Abstract
Autophagy is a lysosomal-dependent degradation process that is highly conserved and maintains cellular homeostasis by sequestering cytosolic material for degradation either non-specifically by non-selective autophagy, or targeting specific proteins aggregates by selective autophagy. Autophagy serves as a protective mechanism defending the cell from stressors and also plays an important role in enabling tumor cells to overcome harsh conditions arising in their microenvironment during growth as well as oxidative and non-oxidative injuries secondary to therapeutic stressors. Recently, autophagy has been implicated to cause tumor resistance to anti-angiogenic therapy, joining an existing literature implicating autophagy in cancer resistance to conventional DNA damaging chemotherapy and ionizing radiation. In this review, we discuss the role of angiogenesis in malignancy, mechanisms of resistance to anti-angiogenic therapy in general, the role of autophagy in driving malignancy, and the current literature in autophagy-mediated anti-angiogenic therapy resistance. Finally, we provide future insight into the current challenges of using autophagy inhibitors in the clinic and provides tips for future studies to focus on to effectively target autophagy in overcoming resistance to anti-angiogenic therapy.
Keywords: Autophagy, Angiogenesis, Anti-angiogenesis, Cancer, VEGF, Drug resistance
1. An introduction to angiogenesis in malignancy
In embryogenesis, wound repair and tumor growth there is a need to supply nutrients to growing tissue [1,2]. However, when tissues are stable and homeostasis has been achieved, it is only necessary to maintain existing vascular supply. Therefore, the process of angiogenesis, the development of new blood vasculature, is carefully regulated so that the generation of new vessels is matched with the tissue’s need for more nutrients. These mechanisms can become activated in the context of an expanding neoplasm which requires additional vascular supply to proliferate. If cancer is unable to induce angiogenesis then the lesion will grow increasingly hypoxic and subsequent necrosis and/or apoptosis may ensue. Therefore, upregulating angiogenesis is considered one of the hallmarks of malignancy [3].
At homeostasis, there is a balance of pro-angiogenic and anti-angiogenic factors present in the extra-cellular space [2]. Endothelial cells are quite stable, dividing once every 1000 days, and therefore do not require quick turnover. However, hypoxic stress can shift the balance to favor pro-angiogenic factors and increase vascular supply. In cancer this process has been especially well studied and a malignant angiogenic cascade has been documented [1]. First, tumor cells secrete paracrine factors that act on local stromal cells to transform them into cancer-associated fibroblasts (CAFs). CAFs then serve a principle role in preparing the local milieu for cancerous growth. CAFs are primarily responsible for breaking down the local extra-cellular matrix (ECM), via metalloproteases; recruiting inflammatory cells, via interleukins; and producing new structural molecules such as fibronectin, laminin and hyaluronate. This produces an environment rich with inflammatory activity and rapid turnover of the ECM. Due to the breakdown of the local stroma, it is possible for budding vessels to push out of pre-established vasculature and perfuse the growing tumor [4]. This budding is largely orchestrated by integrins, which act as anchor points and provide directionality in the context of ECM turnover [5]. All integrins are heterodimers of alpha/beta moieties; and β1, αv, and β8 (present in forming brain blood vessels) have been specifically implicated in tumor angiogenesis [5]. Physiologically, vascular endothelial growth factor (VEGF) is the most important secreted angiogenic factor and this importance carries over to most tumor angiogenesis as well. Many tumors secrete high levels of VEGF to create an angiogenic diffusion gradient. Therefore, the more penetrable ECM established by CAFs and the VEGF gradient produced by tumors are the perfect setup from new vessel budding [4]. As will be discussed below, angiogenesis in tumors is often quite different than healthy tissue. Leaky and atypically branching vessels are a hallmark of malignant angiogenesis and is a reflection that this process is altogether different than what is seen in healthy circumstances.
1.1. Sensing hypoxia and initiating vessel growth
All eukaryotic cells rely on oxygen, and, while cancer cells are more adaptive to low oxygen levels than normal cells, they still need some oxygen to survive. Tumors are able to sense oxygen levels with prolyl hydroxylases and, in times of oxygen abundance, prevent new vessel formation [1]. However, if tumor cells are exposed to hypoxic conditions, then the activity of the Hypoxia-inducible factors (HIF) work to increase local oxygen concentrations [6,7]. As a transcription factor, HIF can translocate to the nucleus and induce transcription of adaptive proteins. HIF activity results in the transcription of proteins that increase local oxygen supply: Some of these, like nitric oxide synthase, are very fast acting and dilate preexisting vasculature to immediately achieve more diffusion. However, HIF transcription factors also increase the amount of VEGF and related proteins, which serve as triggers for angiogenesis. The two major HIF molecules, HIF1α and HIF2α, work in concert to help cancer overcome hypoxia [8]. HIF2α acts to reduce beta-oxidation of fatty acids and increase the production of enzymes capable of dealing with free radicals. HIF1α increases the rate of glycolysis, diverts glycolytic metabolites away from oxidative metabolism, and increase new vessel formation. In times of prolonged hypoxia, angiogenesis is initiated to create a more permanent solution of hypoxia [6]. Although numerous proteins are responsible for angiogenesis, most namely VEGF, HIF is the master regulator of angiogenesis [6].
Blood vessels produced in cancer are not like those produced in healthy tissue [9]. Malignant tumor-induced angiogenesis is defined by the production of leaky and atypically branching vasculature. Therefore, even when angiogenesis is heavily upregulated, large tumors may have pockets of relative hypoperfusion. This is a notable consideration when administering chemotherapy, which requires perfusion for drug delivery; or designing radiation therapy, which requires oxygen for free radical formation [10]. Although upregulated HIF is a sign of hypoxia, which would seem to be associated with poor tumor conditions, it is paradoxically associated with treatment resistant tumors due to impaired therapy delivery.
A number of interesting studies have investigated why tumors develop such atypical vasculature. As with wound healing, tumor angiogenesis appears to be predominately driven by VEGF signaling [9]. It is hypothesized that unabated angiogenic signaling is partially to blame for the unusual vessels in cancer. Specifically, whereas wounds eventually resolve, tumors secrete vascular growth signals for extensive periods of time. Indeed, secretion of pro-angiogenic factors is a hallmark of malignancy and excessive signaling continues even in times of relatively normal perfusion in the tumor [11]. It is also expected that tumors will not express the full spectrum of VEGF subgroups, which are likely responsible for helping vessels mature normally [11]. Additionally, normal angiogenesis requires participation from vessel supporting cells such as pericytes [4]. These cells are dormant, unless acted on by appropriate growth signals, principally platelet-derived growth factor (PDGF). Many cancers do not express these additional angiogenic factors and new blood vessels do not receive developmental support from mural cells. Lastly, upregulation in angiogenic factors is relatively unrefined in cancer, when compared to embryogenesis or wound healing. In physiologic conditions, a concentration gradient of vascular factors is established and later refined to lead budding blood vessels to proper maturity [12]. In summary, the cause of atypical vasculature likely stems from the fact that tumors secrete homogenous amounts of angiogenic signals and are unable to titrate their secretion to the stages of vessel development.
2. Treating cancer by inhibiting angiogenesis
The principle of inhibiting angiogenesis to combat cancer is intuitive: Cancer growth can be stopped by starving a tumor of nutrients. The first application of this approach came from the release of bevacizumab (Avastin) in 2004 – a monoclonal antibody directed against all isoforms of VEGF-A [13]. This drug was shown to have therapeutic effects in a number of different cancers and sparked enthusiasm in this avenue of therapy. Independent clinical trials have validated the role of this drug in colorectal, breast, kidney and ovarian cancer. In 2009, bevacizumab gained accelerated approval for second-line treatment of glioblastoma, but clinical enthusiasm for bevacizumab in glioblastoma has waned due to poor tumor response and application to glioblastoma (GBM) is reserved for recurrent disease [13]. In short, responses to bevacizumab have been varied, but this drug has shown that, for some cancers, anti-angiogenesis could have therapeutic potential.
Enthusiasm for bevacizumab stems from a number of clinical trials. For patients with metastatic (mCRC) colorectal cancer a combination of bevacizumab, irinotecan, fluorouracil, and leucovorin fared better than irinotecan, fluorouracil, and leucovorin alone [14]. Patients additionally treated with bevacizumab for mCRC had a hazard ratio for death of 0.66 (P < 0.001) and 10.2 months median survival (vs. 6.1 months). Similar results were seen in metastatic renal cell carcinoma (mRCC) [15]. When bevacizumab was added to interferon alpha, as opposed to interferon alpha alone, the hazard ratio for death for death was 0.63 (p < 0.001) and patients lived a median 5.2 months longer. For patients that experienced recurrent cervical cancer (despite being treated with chemotherapy initially), bevacizumab boasted a 0.71 hazard ratio for death, and median 3.7 months of addition survival (p = 0.004) when added to combination non-platinum chemotherapy [16]. Lastly, a number of promising results have been seen in ovarian cancer. The overall response depends on cancer subtype and sensitivity to other chemotherapies. Burger et al. showed that a death hazard ratio of 0.71 (p < 0.001) for initial treatment of ovarian cancer with bevacizumab and had a relatively favorable side-effect profile [17].
Following the success of bevacizumab, there was interest in targeting other aspects of the VEGF angiogenic cascade. A number of tyrosine kinase inhibitors (TKIs) have been developed that (nonspecifically) target the VEGF receptor (VEGFR). Instead of removing VEGF from the local tumor milieu, these agents seek to abrogate the downstream reaction of VEGF/VEGFR signaling [18,19] and, in doing so, these agents appear to be mechanistically effective at reducing angiogenesis. The targeting of TKIs by these agents lacks specificity which, although potentially enabling them to broaden their anti-cancer effects, can also create greater toxicity and off-target effects.
3. Resistance to anti-angiogenic agents
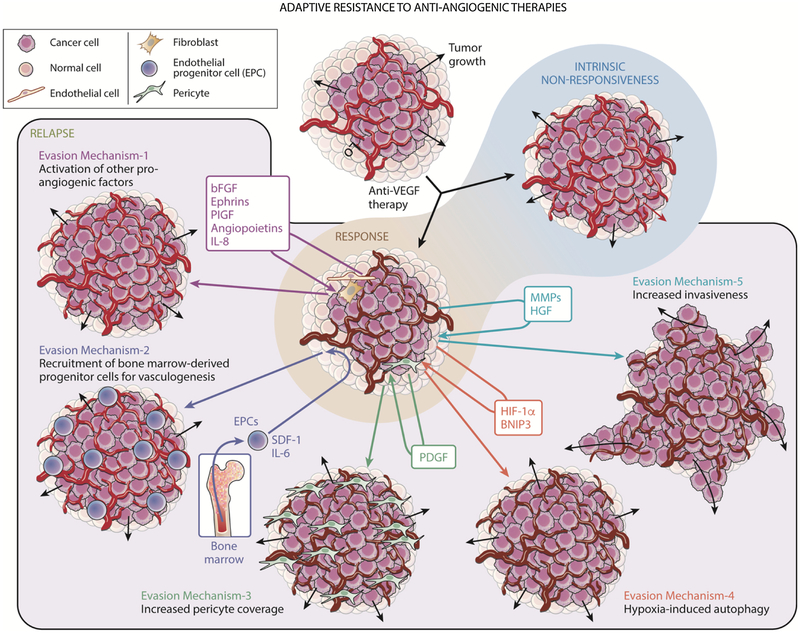
Despite the successes described above, anti-angiogenic therapy for cancer has encountered a number of failures as well. Despite receiving accelerated Food and Drug Administration (FDA) approval for GBM treatment in 2009 on the basis of non-randomized trials in which it was used as monotherapy, randomized trials revealed no impact of bevacizumab on GBM patient survival at recurrence [20] and at diagnosis [21]. Moreover, due to failure to prolong overall survival secondary to a transient therapeutic response, bevacizumab approval was revoked by the FDA for treatment of metastatic breast cancer in 2010 within 2 years of its approval [22]. Tumors have been found to evolve several adaptive responses to overcome the microenvironmental stressors caused by anti-angiogenic therapy (Fig. 1). It is worth noting that some tumors exhibit intrinsic, primary, resistance to anti-angiogenic therapy [23]. Intrinsic resistance is peculiar and likely is present in tumors that:1) have not begun upregulating vascular growth factors, 2) are utilizing vascular growth factors that are not targeted by therapy, or 3) are invading along pre-existing vascular tracks and do not need to produce more vessels [23–25].
Fig. 1. Adaptive mechanisms of evasion to anti-angiogenic therapy in cancer.
While some cancers may be intrinsically resistance to anti-angiogenic therapy, other tumors that are initially responsive may acquire adaptive resistance to anti-angiogenic therapy. These established mechanisms of resistance are as follows. Mechanism 1: Activation of other pro-angiogenic factors like basic fibroblast growth factor (bFGF), ephrins, placental growth factor (PlGF), angiopoietins, or interleukin 8 (IL-8) resulting in tumor vessels to replenish (from brown to red). Mechanism 2: Recruitment of bone marrow-derived progenitor cells for vasculogenesis. This is mediated by secreted factors such as interleukin 6 (IL-6) and stromal derived factor-1α (SDF-1α) which recruit bone marrow-derived endothelial progenitor cells (EPCs) into the tumor for vasculogenesis and replenishing vessels (brown to red). Mechanism 3: Increased pericyte coverage, driven by platelet derived growth factor (PDGF), which also nurtures depleted (brown) vessels. Mechanism 4: Hypoxia-induced autophagy, mediated by Hypoxia Induced Factor-1α and BNIP3, which helps tumor cells thrive in a hypoxic environment. Mechanism 5: Increased invasiveness of the tumor, mediated by matrix metalloproteinases (MMPs) and hepatocyte growth factor (HGF), which helps tumor cells invade despite depleted (brown) vessels and survive.
3.1. Alternative angiogenic pathways
Even for tumors that do initially respond to anti-angiogenic therapy, many will develop resistance over time. The methods of acquired, secondary resistance are varied, but all culminate in treatment failure. The first, and most intuitive, method of overcoming angiogenesis inhibition is upregulation of alternative VEGF-independent angiogenic pathways [26,27]. Because VEGF had been considered to be essential for angiogenesis, it was initially considered surprising that tumors were able to promote angiogenesis without availability of VEGF. Ang2, Bv8, FGF, Il-8, Ephrin, PIGF, and Il-1, to name a few, are some of the molecules associated with these alternative VEGF-independent angiogenic pathways [28]. One of the notable pathways is the Ang/Tie system. When angiogenesis is not indicated, Ang1 binds to Tie2 to promote vascular stability and maturation [29]. However, Ang2 acts as an antagonist to Ang1, and therefore prevents vessel stabilization. Many VEGF resistant tumors have been shown to express high levels of Ang2, which is thought to contribute to vessel expansion [28,30]. Curiously, some pro-angiogenic forces do not stem directly from cancer cells but instead arise from stromal cells in the tumor microenvironment. Due to the pro-inflammatory state present in cancer, there is a wealth of immune cells. Treatment with VEGF-targeted anti-angiogenic agents has been shown to induce immune cell recruitment, and this enrichment is thought to be a component of resistance to VEGF-targeted therapies [31]. Tumor associated neutrophils (TANs) have been shown to produce high quantities of Bv8, a molecule that activates the MAPK/ERK pathway and promotes new vessel formation [32]. Therefore, the inflammatory milieu around tumors is capable of driving VEGF-independent alternative angiogenic pathways and the recruitment of these inflammatory cells to tumors during anti-angiogenic therapy resistance is a mechanism of therapeutic resistance. Lastly, TGF-β, a molecule that has numerous pro-tumoral effects, can also influence angiogenesis [33]. It appears that TGF-β, when interacting with the Neuropilin-1 (nrp-1) receptor inhibits angiogenesis from surrounding endothelium [34]. However, when this receptor is downregulated, as is seen in bevacizumab treated tumors, the endothelium can proliferate to produce new vessels. As these alternative angiogenesis factors have been discovered, specific therapies have been developed to halt the effects of these molecules. Already there have been some promising studies targeting these molecules to potentially restore the therapeutic effect of bevacizumab [35–37], but such an approach may be impractical unless one or two crucial alternative pathways are identified whose targeting alongside VEGF leaves tumor cells with the availability of only minimally effective angiogenic pathways.
3.2. Increased pericyte coverage
The principle mural cell supporting blood vessels are pericytes. These cells are intimately entwined with endothelial cells and provide stability to growing as well as mature vessels [38,39]. It is believed that pericytes can lead advancing blood vessels and serve as a scaffold for further angiogenesis. Some VEGF resistant tumors have been found to be rich with pericytes and these cells appear to be correlated to anti-angiogenic resistance [38,39]. The abundance of pericytes could be due to 1) pericytes directly leading to the maturation of blood vessels without VEGF stimulation, or 2) due to the fact that pericytes have been shown to secrete VEGF themselves and therefore provide the missing angiogenic signal [38]. In short, even though bevacizumab can reduce the local levels of VEGF, the intimate secretion of VEGF by pericytes may not be affected by this agent. However, the extension of pericytes is itself guided by factors in the tumor milieu. PDGF is thought be a principle molecule in promoting pericyte extension and combined PDGF/VEGF has potential to stop this process [40–42]. Therefore, increased pericyte coverage and EPC recruitment are thought to be orchestrated via PDGF secretion and a common PDGF/VEGF therapy may abrogate both of these processes.
3.3. Vascular co-option and perivascular invasion
Early in tumorigenesis, the direct hijacking of blood vessels, termed vascular co-option, is a viable method of acquiring a blood supply [43]. This method of obtaining nutrients can be initiated by primary malignancies or metastases, but usually becomes insufficient as tumors become more massive. As mentioned above, some tumors have an inherent propensity to invade along vascular tracts and are therefore innately resistant to VEGF [24]. However, invasion can also be a form of adaptive resistance – some tumors change their invasiveness to overcome the pressure of VEGF. If tumors cells are always in close proximity to a vascular supply, nutrients can always diffuse into them and there is no need to construct new vessels. This was first demonstrated in a mouse model, where mice treated with anti-VEGF monoclonal antibodies developed tumors that preferentially invaded alongside pre-existing vasculature [44]. This finding has been validated in other studies, and now the mechanism of this resistance is being investigated [45]. Current theories posit that c-Met and/or integrins are responsible for VEGF-resistant-cancer invasion.
4. The autophagy pathway
Autophagy is a highly conserved biological process that allows cells to recycle unneeded or dysfunctional cytosolic elements [46]. This self-catabolic process is highly regulated and is adept at degrading specific intracellular targets. There are three defined types of autophagy: macro-autophagy, micro-autophagy, and chaperone-mediated autophagy (CMA), all of which promote proteolytic degradation of cytosolic components at the lysosome. Macro-autophagy delivers cytoplasmic cargo to the lysosome through the intermediary of a double membrane-bound vesicle, referred to as an autophagosome, that fuses with a lysosome to form an autolysosome [47]. In micro-autophagy, cytosolic components are directly taken up by the lysosome itself through invagination of the lysosomal membrane. Both macro-and micro-autophagy are able to engulf large structures through both selective and non-selective mechanisms. In CMA, targeted proteins are translocated across the lysosomal membrane in a complex with chaperone proteins (such as Hsc-70) that are recognized by the lysosomal membrane receptor lysosomal-associated membrane protein 2A (LAMP-2A), resulting in their unfolding and degradation [48]. Here, we focus on macro-autophagy, which is more implicated in tumor cell survival during therapeutic stressors. Autophagy is a high-regulated adaptive process and has multiple stages: induction, phagopore nucleation, expansion, maturation and fusion.
The first stage of macro-autophagy is induction, whereby the process of autophagy is triggered by cellular stress such as nutrition starvation, oxidative stress, protein aggregation, endoplasmic reticulum (ER) stress, and hypoxia/anoxia [46]. The two most characterized and most studied nutrient deficiency triggers of autophagy are deprivation of amino acids [49] and glucose [50]. Deprivation of amino acids results in the inhibition of mTOR, a serine/threonine kinase that serves as the master cell growth regulator, specifically, in mTORC1 complex, which plays a central role in autophagy. In high-nutrition states, mTORC1 directly binds and phosphorylates autophagy-related protein (Atg) 13 and Unc-51-like kinase 1 (ULK1) to inactivate the two and put the brakes on autophagy [51]. On the contrary, upon starvation, mTORC1 sites on ULK1 are dephosphorylated, allowing ULK1 to dissociate from mTORC1 and undergo autophosphorylation. Subsequently, ULK1 phosphorylates Atg13 and RB1-inducible coiled-coil protein 1 (FIP200) that subsequently activates the ULK1 complex [51]. Another important mediator of autophagy which is regulated by mTORC1 is transcription factor EB (TFEB), which serves as a master transcription factor and controls cellular clearance. This transcription factor is negatively regulated by mTORC1 such that it is released in states of starvation. TFEB controls autophagy by lysosomal recruitment and activity by directly regulating expression of the mTOR-activating Rag GTPase complex component Ras-related GTP-binding protein D (RagD), thereby forming a feedback loop for homeostasis of cellular metabolic state [52]. Autophagy may also be induced during glucose deprivation, which leads to a declining ATP:AMP ratio that is sensed by cell homeostasis regulatory kinase AMPK and serine/threonine-protein kinase STK11 (LKB1) [50]. Activation of autophagy is mediated by LKB1-AMPK-TSC2 axis in which LKB1 via AMPK inhibits mTORC1 directly by phosphorylation of Raptor as well as indirectly through activation of TSC2 complex, that can interact with other molecules in the autophagy pathway [53]. Alternatively, mTORC1 complex is bypassed via AMPK-mediated induction of autophagy by direct phosphorylation of ULK1, VPS34 and Beclin 1 [54].
These cellular stress signals in turn lead to the recruitment of Atgs, a family of proteins essential for formation of the autophagosome, to a specific subcellular site called the phagophore assembly site (PAS), that is present on the ER. Specifically, stress signaling pathways all converge at the ULK1 complex that comprises of ULK1, ATG13, ATG101 and FIP200 [55]. Once this complex is formed, the second stage of autophagy, phagophore nucleation, is initiated which is triggered by phosphorylation of components of the class III PI3K (PI3KC3) complex I that comprises of class III PI3K, vacuolar protein sorting 34 (VPS34), Beclin 1, ATG14, activating molecule in Beclin 1-regulated autophagy protein 1 (AMBRA1) as well as general vesicular transport factor (p115) [46]. This activates local phosphatidylinositol-3-phosphate (PI3P) production at the omegasome [56], an ER-membrane domain where PAS is localized, following which PI3P recruits 3 P effector proteins WD repeat domain phosphoinositide-interacting proteins (WIPI2) and zinc-finger FYVE domain-containing protein 1 (DFCP1) to the omegasome by interacting with their PI3 P-binding domains [57]. At the end of this stage, an isolation membrane is formed.
The third stage, phagophore expansion, is mediated by Ub-like Atg8 family of proteins such as microtubule-associated protein light chain 3 (LC3) proteins and γ-aminobutyric acid receptor-associated proteins (GABARAPs). Following the processing and activation of the nascent Atg8 proteins by Atg4B and Atg7, Atg8 is conjugated to the membrane associated phosphatidylethanolamine (PE) by WIPI2 mediated recruitment of Atg12-Atg5-Atg16L1 complex that propagates the Atg3-mediated conjugation process. Subsequently, this results in formation of conjugated, membrane-bound lipidated forms of Atg8s. Conjugated Atg8s play two prime roles in autophagy: (1) Attract components of the autophagic machinery that contain an LC3-interacting region (LIR) and plays a significant role in mediating selective autophagy where it sequesters specifically labelled cargo or protein aggregates into the autophagolysosomes via LIR-containing cargo receptors, and (2) It is required for elongation and closure of the phagophore membrane. In terms of elongation, the fourth stage of autophagy, in addition to Atg9-mediated delivery of lipid bilayers, several organelles such as ER, mitochondria, the plasma membrane, Golgi complex and recycling endosomes, donate their cellular membranes to the elongating autophagosomal membrane [46,48]. Upon completion of elongation, the membrane encapsules the target and the two ends of the phagophore fuse together to form a double-layered vesicle called the autophago-some [48].
Following formation, the autophagosome undergoes maturation-the next stage of autophagy. During this stage, autophagosome recruits molecules required to be trafficked to lysosomes, specifically, kinesin motor proteins and the proteins needed for fusion with the lysosome, namely, SNARE complex proteins, comprising of synaptosomal-associated protein 29 (SNAP29) and syntaxin 17 on the autophagosome and vesicle associated membrane protein 8 (VAMP8), on the lysosome [58,59]. This process is mediated by Atg8s as well as Atg14, as suggested by recent evidence [59,60]. Following maturation, the autophagosome undergoes fusion with the lysosome to form an autolyso-some. Historically, this process was thought to be only mediated by VTIIB, UVRAG, SNARE machinery such as VAMP7–9, VAMP8, VAMP9, SNAP 29 and syntaxin 17. However, recent reports have also implicated Hippo kinase orthologues serine/threonine-protein kinase 3 (STK3) and STK4 [61] and Atg14 in the fusion process, with Atg14 also playing a role in tethering the autophagosome to the lysosome to enhance the SNARE-mediated fusion process [58,59]. At this stage the targets of the autolysosome are degraded and their components can be recycled into other cellular pathways.
Pharmacologic inhibitors of autophagy are often classified based on whether they inhibit “early” or “late” autophagy, with early autophagy referring to the activities of Atg proteins and signaling complexes necessary for autophagosomal nucleation, elongation, and maturation but not fusion with the lysosome and degradation of the cargo and “late” autophagy referring to the degradation of the autophagosome and its cargo by the lysosome. Table 1 shows a list of autophagy inhibitors identified in preclinical studies that have been classified as early or late inhibitors.
Table 1.
Early and Late inhibitors of autophagy from preclinical studies.
| Class of Inhibitor | Name | Targeted Stage of Autophagy | Target of Inhibitor |
|---|---|---|---|
| Early Inhibitors | 3-methyadine [106] GSK-2126458 [107,108] PT210 [109] Wortmannin [110] |
Induction | PI3K |
| Compound 6 [111] MRT68921 [112] SBI-0206965 [113] |
Induction | ULK | |
| Compound 31 [114] PIK-III [115] Spautin-1 [116,117] SAR405 [118,119] Verteporfin [120,121] VPS34-IN1 [122] |
Phagosome Nucleation/Autophagosome Formation |
Vps34 | |
| NSC185058 [123] | Autophagosome Maturation | Atg4 | |
| Late inhibitors | Bafilomycin A1 [124,125,126] | Acidification of autophagolysosome | V-ATPases |
| E64d [127,128] Leupeptin [129] Pepstatin A [127,128] |
Breakdown of endocytosed product | Proteases | |
| ARN5187 [130] Chloroquine [92,131,132] Clomipramine [133] Hydroxychloroquine [132,134,135,136] Compound 30 [137] Lucanthone [138] Lys05 [139,140,141] ROC325 [142] |
Breakdown of endocytosed product | Lysosomes |
5. Autophagy in malignancy
Various studies [62–65] suggest that autophagy mainly contributes to tumor suppression during the early stage of tumorigenesis and tumor promotion during the late stage of tumorigenesis. During the tumorization of normal cells, autophagy protects genomic stability by slowing cell proliferation enough to allow time for repair of DNA damage, and inhibits the formation of a chronic inflammatory microenvironment, thus protecting normal cell homeostasis and preventing tumor generation. The first link between autophagy and preventing tumor formation was when Beclin-1, a member of the class III PI3K complex, was shown to serve as a tumor suppressor gene by promoting autophagy [66].
On the other hand, once tumorigenesis is initiated, autophagy may help maintain tumor cells and enable them to thrive. For example, the oncogenic mTOR complex is sensitive to the nutritional needs of the cell and as needed will inhibit ULK1, a member of an Atg (autophagy gene) protein complex required for phagophore formation [51]. It is worth noting that autophagy may progress independently of the previously described pathways and thus higher tumor burden caused by tumor suppressor knockouts may not always correlate with a reduction in autophagy. Indeed, targeted knockout of core autophagic factors such as Atg5 and Atg7 show a distinct phenotype that involves increased accumulation of ubiquitinated proteins, but not increased tumor levels [67]. Once tumorigenesis is initiated, autophagy protects tumor cells by providing nutritional resources to meet high metabolic demand via macromolecule digestion [68] and supporting anoikis resistance and dormancy [69]. Tumor suppressors such as LBK1 and PTEN are linked to the mTOR pathway and consequently, their dysregulation can cause downstream inhibition of autophagy [70].
In addition to autophagy promoting the survival of tumor cells after tumorigenesis has initiated, several microenvironmental stressors sometimes seen during anticancer therapies will upregulate autophagy in cancer. Previous research has indicated that increased levels of oxidative stress leading to higher concentrations of reactive oxygen species (ROS), which is a common hallmark of cancer progression and treatment [71], is seen to positively regulate autophagy via redox-dependent and –independent pathways [72]. Autophagy has been shown to work in tandem with processes commonly seen in malignant tumors, such as apoptosis and necrosis, although the exact nature of how they are coordinated is unclear. Multiple proteins such as BCL-2 [73], DRAM1 [74] and PUMA [75] all have been shown to have roles in to co-activation of apoptosis and autophagy in cancers. In addition, studies have shown that the synchronized absence of both apoptosis and autophagy leads to increased tumor growth and necrosis as compared to apoptosis-induced death alone [76]. Autophagy activation can also promote tumor growth via silencing of necrosis-induced inflammatory responses, with a reciprocal response seen in tumor cells evading a necrotic fate by activating the LKB1/AMPK complex, which in turn inhibits mTOR and activates downstream autophagic processes [77].
Taken together, autophagy appears to play a role as a protector for either normal or tumor cells during the early or late stage of tumorigenesis, respectively. One hypothesis to resolve these discordant roles of autophagy is that autophagy always has pro-tumoral consequences, they are just more indirect before tumorigenesis has initiated. Specifically, before tumorigenesis has begun, normal cells that have lower autophagy capacity are slowed in their growth, while normal cells with higher autophagy capacity find it easier to survive internal and external microenvironment stressors and accumulate more mutations to promote malignant progression [78].
6. Autophagy as a resistance mechanism to anti-angiogenic therapy
While initial clinical trials showed promise that anti-angiogenic therapies could halt tumor progression by targeting atypical vasculature in the tumor, the excitement was tempered by failure of angiogenesis inhibitors to produce long-term clinical responses in most patients. For instance, in clinical trials for bevacizumab, an anti-VEGF monoclonal antibody, in glioblastoma patients, while there was an initial termination in tumor progression, over 50% of the tumors progressed with acquiring invasive resistance to anti-angiogenic treatment [79], conferring a poor prognosis to these patients [80]. Several other clinical trials and reports have shown similar outcomes of either non-responsiveness to anti-angiogenic therapies or tumor progression under this therapy in colorectal cancer [81], HER2+ breast cancer [82], HER2+ metastatic or locally recurrent breast cancer [83], esophageal cancer [84] and hepatocellular carcinoma [85].
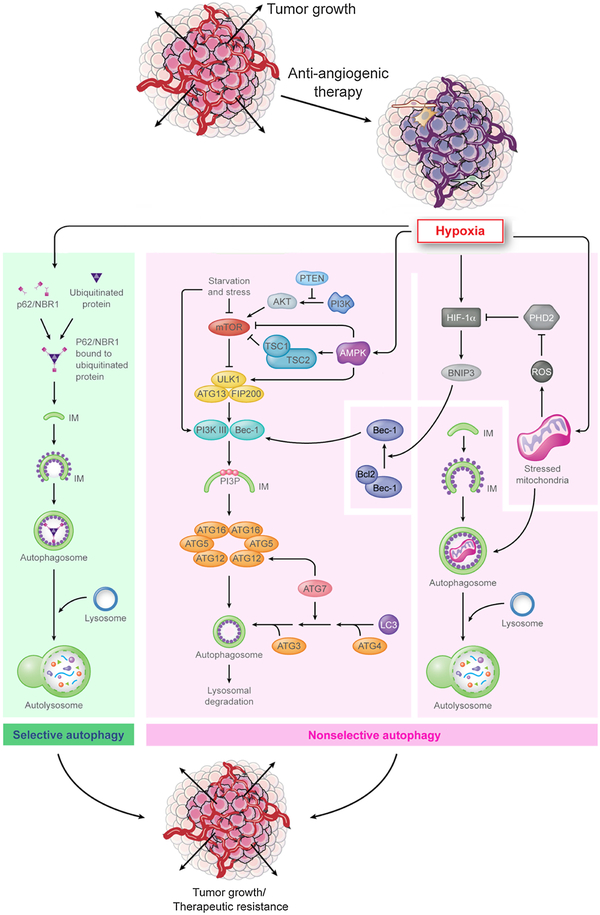
As discussed above, while several mechanisms have been implicated in driving anti-angiogenic resistance in cancer, most of these pathways occur later in tumor evolution. However, hypoxic survival (autophagy) is likely to be the first defense mechanism that occurs at the cellular level without involving tissue or ECM remodeling and thus is important to understand better. Autophagy, a highly conserved and highly regulated adaptive process, has both an apoptotic and pro-survival role in cancer and can lead to anti-angiogenic escape due to its latter role. Anti-angiogenic agents target blood vessel formation in tumors that leads to a hypoxic microenvironment, a critical cellular stressor, that is detrimental to proliferating cells due to lack of effective blood supply to the tumor. However, some tumor cells are able to overcome the harsh microenvironment promoted by anti-angiogenic therapy and actually thrive and proliferate. Studies in the past decade have elucidated two primary mechanisms driving hypoxia-driven autophagy, namely, non-selective and selective autophagy (Fig. 2), with the former catabolizing cytoplasmic components non-selectively and the latter targeting specific protein aggregates designated by the cell for destruction [86]. Non-selective autophagy can be further divided into: (1) HIF-dependent autophagy, induced in hypoxic conditions (3–0.1% O2), is mediated by several genes and proteins such as abnormal BH3-only proteins, specifically, the Bcl-2/E1B 19kDa-interacting protein 3 (BNIP3/BNIP3L (NIX)), sequestosome/p62 [87], DJ-1 [88] and the PDGFR protein family [89]. These proteins also mediate a pro-survival metabolic adaptive process called mitophagy, which modulates oxidative stress, reactive oxygen species (ROS) production and DNA damage [90]. (2) HIF-independent autophagy, induced in anoxic conditions (< 0.1% O2), is driven by the AMPK-mTOR and unfolded protein response, both triggered by severe cellular nutrition deprivation and accumulation of toxic metabolites not cleared due to poor blood flow [90]. In terms of selective autophagy, this process is driven by autophagy and degradation of ubiquitinated protein aggregates that are bound by a molecule, p62, which tags them for autophagic degradation [91]. The hypothesis is that through this process, the tumor cells can target organelles and other proteins associated with oxidative stress and associated damage to the cell, thereby overcoming the secondary effects of hypoxia due to anti-angiogenics, a common end goal of adaptive autophagy.
Fig. 2. Mechanisms underlying autophagy-mediated resistance to anti-angiogenic therapy.
Hypoxia results in anti-angiogenic therapy resistance by either selective autophagy (green), that is mediated by p62/NBR1, which sequesters targeted protein aggregates for autophagy, or by (B) non-selective autophagy (pink), which is mediated by two mechanisms (i) First is driven by HIF-1α that activates downstream mediators such as BNIP3. This mediator interacts with Beclin-1 and activates downstream autophagy pathways. Alternatively, hypoxia-induced mitochondrial stress can lead to autophagy activation. (ii) Second, starvation and cellular stress can modulate the Akt/mTOR pathway that can subsequently activate the autophagy pathway as an adaptive mechanism. Additionally, hypoxia can directly activate AMPK that interacts with the Akt/mTOR pathway and leads to activation of autophagy.
In GBM, anti-angiogenic resistance to bevacizumab has been shown to be driven by BNIP3, a hypoxia-inducible factor-1α (HIF-1α) downstream target gene that is essential for hypoxia-induced autophagy (as described above). In GBM patient specimens, those that were resistant to bevacizumab had a higher level of hypoxia and higher expression of BNIP3 compared to patient specimen that were pretreated with bevacizumab from the same patients. Moreover, pharmacological or genetic inhibition of autophagy in GBM xenograft models increased sensitivity to anti-angiogenic therapy with bevacizumab in these xenografts suggesting autophagy as potential therapeutic target [92]. At the mechanistic level, BNIP3 overexpression leads to autophagy-induced upregulation of AMPK by interacting with Beclin-1/Bcl2 complex that activates mediators of autophagy downstream of AMPK. Activation of AMPK and HIF-1α pathways serves as an adaptive response that maintains energy production in the tumor in the unfavorable hypoxic conditions, thus leading to invasive resistance and tumor progression [92,93]. Interestingly, another report has shown that the activation of autophagy promotes bevacizumab resistance by suppression of mTOR pathway supporting the findings from previous reports [94]. Similar to GBM, in hepatocellular carcinoma, upregulation of AMPK and HIF-1α drives anti-angiogenic therapy resistance [85], while a recent report showed that bevacizumab treatment of colorectal cancers drives autophagy in a HIF-1 dependent manner, that is targetable and can enhance the effects of bevacizumab with an autophagy inhibitor [95].
Additional studies have shown autophagy to be a targetable process in perturbing resistance and enhancing anti-angiogenic therapeutic effects. A recent report in hepatocellular carcinoma found that bevacizumab led to development of hypoxia in the tumor that also led to upregulation of Beclin1 and LC3, autophagy-associated genes, and increased autophagosome formation. Moreover, in hepatocellular carcinoma xenografts, combined treatment of chloroquine, a late inhibitor of autophagy, and bevacizumab was more effective at inhibiting tumor growth, enhanced tumor cell apoptosis, and impaired the proliferation of tumor cells compared with treatment with either drug alone [96]. Similar results were seen in colon cancer models, where pharmacological inhibition with chloroquine and genetic inhibition of using RNA interference of Atg5 and Beclin-1, proteins playing a role in early autophagy, enhanced bevacizumab efficacy in both normoxic and hypoxic environments, while in xenografts, bevacizumab induced hypoxia-driven autophagy and was found to have more potentiated effects on tumor proliferation and growth when xenografts were concomitantly treated with bevacizumab and chloroquine [97].
Studies in other cancers have revealed alternative molecular underpinnings driving autophagy-mediated anti-angiogenic escape. One such finding is in breast adenocarcinoma, in which, using ectopic kinase expression screening for tamoxifen resistance, protein based assays and cell-based assays, a specific kinase, HSPB8, was identified and was found to be upregulated by tumor cells, the role of which was to block autophagy secondary to anti-angiogenic effects of tamoxifen and induced cell proliferation in the presence of tamoxifen via modulation of mTOR pathway [98]. Interestingly, while several different molecules may be implicated in autophagy as a resistance mechanism to anti-angiogenic therapy, most of these studies seem to reveal AMPK/mTOR pathway as a critical node where pathways promoting autophagy intersect, underscoring the relative importance of this pathway in autophagy-mediated anti-angiogenic resistance and pro-survival signals.
At the time of this writing, 22 clinical trials for autophagy inhibitors, primarily, chloroquine or hydroxychloroquine, in cancer have either been completed, are open or active in the United States (www.clinicatrials.gov) Table 3. Based on the pre-clinical insights above suggesting autophagy as a resistance mechanism to anti-angiogenic therapy, of the clinical trials studying autophagy inhibitors in cancer, 23% (n = 5) combine autophagy inhibitors with anti-angiogenic agents (Table 2) with four of the five involving bevacizumab [99–102] and one of the five involving sunitinib [103]. Clinical trials for autophagy inhibitors in various cancers without anti-angiogenics can be found in Table 3.
Table 3.
Active or Completed Clinical Trials of autophagy inhibitors in different cancer types.
| NCT# and Status | Trial Name | Start Date | Participant Number | Study Phase | Indication | Autophagy Inhibitor | Study Arms |
|---|---|---|---|---|---|---|---|
| NCT03598595 Recruiting [143] | Gemcitabine, Docetaxel, and Hydroxychloroquine in Treating Participants With Recurrent or Refractory Osteosarcoma | January 2019 | 31 | I/II | Refractory and Recurrent Osteosarcoma | Hydroxychloroquine | Experimental Arm: (single arm) Hydroxychloroquine, Docetaxel, Gemcitabine |
| NCT03774472 Recruiting [144] | Hydroxychloroquine, Palbociclib, and Letrozole Before Surgery in Treating Participants With Estrogen Receptor Positive, HER2 Negative Breast Cancer | August 2018 | 54 | I/II | ER+/HER2- Breast Cancer | Hydroxychloroquine | Experimental Arm: (single arm) Letrozole, Palbociclib, Hydroxychloroquine |
| NCT03344172 Active/Suspended [145] | Pre-Operative Trial (PGHA vs. PGH) for Resectable Pancreatic Cancer (17134) | December 2017 | 120 | II | Resectable Pancreatic Cancer | Hydroxychloroquine | Experimental Arm: Cohort 1: Gemcitabine, Nab-Paclitaxel, and hydroxychloroquine and Avelumab Cohort 2: Gemcitabine, Nab-Paclitaxel, and hydroxychloroquine |
| NCT03037437 (Recruiting) [146] | Sorafenib Induced Autophagy Using Hydroxychloroquine in Hepatocellular Cancer | June 2017 | 68 | II | Hepatocellular Cancer | Hydroxychloroquine | Experimental Arm: Cohort 1: Sorafenib, Hydroxychloroquine Cohort 2: Sorafenib, Hydroxychloroquine in Sorafenib naive patients |
| NCT02316340 Active [147] | Vorinostat Plus Hydroxychloroquine Versus Regorafenib in Colorectal Cancer | February 2015 | 76 | II | Colorectal Cancer | Hydroxychloroquine | Experimental Arm: Vorinostat, Hydroxychloroquine Control Arm: Regorafenib |
| NCT02257424 Recruiting [148] | Dabrafenib, Trametinib and Hydroxychloroquine in Patients With Advanced BRAF Mutant Melanoma (BAMM) | October 2014 | 53 | I/II | Advanced BRAF Mutant Melanoma | Hydroxychloroquine | Experimental Arm: (single arm) Hydroxychloroquine, Trametinib, dabrafenib |
| NCT01687179 Completed (August 2015) [149] | Safety Study of Sirolimus and Hydroxychloroquine in Women With Lymphangioleiomyomatosis (SAIL) | September 2012 | 14 | I | Lymphangioleiom-yomatosis | Hydroxychloroquine | Experimental Arm: (Single arm) Sirolimus and Hydroxychloroquine |
| NCT01634893 Completed (December 2015) [150] | Oral Hydroxychloroquine Plus Oral Sorafenib to Treat Patients With Refractory or Relapsed Solid Tumors | June 2012 | 28 | I | Refractory or Relapsed Solid Tumors | Hydroxychloroquine | Experimental Arm: (single arm) Sorafenib,Hydroxychloroquine |
| NCT01494155 Active [151] | Short Course Radiation Therapy With Proton or Photon Beam Capecitabine and Hydroxychloroquine for Resectable Pancreatic Cancer | December 2011 | 50 | II | Pancreatic Cancer | Hydroxychloroquine | Experimental Arm: (single arm) Hydroxychloroquine, Capecitabine, Proton or Photon Radiation Therapy |
| NCT01506973 Active [152] | A Phase I/II/Pharmacodynamic Study of Hydroxychloroquine in Combination With Gemcitabine/Abraxane to Inhibit Autophagy in Pancreatic Cancer | December 2011 | 119 | I/II | Advanced and metastatic Pancreatic Cancer | Hydroxychloroquine | Experimental Arm: (single arm) Gemcitabine/Abraxane, Hydroxychloroquine |
| NCT01480154 Active [153] | Akt Inhibitor MK2206 and Hydroxychloroquine in Treating Patients With Advanced Solid Tumors, Melanoma, Prostate or Kidney Cancer | November 2011 | 62 | I | Advanced Malignant Solid Neoplasm, Stage III A-C Cutaneous Melanoma AJCC v7, Stage III/IV Prostate Cancer AJCC v7, Stage III/IV Renal Cell Cancer AJCC v7 | Hydroxychloroquine | Experimental Arm: (single arm) Akt inhibitor MK2206 and hydroxychloroquine |
| NCT01510119 Completed (January 2014) [154] | Autophagy Inhibition to Augment mTOR Inhibition: A Phase I/II Trial of RAD001 and Hydroxychloroquine in Patients With Previously Treated Renal Cell Carcinoma | September 2011 | 40 | I/II | Metastatic Clear Cell Renal Cell Carcinoma | Hydroxychloroquine | Experimental Arm: Everolimus and hydroxychloroquine |
| NCT01266057 Active [155] | Sirolimus or Vorinostat and Hydroxychloroquine in Advanced Cancer | April 2011 | 143 | I | Advanced Cancers | Hydroxychloroquine | Experimental Arm: Sirolimus Hydroxychloroquine; Experimental Arm: Vorinostat, Hydroxychloroquine, |
| NCT00568880 Completed (April 2012) [156] | Hydroxychloroquine and Bortezomib in Treating Patients With Relapsed or Refractory Multiple Myeloma | September 2010 | 25 | I/II | Multiple Myeloma and Plasma Cell Neoplasm | Hydroxychloroquine | Experimental Arm: Bortezomib and hydroxychloroquine |
| NCT00962845 Completed (May 2013) [157] | Hydroxychloroquine in Patients With Stage III or Stage IV Melanoma That Can Be Removed by Surgery | September 2010 | 20 | 0/I | Stage III or IV Resectable Melanoma | Hydroxychloroquine | Experimental Arm: (Single Arm) Hydroxychloroquine pretreatment to biopsy |
| NCT00813423 Active [103] | Sunitinib Malate and Hydroxychloroquine in Treating Patients With Advanced Solid Tumors That Have Not Responded to Chemotherapy | January 2010 | 40 | I | Adult Solid neoplasms | Hydroxychloroquine | Experimental Arm: (single arm) Hydroxychloroquine and Sunitinib Malate |
| NCT01023477 Completed (October 2016) [158] | Study of the Efficacy of Chloroquine in the Treatment of Ductal Carcinoma in Situ (The PINC Trial) | December 2009 | 12 | I/II | Breast Ductal Carcinoma In Situ and Noninfiltrating Intraductal Carcinoma | Chloroquine | Experimental Arm: Cohort 1: Chloroquine (500 mg/week) Cohort 2: Chloroquine (250 mg/week) |
| NCT01023737 Active [159] | Hydroxychloroquine + Vorinostat in Advanced Solid Tumors | November 2009 | 72 | I | Malignant Solid Tumors | Hydroxychloroquine | Experimental Arm: (single arm) Hydroxychloroquine and Vorinostat |
| NCT01978184 Completed (February 2018) [160] | Randomized Phase II Trial of PreOperative Gemcitabine and Nab Paclitaxel With or With Out Hydroxychloroquine | May 2009 | 39 | II | Colorectal Cancer | Hydroxychloroquine | Control Arm: gemcitabine and abraxane Experimental Arm: gemcitabine, abraxane and hydroxychloroquine |
| NCT00909831 Completed (May 2012) [161] | Hydroxychloroquine and Temsirolimus in Treating Patients With Metastatic Solid Tumors That Have Not Responded to Treatment | October 2008 | 40 | I | Adult Solid Tumor | Hydroxychloroquine | Experimental Arm: Temsirolimus, hydroxychloroquine |
| NCT00726596 Active/Unknown [162] | Hydroxychloroquine in Treating Patients With Rising PSA Levels After Local Therapy for Prostate Cancer | August 2008 | 64 | II | Prostate Cancer | Hydroxychloroquine | Experimental Arm: Cohort 1: Hydroxychloroquine (400 mg) Cohort 2: Hydroxychloroquine (600 mg) |
| NCT00486603 Completed (January 2014) [163] | Hydroxychloroquine, Radiation, and Temozolomide Treating Patients With Newly Diagnosed Glioblastoma Multiforme | October 2007 | 92 | I/II | Brain and Central Nervous System Tumors | Hydroxychloroquine | Experimental Arm: Phase I Radiotherapy, temozolomide, hydroxychloroquine (200 mg) Experimental Arm: Phase I Radiotherapy, temozolomide, hydroxychloroquine (400 mg) Experimental Arm: Phase I Radiotherapy, temozolomide, hydroxychloroquine (600 mg) Experimental Arm: Phase I Radiotherapy, temozolomide, hydroxychloroquine (800 mg) Experimental Arm: Phase II Radiotherapy, temozolomide, hydroxychloroquine (maximum tolerable dose) |
Table 2.
Clinical Trials of autophagy inhibitors with anti-angiogenic regimens in different cancer indications.
| NCT# and Status | Trial Name | Start Date | Participant Number | Study Phase | Indication | Autophagy Inhibitor | Anti-angiogeni c agent | Study Arms |
|---|---|---|---|---|---|---|---|---|
| NCT00728845 Completed (December 2010) [99] | Modulation of Autophagy With Hydroxychloroquine in Combination With Carboplatin, Paclitaxel and Bevacizumab in Patients With Advanced/Recurrent Non-Small Cell Lung Cancer - A Phase I/II Study | June 2008 | 8 | I/II | Lung Cancer | Hydroxyc hlor oquine | Bevacizumab | Experimental Arm: Hydroxychloroquine, Carboplatin, Paclitaxel, Bevacizumab Control Arm: Hydroxychloroquine, Carboplatin, Paclitaxel |
| NCT01006369 Completed (August 2015) [100] | Autophagy and Anti-Angiogenesis in Metastatic Colorectal Carcinoma: A Phase II Trial of Hydroxychloroquine to Augment Effectiveness of XELOX-Bevacizumab. A Study of the Cancer Institute of New Jersey Oncology Group (CINJOG) | May 2009 | 39 | II | Colorectal Cancer | Hydroxyc hlor oquine | Bevacizumab | Experimental Arm: FOLFOX6, Bevacizumab, Hydroxyc hlor oquine Control Arm: XELOX, Bevacizumab, Hydroxyc hlor oquine |
| NCT00813423 (Active, not recruiting) [103] | Autophagic Modulation With Anti-angio genic Therapy in Patients With Advanced Malignancies: A Phase I Trial of Sunitinib and Hydroxychloroquine | January 2010 | 40 | I | Adult solid neoplasms | Hydroxyc hlor oquine | Sunitinib | Experimental Arm: (single arm) Sunitinib, Hydroxychloroquine |
| NCT01206530 Completed (November in 2016) [102] | A Phase I/II Pharmacodynamic Study of Hydroxychloroquine in Combination With FOLFOX Plus Bevacizumab to Inhibit Autophagy in Colorectal Cancer | September 2010 | 50 | I/II | Advanced Colorectal and Rectal Cancer, Metastatic Colorectal and Rectal Cancer | Hydroxyc hlor oquine | Bevacizumab | Experimental Arm: (Single arm) FOLFOX, Bevacizumab, Hydroxyc hlor oquine |
| NCT01649947 Completed (June 2015) [101] | Modulation of Autophagy With Hydroxychloroquine in Patients With Advanced/Recurrent Non-small Cell Lung Cancer - a Phase II Study. A Study of The Cancer Institute of New Jersey Oncology Group (CINJOG) | December 2011 | 32 | II | Non-small Cell Lung Cancer, Advanced Non-small Cell Lung Cancer, Recurrent Non-small Cell Lung Cancer | Hydroxyc hlor oquine | Bevacizumab | Experimental Arm: Paclitaxel, Carboplatin, Hydroxychloroquine, Bevacizumab Control Arm: Hydroxychloroquine, Carboplatin, Paclitaxel |
7. Future directions
Autophagy is a high regulated and critical physiological process essential for normal functioning of cells and cell survival in stressful microenvironments. Recent studies have shed light on the role of autophagy in tumorigenesis and resistance of tumors to anticancer therapies like anti-angiogenics. Leveraging this information into translational insights that would benefit patients by prolonging the duration of efficacy of anti-angiogenic therapy will require overcoming challenges related to autophagy and anti-angiogenic therapy.
Regarding autophagy, while several molecules and genes have been implicated in the development of autophagy-mediated anti-angiogenic resistance and may serve as potential targets to overcome this resistance, it is imperative to identify more specific targets to avoid undesirable adverse effects in the clinic. In particular, it remains to be determined whether early versus late inhibitors of autophagy will be best suited to prevent anti-angiogenic therapy resistance with acceptable rates of side effects. Once candidate druggable molecules are identified, further studies may be needed to identify tumor-specific targets on the candidate molecule. For instance, AMPK upregulation has been implicated in anoxia-induced autophagy in GBM. However, targeting AMPK is impractical as this molecule plays a significant role in survival of neurons, glial cells and other cell types in body. To target AMPK, it might be feasible for future preclinical studies to focus on identifying phosphorylation site alterations and/or more downstream molecules that could be targetable in overcoming autophagy-mediated anti-angiogenic resistance. Moreover, simultaneous dysregulation of multiple pathways funneling into autophagy makes targeted therapy challenging which can be overcome with using combination therapies targeting multiple pathways, or a common node to several pathways to prevent any compensatory response to anti-autophagy therapies. Additionally, autophagy inhibition will be ineffective because physiologic autophagy may be cytocidal, whereas stress-induced autophagy as seen in response to chemotherapeutics and anti-angiogenics, may be cytoprotective. In terms of autophagy inhibitors, there is a dire need for developing additional therapeutics targeting autophagy in the context of bevacizumab beyond chloroquine and hydroxychloroquine, given their conflicting treatment effects. While studies have suggested that these agents disrupt autophagy in in vivo models [92], several other reports have shown that chloroquine may potentiate the effects of autophagy-inducing chemotherapies in a non-autophagy dependent manner [104]. While further investigation of these drugs is warranted in the context of identifying sensitive tumors types, temporal onco-genesis stage of optimal therapy and optimal genetical profile leading to susceptible tumors, additional targeted therapies against autophagy need to be developed that could target autophagy specific downstream molecular mediators, proteins involved in regulation of autophagy activation and autophagosome formation which have minimal intracellular roles outside of autophagy. Moreover, combination therapies that could enhance the effects of autophagy inhibitors, chemotherapeutics and anti-angiogenics are imperative to prevent acquired resistance by monotherapy. These discoveries have the potential to improve the efficacy of current anti-angiogenic treatments for malignancies either by identifying new molecular targets or via administration of multi-pathway inhibitors and could unravel the full potential of anti-angiogenic therapies in cancer.
Related to anti-angiogenic therapy, identification of biomarkers predicting when resistance to anti-angiogenic therapy is evolving before it becomes entrenched [105] will be crucial to enable timely intervention with autophagy inhibitors. Studies will also need to determine whether autophagy inhibitors are best used alongside anti-angiogenic therapy from the beginning or added to anti-angiogenic therapy when concerns about resistance arise.
8. Conclusion
Autophagy is a highly conversed cellular process that is essential for degradation of cytosolic protein aggregates and damaged organelles for cellular homeostasis. This process, activated by cellular stress, plays a central role in malignancies and can help tumor cells overcome tissue injury due to radiation therapy and chemotherapeutics causing acquired chemoradiation resistance. Recent literature revealed autophagy as a mechanism of resistance to anti-angiogenic therapy, induced by hypoxia and severe nutrition deficit. While several challenges will need to be overcome to translate these insights into therapeutic gain, studies to date provide a source of optimism that we may someday be able to prolong the efficacy of anti-angiogenic therapy and enable this therapeutic modality to fulfill its promise.
Acknowledgement
We would like to acknowledge Ken Probst for his outstanding art-work.
Funding
Ankush Chandra and Jonathan W. Rick are supported by the Howard Hughes Medical Institute (HHMI). Dr. Garima Yagnik is supported by T32CA151022. Dr. Manish Aghi is supported by 2R01NS079697.
Abbreviations:
- CAF
cancer associated fibroblasts
- ECM
extracellular matrix
- VEGF
vascular endothelial growth factor
- HIF
hypoxia-inducible factor
- PDGF
platelet-derived growth factor
- GBM
glioblastoma
- mRCC
metastatic renal cell carcinoma
- TKI
tyrosine kinase inhibitors
- VEGFR
VEGF receptor
- TAN
tumor associated neutrophils
- EPC
endothelial progenitor cells
- FDA
Food and Drug Administration
- CMA
chaperone-mediated autophagy
- LAMP-2A
lysosomal-associated membrane protein 2A (LAMP-2A)
- Atg
autophagy-related proteins
- ER
endoplasmic reticulum
- ULK1
Unc-51-like kinase 1
- TFEB
transcription factor EB
- AMBRA1
activating molecule in Beclin 1-regulated autophagy protein 1
- GABARAP
γ-aminobutyric acid receptor-associated proteins
- PI3P
phosphatidylinositol-3-phosphate
- PE
phosphatidylethanolamine
- LC3
Mmicrotubule-associated protein light chain 3
- LIR
LC3-interacting region
- SNAP
synaptosomal-associated protein
- VAMP
vesicle associated membrane protein
- STK
serine/threonine-protein kinase
- ROS
reactive oxygen species
Footnotes
Ethics statement
All authors declare no conflict of interest. No studies were performed on humans or animals in the generation of this manuscript. All authors contributed to the composition and intellectual design of this manuscript.
Declaration of Competing Interest
None.
References
- [1].Nishida N, Yano H, Nishida T, Kamura T, Kojiro M, Angiogenesis in cancer, Vasc. Health Risk Manage 2 (2006) 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Adair TH, Montani J-P, Angiogenesis, Angiogenesis, (2010). [Google Scholar]
- [3].Hanahan D, Weinberg RA, Hallmarks of cancer: the next generation, Cell 144 (2011) 646–674. [DOI] [PubMed] [Google Scholar]
- [4].Loizzi V, Del Vecchio V, Gargano G, De Liso M, Kardashi A, Naglieri E, Resta L, Cicinelli E, Cormio G, Biological pathways involved in tumor angiogenesis and bevacizumab based anti-angiogenic therapy with special references to ovarian cancer, Int. J. Mol. Sci 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Avraamides CJ, Garmy-Susini B, Varner JA, Integrins in angiogenesis and lymphangiogenesis, Nat. Rev. Cancer 8 (8) (2008) 604–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Krock BL, Skuli N, Simon MC, Hypoxia-induced angiogenesis: good and evil, Genes Cancer 2 (2011) 1117–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kikuchi H, Pino MS, Zeng M, Shirasawa S, Chung DC, Oncogenic KRAS and BRAF differentially regulate hypoxia-inducible factor-1 and −2 in colon cancer, Cancer Res 69 (2009) 8499–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Majmundar AJ, Wong WJ, Simon MC, Hypoxia-inducible factors and the response to hypoxic stress, Mol. Cell 40 (2) (2010) 294–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Folkman J, Angiogenesis in cancer, vascular, rheumatoid and other disease, Nat. Med 1 (1995) 27–31. [DOI] [PubMed] [Google Scholar]
- [10].Höckel S, Mitze S, Vaupel, Hypoxia and Radiation Response in Human Tumors, Semin. Radiat. Oncol 6 (1996) 3–9. [DOI] [PubMed] [Google Scholar]
- [11].Nagy JA, Chang S-H, Dvorak AM, Dvorak HF, Why are tumour blood vessels abnormal and why is it important to know? Br. J. Cancer 100 (2009) 865–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ozawa CR, Banfi A, Glazer NL, Thurston G, Springer ML, Kraft PE, McDonald DM, Blau HM, Microenvironmental VEGF concentration, not total dose, determines a threshold between normal and aberrant angiogenesis, J. Clin. Invest 113 (2004) 516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Al-Husein B, Abdalla M, Trepte M, Deremer DL, Somanath PR, Antiangiogenic therapy for cancer: an update, Pharmacotherapy 32 (2012) 1095–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S, Holmgren E, Ferrara N, Fyfe G, Rogers B, Ross R, Kabbinavar F, Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer, N. Engl. J. Med 350 (2004) 2335–2342. [DOI] [PubMed] [Google Scholar]
- [15].Escudier B, Pluzanska A, Koralewski P, Ravaud A, Bracarda S, Szczylik C, Chevreau C, Filipek M, Melichar B, Bajetta E, Gorbunova V, Bay J-O, Bodrogi I, Jagiello-Gruszfeld A, Moore N, A.T. investigators, Bevacizumab plus interferon alfa-2a for treatment of metastatic renal cell carcinoma: a randomised, double-blind phase III trial, Lancet 370 (2007) 2103–2111. [DOI] [PubMed] [Google Scholar]
- [16].Tewari KS, Sill MW, Long HJ, Penson RT, Huang H, Ramondetta LM, Landrum LM, Oaknin A, Reid TJ, Leitao MM, Michael HE, Monk BJ, Improved survival with Bevacizumab in advanced cervical cancer, N. Engl. J. Med 370 (2014) 734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Friedman HS, Prados MD, Wen PY, Mikkelsen T, Schiff D, Abrey LE, Yung WKA, Paleologos N, Nicholas MK, Jensen R, Vredenburgh J, Huang J, Zheng M, Cloughesy T, Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma, J. Clin. Oncol 27 (2009) 4733–4740. [DOI] [PubMed] [Google Scholar]
- [18].McTigue M, Murray BW, Chen JH, Deng Y-L, Solowiej J, Kania RS, Molecular conformations, interactions, and properties associated with drug efficiency and clinical performance among VEGFR TK inhibitors, Proc. Natl. Acad. Sci 109 (2012) 18281–18289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Harrison C, A deeper understanding of VEGFR inhibitors, Nat. Rev. Cancer 12 (2012) 735. [DOI] [PubMed] [Google Scholar]
- [20].Kreisl TN, Kim L, Moore K, Duic P, Royce C, Stroud I, Garren N, Mackey M, Butman JA, Camphausen K, Park J, Albert PS, Fine HA, Phase II trial of single-agent bevacizumab followed by bevacizumab plus irinotecan at tumor progression in recurrent glioblastoma, J. Clin. Oncol 27 (5) (2009) 740–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, Colman H, Chakravarti A, Pugh S, Won M, Jeraj R, Brown PD, Jaeckle KA, Schiff D, Stieber VW, Brachman DG, Werner-Wasik M, Tremont-Lukats IW, Sulman EP, Aldape KD, Curran WJ Jr, Mehta MP, A randomized trial of bevacizumab for newly diagnosed glioblastoma, N. Engl. J. Med 370 (8) (2014) 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sasich LD, Sukkari SR, The US FDAs withdrawal of the breast cancer indication for Avastin (bevacizumab), Saudi Pharm. J 20 (4) (2012) 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lu KV, Bergers G, Mechanisms of evasive resistance to anti-VEGF therapy in glioblastoma, CNS Oncol 2 (2013) 49–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kindler HL, Pancreatic cancer: an update, Curr. Oncol. Rep 9 (2007) 170–176. [DOI] [PubMed] [Google Scholar]
- [25].Loges S, Schmidt T, Carmeliet P, Mechanisms of resistance to anti-angiogenic therapy and development of third-generation anti-angiogenic drug candidates, Genes Cancer 1 (2010) 12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Casanovas O, Hicklin DJ, Bergers G, Hanahan D, Drug resistance by evasion of antiangiogenic targeting of VEGF signaling in late-stage pancreatic islet tumors, Cancer Cell 8 (2005) 299–309. [DOI] [PubMed] [Google Scholar]
- [27].Hoeben A, Landuyt B, Highley MS, Wildiers H, Van Oosterom AT, De Bruijn EA, Vascular endothelial growth factor and angiogenesis, Pharmacol. Rev 56 (2004) 549–580. [DOI] [PubMed] [Google Scholar]
- [28].Itatani Y, Kawada K, Yamamoto T, Sakai Y, Resistance to anti-angiogenic therapy in cancer—alterations to anti-VEGF pathway, Int. J. Mol. Sci 19 (2018) 1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Maisonpierre PC, Suri C, Jones PF, Bartunkova S, Wiegand SJ, Radziejewski C, Compton D, McClain J, Aldrich TH, Papadopoulos N, Daly TJ, Davis S, Sato TN, Yancopoulos GD, Angiopoietin-2, a natural antagonist for Tie2 that disrupts in vivo angiogenesis, Science (New York, N.Y.) 277 (1997) 55–60. [DOI] [PubMed] [Google Scholar]
- [30].Rigamonti N, Kadioglu E, Keklikoglou I, Wyser Rmili C, Leow CC, De Palma M, Role of angiopoietin-2 in adaptive tumor resistance to VEGF signaling blockade, Cell Rep 8 (2014) 696–706. [DOI] [PubMed] [Google Scholar]
- [31].Piao Y, Liang J, Holmes L, Zurita AJ, Henry V, Heymach JV, de Groot JF, Glioblastoma resistance to anti-VEGF therapy is associated with myeloid cell infiltration, stem cell accumulation, and a mesenchymal phenotype, Neuro-Oncology 14 (2012) 1379–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].LeCouter J, Lin R, Tejada M, Frantz G, Peale F, Hillan KJ, Ferrara N, The endocrine-gland-derived VEGF homologue Bv8 promotes angiogenesis in the testis: localization of Bv8 receptors to endothelial cells, Proc. Natl. Acad. Sci 100 (2003) 2685–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Massague J, Blain SW, Lo RS, TGFbeta signaling in growth control, cancer, and heritable disorders, Cell 103 (2) (2000) 295–309. [DOI] [PubMed] [Google Scholar]
- [34].Kwiatkowski SC, Guerrero PA, Hirota S, Chen Z, Morales JE, Aghi M, McCarty JH, Neuropilin-1 modulates TGFbeta signaling to drive glioblastoma growth and recurrence after anti-angiogenic therapy, PLoS One 12 (9) (2017) e0185065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Carbone C, Tamburrino A, Piro G, Boschi F, Cataldo I, Zanotto M, Mina MM, Zanini S, Sbarbati A, Scarpa A, Tortora G, Melisi D, Combined inhibition of IL1, CXCR1/2, and TGF$β$ signaling pathways modulates in-vivo resistance to anti-VEGF treatment, Anti-Cancer Drugs 27 (2016) 29–40. [DOI] [PubMed] [Google Scholar]
- [36].Semrad TJ, Kim EJ, Tanaka MS, Sands J, Roberts C, Burich RA, Li Y, Gandara DR, Lara P, Mack PC, Phase II study of dovitinib in patients progressing on anti-vascular endothelial growth factor therapy, Cancer Treat. Res. Commun 10 (2017) 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Curtis VF, Wang H, Yang P, McLendon RE, Li X, Zhou Q-Y, Wang X-F, A PK2/Bv8/PROK2 antagonist suppresses tumorigenic processes by inhibiting angiogenesis in glioma and blocking myeloid cell infiltration in pancreatic cancer, PLoS One 8 (2013) e54916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Baluk P, Hashizume H, McDonald DM, Cellular abnormalities of blood vessels as targets in cancer, Curr. Opin. Genet. Dev 15 (2005) 102–111. [DOI] [PubMed] [Google Scholar]
- [39].Barlow KD, Sanders AM, Soker S, Ergun S, Metheny-Barlow LJ, Pericytes on the Tumor Vasculature: Jekyll or Hyde? Cancer Microenviron 6 (2013) 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Erber R, Thurnher A, Katsen AD, Groth G, Kerger H, Hammes H-P, Menger MD, Ullrich A, Vajkoczy P, Combined inhibition of VEGF and PDGF signaling enforces tumor vessel regression by interfering with pericyte-mediated endothelial cell survival mechanisms, Faseb J 18 (2004) 338–340. [DOI] [PubMed] [Google Scholar]
- [41].Jain RK, Normalization of tumor vasculature: an emerging concept in anti-angiogenic therapy, Science 307 (2005) 58–62. [DOI] [PubMed] [Google Scholar]
- [42].Bergers G, Song S, Meyer-Morse N, Bergsland E, Hanahan D, Benefits of targeting both pericytes and endothelial cells in the tumor vasculature with kinase inhibitors, J. Clin. Invest 111 (2003) 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kuczynski EA, Vermeulen PB, Pezzella F, Kerbel RS, Reynolds AR, Vessel co-option in cancer, Nat. Rev. Clin. Oncol 16 (8) (2019) 469–493. [DOI] [PubMed] [Google Scholar]
- [44].Rubenstein JL, Kim J, Ozawa T, Zhang M, Westphal M, Deen DF, Shuman MA, Anti-VEGF antibody treatment of glioblastoma prolongs survival but results in increased vascular cooption, Neoplasia (New York, N.Y.) 2 (2019) 306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Holash J, Maisonpierre PC, Compton D, Boland P, Alexander CR, Zagzag D, Yancopoulos GD, Wiegand SJ, Vessel cooption, regression, and growth in tumors mediated by angiopoietins and VEGF, Science (New York, N.Y.) 284 (1999) 1994–1998. [DOI] [PubMed] [Google Scholar]
- [46].Dikic I, Elazar Z, Mechanism and medical implications of mammalian autophagy, Nat. Rev. Mol. Cell Biol 19 (2018) 349–364. [DOI] [PubMed] [Google Scholar]
- [47].Hafner Česen M, Pegan K, Špes A, Turk B, Lysosomal pathways to cell death and their therapeutic applications, Exp. Cell Res 318 (2012) 1245–1251. [DOI] [PubMed] [Google Scholar]
- [48].Parzych KR, Klionsky DJ, An overview of autophagy: morphology, mechanism, and regulation, Antioxid. Redox Signal 20 (3) (2014) 460–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Saxton RA, Sabatini DM, mTOR signaling in growth, metabolism, and disease, Cell 169 (2) (2017) 361–371. [DOI] [PubMed] [Google Scholar]
- [50].Gurumurthy S, Xie SZ, Alagesan B, Kim J, Yusuf RZ, Saez B, Tzatsos A, Ozsolak F, Milos P, Ferrari F, Park PJ, Shirihai OS, Scadden DT, Bardeesy N, The Lkb1 metabolic sensor maintains haematopoietic stem cell survival, Nature 468 (7324) (2010) 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N, Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy, Mol. Biol. Cell 20 (7) (2009) 1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Di Malta C, Siciliano D, Calcagni A, Monfregola J, Punzi S, Pastore N, Eastes AN, Davis O, De Cegli R, Zampelli A, Di Giovannantonio LG, Nusco E, Platt N, Guida A, Ogmundsdottir MH, Lanfrancone L, Perera RM, Zoncu R, Pelicci PG, Settembre C, Ballabio A, Transcriptional activation of RagD GTPase controls mTORC1 and promotes cancer growth, Science 356 (6343) (2017) 1188–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Tripathi DN, Chowdhury R, Trudel LJ, Tee AR, Slack RS, Walker CL, Wogan GN, Reactive nitrogen species regulate autophagy through ATM-AMPKTSC2-mediated suppression of mTORC1, Proc. Natl. Acad. Sci. U. S. A 110 (32) (2013) E2950–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kim J, Kundu M, Viollet B, Guan KL, AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1, Nat. Cell Biol 13 (2) (2011) 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Klionsky DJ, Baehrecke EH, Brumell JH, Chu CT, Codogno P, Cuervo AM, Debnath J, Deretic V, Elazar Z, Eskelinen EL, Finkbeiner S, Fueyo-Margareto J, Gewirtz D, Jaattela M, Kroemer G, Levine B, Melia TJ, Mizushima N, Rubinsztein DC, Simonsen A, Thorburn A, Thumm M, Tooze SA, A comprehensive glossary of autophagy-related molecules and processes (2nd edition), Autophagy 7 (11) (2011) 1273–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, Yoshimori T, Autophagosomes form at ER-mitochondria contact sites, Nature 495 (7441) (2013) 389–393. [DOI] [PubMed] [Google Scholar]
- [57].Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA, WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1, Mol. Cell 55 (2) (2014) 238–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Itakura E, Kishi-Itakura C, Mizushima N, The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes, Cell 151 (6) (2012) 1256–1269. [DOI] [PubMed] [Google Scholar]
- [59].Diao J, Liu R, Rong Y, Zhao M, Zhang J, Lai Y, Zhou Q, Wilz LM, Li J, Vivona S, Pfuetzner RA, Brunger AT, Zhong Q, ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes, Nature 520 (7548) (2015) 563–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Olsvik HL, Lamark T, Takagi K, Larsen KB, Evjen G, Overvatn A, Mizushima T, Johansen T, FYCO1 contains a C-terminally extended, LC3A/B-preferring LC3-interacting region (LIR) motif required for efficient maturation of autophagosomes during basal autophagy, J. Biol. Chem 290 (49) (2015) 29361–29374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wilkinson DS, Jariwala JS, Anderson E, Mitra K, Meisenhelder J, Chang JT, Ideker T, Hunter T, Nizet V, Dillin A, Hansen M, Phosphorylation of LC3 by the Hippo kinases STK3/STK4 is essential for autophagy, Mol. Cell 57 (1) (2015) 55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Avalos Y, Canales J, Bravo-Sagua R, Criollo A, Lavandero S, Quest AF, Tumor suppression and promotion by autophagy, Biomed Res. Int 2014 (2014) 603980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Chen ZH, Cao JF, Zhou JS, Liu H, Che LQ, Mizumura K, Li W, Choi AM, Shen HH, Interaction of caveolin-1 with ATG12-ATG5 system suppresses autophagy in lung epithelial cells, Am. J. Physiol. Lung Cell Mol. Physiol 306 (11) (2014) L1016–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, McMahon M, White E, Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors, Cancer Discov 3 (11) (2013) 1272–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Amaravadi R, Kimmelman AC, White E, Recent insights into the function of autophagy in cancer, Genes Dev 30 (17) (2016) 1913–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B, Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene, J. Clin. Invest 112 (12) (2003) 1809–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Mizushima N, Levine B, Autophagy in mammalian development and differentiation, Nat. Cell Biol 12 (9) (2010) 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Mah LY, Ryan KM, Autophagy and cancer, Cold Spring Harb. Perspect. Biol 4 (1) (2012) a008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Yang J, Zheng Z, Yan X, Li X, Liu Z, Ma Z, Integration of autophagy and anoikis resistance in solid tumors, Anat. Rec. (Hoboken) 296 (10) (2013) 1501–1508. [DOI] [PubMed] [Google Scholar]
- [70].Errafiy R, Aguado C, Ghislat G, Esteve JM, Gil A, Loutfi M, Knecht E, PTEN increases autophagy and inhibits the ubiquitin-proteasome pathway in glioma cells independently of its lipid phosphatase activity, PLoS One 8 (12) (2013) e83318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Liou GY, Storz P, Reactive oxygen species in cancer, Free Radic. Res 44 (5) (2010) 479–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Filomeni G, De Zio D, Cecconi F, Oxidative stress and autophagy: the clash between damage and metabolic needs, Cell Death Differ 22 (3) (2015) 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Mukhopadhyay NK, Kim J, You S, Morello M, Hager MH, Huang WC, Ramachandran A, Yang J, Cinar B, Rubin MA, Adam RM, Oesterreich S, Di Vizio D, Freeman MR, Scaffold attachment factor B1 regulates the androgen receptor in concert with the growth inhibitory kinase MST1 and the methyltransferase EZH2, Oncogene 33 (25) (2014) 3235–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM, DRAM, a p53-induced modulator of autophagy, is critical for apoptosis, Cell 126 (1) (2006) 121–134. [DOI] [PubMed] [Google Scholar]
- [75].Yee KS, Wilkinson S, James J, Ryan KM, Vousden KH, PUMA- and Bax-induced autophagy contributes to apoptosis, Cell Death Differ 16 (8) (2009) 1135–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E, Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis, Cancer Cell 10 (1) (2006) 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Buzzai M, Bauer DE, Jones RG, Deberardinis RJ, Hatzivassiliou G, Elstrom RL, Thompson CB, The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid beta-oxidation, Oncogene 24 (26) (2005) 4165–4173. [DOI] [PubMed] [Google Scholar]
- [78].Sun K, Deng W, Zhang S, Cai N, Jiao S, Song J, Wei L, Paradoxical roles of autophagy in different stages of tumorigenesis: protector for normal or cancer cells, Cell Biosci 3 (1) (2013) 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Vredenburgh JJ, Desjardins A, Herndon JE 2nd, Dowell JM, Reardon DA, Quinn JA, Rich JN, Sathornsumetee S, Gururangan S, Wagner M, Bigner DD, Friedman AH, Friedman HS, Phase II trial of bevacizumab and irinotecan in recurrent malignant glioma, Clin. Cancer Res 13 (4) (2007) 1253–1259. [DOI] [PubMed] [Google Scholar]
- [80].Clark AJ, Lamborn KR, Butowski NA, Chang SM, Prados MD, Clarke JL, McDermott MW, Parsa AT, Berger MS, Aghi MK, Neurosurgical management and prognosis of patients with glioblastoma that progresses during bevacizumab treatment, Neurosurgery 70 (2) (2012) 361–370. [DOI] [PubMed] [Google Scholar]
- [81].Mesange P, Poindessous V, Sabbah M, Escargueil AE, de Gramont A, Larsen AK, Intrinsic bevacizumab resistance is associated with prolonged activation of autocrine VEGF signaling and hypoxia tolerance in colorectal cancer cells and can be overcome by nintedanib, a small molecule angiokinase inhibitor, Oncotarget 5 (13) (2014) 4709–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Gianni L, Romieu GH, Lichinitser M, Serrano SV, Mansutti M, Pivot X, Mariani P, Andre F, Chan A, Lipatov O, Chan S, Wardley A, Greil R, Moore N, Prot S, Pallaud C, Semiglazov V, AVEREL: a randomized phase III Trial evaluating bevacizumab in combination with docetaxel and trastuzumab as first-line therapy for HER2-positive locally recurrent/metastatic breast cancer, J. Clin. Oncol 31 (14) (2013) 1719–1725. [DOI] [PubMed] [Google Scholar]
- [83].Lang I, Brodowicz T, Ryvo L, Kahan Z, Greil R, Beslija S, Stemmer SM, Kaufman B, Zvirbule Z, Steger GG, Melichar B, Pienkowski T, Sirbu D, Messinger D, Zielinski C, Central G European Cooperative Oncology, Bevacizumab plus paclitaxel versus bevacizumab plus capecitabine as first-line treatment for HER2-negative metastatic breast cancer: interim efficacy results of the randomised, open-label, non-inferiority, phase 3 TURANDOT trial, Lancet Oncol 14 (2) (2013) 125–133. [DOI] [PubMed] [Google Scholar]
- [84].Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, Han W, Lou F, Yang J, Zhang Q, Wang X, He C, Pan H, Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment, Cell Death Dis 4 (2013) e838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Prieto-Dominguez N, Ordonez R, Fernandez A, Garcia-Palomo A, Muntane J, Gonzalez-Gallego J, Mauriz JL, Modulation of autophagy by sorafenib: effects on treatment response, Front. Pharmacol 7 (2016) 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Hu YL, Jahangiri A, Delay M, Aghi MK, Tumor cell autophagy as an adaptive response mediating resistance to treatments such as antiangiogenic therapy, Cancer Res 72 (17) (2012) 4294–4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, Dipaola RS, Karantza-Wadsworth V, White E, Autophagy suppresses tumorigenesis through elimination of p62, Cell 137 (6) (2009) 1062–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Vasseur S, Afzal S, Tardivel-Lacombe J, Park DS, Iovanna JL, Mak TW, DJ-1/PARK7 is an important mediator of hypoxia-induced cellular responses, Proc. Natl. Acad. Sci. U. S. A 106 (4) (2009) 1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Wilkinson S, O’Prey J, Fricker M, Ryan KM, Hypoxia-selective macroautophagy and cell survival signaled by autocrine PDGFR activity, Genes Dev 23 (11) (2009) 1283–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Mazure NM, Pouyssegur J, Hypoxia-induced autophagy: cell death or cell survival? Curr. Opin. Cell Biol 22 (2) (2010) 177–180. [DOI] [PubMed] [Google Scholar]
- [91].Kirkin V, McEwan DG, Novak I, Dikic I, A role for ubiquitin in selective autophagy, Mol. Cell 34 (3) (2009) 259–269. [DOI] [PubMed] [Google Scholar]
- [92].Hu YL, DeLay M, Jahangiri A, Molinaro AM, Rose SD, Carbonell WS, Aghi MK, Hypoxia-induced autophagy promotes tumor cell survival and adaptation to antiangiogenic treatment in glioblastoma, Cancer Res 72 (7) (2012) 1773–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Paez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Vinals F, Inoue M, Bergers G, Hanahan D, Casanovas O, Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis, Cancer Cell 15 (3) (2009) 220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Huang H, Song J, Liu Z, Pan L, Xu G, Autophagy activation promotes bevacizumab resistance in glioblastoma by suppressing Akt/mTOR signaling pathway, Oncol. Lett 15 (2) (2018) 1487–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Zhao Z, Xia G, Li N, Su R, Chen X, Zhong L, Autophagy inhibition promotes bevacizumab-induced apoptosis and proliferation inhibition in colorectal cancer cells, J. Cancer 9 (18) (2018) 3407–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Guo XL, Li D, Sun K, Wang J, Liu Y, Song JR, Zhao QD, Zhang SS, Deng WJ, Zhao X, Wu MC, Wei LX, Inhibition of autophagy enhances anticancer effects of bevacizumab in hepatocarcinoma, J. Mol. Med. (Berl.) 91 (4) (2013) 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Selvakumaran M, Amaravadi RK, Vasilevskaya IA, O’Dwyer PJ, Autophagy inhibition sensitizes colon cancer cells to antiangiogenic and cytotoxic therapy, Clin. Cancer Res 19 (11) (2013) 2995–3007. [DOI] [PubMed] [Google Scholar]
- [98].Gonzalez-Malerva L, Park J, Zou L, Hu Y, Moradpour Z, Pearlberg J, Sawyer J, Stevens H, Harlow E, LaBaer J, High-throughput ectopic expression screen for tamoxifen resistance identifies an atypical kinase that blocks autophagy, Proc Natl Acad Sci U S A 108 (5) (2011) 2058–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Hydroxychloroquine, Carboplatin, Paclitaxel, and Bevacizumab in Recurrent Advanced Non-Small Cell Lung Cancer, (2019) https://ClinicalTrials.gov/show/NCT00728845.
- [100].Hydroxychloroquine, Capecitabine, Oxaliplatin, and Bevacizumab in Treating Patients With Metastatic Colorectal Cancer, (2019) https://ClinicalTrials.gov/show/NCT01006369.
- [101].Modulation of Autophagy in Patients With Advanced/Recurrent Non-small Cell Lung Cancer - Phase II, (2019) https://ClinicalTrials.gov/show/NCT01649947.
- [102].FOLFOX/Bevacizumab/Hydroxychloroquine (HCQ) in Colorectal Cancer, (2019) https://ClinicalTrials.gov/show/NCT01206530.
- [103].Sunitinib Malate and Hydroxychloroquine in Treating Patients With Advanced Solid Tumors That Have Not Responded to Chemotherapy, (2019) https://ClinicalTrials.gov/show/NCT00813423.
- [104].Maycotte P, Aryal S, Cummings CT, Thorburn J, Morgan MJ, Thorburn A, Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy, Autophagy 8 (2) (2012) 200–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Jahangiri A, Aghi MK, Biomarkers predicting tumor response and evasion to anti-angiogenic therapy, Biochim. Biophys. Acta 1825 (1) (2012) 86–100. [DOI] [PubMed] [Google Scholar]
- [106].Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, Ong CN, Codogno P, Shen HM, Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase, J. Biol. Chem 285 (14) (2010) 10850–10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Basu D, Salgado CM, Bauer B, Khakoo Y, Patel JR, Hoehl RM, Bertolini DM, Zabec J, Brzozowski MR, Reyes-Mugica M, The dual PI3K/mToR inhibitor Omipalisib/GSK2126458 inhibits clonogenic growth in oncogenically-transformed cells from neurocutaneous melanocytosis, Cancer Genomics Proteomics 15 (4) (2018) 239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Avni D, Glucksam Y, Zor T, The phosphatidylinositol 3-kinase (PI3K) inhibitor LY294002 modulates cytokine expression in macrophages via p50 nuclear factor kappaB inhibition, in a PI3K-independent mechanism, Biochem. Pharmacol 83 (1) (2012) 106–114. [DOI] [PubMed] [Google Scholar]
- [109].Miller S, Tavshanjian B, Oleksy A, Perisic O, Houseman BT, Shokat KM, Williams RL, Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34, Science 327 (5973) (2010) 1638–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Hansen SH, Olsson A, Casanova JE, Wortmannin, an inhibitor of phosphoinositide 3-kinase, inhibits transcytosis in polarized epithelial cells, J. Biol. Chem 270 (47) (1995) 28425–28432. [DOI] [PubMed] [Google Scholar]
- [111].Lazarus MB, Novotny CJ, Shokat KM, Structure of the human autophagy initiating kinase ULK1 in complex with potent inhibitors, ACS Chem. Biol 10 (1) (2015) 257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Petherick KJ, Conway OJ, Mpamhanga C, Osborne SA, Kamal A, Saxty B, Ganley IG, Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy, J. Biol. Chem 290 (48) (2015) 28726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Egan DF, Chun MG, Vamos M, Zou H, Rong J, Miller CJ, Lou HJ, Raveendra-Panickar D, Yang CC, Sheffler DJ, Teriete P, Asara JM, Turk BE, Cosford ND, Shaw RJ, Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates, Mol. Cell 59 (2) (2015) 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Pasquier B, El-Ahmad Y, Filoche-Romme B, Dureuil C, Fassy F, Abecassis PY, Mathieu M, Bertrand T, Benard T, Barriere C, El Batti S, Letallec JP, Sonnefraud V, Brollo M, Delbarre L, Loyau V, Pilorge F, Bertin L, Richepin P, Arigon J, Labrosse JR, Clement J, Durand F, Combet R, Perraut P, Leroy V, Gay F, Lefrancois D, Bretin F, Marquette JP, Michot N, Caron A, Castell C, Schio L, McCort G, Goulaouic H, Garcia-Echeverria C, Ronan B, Discovery of (2S)-8-[(3R)-3-methylmorpholin-4-yl]-1-(3-methyl-2-oxobutyl)-2-(trifluoromethyl)- 3,4-dihydro-2H-pyrimido[1,2-a]pyrimidin-6-one: a novel potent and selective inhibitor of Vps34 for the treatment of solid tumors, J. Med. Chem 58 (1) (2015) 376–400. [DOI] [PubMed] [Google Scholar]
- [115].Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E, Menon S, Wang Z, Honda A, Pardee G, Cantwell J, Luu C, Cornella-Taracido I, Harrington E, Fekkes P, Lei H, Fang Q, Digan ME, Burdick D, Powers AF, Helliwell SB, D’Aquin S, Bastien J, Wang H, Wiederschain D, Kuerth J, Bergman P, Schwalb D, Thomas J, Ugwonali S, Harbinski F, Tallarico J, Wilson CJ, Myer VE, Porter JA, Bussiere DE, Finan PM, Labow MA, Mao X, Hamann LG, Manning BD, Valdez RA, Nicholson T, Schirle M, Knapp MS, Keaney EP, Murphy LO, Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo, Nat. Cell Biol 16 (11) (2014) 1069–1079. [DOI] [PubMed] [Google Scholar]
- [116].Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, Jin M, Zhu Z, Wang H, Yu J, Li Y, Hao Y, Choi A, Ke H, Ma D, Yuan J, Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13, Cell 147 (1) (2011) 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Shao S, Li S, Qin Y, Wang X, Yang Y, Bai H, Zhou L, Zhao C, Wang C, Spautin-1, a novel autophagy inhibitor, enhances imatinib-induced apoptosis in chronic myeloid leukemia, Int. J. Oncol 44 (5) (2014) 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Ronan B, Flamand O, Vescovi L, Dureuil C, Durand L, Fassy F, Bachelot MF, Lamberton A, Mathieu M, Bertrand T, Marquette JP, El-Ahmad Y, Filoche-Romme B, Schio L, Garcia-Echeverria C, Goulaouic H, Pasquier B, A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy, Nat. Chem. Biol 10 (12) (2014) 1013–1019. [DOI] [PubMed] [Google Scholar]
- [119].Pasquier B, SAR405, a PIK3C3/Vps34 inhibitor that prevents autophagy and synergizes with MTOR inhibition in tumor cells, Autophagy 11 (4) (2015) 725–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Donohue E, Thomas A, Maurer N, Manisali I, Zeisser-Labouebe M, Zisman N, Anderson HJ, Ng SS, Webb M, Bally M, Roberge M, The autophagy inhibitor verteporfin moderately enhances the antitumor activity of gemcitabine in a pancreatic ductal adenocarcinoma model, J. Cancer 4 (7) (2013) 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Donohue E, Tovey A, Vogl AW, Arns S, Sternberg E, Young RN, Roberge M, Inhibition of autophagosome formation by the benzoporphyrin derivative verteporfin, J. Biol. Chem 286 (9) (2011) 7290–7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Bago R, Malik N, Munson MJ, Prescott AR, Davies P, Sommer E, Shpiro N, Ward R, Cross D, Ganley IG, Alessi DR, Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase, Biochem. J 463 (3) (2014) 413–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Akin D, Wang SK, Habibzadegah-Tari P, Law B, Ostrov D, Li M, Yin XM, Kim JS, Horenstein N, Dunn WA Jr., A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors, Autophagy 10 (11) (2014) 2021–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y, Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells, Cell Struct. Funct 23 (1) (1998) 33–42. [DOI] [PubMed] [Google Scholar]
- [125].Wu YC, Wu WK, Li Y, Yu L, Li ZJ, Wong CC, Li HT, Sung JJ, Cho CH, Inhibition of macroautophagy by bafilomycin A1 lowers proliferation and induces apoptosis in colon cancer cells, Biochem. Biophys. Res. Commun 382 (2) (2009) 451–456. [DOI] [PubMed] [Google Scholar]
- [126].Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC, Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy 4 (7) (2008) 849–850. [DOI] [PubMed] [Google Scholar]
- [127].Muller S, Dennemarker J, Reinheckel T, Specific functions of lysosomal pro-teases in endocytic and autophagic pathways, Biochim. Biophys. Acta 1824 (1) (2012) 34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Ni HM, Bockus A, Wozniak AL, Jones K, Weinman S, Yin XM, Ding WX, Dissecting the dynamic turnover of GFP-LC3 in the autolysosome, Autophagy 7 (2) (2011) 188–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Seguin SJ, Morelli FF, Vinet J, Amore D, De Biasi S, Poletti A, Rubinsztein DC, Carra S, Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly, Cell Death Differ 21 (12) (2014) 1838–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].De Mei C, Ercolani L, Parodi C, Veronesi M, Lo Vecchio C, Bottegoni G, Torrente E, Scarpelli R, Marotta R, Ruffili R, Mattioli M, Reggiani A, Wade M, Grimaldi B, Dual inhibition of REV-ERBbeta and autophagy as a novel pharmacological approach to induce cytotoxicity in cancer cells, Oncogene 34 (20) (2015) 2597–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Cook KL, Warri A, Soto-Pantoja DR, Clarke PA, Cruz MI, Zwart A, Clarke R, Hydroxychloroquine inhibits autophagy to potentiate antiestrogen responsiveness in ER+ breast cancer, Clin. Cancer Res 20 (12) (2014) 3222–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Wang Y, Peng RQ, Li DD, Ding Y, Wu XQ, Zeng YX, Zhu XF, Zhang XS, Chloroquine enhances the cytotoxicity of topotecan by inhibiting autophagy in lung cancer cells, Chin. J. Cancer 30 (10) (2011) 690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Rossi M, Munarriz ER, Bartesaghi S, Milanese M, Dinsdale D, Guerra-Martin MA, Bampton ET, Glynn P, Bonanno G, Knight RA, Nicotera P, Melino G, Desmethylclomipramine induces the accumulation of autophagy markers by blocking autophagic flux, J. Cell. Sci 122 (Pt 18) (2009) 3330–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Pellegrini P, Strambi A, Zipoli C, Hagg-Olofsson M, Buoncervello M, Linder S, De Milito A, Acidic extracellular pH neutralizes the autophagy-inhibiting activity of chloroquine: implications for cancer therapies, Autophagy 10 (4) (2014) 562–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Warhurst DC, Steele JC, Adagu IS, Craig JC, Cullander C, Hydroxychloroquine is much less active than chloroquine against chloroquineresistant Plasmodium falciparum, in agreement with its physicochemical properties, J. Antimicrob. Chemother 52 (2) (2003) 188–193. [DOI] [PubMed] [Google Scholar]
- [136].Manic G, Obrist F, Kroemer G, Vitale I, Galluzzi L, Chloroquine and hydroxychloroquine for cancer therapy, Mol. Cell. Oncol 1 (1) (2014) e29911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Torrente E, Parodi C, Ercolani L, De Mei C, Ferrari A, Scarpelli R, Grimaldi B, Synthesis and in vitro anticancer activity of the first class of dual inhibitors of REVERBbeta and autophagy, J. Med. Chem 58 (15) (2015) 5900–5915. [DOI] [PubMed] [Google Scholar]
- [138].Carew JS, Espitia CM, Esquivel JA 2nd, Mahalingam D, Kelly KR, Reddy G, Giles FJ, Nawrocki ST, Lucanthone is a novel inhibitor of autophagy that induces cathepsin D-mediated apoptosis, J. Biol. Chem 286 (8) (2011) 6602–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Mahalingam D, Mita M, Sarantopoulos J, Wood L, Amaravadi RK, Davis LE, Mita AC, Curiel TJ, Espitia CM, Nawrocki ST, Giles FJ, Carew JS, Combined autophagy and HDAC inhibition: a phase I safety, tolerability, pharmacokinetic, and pharmacodynamic analysis of hydroxychloroquine in combination with the HDAC inhibitor vorinostat in patients with advanced solid tumors, Autophagy 10 (8) (2014) 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB, Histone deacetylases (HDACs): characterization of the classical HDAC family, Biochem. J 370 (Pt 3) (2003) 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].White E, Deconvoluting the context-dependent role for autophagy in cancer, Nat. Rev. Cancer 12 (6) (2012) 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Carew JS, Nawrocki ST, Drain the lysosome: Development of the novel orally available autophagy inhibitor ROC-325, Autophagy 13 (4) (2017) 765–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Gemcitabine, Docetaxel, and Hydroxychloroquine in Treating Participants With Recurrent or Refractory Osteosarcoma, (2019) https://clinicaltrials.gov/ct2/show/NCT03598595.
- [144].Hydroxychloroquine, Palbociclib, and Letrozole Before Surgery in Treating Participants With Estrogen Receptor Positive, HER2 Negative Breast Cancer, (2019).
- [145].] Pre-Operative Trial (PGHA vs. PGH) for Resectable Pancreatic Cancer, (2019), pp. 17–134.
- [146].Sorafenib Induced Autophagy Using Hydroxychloroquine in Hepatocellular Cancer, (2019) https://clinicaltrials.gov/ct2/show/NCT03037437.
- [147].Vorinostat Plus Hydroxychloroquine Versus Regorafenib in Colorectal Cancer, (2019) https://clinicaltrials.gov/ct2/show/NCT02316340. [DOI] [PMC free article] [PubMed]
- [148].Dabrafenib, Trametinib and Hydroxychloroquine in Patients With Advanced BRAF Mutant Melanoma (BAMM), (2019) https://clinicaltrials.gov/ct2/show/NCT02257424.
- [149].Safety Study of Sirolimus and Hydroxychloroquine in Women With Lymphangioleiomyomatosis (SAIL), (2019) https://clinicaltrials.gov/ct2/show/NCT01687179.
- [150].Oral Hydroxychloroquine Plus Oral Sorafenib to Treat Patients With Refractory or Relapsed Solid Tumors, (2019) https://clinicaltrials.gov/ct2/show/NCT01634893.
- [151].Short Course Radiation Therapy With Proton or Photon Beam Capecitabine and Hydroxychloroquine for Resectable Pancreatic Cancer, (2019) https://clinicaltrials.gov/ct2/show/NCT01494155.
- [152].A Phase I/II/Pharmacodynamic Study of Hydroxychloroquine in Combination With Gemcitabine/Abraxane to Inhibit Autophagy in Pancreatic Cancer, (2019) https://clinicaltrials.gov/ct2/show/NCT01506973.
- [153].Akt Inhibitor MK2206 and Hydroxychloroquine in Treating Patients With Advanced Solid Tumors, Melanoma, Prostate or Kidney Cancer, (2019) https://clinicaltrials.gov/ct2/show/NCT01480154.
- [154].Autophagy Inhibition to Augment mTOR Inhibition: A Phase I/II Trial of RAD001 and Hydroxychloroquine in Patients With Previously Treated Renal Cell Carcinoma, (2019) https://clinicaltrials.gov/ct2/show/NCT01510119. [DOI] [PMC free article] [PubMed]
- [155].Sirolimus or Vorinostat and Hydroxychloroquine in Advanced Cancer, (2019) https://clinicaltrials.gov/ct2/show/NCT01266057.
- [156].Hydroxychloroquine and Bortezomib in Treating Patients With Relapsed or Refractory Multiple Myeloma, (2019) https://clinicaltrials.gov/ct2/show/NCT00568880.
- [157].Hydroxychloroquine in Patients With Stage III or Stage IV Melanoma That Can Be Removed by Surgery, (2019) https://clinicaltrials.gov/ct2/show/NCT00962845.
- [158].Study of the Efficacy of Chloroquine in the Treatment of Ductal Carcinoma in Situ (The PINC Trial), (2019) https://clinicaltrials.gov/ct2/show/NCT01023477.
- [159].Hydroxychloroquine + Vorinostat in Advanced Solid Tumors, (2019) https://clinicaltrials.gov/ct2/show/NCT01023737.
- [160].Randomized Phase II Trial of Pre-Operative Gemcitabine and Nab Paclitaxel With or With Out Hydroxychloroquine, (2019) https://clinicaltrials.gov/ct2/show/NCT01978184.
- [161].Hydroxychloroquine and Temsirolimus in Treating Patients With Metastatic Solid Tumors That Have Not Responded to Treatment, (2019) https://clinicaltrials.gov/ct2/show/NCT00909831.
- [162].Hydroxychloroquine in Treating Patients With Rising PSA Levels After Local Therapy for Prostate Cancer, (2019) https://clinicaltrials.gov/ct2/show/NCT00726596.
- [163].Hydroxychloroquine, Radiation, and Temozolomide Treating Patients With Newly Diagnosed Glioblastoma Multiforme, (2019) https://clinicaltrials.gov/ct2/show/NCT00486603.