Abstract
Protein glycosylation is a common post-translational modification that influences the functions and properties of proteins. Despite advances in methods to produce defined glycoproteins by chemoenzymatic elaboration of monosaccharides, the understanding and engineering of glycoproteins remain challenging, in part, due to the difficulty of site-specifically controlling glycosylation at each of several positions within a protein. Here, we address this limitation by discovering and exploiting the unique, conditionally orthogonal peptide acceptor specificities of N-glycosyltransferases (NGTs). We used cell-free protein synthesis and mass spectrometry of self-assembled monolayers to rapidly screen 41 putative NGTs and rigorously characterize the unique acceptor sequence preferences of four NGT variants using 1254 acceptor peptides and 8306 reaction conditions. We then used the optimized NGT-acceptor sequence pairs to sequentially install monosaccharides at four sites within one target protein. This strategy to site-specifically control the installation of N-linked monosaccharides for elaboration to a variety of functional N-glycans overcomes a major limitation in synthesizing defined glycoproteins for research and therapeutic applications.
Short abstract
Discovery and deep characterization of peptide specificity differences in N-glycosyltransferase enzymes enabled the sequential control glycosylation at multiple sites within a single target protein.
Introduction
Protein glycosylation is the attachment of sugar moieties to amino acid side-chains of proteins and is one of the most common post-translational modifications found in nature.1 Glycosylation is important for protein functions and can improve the stability,2 potency,3 and half-life4 of protein therapeutics.5 However, glycoproteins derived from living cells usually contain a complex mixture of glycoforms, with variations in both the sites of glycosylation and oligosaccharide structures.5,6 This lack of control is a key challenge in studies that aim to understand the activity and properties of structurally and site-specifically defined glycoforms, and therefore, the development and optimization of glycoproteins for biotechnological applications.5,7 While significant advances have been made in glycoengineering bacterial,8 yeast,9 and mammalian7,10 cells, a generalizable technique for preparing user-defined glycoforms from cells remains challenging,6,11 and the possibility for understanding or exploiting synergistic interactions between multiple, distinct glycans on a single protein remains largely unexplored.6,11
New developments in chemical and chemoenzymatic methods for in vitro construction of homogeneous glycoproteins have enabled the synthesis and study of diverse glycoproteins with rigorously defined glycan structures.6,11−13 For example, total chemical synthesis has been used to produce human erythropoietin and test the function of each glycan by assembling constituent peptides and glycopeptides.4,14 However, total chemical synthesis has only been successfully applied to a few proteins and is particularly inefficient for larger proteins.11 Davis and co-workers have incorporated noncanonical amino acids and cysteine residues to install monosaccharides or full-length glycans using site-directed mutagenesis.15−17 However, the possible modification of untargeted cysteine residues in proteins and the difficulty of implementing multiple orthogonal noncanonical amino acids may limit its broad application. Finally, the chemoenzymatic method is now important for remodeling glycans or installing defined glycans onto proteins that are first modified with monosaccharides.6,11 We developed a suite of endoglycosidases to remodel glycans,11,18 and this method has been applied to optimize the glycosylation structures of antibody therapeutics for efficient antibody-dependent cell-mediated cytotoxicity.3,19
In practice, the chemoenzymatic method is limited to the synthesis of proteins with one or, at most, two unique carbohydrate structures.11,16,20 In contrast, glycoproteins often contain multiple glycosylation sites, each with distinct glycosylation structures that can synergistically interact to effect protein functions.16,21−25 There is a significant need for methods that can site-specifically control glycosylation at multiple sites so that glycoproteins with defined combinations of glycans and the interactions between them can be studied and optimized to engineer precise or multifunctional glycoprotein therapeutics and vaccines.11 Because recently developed chemoenzymatic techniques can synthesize varieties of complex glycans26,27 and make them into oxazoline donors to elaborate protein-linked monosaccharides to defined full-length glycans,13 the key remaining barrier to the synthesis of proteins with multiple, distinct glycosylation structures at defined locations is the development of strategies to site-specifically control the installation of monosaccharides at each of several positions in a protein.
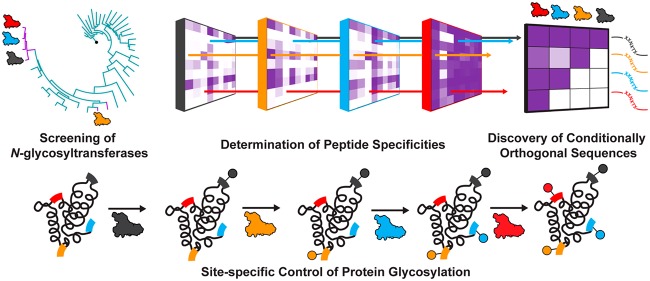
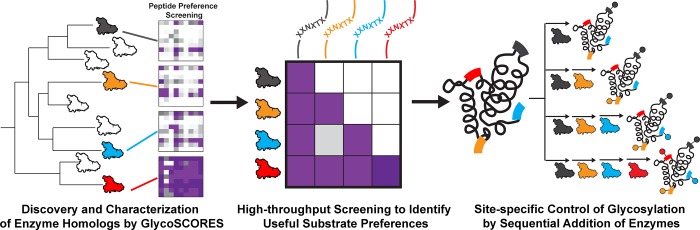
Here, we report a strategy to site-specifically control the glycosylation of four sites within a single protein based on the conditionally orthogonal specificities of N-glycosyltransferase (NGT) variants to install monosaccharides at unique acceptor sites (Figure 1). NGTs are a class of enzymes that post-translationally modify an asparagine residue (at the canonical N-X-S/T acceptor site) with an N-linked glucose from a uracil-diphosphate-glucose (UDP-Glc) sugar donor.28−31 NGTs have several compelling applications in glycoprotein synthesis.30,32−35 However, the substrate specificities for members of the NGT family have largely not been elucidated, making it difficult to identify pairs of enzymes and substrates that are orthogonal and would allow independent glycosylation of distinct sites within a protein. To address this gap, we describe the application of our recently developed method for glycosylation sequence characterization and optimization by rapid expression and screening (GlycoSCORES)36 to characterize 41 putative NGTs—which we prepare with cell-free protein synthesis (CFPS)47—using the self-assembled monolayers for matrix-assisted laser desorption/ionization mass spectrometry (SAMDI-MS) technique. These experiments identified three NGTs that had both high activity and different peptide specificities. We then optimized the sequences of short peptide substrates (which we term GlycTags) for these three enzymes as well as one engineered NGT variant, such that we could sequentially glycosylate each peptide using one of the four enzymes. We show that when these GlycTags are placed into a single target protein, glycosylation can be site-specifically controlled at four sites by the sequential addition of glucose by specific NGTs.
Figure 1.
Site-specific control of glycosylation by sequential enzymatic addition of glycans. The GlycoSCORES workflow was used to screen 41 putative NGTs and discover differences in peptide acceptor specificities. Conditionally orthogonal NGT-substrate sequence pairs were further optimized, such that the first NGT only glycosylates one of the four substrate sequences, the second NGT glycosylates one of the remaining three substrate sequences, and the third NGT only glycosylates one of the remaining two substrate sequences, with the last substrate sequence glycosylated by the fourth NGT. The four acceptor sites were then incorporated as GlycTags into a single protein. Sequential treatment of the protein with the four NGTs gave stepwise glycosylation of each of the four glycosylation sites. Glycans, modification sites, and NGTs have been color coded for illustration purposes.
Results
Discovery and Characterization of Unique NGT Peptide Specificities
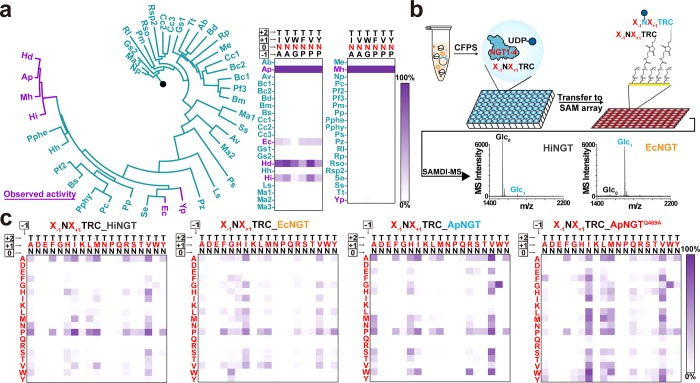
To identify NGTs that may possess different (and therefore conditionally orthogonal) specificities for peptide substrates, we performed a phylogenetic analysis of all bacterial enzymes in family 41 of the Carbohydrate Active enZYmes (CAZY) Database,37 which is known to contain NGTs and O-linked N-acetylglucosaminyltransferases (OGTs). From our initial phylogenetic analysis of 1409 unique protein sequences, we selected 41 putative NGTs (Figure 2a and Supplementary Table 2) for synthesis and characterization. In manually selecting enzymes for screening, we sought to balance sequence diversities with the likelihood of identifying enzymes with NGT activity by selecting enzymes that are either closely or distantly related to previously characterized NGTs. We synthesized the putative NGTs using CFPS (yields and SDS-PAGE autoradiograms are shown in Supplementary Figures 1 and 2) and initially tested the activity by screening for glucose modification against six representative peptides, which are known NGT substrates, using SAMDI-MS (Figure 2a,b).36 Each peptide was incubated with the unpurified NGT and UDP-Glc sugar donor in an in vitro glycosylation (IVG) reaction. The IVG reaction mixtures were then applied to a self-assembled monolayer presenting maleimide groups, where the cysteine-terminated peptides underwent immobilization via a conjugate addition reaction. The monolayer was rinsed and analyzed by matrix-assisted laser desorption/ionization mass spectrometry (i.e., SAMDI-MS). The glycosylated product was detected and quantified using a new peak with a mass increased by 162 Da. The on-chip purification method of SAMDI-MS allowed us to rapidly screen enzymes synthesized in crude lysate CFPS without the need for laborious purification of enzymes.
Figure 2.
GlycoSCORES screening of NGT homologues for unique peptide specificities. (a) Screening for active NGTs from putative bacterial glycosyltransferases. Forty-one putative glycosyltransferases from the CAZY database were screened against six representative peptide substrates for N-glucosyltransferase activity. The phylogenetic tree is rooted with human OGT (black dot) as the outgroup. Six enzymes showed N-glucosyltransferase activity, with strong activity from ApNGT, EcNGT, HdNGT, HiNGT, and MhNGT. Relative MS intensities of peptide substrates and glucose modified peptide products are shown when treated with 5% (v/v) CFPS NGTs and 2.5 mM UDP-Glc at 30 °C for 21 h (n = 1). (b) Scheme for GlycoSCORES workflow for discovery and characterization of the peptide specificities for the NGTs. NGTs were produced in CFPS and mixed with UDP-Glc sugar donor and cysteine-containing peptide substrates, which were then immobilized to a maleimide functionalized self-assembled monolayer and characterized by SAMDI-MS. (c) Peptide acceptor specificity comparison of HiNGT (gray), EcNGT (orange), ApNGT (blue), and ApNGTQ469A (red) using a peptide array of X–1NX+1TRC sequence. Expanded X–1NX+1(T/S)RC substrate libraries with numerical annotation are shown in Supplementary Figure 4. Enzyme concentrations were controlled to obtain a maximum yield of ∼85% in each heatmap to facilitate the comparison of specificities. The heatmap for ApNGT is taken from a previous report.36 Each heatmap shows percentage glucose modification from n = 1 experiment using the conditions: 0.42 μM CFPS HiNGT or 0.75 μM CFPS EcNGT, 30 °C for 21 h; 0.055 μM CFPS ApNGT or 0.014 μM CFPS ApNGTQ469A, 30 °C for 1 h.
In addition to the previously well characterized NGTs from Actinobacillus pleuropneumoniae (ApNGT), Mannheimia haemolytica (MhNGT), and Haemophilus ducreyi (HdNGT) with similar specificity,36 as well as a previously identified NGT from Haemophilus influenzae (HiNGT),38 two previously uncharacterized NGTs from Escherichia coli (EcNGT) and Yersinia pestis (YpNGT) were found to have detectable N-glucosyltransferase activity with representative peptides (Figure 2a and Supplementary Figure 3). The high number of enzymes in CAZY family 41 showing no activity in our tests, even though most of these enzymes expressed well in CFPS, indicates that this family may contain enzymes specific for peptides or sugar donors outside of those tested here. The fact that most of the NGTs that exhibited strong activity in this study were rather closely related to ApNGT supports this hypothesis.
Because EcNGT and HiNGT both showed strong activity and have not been well characterized, we further characterized the substrate specificity of these two NGTs as well as a recently reported, highly active ApNGT mutant, ApNGTQ469A,34,39 using an array of peptides having the sequence X–1-N-X+1-T/S, where X–1 and X+1 are all natural amino acids except Cys (Figure 2c and Supplementary Figure 4). Differences in ionization between modified and unmodified peptides were accounted for using relative ionization factors (RIFs) which were determined as previously described36 (Supplementary Table 4). These data showed that the enzymes EcNGT, HiNGT, ApNGT, and ApNGTQ469A had distinct differences in their specificities.
Inspection of the specificity heatmaps for these four NGTs did not reveal sequences that exhibited absolute orthogonality (the ability to be modified by specific NGTs regardless of the order in which NGTs are added). However, we did find pairs of enzymes and substrates that had “conditional orthogonality”, wherein a first NGT only glycosylates one of the four substrates, the second only glycosylates one of the remaining three substrates (as well as the first, though that substrate was modified in the initial step), and the third NGT only glycosylates one of the remaining two substrates, with the last glycosylated by the fourth NGT. These four pairs of enzymes and substrates could therefore enable a sequential modification scheme in which each substrate is modified only by a single NGT. In other words, it is acceptable for later NGTs to be able to modify sequences that have already been quantitatively modified by previous NGTs. By subtracting peptide specificity maps shown in Supplementary Figure 4 from each other, we found the potential for conditional orthogonality in sequence sets modified by HiNGT after EcNGT; EcNGT after any of the other three NGTs; ApNGT after EcNGT and HiNGT, or EcNGT and ApNGTQ469A; and ApNGTQ469A after any of the three NGTs (Supplementary Figures 5 and 6). Considering both enzyme orthogonality and overall enzyme activity, we reasoned that treatment with HiNGT, EcNGT, ApNGT, and then ApNGTQ469A, in that order, would give the optimal site-specificity for sequential glycosylation at four unique positions.
Optimization of Conditionally Orthogonal GlycTags
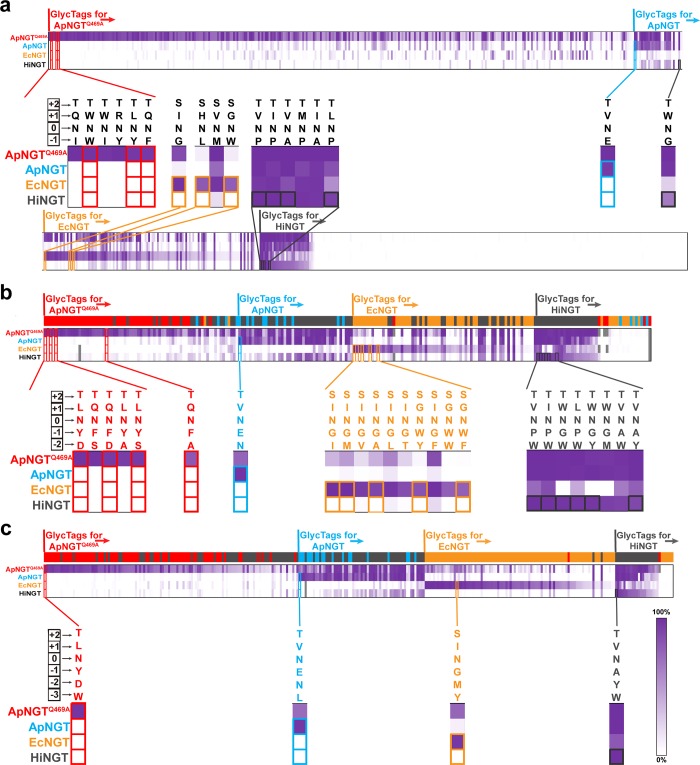
Before attempting the sequential, site-specific glycosylation of a protein having four GlycTags, we first optimized the conditional orthogonality of these GlycTags for the order of NGT treatments. We again used the GlycoSCORES workflow and screened each purified NGT (HiNGT, EcNGT, ApNGT and then ApNGTQ469A) on the full X–1-N-X+1-T/S peptide library to determine modification efficiencies for optimized enzyme concentrations and reaction time (Supplementary Figure 7). To facilitate selection of specifically interacting NGT-GlycTag pairs, we quantified the conditional orthogonality of each peptide sequence as its percent modification when treated with the intended NGT less the sum of its percent modifications when treated with preceding NGTs in our envisioned sequential glycosylation reaction (HiNGT, EcNGT, ApNGT, and then ApNGTQ469A). We organized peptide sequences into regions based on their potential utility as GlycTags for each of the four chosen NGTs. We then arranged the sequences within each region by decreasing conditional orthogonality (Figure 3a). Specifically, we first identified peptides with >5% conditional orthogonality for ApNGTQ469A over the other three NGTs as GlycTags for ApNGTQ469A, and then identified peptides with >5% conditional orthogonality for ApNGT over EcNGT and HiNGT from the remaining sequences as GlycTags for ApNGT, and so on for EcNGT. Remaining sequences modified were identified as GlycTags for HiNGT. We found that each region contained at least one sequence with >94% conditional orthogonality, and we selected 11 GlycTags with particularly high conditional orthogonality in each region for further optimization (Figure 3a). We also selected “GNWT” as a possible core peptide sequence for HiNGT based on our previous finding that this is an optimal sequence for ApNGT when an X–2 residue is present in the peptide.36
Figure 3.
Optimization of conditionally orthogonal GlycTags for NGTs. (a) Screening to improve the conditional orthogonality of NGT-GlycTag pairs for HiNGT, EcNGT, ApNGT, and ApNGTQ469A using an X–1NX+1(T/S)RC peptide library. Peptides were screened with HiNGT, EcNGT, ApNGT, and then ApNGTQ469A, respectively. Sequences are arranged by decreasing conditional orthogonality (differences in modification between specific NGT and the sum of preceding NGTs). The sequences are also divided into regions that are preferred by ApNGTQ469A only (red), ApNGT but not EcNGT or HiNGT (blue), EcNGT but not HiNGT (orange), and HiNGT (gray). GlycTags showing strong conditional orthogonality which were identified in each region used for additional optimization are highlighted (colored boxes). Heatmaps with experimental conditions and numerical annotation from n = 2 separate peptide IVG reactions are shown in Supplementary Figure 7. (b) Conditional orthogonality screening of selected sequences from a which were resynthesized with 19 amino acids in the X–2 position in 5-mer X–2(X–1NX+1T/S)RC library. GlycTags identified from each region and used for additional optimization are highlighted. (c) Conditional orthogonality screening of selected sequences from (b) which were resynthesized with 19 amino acids in the X–3 position. Heatmaps with experimental conditions and numerical annotation from n = 1 separate peptide IVG reactions for (b) and (c) are shown in Supplementary Figures 8 and 9, respectively. The corresponding colors of sequences originating from previous screens are shown above the heatmaps. A set of four GlycTags with >95% conditional orthogonality was selected for site-specific control of sequential glycosylation at four sites within one target protein.
To further increase the conditional orthogonality of the 12 selected peptides, we synthesized a new library wherein the X–2 position was substituted with each of the 19 amino acids (generating 228 peptides), and we again determined their modification efficiencies when treated with HiNGT, EcNGT, ApNGT, or ApNGTQ469A under optimized conditions (Figure 3b). The peptide sequences were arranged similarly as in Figure 3a, and we selected several peptide sequences from each region showing improved conditional orthogonality (Figure 3b). For these 16 peptide candidates, we synthesized a final library of 304 additional peptides wherein the X–3 position was also substituted with each of the 19 amino acids, and we again determined their modification efficiencies when treated with HiNGT, EcNGT, ApNGT, or ApNGTQ469A under optimized conditions (Figure 3c). The sequences WYANVT, YMGNIS, LNENVT, and WDYNLT each showed greater than 95% conditional orthogonality and were selected as optimized GlycTags for insertion into a single protein for sequential, site-specific glycosylation with HiNGT, EcNGT, ApNGT, and then ApNGTQ469A (Figure 3c). These optimized GlycTags were resynthesized and again analyzed with GlycoSCORES (Figure 4a and Supplementary Figure 10) to confirm their conditional orthogonality. In selecting these 6-mer GlycTags for protein engineering, we balanced the increased orthogonality and robustness of a longer GlycTag with the increased flexibility and lessened impact on the protein structure that a shorter GlycTag would allow. We note that the analysis in Figure 3 would likewise identify many other sets of GlycTags or possible optimization paths depending on constraints of protein design.
Figure 4.
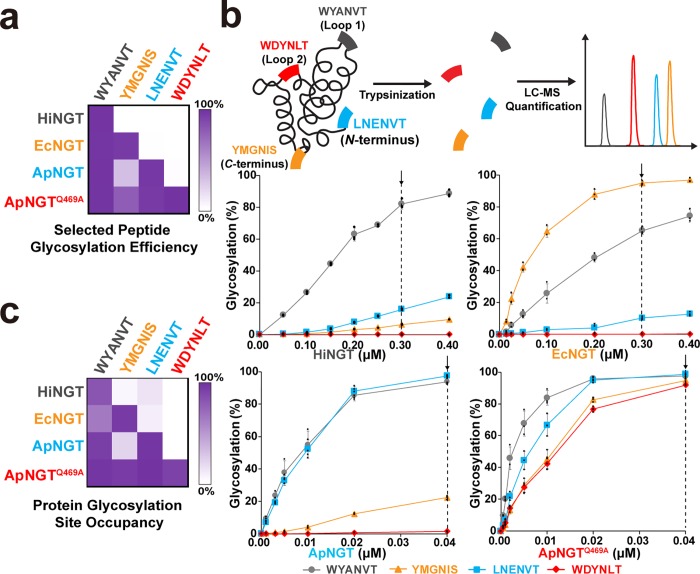
Optimized GlycTag sequences showing conditional orthogonality as peptides and within a single Im7 target protein. (a) The conditional orthogonality of optimized 6-mer GlycTags. HiNGT, EcNGT, ApNGT, and ApNGTQ469A modification of selected GlycTags from Figure 3c under optimized conditions were resynthesized and reanalyzed by GlycoSCORES with purified NGTs. The modification efficiency of each peptide was calculated using individually measured RIFs (Supplementary Table 4). Experimental conditions (n = 3 individual IVGs): 0.2 μM HiNGT or 0.67 μM EcNGT, 30 °C for 21 h; 0.45 μM ApNGT or 0.1 μM ApNGTQ469A, 30 °C for 3 h. (b) Optimized 6-mer GlycTags were inserted into the N-terminus, C-terminus, and two exposed loops of the glycosylation model protein Im7, with flanking sequences of RATT-GlycTag-RAGG to facilitate trypsinization and quantitative LC-qTOF analysis. 10 μM of purified 4gIm7 bearing all four optimized GlycTags was reacted with 2.5 mM UDP-Glc and various concentrations of each purified NGT for 4 h in n = 3 IVG reactions. After modification, 4gIm7 was purified from the reaction, treated with trypsin, and analyzed by LC-qTOF. The modification efficiency (occupancy) of each site was calculated using individually measured RIFs (shown in Supplementary Table 5). A vertical dotted line in each graph denotes optimal conditions for each NGT which provide the greatest conditional orthogonality. Representative LC chromatograms and related MS spectra are presented in Supplementary Figure 13. (c) The conditional orthogonality of each optimized 6-mer GlycTag within 4gIm7 under optimized conditions. Bar graphs with errors for heatmaps in (a) and (c) are shown in Supplementary Figure 10. Similar modification patterns were observed for peptides and GlycTags within engineered 4gIm7.
To evaluate the sugar donor specificity for each of these four NGTs, we screened each enzyme for activity when combined with six representative peptides and UDP-Glc, UDP-Galactose (Gal), UDP-Xylose (Xyl), UDP-Glucosamine (GlcN), GDP-Mannose (Man), UDP-N-acetylglucosamine (GlcNAc), or UDP-N-acetylgalactosamine (GalNAc) (Supplementary Figure 11). We found that all four NGTs could use UDP-Glc, UDP-Gal, and UDP-Xyl and that ApNGT and ApNGTQ469A could also use UDP-GlcN. Overall, these specificities could provide opportunities to install different monosaccharides at the conditionally orthogonal sites designed below.
Site-Specific Control of Protein Glycosylation with Conditionally Orthogonal GlycTags
To show that the GlycTags identified above and their four corresponding NGTs could be used for site-specific control of multiple N-linked glycosylation sequences within a single target protein, we engineered the E. coli immunity protein Im7 to include these four sequences. Im7 has been used previously to study N-linked glycosylation sites.36,40 We have previously found48 that the location of the GlycTag within the protein sequence can impact glycosylation activity, so we first determined the optimal placement of the four GlycTags within the protein. To do this, we placed the promiscuous sequence “IYANVTL”, which our previous analyses indicated could be easily modified by each of the four NGTs, at the N-terminus, the C-terminus, loop 1 (residues N26_D32), or loop 2 (S58_S64). We tested the preference of each NGT for each of these four versions of Im7 by reacting them with each NGT and UDP-Glc, purifying Im7 from the reaction using a C-terminal polyhistidine tag, and quantifying modification using liquid chromatography quadrupole time-of-flight (LC-qTOF) analysis. While we did not observe drastic differences in the preferences of each NGT, we determined that Loop 1, the C-terminus, and the N-terminus were preferred by HiNGT, EcNGT, and ApNGT, respectively (Supplementary Figure 12). As ApNGTQ469A is very active and we use this enzyme as the last treatment in the sequential glycosylation reaction, we did not consider the positional preferences of this enzyme. Based on these data, we placed the GlycTags for HiNGT, EcNGT, ApNGT, and ApNGTQ469A at the Loop 1, C-terminal, N-terminal, and Loop 2 positions, respectively, of Im7 to make 4gIm7 (full sequence available in Supplementary Note 1). We placed three amino acid flanking sequences and an Arg at each side of the GlycTags to facilitate our analytical strategy of trypsin cleavage and site-occupancy analysis by LC-qTOF (Figure 4b).
To test the conditional orthogonality of our optimized GlycTags within a single protein, we reacted purified 4gIm7 with UDP-Glc and various amounts of each NGT, then purified the 4gIm7 (and its glycosylated adducts) from the reaction using a C-terminal polyhistidine tag, digested them with trypsin, and quantified the site-occupancy at each sequence using LC-qTOF (Figure 4b and Supplementary Figure 13). Differences in ionization between the modified and unmodified peptide were accounted for by measuring the RIF of each glycopeptide in LC-qTOF analysis using synthesized peptide standards (Supplementary Table 5). Under optimal conditions, the conditional orthogonality (as defined for peptides above) of all GlycTags remained greater than 75%, with the greatest source of unwanted cross-reactivity observed for glycosylation of the ApNGT sequence by HiNGT. The modest observed decrease in conditional orthogonality or selectivity of NGTs for GlycTags when presented in a protein compared to a peptide may be due to interference from nearby residues, secondary structure, or colocalization of the substrate sequences. Nevertheless, the heatmap of modification for each GlycTag by each NGT in 4gIm7 (Figure 4c and Supplementary Figure 10) showed a similar pattern to the modifications observed at the peptide level in Figure 4a and demonstrates the conditional orthogonality necessary to enable sequential protein modification.
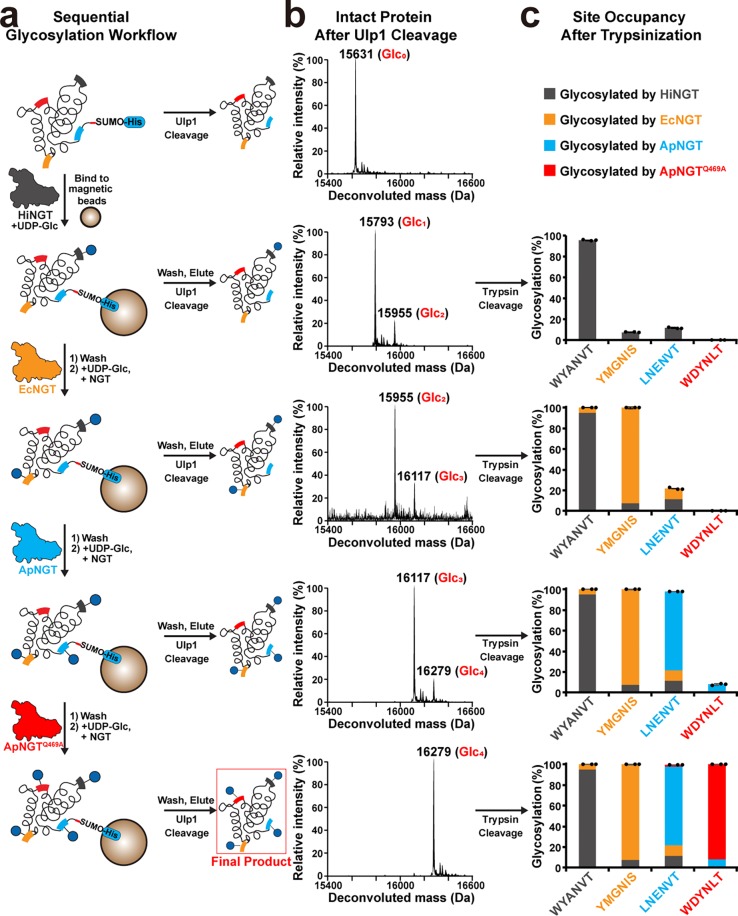
Having validated our GlycTags’ orthogonality at both the peptide and protein level, we finally sought to demonstrate site-specific glycosylation of the engineered 4gIm7 target protein containing all four GlycTags using sequential treatments with HiNGT, EcNGT, ApNGT, and then ApNGTQ469A. To facilitate the sequential reactions of engineered 4gIm7 and increase yields of correctly glycosylated products, we devised a simplified workflow for sequential modification using Ni-NTA functionalized magnetic beads to immobilize the engineered Im7 after the first treatment with HiNGT (Figure 5a). To prevent the bead from interfering with the access of NGTs to the immobilized 4gIm7 substrate, we added a SUMO tag between the N-terminal His-Tag and 4gIm7 to yield SUMO-4gIm7. Purified SUMO-4gIm7 was treated with HiNGT, bound to magnetic beads, and then sequentially reacted with EcNGT, ApNGT, and ApNGTQ469A. The sugar donor UDP-Glc was present in each reaction, and the beads were washed between each NGT treatment. After each step, the sequential elaboration of SUMO-4gIm7 was monitored by eluting the protein from a fraction of the beads. Half of the eluted protein was cleaved from SUMO with Ulp1, and the cleaved 4gIm7 protein was analyzed by LC-qTOF. Deconvoluted spectra from intact 4gIm7 in Figure 5b show that the primary components of the reaction were 4gIm7 with 1, 2, 3, and 4 glucose modifications after treatment with HiNGT, EcNGT, ApNGT, and ApNGTQ469A, respectively. The other half of the eluted protein was treated with trypsin and analyzed with LC-qTOF to quantify the occupancy of each glycosylation site. This analysis showed that glycosylation at each targeted site was specifically controlled by the sequential addition of NGTs, though up to 12% of undesired modification was also observed at the untargeted GlycTags (Figure 5c). Assuming the glycosylation events are not interdependent (supported by Supplementary Figure 14), the site-occupancy data in Figure 5c indicate that at the completion of all four NGT treatment steps, 62 ± 1% of the 4gIm7 was correctly glucosylated in all steps (Supplementary Table 6). While we did not generate a completely homogeneous protein or elaborate these monosaccharides into more complex glycans, this yield of protein correctly modified at all steps represents a more than 150-fold enrichment compared to the 0.39% yield that would be observed if each of the enzymes had no differential specificity for all four sites.
Figure 5.
Site-specific control of sequential glycosylation at four distinct GlycTag sequences within a single Im7 protein. (a) A workflow for site-specific control of glycosylation at four sequences within Im7 by sequential addition of NGT enzymes. (b) Representative deconvoluted MS spectra for intact 4gIm7 of n = 3 IVG reactions showing SUMO-4gIm7 after each NGT treatment, cleaved by UIp1 and analyzed by LC-qTOF. Two μM purified SUMO-4gIm7 was incubated with 0.3 μM HiNGT and then immobilized to magnetic beads and sequentially washed and treated with 0.3 μM EcNGT, 0.4 μM ApNGT, and then 2 μM ApNGTQ469A. Reactions (n = 3) were performed for 4 h at 30 °C containing 2.5 mM UDP-Glc and monitored by elution and LC-qTOF after each wash step. These data show that the dominant forms of 4gIm7 are those modified with one glucose per NGT treatment. The optimization of reaction conditions for SUMO-4gIm7 are shown in Supplementary Figure 15. One representative LC chromatogram and related deconvoluted MS spectra are presented in Supplementary Figure 16. (c) A bar graph of site-occupancy at each of the four GlycTags in the same n = 3 experiments in (b) after each NGT treatment, cleavage by trypsin, and analysis by LC-qTOF. Each GlycTag was mainly glycosylated by its corresponding NGT.
Because many glycoproteins contain fewer than four sites and some applications require more homogeneous samples, we modified the experiments above using a version of Im7 containing only the EcNGT and ApNGTQ469A glycosylation sites (2gIm7) at the C-terminus and N-terminus, respectively (Supplementary Figure 17). We observed complete sequential conversion of the C-terminus site and N-terminus site with almost no off-target modification (98% of the final product was glycosylated in the correct reactions). This demonstrates nearly quantitative control of N-glycan occupancy at two sites.
Next, we investigated the use of recently developed chemoenzymatic transglycosylation strategies to elaborate the monosaccharides site-specifically installed by NGTs.6,11 Specifically, we sought to elaborate the glucose residues installed by EcNGT or ApNGTQ469A to biantennary glycans using endoglycosidases to transfer glycans from chemically synthesized oxazoline donors. We used EcNGT to selectively install a glucose at the C-terminus of 2gIm7 and then used Endo-A to elaborate this glucose to a human-like azido-functionalized trimannose core glycan, AzMan2ManGlcNAcGlc (AzMan3) (see methods). We observed nearly 50% elaboration of the EcNGT-installed glucose to AzMan3 (Supplementary Figure 18). We also investigated the transfer of a human-like biantennary glycan onto the EcNGT-installed glucose to produce Sia2Gal2GlcNAc2Man3GlcNAcGlc (SCT) using EndoCCN180H. We obtained an apparently homogeneous product (nearly 100% modification) with the EcNGT-targeted glycosylation site modified with SCT (Supplementary Figures 19). We also obtained 81% conversion of diglucosylated 2gIm7 to SCT with EndoCCN180H at both sites (Supplementary Figure 20). These data indicate that the combination of site-selective glucose modification with conditionally orthogonal GlycTags and chemoenzymatic elaborations can site-selectively install complex glycans within a target protein bearing multiple glycosylation sites.
Finally, we investigated the use of our sequential glycosylation method using a therapeutically relevant glycoprotein, the constant region of human immunoglobulin G (Fc). We engineered Fc to contain two GlycTags, one at the C-terminus that could be modified by EcNGT and one at the conserved Asn297 glycosylation site that was modified to be specifically preferred by ApNGTQ469A (see Supplementary Note 1 for full sequence of 2gFc). Similar to 2gIm7, we observed nearly quantitative control of sequential glucose modification (Supplementary Figure 21) in 2gFc with 98% of the protein being modified at the correct steps. We then applied our transglycosylation methods to selectively install AzMan3 or SCT onto the EcNGT-targeted glycosylation site at the C-terminus (both with approximately 50% efficiency) (Supplementary Figures 22–23). We then attempted a sequential glycosylation strategy to first install AzMan3 onto the EcNGT site and then install a Sia-Gal-Glc glycan, a simplified glycan similar to the GlycoDelete structure Sia-Gal-GlcNAc7 shown to have desirable properties for IgG, or SCT using a recently discovered glycosyltransferase pathway49 or endoglycosidases (Supplementary Figures 24 and 25) at the second ApNGTQ469A site, respectively. We observed low levels of elaboration at the ApNGTQ469A site, but we did confirm the presence of the intended glycans at the intended sites. While additional work will be required to optimize the efficiency of these elaboration reactions despite differences in protein sequence and structure, these data show proof-of-concept for sequential modification of a therapeutically relevant glycoprotein. With further optimization, such a system may enable the installation of two distinct glycans on Fc. In the future, this workflow could allow the conjugation of a small-molecule cytotoxin or imaging agent (using an azido-functionalized glycan41) at one glycan while maintaining a biantennary human-like glycan at the Asn297 site, which is important for efficient antibody-dependent cell-mediated cytotoxicity.3,19
Discussion
This paper describes the discovery and application of unique peptide acceptor specificities of NGTs to enable the site-specific control of N-linked glycosylation at up to four distinct sites within a single target protein. Our GlycoSCORES approach for efficient glycosyltransferase expression by CFPS and screening by SAMDI-MS allowed us to rapidly test the activities of 41 putative NGTs and rigorously characterize the peptide acceptor and sugar donor specificities of four highly active NGT variants using 1254 peptides and 8306 unique reaction conditions. We developed a set of four conditionally orthogonal NGT-GlycTag pairs, engineered them into a single protein, and demonstrated that these GlycTags retained their conditional orthogonality in the context of the whole protein. The depth of characterization provided and the correlation between peptide and protein glycosylation patterns shown here and elsewhere36,39,40 suggest that this method could be generalized to a variety of proteins. We then demonstrated a magnetic bead-immobilization workflow to facilitate sequential NGT glycosylation steps to site-specifically modify four distinct glycosylation sequences within one target protein.
The sequential glycosylation strategy demonstrated in this work overcomes the critical challenge for achieving the site-specific installation of distinct glycans at multiple positions within one target protein in that it allows for the isolation of glycoprotein with one, two, three, or four monosaccharides at distinct positions. The system demonstrated in Figure 5 enabled 62% of our 4gIm7 target protein with four sites to be glycosylated with glucose at the correct steps. We also showed that nearly 100% of 2gIm7 as well as a therapeutically relevant glycoprotein, 2gFc, with two sites could be glycosylated with glucose at the correct steps (Supplementary Figures 17 and 21). Previous work shows how these monosaccharides can be elaborated at each step to install complex glycosylation structures by treatment with additional glycosyltransferases33,49,50 or by using chemoenzymatic methods with endoglycosidases.13,35,42,43 Using these methods, we elaborated selectively installed glucose residues on 2gIm7 or 2gFc to glycans with biantennary complex glycans, terminal sialic acids, and azido-functionalization (Supplementary Figures 18–20 and 22–25).
However, even with these well-established methods, we were not able to achieve quantitative elaboration of glycans for some glycosylation sites. We are currently working to address this by screening glycosyltransferases and endoglycosidases from different species and subjecting them to further engineering to make these enzymes more efficient and less dependent on target protein sequence/structure, thereby enabling the synthesis of more diverse glycoforms. Future works should also develop and integrate methods to install GlcNAc rather than Glc as the reducing end sugar in order to more closely match naturally occurring human N-glycans. Advances have already been made in this area by using ApNGTQ469A to install a GlcN residue that can be acetylated by GlmA to form GlcNAc34 or by trimming down a longer glycan installed by an oligosaccharyltransferase (OST) to obtain a single N-linked GlcNAc.42 These GlcNAc residues were then elaborated to authentic human N-linked glycans using endoglycosidases.34,42
A key feature of our work was the use of GlycoSCORES to comprehensively map and identify conditionally orthogonal GlycTags. Unlike qualitative screening methods—which are effective at identifying active substrates that serve as starting points for further development—the SAMDI method provides a quantitative measure of the activity of every substrate. Hence, knowledge of which substrates have high activity for certain NGTs and poor activity for others was critical to identifying the pairs of enzyme and substrate that had conditional orthogonality. Directed evolution, an alternative approach for generating orthogonal enzyme–substrate pairs for highly selective positions of peptide substrates,44−46 is best suited to identifying a single orthogonal pair. Another benefit of our approach is that it allowed us to identify multiple, distinct GlycTags within the canonical, eukaryotic glycosylation sequence N-X-S/T, without relaxing specificity such that other protein sites, such as N-X-A, remain aglycosylated. In the future, the set of available conditionally orthogonal GlycTags may be expanded, and the need to engineer target protein sequences may be alleviated by engineering the peptide binding sites of known NGTs for alternative specificities.
In summary, this work describes a systematic method to site-specifically control protein glycosylation at multiple sequences within a single target protein, overcoming a major limitation in the synthesis of defined glycoforms of proteins with multiple glycosylation sites for basic science and biotechnological applications. We expect this method, and further improvements, will find use in preparing glycoproteins for mechanistic studies of the roles of carbohydrates in protein function as well as to explore new paradigms for multifunctional therapeutic molecules based on synergistic glycan interactions.
Acknowledgments
The authors acknowledge A. Ramesh, K. Duncker, and A. Thames for assistance with molecular cloning, enzyme expression and purification, and LC-qTOF data analysis; M. P. DeLisa, A. S. Karim, and J. C. Stark for discussions; J. Hershewe and J. Kath for sharing reagents; and S. Habibi for assistance with LC-qTOF instrumentation. This work made use of IMSERC at Northwestern University, which has received support from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF ECCS-1542205), the State of Illinois, and International Institute for Nanotechnology (IIN). This material is based upon work supported by the Defense Threat Reduction Agency (HDTRA1-15-10052/P00001), the David and Lucile Packard Foundation, the Dreyfus Teacher-Scholar Program, and the National Science Foundation (Graduate Research Fellowship under Grant No. DGE-1324585) and MCB-1413563.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.9b00021.
Additional methods, data, and figures including DNA constructs used in this study, theoretical intact protein masses and observed errors, yield and visualization of CFPS products, supporting and numerically annotated NGT specificity heatmaps, representative LC-qTOF chromatograms and deconvolutions, optimization of GlycTag locations and modification conditions, calculation of RIFs and sequential glycosylation yields, and sequential glycosylation of Im7 and Fc with two sites and subsequent chemoenzymatic elaboration (PDF)
Author Contributions
∇ L.L. and W.K. contributed equally to the work. L.L. and W.K. designed, performed, and analyzed experiments. S.K.P. performed experiments and analyzed the data for chemoenzymatic transglycosylation reactions with the help of C.L. A.J.H. performed phylogenetic analyses of putative NGTs and helped choose NGTs for screening. M.C.J. and M.M. designed the experiments, directed the studies, and interpreted the data. L.L., W.K., S.K.P., L.-X.W., M.C.J., and M.M. wrote the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Khoury G. A.; Baliban R. C.; Floudas C. A. Proteome-wide post-translational modification statistics: Frequency analysis and curation of the swiss-prot database. Sci. Rep. 2011, 1, 90. 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott S.; Lorenzini T.; Asher S.; Aoki K.; Brankow D.; Buck L.; Busse L.; Chang D.; Fuller J.; Grant J.; Hernday N.; Hokum M.; Hu S.; Knudten A.; Levin N.; Komorowski R.; Martin F.; Navarro R.; Osslund T.; Rogers G.; Rogers N.; Trail G.; Egrie J. Enhancement of therapeutic protein in vivo activities through glycoengineering. Nat. Biotechnol. 2003, 21 (4), 414–421. 10.1038/nbt799. [DOI] [PubMed] [Google Scholar]
- Lin C.-W.; Tsai M.-H.; Li S.-T.; Tsai T.-I.; Chu K.-C.; Liu Y.-C.; Lai M.-Y.; Wu C.-Y.; Tseng Y.-C.; Shivatare S. S.; Wang C.-H.; Chao P.; Wang S.-Y.; Shih H.-W.; Zeng Y.-F.; You T.-H.; Liao J.-Y.; Tu Y.-C.; Lin Y.-S.; Chuang H.-Y.; Chen C.-L.; Tsai C.-S.; Huang C.-C.; Lin N.-H.; Ma C.; Wu C.-Y.; Wong C.-H. A common glycan structure on immunoglobulin g for enhancement of effector functions. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (34), 10611–10616. 10.1073/pnas.1513456112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M.; Kiuchi T.; Nishihara M.; Tezuka K.; Okamoto R.; Izumi M.; Kajihara Y. Chemical synthesis of erythropoietin glycoforms for insights into the relationship between glycosylation pattern and bioactivity. Sci. Adv. 2016, 2 (1), e1500678. 10.1126/sciadv.1500678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethuraman N.; Stadheim T. A. Challenges in therapeutic glycoprotein production. Curr. Opin. Biotechnol. 2006, 17 (4), 341–346. 10.1016/j.copbio.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Rich J. R.; Withers S. G. Emerging methods for the production of homogeneous human glycoproteins. Nat. Chem. Biol. 2009, 5 (4), 206–215. 10.1038/nchembio.148. [DOI] [PubMed] [Google Scholar]
- Meuris L.; Santens F.; Elson G.; Festjens N.; Boone M.; Dos Santos A.; Devos S.; Rousseau F.; Plets E.; Houthuys E.; Malinge P.; Magistrelli G.; Cons L.; Chatel L.; Devreese B.; Callewaert N. Glycodelete engineering of mammalian cells simplifies n-glycosylation of recombinant proteins. Nat. Biotechnol. 2014, 32 (5), 485–489. 10.1038/nbt.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valderrama-Rincon J. D.; Fisher A. C.; Merritt J. H.; Fan Y. Y.; Reading C. A.; Chhiba K.; Heiss C.; Azadi P.; Aebi M.; DeLisa M. P. An engineered eukaryotic protein glycosylation pathway in escherichia coli. Nat. Chem. Biol. 2012, 8 (5), 434–436. 10.1038/nchembio.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton S. R.; Gerngross T. U. Glycosylation engineering in yeast: The advent of fully humanized yeast. Curr. Opin. Biotechnol. 2007, 18 (5), 387–392. 10.1016/j.copbio.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Yang Z.; Wang S.; Halim A.; Schulz M. A.; Frodin M.; Rahman S. H.; Vester-Christensen M. B.; Behrens C.; Kristensen C.; Vakhrushev S. Y.; Bennett E. P.; Wandall H. H.; Clausen H. Engineered cho cells for production of diverse, homogeneous glycoproteins. Nat. Biotechnol. 2015, 33 (8), 842–844. 10.1038/nbt.3280. [DOI] [PubMed] [Google Scholar]
- Wang L.-X.; Amin M. N. Chemical and chemoenzymatic synthesis of glycoproteins for deciphering functions. Chem. Biol. 2014, 21 (1), 51–66. 10.1016/j.chembiol.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.-X.; Davis B. G. Realizing the promise of chemical glycobiology. Chem. Sci. 2013, 4 (9), 3381–3394. 10.1039/c3sc50877c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.; Wang L.-X. Chemoenzymatic methods for the synthesis of glycoproteins. Chem. Rev. 2018, 118 (17), 8359–8413. 10.1021/acs.chemrev.8b00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Tejada A.; Brailsford J.; Zhang Q.; Shieh J.-H.; Moore M. A. S.; Danishefsky S. J. Total synthesis of glycosylated proteins. Top. Curr. Chem. 2014, 362, 1–26. 10.1007/128_2014_622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kasteren S. I.; Kramer H. B.; Gamblin D. P.; Davis B. G. Site-selective glycosylation of proteins: Creating synthetic glycoproteins. Nat. Protoc. 2007, 2 (12), 3185–3194. 10.1038/nprot.2007.430. [DOI] [PubMed] [Google Scholar]
- van Kasteren S. I.; Kramer H. B.; Jensen H. H.; Campbell S. J.; Kirkpatrick J.; Oldham N. J.; Anthony D. C.; Davis B. G. Expanding the diversity of chemical protein modification allows post-translational mimicry. Nature 2007, 446 (7139), 1105–1109. 10.1038/nature05757. [DOI] [PubMed] [Google Scholar]
- Wright T. H.; Bower B. J.; Chalker J. M.; Bernardes G. J.; Wiewiora R.; Ng W. L.; Raj R.; Faulkner S.; Vallee M. R.; Phanumartwiwath A.; Coleman O. D.; Thezenas M. L.; Khan M.; Galan S. R.; Lercher L.; Schombs M. W.; Gerstberger S.; Palm-Espling M. E.; Baldwin A. J.; Kessler B. M.; Claridge T. D.; Mohammed S.; Davis B. G. Posttranslational mutagenesis: A chemical strategy for exploring protein side-chain diversity. Science 2016, 354 (6312), aag1465. 10.1126/science.aag1465. [DOI] [PubMed] [Google Scholar]
- Huang W.; Giddens J.; Fan S. Q.; Toonstra C.; Wang L. X. Chemoenzymatic glycoengineering of intact igg antibodies for gain of functions. J. Am. Chem. Soc. 2012, 134 (29), 12308–12318. 10.1021/ja3051266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.; DiLillo D. J.; Bournazos S.; Giddens J. P.; Ravetch J. V.; Wang L. X. Modulating igg effector function by fc glycan engineering. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (13), 3485–3490. 10.1073/pnas.1702173114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q.; An Y.; Zhu S.; Zhang R.; Loke C. M.; Cipollo J. F.; Wang L.-X. Glycan remodeling of human erythropoietin (epo) through combined mammalian cell engineering and chemoenzymatic transglycosylation. ACS Chem. Biol. 2017, 12 (6), 1665–1673. 10.1021/acschembio.7b00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang I.; Lin C.-w.; Grant O. C.; Fleurkens S.; Villiger T. K.; Soos M.; Morbidelli M.; Woods R. J.; Gauss R.; Aebi M. Analysis of site-specific n-glycan remodeling in the endoplasmic reticulum and the golgi. Glycobiology 2015, 25 (12), 1335–1349. 10.1093/glycob/cwv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losfeld M.-E.; Scibona E.; Lin C.-W.; Villiger T. K.; Gauss R.; Morbidelli M.; Aebi M. Influence of protein/glycan interaction on site-specific glycan heterogeneity. FASEB J. 2017, 31 (10), 4623–4635. 10.1096/fj.201700403R. [DOI] [PubMed] [Google Scholar]
- Go E. P.; Irungu J.; Zhang Y.; Dalpathado D. S.; Liao H.-X.; Sutherland L. L.; Alam S. M.; Haynes B. F.; Desaire H. Glycosylation site-specific analysis of hiv envelope proteins (jr-fl and con-s) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopesʼ accessibility. J. Proteome Res. 2008, 7 (4), 1660–1674. 10.1021/pr7006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y.; Raghwani J.; Allen J. D.; Seabright G. E.; Li S.; Moser F.; Huiskonen J. T.; Strecker T.; Bowden T. A.; Crispin M. Structure of the lassa virus glycan shield provides a model for immunological resistance. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (28), 7320–7325. 10.1073/pnas.1803990115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schriebl K.; Trummer E.; Lattenmayer C.; Weik R.; Kunert R.; Muller D.; Katinger H.; Vorauer-Uhl K. Biochemical characterization of rhepo-fc fusion protein expressed in cho cells. Protein Expression Purif. 2006, 49 (2), 265–275. 10.1016/j.pep.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Liu L.; Prudden A. R.; Capicciotti C. J.; Bosman G. P.; Yang J.-Y.; Chapla D. G.; Moremen K. W.; Boons G.-J. Streamlining the chemoenzymatic synthesis of complex n-glycans by a stop and go strategy. Nat. Chem. 2019, 11 (2), 161–169. 10.1038/s41557-018-0188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.; Liu L.; Wei N.; Yang J.-Y.; Chapla D. G.; Moremen K. W.; Boons G.-J. An automated platform for the enzyme-mediated assembly of complex oligosaccharides. Nat. Chem. 2019, 11, 229–236. 10.1038/s41557-019-0219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz F.; Fan Y. Y.; Schubert M.; Aebi M. Cytoplasmic n-glycosyltransferase of actinobacillus pleuropneumoniae is an inverting enzyme and recognizes the nx(s/t) consensus sequence. J. Biol. Chem. 2011, 286 (40), 35267–35274. 10.1074/jbc.M111.277160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naegeli A.; Michaud G.; Schubert M.; Lin C. W.; Lizak C.; Darbre T.; Reymond J. L.; Aebi M. Substrate specificity of cytoplasmic n-glycosyltransferase. J. Biol. Chem. 2014, 289 (35), 24521–24532. 10.1074/jbc.M114.579326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuccui J.; Terra V. S.; Bossé J. T.; Naegeli A.; Abouelhadid S.; Li Y.; Lin C.-W.; Vohra P.; Tucker A. W.; Rycroft A. N.; Maskell D. J.; Aebi M.; Langford P. R.; Wren B. W. The n-linking glycosylation system from actinobacillus pleuropneumoniae is required for adhesion and has potential use in glycoengineering. Open Biol. 2017, 7 (1), 160212. 10.1098/rsob.160212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.-J.; Grass S.; Paek S.; St. Geme J. W. III; Yeo H.-J. The actinobacillus pleuropneumoniae hmw1c-like glycosyltransferase mediates n-linked glycosylation of the Haemophilus influenzae hmw1 adhesin. PLoS One 2010, 5 (12), e15888. 10.1371/journal.pone.0015888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys T. G.; Aebi M. Engineering protein glycosylation in prokaryotes. Curr. Opin. Syst. Biol. 2017, 5, 23–31. 10.1016/j.coisb.2017.05.016. [DOI] [Google Scholar]
- Keys T. G.; Wetter M.; Hang I.; Rutschmann C.; Russo S.; Mally M.; Steffen M.; Zuppiger M.; Müller F.; Schneider J.; Faridmoayer A.; Lin C.-w.; Aebi M. A biosynthetic route for polysialylating proteins in escherichia coli. Metab. Eng. 2017, 44, 293–301. 10.1016/j.ymben.2017.10.012. [DOI] [PubMed] [Google Scholar]
- Xu Y.; Wu Z.; Zhang P.; Zhu H.; Zhu H.; Song Q.; Wang L.; Wang F.; Wang P. G.; Cheng J. A novel enzymatic method for synthesis of glycopeptides carrying natural eukaryotic n-glycans. Chem. Commun. 2017, 53 (65), 9075–9077. 10.1039/C7CC04362G. [DOI] [PubMed] [Google Scholar]
- Lomino J. V.; Naegeli A.; Orwenyo J.; Amin M. N.; Aebi M.; Wang L.-X. A two-step enzymatic glycosylation of polypeptides with complex n-glycans. Bioorg. Med. Chem. 2013, 21 (8), 2262–2270. 10.1016/j.bmc.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kightlinger W.; Lin L.; Rosztoczy M.; Li W.; DeLisa M. P.; Mrksich M.; Jewett M. C. Design of glycosylation sites by rapid synthesis and analysis of glycosyltransferases. Nat. Chem. Biol. 2018, 14 (6), 627–635. 10.1038/s41589-018-0051-2. [DOI] [PubMed] [Google Scholar]
- Silverman A. D.; Karm A. S.; Jewett M. C.. Nat. Rev. Genet. 2019 10.1038/s41576-019-0186-3. [DOI] [PubMed] [Google Scholar]
- Lombard V.; Golaconda Ramulu H.; Drula E.; Coutinho P. M.; Henrissat B. The carbohydrate-active enzymes database (cazy) in 2013. Nucleic Acids Res. 2014, 42 (D1), D490–495. 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass S.; Lichti C. F.; Townsend R. R.; Gross J.; St. Geme J. W. III The Haemophilus influenzae hmw1c protein is a glycosyltransferase that transfers hexose residues to asparagine sites in the hmw1 adhesin. PLoS Pathog. 2010, 6 (5), e1000919. 10.1371/journal.ppat.1000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q.; Wu Z.; Fan Y.; Song W.; Zhang P.; Wang L.; Wang F.; Xu Y.; Wang P. G.; Cheng J. Production of homogeneous glycoprotein with multisite modifications by an engineered n-glycosyltransferase mutant. J. Biol. Chem. 2017, 292 (21), 8856–8863. 10.1074/jbc.M117.777383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. M.; Glover K. J.; Imperiali B. From peptide to protein: Comparative analysis of the substrate specificity of n-linked glycosylation in c. Jejuni. Biochemistry 2007, 46 (18), 5579–5585. 10.1021/bi602633n. [DOI] [PubMed] [Google Scholar]
- Techner J.-M.; Kightlinger W.; Lin L.; Hershewe J.; Ramesh A.; DeLisa M. P.; Jewett M. C.; Mrksich M. High-Throughput Synthesis and Analysis of Intact Glycoproteins Using SAMDI-MS. Anal. Chem. 2020, 92 (2), 1963–1971. 10.1021/acs.analchem.9b04334. [DOI] [PubMed] [Google Scholar]
- Tytgat H. L. P.; Lin C.-w.; Levasseur M. D.; Tomek M. B.; Rutschmann C.; Mock J.; Liebscher N.; Terasaka N.; Azuma Y.; Wetter M.; Bachmann M. F.; Hilvert D.; Aebi M.; Keys T. G. Cytoplasmic glycoengineering enables biosynthesis of nanoscale glycoprotein assemblies. Nat. Commun. 2019, 10 (1), 5403. 10.1038/s41467-019-13283-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Geel R.; Wijdeven M. A.; Heesbeen R.; Verkade J. M. M.; Wasiel A. A.; van Berkel S. S.; van Delft F. L. Chemoenzymatic conjugation of toxic payloads to the globally conserved n-glycan of native mabs provides homogeneous and highly efficacious antibody–drug conjugates. Bioconjugate Chem. 2015, 26 (11), 2233–2242. 10.1021/acs.bioconjchem.5b00224. [DOI] [PubMed] [Google Scholar]
- Kightlinger W.; Duncker K. E.; Ramesh A.; Thames A. H.; Natarajan A.; Stark J. C.; Yang A.; Lin L.; Mrksich M.; DeLisa M. P.; Jewett M. C. A cell-free biosynthesis platform for modular construction of protein glycosylation pathways. Nat. Commun. 2019, 10 (1), 5404. 10.1038/s41467-019-12024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz F.; Huang W.; Li C.; Schulz B. L.; Lizak C.; Palumbo A.; Numao S.; Neri D.; Aebi M.; Wang L.-X. A combined method for producing homogeneous glycoproteins with eukaryotic n-glycosylation. Nat. Chem. Biol. 2010, 6 (4), 264–266. 10.1038/nchembio.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.; Tong X.; Yang Q.; Giddens J. P.; Wang L.-X. Glycosynthase mutants of endoglycosidase s2 show potent transglycosylation activity and remarkably relaxed substrate specificity for antibody glycosylation remodeling. J. Biol. Chem. 2016, 291 (32), 16508–16518. 10.1074/jbc.M116.738765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L.; Gebhard M. C.; Li Q.; Taft J. M.; Georgiou G.; Iverson B. L. Engineering of tev protease variants by yeast er sequestration screening (yess) of combinatorial libraries. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (18), 7229–7234. 10.1073/pnas.1215994110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill M. E.; MacPherson D. J.; Wu P.; Julien O.; Wells J. A.; Hardy J. A. Reprogramming caspase-7 specificity by regio-specific mutations and selection provides alternate solutions for substrate recognition. ACS Chem. Biol. 2016, 11 (6), 1603–1612. 10.1021/acschembio.5b00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight Z. A.; Garrison J. L.; Chan K.; King D. S.; Shokat K. M. A remodelled protease that cleaves phosphotyrosine substrates. J. Am. Chem. Soc. 2007, 129 (38), 11672–11673. 10.1021/ja073875n. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.