Abstract
Knotted1-like Homeobox (KNOX) proteins play important roles in regulating plant growth, development, and other biological processes. However, little information is available on the KNOX gene family in apple (Malus domestica Borkh.). In this study, 22 KNOX genes were identified in the apple genome. The gene structure, protein characteristics, and promoter region were characterized. The MdKNOX family members were divided into three classes based on their phylogenetic relationships. Quantitative real-time PCR analysis revealed that the majority of MdKNOX genes exhibited strongly preferential expression in buds and were significantly up-regulated during the flower induction period. The transcript levels of MdKNOX genes were responsive to treatments with flowering- and stress-related hormones. The putative upstream regulation factor MdGRF could directly bind to the promoter of MdKNOX15 and MdKNOX19, and inhibit their transcriptional activities, which were confirmed by yeast one-hybrid and dual-luciferase assays. The results provide an important foundation for future analysis of the regulation and functions of the MdKNOX gene family.
Keywords: Malus domestica, Knotted1-like Homeobox, flower induction, growth-regulating factor, yeast one-hybrid
Introduction
Homeobox proteins are considered to act as sequence-specific DNA-binding proteins and contain a 60 amino-acid-long DNA-binding domain termed a homeodomain (HD) that directly regulates the expression of specific groups of target genes (Hayashi and Scott, 1990). Different HD proteins have been grouped into separate families (or classes) based on either sequence identity within the HD or conserved protein motifs outside of the HD (Bürglin and Affolter, 2016). Although their structures are similar, different homeodomains are able to recognize diverse DNA binding sites (Berger et al., 2008). KNOTTED1-LIKE HOMEOBOX (KNOX) genes belong to the three amino acid loop extension (TALE) homeodomain superfamily and are generally distinguished by four characteristic domains—KNOXI, KNOXII, ELK, and HD—although some genes lack the ELK and HD domains (Gao et al., 2015). The first homeobox gene reported in a plant species was ZmKN1 from maize (Vollbrecht et al., 1991). Subsequently, a number of KNOX proteins have been characterized in many plant species (Hay and Tsiantis, 2010). Arabidopsis KNOX proteins can be divided into Classes I and II based on sequence similarity conventionally. Further, KNOX lost the HD domain was found in Arabidopsis, which defined a novel class, named as the KNATM (Magnani and Hake, 2008). Four Class I (SHOOT-MERISTEMLESS (STM), KNAT1, KNAT2, and KNAT6) and Four Class II KNOX (KNAT3, KNAT4, KNAT5, and KNAT7) genes were identified from Arabidopsis. Class I genes have been intensively studied and shown to play important roles in meristem maintenance, control of leaf blade shape, internode elongation, hormone homeostasis, and establishment of inflorescence architecture (Tsuda et al., 2011; Tsuda and Hake, 2015). Loss-of-function mutations in the Arabidopsis STM resulted in embryos that lack a SAM (Barton and Poethig, 1993). KNAT1 transcripts are detected in whole-shoot and inflorescence tissue but not in leaves, and KNAT2 transcripts are present at high levels in shoot and inflorescence tissue as expected but are of low abundance in leaves, which affects leaf morphological development (Byrne et al., 2000). KNAT6 is expressed at the site of lateral root initiation, and is involved in meristem activity and organ separation (BellesBoix et al., 2006). With regard to Class II KNOX genes, expression patterns have been characterized in maize by RNA gel-blot analysis (Kerstetter et al., 1994). Serikawa et al. (1997) detected Arabidopsis KNAT3 expression patterns through the use of promoter-GUS (β-glucuronidase) fusion analysis and in situ hybridization. The varied expression patterns indicate that KNAT3 plays several different roles in plants, depending on when and where it is expressed. Despite several reports of expression patterns, comparatively little is known about the function of Class II KNOX genes in plants. In Arabidopsis, domain exchange and phenotypes analysis suggest that the sequences outside of the third helix and N-terminal arm of the homeodomain endow the specificity of KNAT3 and KNAT1 (Serikawa and Zambryski, 1997). The Class II genes KNAT3, KNAT4 and KNAT5 perform redundant and important functions in root (Truernit and Haseloff, 2007) and lateral organ differentiation (Furumizu et al., 2015). Promoter-GUS and fluorescent protein analysis have demonstrated the transcriptional regulation and protein products localization of KNAT3, KNAT4, and KNAT5 in specific domains and cell types of the Arabidopsis root (Truernit et al., 2006). KNAT3 may also modulate abscisic acid (ABA) responses to regulate germination and early seedling development (Kim et al., 2013). KNAT3 and KNAT7 are involved in secondary cell wall biosynthesis in Arabidopsis and Populus (Li et al., 2012; Wang et al., 2020) and GhKNL1 participates in the regulation of fiber development in cotton (Gong et al., 2014). Three Class II KNOX genes, MtKNAT3/4/5-like, from Medicago truncatula regulate legume nodule boundaries and shape development (Di Giacomo et al., 2017).
Additional research has revealed that KNOX genes are involved in diverse developmental processes mainly by affecting the metabolism and signaling pathway of hormones (Chan et al., 1998; Himmelbach et al., 2002; Bolduc and Hake, 2009). KNOX genes activate cytokinin biosynthesis (Jasinski et al., 2005; Yanai et al., 2005). For example, in M. truncatula, MtKNOX3 activates the cytokinin biosynthesis ISOPENTENYL TRANSFERASE (IPT) genes, regulates nodule development, and activates cytokinin biosynthesis upon nodulation (Azarakhsh et al., 2015). KNOX proteins have been reported to repress the production of gibberellins (GAs). On the one hand, KNOX negatively modulates the accumulation of GAs by controlling the abundance of GA2-oxidase, by binding to an intron of ga2ox1 and up-regulating the metabolic gene (Bolduc and Hake, 2009). On the other hand, KNOX inhibits GA biosynthesis by down-regulation of the key biosynthetic gene GA20-oxidase (Kusaba et al., 1998; Rosin et al., 2003). In addition, KNOX proteins are involved in other hormonal signaling pathways. KNOX change the abundance of proteins associated with auxin transporter signaling components to regulate abscission in tomato (Ma et al., 2015). Rice HOMEOBOX 1 (OSH1) represses the brassinosteroid phytohormone pathway through activation of brassinosteroid catabolism genes (CYP734A2, CYP734A4, and CYP734A6) and then arrests the growth of the SAM (Tsuda et al., 2014). KNAT3 interacts with a BELL-like homeodomain (BLH) protein and synergistically modulates ABA responses during germination and early seedling development in Arabidopsis (Dachan et al., 2013).
In addition to being a transcriptional regulator, KNOX genes are regulated by other protein factors to prevent misexpression. Arabidopsis BELL-like homeodomain proteins BLH2/SAW1 and BLH4/SAW2 act redundantly to regulate expression of one or more KNOX genes and to establish leaf shape (Kumar et al., 2007). NtSVP, a MADS-box transcription factor from tobacco, acts as a repressor of the BP-like Class I KNOX gene NtBPL by directly binding to the NtBPL promoter, causing shortened pedicels (Wang et al., 2015). YABBY contributes to the repression of KNOX genes (STM, KNAT1/BP and KNAT2) to prevent development of ectopic meristems in Arabidopsis (Kumaran et al., 2002). The Arabidopsis polycomb group (PcG) protein FERTILIZATION-INDEPENDENT ENDOSPERM (FIE) and CURLY LEAF (CLF) could also repress expression of KNOX genes (Katz et al., 2004). In particular, transcription of KNOX genes is indicated to be suppressed by a growth-regulating factor (GRF), and such interactions have been confirmed in several species, including barley (Osnato et al., 2010), Arabidopsis and rice (Kuijt et al., 2014). This result implies that the GRF–KNOX regulatory module was relatively conservative.
Although roles of KNOX genes in plant development have been partly elucidated in Arabidopsis and other species, little information is available about the possible roles of these genes in fruit crops. Apple (Malus domestica) is one of the most widely cultivated fruit trees. To date, only two studies have been reported concerning KNOX genes from apple (Watillon et al., 1997; Gao et al., 2015). Here, we conducted the genome-wide identification of members of the KNOX gene family in M. domestica. The expression profiles in various tissues and in response to exogenous hormone treatment as well as during the floral development period were explored. In addition, the regulatory interaction between MdKNOX and the putative upstream regulator MdGRF was tested. This study may provide a foundation for further investigation of the regulation and functions of the MdKNOX gene family.
Materials and Methods
Identification of KNOX Encoding Genes in the Apple (Malus domestica Borkh.) Genome
Arabidopsis thaliana and Oryza sativa KNOX protein sequences were downloaded from The Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/) and the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/cgi-bin/ORF_infopage.cgi) databases. To identify the genes encoding KNOX proteins in the apple (M. domestica) genome, the BLASTP program was used to search for potential KNOX-encoding genes in the complete genome, using the known KNOX sequences from Arabidopsis and rice as queries. All non-redundant putative protein sequences were manually checked with the Pfam database (http://pfam.xfam.org/search/sequence) and the NCBI Conserved Domains database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The 22 MdKNOX genes obtained were designated MdKNOX1 to MdKNOX22 based on their chromosomal locations. Protein physicochemical characteristics were predicted with the ExPASy program (http://web.expasy. org/protparam/).
Multiple Sequence Alignment and Phylogenetic Analysis
The full-length amino acid sequences of KNOX proteins from Arabidopsis, rice, and apple were used for multiple alignment performed with DNAMAN software. We chose the following parameter settings—substitution matrix: Blosum62, mismatch score: −15, and gap open/extend penalty: 10/5. Phylogenetic trees were constructed using the MEGA 7.0 program (Kumar et al., 2016). Sequence alignment was carried out using MUSCLE (Edgar, 2004) program with default parameters. The optimal protein substitution model was the Jones–Taylor–Thornton (JTT) model with gamma distribution. The evolutionary history was inferred using the neighbor-joining method based on the JTT matrix-based model and gamma distribution. Support for the phylogeny topology was assessed by means of a bootstrap analysis with 500 replications. Sequence logos were generated using the Weblogo online platform (http://weblogo.berkeley.edu/logo.cgi).
Gene Structure, Conserved Motif, and Promoter Sequence Analysis
A gene structures map was obtained with the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn). Conserved motifs in MdKNOX protein sequences were elucidated with the MEME platform (http://meme-suite.org/) (Bailey et al., 2006). The dimensional structure of MdKNOX proteins was predicted with the PHYRE server v2.0 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index). The 1,500-bp genomic DNA sequence upstream of the start codon (ATG) of each MdKNOX gene was obtained from the apple genome sequence. Cis-elements in the promoters were identified using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Homologous Gene Pairs and Synteny Analysis
Information on syntenic blocks within the apple genome and between the Arabidopsis and apple genomes were downloaded from the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/). The genome sequences of homologous KNOX proteins from Arabidopsis and apple were assessed using BLASTP. The OrthoMCL algorithm was used to identify paralogous genes within the apple genome as well as between the apple and Arabidopsis genomes (Li et al., 2003). The MCScan algorithm (Wang et al., 2012) was applied to detect syntenic blocks containing apple KNOX genes. Circos (Krzywinski and Schein, 2009) was used to visualize the syntenic relationships between the genomes.
Plant Materials and Treatments
Samples were collected from six-year-old apple Fuji/T337/Malus robusta Rehd. trees for tissue-specific expression analysis, comprising roots, stems, leaves, buds, flowers, and fruits. Newly developed lateral roots of 1–2 mm diameter, stems of 2–3 mm diameter near to the shoot apices, fully expanded leaves adjacent to buds, flower buds, flowers at anthesis, and young fruits were collected, immediately frozen in liquid nitrogen, and stored at −80°C until use.
For hormone treatments, apple trees of uniform growth in the experimental orchard of the College of Horticulture, Northwest A&F University, Yangling, China (108°04′ E, 34°16′ N) were chosen and randomly divided into six groups. Each group was treated with 4 mmol/L salicylic acid (SA), 150 μmol/L ABA, 700 mg/L gibberellin (GA3), 300 mg/L 6-Benzylaminopurine (6-BA) or water (control). Solutions were applied by spraying the leaves with a low-pressure hand-wand sprayer, followed by sampling at 30, 50, and 70 days after full bloom (DAFB). Plant samples were immediately frozen in liquid nitrogen after collection and stored at −80°C until use.
RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR
Total RNA was isolated using a RNA extraction kit (OMEGA, Doraville, GA, USA). RNA integrity was verified by electrophoresis and RNA concentration was determined using a Nanodrop 2000 spectrophotometer. One microgram of total RNA was used as the template for first-strand cDNA synthesis, using the PrimeScript™ RT Reagent kit (Takara, Shiga, Japan) following the manufacturer's instructions.
Primer pairs for quantitative real-time PCR (qRT-PCR) were designed using Primer Premier 6.0 (Premier Biosoft, Palo Alto, CA, USA) ( Supplementary Table 1 ). It was difficult to distinguish the amplification products because of the high similarity in coding regions among several MdKNOX genes. Therefore, the same primer pair was used to analyze the expression of both MdKNOX1 and MdKNOX20, MdKNOX2 and MdKNOX5, MdKNOX4 and MdKNOX12, MdKNOX10 and MdKNOX22. Consequently, 18 pairs of primers were designed for 22 MdKNOX genes. Each primer pair was checked via RT-PCR followed by 1.2% agarose gel electrophoresis to verify the specificity of the amplification products.
Real-time RT-PCR was performed in a total volume of 20 μl containing 2 μl cDNA, 10 μl of 2×SYBR® Green II Mix, 0.5 μM of each primer, and distilled deionized H2O. Analyses were conducted with the Bio-Rad CFX Connect™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The PCR protocol was as follows: 94°C pre-incubation for 3 min; followed by 40 cycles of denaturation at 94°C for 10 s, and annealing at 60°C for 30 s. At the end of the amplification, a melting curve from 65 to 95°C with 0.5°C increments was performed to verify the presence of gene-specific PCR products. Apple Actin (MD04G1127400) and HistoneH3 (MD01G1035300) were used as internal standard genes. Three biological replicates for each sample and three technical replicates for each biological replicate were analyzed. The relative expression levels were calculated using the relative 2−△△ Ct method (Livak and Schmittgen, 2001).
Yeast One-Hybrid (Y1H) and Dual-Luciferase Assays
To clone the promoters of MdKNOX15 and MdKNOX19, genomic DNA was isolated from fresh young leaves of apple ‘Fuji' and used as the DNA template. Each PCR system contained the Phusion® High-Fidelity PCR Master Mix, 0.5 μM primer pairs (proKNOX15-F/proKNOX15-R for MdKNOX15 promoter, and proKNOX19-F/proKNOX19-R for MdKNOX19 promoter) and 1 ng/μl genomic DNA. The standard thermal profile was as follows: 95°C for 2 min; 30 cycles of 95°C for 10 s, 57°C for 20 s, and 72°C for 1 min; followed by a final extension for 10 min at 72°C. The PCR products were cloned into the pBlunt vector (CloneSmart, USA) for sequencing.
A yeast one-hybrid (Y1H) assay was performed using the Gold Matchmaker™ Gold Yeast One-Hybrid System (Clontech, Mountain View, CA, USA). The open reading frame of MdGRF was cloned using the primers MdGRF-F1 and MdGRF-R1, and inserted into the pGADT7 vector. Then the 277-bp or 434-bp promoter fragment of the MdKNOX gene (proKNOX) was inserted into the pAbAi vector. After linearization, the constructs were transformed into the yeast cells, which were plated on SD/−Ura media supplemented with aureobasidin A (AbA) to determine the minimal inhibitory concentration of AbA. Growth of the co-transformant yeast cells (harboring pGADT7-MdGRF and pAbAi-proMdKNOX) was detected on SD/−Ura medium supplemented with AbA.
For dual-luciferase assays, the complete expression units of improved firefly (coleopteran) luciferase (FLuc) and Renilla (Renilla reniformis) luciferase (RLuc) were cloned from pGL3 basic-2X35S-Rluc-2X35S-Fluc plasmid (Gu et al., 2013), and inserted into the multiple cloning site (MCS) of pCAMBIA0309 vector to generate the dual reporter expression vector. To detect the effect of MdGRF on the promoter activities of MdKNOX, the 2× 35S promoter upstream of the luciferase gene was replaced by the promoter of the KNOX gene to generate the reporter, and the open reading frame of MdGRF was cloned using the primers MdGRF-F2/MdGRF-R2 and inserted into pRI101-AN vector o generate the effector (MdGRF-OE) plasmid. The recombinant vectors were transformed into Agrobacterium strain GV3103. Tobacco leaves were infected with the mixed Agrobacterium cells by means of Agrobacterium-mediated transient injection (Krenek et al., 2015). RLuc/FLuc activity was assessed using the Dual-Luciferase® Reporter Assay System (Promega, USA).
Statistical Analysis
Data were subjected to analysis of variance and the means were compared using Student's t-test at the 5% significance level using SPSS 11.5 software (SPSS, Chicago, IL, USA).
Results
Genome-Wide Identification of Arabidopsis and Apple KNOX Genes
Nine KNOX genes were previously identified and reported in the A. thaliana genome, named KNATM, KNAT6, STM, KNAT7, KNAT2, KNAT1/BP, KNAT5, KNAT4, and KNAT3. To identify apple KNOX genes, a BLASTP search of the apple genome was conducted with the nine AtKNOX protein sequences as queries. After manual checking and confirmation using the NCBI Conserved Domains database, 22 candidate MdKNOX genes were obtained ( Table 1 ). The MdKNOX genes were named in accordance with their chromosomal locations (MdKNOX1–MdKNOX22). The 22 MdKNOX genes were located on 12 chromosomes in the apple genome. The chromosomes 6 and 15 harbored the highest number of genes (three genes each), the chromosomes 4, 8, 10, 14 and 15 contained two genes, and chromosome 3, 5, 9,12, 16, and 17 each carried a single gene ( Table 1 ).
Table 1.
Arabidopsis thaliana and apple KNOX gene families.
| Name | Gene ID | Location | CDS(bp) | Peptide(aa) |
|---|---|---|---|---|
| KNATM | AT1G14760 | chr1:5,084,315..5,084,315 | 429 | 142 |
| STM | AT1G62360 | chr1:23,058,582..23,058,582 | 1,149 | 382 |
| KNAT1/BP | AT4G08150 | chr4:5,147,699..5,147,699 | 1,197 | 398 |
| KNAT2 | AT1G70510 | chr1:26,576,486..26,576,486 | 933 | 310 |
| KNAT6 | AT1G23380 | chr1:8,297,241..8,297,241 | 990 | 329 |
| KNAT3 | AT5G25220 | chr5:8,735,944..8,735,944 | 1,296 | 431 |
| KNAT4 | AT5G11060 | chr5:3,509,833..3,509,833 | 1,182 | 393 |
| KNAT5 | AT4G32040 | chr4:15,493,989..15,493,989 | 1,152 | 383 |
| KNAT7 | AT1G62990 | chr1:23,337,167..23,337,167 | 876 | 291 |
| KNOX01 | MD02G1012900 | Chr03:821,764..821,764 | 1,083 | 360 |
| KNOX02 | MD04G1069700 | Chr04:9,546,107..9,546,107 | 765 | 254 |
| KNOX03 | MD04G1215500 | Chr04:29,846,452..29,846,452 | 993 | 330 |
| KNOX04 | MD05G1352500 | Chr05:46,891,824..46,891,824 | 1,161 | 386 |
| KNOX05 | MD06G1071100 | Chr06:17,202,997..17,202,997 | 867 | 288 |
| KNOX06 | MD06G1171700 | Chr06:31,218,154..31,218,154 | 1,056 | 351 |
| KNOX07 | MD06G1232400 | Chr06:36,328,062..36,328,062 | 426 | 141 |
| KNOX08 | MD08G1075200 | Chr08:6,111,898..6,111,898 | 750 | 249 |
| KNOX09 | MD08G1153600 | Chr08:16,709,140..16,709,140 | 1,293 | 430 |
| KNOX10 | MD09G1112500 | Chr09:8,548,362..8,548,362 | 1005 | 334 |
| KNOX11 | MD10G1276200 | Chr10:36,731,509..36,731,509 | 621 | 206 |
| KNOX12 | MD10G1326500 | Chr10:40,664,274..40,664,274 | 1,182 | 393 |
| KNOX13 | MD12G1205700 | Chr12:28,622,790..28,622,790 | 990 | 329 |
| KNOX14 | MD13G1018900 | Chr13:1,182,375..1,182,375 | 330 | 109 |
| KNOX15 | MD13G1095800 | Chr13:6,760,799..6,760,799 | 1,059 | 352 |
| KNOX16 | MD14G1177200 | Chr14:27,054,900..27,054,900 | 933 | 310 |
| KNOX17 | MD14G1239200 | Chr14:31,827,706..31,827,706 | 426 | 141 |
| KNOX18 | MD15G1062700 | Chr15:4,315,813..4,315,813 | 753 | 250 |
| KNOX19 | MD15G1130800 | Chr15:9,443,279..9,443,279 | 1,314 | 437 |
| KNOX20 | MD15G1159800 | Chr15:11,950,827..11,950,827 | 1,107 | 368 |
| KNOX21 | MD16G1097200 | Chr16:6,783,377..6,783,377 | 1,077 | 358 |
| KNOX22 | MD17G1102600 | Chr17:8,723,554..8,723,554 | 987 | 328 |
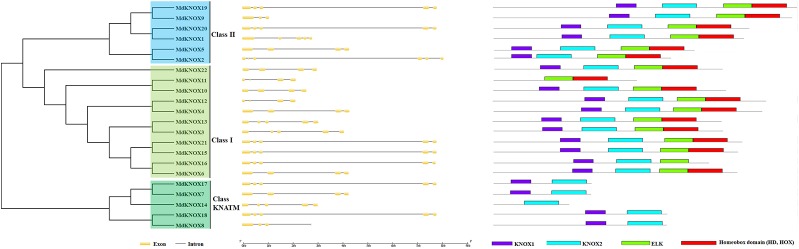
Multiple sequence alignment showed that the majority of the MdKNOX proteins shared four conserved domains: KNOXI, KNOXII, ELK, and HOX domain ( Figure 1 ). The ELK and HOX domains were located at the C terminus of the MdKNOX protein, whereas the KNOXI and KNOXII domains were located at the N terminus. Among the proteins, ELK and HOX domains were absent in five MdKNOX proteins (MdKNOX7, MdKNOX8, MdKNOX14, MdKNOX17, and MdKNOX18) ( Figure 1 ).
Figure 1.
Multiple sequences alignment of MdKNOX proteins.
Gene Characterization and Structure Analysis of MdKNOX
KNOX protein characteristics were analyzed using the ExPASy portal, including molecular weight, isoelectric point, grand average of hydropathicity, instability index, major amino acid content, and aliphatic index ( Table 2 ). The molecular weight of the analyzed MdKNOX proteins ranged from 11.99 (MdKNOX14) to 40.81 kDa (MdKNOX1). The molecular weight of the MdKNOX proteins was greater than 13 except for MdKNOX14, which indicated that MdKNOX is a group of macromolecular proteins. The isoelectric point ranged from 4.66 (MdKNOX7) to 6.89 (MdKNOX11) ( Table 2 ). Given that the instability index values were greater than 40, all KNOX proteins were considered to be unstable except for KNOX7. Grand average of hydropathicity values indicated that the MdKNOX proteins were hydrophilic. The aliphatic index values ranged from 54.15 (MdKNOX4) to 85.18 (MdKNOX17). Amino acid content analysis showed that Lys and Ser were the predominant residues, and that Glu, Ala, Gln, and Lys also accounted for a large proportion of the proteins. Alpha helices, β sheets, extended strands, and random coils were present in the predicted protein structures of all the MdKNOX proteins except for MdKNOX16 ( Supplementary Figure 1 ). The Gene Structure Display Server was used to display the exon–intron structure based on the annotated apple genome. All MdKNOX family members contained 3–6 introns. The number and distribution of introns for MdKNOX genes was rather conserved within each cluster ( Figure 2 ). For example, the KNAT2/6 cluster, including MdKNOX15, MdKNOX16, and MdKNOX21, was highly conserved and comprised four introns and five exons. However, although the genes MdKNOX2 and MdKNOX5 showed high similarity in protein sequences, the distribution and location of exons were distinct. These differences suggested that the two genes have functionally diverged during evolution.
Table 2.
Amino acid compositions as well as physical and chemical characteristics of KNOX proteins.
| Name | pI | MW | Instability Index | GRAVY | Major Amino Acid | Aliphatic Index |
|---|---|---|---|---|---|---|
| MdKNOX01 | 5.19 | 40.81 | 53.81 | −0.747 | S(9.4%)L(9.4%)E(8.6%)D(7.5%) | 72.03 |
| MdKNOX02 | 6.27 | 29.08 | 58.41 | −0.729 | L(10.2%)E(9.0%)S(7.4%)G(7.0%) | 74.45 |
| MdKNOX03 | 5.14 | 36.83 | 41.83 | −0.547 | G(10.3%)E(9.6%)S(8.7%)L(8.4%) | 67.36 |
| MdKNOX04 | 6.3 | 43.04 | 44.81 | −0.726 | S(11.9%)G(7.5%)L(6.9%)A(6.7%) | 54.15 |
| MdKNOX05 | 6.31 | 32.85 | 59.96 | −0.739 | L(10.7%)Q(9.0%)S(8.3%)E(7.9%) | 76.18 |
| MdKNOX06 | 5.15 | 39.57 | 46.5 | −0.636 | S(10.2%)L(7.9%)A(7.4%)D(7.1%) | 65.38 |
| MdKNOX07 | 4.66 | 15.73 | 36.75 | −0.655 | L(13.4%)E(11.3%)K(8.5%)S(8.5%) | 83.05 |
| MdKNOX08 | 5.87 | 27.70 | 49.68 | −0.895 | S(9.6%)N(9.2%)G(8.8%)Q(7.6%) | 57.19 |
| MdKNOX09 | 5.95 | 48.31 | 47.72 | −0.788 | L(9.3%)S(8.8%)A(7.2%)N(7.2%) | 71.02 |
| MdKNOX10 | 6.32 | 37.72 | 48.13 | −0.61 | L(10.1%)S(9.8%)A(7.4%)E(7.1%) | 68.71 |
| MdKNOX11 | 6.89 | 24.08 | 45.14 | −0.723 | L(11.6%)K(9.7%)E(7.7%)S(7.2%) | 71.99 |
| MdKNOX12 | 6.32 | 43.91 | 48.22 | −0.734 | S(11.1%)L(7.6%)A(7.1%)G(6.8%) | 56.41 |
| MdKNOX13 | 5.14 | 36.82 | 41.95 | −0.663 | S(9.4%)E(9.1%)G(9.1%)L(8.2%) | 63.74 |
| MdKNOX14 | 4.88 | 12.00 | 40.56 | −0.492 | S(9.1%)A(8.2%)D(8.2%)K(8.2%) | 64.5 |
| MdKNOX15 | 5.17 | 39.87 | 46.5 | −0.679 | S(10.7%)L(8.8%)E(8.5%)D(7.1%) | 65.2 |
| MdKNOX16 | 4.94 | 34.65 | 50.12 | −0.553 | S(10.9%)L(8.7%)A(8.3%)D(7.0%) | 67.45 |
| MdKNOX17 | 4.72 | 15.78 | 49.08 | −0.561 | L(12.7%)E(11.3%)S(9.2%)K(7.8%) | 85.18 |
| MdKNOX18 | 5.85 | 27.74 | 46.13 | −0.857 | S(10.0%)G(9.2%)N(9.2%)Q(7.6%) | 57.76 |
| MdKNOX19 | 6.01 | 48.66 | 49.41 | −0.762 | S(8.9%)L(8.6%)A(7.3%)Q(7.0%) | 68.99 |
| MdKNOX20 | 5.43 | 41.65 | 47 | −0.799 | S(8.9%)L(8.6%)D(7.8%)E(7.6%) | 68.61 |
| MdKNOX21 | 5.62 | 40.88 | 43.57 | −0.643 | S(10.3%)L(8.9%)E(7.8%)A(7.2%) | 67.37 |
| MdKNOX22 | 6.16 | 37.11 | 47.42 | −0.564 | L(10.3%)S(10.3%)A(7.9%)E(7.3%) | 71.46 |
| KNATM | 5.64 | 16.44 | 56.91 | −0.349 | L(14.0%)S(11.2%)K(9.1%)E(8.4%) | 87.89 |
| STM | 6.19 | 42.75 | 55.48 | −0.652 | S(12.0%)A(7.8%)L(7.0%)E(6.8%) | 55.99 |
| KNAT1/BP | 6.02 | 45.84 | 50.08 | −1.113 | N(10.5%)S(10.0%)L(7.5%)E(7.2%) | 56.41 |
| KNAT2 | 4.9 | 35.64 | 48.97 | −0.705 | D(9.6%)L(9.6%)S(8.3%)E(8.0%) | 70.87 |
| KNAT6 | 4.92 | 37.19 | 53.6 | −0.542 | S(10.0%)L(9.1%)D(8.5%)E(8.5%) | 73.56 |
| KNAT3 | 5.86 | 47.60 | 57.27 | −0.696 | A(9.2%)L(8.8%)S(8.5%)Q(7.6%) | 69.56 |
| KNAT4 | 5.87 | 44.39 | 67.02 | −0.856 | S(9.6%)L(8.9%)E(8.3%)Q(8.3%) | 66.82 |
| KNAT5 | 6.03 | 43.28 | 55.48 | −0.651 | L(9.3%)S(9.3%)E(7.5%)T(6.7%) | 73.79 |
| KNAT7 | 6.1 | 32.91 | 47.98 | −0.61 | L(10.3%)E(8.2%)A(7.2%)G(6.8%) | 76.43 |
pI, isoelectric point; MW, molecular weight; kDa, GRAVY: grand average of hydropathicity.
Figure 2.
Analysis of MdKNOX gene structure. An unrooted neighbor-joining tree was constructed derived from MdKNOX protein sequences (left) and exon–intron composition analysis (right).
Synteny Analysis and Phylogenetic Relationships Among KNOX Genes
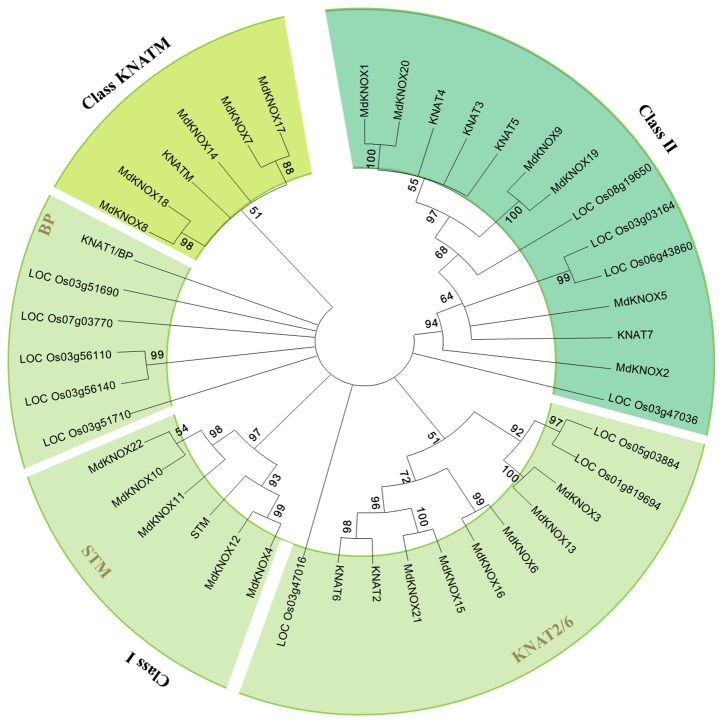
To clarify the evolutionary relationships among KNOX proteins, a neighbor-joining tree was constructed derived from Arabidopsis, rice, and apple KNOX protein sequences. According to the phylogenetic tree ( Figure 3 ), the KNOX proteins were clustered into three groups, designated Class I, Class II and Class KNATM. Class I was further divided into four subgroups: STM, KNAT2, KNAT6, and BP. Six apple proteins (MdKNOX1, MdKNOX2, MdKNOX5, MdKNOX9, MdKNOX19, and MdKNOX20) were clustered in Class II, and eleven apple proteins were clustered in the Class I. MdKNOX7, MdKNOX8, MdKNOX14, MdKNOX17, and MdKNOX18 were clustered in Class KNATM ( Figure 3 ), which lakes the HOX domain ( Figure 1 )
Figure 3.
Neighbor-joining tree representing phylogenetic relationships among KNOX genes from apple, Arabidopsis, and rice.
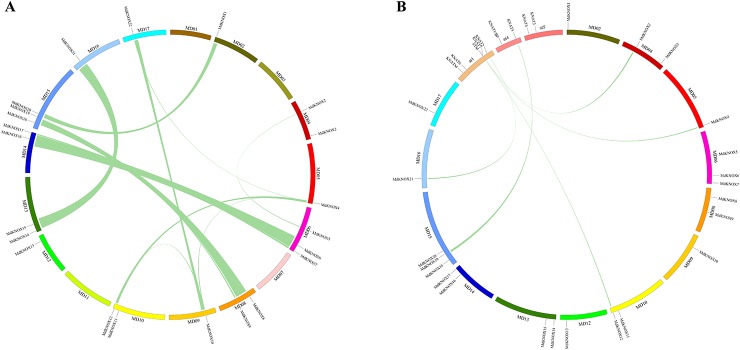
Segmental and tandem duplications are reported to be the predominant mechanisms of diversification of the KNOX gene family (Cannon et al., 2004). To analyze MdKNOX gene duplication events, the Circos software was used to detect duplicated blocks in the apple genome. More than ten pairs of MdKNOX genes, such as MdKNOX1/MdKNOX20, MdKNOX2/MdKNOX5, MdKNOX4/MdKNOX12, MdKNOX7/MdKNOX17, MdKNOX8/MdKNOX18, MdKNOX10/MdKNOX22, MdKNOX12/MdKNOX21, were located in duplicated genomic regions. Chromosomes 1, 3, 5, 7, and 12 did not contain any duplicated genes, whereas Chromosomes 6 and 15 contained the highest number of duplications ( Figure 4A ). Given that Arabidopsis is a well-characterized model plant species, we generated a comparative KNOX synteny map between Arabidopsis and apple to investigate orthologous genes and extract information on evolutionary relationships between the two species. Five pairs of syntenic orthologous genes were matched between the two species, including KNAT5/MdKNOX19, KNAT2/MdKNOX21, STM/MdKNOX12, STM/MdKNOX4, and KNAT7/MdKNOX2 ( Figure 4B ).
Figure 4.
Analysis of evolutionary relationships among KNOX gene family members. Relative positive positions are depicted according to apple chromosomes, colored lines indicate syntenic regions of the apple genome (A). Synteny analysis of KNOX genes between Arabidopsis and apple; relative positive positions are depicted according to apple and Arabidopsis chromosomes, colored lines indicate syntenic regions of the apple and Arabidopsis genomes (B).
MdKNOX Expression Patterns in Different Tissues
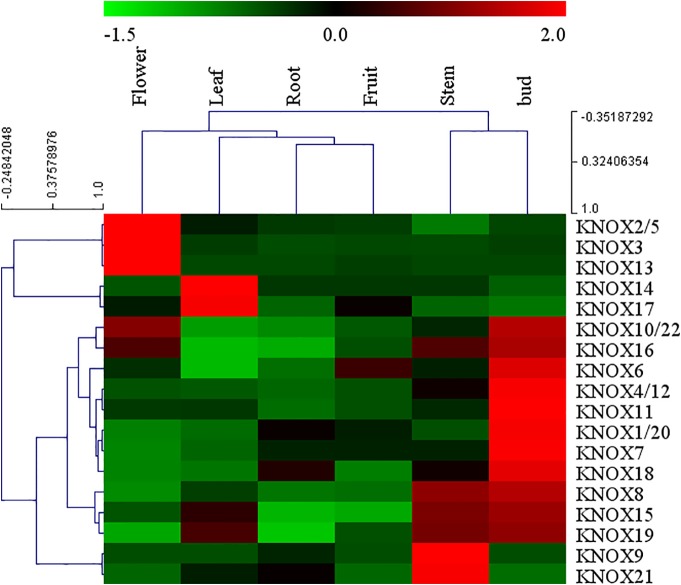
Arabidopsis KNOX genes have been well characterized, whereas little information on expression of apple KNOX genes is available. To elucidate the expression patterns of MdKNOX genes in apple, the expression patterns in a variety of tissues were analyzed by qRT-PCR. A heat map was drawn to visualize the expression profiles of individual MdKNOX genes based on the qRT-PCR data ( Figure 5 ). The majority of (fourteen) MdKNOX genes exhibited strongly preferential expression in the floral bud. MdKNOX8, MdKNOX15, MdKNOX16, and MdKNOX19 were highly expressed in the floral bud and stem. Low expression levels in the root and fruit were recorded for all MdKNOX genes except for MdKNOX16 and MdKNOX12. Only MdKNOX2/5, MdKNOX3, MdKNOX10/22, MdKNOX13, and MdKNOX16 were highly expressed in the flower, and MdKNOX14, MdKNOX15, MdKNOX17, and MdKNOX19 were highly expressed in the leaf.
Figure 5.
MdKNOX gene expression profiles in different tissues. The heat map was generated using MEV software. Relative expression profiles are based on quantitative real-time PCR data.
MdKNOX Expression Patterns During the Flower Induction Period
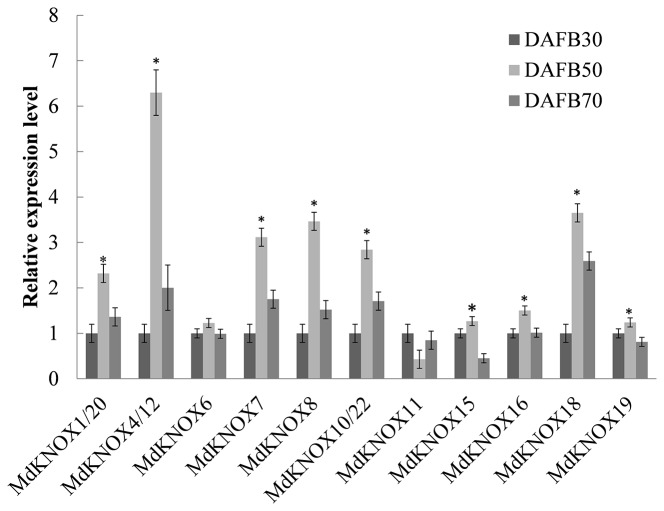
Fourteen MdKNOX genes that exhibited strongly preferential expression in the floral bud were chosen to detect MdKNOX expression patterns during the flower induction period ( Figure 6 ). The transcript level of the majority of these MdKNOX genes was increased, including MdKNOX1/20, MdKNOX4/12, MdKNOX7, MdKNOX8, MdKNOX10/22, MdKNOX15, MdKNOX18, and MdKNOX19. However, the gene expression level of MdKNOX6 was not significantly up-regulated and that of MdKNOX11 showed a downward trend.
Figure 6.
MdKNOX gene expression profiles during the flower induction period. Samples were collected at 30, 50, and 70, days after full bloom (DAFB). Each value represents the mean ± standard error of three replicates. Asterisks (*) means significant difference at the 0.05 level.
Effect of Phytohormone Treatments on MdKNOX Expression During the Flower Induction Period
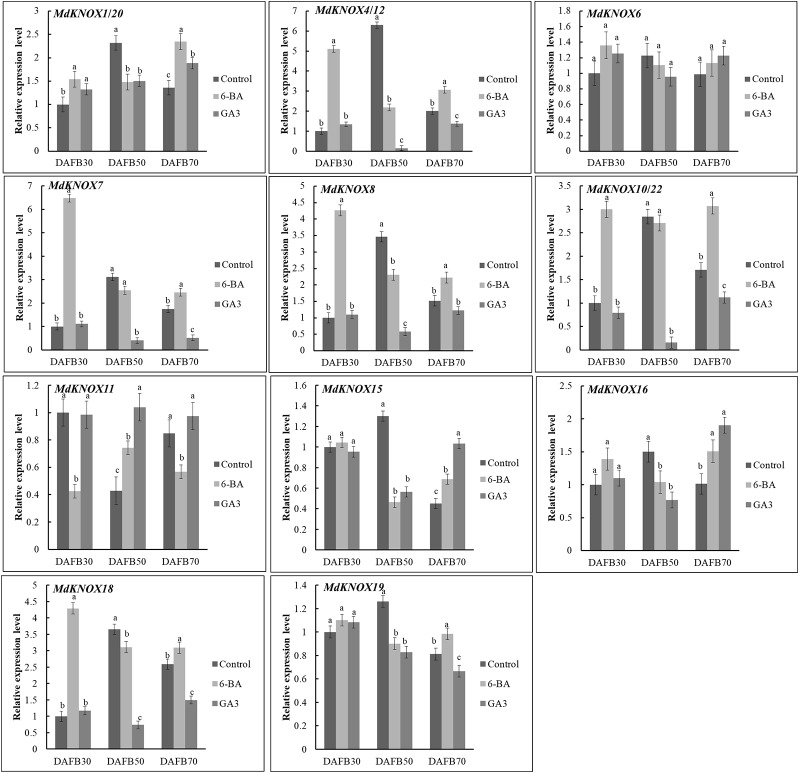
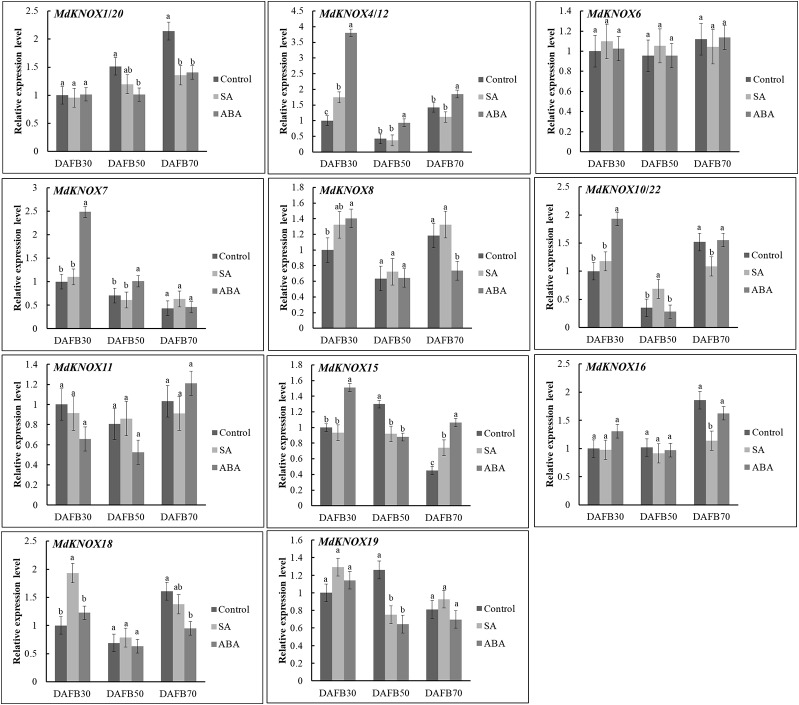
To assess the potential effects of phytohormones on MdKNOX expression during the flower induction period, the transcript levels were estimated after treatment with 6-BA, GA3, ABA, or SA. Treatment with 6-BA significantly increased the transcript levels of MdKNOX4/12, MdKNOX7, MdKNOX8, MdKNOX10/22, and MdKNOX18 at DAFB30 ( Figure 7 ). At DAFB50, all MdKNOX genes showed lower transcript levels compared with that of the control except for MdKNOX11. MdKNOX11 was down-regulated in response to 6-BA treatment in both the early and late sampling periods but was significantly induced at DAFB50. MdKNOX1/20, MdKNOX4/12, MdKNOX7, MdKNOX8, MdKNOX10/22, MdKNOX15, MdKNOX16, MdKNOX18, and MdKNOX19 also showed higher transcript levels than that of the control at DAFB70. In response to exogenously applied GA3 ( Figure 7 ), MdKNOX expression was unaffected at DAFB30, whereas all MdKNOX genes except MdKNOX11 were down-regulated at DAFB50, especially MdKNOX4/12, MdKNOX7, MdKNOX8, MdKNOX10/22, and MdKNOX18. The transcript abundance of MdKNOX4/12, MdKNOX7, MdKNOX8, MdKNOX10/22, and MdKNOX18 remained at lower levels than that of the control at DAFB70. MdKNOX expression patterns varied over time in response to SA and ABA treatment ( Figure 8 ). MdKNOX4/12, MdKNOX15, and MdKNOX18 were initially up-regulated in response to SA treatment and subsequently showed no significant difference at DAFB50 and DAFB70. MdKNOX15 transcription was suppressed at DAFB50. MdKNOX1/20 was significantly inhibited by SA at DAFB70 and MdKNOX19 was significantly inhibited at DAFB50. MdKNOX10/22 was up-regulated by SA at DAFB50, while down-regulated at DAFB70. With regard to ABA ( Figure 8 ), MdKNOX4/12, MdKNOX7, MdKNOX8, MdKNOX10/22, MdKNOX15, and MdKNOX16 were up-regulated at DAFB30. However, no significant difference was observed for all the MdKNOX at subsequent time points between control and ABA-treated group except for MdKNOX15 and MdKNOX19.
Figure 7.
MdKNOX transcript levels in response to 6-benzylaminopurine (6-BA) and gibberellic acid (GA3) treatments. Samples were collected at 30, 50, 70 days after full bloom (DAFB) after 6-BA or GA3 treatment, with water used as a control. Each value represents the mean± standard error of three replicates. Different letters means significant difference at the 0.05 level.
Figure 8.
MdKNOX expression levels in response to SA and ABA treatments. Samples were collected at 30, 50, 70 days after full bloom (DAFB) after SA or ABA treatment, with water used as a control. Each value represents the mean ± standard error of three replicates. Different letters means significant difference at the 0.05 level.
Analysis of the cis-Elements in the MdKNOX Promoters
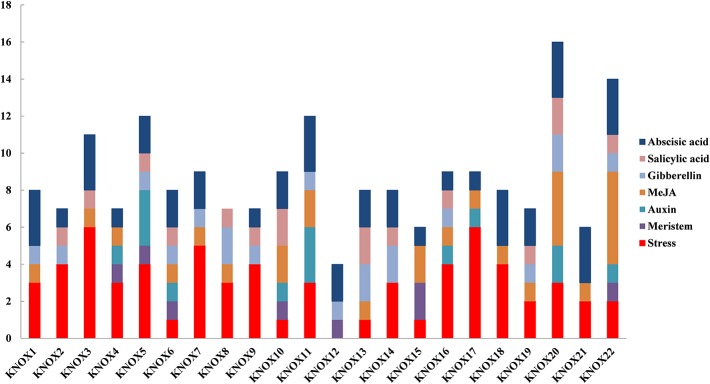
To further investigate the regulatory mechanisms and potential functions of MdKNOX genes, cis-element motifs associated with responses to environmental factors and phytohormones were detected in the 1.5-kb promoter region upstream of the start codon (ATG) ( Figure 9 ). Stress-related elements were detected in the promoters of all MdKNOX genes except for MdKNOX2. Meristem-related cis-elements were also identified in the MdKNOX4, MdKNOX5, MdKNOX6, MdKNOX10, MdKNOX15, and MdKNOX22 promoters. Several hormone-related cis-elements were detected in all MdKNOX genes, including ABA-, SA-, GA-, methyl jasmonate-, and auxin-responsive elements. Among the cis-acting elements involved in hormone-related responses, the ABA-responsive element was present (as one to three copies) in all studied promoters except for MdKNOX8.
Figure 9.
Predicted cis-elements in the MdKNOX promoters. The 1.5 kb sequence upstream from the start codon of MdKNOX genes was analyzed using the PlantCARE database.
MdGRF Directly Binds to the MdKNOX15 and KNOX19 Promoter
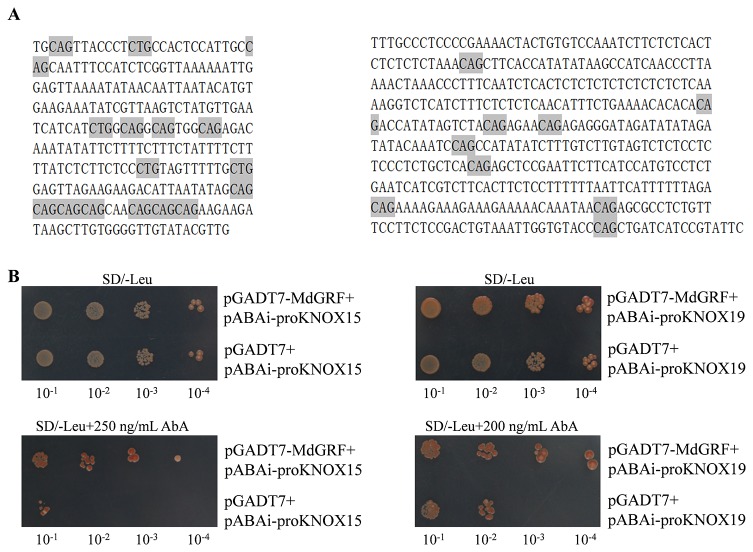
We conducted Y1H assays to test the interaction between the MdGRF protein and MdKNOX15 and MdKNOX19 promoters. The open reading frame of MdGRF was cloned into pGADT7 vector. The promoter fragments of MdKNOX15 and MdKNOX19 ( Figure 10A ) were inserted into the pAbAi vector, respectively. Yeast strains carrying the pGADT7-MdGRF and pAbAi-proKNOX constructs grew normally on selective medium supplemented with AbA (200 ng/ml for KNOX15 and 250 ng/ml for KNOX19), whereas the pGADT7 empty vector control did not grow ( Figure 10B ). These results suggested that MdGRF directly interacted with the MdKNOX promoter.
Figure 10.
MdGRF binds to MdKNOX15 and MdKNOX19 promoters. (A) Promoter sequences of MdKNOX15 and MdKNOX19 genes used for yeast one-hybrid (Y1H) assay. The putative GRF binding sites, core CAG repeats or its reverse complementary sequence CTG were marked with gray shadow. (B) The yeast strains were grown on SD/-Leu and SD/-Leu/+ AbA medium for 3 d.
MdGRF Inhibited the Promoter Activities of MdKNOX15 and MdKNOX19
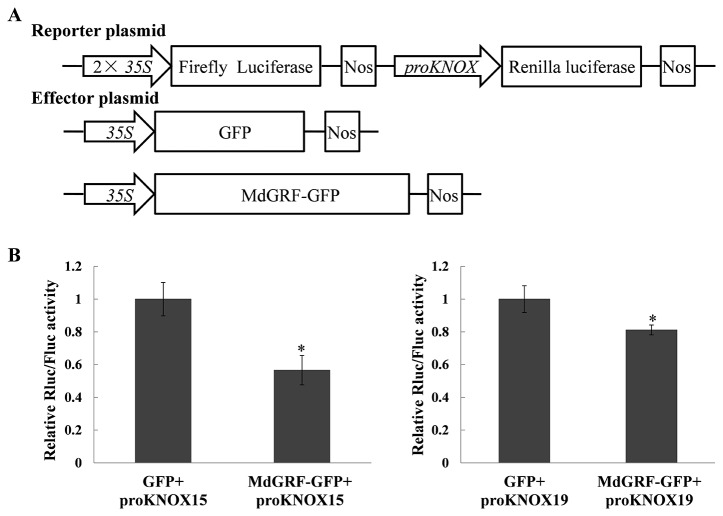
To test whether MdGRF regulated the transcription of MdKNOX genes, a transient transformation assay was conducted. A dual effector–reporter system was established using MdGRF as the effector and the RLuc gene under the control of the MdKNOX promoter as the reporter ( Figure 11A ). The Rluc/Fluc activity was decreased under co-transformation with 35S:GRF-GFP and proKNOX compared with that under co-transformation with 35S:GFP and proKNOX ( Figure 11B ). These results suggested that MdGFR negatively regulated the expression level of MdKNOX15 and MdKNOX19.
Figure 11.
MdGRF inhibited the promoter activities of MdKNOX15 and MdKNOX19. (A) Schematic diagram of the reporter vector and effector vector. (B) Dual-luciferase assays showing that MdGRF inhibits the transcription MdKNOX15 and MdKNOX19 in tobacco leaves. The MdGRF effector vector (MdGRF-GFP) or the control effector vector (GFP) with the reporter vector containing the MdKNOX promoter (proKNOX15 and proKNOX19) were infiltrated into tobacco leaves for analysis of Rluc/Fluc activity. Each experiment was performed in three replicates. Asterisks (*) indicates a significant difference (p <0.05) compared with the control.
Discussion
Identification of Apple KNOX Genes
We identified 22 MdKNOX genes in the apple genome, which is greater than the number of KNOX genes identified in Arabidopsis and rice, and may reflect that the apple genome (881 Mb) is larger than those of rice (466 Mb) and Arabidopsis (12 Mb). The identified MdKNOX genes were unevenly distributed on 12 of the 17 apple chromosomes ( Table 1 ).
Multiple sequence alignment showed that the majority of MdKNOX proteins contained a series of conserved domains: KNOXI, KNOXII, ELK, and HOX domains ( Figure 1 ). The HOX domain is located in the C-terminal portion of the protein and is involved in DNA binding and possibly in homodimer formation (Scofield and Murray, 2006). The ELK domain is located adjacent to the HD domain, spans about 21 amino acids, and is composed of a conserved series of Glu (E), Leu (L), and Lys (K) amino acids. The ELK domain may function as a nuclear localization signal and also is considered to be involved in transcriptional repression, but the precise role of this domain has not been determined (Kerstetter et al., 1994; Sakamoto et al., 1999; Nagasaki et al., 2001). The KNOXI and KNOXII domains are located in the N-terminal half of the protein. KNOXI plays a role in suppressing target gene expression and KNOXII is considered to be necessary for homodimerization (Nagasaki et al., 2001) and transactivation (Scofield and Murray, 2006).
Phylogenesis, Evolution, and Expansion of MdKNOX Gene
An unrooted neighbor-joining tree was constructed from a multiple alignment of the KNOX protein sequences from apple, rice, and Arabidopsis to investigate evolutionary relationships. The analysis separated the KNOX proteins into three groups. MdKNOX1, MdKNOX2, KNOX5, MdKNOX9, MdKNOX19, and MdKNOX20 were clustered with KNAT3, KNAT4, KNAT5, and KNAT7, which belong to the Class II group ( Figure 3 ). The majority of MdKNOX genes were clustered into the Class I group, which consisted of four subfamilies. MdKNOX7, MdKNOX8, MdKNOX14, MdKNOX17, and MdKNOX18 lost the ELK and HOX domains, which clustered in the Class KNATM ( Figure 1 and 3 ). KNOX genes with similar functions and structural motifs showed a tendency to cluster in the same subgroup, which provided a foundation to explore the functions of each MdKNOX gene.
Previous research has shown that gene duplications are important in the evolution of species. Genome-wide duplication events occurred in apple about 60 million years ago, resulting in expansion from nine to 17 chromosomes and diversification of some gene families (Velasco et al., 2010). A number of apple gene duplications have been reported, such as the CCOs (Chen et al., 2018), IDD (Fan et al., 2017a), and GASA families (Fan et al., 2017b). In the present study, eight gene pairs were tentatively identified as duplicated genes ( Figure 4A ). Gene duplications and expansion resulted in MdKNOX gene clusters and increased the diversification of MdKNOX gene structures and functions.
Genomic comparisons with orthologous genes from well-studied plant species may provide a valuable reference for newly identified genes (Koonin, 2005). Thus, the functions of MdKNOX were inferred by comparative genomic analyses with the KNOX genes from Arabidopsis. Five orthologous gene pairs between Arabidopsis and apple were identified ( Figure 4B ), which suggested that the genes in question may share a common ancestor their functions have been conserved during evolution. Although many genetic prediction resources are available, additional research is needed to determine the specific function of each gene.
MdKNOX Gene Expression Profiles and Potential Functions
Given the gene functional diversity, all members of the MdKNOX gene family need to be further functionally characterized. Analysis of tissue expression patterns of MdKNOX genes may provide insights into their possible functions. The majority of MdKNOX family members showed high transcript levels in floral buds, whereas extremely low transcript levels were detected in roots ( Figure 5 ). We observed some differences between the present results and previous reports for other plant species that Class II KNOX genes in angiosperms are expressed in differentiating organs, including leaves, stems, flowers, and roots (Kerstetter et al., 1994). On the other hand, according to the more accurate KNAT3 promoter-driven GUS staining patterns, the Class II KNOX gene KNAT3 is highly expressed in cotyledons, and apical and floral tissues, and is moderately expressed in roots. Moreover, light has a significant effect on the expression profile of KNAT3 (Serikawa et al., 1997). Therefore, we inferred that the developmental stage, sampling method, and species specificity may affect the experimental results. Despite these differences, detection of high transcript levels in floral buds implied that the majority of genes (MdKNOX1/20, MdKNOX4/12, MdKNOX6, MdKNOX7, MdKNOX8, MdKNOX10/22, MdKNOX11, MdKNOX15, MdKNOX16, MdKNOX18, and MdKNOX19) were involved in the regulation of flowering.
Insufficient production of flower buds is an intractable problem in the apple industry. The physiological differentiation of apple flower buds is essential for flowering and fruiting. Therefore, we analyzed the expression of the MdKNOX genes that were highly expressed in floral buds during the flower induction period (floral bud physiological differentiation, at DAFB30, DAFB50, and DAFB70). The genes were highly induced at 50 DAFB ( Figure 6 ), suggesting that these genes may play an active role in floral induction. Several phytohormone-associated cis-element motifs were predicted within the MdKNOX promoters ( Figure 9 ). In addition, we analyzed the expression profiles under different hormone treatments. The exogenous plant hormone 6-BA promotes flower bud formation (Li et al., 2019), whereas GA3 reduces flowering rates in apple (Zhang et al., 2016). MdKNOX1/20 showed identical expression patterns in response to 6-BA and GA3 treatments. The transcription of MdKNOX1/20 was strongly induced by both 6-BA and GA3 at DAFB30 and DAFB70, but was inhibited at DAFB50. MdKNOX4/12 showed a similar expression pattern to that of MdKNOX1/20 in response to 6-BA treatment. Treatment with 6-BA also increased the transcript levels of MdKNOX4/12, MdKNOX7, MdKNOX8, MdKNOX10/22, and MdKNOX18 in the initial stage of flower induction (DAFB30), whereas transcription of the genes was suppressed at the intermediate stage of flower induction (DAFB50). Only MdKNOX11 was down-regulated by 6-BA (in the early and late sampling stages), but was induced at 50 DAFB. With regard to MdKNOX15, MdKNOX16, and MdKNOX19, 6-BA treatment affected their expression at 30 DAFB, and each gene was suppressed at 50 DAFB by 6-BA. This finding is similar to previously reported results, for example, KNAT3 transcript levels are decreased in response to exposure to kinetin (Truernit et al., 2006). These results implied that MdKNOX genes were regulated by 6-BA and might also regulate the 6-BA hormone signal. All MdKNOX genes did not show a significant difference in transcript level at DAFB30 in response to GA3 treatment, which is consistent with a previous report that GA does not influence KNAT promoter activity (Truernit et al., 2006). The transcription of MdKNOX1/20, MdKNOX4/12, MdKNOX7, MdKNOX8, MdKNOX10/22, MdKNOX15, MdKNOX16, MdKNOX18, and MdKNOX19 was inhibited at DAFB50 ( Figure 7 ). These results suggested that GA3 inhibited floral induction in apple, which might be partly achieved by inhibiting the expression of MdKNOX genes. Environmental factors, such as drought (which is common on the Loess Plateau), stimulates ABA accumulation and triggers an early flowering response (Verslues and Juenger, 2011). Only MdKNOX4/12, MdKNOX7, MdKNOX10/22, and MdKNOX15 were induced by exogenous ABA treatment. This was especially the case for MdKNOX4/12, for which the transcript level increased over time in response to ABA treatment. Exogenous SA treatment induces accumulation of SA and accelerates the transition to flowering (MartãNez et al., 2004). MdKNOX18 was induced by exogenous SA at 30 DAFB, whereas all other MdKNOX genes were not affected. The transcript levels of MdKNOX15 and MdKNOX19 were significantly suppressed at the intermediate stage of flower induction (50 DAFB) ( Figure 8 ).
MdKNOX Gene Under the Transcriptional Regulation of MdGRF
KNOX gene expression is regulated at multiple levels to prevent misexpression. Several regulators of KNOX gene expression have been identified, including MYB domain transcription factors (Waites et al., 1998), ASYMETRIC LEAVES1 (AS1) (Byrne et al., 2002), CUP-SHAPED COTYLEDON (CUC) (Hibara et al., 2003), and GROWTH-REGULATING FACTOR (GRF) (Kuijt et al., 2014), YABBY (Kumaran et al., 2002), and FIE and CLF (Katz et al., 2004). The plant-specific GRF transcription factors, which are negative regulators of KNOX genes, were identified for their roles in developmental processes, including root, stem, and leaf development, flower and seed formation, and coordination of growth processes under adverse environmental conditions (Omidbakhshfard et al., 2015). The MdGRF genes in the apple genome were identified in previous work in our laboratory (Zheng et al., 2018). The MdGRF gene (MD00G1142400) used in the present research was homologous to Arabidopsis AtGRF5 (full-length sequence identity was 30%, and the characteristic WRC and QLQ domains were highly conserved), and showed a negative correlation with MdKNOX15 and MdKNOX19 at the transcript level in transcriptome data (data not shown). Therefore, the interaction between MdKNOX and MdGR was evaluated in the current study. As a transcriptional regulator, MdGRF protein could direct localization of the GFP marker protein to the nucleus ( Supplementary Figure 2 ). As shown in Figure 10 , the promoter regions of MdKNOX15 and MdKNOX19 contained the putative GRF binding sites, core ‘CAG' repeats or its reverse complementary sequence ‘CTG' (Kuijt et al., 2014). Therefore, MdKNOX15 and MdKNOX19 were chosen to detect the interaction. MdGRF protein interacted with the MdKNOX15 and MdKNOX19 promoter in yeast. In rice, KNOX gene expression is down-regulated by GRF overexpression and is up-regulated by RNA interference (RNAi)-mediated GRF silencing. In the present study, the promoter activities of MdKNOX15 and MdKNOX19 were inhibited by MdGRF ( Figure 11 ). Taken together, these results suggest that GRF–KNOX interactions might be conserved both in herbaceous and woody plants.
Conclusion
Twenty-two KNOX genes were identified in the apple (M. domestica) genome. The MdKNOX members were divided into three subfamilies based on their phylogenetic relationships. Duplications have likely been important for the expansion and evolution of MdKNOX genes. The majority of MdKNOX genes exhibit strongly preferential expression in floral buds and are significantly up-regulated during the flower induction period, implying that they perform specific roles in floral induction. Furthermore, most MdKNOX genes are responsive to flowering-related and stress-related hormone treatments, suggesting that the genes are involved in flowering and stress response processes. The putative upstream regulatory factor MdGRF is able to bind directly to the promoter of MdKNOX15 and MdKNOX19, and inhibits their transcriptional activities, as confirmed by Y1H and dual-luciferase assays. To our knowledge, this study is the first systematic and in-depth analysis of apple KNOX genes. The data provide useful information for future functional characterization of apple KNOX genes.
Data Availability Statement
All datasets generated for this study are included in the article/ Supplementary Material .
Author Contributions
PJ, XR, and MH designed the experiments and analyzed the data. PJ, CZ, LX, XZ, YL, and KS collected the sample materials and completed the field and laboratory measurements. PJ, DZ, NA, and XR wrote the manuscript. All authors have read and approved the manuscript.
Funding
This study was sponsored by the Chinese Postdoctoral Science Foundation (2018M631207), the Ecological Adaptability Selection of Apple Superior Stock and Scion Combinations in the Loess Plateau (A2990215082), the Screening and Interaction Molecular Mechanism of Apple Stock and Scion Combinations (K3380217027), and the National Apple Industry Technology System of the Agriculture Ministry of China (CARS-27).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00128/full#supplementary-material
Predicted dimensional structures of MdKNOX proteins.
Subcellular localization assays of MdGRF in Nicotiana benthamiana leaves. Leaves from 5-week-old tobacco plants were infiltrated with Agrobacterium strain GV3103 harboring the 35S:GRF-GFP or 35S:GFP expression cassette, as described in section 2.7. DAPI (4',6-diamidino-2-phenylindole) was used to stain the nucleus. Green fluorescent protein (GFP) signals in transformed tobacco leaves were detected with a LEICA TCS SP8 confocal microscope (Wetzlar, Germany).
Abbreviations
KNOX, Knotted1-like Homeobox; GRF, Growth-regulating factor; 6-BA, 6-Benzylaminopurine; GA3, gibberellic acid; ABA, abscisic acid; SA, salicylic acid; AbA, Aureobasidin A; DAFB, days after full bloom.
References
- Azarakhsh M., Kirienko A. N., Zhukov V. A., Lebedeva M. A., Dolgikh E. A., Lutova L. A. (2015). KNOTTED1-LIKE HOMEOBOX 3: a new regulator of symbiotic nodule development. J. Exp. Bot. 66, 7181–7195. 10.1093/jxb/erv414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin T. R., Affolter M. (2016). Homeodomain proteins: an update. Chromosoma 125, 497–521. 10.1007/s00412-015-0543-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Williams N., Misleh C., Li W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. 10.1093/nar/gkl198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M. K., Poethig R. S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development 119, 823–831. [Google Scholar]
- BellesBoix E., Hamant O., Witiak S. M., Morin H., Traas J., Pautot V. (2006). KNAT6: An arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell. 18, 1900. 10.1105/tpc.106.041988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M. F., Badis G., Gehrke A. R., Talukder S., Philippakis A. A., Peña-Castillo L., et al. (2008). Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133, 1266–1276. 10.1016/j.cell.2008.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N., Hake S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell. 21, 1647–1658. 10.1105/tpc.109.068221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M. E., Barley R., Curtis M., Arroyo J. M., Dunham M., Hudson A., et al. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. 10.1038/35050091 [DOI] [PubMed] [Google Scholar]
- Byrne M. E., Joseph S., Martienssen R. A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis . Development 129, 1957–1965. Retrieved from https://dev.biologists.org/content/129/8/1957 [DOI] [PubMed] [Google Scholar]
- Cannon S. B., Mitra A., Baumgarten A., Young N. D., May G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana . BMC Plant Biol. 4, 10–10. 10.1186/1471-2229-4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R. L., Gago G. M., Palena C. M., Gonzalez D. H. (1998). Homeoboxes in plant development. Biochim. Biophys. Acta 1442, 1–19. 10.1016/S0167-4781(98)00119-5 [DOI] [PubMed] [Google Scholar]
- Chen H., Zuo X., Shao H., Fan S., Ma J., Zhang D., et al. (2018). Genome-wide analysis of carotenoid cleavage oxygenase genes and their responses to various phytohormones and abiotic stresses in apple (Malus domestica). Plant Physiol. Biochem. 123, 81–93. 10.1016/j.plaphy.2017.12.001 [DOI] [PubMed] [Google Scholar]
- Dachan K., Young-Hyun C., Hojin R., Yoonhee K., Tae-Houn K., Ildoo H. (2013). BLH1 and KNAT3 modulate ABA responses during germination and early seedling development in Arabidopsis. Plant J. 75, 755–766. 10.1111/tpj.12236 [DOI] [PubMed] [Google Scholar]
- Di Giacomo E., Laffont C., Sciarra F., Iannelli M. A., Frugier F., Frugis G. (2017). KNAT3/4/5‐like class 2 KNOX transcription factors are involved in Medicago truncatula symbiotic nodule organ development. New Phytol. 213, 822–837. 10.1111/nph.14146 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S., Zhang D., Xing L., Qi S., Du L., Wu H., et al. (2017. a). Phylogenetic analysis of IDD gene family and characterization of its expression in response to flower induction in Malus . Mol. Genet. Genomics 292, 755–771. 10.1007/s00438-017-1306-4 [DOI] [PubMed] [Google Scholar]
- Fan S., Zhang D., Zhang L., Gao C., Xin M., Tahir M. M., et al. (2017. b). Comprehensive analysis of GASA family members in the Malus domestica genome: identification, characterization, and their expressions in response to apple flower induction. BMC Genomics 18, 827. 10.1186/s12864-017-4213-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumizu C., Alvarez J. P., Sakakibara K., Bowman J. L. (2015). Antagonistic roles for KNOX1 and KNOX2 genes in patterning the land plant body plan following an ancient gene duplication. PloS Genet. 11, e1004980. 10.1371/journal.pgen.1004980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Yang X., Zhao W., Lang T., Samuelsson T. (2015). Evolution, diversification, and expression of KNOX proteins in plants. Front. Plant Sci. 6, 882. 10.3389/fpls.2015.00882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S.-Y., Huang G.-Q., Sun X., Qin L.-X., Li Y., Zhou L., et al. (2014). Cotton KNL1, encoding a class II KNOX transcription factor, is involved in regulation of fibre development. J. Exp. Bot. 65, 4133–4147. 10.1093/jxb/eru182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L., Han Z., Zhang L., Downie B., Zhao T. (2013). Functional analysis of the 5′ regulatory region of the maize GALACTINOL SYNTHASE2 gene. Plant Sci. 213, 38–45. 10.1016/j.plantsci.2013.09.002 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2010). KNOX genes: versatile regulators of plant development and diversity. Development 137, 3153–3165. 10.1242/dev.030049 [DOI] [PubMed] [Google Scholar]
- Hayashi S., Scott M. P. (1990). What determines the specificity of action of Drosophila homeodomain proteins? Cell 63, 883–894. 10.1016/0092-8674(90)90492-W [DOI] [PubMed] [Google Scholar]
- Hibara K., Takada S., Tasaka M. (2003). CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J. 36, 687–696. 10.1046/j.1365-313X.2003.01911.x [DOI] [PubMed] [Google Scholar]
- Himmelbach A., Hoffmann T., Leube M., Höhener B., Grill E. (2002). Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 21, 3029. 10.1093/emboj/cdf316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., et al. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol.: CB. 15, 1560–1565. 10.1016/j.cub.2005.07.023 [DOI] [PubMed] [Google Scholar]
- Katz A., Oliva M., Mosquna A., Hakim O., Ohad N. (2004). FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 37, 707–719. 10.1111/j.1365-313X.2003.01996.x [DOI] [PubMed] [Google Scholar]
- Kerstetter R., Vollbrecht E., Lowe B., Veit B., Yamaguchi J., Hake S. (1994). Sequence analysis and expression patterns divide the maize knotted1-like homeobox genes into two classes. Plant Cell. 6, 1877–1887. 10.1105/tpc.6.12.1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Cho Y.-H., Ryu H., Kim Y., Kim T.-H., Hwang I. (2013). BLH1 and KNAT3 modulate ABA responses during germination and early seedling development in Arabidopsis. Plant J. 75, 755–766. 10.1111/tpj.12236 [DOI] [PubMed] [Google Scholar]
- Koonin E. V. (2005). Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 39, 309–338. 10.1146/annurev.genet.39.073003.114725 [DOI] [PubMed] [Google Scholar]
- Krenek P., Samajova O., Luptovciak I., Doskocilova A., Komis G., Samaj J. (2015). Transient plant transformation mediated by Agrobacterium tumefaciens: Principles, methods and applications. Biotechnol. Adv. 33, 1024–1042. 10.1016/j.biotechadv.2015.03.012 [DOI] [PubMed] [Google Scholar]
- Krzywinski M., Schein J. I. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijt S. J., Greco R., Agalou A., Shao J., Cc T. H., Overnäs E., et al. (2014). Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant Physiol. 164, 1952. 10.1104/pp.113.222836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Kushalappa K., Godt D., Pidkowich M. S., Pastorelli S., Hepworth S. R., et al. (2007). The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. Plant Cell. 19, 2719–2735. 10.1105/tpc.106.048769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran M. K., Bowman J. L., Sundaresan V. (2002). YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell. 14, 2761–2770. 10.1105/tpc.004911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba S., Fukumoto M., Honda C., Yamaguchi I., Sakamoto T., Kanomurakami Y. (1998). Decreased GA1 content caused by the overexpression of OSH1 is accompanied by suppression of GA 20-oxidase gene expression. Plant Physiol. 117, 1179–1184. 10.1104/pp.117.4.1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C. J., Jr., Roos D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E., Bhargava A., Qiang W., Friedmann M. C., Forneris N., Savidge R. A., et al. (2012). The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytologist 194, 102–115. 10.1111/j.1469-8137.2011.04016.x [DOI] [PubMed] [Google Scholar]
- Li Y., Zhang D., An N., Fan S., Zuo X., Zhang X., et al. (2019). Transcriptomic analysis reveals the regulatory module of apple (Malus × domestica) floral transition in response to 6-BA. BMC Plant Biol. 19, 93. 10.1186/s12870-019-1695-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego Calif.) 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Ma C., Meir S., Xiao L., Tong J., Liu Q., Reid M. S., et al. (2015). A KNOTTED1-LIKE HOMEOBOX protein regulates abscission in tomato by modulating the auxin pathway. Plant Physiol. 167, 844–853. 10.1104/pp.114.253815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani E., Hake S. (2008). KNOX lost the OX: the arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell. 20, 875–887. 10.1105/tpc.108.058495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MartãNez C., Pons E., Prats G., Leã³ N. ,. J. (2004). Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J. 37, 209–217. 10.1046/j.1365-313X.2003.01954.x [DOI] [PubMed] [Google Scholar]
- Nagasaki H., Sakamoto T., Sato Y., Matsuoka M. (2001). Functional analysis of the conserved domains of a rice KNOX homeodomain protein, OSH15. Plant Cell. 13, 2085–2098. 10.1105/TPC.010113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidbakhshfard M. A., Proost S., Fujikura U., Mueller-Roeber B. (2015). Growth-regulating factors (GRFs): a small transcription factor family with important functions in plant biology. Mol. Plant 8, 998–1010. 10.1016/j.molp.2015.01.013 [DOI] [PubMed] [Google Scholar]
- Osnato M., Stile M. R., Wang Y., Meynard D., Curiale S., Guiderdoni E., et al. (2010). Cross talk between the KNOX and ethylene pathways is mediated by intron-binding transcription factors in barley. Plant Physiol. 154, 1616. 10.1104/pp.110.161984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin F. M., Hart J. K., Horner H. T., Davies P. J., Hannapel D. J. (2003). Overexpression of a knotted-like homeobox gene of potato alters vegetative development by decreasing gibberellin accumulation. Plant Physiol. 132, 106–117. 10.1104/pp.102.015560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Nishimura A., Tamaoki M., Kuba M., Tanaka H., Iwahori S., et al. (1999). The conserved KNOX domain mediates specificity of tobacco KNOTTED1-type homeodomain proteins. Plant Cell. 11, 1419–1432. 10.1105/tpc.11.8.1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield S., Murray J. A. H. (2006). KNOX gene function in plant stem cell niches. Plant Mol. Biol. 60, 929–946. 10.1007/s11103-005-4478-y [DOI] [PubMed] [Google Scholar]
- Serikawa K. A., Zambryski P. C. (1997). Domain exchanges between KNAT3 and KNAT1 suggest specificity of the kn1-like homeodomains requires sequences outside of the third helix and N-terminal arm of the homeodomain. Plant J.: Cell Mol. Biol. 11, 863–869. 10.1046/j.1365-313X.1997.11040863.x [DOI] [PubMed] [Google Scholar]
- Serikawa K. A., Martinez-Laborda A., Kim H. S., Zambryski P. C. (1997). Localization of expression of KNAT3, a class 2 knotted1-like gene. Plant J.: Cell Mol. Biol. 11, 853–861. 10.1046/j.1365-313X.1997.11040853.x [DOI] [PubMed] [Google Scholar]
- Truernit E., Haseloff J. (2007). A role for KNAT class II genes in root development. Plant Signal. Behav. 2, 10–12. 10.4161/psb.2.1.3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Siemering K. R., Hodge S., Grbic V., Haseloff J. (2006). A map of KNAT gene expression in the Arabidopsis root. Plant Mol. Biol. 60, 1–20. 10.1007/s11103-005-1673-9 [DOI] [PubMed] [Google Scholar]
- Tsuda K., Hake S. (2015). Diverse functions of KNOX transcription factors in the diploid body plan of plants. Curr. Opin. Plant Biol. 27, 91–96. 10.1016/j.pbi.2015.06.015 [DOI] [PubMed] [Google Scholar]
- Tsuda K., Ito Y., Sato Y., Kurata N. (2011). Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell. 23, 4368–4381. 10.1105/tpc.111.090050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Kurata N., Ohyanagi H., Hake S. (2014). Genome-wide study of KNOX regulatory network reveals brassinosteroid catabolic genes important for shoot meristem function in rice. Plant Cell. 26, 3488–3500. 10.1105/tpc.114.129122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R., Zharkikh A., Affourtit J., Dhingra A., Cestaro A., Kalyanaraman A., et al. (2010). The genome of the domesticated apple (Malus x domestica Borkh.). Nat. Genet. 42, 833–839. 10.1038/ng.654 [DOI] [PubMed] [Google Scholar]
- Verslues P. E., Juenger T. E. (2011). Drought, metabolites, and Arabidopsis natural variation: a promising combination for understanding adaptation to water-limited environments. Curr. Opin. Plant Biol. 14, 240–245. 10.1016/j.pbi.2011.04.006 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E., Veit B., Sinha N., Hake S. (1991). The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350, 241–243. 10.1038/350241a0 [DOI] [PubMed] [Google Scholar]
- Waites R., Selvadurai H. R., Oliver I. R., Hudson A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum . Cell 93, 779–789. 10.1016/S0092-8674(00)81439-7 [DOI] [PubMed] [Google Scholar]
- Wang Y., Tang H., Debarry J. D., Tan X., Li J., Wang X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49–e49. 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Chen X., Zhang Z., Liu D., Song G., Kong X., et al. (2015). A MADS-box gene NtSVP regulates pedicel elongation by directly suppressing a KNAT1-like KNOX gene NtBPL in tobacco (Nicotiana tabacum L.). J. Exp. Bot. 66, 6233–6244. 10.1093/jxb/erv332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Yamaguchi M., Grienenberger E., Martone P. T., Samuels A. L., Mansfield S. D. (2020). The Class II KNOX genes KNAT3 and KNAT7 work cooperatively to influence deposition of secondary cell walls that provide mechanical support to Arabidopsis stems. Plant J. 101 (2), 293–309. 10.1111/tpj.14541 [DOI] [PubMed] [Google Scholar]
- Watillon B., Kettmann R., Boxus P., Burny A. (1997). Knotted1-like homeobox genes are expressed during apple tree (Malus domestica [L] Borkh) growth and development. Plant Mol. Biol. 33, 757–763. 10.1023/A:1005708700636 [DOI] [PubMed] [Google Scholar]
- Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., et al. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol.: CB. 15, 1566–1571. 10.1016/j.cub.2005.07.060 [DOI] [PubMed] [Google Scholar]
- Zhang S., Zhang D., Fan S., Du L., Shen Y., Xing L., et al. (2016). Effect of exogenous GA3 and its inhibitor paclobutrazol on floral formation, endogenous hormones, and flowering-associated genes in ‘Fuji' apple (Malus domestica Borkh.). Plant Physiol. Biochem. 107, 178–186. 10.1016/j.plaphy.2016.06.005 [DOI] [PubMed] [Google Scholar]
- Zheng L., Ma J., Song C., Zhang L., Gao C., Zhang D., et al. (2018). Genome-wide identification and expression analysis of GRF genes regulating apple tree architecture. Tree Genet. Genomes 14, 54. 10.1007/s11295-018-1267-8 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Predicted dimensional structures of MdKNOX proteins.
Subcellular localization assays of MdGRF in Nicotiana benthamiana leaves. Leaves from 5-week-old tobacco plants were infiltrated with Agrobacterium strain GV3103 harboring the 35S:GRF-GFP or 35S:GFP expression cassette, as described in section 2.7. DAPI (4',6-diamidino-2-phenylindole) was used to stain the nucleus. Green fluorescent protein (GFP) signals in transformed tobacco leaves were detected with a LEICA TCS SP8 confocal microscope (Wetzlar, Germany).
Data Availability Statement
All datasets generated for this study are included in the article/ Supplementary Material .