Abstract
Escherichia coli is a common inhabitant of the human microbiota and a beacon model organism in biology. However, an understanding of its signaling systems that regulate population-level phenotypes known as quorum sensing remain incomplete. Here, we define the structure and biosynthesis of autoinducer-3 (AI-3), a metabolite of previously unknown structure involved in the pathogenesis of enterohemorrhagic E. coli (EHEC). We demonstrate that novel AI-3 analogs are derived from threonine dehydrogenase (Tdh) products and “abortive” tRNA synthetase reactions, and they are distributed across a variety of Gram-negative and Gram-positive bacterial pathogens. In addition to regulating virulence genes in EHEC, we show that the metabolites exert diverse immunological effects on primary human tissues. The discovery of AI-3 metabolites and their biochemical origins now provides a molecular foundation for investigating the diverse biological roles of these elusive yet widely distributed bacterial signaling molecules.
Short abstract
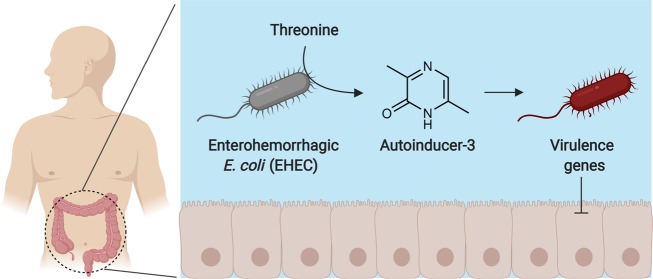
Structure and biosynthesis of autoinducer-3 (AI-3), a virulence regulator in enterohemorrhagic E. coli (EHEC).
Introduction
Natural variants of Escherichia coli are among the first to colonize the human intestinal tract after birth and are estimated to reside in about 90% of the population.1 Pathogenic strains fall within eight known pathotypes2 and cause a variety of severe infections, such as meningitis, hemorrhagic colitis, pneumonia, urinary tract infections, hemolytic uremic syndrome, and others with greater than 200 000 infections being reported annually in the United States alone.3 Additionally, some strains are oncogenic risk factors and are thought to significantly contribute to the initiation of colorectal cancer.4−7 Despite the bacterium’s biomedical importance, a molecular understanding of the signaling systems that E. coli use to regulate virulence and quorum sensing remain undefined. In quorum sensing, bacteria produce small molecule autoinducers that accumulate during growth, facilitating concentration-dependent signaling as a function of population density.8−10E. coli has three major quorum sensing systems,8−10 but extensive genetic efforts over the last two decades have failed to identify the biosynthetic pathway and chemical structure of autoinducer-3 (AI-3). Early efforts defined the QseBC two-component signal transduction pathway of AI-311,12 and established that the QseC receptor responds to the host-derived adrenergic signals epinephrine and norepinephrine, supporting an interkingdom sensing mechanism.13 In the enterohemorrhagic E. coli (EHEC) pathotype, stimulation of this pathway upregulates the locus of enterocyte effacement (LEE), which encodes a type III secretion system (T3SS) and effectors responsible for hemorrhagic colitis and the formation of attaching and effacing lesions in the intestinal tract.14 However, in the absence of accessible small molecule signals, elucidation of AI-3 quorum sensing at the molecular level across E. coli strains has been intractable. Through the use of cellular stress-induced metabolite stimulation techniques,15,16 here we were able to isolate sufficient quantities of select AI-3 analogs, which facilitated the broader structural, biochemical, and functional characterization of this chemical signaling family.
Results
Structural Characterization of AI-3 Analogs
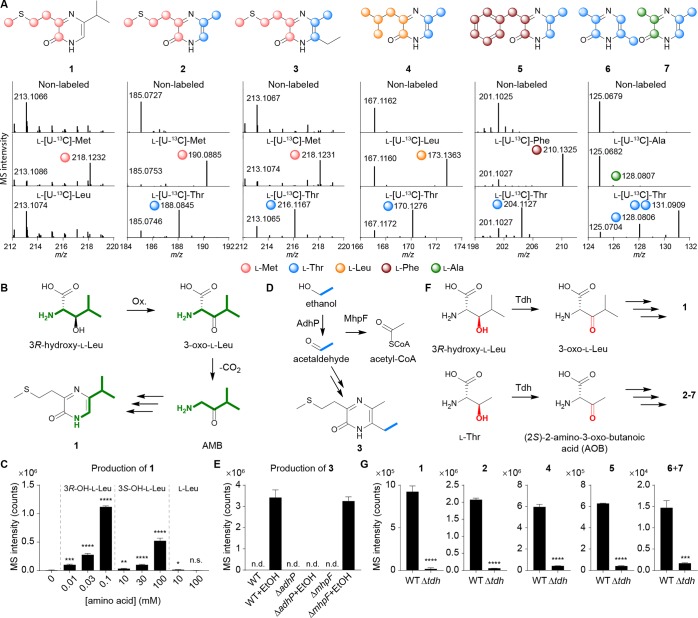
As a representative strain, we first treated the human probiotic E. coli Nissle 1917 with sublethal levels of a representative ribosome inhibitor, erythromycin (Figure 1A,B), and noticed the dose-dependent stimulation of a metabolite with a protonated m/z of 213.1066 (Figure 1C, structure 1, proposed formula C10H17N2OS+, calc 213.1056) by high-resolution liquid chromatography–mass spectrometry (LC-MS). While the core molecular mechanisms remain undefined, increasing evidence suggests that cellular stress responses are integrated with quorum sensing,17−21 and preliminary isolation efforts of AI-3 had identified a signal with a protonated mass of 213.1.12 Given the stress rationale and spectral similarities, we isolated this molecule (1) from an 18 L culture of E. coli Nissle 1917 under drug stress and established its structure using one- and two-dimensional NMR (1H, COSY, HSQC, and HMBC, see Figure 1C) and chemical synthesis (Supporting Information). The new metabolite belonged to the pyrazinone class of natural products (keto-form favored versus enol-form, Figures 1D and S1) that can inhibit human proteases22−24 and trigger inflammatory responses.25 During the course of our efforts, we also characterized a family of structurally related metabolites with protonated masses of 185.0727 (2, C8H13N2OS+, calc 185.0743), 213.1067 (3, C10H17N2OS+, calc 213.1056), 167.1162 (4, C9H15NO+, calc 167.1179), 201.1025 (5, C12H13N2O+, calc 201.1022), and 125.0679 (6, C6H9N2O+, calc 125.0709). Their structures were similarly established via multidimensional NMR (isolation) or synthesis or both (Figure S2 and Supporting Information), leading to the collective identification of six new metabolites (1–6, Figure 1E). We noticed that the novel metabolite 6 coeluted with a trace amount (2.5% relative, Figures S2C and S3) of 3,5-dimethylpyrazine-2-ol (DPO; keto-form 7 is shown), which was recently proposed to be an autoinducer in Vibrio cholerae quorum sensing and a bacteriophage lytic signal.26,27 We also identified these metabolites (1, 2, and 4–7) in other selected pathogenic and commensal model E. coli strains, including enterohemorrhagic E. coli (EHEC) O157:H7, adherent invasive E. coli (AIEC) LF82, E. coli MG1655, and E. coli BW25113 (Figure S4).
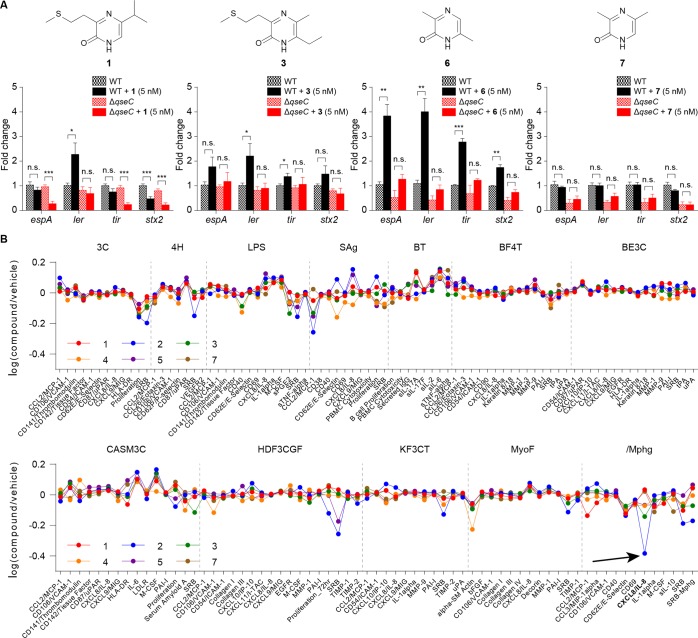
Figure 1.
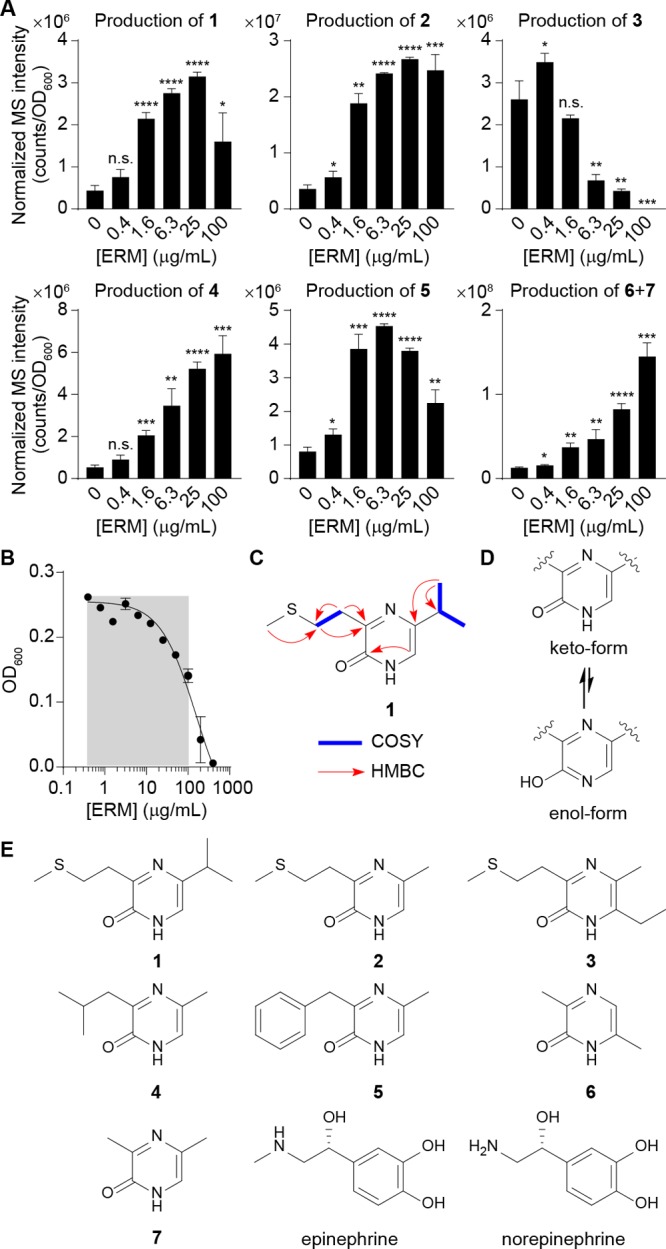
Characterization of pyrazinone metabolites differentially regulated by ribosomal inhibitory stress in E. coli. (A) Dose-dependent production of 1–7 in E. coli Nissle 1917 by erythromycin, as established by LC-MS. Compounds 6 and 7 are indistinguishable by the standard LC-MS method. ERM, erythromycin. (B) Half-maximal inhibitory concentration (IC50) analysis of E. coli Nissle 1917 in response to erythromycin. Gray area represents sublethal antibiotic range examined for metabolite production. (C) Key NMR correlations for representative pyrazinone 1. NMR spectra and analysis for all metabolites can be found in the Supporting Information. COSY, blue bold. HMBC, red arrows. (D) Keto-form is favored versus enol-form. (E) Structures of characterized bacterial metabolites (1–7) and human-derived epinephrine and norepinephrine. Revised pyrazinone tautomeric structure of DPO (7) is shown. n = 3 biological replicates. Data are mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Two-tailed t-test. ns, not significant.
Identification of Biosynthetic Origins of AI-3 Analogs
To establish substrate origins for the new pyrazinone family, we individually supplemented E. coli Nissle 1917 cultures with universally 13C-labeled amino acids l-Met, l-Leu, l-Thr, l-Phe, and l-Ala (10 mM) and analyzed their metabolite extracts by LC-MS. Expected 13C-mass shifts were observed for all metabolites except 1, establishing their amino acid-derived origins (Figure 2A). While we initially anticipated that 1 would be derived from l-Leu, we only observed dose-dependent enhancement of 1 when E. coli Nissle 1917 was cultivated with increasing concentrations of 3R-hydroxy-l-Leu (Figure 2B,C). l-Leu supplementation did not enhance the production of 1, and epimeric 3S-hydroxy-l-Leu exhibited an ∼2000-fold lower enhancement relative to substrate 3R-hydroxy-l-Leu (Figure 2C). These data are consistent with the lack of l-[U-13C]-Leu labeling of 1 (Figure 2A). In the proposed DPO (7) biosynthesis, l-Ala is condensed with aminoacetone (AA), which is derived from l-Thr via threonine dehydrogenase (Tdh) oxidation and subsequent decarboxylation.26l-Thr-derived 13C-labeling of 2–7 was consistent with this proposal; however, 6 was derived from two substrate units instead of one. Based on these studies, we reasoned that analogous dehydrogenation of 3R-hydroxy-l-Leu and decarboxylation to generate 1-amino-3-methylbutan-2-one (AMB) would account for the production of 1 (Figure 2B). Indeed, we observed an ∼1500-fold increase in the production of 1 in AMB-treated (1 mM) compared to untreated culture samples (Figure S5). Pyrazinone 3 production required ethanol culture supplementation, which was used as a solvent vehicle in the drug-stress studies, suggesting that its ethyl substituent is derived from ethanol (Figure 2D). We propose that acetaldehyde is the specific substrate, as alcohol dehydrogenase (AdhP), which oxidizes ethanol to acetaldehyde (Figure 2D), was required for the production of 3 in E. coli BW25113 (Figure 2E). Additionally, acetaldehyde dehydrogenase (MhpF), which converts acetaldehyde to acetyl-CoA, was not required, supporting the intermediary reactive carbonyl species (Figure 2D,E). Finally, deletion of threonine dehydrogenase tdh impaired the production of all the pyrazinones, suggesting that Tdh initiates the pathway from l-Thr or noncanonical 3R-hydroxy-l-Leu (Figure 2F,G).
Figure 2.
Identification of pyrazinone biosynthetic origins. (A) Proposed 13C-isotopic labeling patterns of 1–7 and their HRMS spectra from organic extracts of E. coli Nissle 1917 individually cultivated with universal 13C-labeled amino acids. Co-eluting 6 and 7 could be distinguished using this labeling strategy. (B, C) Individual 3R- and 3S-hydroxy-l-Leu and l-Leu feeding experiments support 3R-hydroxy-l-Leu as a dominant precursor of 1. (D, E) Acetaldehyde derived from ethanol by AdhP represents the observed origin for the ethyl substituent of 3. WT, wild type. (F, G) Comparison of the metabolite production levels between WT E. coli BW25113 and its tdh mutant strain. Tdh was found to dehydrogenate noncanonical 3R-hydroxy-l-Leu in addition to l-Thr in these studies. n = 3 biological replicates. Data are mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Two-tailed t-test. ns, not significant; nd, not detected.
Reconstitution of AI-3 Analog Production in Vitro
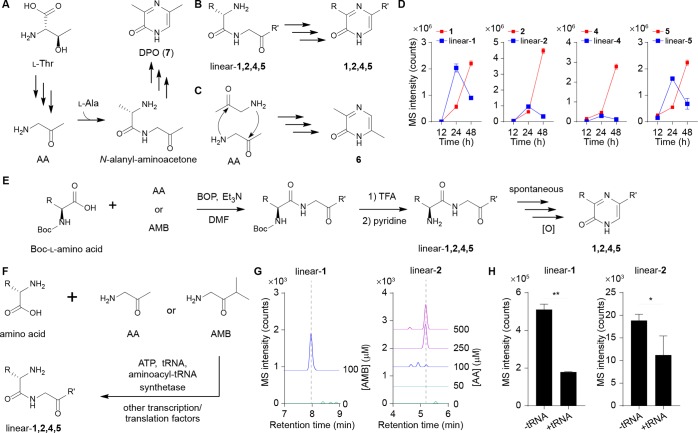
Dipeptide aldehydes have been characterized as intermediates in pyrazinone biosynthesis,24 and for DPO, a dipeptide ketone intermediate, N-alanyl-aminoacetone, was proposed (Figure 3A).26 We propose that the new pyrazinones (1, 2, 4, and 5) would be generated from analogous dipeptide ketones (Figure 3B). However, the formation of 6 from two units of l-Thr requires a different mechanism, invoking head-to-tail condensation of AA, oxidation, and tautomerization (Figure 3C and see below). Indeed, we only detected the linear dipeptide precursors for 1, 2, 4, and 5 (hereafter, linear-1, -2, -4, and -5) in E. coli Nissle 1917 extracts by LC-MS (Figure 3B,D). Their structures were confirmed by universal 13C-isotopic amino acid labeling and tandem MS analysis (Figure S6). Production of linear-1, -2, -4, and -5 increased over 12–24 h, and as expected, they then decreased with an inverse relationship to pyrazinones 1, 2, 4, and 5 in stationary phase (24–48 h), supporting their intermediacy (Figure 3D). Through biomimetic synthetic studies of linear-1, -2, -4, and -5, we found that they underwent spontaneous cyclization, dehydration, tautomerization, and oxidation to produce 1, 2, 4, and 5 on a similar multihour time scale as in cultures (Figure 3E), supporting a spontaneous chemical process that proceeds from the linear precursors.
Figure 3.
Formation of dipeptide ketone intermediates. (A) Proposed DPO (7) pathway. (B) Pyrazinones 1, 2, 4, and 5 would be generated from their corresponding dipeptide ketone precursors. (C) Proposed pathway of 6 from two molecules of AA. See Figure 4D for full pathway. (D) Time-course analysis for production of metabolites 1, 2, 4, and 5, and their linear precursors in E. coli Nissle 1917. n = 3 biological replicates. Data are mean ± SD. (E) Biomimetic synthesis of 1, 2, 4, and 5. Linear-1, -2, -4, and -5 spontaneously converted to 1, 2, 4, and 5, respectively. (F) Selective in vitro synthesis of linear-1, -2, -4, and -5 using PURExpress in vitro protein synthesis technologies. (G) Aminoketone (AMB/AA) dose-dependent production of linear-1 and -2 was observed. The Extracted Ion Chromatograms (EICs) were generated with a 10 ppm error window. (H) Presence of tRNAs decreased production of linear-1 and -2. n = 2 (linear-1) or 3 (linear-2) biological replicates. Data are mean ± SD; *P < 0.05, **P < 0.01. Two-tailed t-test.
While we demonstrated that the dipeptide ketones convert to pyrazinones spontaneously, formation of their peptide bond remained undefined. Prior transposon mutagenesis studies in V. cholerae failed to establish genes beyond tdh that are responsible for DPO (7) biosynthesis.26 After similar failures to identify mutant strains exhibiting disrupted AI-3 biosynthesis in E. coli, we were eventually able to reconstitute AI-3 analog production using in vitro protein synthesis technologies. Supplementation of AMB and AA in protein synthesis reaction mixtures, which contain the 20 E. coli aminoacyl-tRNA synthetases as well as additional transcription and translation factors, led to the selective formation of linear-1, -2, -4, and -5 and pyrazinone 6 in a substrate-dependent manner (Figures 3F,G and S7A). However, tRNAs were not required, and in fact, their elimination from the reaction mixtures significantly enhanced the production of linear-1, -2, -4, and -5 (Figures 3H and S7B). Additionally, the ribosome was not required for their production (Figure S7C), and 6 could be formed spontaneously in buffers from AA alone (Figure S8).
Biosynthetic Proposal of AI-3 Analog Formation
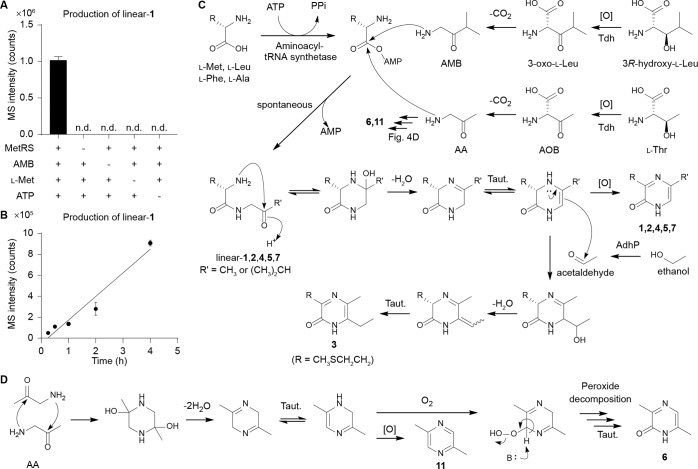
Based on our in vitro protein synthesis technologies studies, we hypothesized that the Tdh product and aminoacyl-tRNA synthetases were directly or indirectly responsible for AI-3 family biosynthesis. To test this possibility, we first incubated representative methionyl-tRNA synthetase (MetRS) with AMB, l-Met, and ATP in the absence of tRNAs. Under these conditions, we detected linear-1, and consistent with in vitro protein synthesis technology results, its production was abolished when MetRS, AMB, l-Met, or ATP were omitted in the reaction mixtures (Figure 4A). Importantly, production occurred on the multihour-long time scale (Figure 4B), suggesting that activated methionyl-AMP reacts spontaneously with AMB. Similar results were observed when we used AA instead of AMB for the production of linear-2 (Figure S9). Based on these data, we propose that AI-3 analogs 1–5 and minor 7 are derived from “abortive” tRNA synthetase charging reactions (release of an aminoacyl-AMP prior to tRNA charging) and spontaneous condensation of aminoacyl-AMP and aminoketone (AMB or AA) substrates (Figure 4C). On the other hand, spontaneous condensation of two units of AA lead to pyrazine 11 and, in the presence of oxygen, pyrazinone 6 (Figures 4D and S8). Oxygen is available at the gut host–bacteria interface, which E. coli use to sense their environment in the gut.14 Potential differential signaling effects between 6 and 11 will be determined in future studies.
Figure 4.
Tdh- and tRNA synthetases-derived origins of AI-3 analogs. (A) Results for in vitro synthesis of linear-1 by isolated MetRS. MetRS, AMB, l-Met, and ATP are required for the in vitro synthesis of linear-1. (B) Time-course analysis results of linear-1 production from in vitro assays. (C) Proposed biosynthesis of pyrazinones 1–7 and pyrazine 11. (D) Biosynthetic pathway of pyrazinone 6 and pyrazine 11. Taut., tautomerization. n = 3 biological replicates. Data are mean ± SD. nd, not detected.
Cellular Responses to AI-3 Analogs
Pyrazines and pyrazine-containing molecules, including pterins, have been linked to chemical signaling in bacteria.20,21,26,28,29 Additionally, pyrazinones 1 and 3 have the mass (m/z 213.1) previously reported for a structurally uncharacterized molecule in an active AI-3 fraction.12 Thus, we analyzed the new family of metabolites for their transcriptional effects on genes in the AI-3 regulon in EHEC. Although 1 enhanced expression of ler (the transcriptional activator of all LEE genes) in a qseC receptor-dependent manner, its effect on the expression of virulence factors espA, tir, and stx2a were independent of qseC (Figure 5A). Expression of effector protein gene espA, translocated intimin receptor gene tir and shiga toxin type 2a gene stx2a are subject to a high level of post-transcriptional control,30−35 suggesting that this compound is also affecting other signaling pathways. Pyrazinone 3 activated expression of ler and tir in a qseC-dependent manner, consistent with the previously reported AI-3 activity.36 However, it did not affect espA and stx2a expression, suggesting that this compound has limited AI-3 activity. Pyrazinone 6 increased expression of all LEE (ler, espA, and tir) and stx2a genes in a qseC-dependent fashion. It had the most robust activity of all the compounds, and it exhibited the same activity spectrum as AI-3.36 These data suggest that new metabolite 6 is the primary form of AI-3 activity and that AI-3 analogs can modulate this activity. This compound is very potent and activates AI-3 genes at a 5 nM concentration. Importantly, 6 can be found in isolations and commercial preparations of DPO (7) (3.1% relative, Figure S10); preparations of which regulate biofilm formation in V. cholerae at 100 μM.26 In our EHEC studies, DPO (7) at a 5 nM dose had no activity in the expression of any of the virulence genes tested. Pyrazinones 2, 4, and 5 had marginal or no activation of the tested genes in WT EHEC, but some of them exerted effects in the qseC mutant, suggesting that they have different activities than AI-3 (Figure S11).
Figure 5.
Evaluation of the biological activities of metabolites 1–7. (A) qRT-PCR analysis for the expression of virulence genes from WT EHEC and ΔqseC EHEC with pyrazinones 1, 3, 6, or 7 (5 nM). n = 6 (three biological replicates and two technical replicates). Fold change was calculated relative to rpoA as an internal control. Data are mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant. Two-tailed t-test. See Figure S11 for marginal 2, 4, and 5 responses. (B) BioMAP Phenotypic Profiling assay with pyrazinones 1–5 and 7 (36 μM). Cell types and stimuli used in each system are as follows: 3C system [HUVEC + (IL-1β, TNFα, and IFNγ)], 4H system [HUVEC + (IL-4 and histamine)], LPS system [PBMC and HUVEC + LPS (TLR4 ligand)], SAg system [PBMC and HUVEC + TCR ligands (1×)], BT system [CD19+ B cells and PBMC + (α-IgM and TCR ligands (0.001×))], BF4T system [bronchial epithelial cells and HDFn + (TNFα and IL-4)], BE3C system [bronchial epithelial cells + (IL-1β, TNFα, and IFNγ)], CASM3C system [coronary artery smooth muscle cells + (IL-1β, TNFα, and IFNγ)], HDF3CGF system [HDFn + (IL-1β, TNFα, IFNγ, EGF, bFGF, and PDGF-BB)], KF3CT system [keratinocytes and HDFn + (IL-1β, TNFα, and IFNγ)], MyoF system [differentiated lung myofibroblasts + (TNFα and TGFβ)] and /Mphg system [HUVEC and M1 macrophages + zymosan (TLR2 ligand)].
Given that bacterial QseC also responds to two human adrenergic signaling molecules, epinephrine and norepinephrine (Figure 1E),13 we also tested whether the AI-3 analogs could regulate human adrenergic signaling through ADRA1A, ADRA1B, ADRA1D, ADRA2A, ADRA2B, ADRA2C, ADRB1, and ADRB2 in HEK293 cells via the parallel receptor-ome expression and screening via transcriptional output (PRESTO-Tango) assay.37 The pyrazinones (1–7) and pyrazine (11) neither activated nor suppressed human adrenergic signaling relative to epinephrine, norepinephrine, and DMSO vehicle controls (Figures S12 and S13), indicating that bacteria can eavesdrop on human adrenergic signaling but not vice versa.
To further evaluate the metabolites for human cell responses, metabolites 1–5 and 7 (36 μM) were screened against a panel of 12 human primary cell-based coculture systems, including venular endothelial cells, lung fibroblasts, and peripheral blood mononuclear cells (PBMCs), that model several tissues and disease states (BioMAP Phenotypic Profiling Assay). Broad immunological effects of the pyrazinones were observed across the panel (Figure 5B). Specifically, protein biomarker readouts allowed for the effects of the metabolites on the different cell systems to be quantified. The structurally related metabolites 1–5 and 7 had a similar effect on most of the protein biomarkers measured in these cell systems. Each of the metabolites reduced proliferation in venular endothelial cells (3C). In addition, each of the metabolites caused an increase in protein biomarker interleukin-6 (IL-6) in coronary artery smooth muscle cells (CASM3C) and a decrease in protein biomarker α-SM actin in lung fibroblasts (MyoF). However, metabolite 2 affected several protein biomarkers differently compared to the other metabolites, including a decrease in interleukin-8 (IL-8) in the venular endothelial cell–macrophage coculture system (/Mphg).
To compare the primary cell systems with laboratory tissue culture, we selected the IL-8 response for further dose response and validation. Although IL-8 secretion was downregulated in the BioMAP mixed cell system results (see black arrow in Figure 5B), we observed increased IL-8 secretion in macrophages differentiated from human THP-1 cells when stimulated by 2 at doses of 100 nM or higher (Figure S14). This suggests that 2 in a context-dependent manner could contribute to E. coli-mediated IL-8 secretion, a known signal of macrophage phagocytosis and neutrophil chemotaxis. Combined with prior pyrazinone NF-κB signaling studies,25 this also suggests that the host can recognize select pyrazinones as microbe-associated molecular patterns, but the host sensory system(s) remain unknown at this time.
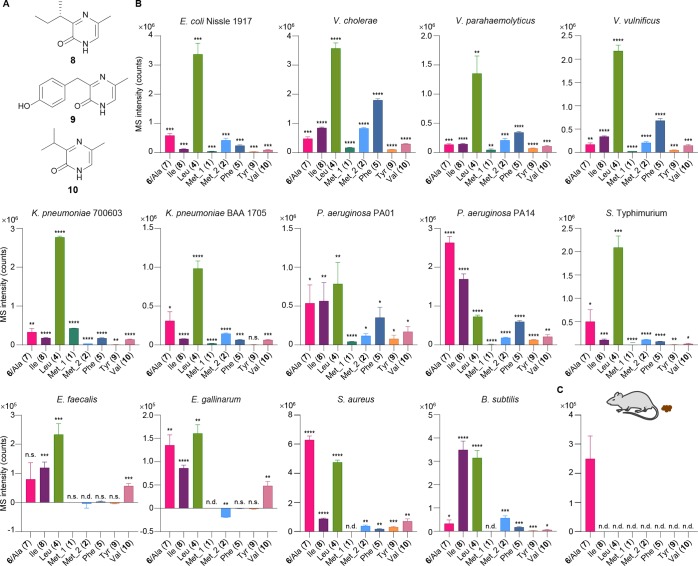
AI-3 Analogs Are Found in Gram-Negative and Gram-Positive Bacteria and Mouse Fecal Samples
Because the AI-3 variants above were derived from threonine dehydrogenase products and a biologically universal metabolic pathway, tRNA synthetases, we reasoned that similar metabolites could be detected in other Gram-negative and Gram-positive bacteria (Figure 6). Additionally, differential patterns of AI-3 production could suggest variations in abortive tRNA synthetase activity as a potential signaling strategy. Consequently, we first analyzed well-known Gram-negative pathogens for AI-3 analog production, including V. cholerae, Vibrio parahaemolyticus, Vibrio vulnificus, carbapenem-resistant Klebsiella pneumoniae, Pseudomonas aeruginosa, and Salmonella enterica serovar Typhimurium (Figure 6B). During the course of these efforts, we characterized three additional new pyrazinone metabolites 8–10 (Figures 6A and S2, Table S6). We also analyzed well-known Gram-positive bacteria, including pathogenic vancomycin-resistant Enterococcus faecalis (VRE), Enterococcus gallinarum,38 methicillin-resistant Staphylococcus aureus (MRSA), and nonpathogenic Bacillus subtilis. The patterns of AI-3 analog production were consistent with taxonomic similarities across the selected strains. These data demonstrate that diverse bacteria are capable of synthesizing AI-3 analogs and suggest that analog patterns may facilitate differential signaling effects among species. Production and sensing of these signaling molecules likely go well beyond that of E. coli described here and now represent exciting questions for future studies. Indeed, homologues of the AI-3 receptor QseC can be found in many Gram-negative bacterial species, including the Gram-negative bacteria in this study, in addition to Shigella flexneri, Citrobacter koseri, Enterobacter sp. 638, Yersinia mollaretii ATCC 43969, Y. pestis CO92, Y. pseudotuberculosis IP 32953, Y. enterocolitica subsp. enterocolitica 8081, Haemophilus influenzae PittGG, Pasteurella multocida subsp. multocida str. Pm70, Coxiella burnetii RSA 493, Burkholderia phymatum STM815, Ralstonia eutropha H16, Legionella pneumophila str. Paris, Bordetella parapertussis 12822, Francisella tularensis subsp. tularensis SCHU S4, Pseudomonas fluorescens Pf-5, Erwinia carotovora subsp. atroseptica SCRI1043, Actinobacillus pleuropneumoniae serovar 1 str. 4074, and Chromobacterium violaceum ATCC 12472, among others.36
Figure 6.
Pyrazinone profiling in different bacterial strains. (A) Structures of pyrazinones derived from l-Ile (8), l-Tyr (9), and l-Val (10). (B) Production of the pyrazinones derived from l-Thr (right-hand side) and l-Ala (7, minor in E. coli, which is indistinguishable with major 6), l-Ile (8), l-Leu (4), l-Met (2), l-Phe (5), l-Tyr (9), or l-Val (10) (left-hand side), and 3R-hydroxy-l-Leu (right-hand side) and l-Met (1) (left-hand side) from E. coli Nissle 1917, V. cholerae, V. parahaemolyticus, V. vulnificus, K. pneumonia 700603, K. pneumonia BAA 1705, P. aeruginosa PA01, P. aeruginosa PA14, S. enterica serovar Typhimurium, E. faecalis, E. gallinarum, S. aureus, and B. subtilis. Compounds 6 and 7 are indistinguishable in these studies (Figure S2C). Data are presented by mean ± SD of MS intensities (counts) observed in bacterial culture extracts subtracted by the mean of the media background. Statistical analysis was performed between the bacterial culture extracts (n = 3) and media control extracts (n = 3) using two-tailed t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant; nd, not detected. (C) A major peak corresponding to pyrazinones 6/7 was detected in C57BL/6 mouse fecal samples. n = 5 biological replicates. Data are mean ± SD.
To test whether the pyrazinone family could be detected in vivo, we examined fecal samples from C57BL/6 specific pathogen free mice.39 The mice were first treated with kanamycin to improve subsequent supplemental colonization. After treatment, the mice were inoculated with a kanamycin-resistant variant of the commensal model E. coli BW25113. Cellular stressors were not included during the colonization period. Under these conditions, a major peak corresponding to pyrazinones 6/7 was readily detected (Figure 6C), suggesting a likely functional role in vivo. However, the other pyrazinones were undetectable under the conditions of our studies. Given the robust production of the broader pryazinone family in cell culture, tracking their regulation, trafficking, fate, and function in vivo remain exciting future directions.
Discussion
Spontaneous chemical reaction sequences in cellular metabolism and signaling regulate diverse physiological responses in humans and in bacteria.40,41 However, this class of biosynthetic pathways is often challenging to characterize as genetic methods are limited to genes that are involved in substrate supply (i.e., tdh for E. coli and V. cholerae). We show here that tdh and essential tRNA synthetases are responsible for generating the substrates that react spontaneously to form the AI-3 family. Functional redundancies in amino acid activation may also contribute to these processes; however, the selectivity observed in our in vitro tRNA synthetase studies and cellular metabolite analyses support Tdh and tRNA synthetases as the dominant AI-3 biochemical origins. Our ribosome inhibitor-induced stress studies enhanced AI-3 family production levels, which facilitated isolation and characterization of these potent small molecules from laboratory-scale cultures. Strikingly, our studies support two mechanisms of AI-3 signal modulation. One, the Tdh product AA spontaneously dimerizes, oxidizes, and tautomerizes to form new metabolite 6 (Figure 4D), which is detected in vivo and accounts for the spectrum of AI-3 activities previously observed. And two, abortive tRNA synthetase products (aminoacyl-AMPs) spontaneously react with the aminoketones (AMB or AA) to modulate AI-3 transcriptional effects, as observed in cell culture. AI-3 6 is likely the major signal in vivo in the absence of cellular stressors, as the abortive products (i.e., 1–5, 7–10) were suppressed in fecal samples relative to cell cultures. In the latter, the resulting unstable dipeptide ketones, linear-1, -2, -4, or -5, undergo further oxidative spontaneous reactions to establish pyrazinone signals, 1, 2, 4, or 5, in E. coli. These AI-3 signals thus act as markers of cellular stress resulting from abortive aminoacyl-tRNA synthetase charging to enhance quorum sensing responses. Our results underscore the role of aminoacyl-tRNA charging fidelity as a common signal of cellular stress and starvation across multiple kingdoms. Indeed, aminoacyl-tRNA charging is monitored in humans by GCN242 and in bacteria by RelA and Hfq,43−46 which regulate the stringent response and the general stress response, respectively, in bacteria like E. coli. In the bacterial stringent response, RelA binds to uncharged tRNAs and the ribosome to initiate biosynthesis of the major stress signal (p)ppGpp.45,46 In the general stress response, Hfq represses RNA polymerase stress response sigma factor rpoS. Hfq binds to a variety of noncoding RNAs and appears to have a preference for uncharged tRNAs over charged tRNAs, and binding derepresses rpoS expression.43,44 Our cell culture studies here now integrate quorum sensing and cellular stress response behaviors at the molecular level, identify Tdh metabolism and abortive aminoacyl-tRNA synthetase charging as a biochemical basis for AI-3 family production, and define the structures of AI-3 and pathway intermediates that regulate immunological responses in the host and virulence-associated phenotypes in a broadly studied model organism and clinically relevant pathogen.
Acknowledgments
This work was supported by the Burroughs Wellcome Foundation (No. 1016720 to J.M.C.), the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2019R1A6A3A12033304 to C.S.K.), and the National Institutes of Health (No. AI053067 to V.S.). We also acknowledge prior support of our general human microbiota metabolism work from the National Institutes of Health (Nos. 1DP2-CA186575 and R01CA215553 to J.M.C.), the Camille and Henry Dreyfus Foundation (No. TC-17-011 to J.M.C.), and Yale University. The TOC image was created with BioRender.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.9b01076.
Materials and Methods, additional figures and tables, synthetic procedures, and NMR spectra of characterized compounds (PDF)
Author Contributions
C.S.K. characterized the structures and the biosynthetic pathway of AI-3 and wrote the manuscript. A.G. synthesized AI-3 analogs and performed metabolite profiling in Gram-negative and Gram-positive bacteria. S.C. performed the qRT-PCR experiments in WT and ΔqseC EHEC strains and analyzed the data. Y.C.L. contributed to MetRS studies. Z.W. validated the IL-8 secretion activity of 2 from THP-1 cells. H.C. performed the PRESTO-Tango assays. R.M.R. and E.E.S. performed preliminary qRT-PCR and EHEC cultivation experiments. R.W., T.P.W., and G.P. contributed to BioMAP Phenotypic Profiling Assay analysis. R.A.F. oversaw the IL-8 validation experiments. N.W.P. oversaw the PRESTO-Tango studies. V.S. oversaw the qRT-PCR experiments in WT and ΔqseC EHEC strains. J.M.C. conceived the study, oversaw experiments, and wrote the manuscript. All authors reviewed and contributed to the editing of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Secher T.; Brehin C.; Oswald E. Early settlers: which E. coli strains do you not want at birth?. Am. J. Physiol.-Gastroint. Liver Physiol. 2016, 311 (1), G123–G129. 10.1152/ajpgi.00091.2016. [DOI] [PubMed] [Google Scholar]
- Clements A.; Young J. C.; Constantinou N.; Frankel G. Infection strategies of enteric pathogenic Escherichia coli. Gut Microbes 2012, 3 (2), 71–87. 10.4161/gmic.19182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E.; Hoekstra R. M.; Angulo F. J.; Tauxe R. V.; Widdowson M.-A.; Roy S. L.; Jones J. L.; Griffin P. M. Foodborne illness acquired in the United States—major pathogens. Emerging Infect. Dis. 2011, 17 (1), 7–15. 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur J. C.; Perez-Chanona E.; Mühlbauer M.; Tomkovich S.; Uronis J. M.; Fan T.-J.; Campbell B. J.; Abujamel T.; Dogan B.; Rogers A. B.; et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338 (6103), 120–123. 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougnoux A.; Dalmasso G.; Martinez R.; Buc E.; Delmas J.; Gibold L.; Sauvanet P.; Darcha C.; Déchelotte P.; Bonnet M.; et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 2014, 63 (12), 1932–1942. 10.1136/gutjnl-2013-305257. [DOI] [PubMed] [Google Scholar]
- Tomkovich S.; Yang Y.; Winglee K.; Gauthier J.; Mühlbauer M.; Sun X.; Mohamadzadeh M.; Liu X.; Martin P.; Wang G. P. Locoregional effects of microbiota in a preclinical model of colon carcinogenesis. Cancer Res. 2017, 77 (10), 2620–2632. 10.1158/0008-5472.CAN-16-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M.; Kim C. S.; Healy A. R.; Wernke K. M.; Wang Z.; Frischling M. C.; Shine E. E.; Wang W.; Herzon S. B.; Crawford J. M. Structure elucidation of colibactin and its DNA cross-links. Science 2019, 365, eaax2685 10.1126/science.aax2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W.-L.; Bassler B. L. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009, 43, 197–222. 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S. T.; Bassler B. L. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harbor Perspect. Med. 2012, 2 (11), a012427. 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K.; Bassler B. L. Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14 (9), 576–588. 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio V.; Torres A. G.; Kaper J. B. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 2002, 43 (3), 809–821. 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- Sperandio V.; Torres A. G.; Jarvis B.; Nataro J. P.; Kaper J. B. Bacteria–host communication: the language of hormones. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (15), 8951–8956. 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M. B.; Hughes D. T.; Zhu C.; Boedeker E. C.; Sperandio V. The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U. S. A. 2006, 103 (27), 10420–10425. 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson-Banning K. M.; Sperandio V. Enterohemorrhagic Escherichia coli outwits hosts through sensing small molecules. Curr. Opin. Microbiol. 2018, 41, 83–88. 10.1016/j.mib.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford J. M.; Kontnik R.; Clardy J. Regulating alternative lifestyles in entomopathogenic bacteria. Curr. Biol. 2010, 20 (1), 69–74. 10.1016/j.cub.2009.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao D.; Okada B. K.; Wu Y.; Xu F.; Seyedsayamdost M. R. Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr. Opin. Microbiol. 2018, 45, 156–163. 10.1016/j.mib.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal I.; Hazan R.; Gaathon A.; Carmeli S.; Engelberg-Kulka H. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science 2007, 318 (5850), 652–655. 10.1126/science.1147248. [DOI] [PubMed] [Google Scholar]
- Lee J.; Wu J.; Deng Y.; Wang J.; Wang C.; Wang J.; Chang C.; Dong Y.; Williams P.; Zhang L.-H. A cell-cell communication signal integrates quorum sensing and stress response. Nat. Chem. Biol. 2013, 9 (5), 339–343. 10.1038/nchembio.1225. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Kolodkin-Gal I.; Vesper O.; Alam N.; Schueler-Furman O.; Moll I.; Engelberg-Kulka H. Escherichia coli quorum-sensing EDF, a peptide generated by novel multiple distinct mechanisms and regulated by trans-translation. mBio 2016, 7 (1), e02034-15. 10.1128/mBio.02034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. B.; Perez C. E.; Barber K. W.; Rinehart J.; Crawford J. M. Genome mining unearths a hybrid nonribosomal peptide synthetase-like-pteridine synthase biosynthetic gene cluster. eLife 2017, 6, e25229 10.7554/eLife.25229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez C. E.; Park H. B.; Crawford J. M. Functional characterization of a condensation domain that links nonribosomal peptide and pteridine biosynthetic machineries in Photorhabdus luminescens. Biochemistry 2018, 57 (3), 354–361. 10.1021/acs.biochem.7b00863. [DOI] [PubMed] [Google Scholar]
- Alvarez M. E.; White C. B.; Gregory J.; Kydd G. C.; Harris A.; Sun H. H.; Gillum A. M.; Cooper R. Phevalin, a new calpain inhibitor, from a Streptomyces sp. J. Antibiot. 1995, 48 (10), 1165–1167. 10.7164/antibiotics.48.1165. [DOI] [PubMed] [Google Scholar]
- Park H. B.; Crawford J. M. Pyrazinone protease inhibitor metabolites from Photorhabdus luminescens. J. Antibiot. 2016, 69 (8), 616–621. 10.1038/ja.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C.-J.; Chang F.-Y.; Wyche T. P.; Backus K. M.; Acker T. M.; Funabashi M.; Taketani M.; Donia M. S.; Nayfach S.; Pollard K. S.; et al. Discovery of reactive microbiota-derived metabolites that inhibit host proteases. Cell 2017, 168 (3), 517–526. 10.1016/j.cell.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J.; Patel J.; Park H. B.; Crawford J. M. β-lactam biotransformations activate innate immunity. J. Org. Chem. 2018, 83 (13), 7173–7179. 10.1021/acs.joc.8b00241. [DOI] [PubMed] [Google Scholar]
- Papenfort K.; Silpe J. E.; Schramma K. R.; Cong J.-P.; Seyedsayamdost M. R.; Bassler B. L. A Vibrio cholerae autoinducer–receptor pair that controls biofilm formation. Nat. Chem. Biol. 2017, 13 (5), 551–557. 10.1038/nchembio.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silpe J. E.; Bassler B. L. A host-produced quorum-sensing autoinducer controls a phage lysis-lysogeny decision. Cell 2019, 176 (1–2), 268–280. 10.1016/j.cell.2018.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feirer N.; Xu J.; Allen K. D.; Koestler B. J.; Bruger E. L.; Waters C. M.; White R. H.; Fuqua C. A pterin-dependent signaling pathway regulates a dual-function diguanylate cyclase-phosphodiesterase controlling surface attachment in Agrobacterium tumefaciens. mBio 2015, 6 (4), e00156-15. 10.1128/mBio.00156-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Junior E. A.; Ruzzini A. C.; Paludo C. R.; Nascimento F. S.; Currie C. R.; Clardy J.; Pupo M. T. Pyrazines from bacteria and ants: convergent chemistry within an ecological niche. Sci. Rep. 2018, 8 (1), 2595. 10.1038/s41598-018-20953-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato P. B.; Kaper J. B. Post-transcriptional processing of the LEE4 operon in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 2009, 71 (2), 273–290. 10.1111/j.1365-2958.2008.06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakhnovich E. A.; Davis B. M.; Waldor M. K. Hfq negatively regulates type III secretion in EHEC and several other pathogens. Mol. Microbiol. 2009, 74 (2), 347–363. 10.1111/j.1365-2958.2009.06856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall M. M.; Gruber C. C.; Rasko D. A.; Hughes D. T.; Sperandio V. Hfq virulence regulation in enterohemorrhagic Escherichia coli O157:H7 strain 86–24. J. Bacteriol. 2011, 193 (24), 6843–6851. 10.1128/JB.06141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber C. C.; Sperandio V. Posttranscriptional control of microbe-induced rearrangement of host cell actin. mBio 2014, 5 (1), e01025-13. 10.1128/mBio.01025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tree J. J.; Granneman S.; McAteer S. P.; Tollervey D.; Gally D. L. Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol. Cell 2014, 55 (2), 199–213. 10.1016/j.molcel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber C. C.; Sperandio V. Global analysis of posttranscriptional regulation by GlmY and GlmZ in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 2015, 83 (4), 1286–1295. 10.1128/IAI.02918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko D. A.; Moreira C. G.; Li D. R.; Reading N. C.; Ritchie J. M.; Waldor M. K.; Williams N.; Taussig R.; Wei S.; Roth M.; Hughes D. T.; Huntley J. F.; Fina M. W.; Falck J. R.; Sperandio V. Targeting QseC signaling and virulence for antibiotic development. Science 2008, 321 (5892), 1078–1080. 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeze W. K.; Sassano M. F.; Huang X.-P.; Lansu K.; McCorvy J. D.; Giguère P. M.; Sciaky N.; Roth B. L. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 2015, 22 (5), 362–369. 10.1038/nsmb.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredo Vieira S.; Hiltensperger M.; Kumar V.; Zegarra-Ruiz D.; Dehner C.; Khan N.; Costa F.; Tiniakou E.; Greiling T.; Ruff W.; Barbieri A.; Kriegel C.; Mehta S. S.; Knight J. R.; Jain D.; Goodman A. L.; Kriegel M. A. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018, 359 (6380), 1156–1161. 10.1126/science.aar7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. B.; Wei Z.; Oh J.; Xu H.; Kim C. S.; Wang R.; Wyche T. P.; Piizzi G.; Flavell R. A.; Crawford J. M.. Sulfamethoxazole drug stress upregulates redox active immunomodulatory metabolites in E. coli. 2019, submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J. W. Nitric oxide in immunity and inflammation. Int. Immunopharmacol. 2001, 1 (8), 1397–1406. 10.1016/S1567-5769(01)00086-8. [DOI] [PubMed] [Google Scholar]
- Pereira C. S.; Thompson J. A.; Xavier K. B. AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 2013, 37 (2), 156–181. 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- He F.; Wu C.; Li P.; Li N.; Zhang D.; Zhu Q.; Ren W.; Peng Y. Functions and signaling pathways of amino acids in intestinal inflammation. BioMed Res. Int. 2018, 2018, 9171905. 10.1155/2018/9171905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.; Feig A. L. The RNA binding protein Hfq interacts specifically with tRNAs. RNA 2008, 14 (3), 514–523. 10.1261/rna.531408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson K. M.; Gottesman S. The MiaA tRNA modification enzyme is necessary for robust RpoS expression in Escherichia coli. J. Bacteriol. 2014, 196 (4), 754–761. 10.1128/JB.01013-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther K. S.; Roghanian M.; Gerdes K. Activation of the stringent response by loading of RelA-tRNA complexes at the ribosomal A-site. Mol. Cell 2018, 70 (1), 95–105. 10.1016/j.molcel.2018.02.033. [DOI] [PubMed] [Google Scholar]
- Wang B.; Dai P.; Ding D.; Del Rosario A.; Grant R. A.; Pentelute B. L.; Laub M. T. Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 2019, 15 (2), 141–150. 10.1038/s41589-018-0183-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.