Abstract
Purpose:
Cell size is a fundamental characteristic of all tissues, and changes in cell size in cancer reflect tumor status and response to treatments, such as apoptosis and cell cycle arrest. Unfortunately, cell size can currently be obtained only by pathologic evaluation of tumor tissue samples obtained invasively. Previous imaging approaches are limited to preclinical MRI scanners or require relatively long acquisition times that are impractical for clinical imaging. There is a need to develop cell size imaging for clinical applications.
Methods:
We propose a clinically feasible IMPULSED (Imaging Microstructural Parameters Using Limited Spectrally Edited Diffusion) approach that can characterize mean cell sizes in solid tumors. We report the use of a combination of pulse sequences, using different gradient waveforms implemented on clinical MRI scanners and analytical equations based on these waveforms to analyze diffusion-weighted MRI signals and derive specific microstructural parameters such as cell size. We also describe comprehensive validations of this approach using computer simulations, cell experiments in vitro, and animal experiments in vivo and demonstrate applications in pre-operative breast cancer patients.
Results:
With fast acquisitions (~ 7 mins), IMPULSED can provide high-resolution (1.3 mm in-plane) mapping of mean cell size of human tumors in vivo on clinical 3T MRI scanners. All validations suggest IMPULSED provides accurate and reliable measurements of mean cell size.
Conclusion:
The proposed IMPULSED method can assess cell size variations in the tumor of breast cancer patients, which may have the potential to assess early response to neoadjuvant therapy.
Keywords: MRI, cell size, density, diameter, diffusion, IMPULSED, OGSE, oscillating gradient
Introduction
Cell size is a basic feature of living cells that plays an important role from the molecular to the organ level, including cellular metabolism, (1) proliferation, (2) and tissue growth (3). Cell size may vary significantly during disease progression or after therapy. For example, cells swell significantly after acute stroke (4,5) and cell shrinkage is a hallmark of apoptotic cell death (6). For cancer diagnosis and prognosis, cell size is of interest because it varies during mitosis and before death, and thus may provide a unique means to evaluate tumor progression and response to treatments. Measurements of cell sizes are reportedly capable of differentiating cancer types (7) and monitoring tumor early therapeutic response by detecting treatment-induced apoptosis (8,9) or mitotic-arrest (10). Therefore, quantitative microstructural measurements such as cell size may provide specific means to probe the status of cancerous tissues and would be of potential value in preclinical and clinical applications. Currently, such microstructural information is obtained during conventional clinical care only via invasive biopsies, which are limited not only by the potential to miss important changes due to tumor heterogeneity and the small sample size of each specimen, but also may introduce various clinical complications, including pain, hemorrhage, infection, and even death (11). Therefore, a non-invasive imaging technique capable of characterizing tissue microstructural information would be of great interest to clinicians.
Diffusion-weighted magnetic resonance imaging (DWI) is an exogenous-agent-free non-invasive imaging technique that provides unique capabilities to probe biological tissue microstructure by evaluating the degree of restriction and hindrance to the free motion of randomly diffusing water molecules. Values of the apparent diffusion coefficient (ADC), a metric obtained using DWI, have been found to be sensitive to cell density and hence are widely used to evaluate cellularity changes after anti-cancer treatment (12–15). However, ADC represents an overall diffusion property of water molecules inside each image voxel, and ADC values are influenced by several tissue parameters simultaneously, including but not limited to cell size (16), cell membrane permeability (17), intra- and extracellular diffusion coefficients (18), and intracellular volume fraction (19). As a result, ADC and tumor cellularity are not always strongly correlated (20–22) so that ADC does not reliably provide specific information on cell size and density. Recently, numerous attempts have been made to enhance the specificity of DWI measurements, such as the DDR (23), VERDICT (24–26), qTDS and IMPULSED (27,28), and POMACE approaches (29). Some of these e.g. qTDS and IMPULSED, exploit the dependence of ADC on the time scale (the diffusion time) over which diffusion affects the measured signals, which in principle enables the derivation of the spatial scales of restrictions to free displacements (27,28). However, previous reports of their applications were either implemented on animal MRI scanners only and used much stronger diffusion gradient amplitudes than are available on clinical MRI scanners, or the total acquisition times were long for clinical applications. Therefore, there is still a need to develop a fast (< 10 mins) and quantitative DWI method that is capable of measuring cell size on clinical MRI machines.
Here, we introduce a modified implementation of IMPULSED for clinical MR imaging. Instead of using apodised cosine oscillating diffusion gradients as reported earlier (27,28), we used IMPULSED acquisitions that incorporate cosine-modulated oscillating trapezoidal diffusion gradients (30–32) that are readily capable of running on clinical MRI machines as shown in Figure 1. An IMPULSED protocol incorporates a set of diffusion weighted imaging sequences, each of which uses a different diffusion time, with a diffusion time range that makes the DWI images highly sensitive to variations in cell size in human tissues, and from which mapping cell size and density can be derived. In the current work, we demonstrate a practical imaging protocol that combines different DWI sequences along with new analytical equations that link the DWI signals using the real gradient waveforms to specific microstructural parameters such as cell size. We also provide validations of the IMPULSED method using computer simulations and experimental measurements of cells in vitro and animal models in vivo, along with practical demonstrations of clinical applications in breast cancer patients.
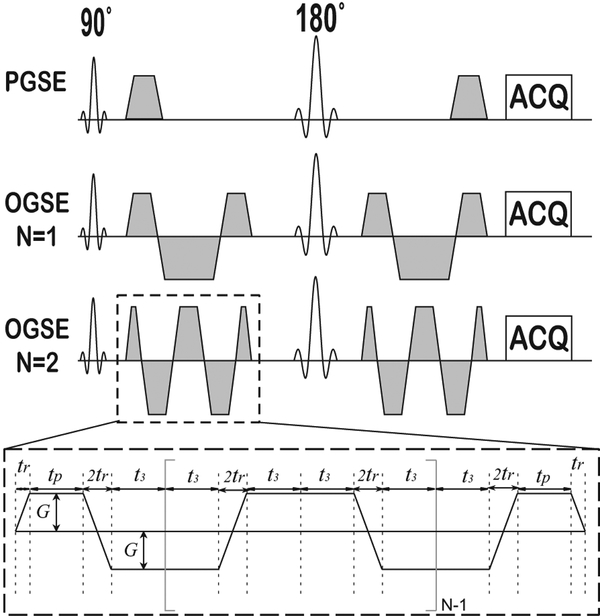
Figure 1.
Diagram of the pulse sequences used in the IMPULSED method. In addition to conventional PGSE acquisitions, OGSE acquisitions with two frequencies (N = 1 and 2) are used.
Methods
Pulse sequence
Figure 1 shows the pulse sequences used to acquire DWI data for the IMPULSED method on clinical 3T scanners. In addition to a conventional pulsed gradient spin echo (PGSE) sequence which measures ADC over longer diffusion times, IMPULSED also uses cosine-modulated, trapezoidal, oscillating gradients in spin echo sequences (OGSE) to measure ADC over different, shorter diffusion times. The combination of longer and shorter diffusion times ensures that ADC values from each sequence will differ, and these differences then reflect the length scales of major restrictions to diffusion, which in tumors correspond to cell sizes (33). This combination enables detection of a broad range of length scales, providing more comprehensive information on tissue microstructure than single measurements of ADC (27,28). For all diffusion sequences, G is the gradient strength, δ is the duration of each diffusion gradient, Δ is the separation of two gradients, tr is the gradient rise time (= 0.9 ms on Philips Achieva 3T MRI scanner), tp is the duration time of the first gradient plateau, and t3 = tp + tr/2 for OGSE sequences. N is the number of cycles in each diffusion gradient in the OGSE sequence. for the PGSE sequence and for the OGSE sequences (30,31) shown in Figure 1.
Theory
Quantitative information on cell size and density are obtained by fitting a simple model of tissue water to the DWI experimental data. Like previous reports (24,27,29,34), DWI signals in IMPULSED are modeled as the sum of signals arising from two compartments, i.e., intracellular and extracellular spaces. The measured DWI signal S is
| [1] |
where Sin, and Sex are the signal magnitudes per volume from the intracellular and extracellular spaces, respectively, and vin is intracellular water fraction. Note that water exchange between intra- and extracellular spaces is ignored but does not affect the estimation of mean cancer cell size although it may bias the estimation of cell density (35).
Modeling intracellular diffusion (the 1st term in Eq.[1]):
Water molecules are restricted inside cells because cell membranes have only finite permeabilities. For simplification, cancer cells are usually modeled as impermeable spheres and analytical expressions similar to previous reports (34) may be derived to link DWI signals and underlying microstructural parameters such as cell size. The derived analytical equations for cosine-modulated, trapezoidal-shaped OGSE sequences can be found in the Supporting Information Eq. S5 and S6. The computer simulation validation of the accuracy of PGSE and OGSE equations using real gradient waveforms are shown in Supporting Information Figure S1 and S2.
Modeling extracellular diffusion (the 2nd term in Eq.[1]).
It is challenging to obtain an explicit analytical form to describe extracellular diffusion. When the diffusion time range is limited, previous studies have suggested the extracellular diffusion coefficient shows an approximately linear dependence on the gradient frequency (proportional to the inverse of diffusion time) (23,36). However, partly due to hardware limitations and by the nature of the cell sizes found in real tissues, the frequency range acquired or available on clinical MRI scanners is narrow, so that the dependency of extracellular diffusion on frequency is minor. All our investigations using simulations in silico, cell lines in vitro, and animals in vivo have suggested the extracellular diffusion coefficient obtained using the IMPULSED method is largely insensitive to diffusion times so that the extracellular diffusion coefficient is modeled as a constant Dex.
Diffusion anisotropy.
Except for some ex vivo investigations of fixed breast tissues with strong gradients (37), a previous in vivo study involving 81 patients reported that water diffusion is nearly isotropic in various human breast tumors (38). Therefore, the DWI signals for IMPULSED acquisitions are obtained by averaging three acquisitions with diffusion gradients along three orthogonal oblique directions ([gx, gy, gz] = [1, 1, −0.5], [1, −0.5, 1], and [−0.5, 1, 1]), corresponding to the trace of a diffusion tensor.
IMPULSED outcomes:
Table 1 lists tissue parameters that can be determined by analyses of DWI data. The estimates of d are actually volume-weighted mean cell sizes (39) which, for a population of cells, is given by , where dn is the cell size of the nth cell. Note that, unlike our pre-clinical animal studies (27,28), Din is fixed as 1.58 μm2/ms (10) which better stabilizes the fittings of IMPULSED data. Supporting Information Figure S4 shows the fitting precisions of d and vin significantly decrease if Din is a free fitting parameter. Our simulations suggest that difference choices of Din in the data fitting have little influences on the fitted IMPULSED metrics (see Supporting Information Figure S5).
Table 1.
Summary of IMPULSED derived parameters and corresponding biophysical features.
| IMPULSED parameters | Biophysical features |
|---|---|
| d | Volume-weighted mean cell size |
| vin | Intracellular volume fraction |
| Dex | Extracellular diffusion coefficient |
Experimental diffusion parameters in IMPULSED
It is desirable to use higher gradient strengths and slew rates in diffusion measurements to achieve greater diffusion weighting (b values) and a range of diffusion times appropriate for each sample of interest (40). However, due to human physiological thresholds and hardware limitations, the gradient strength and slew rate are limited on clinical MRI scanners, which makes it challenging to implement some quantitative DWI methods. To ensure IMPULSED can be translated clinically, a maximum gradient strength of 80 mT/m and a slew rate < 100 mT/m was assumed, and these limits were imposed on our simulations and experiments including cells, animals, and humans. Table 2 shows diffusion parameters for IMPULSED measurements that are readily available on human 3T MRI systems and were selected for practical implementation. All studies acquired two opposite diffusion gradient directions for each axis and the geometric means were used as final images to mitigate cross-terms between diffusion and background gradients (41).
Table 2.
Summary of diffusion parameters used in IMPULSED measurements which are readily available for human 3T MRI systems. Gmax is the maximum gradient strength used with all gradients on.
| δ /Δ [ms] | N | f [Hz] | b [s/mm2] | Gmax [mT/m] | |
|---|---|---|---|---|---|
| PGSE | 12/74 | N/A | N/A | 0,250,500,750,1000, 1400, 1800 | 50.0 |
| OGSE | 40.9/51.4 | 1 | 25 | 0,250,500,750,1000 | 72.7 |
| 2 | 50 | 0,100,200,300 | 80.5 |
Validation using simulations in silico
Computer simulations were performed to evaluate both the accuracy and precision of IMPULSED derived parameters obtainable for signal to noise ratios (SNRs) practically available on clinical 3T MRI scanners. A finite difference method was used to simulate DWI signals as reported previously (42). Tumors were modeled as tightly-packed spherical cells on a face-centered-cubic lattice (43) with vin = 61.8%, Din = 1.58 μm2/ms (10), Dex = 2 μm2/ms, and homogeneous relaxation times for simplicity. Eight different values of cell diameter d evenly distributed from 6 to 20 μm were evaluated, covering the cell sizes typical of lymphocytes to cancer cells. After noise-free DWI signals were calculated for each cell diameter, Rician noise equivalent to achieve an SNR = 20 was added, and then the noisy signals were used for data fitting. This process was repeated 100 times to evaluate both accuracy and precision of IMPULSED derived metrics.
Validation using cell lines in vitro
Cells.
Three types of breast cancer cell lines, MDA-MB-231, MCF7, and MDA-MB-453, were purchased from American Type Culture Collection (Manassas, Virginia, USA). In addition to cancer cell lines, the Jurkat acute T cell leukemia cell line and lymphocytes were used to mimic smaller cells. Jurkat cells were generously provided by Dr. James Thomas at Vanderbilt University. Lymphocytes were extracted from human peripheral blood by using the Ficoll method (44). The details of cell preparation can be found in the Supporting Information.
For MR experiments, cells were washed with PBS after fixation, about 3 × 107 cultured cells (or 1×109 lymphocytes) were centrifuged at 2000g for 2 minutes in a 0.65ml of Eppendorf tube to obtain a tight cell pellet. The supernatant was carefully removed for MRI measurements. A small aliquot of cells from each sample was spotted on a glass slide and covered by a coverslip. Digital images of the cells were recorded at both 20X and 40X amplification. A stage micrometer was used for size calibration.
MRI experiments.
All MRI measurements of cells in vitro were performed on a Varian/Agilent 4.7T MRI spectrometer similar to the approach described previously (35). A 2-mm thick slice crossing the center of each cell pellet was imaged with a field-of-view 16×16mm and a matrix size 64×64, yielding a spatial resolution of 250μm. All diffusion sequence parameters were the same as in Table 2, and other acquisition parameters are the same as in the animal study in vivo.
Light microscopy.
Bright field images were captured using a Zeiss Axio Observer microscope. Two representative fields of view were chosen for each slide. Each field of view was imaged at 40X magnification with the focus set slightly above, below, and equal to the optimal focal plane. Differences between two out-of-focus images resulted in enhanced contrast of the cell boundaries (45). The area of each cell was then calculated from these microscopic images using an auto-segmentation program written in Matlab, and these measurements were converted to a diameter assuming each cell is a sphere. The detailed procedures are provided in the Supporting Information and representative raw and segmented light microscopy images are shown in Supporting Information Figure S6.
Validation using animals in vivo
Animal preparation.
All procedures were approved by the Vanderbilt University Institutional Animal Care and Usage Committee. MDA-MB-231(n=6) and MCF7 (n=4) xenografts were generated following subcutaneous injection of 1–2×106 cells in female athymic nude mice (Harlan Laboratories, Inc., Indianapolis, IN). When each tumor reached a size of 200–300 mm3, MR imaging was performed as described below, and each mouse was euthanized for histology immediately afterward.
MRI experiments.
All MR images of animals were acquired on a 4.7 T Varian/Agilent horizontal small animal scanner using a Litz38 volume coil for both transmission and reception. A single-shot echo-planar imaging (EPI) diffusion sequence with fat suppression was used for all diffusion measurements to minimize motion artefacts. Axial slices with 1 mm thickness were acquired to cover the entire tumors, with in-plane matrix size 128 × 64 and field-of-view =40 × 20 mm yielding an in-plane resolution of 312.5 × 312.5 μm. All diffusion sequence parameters are outlined in Table 2. TR/TE=4500/104 ms; receiver spectral width 250 kHz; half scan factor = 0.875.
Histology.
Animals were euthanized immediately following MRI scans. Tumors were collected, cut into 2mm thick pieces and fixed in 10% formalin for 24 hours. Tissues were transferred to 70% ethanol prior to paraffin embedding. Tumors were sectioned at 8 μm thickness and stained with hematoxylin and eosin (H&E) for cellularity, and Na+/K+-ATPase (ab76020, Abcam).
Applications in patients
Breast cancer patients.
The human imaging study was approved by the Institutional Review Board at Vanderbilt University Medical Center. Seven women (age 55.3±8.0) diagnosed with breast cancer with tumors 1cm or greater were recruited in this study. Written informed consents were received from participants prior to inclusion in the study. The detailed patient information and tumor types can be found in Supporting Information Table S1.
Human MR imaging.
IMPULSED imaging was performed using a Philips Achieva 3T scanner with a 16-channel breast coil. Acquisition sequence parameters were TR/TE=4500/103ms; FOV=192×192mm; acquisition matrix size 96×96; reconstructed in-plane resolution = 1.3×1.3 mm; 10 or 20 slices; slice thickness=5 mm; single shot EPI; SENSE factor=3; half scan factor 0.64; WFS(pix) / BW (Hz) = 12.181/35.7; fat suppression with SPAIR; and dynamic stabilization was used to minimize DWI signal drifts during scanning. All diffusion sequence parameters were the same as in Table 2. The total scan time ≈ 7 mins. In addition, ADC measurements using PGSE acquisitions with Δ = 54 and 34 ms were performed to further investigate the ADC dependence on diffusion times.
Data analyses
All diffusion images were co-registered to the corresponding T2-weighted S(b=0) image to correct for subject motion. For cell experiments in vitro, an ROI was manually drawn on each image and the total DWI signals from the ROI were used in the data fitting. The light microcopy images of cells were analyzed using a locally developed pipeline with details provided in the Supporting Information. For in vivo experiments, tumor ROIs were manually drawn based on PGSE diffusion weighted images with b = 1000 s/mm2. All ROIs were drawn without boundary voxels to avoid partial volume effects. The IMPULSED fitting was performed only inside ROIs. The histology images were analyzed using CellProfiler™ and the details are provided in the Supporting Information.
All data fittings were performed using Matlab (Mathworks, Natick, Massachusetts) to generate DWI parametric maps on a voxel-wise basis (39). The fitting parameter ranges were limited by possible physiologically relevant values, i.e., 0.2 ≤ d ≤ 25 μm (i.e., typical breast cancer cell size range. Note that the upper limit is determined by the root mean square displacement of free water diffusion at 37°C), 0 ≤ vin ≤ 1 (max volume fraction 100%), and 0 ≤ Dex ≤ 3.1 μm2/ms (the free water diffusion coefficient is 3.07 μm2/ms at 37°C). Din was fixed as 1.58 μm2/ms (10) in the data analysis to stabilize fittings as shown in Figure S5. Note that fitting results are insensitive to the choices of Din. The fittings were to maximize the log likelihood function with Rician noise, i.e., , where Sn and Mn are the model-predicted and measured signals of the nth measurements, respectively, I0 is the modified Bessel function of the first kind with order zero, and σ2 is the noise variance. In the human study, all diffusion images and IMPULSED derived parametric images were co-registered to corresponding high-resolution T1-weighted anatomical images using the FMRIB’s Linear Image Registration Tool (FLIRT) (46) in the FSL toolbox (47).
The Spearman correlation was calculated to determine the relationship between fitted cell size and histology values at the ROI level. The Bland Altman plot was used to evaluate the agreement of cell size values between those from fitted and those derived from histology.
Results
Computer simulations in silico
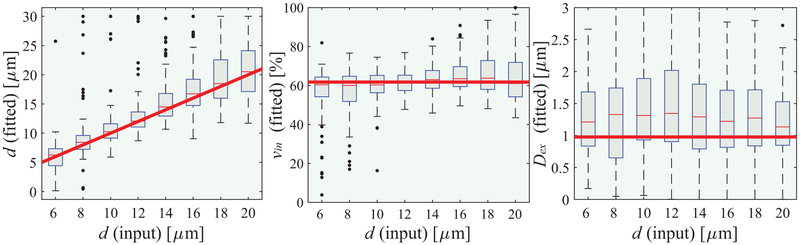
Figure 2 shows the simulated dependence of IMPULSED derived metrics on input values with a signal to noise ratio (SNR) of 20, typical of human MRI. In the range 6 – 20 μm, the fitted cell size d shows a clear linear dependence on ground-truth values. A mixed linear model reports p<0.01. The fitted intracellular water fraction vin shows a good match to any ground-truth value with a Bonferroni adjusted p < 0.05. By contrast, the fitted extracellular diffusion coefficient Dex shows significant uncertainties, indicating the precision of fitted Dex is limited when SNR is low. Note that the fitted Dex is expected to be lower than the intrinsic extracellular diffusion coefficient due to restriction effects. If SNR increases to 50, the precisions of fitted d, vin and Dex can be dramatically improved (see Supporting Information Figure S3). In conclusion, although limited in practice by the available gradient strength and slew rate, IMPULSED can measure mean cancer cell size reliably when 6 < d < 20 μm with practical SNRs, but the estimation of extracellular diffusion coefficient is not reliable with low SNRs.
Figure 2.
Simulated influence of noise with SNR = 20 on fitted d (left), vin (middle), and Dex (rigth) using IMPULSED. For each real input d, the fittings were repeated 100 times each with different noise samples but with the same SNR level. The red solid lines represent the ground-truth values, boxes represent ranges between the 25th and 75th percentiles, and dots are outliers.
Imaging cells in vitro
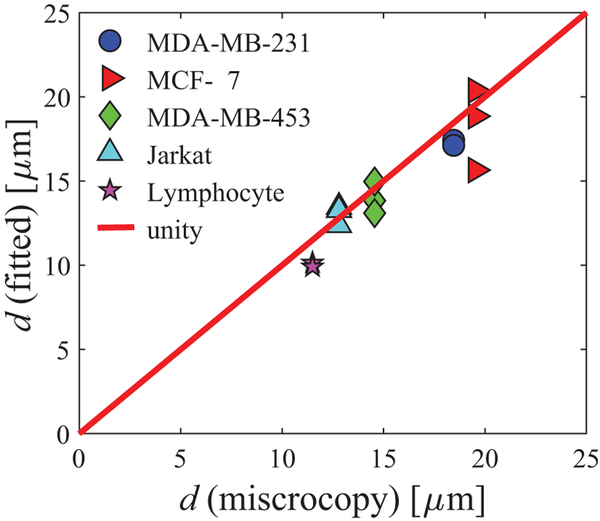
Supporting Information Figure S6 shows an example of the cell segmentation of light microscopy images. The cell size information obtained from microscopy was assumed to be the ground truth for validating the cell sizes fitted using IMPULSED. Figure 3 compares the mean cell sizes obtained from IMPULSED fitting and light microscopy for three breast cancer cell lines, the Jarkat cell line, and lymphocytes. Over a broad range of cell sizes (11 – 18 μm) and cell types (breast cancer, leukemia, and lymphocytes), IMPULSED fitted d values show a strong correlation (r = 0.92 and p < 0.001 provided by the Pearson correlation) with the values obtained from light microscopy.
Figure 3.
Comparison of mean cell sizes obtained from IMPULSED fitting and light microscopy.
Imaging animals in vivo
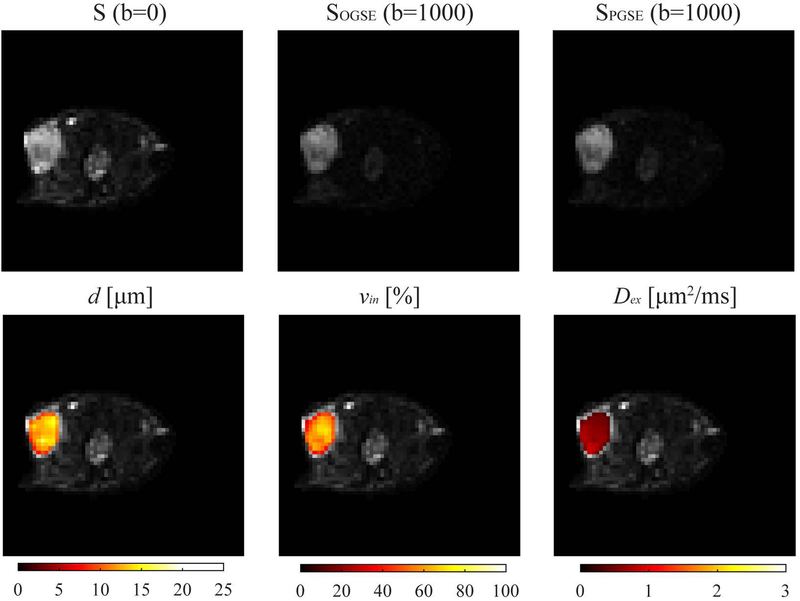
Figure 4 shows representative multi-parametric images of a mouse MDA-MB-231 xenograft. The T2-weighted S(b=0), diffusion-weighted OGSE with N=1 and b=1000 μm2/ms and PGSE with b=1000 μm2/ms images are shown in the top row. IMPULSED-derived maps of d, vin, and Dex were overlaid on the corresponding diffusion-weighted image shown in the bottom row. There is significant inhomogeneity within the tumor.
Figure 4.
Representative multi-parametric images of a mouse bearing MDA-MB-231 xenograft. (Top) T2-weighted S(b=0), OGSE with N=1 and b=1000 μm2/ms and PGSE with b=1000 μm2/ms. (Bottom) IMPULSED-derived maps of mean cell size d, intracellular volume fraction vin, and extracellular diffusion coefficient Dex overlaid on the corresponding S0 image.
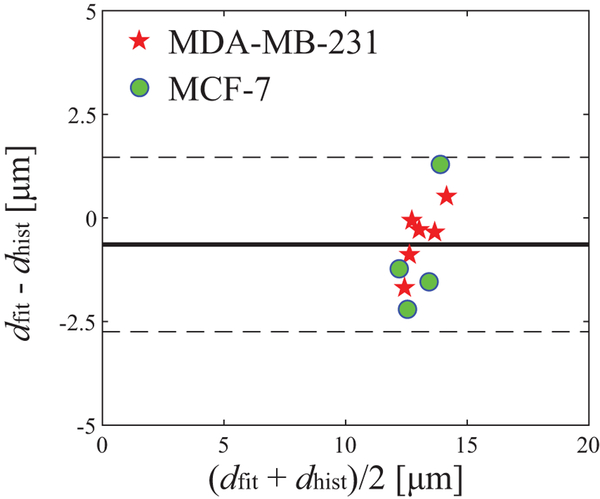
Figure 5 is a Bland Altman plot of fitted and histology derived mean cell sizes for two types of breast cancer xenografts, MDA-MB-231 (n=6) and MCF7 (n=4), imaged in mice in vivo. The IMPULSED slightly underestimates d (mean difference = 0.6 μm) and 95% limits of agreement is 2.1 μm. This suggests good agreement between the IMPULSED and histology derived mean cell sizes in animals in vivo.
Figure 5.
Bland Altman plot of fitted and histologically obtained mean cell sizes of two types of breast cancer xenografts. The solid line represents the mean difference and two dashed lines represent 95% of limits of agreement (i.e., 1.96 SD).
Imaging breast cancer patients in vivo
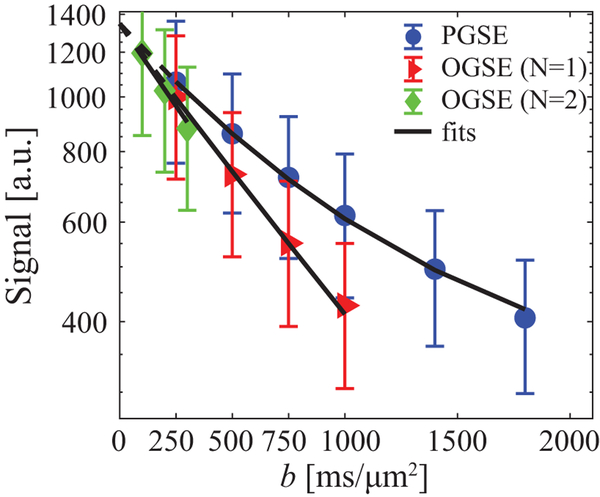
The Supporting Information Table S1 summarizes the information on breast cancer patients and breast tumors. The SNR was ~45 in the tumors. Figure 6 shows IMPULSED signals (symbols) from a tumor region of interest (ROI) and the IMPULSED fits (solid lines) of a representative human breast tumor. The fitted overall average cell size d = 14.88 ± 4.39 μm, vin = 38.87 ± 7.95%, and Dex = 1.81 ± 0.46 μm2/ms, yielding a cell density = 2.86 ± 1.31 × 108 cells/cm3, which is consistent with previous reports of ~108 cells/cm3 for tumors of epithelial origin (such as breast tumors) (48). Note that for the same b values, signals acquired with OGSE with an effective diffusion time tdiff of 10 ms decay significantly more than those obtained with PGSE with tdiff = 70 ms. This increase in ADC at shorter diffusion times obtained provides the contrast that enables the possibility to measure cancer cell size and density. An example of ADC dependence on diffusion times can be found in Supporting Information Figure S8. For the IMPULSED fitting, all b = 0 images were excluded from the fittings in order to minimize the influences of blood perfusion.
Figure 6.
The ROI-based diffusion-weighted signal attenuations of a representative human breast tumor. Markers are mean signals and the error bars represent standard deviations. The solid lines are fitted results using Eq.[1], and dashed lines with low b values indicate b=0 images were excluded from fittings to minimize perfusion effects.
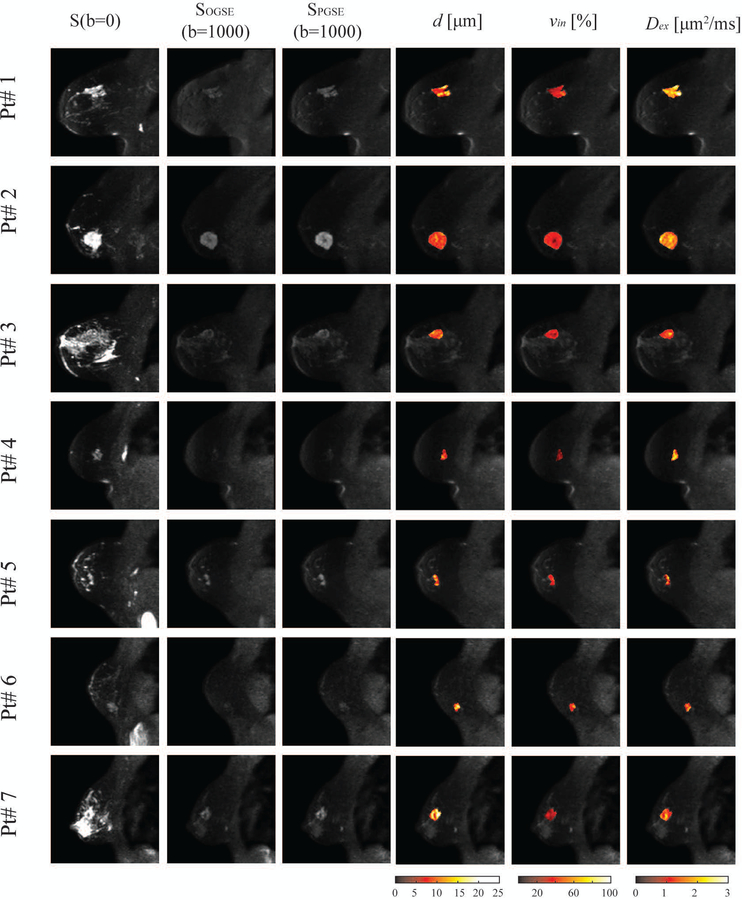
Figure 7 shows multi-parametric images of all seven patients. In addition to the large variations of tumor sizes across all patients, the parametric maps are significantly inhomogeneous within each tumor. For example, there are significantly lower intracellular volume fraction and increased extracellular diffusion coefficient at the center of the tumor of patient# 2, suggesting an acellular, necrotic core has developed. Note that the parametric map of Dex is more heterogeneous inside tumors which may be due to the lower fitting precision (see Figure 1).
Figure 7.
Representative multi-parametric images of all seven patients. (From left to right) T2-weighted S(b=0), OGSE with N=1 and b=1000 μm2/ms, PGSE with b=1400 μm2/ms, IMPULSED-derived maps of mean cell size d, intracellular volume fraction vin, and extracellular diffusion coefficient Dex overlaid on the corresponding diffusion-weighted image. .
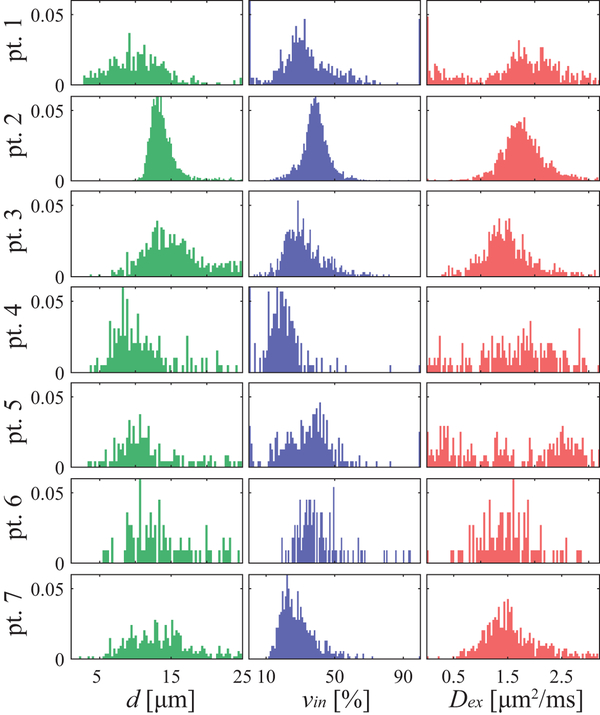
Figure 8 summarizes the histograms of all fitted IMPULSED metrics for seven patients. Although different patients and breast tumors show different histograms of the IMPULSED metrics, the peaks of d are all in the range of 12 – 18 μm and vin are in the range of 25 – 40 %. Values of vin may be underestimated due to transcytolemmal water change, but this does not affect d (35). Dex show broader ranges of distribution compared with those of d and vin. This may be due to their larger fitting variations as predicted by the simulation results (Figure 1). Note that IMPULSED derived parameters do not correlate with each other (data not shown), which is not surprising since they represent independent tissue features.
Figure 8.
A summary of histograms of all fitted IMPULSED metrics (columns) of all seven patients (rows).
Discussion
We have implemented a clinically feasible non-invasive imaging method termed IMPULSED to measure mean cancer cell size of tumors using clinical MRI scanners in vivo. The potential clinical relevance of the IMPULSED method is based on the important role of cell size in cancer. For example, one potential clinical application is to use cell size variations to monitor tumor early response to anti-cancer treatment. As shown in our previous work using animal models, cell size variation is an early and specific biomarker to report the effectiveness of anti-cancer treatment, such as in treatment-induced mitotic arrest (10) and apoptosis (9). The current work provides a reliable technique that allows measurements of mean cell sizes of human breast tumors in a clinical setting, which is an essential step to translate our previous preclinical findings to clinical practice. Moreover, it would be interesting to correlate IMPULSED derived parameters with other pathohistological and/or genetic features of tumors to assess tumor status, perhaps as part of a “radiomics” characterization of cancers. However, this is out of the scope of this paper.
Despite numerous attempts at using diffusion MRI to map microstructural information in tumors in vivo, successful applications to date have so far been very limited. Although providing similar information such as cell size, IMPULSED and other diffusion-based methods such as VERDICT and POMACE use different acquisitions and methods of analysis. It is therefore of interest to compare the accuracy and precision of different methods. The VERDICT approach attemps to map mean cell size in human tumors, but to date has been exclusively used in human prostate cancers (24–26). The VERDICT approach is different from IMPULSED in that though VERDICT also employs multiple diffusion times, it uses conventional PGSE sequences with higher b values. Consequently, the range of diffusion times probed is narrower than for IMPULSED. Moreover, VERDICT uses multiple acquisitions with different echo times, which may introduce biases in fitting from relaxation time effects. IMPULSED incorporates cosine-modulated trapezoidal oscillating gradients to significantly increase the range of effective diffusion times in the short time range. Supporting Information Figure S8 shows the diffusion time dependence of ADC of a human breast tumor. Over the range tdiff 30 – 70 ms achievable using PGSE sequences and realistic gradient strengths only (such as those used in VERDICT), the ADC does not change significantly. The most significant ADC change occurs between 30 ms and 5 ms, the range used in IMPULSED. This provides good sensitivity to cellular properties, which forms the foundation of IMPULSED in measuring cell size.
Because this is the first demonstration of IMPULSED in human breast cancer imaging, the experimental parameters were not all optimized, and the protocol was chosen to include more acquisitions to maximize the accuracy. For example, the current IMPULSED method acquires both positive and negative gradients in each direction to remove cross-terms, acquires three orthogonal gradient directions to account for any small diffusion anisotropy in breast tumors, and uses a much longer TR (4.5 sec) to reduce any possible influences from the relatively long T1 relaxation times in tumors. All these strategies are theoretically valid for any diffusion method. However, their exact influence on IMPULSED derived parameters remains unclear. Moreover, an optimization of experimental parameters could maintain the precision while minimizing scan time, such as employed in our previous work using the Cramér–Rao lower bound (CRLB) approach for optimizing T1 and qMT protocols (49). Recent optimization efforts have shortened VERDICT acquisition time from 35 minutes (25) to 12 minutes (26)., suggesting there is much room to accelerate the acquisition protocol of diffusion MRI based methods.
Although IMPULSED detects the mean cell size within each image voxel, it is plausible to obtain the distribution of cell sizes inside each voxel as well. However, it is challenging to fit cell size distributions without a priori knowledge of their nature (50). Most quantitative DWI studies to date have modeled cancer cells as spheres (10,27) or neural axons as cylinders (39) with uniform cell sizes to simplify the mathematical complexity yet preserve basic microstructural features. Although a mean cell size and density can be measured inside each imaging voxel, the voxel-by-voxel heterogeneous spatial distributions of cell size and cell density across whole tumors can still be obtained as shown in Figure 8.
Not only mean cell size d but also the intracellular volume fraction vin can be derived from IMPULSED. The apparent cell density can be calculated from d and vin (28). However, our previous study (35) found that, if the transcytolemmal water exchange cannot be ignored, the intracellular volume fraction is intrinsically underestimated for any biophysical diffusion model that assumes no water exchange (such as IMPULSED and VERDICT). Because cell membrane permeability is likely to increase during treatment-induced apoptosis, the accuracy of cell density derived from IMPULSED may be compromised. Therefore, caution should be expressed in the interpretation of fitted vin values as intracellular volume fraction. Interestingly, vin has been found to correlate well with ground truth cell density in simulations (27), which suggests vin might be still a good indicator of cell density although its absolute accuracy is uncertain.
Although only breast cancer was investigated in the current study, IMPULSED can be used to assess other extracranial tumors such as head and neck tumors, wherever a two-compartment model is valid. Note that IMPULSED cannot specifically differentiate cancer cells from other cells. Therefore, the mean cell size obtained using IMPULSED includes all cell types (e.g., cancer cells, stromal cells, and lymphocytes). Our in vitro cell studies (Figure 3) suggest IMPULSED has sufficient sensitivity to differentiate small lymphocytes, relatively small cancer cells (leukemia Jarkat), and relatively large breast cancer cells. The relative fraction of these cells will change the mean cell size. If more detailed information on specific types of cells is needed, the two-compartment model may need to be modified.
To ensure clinical translation, the IMPULSED method presented in the current study was strictly limited to diffusion parameters that are achievable on clinical 3T MRI scanners, such as a maximum gradient strength < 80 mT/m and maximum gradient slew rate < 100 mT/m/sec. However, the ability of IMPULSED is not limited by these standard parameters. For example, more advanced MRI hardware such as the Human Connectome gradient coil with a maximum gradient strength 300 mT/m and maximum slew rate 200 mT/m/sec can remarkably improve the ability of diffusion MRI to probe brain microstructure (40). Different from fixing intracellular diffusivity Din in the fittings in the current work, we previously found Din can be fit reliably with a gradient strength up to 360 mT/m using the IMPULSED method (35). Because Din is an important indicator of overall intracellular microenvironment, high performance gradient systems can remarkably enhance the ability of IMPULSED to characterize intracellular space in human imaging.
Conclusions
A clinically feasible IMPULSED imaging method has been successfully developed and validated on clinical MRI scanners for in vivo imaging of mean cell sizes of solid tumors in breast cancer patients. To the best of our knowledge, this is the first clinical study that uses a non-invasive imaging method for spatially mapping distributions of mean cancer cell size of heterogeneous human breast tumors in vivo.
Supplementary Material
Supporting Information Figure S1 Comparison of intracellular PGSE DWI signals obtained from simulation vs analytical forms with trapezoid waveform (Eq.[S3]) and square waveform (Eq.[S4])
Supporting Information Figure S2 Comparison of intracellular DWI signals obtained from simulation vs analytical expressions with trapezoidal OGSE sequences and N=1 (Eq.[S5]) and N=2 (Eq.[S6])
Supporting Information Figure S3 Simulated influence of noise with SNR = 50 on IMPULSED derived metrics. For each real input d, the fittings were repeated 100 times each with different noises but with the same SNR level. The red solid lines represent the ground truth values inputted in the simulations, boxes represent ranges between the 25th and 75th percentiles, and dots are outliers.
Supporting Information Figure S4 Simulated IMPULSED derived metrics with SNR = 20 if Din is a free fitting parameter.
Supporting Information Figure S5 Simulated influence of noise with SNR = 20 on IMPULSED derived metrics with different choices of fixed Din values in the data analyses. From the top to bottom, fixed Din = 1.0, 1.4, 1.58 (ground truth), 1.8, 2.2 μm2/ms.
Supporting Information Figure S6 A representative FOV of light microscopy of cells in vitro. The left is the raw image and the right shows the segmented images with identified cells highlighted in green.
Supporting Information Figure S7 A representative image of an MDA-MB-231 breast tumor in mouse. The top is the raw image, and the bottom is the images after the processing of CellProfiler.
Supporting Information Figure S8 Tumor ADC dependence on diffusion time. The error-bars represent the standard deviations over the tumor region shown in Figure 4.
Acknowledgements
The authors thank MR technologists Clair Jones, Leslie McIntosh, Christopher Thompson, and Fuxue Xin for assistance in data acquisitions, Drs. Katy Beckermann and Kirsten Young for collecting lymphocytes. This work was funded by NIH grants K25CA168936, R01CA109106, R01CA173593, UL1TR002243, S10OD021771, U01CA142565, F32CA216942, UL1TR000445, P30 CA068485, and American Cancer Society grant IRG#58-009-56.
Footnotes
COI statement: The authors declare no potential conflicts of interest.
References
- 1.Kozlowski J, Konarzewski M, Gawelczyk AT. Cell size as a link between noncoding DNA and metabolic rate scaling. P Natl Acad Sci USA 2003;100(24):14080–14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baserga R. Is cell size important? Cell cycle 2007;6(7):814–816. [DOI] [PubMed] [Google Scholar]
- 3.Savage VM, Allen AP, Brown JH, Gillooly JF, Herman AB, Woodruff WH, West GB. Scaling of number, size, and metabolic rate of cells with body size in mammals. P Natl Acad Sci USA 2007;104(11):4718–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia JH, Yoshida Y, Chen H, Li Y, Zhang ZG, Lian J, Chen S, Chopp M. Progression from ischemic injury to infarct following middle cerebral artery occlusion in the rat. The American Journal of Pathology 1993;142(2):623–635. [PMC free article] [PubMed] [Google Scholar]
- 5.Liang D, Bhatta S, Gerzanich V, Simard JM. Cytotoxic edema: mechanisms of pathological cell swelling. Neurosurgical focus 2007;22(5):E2–E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saraste A, Pulkki K. Morphologic and biochemical hallmarks of apoptosis. Cardiovascular research 2000;45:528–537. [DOI] [PubMed] [Google Scholar]
- 7.Sun L, Sakurai S, Sano T, Hironaka M, Kawashima O, Nakajima T. High-grade neuroendocrine carcinoma of the lung: Comparative clinicopathological study of large cell neuroendocrine carcinoma and small cell lung carcinoma. Pathol Int 2009;59(8):522–529. [DOI] [PubMed] [Google Scholar]
- 8.Brauer M. In vivo monitoring of apoptosis. Prog Neuro-Psychoph 2003;27(2):323–331. [DOI] [PubMed] [Google Scholar]
- 9.Jiang X, McKinley ET, Xie J, Li H, Xu J, Gore JC. In vivo magnetic resonance imaging of treatment-induced apoptosis. Scientific reports 2019;9(1):9540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang X, Li H, Zhao P, Xie J, Khabele D, Xu J, Gore JC. Early detection of treatment-induced mitotic arrest using temporal diffusion magnetic resonance spectroscopy. Neoplasia 2016;18(6):387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tobkes AI, Nord HJ. Liver-Biopsy - Review of Methodology and Complications. Digest Dis 1995;13(5):267–274. [DOI] [PubMed] [Google Scholar]
- 12.Zhao M, Pipe JG, Bonnett J, Evelhoch JL. Early detection of treatment response by diffusion-weighted 1H-NMR spectroscopy in a murine tumour in vivo. Br J Cancer 1996;73(1):61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugahara T, Korogi Y, Kochi M, Ikushima I, Shigematu Y, Hirai T, Okuda T, Liang L, Ge Y, Komohara Y, Ushio Y, Takahashi M. Usefulness of diffusion-weighted MRI with echo-planar technique in the evaluation of cellularity in gliomas. J Magn Reson Imaging 1999;9(1):53–60. [DOI] [PubMed] [Google Scholar]
- 14.Gauvain KM, McKinstry RC, Mukherjee P, Perry A, Neil JJ, Kaufman BA, Hayashi RJ. Evaluating pediatric brain tumor cellularity with diffusion-tensor imaging. AJR Am J Roentgenol 2001;177(2):449–454. [DOI] [PubMed] [Google Scholar]
- 15.Ross BD, Moffat BA, Lawrence TS, Mukherji SK, Gebarski SS, Quint DJ, Johnson TD, Junck L, Robertson PL, Muraszko KM, Dong Q, Meyer CR, Bland PH, McConville P, Geng H, Rehemtulla A, Chenevert TL. Evaluation of cancer therapy using diffusion magnetic resonance imaging. Mol Cancer Ther 2003;2(6):581–587. [PubMed] [Google Scholar]
- 16.Cory DG, Garroway AN. Measurement of translational displacement probabilities by NMR: an indicator of compartmentation. Magn Reson Med 1990;14(3):435–444. [DOI] [PubMed] [Google Scholar]
- 17.Tanner JE. Transient diffusion in a system partitioned by permeable barriers - application to NMR measurements with a pulsed field gradient. J Chem Phys 1978;69(4):1748–1754. [Google Scholar]
- 18.Szafer A, Zhong J, Gore JC. Theoretical model for water diffusion in tissues. Magn Reson Med 1995;33(5):697–712. [DOI] [PubMed] [Google Scholar]
- 19.van der Toorn A, Syková E, Dijkhuizen RM, Vorísek I, Vargová L, Skobisová E, van Lookeren Campagne M, Reese T, Nicolay K. Dynamic changes in water ADC, energy metabolism, extracellular space volume, and tortuosity in neonatal rat brain during global ischemia. Magnetic resonance in medicine 1996;36:52–60. [DOI] [PubMed] [Google Scholar]
- 20.Padhani AR, Liu G, Koh DM, Chenevert TL, Thoeny HC, Takahara T, Dzik-Jurasz A, Ross BD, Van Cauteren M, Collins D, Hammoud DA, Rustin GJ, Taouli B, Choyke PL. Diffusion-weighted magnetic resonance imaging as a cancer biomarker: consensus and recommendations. Neoplasia 2009;11(2):102–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshikawa MI, Ohsumi S, Sugata S, Kataoka M, Takashima S, Mochizuki T, Ikura H, Imai Y. Relation between cancer cellularity and apparent diffusion coefficient values using diffusion-weighted magnetic resonance imaging in breast cancer. Radiat Med 2008;26(4):222–226. [DOI] [PubMed] [Google Scholar]
- 22.Squillaci E, Manenti G, Cova M, Di Roma M, Miano R, Palmieri G, Simonetti G. Correlation of diffusion-weighted MR imaging with cellularity of renal tumours. Anticancer Res 2004;24(6):4175–4179. [PubMed] [Google Scholar]
- 23.Xu J, Li K, Smith RA, Waterton JC, Zhao P, Chen H, Does MD, Manning HC, Gore JC. Characterizing tumor response to chemotherapy at various length scales using temporal diffusion spectroscopy. PLoS One 2012;7(7):e41714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panagiotaki E, Walker-Samuel S, Siow B, Johnson SP, Rajkumar V, Pedley RB, Lythgoe MF, Alexander DC. Noninvasive quantification of solid tumor microstructure using VERDICT MRI. Cancer Res 2014;74(7):1902–1912. [DOI] [PubMed] [Google Scholar]
- 25.Panagiotaki E, Chan RW, Dikaios N, Ahmed HU, O’Callaghan J, Freeman A, Atkinson D, Punwani S, Hawkes DJ, Alexander DC. Microstructural characterization of normal and malignant human prostate tissue with vascular, extracellular, and restricted diffusion for cytometry in tumours magnetic resonance imaging. Investigative radiology 2015;50(4):218–227. [DOI] [PubMed] [Google Scholar]
- 26.Bonet-Carne E, Johnston E, Daducci A, Jacobs JG, Freeman A, Atkinson D, Hawkes DJ, Punwani S, Alexander DC, Panagiotaki E. VERDICT-AMICO: Ultrafast fitting algorithm for non-invasive prostate microstructure characterization. NMR in biomedicine 2019;32(1):e4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang X, Li H, Xie J, Zhao P, Gore JC, Xu J. Quantification of cell size using temporal diffusion spectroscopy. Magn Reson Med 2016;75(3):1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang X, Li H, Xie J, McKinley ET, Zhao P, Gore JC, Xu J. In vivo imaging of cancer cell size and cellularity using temporal diffusion spectroscopy. Magn Reson Med 2017;78(1):156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynaud O, Winters KV, Hoang DM, Wadghiri YZ, Novikov DS, Kim SG. Pulsed and oscillating gradient MRI for assessment of cell size and extracellular space (POMACE) in mouse gliomas. NMR in biomedicine 2016;29(10):1350–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van AT, Holdsworth SJ, Bammer R. In vivo investigation of restricted diffusion in the human brain with optimized oscillating diffusion gradient encoding. Magn Reson Med 2014;71(1):83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baron CA, Beaulieu C. Oscillating gradient spin-echo (OGSE) diffusion tensor imaging of the human brain. Magn Reson Med 2014;72(3):726–736. [DOI] [PubMed] [Google Scholar]
- 32.Ianus A, Siow B, Drobnjak I, Zhang H, Alexander DC. Gaussian phase distribution approximations for oscillating gradient spin echo diffusion MRI. J Magn Reson 2013;227:25–34. [DOI] [PubMed] [Google Scholar]
- 33.Gore JC, Xu J, Colvin DC, Yankeelov TE, Parsons EC, Does MD. Characterization of tissue structure at varying length scales using temporal diffusion spectroscopy. NMR Biomed 2010;23(7):745–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J, Does MD, Gore JC. Quantitative characterization of tissue microstructure with temporal diffusion spectroscopy. J Magn Reson 2009;200(2):189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Jiang X, Xie J, Gore JC, Xu J. Impact of transcytolemmal water exchange on estimates of tissue microstructural properties derived from diffusion MRI. Magn Reson Med 2017;77(6):2239–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aggarwal M, Jones MV, Calabresi PA, Mori S, Zhang J. Probing mouse brain microstructure using oscillating gradient diffusion MRI. Magn Reson Med 2012;67(1):98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bailey C, Siow B, Panagiotaki E, Hipwell JH, Mertzanidou T, Owen J, Gazinska P, Pinder SE, Alexander DC, Hawkes DJ. Microstructural models for diffusion MRI in breast cancer and surrounding stroma: an ex vivo study. NMR in biomedicine 2017;30(2):e3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Partridge SC, Ziadloo A, Murthy R, White SW, Peacock S, Eby PR, DeMartini WB, Lehman CD. Diffusion tensor MRI: Preliminary anisotropy measures and mapping of breast tumors. Journal of Magnetic Resonance Imaging 2010;31:339–347. [DOI] [PubMed] [Google Scholar]
- 39.Xu J, Li H, Harkins KD, Jiang X, Xie J, Kang H, Does MD, Gore JC. Mapping mean axon diameter and axonal volume fraction by MRI using temporal diffusion spectroscopy. Neuroimage 2014;103:10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Setsompop K, Kimmlingen R, Eberlein E, Witzel T, Cohen-Adad J, McNab JA, Keil B, Tisdall MD, Hoecht P, Dietz P, Cauley SF, Tountcheva V, Matschl V, Lenz VH, Heberlein K, Potthast A, Thein H, Van Horn J, Toga A, Schmitt F, Lehne D, Rosen BR, Wedeen V, Wald LL. Pushing the limits of in vivo diffusion MRI for the Human Connectome Project. NeuroImage 2013;80:220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neeman M, Freyer JP, Sillerud LO. A simple method for obtaining cross-term-free images for diffusion anisotropy studies in NMR microimaging. Magnetic Resonance in Medicine 1991;21:138–143. [DOI] [PubMed] [Google Scholar]
- 42.Xu J, Does MD, Gore JC. Numerical study of water diffusion in biological tissues using an improved finite difference method. Phys Med Biol 2007;52(7):N111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu J, Does MD, Gore JC. Sensitivity of MR diffusion measurements to variations in intracellular structure: effects of nuclear size. Magn Reson Med 2009;61(4):828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyle W, Chow A. Isolation of human lymphocytes by a Ficoll barrier method. Transfusion 1969;9(3):151–155. [DOI] [PubMed] [Google Scholar]
- 45.Ali R, Gooding M, Szilágyi T, Vojnovic B, Christlieb M, Brady M. Automatic segmentation of adherent biological cell boundaries and nuclei from brightfield microscopy images. Machine Vision and Applications 2012;23(4):607–621. [Google Scholar]
- 46.Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Medical image analysis 2001;5(2):143–156. [DOI] [PubMed] [Google Scholar]
- 47.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. Neuroimage 2012;62(2):782–790. [DOI] [PubMed] [Google Scholar]
- 48.Del Monte U. Does the cell number 10(9) still really fit one gram of tumor tissue? Cell cycle 2009;8(3):505–506. [DOI] [PubMed] [Google Scholar]
- 49.Li K, Zu Z, Xu J, Janve VA, Gore JC, Does MD, Gochberg DF. Optimized inversion recovery sequences for quantitative T1 and magnetization transfer imaging. Magn Reson Med 2010;64(2):491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Assaf Y, Blumenfeld-Katzir T, Yovel Y, Basser PJ. AxCaliber: a method for measuring axon diameter distribution from diffusion MRI. Magn Reson Med 2008;59(6):1347–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szczepankiewicz F, van Westen D, Englund E, Westin CF, Stahlberg F, Latt J, Sundgren PC, Nilsson M. The link between diffusion MRI and tumor heterogeneity: Mapping cell eccentricity and density by diffusional variance decomposition (DIVIDE). Neuroimage 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1 Comparison of intracellular PGSE DWI signals obtained from simulation vs analytical forms with trapezoid waveform (Eq.[S3]) and square waveform (Eq.[S4])
Supporting Information Figure S2 Comparison of intracellular DWI signals obtained from simulation vs analytical expressions with trapezoidal OGSE sequences and N=1 (Eq.[S5]) and N=2 (Eq.[S6])
Supporting Information Figure S3 Simulated influence of noise with SNR = 50 on IMPULSED derived metrics. For each real input d, the fittings were repeated 100 times each with different noises but with the same SNR level. The red solid lines represent the ground truth values inputted in the simulations, boxes represent ranges between the 25th and 75th percentiles, and dots are outliers.
Supporting Information Figure S4 Simulated IMPULSED derived metrics with SNR = 20 if Din is a free fitting parameter.
Supporting Information Figure S5 Simulated influence of noise with SNR = 20 on IMPULSED derived metrics with different choices of fixed Din values in the data analyses. From the top to bottom, fixed Din = 1.0, 1.4, 1.58 (ground truth), 1.8, 2.2 μm2/ms.
Supporting Information Figure S6 A representative FOV of light microscopy of cells in vitro. The left is the raw image and the right shows the segmented images with identified cells highlighted in green.
Supporting Information Figure S7 A representative image of an MDA-MB-231 breast tumor in mouse. The top is the raw image, and the bottom is the images after the processing of CellProfiler.
Supporting Information Figure S8 Tumor ADC dependence on diffusion time. The error-bars represent the standard deviations over the tumor region shown in Figure 4.