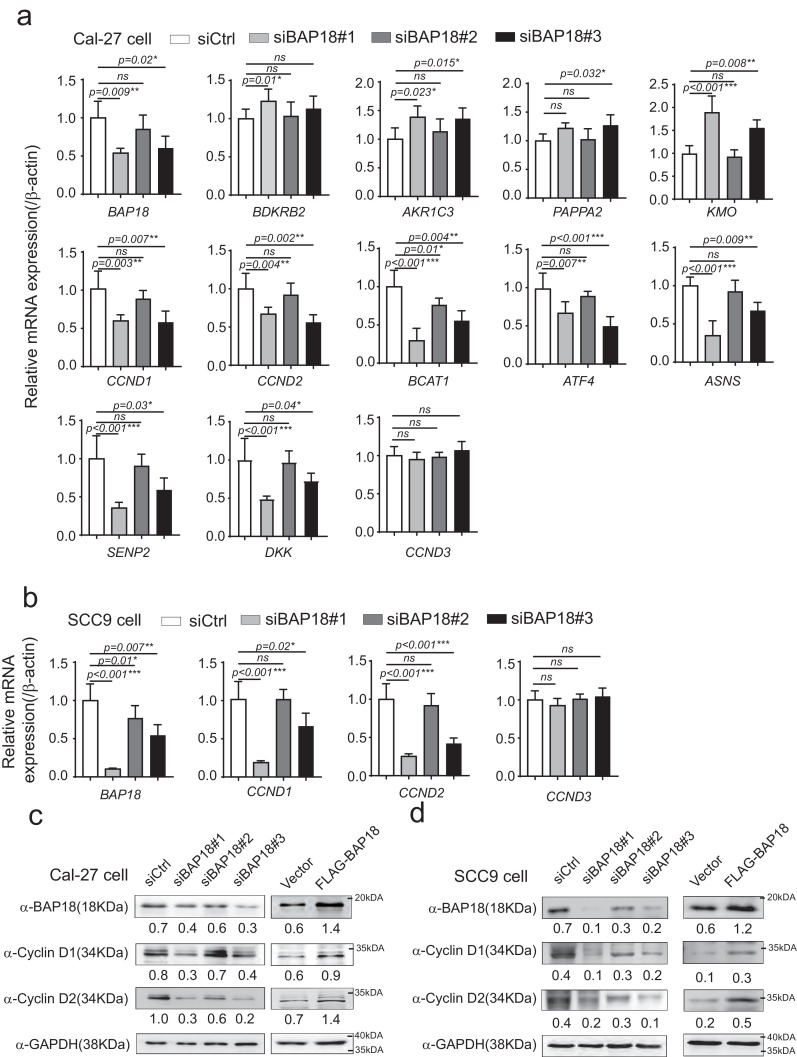
To validate the molecular mechanism underlying the modulation of BAP18 on cancer-related genes, we performed the quantitative PCR (qPCR) experiments to determine the effect of BAP18 on the transcription of a series of genes as shown in Fig. 2. Three independent siRNAs against BAP18 (siBAP18s) were generated for knocking down BAP18 in OSCC-derived cell line (Cal-27 cell). Among them, no.1 and no. 3 siBAP18s (siBAP18#1 and siBAP18#3) significantly decreased BAP18 mRNA expression. Our results have demonstrated that knockdown of BAP18 increased BDKRB2, AKR1C3, PAPPA2, and KMO mRNA expression; whereas the depletion of BAP18 obviously decreased a series of gene transcription, including CCND1 and CCND2. However, BAP18 had no effect on gene transcription of CCND3, which is also a member of Cyclin-Ds family in Cal-27 cell line (Fig. 2a). We further provided the evidence to show that BAP18 depletion decreased CCND1 and CCND2 transcription in another OSCC-derived cell line, SCC9 (Fig. 2b). Moreover, the results from qPCR experiments performed in Cal-27 cells or SCC9 cells carrying ectopic expression of BAP18 (FLAG-BAP18) were in agreement with those from qPCR experiments with BAP18 depletion (Supplementary Fig. S1a and b). We further examined the influence of BAP18 on protein expression of Cyclin D1/D2 in OSCC-derived cell lines. The results showed that ectopic expression of BAP18 enhanced Cyclin D1/D2 protein expression, whereas the depletion of BAP18 decreased the expression of those proteins in Cal-27 cells and SCC9 cells (Fig. 2c and d). Collectively, the results suggest that BAP18 is involved in modulation of the transcription of a series of genes, including CCND1 and CCND2 in OSCC-derived cell lines.
Fig. 2.
BAP18 significantly regulates the transcription of a series of genes in OSCC-derived cell lines. (a) The effects of BAP18 depletion on a series of genes in Cal-27 cells by real-time quantitative PCR. Three independent siRNAs against BAP18 (siBAP18s) were designed and generated for BAP18 depletion. Levels of all kinds of RNAs as indicated were normalized to that of β-Actin. Student t-test were used, ns stands for no significant, ***p<0.001, **p<0.01, *p<0.05. (b) Effect of knockdown of BAP18 on mRNA expression of CCND1, CCND2, or CCND3 in SCC9 cells. Student t-test were performed, ns stands for no significant, ***p<0.001, **p<0.01, *p<0.05. (c,d) Effects of depletion of BAP18 or ectopic expression of BAP18 on protein expression of cyclin D1 and cyclin D2 by western blotting in Cal-27 cells (c) or SCC9 cells (d) with indicated antibodies.
3.3. BAP18 depletion reduces the recruitment of the core subunits of MLL1 complex to the promoter regions of CCND1/2
It has been previously demonstrated that BAP18 as a reader of histone H3K4me3 is a subunit of MLL1/WDR5 complex, which triggers histone H3K4me3 to be involved in maintenance of active transcription. Having demonstrated that BAP18 enhances the gene transcription of CCND1/2 as shown in Fig. 3, we thus turn to ask whether BAP18 could be recruited to the promoter regions of CCND1/2, and what is the molecular mechanism underlying the modulation of BAP18 on CCND1/2 transcription. We then examined the sequences of CCND1/2 and designed two pairs of primers within 1000 bps to their transcription start sites (TSS) for chromatin immunoprecipitation (ChIP) assay (Fig. 4a). ChIP assay was performed to determine the recruitment of BAP18to the promoter regions of CCND1/2 in Cal-27 cells. The results have demonstrated that BAP18 or the core subunit of MLL1/WDR5 complex, including WDR5, ASH2 and DPY30, was recruited to the promoter regions of CCND1/2. Meanwhile, ChIP assay was further performed in Cal-27 cells with BAP18 depletion induced by shRNA against BAP18 (shBAP18). The results have shown that BAP18 depletion significantly reduced the recruitment of the core subunit of MLL1/WDR5 complex to the same regions. In addition, we examined the effect of BAP18 on modulation of histone modification levels at two promoter regions. The results have shown that BAP18 knockdown obviously diminished the histone H3K4me3 level at the promoter regions of CCND1/2, whereas H3Ac and H4Ac levels were differently influenced by BAP18 as indicated (Fig. 4b–e). Taken together, our results suggest that BAP18 depletion reduces the recruitment of the core subunit of MLL1/WDR5 complex, thereby influencing histone H3K4me3 or H3/H4Ac levels on the promoter regions of CCND1/2 in Cal-27 cells.
Fig. 3.
BAP18 is recruited to the promoter regions of CCND1/2. (a) Schematic representation of primers designed for CCND1 and CCND2 gene promoter-TSS regions. BBS is an abbreviation of BAP18 binding sites. (b–e) ChIP assays performed on the promoter regions of CCND1/2 with the indicated antibodies in Cal-27 cells with shCtrl or shBAP18. Student t-test was performed, ns stands for no significant, ***p<0.001, **p<0.01, *p<0.05.
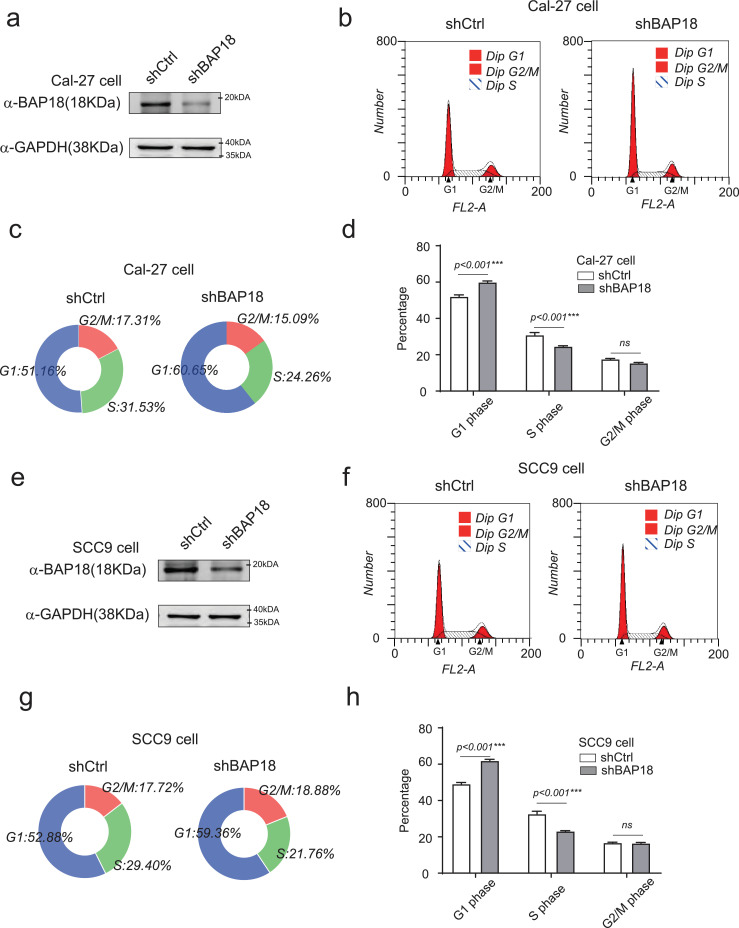
Fig. 4.
BAP18 depletion influences the G1-S phase in OSCC-derived cell lines. (a) Cal-27 cells were infected with lentivirus against BAP18 (shBAP18) and its negative control (shCtrl). The efficiency of the shBAP18 detected by western blotting in Cal-27 cells. (b–d) The impact of BAP18 on cell cycle progression by flow cytometry assay in Cal-27 cells. (e) SCC9 cells were infected with lentivirus against BAP18 (shBAP18) and its negative control (shCtrl). The efficiency of the shBAP18 detected by western blotting in SCC9 cells. (f–h) The impact of BAP18 on cell cycle progression by flow cytometry assay in SCC9 cells. All statistics were performed by Graphpad7.0 with Student t-test, ns stands for no significant, ***p<0.001, **p<0.01, *p<0.05.
3.4. BAP18 promotes cell growth/proliferation in OSCC-derived cell lines
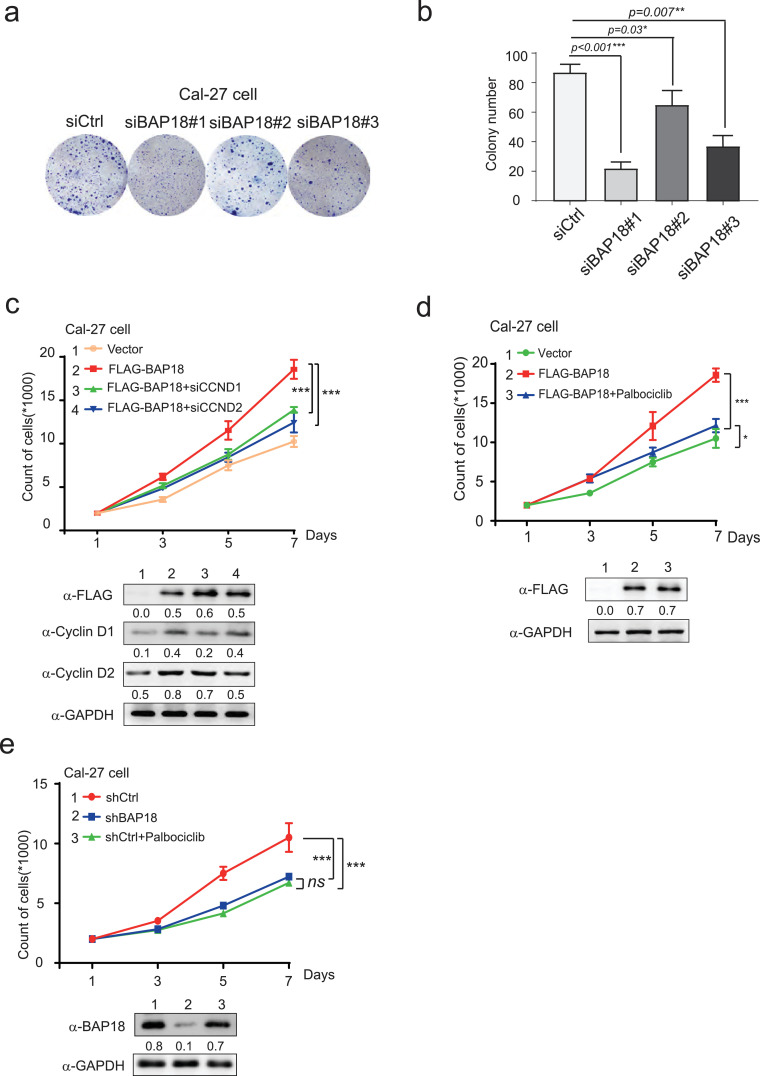
Cyclin D1 and Cyclin D2 are the members of Cyclin-Ds family, controlling cell cycle G1 to S phase transition to be essential for cell mitotic process. Having shown that BAP18 was involved in modulation of CCND1/2 mRNA expression, indicating that BAP18 might influence cellular mitotic G1/S phases in OSCC cells, flow cytometry analysis was performed to detect the effect of BAP18 on cell cycle process. The results demonstrated that the depletion of BAP18 significantly delayed cell G1-S phase transition and the number of Cal-27 cells which lagged into G1 phase increased from 51% to 60%, and S phase decreased from 31.53% to 24.26% (Fig. 4a–d). The similar experiments were further performed in another OSCC-derived cell line, SCC9 cells. In agreement with the results generated from Cal-27 cells, the data from SCC9 demonstrated that BAP18 depletion obviously delayed cell G1-S phase transition (Fig. 4e–h). Moreover, colony formation experiments were performed to examine the influence of BAP18 on cell growth with three independent siRNAs against BAP18 (siBAP18#1, #2, #3). The results showed that BAP18 depletion by siBAP18#1 and siBAP18#3 obviously decreased the colony numbers in Cal-27 cells, suggesting that BAP18 depletion inhibits cell growth in OSCC-derived cell line (Fig. 5a and b). In addition, in order to examine the effect of BAP18 on cell growth is related with CCND1/2 signaling pathway, siRNA against CCND1 or CCND2 (siCCND1/2) or Palbociclib, a selective CDK4/6 inhibitor was used for cell growth experiments. Cal-27 cells with ectopic expression of BAP18 (FLAG-BAP18) were treated with siCCND1/2 or Palbociclib. The results demonstrated that the enhancement of cell growth by ectopic expression of BAP18 was decreased by siCCND1/2 or Palbociclib (Fig. 5c and d). Meanwhile, we analyzed the function of BAP18 depletion or CDKs inhibitor on cell growth, the results showed that BAP18 depletion has the similar effect as that of Palbociclib on suppression of cell growth (Fig. 5e). Taken together, the results suggest that BAP18 is involved in promotion of cell growth, if not all, at least partially through Cyclin D1/2-CDKs pathway in OSCC.
Fig. 5.
BAP18 promotes the cell proliferation in OSCC-derived cell lines. (a) Colony formation assays in Cal-27 cells carrying siCtrl or siBAP18s (siBAP18#1, 2, 3). Amount of 400 cells were seeded into 35 mm dishes for 14 days and stained by crystal violet. (b) Representation of histogram for colony formation experiments. Standard of account is one cell growth colony contains more than 50 cloned cells. (c) siRNAs against CCND1/CCND2 (siCCND1/2) were transfected into Cal-27 cells with overexpression plasmid of BAP18 tagged with FLAG or vector plasmids for Growth curve analysis. (d) Growth curve analysis for Cal-27 cells with overexpression plasmids of BAP18 tagged with FLAG or vector plasmids were treated with CDK4/6 inhibitor (Palbociclib, 25 nM). (e) Growth curve analysis for Cal-27 cells with depletion of BAP18 (shBAP18) or shCtrl were treated with CDK4/6 inhibitor (Palbociclib, 25 nM). The expression levels of proteins as indicated were detected by western blotting shown at the bottom of panel c, d, e. Error bars represent mean ± SD. Student t-test was used, ns stands for no significant. ***p<0.001, *p<0.05.
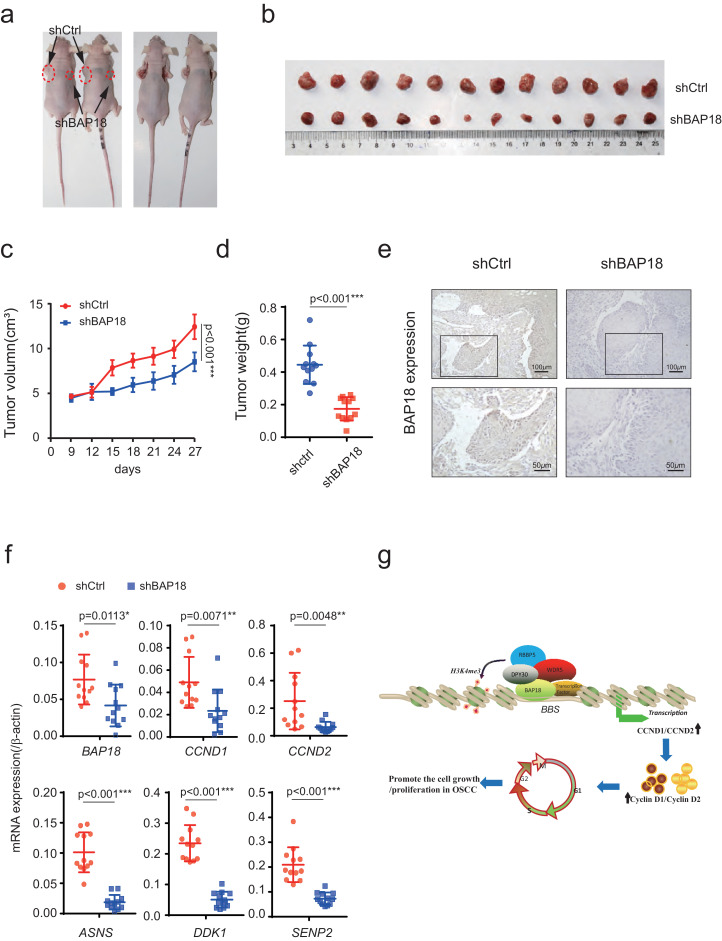
3.5. Knockdown of BAP18 inhibits the cell growth of OSCC in vivo
We next intend to explore the biological function of BAP18 on cell growth of OSCC in vivo. Firstly, we constructed the lentivirus-mediated stable BAP18 knockdown (shBAP18) and control (shCtrl) Cal-27 OSCC cells for xenografts in4-week-old BALB/c nude mice. The results have demonstrated that the depletion of BAP18 forms smaller xenograft tumor than that of control (Fig. 6a and b). Measuring the tumor volume and tumor weight, we found BAP18 knockdown cells had slower growth rate, and the average tumor weight was lower than that of control (Fig. 6c and d). Next, we detected BAP18 protein expression in two kinds of xenograft tumors by IHC assays. The immunohistochemical results showed that BAP18 was lower expressed in BAP18 knockdown xenograft tumors (Fig. 6e). Meanwhile, we detected the mRNA expression in xenograft tumor. The results showed that BAP18, CCND1, CCND2, ASNS, DDK1, and SENP2 mRNA were significantly lower in shBAP18 xenograft tumors than that in shCtrl xenograft tumors (Fig. 6f). Our data demonstrated that BAP18 knockdown suppressed the cell growth of OSCC in vivo.
Fig. 6.
BAP18 depletion inhibits the OSCC-derived cell growth in xenograft mice. (a) Representative photos were pictured of mice xenograft tumor. shCtrl (left) and shBAP18 (right) were stably expression in Cal-27 cells and injected into male mice at 4 weeks old. (b) Xenograft tumors were shown after mice were killed. (c) The average tumor volume of shCtrl and shBAP18 were measured every six days. Bars represented standard alteration of the mean, ***p<0.001. (d) Tumor weights were measured and statistically calculated with Student t-test, ***p<0.001. (e) Expressions of BAP18 proteins in xenograft tumors were detected by IHC assays. Magnification: 10*; Scale bars: 100 μm. (f) RNA isolated from xenograft tumors and the mRNA expression of BAP18, CCND1, CCND2, ASNS, DDK1andSENP2 was shown as indicated. Student t-test was used, *p<0.05, **p<0.01. (g) Schematic representation of the function of BAP18on up-regulation of CCND1/2 transcription. BAP18 is recruited to the promoter regions of CCND1/2, thereby facilitating the recruitment of the core subunits of MLL1 complex to enhance CCND1/2 transcription, to promote the cell growth and proliferation in OSCC progression.
4. Discussion
We have previously identified that BPTF associated protein of 18 kDa (BAP18) as a key subunit of multimeric histone methyltransferase complexes participates in modulation of androgen receptor-induced transactivation and plays a crucial role in the progression of prostate cancer. However, the biological function of BAP18 in OSCC is largely unclear. In this study, we provided the evidence to show that BAP18 is highly expressed in clinical OSCC samples compared with that in the non-cancerous oral epithelial tissues (non-cancerous tissues, NC). Using quantitative RT-PCR (qPCR) experiments, we have demonstrated BAP18 depletion up-regulates or down-regulates a series of genes in OSCC cells. Focusing on the regulation of cell cycle pathway, our results showed that BAP18 is significantly modulate the transcription of CCND1 and CCND2. Moreover, BAP18 was recruited to the promoter regions of CCND1 and CCND2, and BAP18 facilitates the recruitment of the core subunits of MLL1/WDR5 complex, thereby regulating the histone H3K4me3 level to enhance CCND1 and CCND2 transcription. Finally, BAP18 depletion inhibits cell growth and proliferation in OSCC-derived cell lines (Fig. 6g).
Oral squamous cell carcinoma is considered to be a rapid proliferation malignant tumor. Patients with T1-2N0M0 OSCC can be treated with surgery, whereas T3+ OSCC patients undergoing radiotherapy or immunotherapy still exhibits poor outcomes [32]. Although the five-year survival rate of oral cancer is 62.1% (2003–2009), survival rates aggravate with advancement in clinical stages [33], [34], [35]. Terminal OSCC patients (stage III or IV) always show malignant proliferation, deadly lymphatic metastasis and low reaction to drug therapy. Therefore, it is necessary to identify the novel biomarkers for OSCC early diagnosis. In this study, we identified BAP18, a reader of histone H3K4me3, was significantly highly expressed in OSCC tumor tissue, compared with that in non-cancerous tissues. In the immunohistochemistry (IHC) experiments with OSCC tissue sections, the results demonstrated that BAP18 exhibited abundantly expression in evolved stages (stage III/IV), while relative low-level expression in early stages (stage I/II). All these analyses demonstrated that BAP18 may act as a potential biomarker for OSCC early diagnosis and tumor rating standard. Thus, qPCR experiments were further performed to try to clarify the molecular mechanism underlying the biological function of BAP18 on OSCC development and progression. Our results have demonstrated that BAP18 participates in modulation of CCND1 and CCND2 gene transcription. Several proteins have been reported to be highly expressed in OSCC and become potential biomarkers in OSCC. Toll-like receptor 2 (TLR2) is highly expressed in OSCC, and ligand-activated TLR2 induced miR-146a expression and downregulated the caspase recruitment domain-containing protein 10 (CARD10) mRNA to be implicated in OSCC development [36]. Protein regulator of cytokinesis 1 participates in cell proliferation regulation and cell cycle in OSCC. Salvianolic acid B is involved in glycolysis inhibition through targeting PI3K/Akt/HIF-1α in OSCC. Other highly expressed proteins in OSCC, have also been reported that notable influence the signaling pathways, such as EMT, mTOR, and GS3K, which are involved in OSCC rapid proliferation and tumor growth [37], [38], [39], [40], [41].
Cell proliferation is supposed to primarily regulated in the G1 phase of the cell cycle, and the abnormal regulation of the transition from G1 to S phase can cause neoplastic transformation [13,14,42]. Cyclin-Ds and their CDKs have been reported to delay early genes in response to growth factor stimulation of cell proliferation, while protein expression accompanied with CDK4/6 activities peak at G1/S transition of cell cycle. Therapies based on cell cycle regulation by Cyclin-Ds family members were discovered in many ways [43]. For instant, targeting cyclin-dependent kinase (CDK4/6), Palbociclib has found to promote cancer cell senescence through G1 arresting [44,45]. Meanwhile, CyclinD1-CDK4 complex exerts the function on controlling glucose metabolism independently of cell division [20,45]. Besides cell growth delay and senescence, Cyclin-Ds members and CDKs (Cyclin-Ds/CDKs) are reported to regulate PD-L1 protein to administer immunotherapy efficacy [46]. Therefore, the study of Cyclin-Ds/CDKs would be important for cancer therapy. In our study, we provided the evidence that BAP18 is involved in up-regulation of CCND1 and CCND2 gene transcription. In addition, BAP18 was recruited to the promoter regions of CCND1 and CCND2, facilitating the recruitment of WDR5, ASH2 and DPY30 to the same regions. Knockdown of BAP18 in OSCC-derived cells exhibited remarkable cell growth delay. In addition, the promotion of cell proliferation induced by ectopic expression of BAP18 was obviously suppressed by CCND1/2 depletion or CDK4/6 inhibitor (Palbociclib), suggesting that BAP18 promotes the cell growth/proliferation in OSCC, if not all, at least partially through the regulation of CCND1/2 transcription. It remains to be elusive whether BAP18 would be involved in the regulation of glucose metabolism or PD-L1 protein, which was related with Cyclin-Ds/CDKs.
Declaration of Competing Interest
The authors declare no conflict of interests.
Acknowledgments
Acknowledgments
We appreciate Dr. Yunlong Huo (Shengjing Hospital, China Medical University) and Dr. Hongyan Zhang (Department of Molecular Cellular Biology, China Medical University) for helpful technique support. We thank Dr. Yanshu Li (Department of Molecular Cellular Biology, China Medical University) for kindly providing the OSCC-derived cell line (Cal-27 cells and SCC9 cells).
Funding sources
This study was supported by the grants from National Natural Science Foundation of China [31871286 for Yue Zhao, 81872015 for Chunyu Wang, 31701102 for Shengli Wang, 81702800 for Renlong Zou, 81902889 for Wensu Liu]; Foundation for Special Professor of Liaoning Province (the 5thbatch) for Yue Zhao; Supported project for Young Technological Innovation-Talents in Shenyang (No. RC170541) for Chunyu Wang.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102685.
Appendix. Supplementary materials
References
- 1.Chuang S.C., Scelo G., Tonita J.M., Tamaro S., Jonasson J.G., Kliewer E.V. Risk of second primary cancer among patients with head and neck cancers: a pooled analysis of 13 cancer registries. Int J Cancer. 2008;123(10):2390–2396. doi: 10.1002/ijc.23798. [DOI] [PubMed] [Google Scholar]
- 2.Sant M., Aareleid T., Berrino F., Bielska Lasota M., Carli P.M., Faivre J. EUROCARE-3: survival of cancer patients diagnosed 1990-94–results and commentary. Ann Oncol. 2003;14(Suppl 5):v61–118. doi: 10.1093/annonc/mdg754. [DOI] [PubMed] [Google Scholar]
- 3.Patel S.C., Carpenter W.R., Tyree S., Couch M.E., Weissler M., Hackman T. Increasing incidence of oral tongue squamous cell carcinoma in young white women, age 18 to 44 years. J Clin Oncol. 2011;29(11):1488–1494. doi: 10.1200/JCO.2010.31.7883. [DOI] [PubMed] [Google Scholar]
- 4.Ng J.H., Iyer N.G., Tan M.H., Edgren G. Changing epidemiology of oral squamous cell carcinoma of the tongue: a global study. Head Neck. 2017;39(2):297–304. doi: 10.1002/hed.24589. [DOI] [PubMed] [Google Scholar]
- 5.van Dijk B.A., Brands M.T., Geurts S.M., Merkx M.A., Roodenburg J.L. Trends in oral cavity cancer incidence, mortality, survival and treatment in the Netherlands. Int J Cancer. 2016;139(3):574–583. doi: 10.1002/ijc.30107. [DOI] [PubMed] [Google Scholar]
- 6.Frame F.M., Maitland N.J. Cancer stem cells, models of study and implications of therapy resistance mechanisms. Adv Exp Med Biol. 2011;720:105–118. doi: 10.1007/978-1-4614-0254-1_9. [DOI] [PubMed] [Google Scholar]
- 7.Amit M., Yen T.C., Liao C.T., Chaturvedi P., Agarwal J.P., Kowalski L.P. The origin of regional failure in oral cavity squamous cell carcinoma with pathologically negative neck metastases. JAMA Otolaryngol Head Neck Surg. 2014;140(12):1130–1137. doi: 10.1001/jamaoto.2014.1539. [DOI] [PubMed] [Google Scholar]
- 8.da Silva S.D., Hier M., Mlynarek A., Kowalski L.P., Alaoui-Jamali M.A. Recurrent oral cancer: current and emerging therapeutic approaches. Front Pharmacol. 2012;3:149. doi: 10.3389/fphar.2012.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vermorken J.B., Remenar E., van Herpen C., Gorlia T., Mesia R., Degardin M. Cisplatin, fluorouracil, and docetaxel in unresectable head and neck cancer. N Engl J Med. 2007;357(17):1695–1704. doi: 10.1056/NEJMoa071028. [DOI] [PubMed] [Google Scholar]
- 10.Greenberg J.S., El Naggar A.K., Mo V., Roberts D., Myers J.N. Disparity in pathologic and clinical lymph node staging in oral tongue carcinoma. Implication for therapeutic decision making. Cancer. 2003;98(3):508–515. doi: 10.1002/cncr.11526. [DOI] [PubMed] [Google Scholar]
- 11.Alaoui-Jamali M.A., Dupre I., Qiang H. Prediction of drug sensitivity and drug resistance in cancer by transcriptional and proteomic profiling. Drug Resist Updat. 2004;7(4–5):245–255. doi: 10.1016/j.drup.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Messadi D.V., Sato K. Oral cancer chernoprevention: current status and future direction. J Calif Dent Assoc. 2016;44(2):101–111. [PubMed] [Google Scholar]
- 13.Hosokawa Y., Onga T., Nakashima K. Induction of D2 and D3 cyclin-encoding genes during promotion of the G1/S transition by prolactin in rat Nb2 cells. Gene. 1994;147(2):249–252. doi: 10.1016/0378-1119(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 14.Cole A.M., Myant K., Reed K.R., Ridgway R.A., Athineos D., Van den Brink G.R. Cyclin D2-cyclin-dependent kinase 4/6 is required for efficient proliferation and tumorigenesis following Apc loss. Cancer Res. 2010;70(20):8149–8158. doi: 10.1158/0008-5472.CAN-10-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calegari F. CyclinD2 at the edge: splitting up cell fate. EMBO J. 2012;31(8):1850–1852. doi: 10.1038/emboj.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fatrai S., Elghazi L., Balcazar N., Cras-Meneur C., Krits I., Kiyokawa H. Akt induces beta-cell proliferation by regulating cyclin D1, cyclin D2, and p21 levels and cyclin-dependent kinase-4 activity. Diabetes. 2006;55(2):318–325. doi: 10.2337/diabetes.55.02.06.db05-0757. [DOI] [PubMed] [Google Scholar]
- 17.Nath K., Fisher C., Elinson R.P. Expression of cyclin D1, cyclin D2, and N-myc in embryos of the direct developing frog eleutherodactylus coqui, with a focus on limbs. Gene Expr Patterns. 2013;13(5–6):142–149. doi: 10.1016/j.gep.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shan L., Li X., Liu L., Ding X., Wang Q., Zheng Y. GATA3 cooperates with PARP1 to regulate CCND1 transcription through modulating histone H1 incorporation. Oncogene. 2014;33(24):3205–3216. doi: 10.1038/onc.2013.270. [DOI] [PubMed] [Google Scholar]
- 19.Jena N., Deng M., Sicinska E., Sicinski P., Daley G.Q. Critical role for cyclin D2 in BCR/ABL-induced proliferation of hematopoietic cells. Cancer Res. 2002;62(2):535–541. [PubMed] [Google Scholar]
- 20.Lee Y., Dominy J.E., Choi Y.J., Jurczak M., Tolliday N., Camporez J.P. Cyclin D1-Cdk4 controls glucose metabolism independently of cell cycle progression. Nature. 2014;510(7506):547–551. doi: 10.1038/nature13267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bienvenu F., Jirawatnotai S., Elias J.E., Meyer C.A., Mizeracka K., Marson A. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature. 2010;463(7279):374–378. doi: 10.1038/nature08684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poudel S., Song J., Jin E.J., Song K. Sulfuretin-induced miR-30C selectively downregulates cyclin D1 and D2 and triggers cell death in human cancer cell lines. Biochem Biophys Res Commun. 2013;431(3):572–578. doi: 10.1016/j.bbrc.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 23.Tamamori-Adachi M., Goto I., Yamada K., Kitajima S. Differential regulation of cyclin D1 and D2 in protecting against cardiomyocyte proliferation. Cell Cycle. 2008;7(23):3768–3774. doi: 10.4161/cc.7.23.7239. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt B.A., Rose A., Steinhoff C., Strohmeyer T., Hartmann M., Ackermann R. Up-regulation of cyclin-dependent kinase 4/cyclin D2 expression but down-regulation of cyclin-dependent kinase 2/cyclin e in testicular germ cell tumors. Cancer Res. 2001;61(10):4214–4221. [PubMed] [Google Scholar]
- 25.Yu Q., Geng Y., Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411(6841):1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- 26.Kerkhoff E., Ziff E.B. Cyclin D2 and Ha-Ras transformed rat embryo fibroblasts exhibit a novel deregulation of cell size control and early S phase arrest in low serum. EMBO J. 1995;14(9):1892–1903. doi: 10.1002/j.1460-2075.1995.tb07181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tremblay V., Zhang P., Chaturvedi C.P., Thornton J., Brunzelle J.S., Skiniotis G. Molecular basis for DPY-30 association to COMPASS-like and NURF complexes. Structure. 2014;22(12):1821–1830. doi: 10.1016/j.str.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H., Ilin S., Wang W., Duncan E.M., Wysocka J., Allis C.D. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442(7098):91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vermeulen M., Eberl H.C., Matarese F., Marks H., Denissov S., Butter F. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142(6):967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 30.Sun S., Zhong X., Wang C., Sun H., Wang S., Zhou T. BAP18 coactivates androgen receptor action and promotes prostate cancer progression. Nucleic Acids Res. 2016;44(17):8112–8128. doi: 10.1093/nar/gkw472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Y., Takeyama K., Sawatsubashi S., Ito S., Suzuki E., Yamagata K. Corepressive action of CBP on androgen receptor transactivation in pericentric heterochromatin in a drosophila experimental model system. Mol Cell Biol. 2009;29(4):1017–1034. doi: 10.1128/MCB.02123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fridman E., Na'ara S., Gil Z. Reply to are adjuvant radiotherapy outcomes really better in patients with early-stage oral cavity squamous cell carcinoma? Cancer. 2018;124(23):4578–4579. doi: 10.1002/cncr.31796. [DOI] [PubMed] [Google Scholar]
- 33.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 34.Torre L.A., Siegel R.L., Ward E.M., Jemal A. Global cancer incidence and mortality rates and trends–an update. Cancer Epidemiol Biomark Prev. 2016;25(1):16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 35.Ferlay J., Colombet M., Soerjomataram I., Mathers C., Parkin D.M., Pineros M. Estimating the global cancer incidence and mortality in 2018: globocan sources and methods. Int J Cancer. 2019;144(8):1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 36.Ikehata N., Takanashi M., Satomi T., Watanabe M., Hasegawa O., Kono M. Toll-like receptor 2 activation implicated in oral squamous cell carcinoma development. Biochem Biophys Res Commun. 2018;495(3):2227–2234. doi: 10.1016/j.bbrc.2017.12.098. [DOI] [PubMed] [Google Scholar]
- 37.Lu M., Chen W.H., Wang C.Y., Mao C.Q., Wang J. Reciprocal regulation of miR-1254 and c-Myc in oral squamous cell carcinoma suppresses EMT-mediated metastasis and tumor-initiating properties through MAPK signaling. Biochem Biophys Res Commun. 2017;484(4):801–807. doi: 10.1016/j.bbrc.2017.01.170. [DOI] [PubMed] [Google Scholar]
- 38.Wu F., Shi X., Zhang R., Tian Y., Wang X., Wei C. Regulation of proliferation and cell cycle by protein regulator of cytokinesis 1 in oral squamous cell carcinoma. Cell Death Dis. 2018;9(5):564. doi: 10.1038/s41419-018-0618-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai D., Chen H., Tang J., Tang Y. Inhibition of mTOR/eIF4E by anti-viral drug ribavirin effectively enhances the effects of paclitaxel in oral tongue squamous cell carcinoma. Biochem Biophys Res Commun. 2017;482(4):1259–1264. doi: 10.1016/j.bbrc.2016.12.025. [DOI] [PubMed] [Google Scholar]
- 40.Chien C.S., Wang M.L., Chu P.Y., Chang Y.L., Liu W.H., Yu C.C. Lin28B/Let-7 regulates expression of oct4 and sox2 and reprograms oral squamous cell carcinoma cells to a stem-like state. Cancer Res. 2015;75(12):2553–2565. doi: 10.1158/0008-5472.CAN-14-2215. [DOI] [PubMed] [Google Scholar]
- 41.Wei J., Wu J., Xu W., Nie H., Zhou R., Wang R. Salvianolic acid b inhibits glycolysis in oral squamous cell carcinoma via targeting PI3K/AKT/HIF-1alpha signaling pathway. Cell Death Dis. 2018;9(6):599. doi: 10.1038/s41419-018-0623-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doerner P., Jorgensen J.E., You R., Steppuhn J., Lamb C. Control of root growth and development by cyclin expression. Nature. 1996;380(6574):520–523. doi: 10.1038/380520a0. [DOI] [PubMed] [Google Scholar]
- 43.Yoshiba S., Ito D., Nagumo T., Shirota T., Hatori M., Shintani S. Hypoxia induces resistance to 5-fluorouracil in oral cancer cells via G(1) phase cell cycle arrest. Oral Oncol. 2009;45(2):109–115. doi: 10.1016/j.oraloncology.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 44.Leite de Oliveira R., Bernards R. Anti-cancer therapy: senescence is the new black. EMBO J. 2018;37(10):e99386. doi: 10.15252/embj.201899386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sherr C.J., Roberts J.M. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13(12):1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J., Bu X., Wang H., Zhu Y., Geng Y., Nihira N.T. Cyclin d-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553(7686):91–95. doi: 10.1038/nature25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.