Abstract

We have developed a syringolin-based chemical probe and explored its utility for the profiling of metabolite extracts as potent inhibitors of the 20S proteasome. Activity-guided fractionation by competitive labeling allowed us to isolate and identify glidobactin A and C as well as luminmycin A from a Burkholderiales strain. The natural products exhibited unique subunit specificities for the proteolytic subunits of human and mouse constitutive and immunoproteasome in the lower nanomolar range. In particular, glidobactin C displayed an unprecedented β2/β5 coinhibition profile with single-digit nanomolar potency in combination with sufficiently high cell permeability. These properties render glidobactin C a promising live cell proteasome inhibitor with potent activity against human breast cancer cell lines and comparably low immunotoxicity.
Short abstract
Competitive profiling with a syringolin A probe allows us to find subunit specific proteasome inhibitors “in the haystack” of complex metabolite extracts.
Introduction
The ubiquitin-proteasome system is the central mechanism responsible for the dynamic regulation of protein turnover and the degradation of damaged and misfolded proteins in the eukaryotic cell.1,2 The barrel-shaped 20S core of the constitutive proteasome contains three proteolytic subunits with distinct caspase-like (β1), trypsin-like (β2), and chymotrypsin-like (β5) activities. In response to immune and inflammatory stimuli, these subunits get replaced by their isoforms β1i (LMP2), β2i (MECL-1), and β5i (LMP7) to constitute the 20S immunoproteasome which has importance for antigen processing and major relevance for cancer and immune diseases.3 Subunit specific inhibitors are thus much sought after and a major goal in synthetic approaches of scaffold derivatization and development of chemical probes.4,5 In particular, recent insights into the requirement of coinhibition of β2 and β5 for inhibiting the growth of solid tumors6 and coinhibition of β2i and β5i for the therapy of multiple myeloma7 and autoimmune diseases8 pose challenges for subunit-specific coinhibition of constitutive proteasomes and immunoproteasomes. In addition, the proteasome is an attractive target for the development of anti-malarial and anti-mycobacterial drugs.9,10 A large number of existing proteasome inhibitor scaffolds are based on natural products of microbial origin including syrbactins, epoxyketones, and β-lactones.11 However, it remains challenging to detect activity of natural products directly in crude metabolite extracts with thousands of potentially interfering substances including salts and lipids.
In order to meet this challenge, we have envisioned the application of active site-directed chemical probes for the competitive screening of inhibitors. We have recently demonstrated the power of such an approach for the discovery of synthetic inhibitors of quinolone biosynthesis of the human pathogen Pseudomonas aeruginosa.12,13 Hereby, an active site-directed probe was applied as a tool to profile the inhibition of PqsD, a central enzyme responsible for the biosynthesis of quinolones. Competition experiments with a library of synthetic compounds allowed the detection of those which blocked the probe from accessing and binding to the active site of the enzyme. Since competitive profiling yielded potent inhibitors against PqsD that successfully abrogated quinolone production,12 we now aimed to explore the applicability of this strategy for the discovery of natural products directly from bacterial culture extracts. We hereby aimed to exploit a chemical probe as a tool for detection and activity-guided purification of natural products as potent proteasome inhibitors. As a proof-of-principle, we selected a Burkholderiales strain which is known to possess a biosynthetic gene cluster for syrbactin production14 and report here the application of the competitive profiling strategy with an active site-directed probe for the isolation of the three syrbactins luminmycin A, glidobactin A, and glidobactin C. The full potential of this strategy is highlighted by the discovery of a surprising subunit-specific inhibition profile of glidobactin C.
Results and Discussion
Active Site-Directed Proteasome Probe
We selected syringolin A (SylA),15 a covalently binding Michael-acceptor-type proteasome inhibitor as probe scaffold which is known to target all three proteolytic subunits of the 20S proteasome.16 To this aim, we isolated SylA from an overproducing strain of Pseudomonas syringae pv syringae (Figure 1A and B, Figure S1).15
Figure 1.
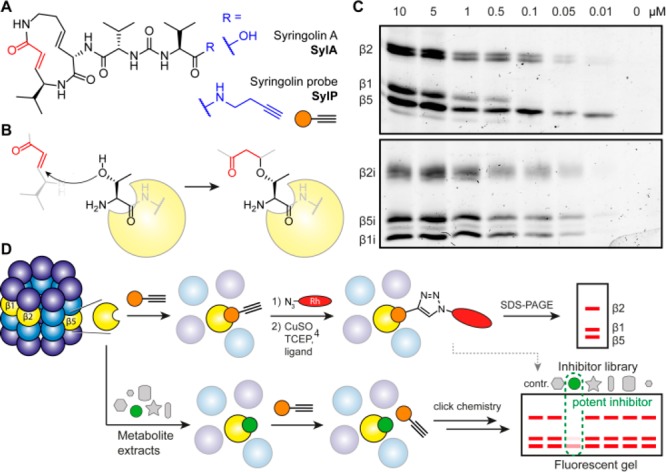
Syringolin and competitive profiling strategy. (A) Structures of Syringolin A (SylA) and the corresponding SylP probe. (B) Covalent Michael-acceptor binding mode of syrbactins on the active site Thr of the proteolytic subunits of the proteasome. (C) Concentration-dependent labeling of constitutive and immunoproteasome. (D) Competitive profiling strategy of natural product extracts for proteasome inhibitors.
Chemical modification of the compound at its free carboxyl group led to a syringolin probe (SylP) comprising a terminal alkyne handle for bioorthogonal modification with a fluorescent reporter tag via the copper(I)-catalyzed alkyne–azide cycloaddition (click chemistry). To test the ability of the SylP probe to label the proteolytic subunits of the proteasome, we incubated a purified constitutive proteasome (CP) and immunoproteasome (IP) of human and mouse origin with titrated dose down of SylP for 1 h and appended a fluorescent tag via click chemistry with a tetramethylrhodamine (TAMRA) azide. SDS-PAGE and fluorescence scanning revealed labeling of β2/β2i, β5/β5i, and β1i down to 50 nM, while β1 was only labeled down to 0.5 μM (Figure 1C). The subunits were identified according to their molecular weights and previous assignments.17−19
Competitive Profiling Assays
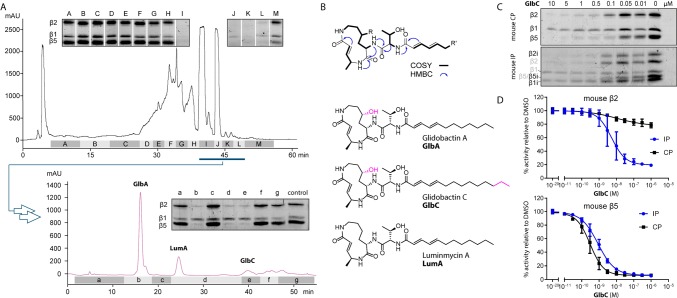
Since labeling of the proteasome was successful, we investigated whether a competitive labeling strategy allowed the detection of the natural product syringolin A from culture supernatants. In short, the purified 20S proteasome was preincubated with the extracts for 1 h followed by treatment with the SylP probe and fluorescent labeling via click chemistry. A potent proteasome inhibitor would block the active sites of the proteolytic subunits and exclude probe binding and consequently abolish fluorescent labeling (Figure 1D). First, we used purified SylA to determine the limit of detection, which was between 5 and 1 μM (Figure S2). Since extracts of spent culture media would be concentrated by evaporating the organic solvent, we reasoned that this would allow the detection of syringolin-like proteasome inhibitors in native production levels in the extracts of 6 L culture scale. Indeed, we could confirm this with crude extracts from our syringolin overproducing strain (Figure S3). We next aimed to apply our competitive labeling strategy for natural product discovery using a wild-type strain (Figure 1D). We selected the Burkholderiales strain DSM 7029 (previously named Polyangium brachysporum) which is known to produce glidobactins via a syrbactin biosynthetic gene cluster (BGC) related to the BGC for syringolin biosynthesis of P. syringae.14 Culture supernatants were extracted with ethyl acetate, and competitive screening of RP-HPLC fractions identified multiple fractions that inhibited probe labeling (Figure 2A, Figures S4 and S5). Preparative scale fractionation in an activity-guided manner allowed purification of the compounds to homogeneity and elucidation of the chemical structures of the active metabolites by NMR spectroscopy in combination with high-resolution mass spectrometry (Figure S6). Three members of the syrbactin family were identified including luminmycin A (LumA),20−22 glidobactin A (GlbA), and glidobactin C (GlbC) (Figure 2B).23 Each of the pure compounds was subjected to a dose-down competitive labeling experiment with the probe SylP against mouse CP and IP (Figure 2C, Figures S7 and S8). For some of the compounds, we hereby noticed a surprising subunit selective competition which was particularly interesting for β2 and β5.
Figure 2.
Competitive profiling and identification of proteasome inhibitors. (A) Fractionation of crude metabolite extracts by preparative HPLC and competitive labeling was performed with mouse CP against 1 μM SylP. Representative fluorescent gel results of the competitive labeling of fractions A–M are displayed. Active fractions (I–L) were further purified by several rounds of semipreparative HPLC runs and activity was assayed in SylP probe competition experiments with mouse CP. Fractions b, d, and e were purified to homogeneity and (B) identified by NMR spectroscopy and mass spectrometry as glidobactin A (GlbA), glidobactin C (GlbC), and luminmycin A (LumA). (C) Dose-down competition experiment of GlbC against 1 μM of probe SylP with mouse CP and IP. (D) Subunit specific inhibition assays with fluorogenic peptide substrates shown as examples for β2 and β5 inhibition by GlbC in mouse CP and IP.
Subunit-Selectivity of Isolated Inhibitors
These selectivities warranted a more detailed investigation of subunit specific inhibition using enzyme assays with fluorogenic peptide substrates. Different syrbactins exhibited interesting selectivities for various subunits of constitutive and immunoproteasome with individual species preferences for proteasomes of mouse or human origin (Figure 2D, Figure S9, and Table 1). Most importantly, however, glidobactins GlbA and GlbC inhibited β2 and β5, as well as the corresponding immunoproteasome subunits β2i and β5i with exceptional potency largely in the single-digit nanomolar range (Table 1 and Figure S9). This β2/β5 coinhibition profile is of particular interest, since it has been shown to suppress Nrf1 activation preventing the recovery of proteasome activity. Dual β2/β5 inhibitors are thus considered a hallmark for the treatment of breast cancer and multiple myeloma.6,7,24 While unique structural features of the subunits β1, β1i, β5, and β5i have allowed the development of selective inhibitors,25−27 discriminating β2 and β2i has remained a challenge due to their high structural similarity.28 Just recently, the first synthetic inhibitors based on epoxyketones with up to 45-fold selectivity for β2i were reported.29 To our surprise, we found that GlbA and GlbC displayed the highest selectivity for mouse β2i with IC50 values of 1.4 and 15 nM, respectively, while β2 of the constitutive proteasome was almost unaffected at concentrations up to 1 μM (β2/β2i > 714, GlbA; β2/β2i > 66, GlbC).
Table 1. IC50 Values of Isolated Compounds in mol/L Were Determined Using the Hydrolysis of Fluorogenic Substrates for the 20S Immunoproteasomes (IP) or Constitutive Proteasomes (CP) of Human or Mouse Origina.
| activity/species | proteasome | SylA | SylP | LumA | GlbA | GlbC |
|---|---|---|---|---|---|---|
| Chymotrypsin-Like Activity (β5/β5i)b | ||||||
| human | CP | (1.8 ± 1.1) × 10–7 | (1.0 ± 0.8) × 10–8 | (3.9 ± 0.2) × 10–8 | (3.6 ± 1.1) × 10–9 | (2.9 ± 2.2) × 10–9 |
| IP | (2.4 ± 1.9) × 10–6 | (3.6 ± 1.8) × 10–8 | (1.6 ± 0.6) × 10–6 | (2.5 ± 1.9) × 10–8 | (7.1 ± 5.3) × 10–9 | |
| mouse | CP | (5.9 ± 1.6) × 10–8 | (2.1 ± 0.0) × 10–9 | (3.2 ± 2.9) × 10–8 | (6.5 ± 0.8) × 10–10 | (2.7 ± 3.4) × 10–9 |
| IP | (1.8 ± 2.0) × 10–7 | (3.2 ± 1.9) × 10–9 | (8.9 ± 0.6) × 10–9 | (1.1 ± 0.1) × 10–9 | (1.3 ± 0.7) × 10–9 | |
| Trypsin-like activity (VGR) (β2/β2i)b | ||||||
| human | CP | n.i. | (1.4 ± 1.8) × 10–7 | (2.6 ± 0.8) × 10–7 | (1.5 ± 1.5) × 10–8 | (2.4 ± 2.8) × 10–8 |
| IP | (6.7 ± 7.5) × 10–7 | (1.6 ± 0.2) × 10–8 | (1.7 ± 1.6) × 10–7 | (4.2 ± 0.8) × 10–8 | (2.5 ± 2.0) × 10–9 | |
| mouse | CP | n.i. | n.i. | n.i. | n.i. | n.i. |
| IP | n.i | (1.1 ± 1.4) × 10–7 | n.i. | (1.4 ± 1.6) × 10–9 | (1.5 ± 1.3) × 10–8 | |
| Caspase-Like Activity (β1)b | ||||||
| human | CP | n.i. | n.i. | n.i. | n.i. | n.i. |
| mouse | CP | (5.0 ± 7.0) × 10–5 | (2.1 ± 0.5) × 10–7 | n.i. | n.i. | n.i. |
| LMP2 Activity (β1i)b | ||||||
| human | IP | (7.6 ± 0.0) × 10–7 | (1.7 ± 0.6) × 10–7 | n.i. | n.i. | n.i. |
| mouse | IP | (2.8 ± 1.0) × 10–8 | (2.2 ± 1.5) × 10–8 | n.i. | (2.5 ± 0.8) × 10–7 | n.i. |
IC50 values ± SD of two series of four independent experiments are indicated.
Chymotrypsin-like (Suc-LLVY-AMC), trypsin-like (Bz-VGR-AMC), caspase-like (Z-LLE-βNA), or LMP2 (Ac-PAL-AMC) activity. n.i.: inhibition below 50% at 1 μM.
This selectivity in combination with the high potency is unprecedented and even more surprising to be found for microbial natural products. In contrast to the mouse proteasome, no such selectivity was found between human β2 and β2i, indicating distinct substrate binding affinities of these proteasome subunits in mouse and human. Interestingly, LumA, which corresponds to GlbA lacking a hydroxyl group at the ring, poorly inhibited β2 and β2i of human and mouse at orders of magnitude higher concentrations.
Cell Permeability
Finally, we were interested in the cell permeability of the different syrbactins which we investigated in live LCL721.145 cells using the cell-permeable substrate MeO-Suc-GLF-AMC to quantify the chymotrypsin-like activity. Hereby, SylA failed to inhibit chymotrypsin-like activity up to 1 μM, indicating poor cell permeability. Surprisingly, the more hydrophobic LumA and GlbA also remained inactive in live cell experiments.
However, GlbC which differs from GlbA only by encompassing two additional methylene groups in its acyl-chain considerably inhibited substrate cleavage in live cells, suggesting that its slightly increased chain length sufficiently enhanced membrane penetration (Figure S10).
Activity in Cell-Based Assays
Consequently, we further explored the activity of GlbC in live cells. Mouse fibrosarcoma MC57 cells treated for 6 h with GlbC accumulated ubiquitin conjugates in concentration dependence with even slightly better efficiency as compared to the synthetic aldehyde proteasome inhibitor MG132 (Figure 3A and B). In addition, the pro-inflammatory cytokine response upon lipopolysaccharide (LPS) stimulation of mouse splenocytes was inhibited by GlbC treatment (Figure 3C). These results indicate potent inhibition of CP and IP in live cells. Finally, we also investigated the anticancer potential of GlbC in the human breast cancer cell lines BT-549, MDA-MB-231, and MDA-MB-361 for which β2/β5 coinhibition is particularly required. Indeed, 0.25 μM of GlbC blocked proliferation, and higher concentrations led to cancer cell death (Figure 3D, Figure S11). In order to test whether GlbC alters T cell proliferation differently in immunoproteasome-proficient wild-type T cells and immunoproteasome-deficient MECL-1–/–/LMP7–/– T cells, cell division of CD4+ T cells was analyzed after T cell receptor (TCR) stimulation (Figure S12). GlbC inhibited T cell proliferation in a dose-dependent manner. However, no difference between immunoproteasome-proficient and -deficient T cells could be observed. Although GlbC showed selectivity for mouse β2i (Table 1), this result is not unexpected since β2i inhibition by GlbC is less potent (IC50 = (1.5 ± 1.3) × 10–8) and since it has been shown that inhibition of LMP7 together with LMP2 or MECL-1 is important to alter T cell differentiation and cytokine secretion.8,30 However, similar selectivity of GlbC for LMP7 and β5 as well as for LMP2 and β1 was observed in this study (Table 1). Nevertheless, it is remarkable that the proliferation inhibition of CD4+ T helper cells by GlbC is much lower than the inhibition by bortezomib, although the proliferation inhibition of the breast carcinoma line MDA-MB-231 by GlbC (Figure S11) is similar to that of bortezomib.6 This result suggests that the β2/β5 selectivity of GlbC may enable a strong anticancer effect accompanied by less immunotoxicity than bortezomib.
Figure 3.
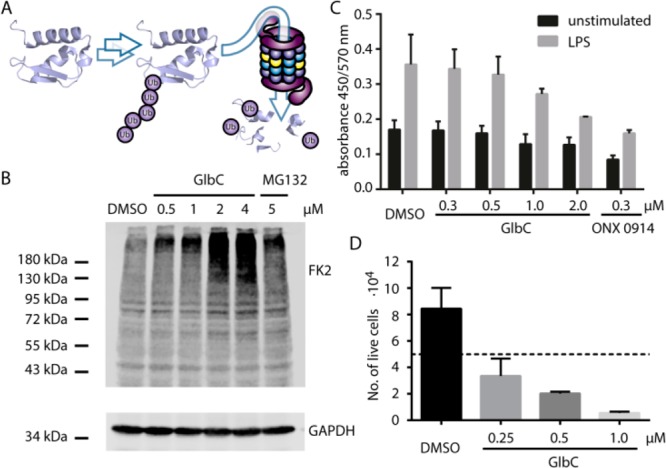
Live cell activity assays with GlbC. (A) The proteasome degrades ubiquitinated proteins. (B) These ubiquitin–protein conjugates accumulate upon GlbC treatment as visualized by Western blot analysis with mAb FK2. Loading control: GADPH; positive control: proteasome inhibitor MG132. (C) ELISA analysis of IL-6 in the culture supernatant of C57BL/6 mouse splenocytes treated with GlbC. Positive control: β1i/β5i specific inhibitor ONX 0914. (D) Cell count after 2-day treatment of breast cancer cell line BT-549 with GlbC. The dotted line corresponds to the number of cells seeded on day 0.
These results demonstrate the strength of competitive profiling for the discovery of proteasome inhibitors and activity-guided fractionation of crude metabolite extracts. The value of direct assays to discover proteasome inhibitors from complex microbial metabolite mixtures has also been shown by an NMR-based technique using a 13C-labeled substrate that led to the isolation of the syrbactin cepafungin I.31 Although the family of syrbactins has been known for decades,20,23,32 our understanding of their specificity for subunits of the 20S proteasome has so far been incomplete.33 A syringolin-based probe directly linked with fluorophore has been useful for 20S proteasome labeling in leukemia cell lines,18 as well as for characterizing the subunit-selectivity of SylA and for noninvasive imaging in live cells of Arabidopsis.16 A synthetic SylA-GlbA hybrid has been reported which inhibited all three subunits of the proteasome of tumor cell lines.34,35 This chimera consisted of a SylA core combined with the tail of GlbA. Our results show that the ring hydroxyl group of the GlbA/C core is important for efficacy in β2/β2i inhibition, and efficacy is reduced when the hydroxyl group is missing (LumA). In contrast, the SylA core exhibits increased β1/β1i binding that can be partially modulated by the hydrophobicity of the tail (SylP). Thus, the selectivity of GlbA/C has so far been missed and we present here the first account demonstrating that the inhibition profile and efficacy of GlbA and GlbC are markedly different from those of SylA and LumA.
Conclusions
In summary, glidobactin C exhibits a highly interesting β2/β5 subunit coinhibition profile that in combination with its cell permeability makes glidobactin C a promising anticancer drug candidate and a tool for subunit-specific proteasome inhibition in live cells. So far, it has been shown that the combination of β5/β5i inhibitors with a β2/β2i inhibitor inhibits the growth of cancer cell lines of breast, lung, kidney, and ovarian carcinoma,6 but to the best of our knowledge, an inhibitor like GlbC that inhibits the active centers of both human β5/β5i and β2/β2i subunits at single-digit nanomolar potency has not been reported to date and holds great potential for cancer therapy. Hence, it will be pertinent to test the effect of GlbC on the growth of cancer cell lines derived from other solid tumors of humans. These results demonstrate that competitive profiling using active site-directed chemical probes is a suitable strategy to identify and isolate proteasome inhibitors from complex microbial metabolite mixtures. Furthermore, minor structural differences in native syrbactins have a major impact on subunit and species selectivity of these proteasome inhibitors. A major advantage of the SylP probe developed herein is that, compared to the natural proteasome inhibitor SylA, it is only minimally extended by introduction of the alkyne moiety affording facilitated entry into the proteolytic central chamber of the 20S proteasome. Nevertheless, bulky fluorescent or other labels can subsequently be attached via click reaction after the proteasome subunits have been disassembled by denaturation. Our results indicate that the syrbactin core largely determines subunit selectivity, while the tail modulates efficacy and cell permeability. These results indicate that structural diversification of syrbactins within a single producer species may be driven by functional specialization. This again may represent a strategy for evolutionary adaptation to complex ecological interactions with multiple different eukaryotic hosts, competitors, or predators.
Acknowledgments
We thank Prof. Dr. Andreas Marx and his group for generous support. We gratefully acknowledge funding by SFB969 project C06, the Emmy Noether program of the Deutsche Forschungsgemeinschaft (DFG), EU FP7Marie Curie Zukunftskolleg Incoming Fellowship Program, Fonds der Chemischen Industrie (FCI), and Konstanz Research School Chemical Biology (KoRS-CB). We thank Prof. Dr. Robert Dudler and Zsuzsanna Hasenkamp for kindly providing P. syringae pv. syringae B301D-R with the pOEAC plasmid construct. Flow cytometry experiments were performed at the flow cytometry facility FlowKon of the University of Konstanz.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.9b01170.
Additional data, methods, and supplementary figures including probe synthesis, fluorescent gels of proteasome labeling experiments, competitive profiling, isolation methods, characterization of isolated natural products, NMR spectra, proteasome inhibition assays, Western blot analysis of ubiquitin accumulation, ELISA assay of cytokine response upon lipopolysaccharide (LPS), cancer cell viability assays, and T-cell proliferation assays (PDF)
Author Contributions
# A.P. and M.B. contributed equally to this work. T.B. and M.G. conceived of the project. A.P. designed and generated the probe, conducted protein profiling experiments and isolated and characterized syrbactins. M.B. performed the detailed biochemical investigation of proteasome inhibitors. G.O.A.S. purified proteasomes from mouse and human. H.G. conducted proteasome inhibition assays. A.P., M.B., M.G., and T.B. wrote the manuscript. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Collins G. A.; Goldberg A. L. The Logic of the 26S Proteasome. Cell 2017, 169 (5), 792–806. 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O.; Felberbaum R.; Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006, 22, 159–80. 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Murata S.; Takahama Y.; Kasahara M.; Tanaka K. The immunoproteasome and thymoproteasome: functions, evolution and human disease. Nat. Immunol. 2018, 19 (9), 923–931. 10.1038/s41590-018-0186-z. [DOI] [PubMed] [Google Scholar]
- Hewings D. S.; Flygare J. A.; Wertz I. E.; Bogyo M. Activity-based probes for the multicatalytic proteasome. FEBS J. 2017, 284 (10), 1540–1554. 10.1111/febs.14016. [DOI] [PubMed] [Google Scholar]
- Gu C.; Kolodziejek I.; Misas-Villamil J.; Shindo T.; Colby T.; Verdoes M.; Richau K. H.; Schmidt J.; Overkleeft H. S.; van der Hoorn R. A. Proteasome activity profiling: a simple, robust and versatile method revealing subunit-selective inhibitors and cytoplasmic, defense-induced proteasome activities. Plant J. 2010, 62 (1), 160–70. 10.1111/j.1365-313X.2009.04122.x. [DOI] [PubMed] [Google Scholar]
- Weyburne E. S.; Wilkins O. M.; Sha Z.; Williams D. A.; Pletnev A. A.; de Bruin G.; Overkleeft H. S.; Goldberg A. L.; Cole M. D.; Kisselev A. F. Inhibition of the Proteasome beta2 Site Sensitizes Triple-Negative Breast Cancer Cells to beta5 Inhibitors and Suppresses Nrf1 Activation. Cell chemical biology 2017, 24 (2), 218–230. 10.1016/j.chembiol.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey-Kopyscinski S.; Daily E. W.; Gautier M.; Bhatt A.; Florea B. I.; Mitsiades C. S.; Richardson P. G.; Driessen C.; Overkleeft H. S.; Kisselev A. F. An inhibitor of proteasome beta2 sites sensitizes myeloma cells to immunoproteasome inhibitors. Blood Adv. 2018, 2 (19), 2443–2451. 10.1182/bloodadvances.2018016360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H. W. B.; Lowe E.; Anderl J. L.; Fan A.; Muchamuel T.; Bowers S.; Moebius D. C.; Kirk C.; McMinn D. L. Required Immunoproteasome Subunit Inhibition Profile for Anti-Inflammatory Efficacy and Clinical Candidate KZR-616 ((2 S,3 R)- N-((S)-3-(Cyclopent-1-en-1-yl)-1-((R)-2-methyloxiran-2-yl)-1-oxopropan-2-yl)-3-hydroxy-3-(4-methoxyphenyl)-2-((S)-2-(2-morpholinoacetamido)propanamido)propenamide). J. Med. Chem. 2018, 61 (24), 11127–11143. 10.1021/acs.jmedchem.8b01201. [DOI] [PubMed] [Google Scholar]
- Darwin K. H.; Ehrt S.; Gutierrez-Ramos J. C.; Weich N.; Nathan C. F. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 2003, 302 (5652), 1963–6. 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- Li H.; O’Donoghue A. J.; van der Linden W. A.; Xie S. C.; Yoo E.; Foe I. T.; Tilley L.; Craik C. S.; da Fonseca P. C.; Bogyo M. Structure- and function-based design of Plasmodium-selective proteasome inhibitors. Nature 2016, 530 (7589), 233–6. 10.1038/nature16936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev A. F.; van der Linden W. A.; Overkleeft H. S. Proteasome inhibitors: an expanding army attacking a unique target. Chem. Biol. 2012, 19 (1), 99–115. 10.1016/j.chembiol.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prothiwa M.; Englmaier F.; Böttcher T. Competitive Live-Cell Profiling Strategy for Discovering Inhibitors of the Quinolone Biosynthesis of Pseudomonas aeruginosa. J. Am. Chem. Soc. 2018, 140 (43), 14019–14023. 10.1021/jacs.8b07629. [DOI] [PubMed] [Google Scholar]
- Prothiwa M.; Szamosvari D.; Glasmacher S.; Böttcher T. Chemical probes for competitive profiling of the quorum sensing signal synthase PqsD of Pseudomonas aeruginosa. Beilstein J. Org. Chem. 2016, 12, 2784–2792. 10.3762/bjoc.12.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg B.; Bigler L.; Dudler R. Identification of genes involved in the biosynthesis of the cytotoxic compound glidobactin from a soil bacterium. Environ. Microbiol. 2007, 9 (7), 1640–50. 10.1111/j.1462-2920.2007.01278.x. [DOI] [PubMed] [Google Scholar]
- Groll M.; Schellenberg B.; Bachmann A. S.; Archer C. R.; Huber R.; Powell T. K.; Lindow S.; Kaiser M.; Dudler R. A plant pathogen virulence factor inhibits the eukaryotic proteasome by a novel mechanism. Nature 2008, 452 (7188), 755–8. 10.1038/nature06782. [DOI] [PubMed] [Google Scholar]
- Kolodziejek I.; Misas-Villamil J. C.; Kaschani F.; Clerc J.; Gu C.; Krahn D.; Niessen S.; Verdoes M.; Willems L. I.; Overkleeft H. S.; Kaiser M.; van der Hoorn R. A. Proteasome activity imaging and profiling characterizes bacterial effector syringolin A. Plant Physiol. 2011, 155 (1), 477–89. 10.1104/pp.110.163733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidtke G.; Schregle R.; Alvarez G.; Huber E. M.; Groettrup M. The 20S immunoproteasome and constitutive proteasome bind with the same affinity to PA28alphabeta and equally degrade FAT10. Mol. Immunol. 2019, 113, 22–30. 10.1016/j.molimm.2017.11.030. [DOI] [PubMed] [Google Scholar]
- Clerc J.; Florea B. I.; Kraus M.; Groll M.; Huber R.; Bachmann A. S.; Dudler R.; Driessen C.; Overkleeft H. S.; Kaiser M. Syringolin A selectively labels the 20 S proteasome in murine EL4 and wild-type and bortezomib-adapted leukaemic cell lines. ChemBioChem 2009, 10 (16), 2638–43. 10.1002/cbic.200900411. [DOI] [PubMed] [Google Scholar]
- Groettrup M.; Standera S.; Stohwasser R.; Kloetzel P. M. The subunits MECL-1 and LMP2 are mutually required for incorporation into the 20S proteasome. Proc. Natl. Acad. Sci. U. S. A. 1997, 94 (17), 8970–5. 10.1073/pnas.94.17.8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian X.; Plaza A.; Zhang Y.; Müller R. Luminmycins A-C, cryptic natural products from Photorhabdus luminescens identified by heterologous expression in Escherichia coli. J. Nat. Prod. 2012, 75 (9), 1652–5. 10.1021/np300444e. [DOI] [PubMed] [Google Scholar]
- Bian X.; Huang F.; Wang H.; Klefisch T.; Müller R.; Zhang Y. Heterologous production of glidobactins/luminmycins in Escherichia coli Nissle containing the glidobactin biosynthetic gene cluster from Burkholderia DSM7029. ChemBioChem 2014, 15 (15), 2221–4. 10.1002/cbic.201402199. [DOI] [PubMed] [Google Scholar]
- Fu J.; Bian X.; Hu S.; Wang H.; Huang F.; Seibert P. M.; Plaza A.; Xia L.; Müller R.; Stewart A. F.; Zhang Y. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat. Biotechnol. 2012, 30 (5), 440–6. 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- Oka M.; Nishiyama Y.; Ohta S.; Kamei H.; Konishi M.; Miyaki T.; Oki T.; Kawaguchi H. Glidobactins A, B and C, new antitumor antibiotics. I. Production, isolation, chemical properties and biological activity. J. Antibiot. 1988, 41 (10), 1331–7. 10.7164/antibiotics.41.1331. [DOI] [PubMed] [Google Scholar]
- Besse A.; Besse L.; Kraus M.; Mendez-Lopez M.; Bader J.; Xin B. T.; de Bruin G.; Maurits E.; Overkleeft H. S.; Driessen C. Proteasome Inhibition in Multiple Myeloma: Head-to-Head Comparison of Currently Available Proteasome Inhibitors. Cell chemical biology 2019, 26 (3), 340–351. 10.1016/j.chembiol.2018.11.007. [DOI] [PubMed] [Google Scholar]
- Huber E. M.; de Bruin G.; Heinemeyer W.; Paniagua Soriano G.; Overkleeft H. S.; Groll M. Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors. J. Am. Chem. Soc. 2015, 137 (24), 7835–42. 10.1021/jacs.5b03688. [DOI] [PubMed] [Google Scholar]
- de Bruin G.; Huber E. M.; Xin B. T.; van Rooden E. J.; Al-Ayed K.; Kim K. B.; Kisselev A. F.; Driessen C.; van der Stelt M.; van der Marel G. A.; Groll M.; Overkleeft H. S. Structure-based design of beta1i or beta5i specific inhibitors of human immunoproteasomes. J. Med. Chem. 2014, 57 (14), 6197–209. 10.1021/jm500716s. [DOI] [PubMed] [Google Scholar]
- Xin B. T.; de Bruin G.; Huber E. M.; Besse A.; Florea B. I.; Filippov D. V.; van der Marel G. A.; Kisselev A. F.; van der Stelt M.; Driessen C.; Groll M.; Overkleeft H. S. Structure-Based Design of beta5c Selective Inhibitors of Human Constitutive Proteasomes. J. Med. Chem. 2016, 59 (15), 7177–87. 10.1021/acs.jmedchem.6b00705. [DOI] [PubMed] [Google Scholar]
- Huber E. M.; Basler M.; Schwab R.; Heinemeyer W.; Kirk C. J.; Groettrup M.; Groll M. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell 2012, 148 (4), 727–38. 10.1016/j.cell.2011.12.030. [DOI] [PubMed] [Google Scholar]
- Xin B. T.; Huber E. M.; de Bruin G.; Heinemeyer W.; Maurits E.; Espinal C.; Du Y.; Janssens M.; Weyburne E. S.; Kisselev A. F.; Florea B. I.; Driessen C.; van der Marel G. A.; Groll M.; Overkleeft H. S. Structure-Based Design of Inhibitors Selective for Human Proteasome beta2c or beta2i Subunits. J. Med. Chem. 2019, 62 (3), 1626–1642. 10.1021/acs.jmedchem.8b01884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M.; Lindstrom M. M.; LaStant J. J.; Bradshaw J. M.; Owens T. D.; Schmidt C.; Maurits E.; Tsu C.; Overkleeft H. S.; Kirk C. J.; Langrish C. L.; Groettrup M. Co-inhibition of immunoproteasome subunits LMP2 and LMP7 is required to block autoimmunity. EMBO Rep. 2018, 19 (12), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M. L.; Beck P.; Kaiser M.; Dudler R.; Becker C. F.; Groll M. One-shot NMR analysis of microbial secretions identifies highly potent proteasome inhibitor. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (45), 18367–71. 10.1073/pnas.1211423109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wäspi U.; Blanc D.; Winkler T.; Rüedi P.; Dudler R. Syringolin, a novel peptide elicitor from Pseudomonas syringae pv. syringae that induces resistance to Pyricularia oryzae in rice. Mol. Plant-Microbe Interact. 1998, 11 (8), 727–733. 10.1094/MPMI.1998.11.8.727. [DOI] [Google Scholar]
- Krahn D.; Ottmann C.; Kaiser M. The chemistry and biology of syringolins, glidobactins and cepafungins (syrbactins). Nat. Prod. Rep. 2011, 28 (11), 1854–67. 10.1039/c1np00048a. [DOI] [PubMed] [Google Scholar]
- Archer C. R.; Groll M.; Stein M. L.; Schellenberg B.; Clerc J.; Kaiser M.; Kondratyuk T. P.; Pezzuto J. M.; Dudler R.; Bachmann A. S. Activity enhancement of the synthetic syrbactin proteasome inhibitor hybrid and biological evaluation in tumor cells. Biochemistry 2012, 51 (34), 6880–8. 10.1021/bi300841r. [DOI] [PubMed] [Google Scholar]
- Clerc J.; Li N.; Krahn D.; Groll M.; Bachmann A. S.; Florea B. I.; Overkleeft H. S.; Kaiser M. The natural product hybrid of Syringolin A and Glidobactin A synergizes proteasome inhibition potency with subsite selectivity. Chem. Commun. 2011, 47 (1), 385–7. 10.1039/C0CC02238A. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.