Abstract
Sensing bacterial infections and monitoring drug resistance are very important for the selection of treatment options. However, the common methods of sensing resistance are limited by time-consuming, the requirement for professional personnel, and expensive instruments. Moreover, the abuse of antibiotics causes the accelerated process of bacterial resistance. Herein, we construct a portable paper-based band-aid (PBA) which implements a selective antibacterial strategy after sensing of drug resistance. The colors of PBA indicate bacterial infection (yellow) and drug resistance (red), just like a bacterial resistance colorimetric card. On the basis of color, antibiotic-based chemotherapy and Zr-MOF PCN-224-based photodynamic therapy (PDT) are used on site to treat sensitive and resistant strains, respectively. Eventually, it takes 4 h to sense, and the limit of detection is 104 CFU/mL for drug-resistant E. coli. Compared with traditional PDT-based antibacterial strategies, our design can alleviate off-target side effects, maximize therapeutic efficacy, and track the drug resistance in real time with the naked eye. This work develops a new way for the rational use of antibiotics. Given the low cost and easy operation of this point-of-care device, it can be developed for practical applications.
Short abstract
A drug-resistant visualizing paper-based band-aid has been constructed for point-of-care sensing and selectively treating drug-resistant bacteria.
Introduction
Currently, 700 000 deaths are associated with antimicrobial resistance every year.1 The accelerated emergence and widespread development of antibiotic-resistant bacterial strains have been one of the most serious threats to human health worldwide.2−5 However, commercial antibiotics, with their clear antibacterial mechanisms, are still the most widely accepted treatment paradigms.6,7 Because of the “auto-obsolescence” of antibacterial treatments, it is an important issue in the current antibacterial field how to rationally use of existing antibiotics and overcome tolerance.
Early sensing of bacterial infections and tracing the emergence of drug resistance are essential prerequisites for the selection of treatment regimens. On one hand, for sensing bacterial infection, the unique microenvironment (pH, toxins, enzymes, etc.) of bacteria is an excellent inspiration for researchers.8−12 Among them, the acidity is a result of the glycometabolism of most pathogens and has been widely used for sensing of bacterial infections.13,14 On the other hand, for tracking drug resistance, specific enzymes which are actively produced by drug-resistant (DR) bacteria are often considered as markers, especially β-lactamase.15−17 Because of the most extensive use of β-lactam antibiotics, resistance toward them is considered a serious threat. Therefore, β-lactamase has been widely used as a marker to identify DR bacteria.
For weakening resistance, reactive oxygen species (ROS)-based strategies, including photodynamic therapy (PDT) and chemodynamic therapy (CDT), are expected to be excellent candidates.18−21 Their main advantage is the multitarget effect, that is, factors associated with bacterial resistance, such as bacterial cell walls, nucleic acids, and proteins, can be destroyed. Noticeably, ROS lacks specific targeting, and its abuse can cause undesirable adverse effects on healthy tissue.22 Therefore, rational use of ROS-based strategy is an effective measure to combat drug resistance.
Inspired by World Health Organization (WHO)’s need for portable and inexpensive devices to combat Antimicrobial Resistance in the Global Action Plan, paper-based devices (PBD) are promising platforms for antibacterial therapy.23 Because of the advantages of sustainability, biosafety, low cost and easy modification, PBD have been widely used for portable biosensors and sterilization paper.23−27 However, it has not been constructed for PBD to achieve “sense-and-treat” function.28−30 Thus, we aim to develop a portable paper-based band-aid (PBA) for sensing and treating drug resistance.
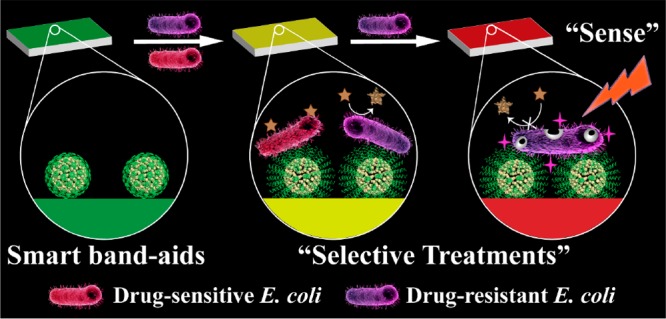
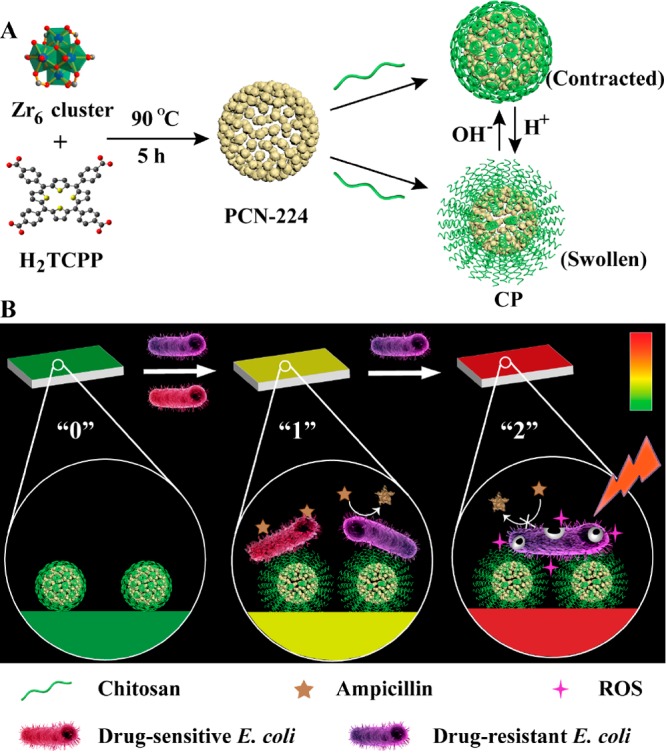
Herein, we developed a drug-resistant visualizing PBA which implemented a selective antibacterial strategy after sensing of drug resistance (Scheme 1). For drug-sensitive (DS) bacteria, the acidic nature of bacterial microenvironment was utilized. On one hand, bromothymol blue (BTB) responded to the acid environment at infectious sites, accompanied by a color change from green to yellow. On the other hand, ampicillin-loaded nanomaterials were coated with chitosan to lure negatively charged bacteria and achieved acid-responsive drug release, thereby killing DS bacteria. For DR bacteria, nitrocefin was changed from yellow to red by the action of β-lactamase, which was secreted by many resistant bacteria. We synthesized PCN-224, a porphyrin-based metal–organic framework (MOF) that has superior PDT capabilities compared with free porphyrin molecules.31 With the irradiation of light, ROS produced by MOF caused great damage to bacteria and weakened resistance. Through this elaborate design, we integrated the above elements into cellulose paper and prepared PBA. Eventually, we can judge whether there exists resistant bacteria by the color changes and distinguish the treatment. As a proof-of-concept, our strategy enabled the fast sensing (4 h) and effective treatment of DS and DR strains of Escherichia coli (E. coli) as model examples.
Scheme 1. (A) Preparation Routes of CP and pH-Responsive Transformation between Contracted State and Swollen State of Chitosan. (B) Schematic Illustration of PBA (“0”, Green) for Sensing Bacterial Infection (“1”, Yellow) and Drug Resistance (“2”, Red), and Implementing Antibiotic-Based Chemotherapy and PCN-224-Based PDT, Respectively.
Results and Discussion
Design and Preparation of PBA
In our design, based on the Zr6 cluster and the H2TCPP ligand (TCPP = tetrakis(4-carboxyphenyl)-porphyrin), porphyrin-based MOF nanoparticles named as PCN-224 were synthesized at 90 °C first. An average diameter of about 58 nm was revealed by scanning electron microscopy (SEM) (Figure S1). The X-ray diffraction (XRD) pattern clearly showed a crystalline structure of PCN-224 (Figure S2). Next, for realizing acid-responsive character, chitosan (CS) was attached on the surface of PCN-224 (PCN-224@CS, thus denoted as CP) using the postsynthesis modification method.32−34 SEM and transmission electron microscopy (TEM) images (Figure S3A and Figure 1A) revealed that the modification of CS did not influence the structural integrity of the MOF. And TEM elemental mappings (Figure 1B) exhibited the composition of CP. Meanwhile, the increased hydrodynamic diameter also indicated the encapsulation of CS (Figure S3B). Furthermore, the formation of CP was proved by the change of vibration frequency of carbonyl group in Fourier transform infrared spectra (FT-IR) (Figure S4).
Figure 1.
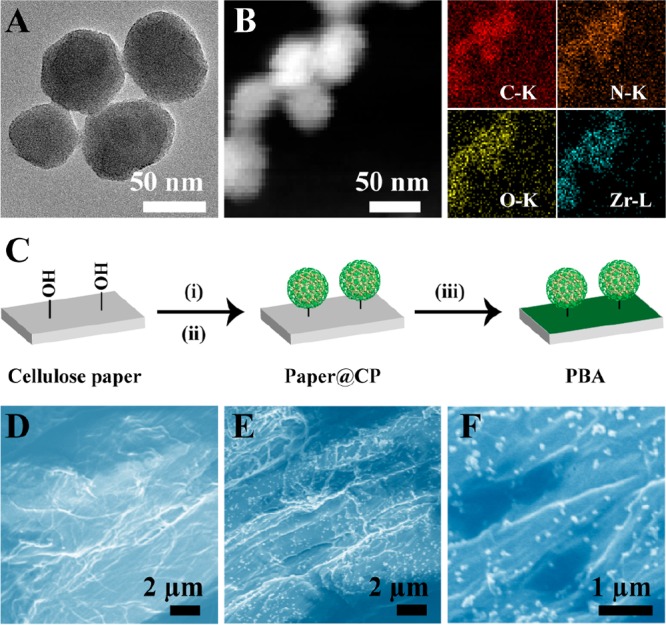
(A) TEM image of CP. (B) Dark-field TEM image of CP and its corresponding TEM elemental mappings of C–K, N–K, O–K, and Zr–L signals. (C) The preparation route of PBA: i, LiCl and NaIO4; ii, NaCNBH3 and CP; iii, BTB solution (5% PEG, pH 7.4). SEM graphs of cellulose paper (D) and PBA (E, F).
Subsequently, CP was immobilized onto sterile cellulose paper (paper@CP) for portability. As illustrated in Figure 1C, hydroxyl groups of cellulose paper were oxidized by periodate to aldehyde groups. Then CP was immobilized onto the paper through the formation of secondary amines. As shown in Figure 1D–F, CP was evenly dispersed on the paper. The porous microstructure of cellulose paper and uniform porosity of PCN-224 were beneficial for the efficient diffusion of molecular oxygen (Figure S5). The appearance of characteristic peaks of Zr 3d and N 1s was observed by X-ray photon spectroscopy (XPS) analysis (Figure S6), indicating that CP was modified on the surface of the paper. Additionally, FT-IR spectra were recorded to further demonstrate the successful modification of CP on the paper (Figure S4). The 1.89 μg/cm2 of Zr6+ ion amounts were measured by inductively coupled plasma (ICP) analysis. Furthermore, the modification of CP was characterized by the corresponding thermogravimetric analysis (TGA) (Figure S7), Thus, CP-modified cellulose paper was prepared successfully.
After that, BTB and nitrocefin were used as indicators of bacterial infection and drug resistance, respectively. To combine them into one system, nitrocefin and antibiotics were coencapsulated into MOF (Figure S8), and the polyethylene glycol (PEG) solution of BTB was used to rinse the entire paper surface to act as a chromogenic layer (Figure S9). Such a design not only helps the bacterial microenvironment to fully infiltrate the paper but also realizes the cascade reaction. After these steps, the preparation of PBA was completed.
Use of PBA for Sensing Bacterial Infection
The acid responsiveness of PBA includes the acid-triggered color change and acid-responsive payload release (Figure 2A). In the first place, the spectral and color changes of BTB were examined under different levls of pH. When pH was decreased from 8.0 to 6.0, the BTB solution changed from blue to green and then yellow with a significant absorbance decrease at 615 nm (Figure S10). When E. coli was coincubated with the mixture of CP and BTB, the absorbance value at 615 nm decreased with the increase of bacterial concentration (Figure S11). Accordingly, regardless of adding DS or DR E. coli, PBAno nitrocefin was also turned yellow, and a slight but observable color change occurred at the concentration of 103 CFU/mL (Figure 2B). In the second place, the acid-responsive drug release properties of CP were investigated (Figure S12). Rhodamine B (RhB) was loaded into the MOF (CP-RhB) as a model drug with a loading efficiency of 68.9%. After dissolving CP-RhB in a buffer (pH 6.0) or bacterial solution, RhB was rapidly released within hours, demonstrating that CP possessed the ability of acid-responsive payload release.
Figure 2.
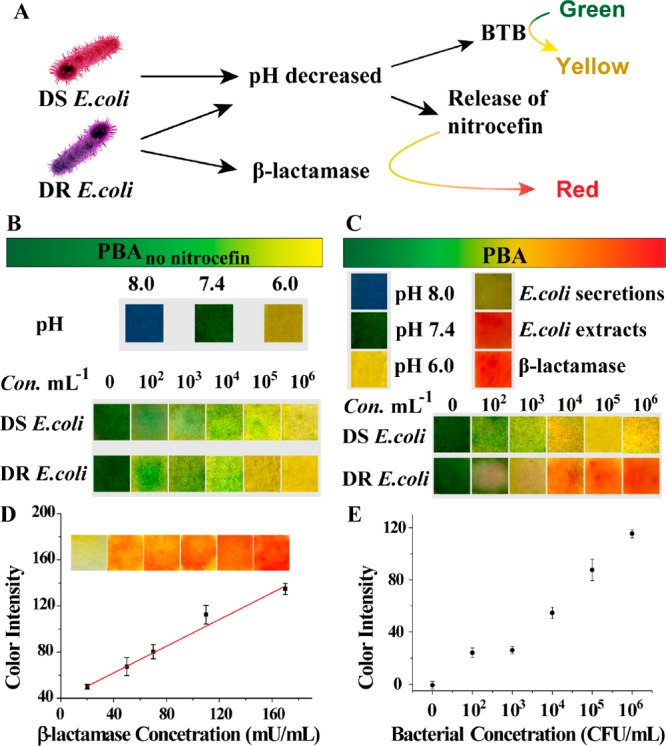
Use of PBA for sensing drug resistance. (A) The schematic diagram of PBA (green) for sensing bacterial infection (yellow) and drug resistance (red). The typical pictures of (B) PBAno nitrocefin and (C) PBA after incubation with buffer (pH 8.0–6.0), E. coli solution, bacterial secretions, and extracts for 4 h. (D) Quantitative analysis of PBA with different concentrations of β-lactamase ranging from 0.02 to 0.18 U/mL. The inset includes the typical images of PBA after reaction with β-lactamase. (E) Quantitative analysis of PBA with different concentrations of DR E. coli. The color intensity was calibrated by subtracting the mean intensity of DS E. coli group on each side of the DR E. coli group.
Prior to explore the sensing β-lactamase ability of PBA, the feasibility of the mixture solution to respond to β-lactamase was investigated. First, reaction between β-lactamase and nitrocefin was monitored by UV–vis spectra. The red products were gradually formed, and the absorbance at 486 nm was increased (Figure S13). Under different pH levels, β-lactamase still retained catalytic activity (Figure S14). UV–vis spectra of CP were almost unchanged (Figure S15), so the absorbance at 486 nm was used to monitor the reaction of nitrocefin. Second, the mixture solution of CP-N and BTB was treated with DR E. coli, and the absorbance values at 486 nm increased with bacteria concentration ranging from 102 to 108 CFU/mL (Figure S16), indicating that our designed mixture system could sense DR E. coli secreting β-lactamase.
Furthermore, 50 μL of E. coli (DR or DE) solution was dripped onto the test paper. PBA turned red from green in the presence of DR E. coli, and the higher the concentration of bacteria, the stronger the red intensity of PBA. As shown in Figure 2C, a perceivable red signal was observed at the concentration of 104 CFU/mL, which meets the need of clinical diagnosis. Besides, bacterial secretions and extracts were used to test the specificity of PBA. Results showed that only bacterial secretions did not cause red, which was due to the secretion of β-lactamase from E. coli to the periplasm, rather than the outside of the whole cell.35,36 Therefore, the color of PBA could indicate both bacterial infection and drug resistance with a low detection limit and high specificity.
In addition, we further estimated bacterial infection levels on the basis of the relationship between the color intensity change and the concentration of β-lactamase itself. As shown in Figure 2D, the color intensity was enhanced with the increase of β-lactamase concentration. A linear dependence was obtained when the β-lactamase concentration range was 0.02–0.18 U/mL. The detectable increase of color intensity was observed at the concentration of 104 CFU/mL (Figure 2E), which was comparable to the level of 0.02 U/mL of β-lactamase. Therefore, under our experiment conditions, concentration-dependent colorimetric detection of β-lactamase may act as a reference to evaluate bacterial infection levels.
Use of PBA for Selective Treatment on the Basis of Drug Resistance
We optimized the selective antimicrobial experiments (Figure 3A). The ROS generation ability of CP was proved under visible light irradiation of 638 nm (Figure S17 and S18), and four PBA were prepared with different amounts of CP (Figure 3B). For DS E. coli, the survival rates obviously decreased with the increase of the amount of CP (Figure 3C). For DR E. coli, the increase of the concentration of CP did not significantly reduce the viability, while their viability became extremely low after light irradiation (Figure 3D and Figure S19). These results indicated that chemotherapy alone could eradicate DS E. coli while killing DR E. coli required the combination of chemotherapy and PDT. Moreover, compared with the control groups, there was a considerable synergistic effect between PDT (45.3%) and chemotherapy (72.4%) on killing DR E. coli, which was proved by the obviously lower viability than the projected additive value (32.8%) (Figure 3D). Noticeably, considering the side effects of PDT on normal cells, we chose the appropriate density (denoted as c) to balance the performance and biosafety. Thus, PBA was a promising candidate to realize the selective antimicrobial activity against DS and DR E. coli.
Figure 3.
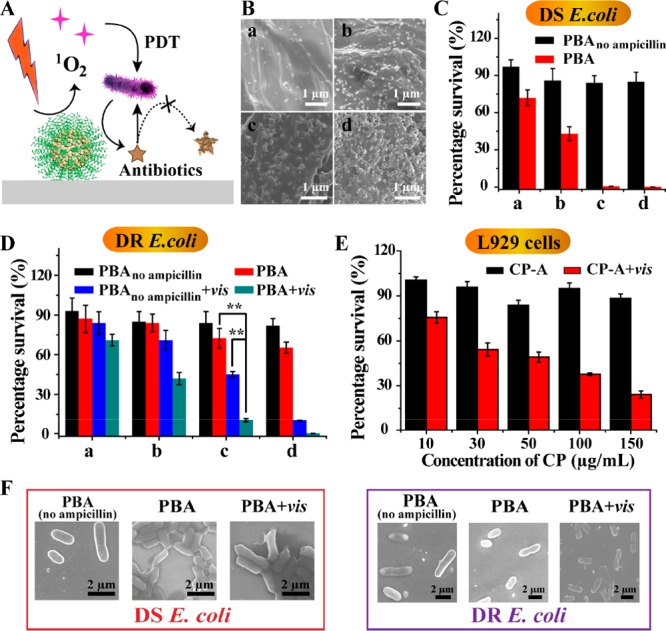
Use of PBA for selective treatment on the basis of drug resistance. (A) Schematic illustration of PBA combined with chemotherapy and PDT to kill DR E. coli. (B) Typical SEM images of PBA modified with different amounts of CP. Viability of DS E. coli (C) and DR E. coli (D) incubated on PBA or PBAno ampicillin with or without light irradiation. Statistical analysis was performed using the student’s two-tailed t test (**p < 0.01). (E) L929 cell viability after incubation with different concentrations of CP for 12 h. (F) The typical SEM images of E. coli after incubation with PBA.
Bacterial cell walls are closely related to drug resistance, and the change of cell morphology was monitored by SEM. When treated with PBA, the cell wall was wrinkled for DS E. coli, whereas it was almost intact for DR E. coli (Figure 3F). That may be the result of ampicillin, which prevented bacterial cell wall synthesis. However, when being supplemented by light, the cell wall was significantly damaged, demonstrating that ROS-based therapy had great harm to the bacterial cell wall. It was reported that cell walls are a basic permeability barrier and carry proteins related to efflux pumps and hydrolases.37 The oxidative damage of the cell wall may impair the function of the cell wall and its adjacent proteins, which contributed to the enhancement of chemotherapy.
Investigation of Validity Period
The validity period was an important index in practical application. PBA was stored hermetically at 4 °C and then taken out for sensing and antimicrobial experiments (Figure 4). A month later, PBA still responded to DS and DR E. coli, showing the corresponding color. At the same time, the effective killing ability of E. coli was also maintained. Thus, the validity period of PBA was at least one month, which was promising for use in practical applications.
Figure 4.
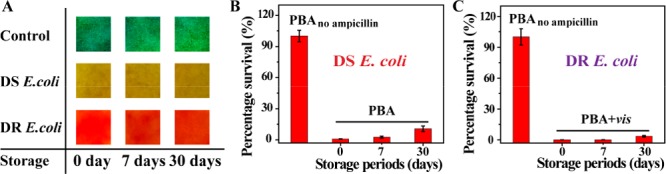
Investigation of validity periods. (A) Typical pictures of PBA when adding bacterial solutions. The reaction time is 4 h. Survival percentages of DS E. coli (B) and DR E. coli (C). Bacterial concentration is 107 CFU/mL.
Use of PBA for Wound Disinfection in Mice
Finally, the potential of PBA for sensing and treating resistance in mice with wounds on their back was evaluated. The wounds were, respectively, infected by DS E. coli and DR E. coli, and PBA was attached to the wound for 4 h. As shown in Figure 5A, for the infection by DS E. coli, all test paper exhibited yellow, proving the feasibility of biosensors for bacterial infection. The wound treated with PBA was almost healed on the third day, and hematoxylin and eosin (H&E) stain of wound tissues also revealed the intact epidermal layer (Figure 5B), suggesting that antibiotics were sufficient for wound healing caused by DS E. coli. However, when irradiated with light, the wound was swollen indicating that ROS-based therapy caused side effects. Accordingly, for infection by DR E. coli, PBA turned red, demonstrating the feasibility of biosensors for drug resistance. As shown in Figure 5C, when being treated with the combination of light and PBA, the wound was healed better, and a nascent epidermal layer was observed, showing that the combination of PDT and chemotherapy required for wound healing when infected by DR E. coli under this condition. Additionally, wound tissues around infected sites were taken out to quantify the survival bacteria (Figure 5C). The obvious antibacterial effects were displayed with the treatment of PBA for DS E. coli, and the combination of light and PBA for DR E. coli. Considering the antimicrobial effects and biosafety (Figure S20 and S21), PBA was good enough for treating DS E. coli-infection, and the combination of PBA and light was more suitable for combating DR E. coli-infection. Therefore, PBA can be a promising candidate for treating wound infection.
Figure 5.
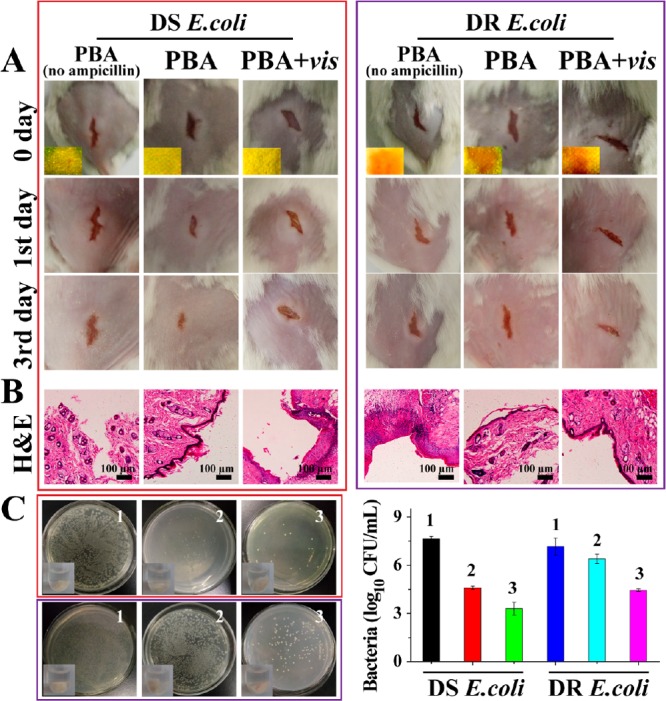
Use of PBA for wound disinfection in mice. (A) Photographs of wounds on the mice. Inset images in the first row revealed the color on PBA. (B) Photomicrographs showing section of skin tissues with H&E staining. (C) Bacteria was separated from wound tissue and then cultured on agar plates. The upper row belongs to DS E. coli, and the lower row belongs to DR E. coli. 1, PBAno ampicillin; 2, PBA; 3, PBA+vis.
Besides, the potential of PBA for sensing and treating resistance was further shown by a fruit preservation model. As an example, the infected tomato model was constructed, and PBA was attached on the infectious sites (Figure S22). In the group of mixed strains, the red color of PBA appeared on the second day, indicating that PBA could monitor drug resistance in real time. If no light treatment was supplemented, the infection site of tomatoes became soft and sunken, and thousands of bacteria appeared on the surface after a 3-day treatment, indicating the necessity of PDT. This model also confirmed that PBA could sense the bacterial infection and monitor the drug resistance in real time and subsequently eradicate the drug resistance by PDT.
Conclusions
In summary, we constructed a drug-resistant visualizing PBA which implemented a selective antibacterial strategy after sensing of drug resistance. For the DS E. coli-induced infection, PBA turned from green to yellow, and antibiotics were released to eradicate DS E. coli (antibiotic-based chemotherapy). For DR bacteria-induced infection, PBA turned red, and the supplement of light was taken to eradicate resistance. Compared with traditional PDT-based antibacterial strategies, drug resistance can be tracked in real time with the naked eye, and off-target side effects are alleviated. Eventually, it took only 2–4 h, and the limit of detection was 104 CFU/mL for DR E. coli, which meets the need for clinical diagnosis. In consideration of PBA with excellent performance, low cost, and easy operation, it can be developed for practical point-of-care application.
Acknowledgments
The authors are grateful for support from NSFC (21431007, 21533008, 21820102009, 21871249, 91856205, 21877105), Key Program of Frontier of Sciences, CAS QYZDJ-SSW-SLH052.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.9b01104.
Materials, measurements, experimental section, and additional data (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Mckenna M. How to fight superbugs: start spending money. https://www.wired.com/2015/02/oneill-amr-2/.
- O’Connell K. M. G.; Hodgkinson J. T.; Sore H. F.; Welch M.; Salmond G. P. C.; Spring D. R. Combating multidrug-resistant bacteria: current strategies for the discovery of novel antibacterials. Angew. Chem., Int. Ed. 2013, 52, 10706–10733. 10.1002/anie.201209979. [DOI] [PubMed] [Google Scholar]
- Soh M.; Kang D.-W.; Jeong H.-G.; Kim D.; Kim D. Y.; Yang W.; Song C.; Baik S.; Choi I.-Y.; Ki S.-K.; Kwon H. J.; Kim T.; Kim C. K.; Lee S.-H.; Hyeon T. Ceria–zirconia nanoparticles as an enhanced multi-antioxidant for sepsis treatment. Angew. Chem., Int. Ed. 2017, 56, 11399–11403. 10.1002/anie.201704904. [DOI] [PubMed] [Google Scholar]
- Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature 2000, 406, 775–781. 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- Coates A.; Hu Y.; Bax R.; Page C. The future challenges facing the development of new antimicrobial drugs. Nat. Rev. Drug Discovery 2002, 1, 895. 10.1038/nrd940. [DOI] [PubMed] [Google Scholar]
- Chellat M. F.; Raguž L.; Riedl R. Targeting antibiotic resistance. Angew. Chem., Int. Ed. 2016, 55, 6600–6626. 10.1002/anie.201506818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarlagadda V.; Sarkar P.; Samaddar S.; Haldar J. A vancomycin derivative with a pyrophosphate-binding group: a strategy to combat vancomycin-resistant bacteria. Angew. Chem., Int. Ed. 2016, 55, 7836–7840. 10.1002/anie.201601621. [DOI] [PubMed] [Google Scholar]
- Chen J.; Andler S. M.; Goddard J. M.; Nugen S. R.; Rotello V. M. Integrating recognition elements with nanomaterials for bacteria sensing. Chem. Soc. Rev. 2017, 46, 1272–1283. 10.1039/C6CS00313C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gram L.; Ravn L.; Rasch M.; Bruhn J. B.; Christensen A. B.; Givskov M. Food spoilage—interactions between food spoilage bacteria. Int. J. Food Microbiol. 2002, 78, 79–97. 10.1016/S0168-1605(02)00233-7. [DOI] [PubMed] [Google Scholar]
- Pornpattananangkul D.; Zhang L.; Olson S.; Aryal S.; Obonyo M.; Vecchio K.; Huang C.-M.; Zhang L. Bacterial toxin-triggered drug release from gold nanoparticle-stabilized liposomes for the treatment of bacterial infection. J. Am. Chem. Soc. 2011, 133, 4132–4139. 10.1021/ja111110e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H.; Dong K.; Yan Z.; Ding C.; Chen Z.; Ren J.; Qu X. Bacterial hyaluronidase self-triggered prodrug release for chemo-photothermal synergistic treatment of bacterial infection. Small 2016, 12, 6200–6206. 10.1002/smll.201601729. [DOI] [PubMed] [Google Scholar]
- Gupta A.; Mumtaz S.; Li C.-H.; Hussain I.; Rotello V. M. Combatting antibiotic-resistant bacteria using nanomaterials. Chem. Soc. Rev. 2019, 48, 415–427. 10.1039/C7CS00748E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-d.; Meier R. J.; Wolfbeis O. S. Fluorescent pH-sensitive nanoparticles in an agarose matrix for imaging of bacterial growth and metabolism. Angew. Chem., Int. Ed. 2013, 52, 406–409. 10.1002/anie.201205715. [DOI] [PubMed] [Google Scholar]
- Yan Z.; Shi P.; Ren J.; Qu X. A. Sense-and-treat” hydrogel used for treatment of bacterial infection on the solid matrix. Small 2015, 11, 5540–5544. 10.1002/smll.201501958. [DOI] [PubMed] [Google Scholar]
- Li Y.; Liu G.; Wang X.; Hu J.; Liu S. Enzyme-responsive polymeric vesicles for bacterial-strain-selective delivery of antimicrobial agents. Angew. Chem., Int. Ed. 2016, 55, 1760–1764. 10.1002/anie.201509401. [DOI] [PubMed] [Google Scholar]
- Aw J.; Widjaja F.; Ding Y.; Mu J.; Liang Y.; Xing B. Enzyme-responsive reporter molecules for selective localization and fluorescence imaging of pathogenic biofilms. Chem. Commun. 2017, 53, 3330–3333. 10.1039/C6CC09296A. [DOI] [PubMed] [Google Scholar]
- Varadi L.; Luo J. L.; Hibbs D. E.; Perry J. D.; Anderson R. J.; Orenga S.; Groundwater P. W. Methods for the detection and identification of pathogenic bacteria: past, present, and future. Chem. Soc. Rev. 2017, 46, 4818–4832. 10.1039/C6CS00693K. [DOI] [PubMed] [Google Scholar]
- Chen Z.; Wang Z.; Ren J.; Qu X. Enzyme mimicry for combating bacteria and biofilms. Acc. Chem. Res. 2018, 51, 789–799. 10.1021/acs.accounts.8b00011. [DOI] [PubMed] [Google Scholar]
- Jia H.-R.; Zhu Y.-X.; Chen Z.; Wu F.-G. Cholesterol-assisted bacterial cell surface engineering for photodynamic inactivation of Gram-positive and Gram-negative bacteria. ACS Appl. Mater. Interfaces 2017, 9, 15943–15951. 10.1021/acsami.7b02562. [DOI] [PubMed] [Google Scholar]
- Ristic B. Z.; Milenkovic M. M.; Dakic I. R.; Todorovic-Markovic B. M.; Milosavljevic M. S.; Budimir M. D.; Paunovic V. G.; Dramicanin M. D.; Markovic Z. M.; Trajkovic V. S. Photodynamic antibacterial effect of graphene quantum dots. Biomaterials 2014, 35, 4428–4435. 10.1016/j.biomaterials.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Yang B.; Chen Y.; Shi J. Reactive xxygen species (ROS)-based nanomedicine. Chem. Rev. 2019, 119, 4881–4985. 10.1021/acs.chemrev.8b00626. [DOI] [PubMed] [Google Scholar]
- Linden G.; Zhang L.; Pieck F.; Linne U.; Kosenkov D.; Tonner R.; Vazquez O. Conditional singlet oxygen generation via DNA-targeted tetrazine bioorthogonal reaction. Angew. Chem., Int. Ed. 2019, 58, 12868–12873. 10.1002/anie.201907093. [DOI] [PubMed] [Google Scholar]
- Boehle K. E.; Gilliand J.; Wheeldon C. R.; Holder A.; Adkins J. A.; Geiss B. J.; Ryan E. P.; Henry C. S. Utilizing paper-based devices for antimicrobial resistant bacteria detection. Angew. Chem., Int. Ed. 2017, 56, 6886–6890. 10.1002/anie.201702776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tram K.; Kanda P.; Salena B. J.; Huan S.; Li Y. Translating bacterial detection by DNAzymes into a litmus test. Angew. Chem., Int. Ed. 2014, 53, 12799–12802. 10.1002/anie.201407021. [DOI] [PubMed] [Google Scholar]
- Carpenter B. L.; Scholle F.; Sadeghifar H.; Francis A. J.; Boltersdorf J.; Weare W. W.; Argyropoulos D. S.; Maggard P. A.; Ghiladi R. A. Synthesis, characterization, and antimicrobial efficacy of photomicrobicidal cellulose paper. Biomacromolecules 2015, 16, 2482–2492. 10.1021/acs.biomac.5b00758. [DOI] [PubMed] [Google Scholar]
- Deiss F.; Funes-Huacca M. E.; Bal J.; Tjhung K. F.; Derda R. Antimicrobial susceptibility assays in paper-based portable culture devices. Lab Chip 2014, 14, 167–171. 10.1039/C3LC50887K. [DOI] [PubMed] [Google Scholar]
- Hu W.; Peng C.; Luo W.; Lv M.; Li X.; Li D.; Huang Q.; Fan C. Graphene-based antibacterial paper. ACS Nano 2010, 4, 4317–4323. 10.1021/nn101097v. [DOI] [PubMed] [Google Scholar]
- van Oosten M.; Schäfer T.; Gazendam J. A. C.; Ohlsen K.; Tsompanidou E.; de Goffau M. C.; Harmsen H. J. M.; Crane L. M. A.; Lim E.; Francis K. P.; Cheung L.; Olive M.; Ntziachristos V.; van Dijl J. M.; van Dam G. M. Real-time in vivo imaging of invasive- and biomaterial-associated bacterial infections using fluorescently labelled vancomycin. Nat. Commun. 2013, 4, 2584. 10.1038/ncomms3584. [DOI] [PubMed] [Google Scholar]
- Li Y.-Q.; Zhu B.; Li Y.; Leow W. R.; Goh R.; Ma B.; Fong E.; Tang M.; Chen X. A Synergistic capture strategy for enhanced detection and elimination of bacteria. Angew. Chem., Int. Ed. 2014, 53, 5837–5841. 10.1002/anie.201310135. [DOI] [PubMed] [Google Scholar]
- López-Igual R.; Bernal-Bayard J.; Rodríguez-Patón A.; Ghigo J.-M.; Mazel D. Engineered toxin–intein antimicrobials can selectively target and kill antibiotic-resistant bacteria in mixed populations. Nat. Biotechnol. 2019, 37, 755–760. 10.1038/s41587-019-0105-3. [DOI] [PubMed] [Google Scholar]
- Park J.; Jiang Q.; Feng D.; Mao L.; Zhou H.-C. Size-controlled synthesis of porphyrinic metal–organic framework and functionalization for targeted photodynamic therapy. J. Am. Chem. Soc. 2016, 138, 3518–3525. 10.1021/jacs.6b00007. [DOI] [PubMed] [Google Scholar]
- Deria P.; Mondloch J. E.; Karagiaridi O.; Bury W.; Hupp J. T.; Farha O. K. Beyond post-synthesis modification: evolution of metal-organic frameworks via building block replacement. Chem. Soc. Rev. 2014, 43, 5896–5912. 10.1039/C4CS00067F. [DOI] [PubMed] [Google Scholar]
- Cohen S. M. Postsynthetic methods for the functionalization of metal–organic frameworks. Chem. Rev. 2012, 112, 970–1000. 10.1021/cr200179u. [DOI] [PubMed] [Google Scholar]
- Tanabe K. K.; Cohen S. M. Postsynthetic modification of metal–organic frameworks—a progress report. Chem. Soc. Rev. 2011, 40, 498–519. 10.1039/C0CS00031K. [DOI] [PubMed] [Google Scholar]
- Koshland D.; Botstein D. Evidence for posttranslational translocation of β-lactamase across the bacterial inner membrane. Cell 1982, 30, 893–902. 10.1016/0092-8674(82)90294-X. [DOI] [PubMed] [Google Scholar]
- Minsky A.; Summers R. G.; Knowles J. R. Secretion of beta-lactamase into the periplasm of Escherichia coli: evidence for a distinct release step associated with a conformational change. Proc. Natl. Acad. Sci. U. S. A. 1986, 83, 4180–4184. 10.1073/pnas.83.12.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Ding S.; Dietrich R.; Märtlbauer E.; Zhu K. A biosurfactant-inspired heptapeptide with improved specificity to kill MRSA. Angew. Chem., Int. Ed. 2017, 56, 1486–1490. 10.1002/anie.201609277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


