Abstract
The ribose of protein-bound nucleosides and nucleotides displays preferred conformations (usually either North or South), which can be exploited to design enhanced analogues having chemically fixed conformations. We introduce a computational protocol for assembling data from the protein database (PDB) on the ribose and ribose-like conformation of small molecule ligands when complexed with purinergic signaling proteins (including receptors, enzymes and transporters, and related intracellular pathways). Some targets prefer exclusively North (adenosine and P2Y1 receptors, CD73, adenosine kinase ATP/ADP-binding site, adenosine deaminase), others prefer South (P2Y12 receptor, E-NTPDase2) or East (adenosine kinase substrates), while others (P2XRs) allow various conformations.
Keywords: nucleoside, nucleotide, G protein-coupled receptor, enzyme, transporter
INTRODUCTION
Small nucleoside and nucleotide derivatives have important biological signaling roles [1,2], in addition to the central role of nucleic acids in genetics, ribozymes and protein synthesis. Both extracellular and intracellular signaling and metabolism involve various endogenous purine and pyrimidine nucleoside derivatives [3-5]. Adenosine, ATP and various nucleotide derivatives are key extracellular mediators in living systems, by virtue of the activation of four subtypes of adenosine receptors (ARs), eight P2Y receptors (P2YRs) and seven ATP-gated P2XRs on the cell surface. Other functions of adenine nucleotides include ATP as the energy currency of the cell, cAMP as a second messenger, NAD+ as a coenzyme, AMP as metabolic regulator and other roles [1-8]. Similarly, derivatives of guanosine and uridine are important in intracellular signaling through G proteins and other GTPases, in the posttranslational modification of proteins (e.g. intracellular UDP-sugars). Also, extracellular UDP, UTP and UDP-glucose activate four of the eight P2YR subtypes [1].
Purines and pyrimidine nucleosides and nucleotides are present as ligands in the X-ray or cryo-electron microscopy (cryo-EM) structures of an increasing number of proteins in the Protein Data Bank (PDB) [9]. Thus, it is feasible to study the detailed molecular recognition of both naturally occurring and synthetic nucleosides and nucleotides by their macromolecular targets. The conformation of the ribose moiety of the nucleosides and nucleotides determines the angle of the projection of the nucleobase, phosphate group (if present) and other substituents, essential for recognition by the target protein. Therefore, we have collected and categorized quantitative data on the ribose ring pucker of small molecular nucleosides and nucleotides in complex with proteins of the purinome and related intracellular signaling pathways. These signaling proteins include receptors, enzymes and transporters.
Our interest in the ribose ring conformation derives from the ability to pre-establish by chemical modification a preferred conformation of this ring, to increase energetic stabilization when protein-bound, and thus enhance the potency or selectivity of these analogues in signaling pathways [10]. Ribose in nature approximates one of two opposite conformational states: North (N) and South (S). A mathematical description of the ribose ring pucker is the pseudorotational cycle [11,21], which provides parameters for both the angular twist, with the phase angle of pseudorotation (P), and the degree of deformation from the plane (νmax). The regions of (N) and (S) conformations on the pseudorotational cycle correspond to a P range centered at 0° (−45° < P ≤ 45°) and 180° (135° < P ≤ 225°), respectively [11], and they are intermediated by the East (E, 45° < P ≤ 135°) and West (W, 225° < P ≤ 315°) conformations, around 90° and 270°, respectively. Values of νmax lower than 20 ° are associated with a flatter conformation of the ribose ring that could be termed Central (C) [25].
Information on the ribose ring pucker can suggest the design and synthesis of novel nucleoside and nucleotide analogues as pharmacological probes of these pathways that might display an increased selectivity for a particular target. Methods for chemically limiting the conformations of a ribose or a pseudoribose moiety to favor either a (N) or (S) conformation include methyl groups at the 2′ or 3′ position and the sterically constrained methanocarba (bicyclo[3.1.0]hexane) nucleosides, introduced by Marquez and colleagues for anticancer and antiviral applications [11,12]. These modifications, in some cases, not only increase the affinity or potency at the target protein, but also the selectivity. While 2′ or 3′-methyl analogues have a preference for either a (N) or (S) conformation, respectively, a methanocarba modification enforces a rigid conformation. The two methanocarba isomers provide either a (N, 3′-endo) or (S, 2′-endo) envelope conformation of the pseudoribose depending on the cyclopropane fusion bond of the cyclopentane ring. We and others have explored the influence of methanocarba modifications within the signaling purinome, i.e. on affinity at ARs, P2YRs, P2XRs, nucleoside transporters and deaminases, and ribokinases [10,13-19].
COMPUTATIONAL METHODS
We have developed an automatic procedure to look for experimental structures of selected proteins in complex with nucleosides/nucleotides through the PDB website and compute the puckering parameters of the ribose ring. The input file consists of a text file with the list of the protein gene names of interest, with the optional addition of the protein name. Each gene is searched through the UniProt database, and the UniProt code is collected for the human, mouse and rat proteins corresponding to the gene, if a 3D structure is available for that protein. At this point, the collected UniProt code is used as search string in the PDB website, and all the 3D structures (X-ray, NMR, cryo-EM) are collected for that protein. All ligands in complex with the protein are identified in the PDB file, searching the string “HETNAM”, and the corresponding sdf files are retrieved from the PDB website. The compounds are searched for the presence of a nucleosidic substructure by employing a SMARTS [35] string encoding the structures shown in Figure 1A. The criteria to define a nucleoside/nucleotide were inspired by the PROSIT web-based calculator of parameters of the pseudorotational cycle established by Sun et al. [25], and a few modifications were made mainly to include 5′-truncated nucleosides. The P and νmax are computed for each identified ribose-like substructure belonging to a nucleoside/nucleotide moiety, using equations (1,2,3) [25] (see Figure 1B for the definition of the dihedral angles ν0, ν1, ν2, ν3, ν4).
Figure 1.
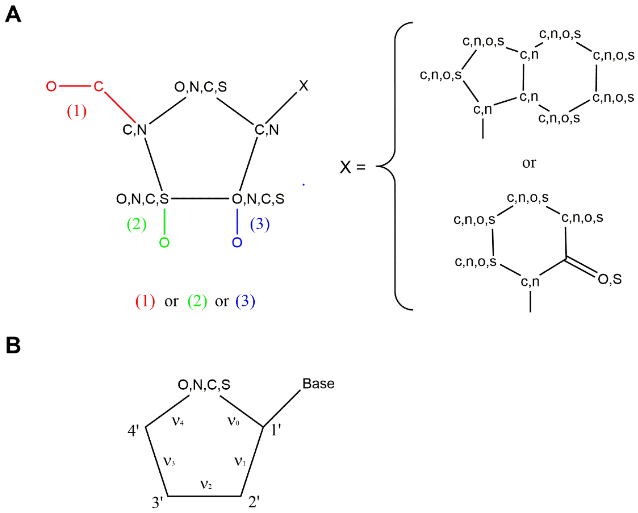
A. Schematic representation of the nucleosidic substructure used as query by the script, in the form of SMARTS string. Six substructures are searched, involving three possibilities for the ribose-like ring (identified by three different colors in the image) combined to a purine-like or pyrimidine-like scaffold. B. Schematic representation of the dihedral angles ν0, ν1, ν2, ν3, ν4.
| Equation 1. |
| Equation 2. |
| Equation 3. |
P and νmax are finally plotted in two polar axis graphs, where the spots are colored on a per-ligand or per-protein subtype basis, respectively.
Given an input gene name, the script (Supplemental file geneToPDB_puckering.txt) identifies and computes the pseudorotational parameters for all the nucelosidic/nucleotidic moieties present in all the chains of each PDB structure associated with that gene and it is also able to consider multiple substructures belonging to the same compound.
Since the procedure is completely automatic, some anomalies may appear in the results. The user should be aware of this and carefully analyze the results. In SI (Notes) we have reported all the anomalies we found in our results and the modifications we applied to the output.
The script is written in Python2.7 and exploits the modules: requests and urllib for the interaction with PDB and UniProt websites, rdkit [36] for the chemoinformatic part (SMARTS, substructure managing, dihedral computation), together with the software OpenBabel [37, 38] for sdf-file management, and matplotlib [39] for the graphs.
RESULTS AND DISCUSSION
Here we introduce a computational script (reported in SI, geneToPDB_puckering.txt) with the aim to extract ribose and ribose-like conformations from the structures of nucleosides and nucleotides in complex with proteins starting from their gene names. We have initially applied this method to proteins of the broad signaling purinome, including receptors, enzymes and transporters, and including various intracellular pathways (Table 1). Data from human (h), mouse (m) and rat (r) protein structures were included in the searches, except for P2XRs in which all species were considered. Puckering information of all nucleosidic/nucleotidic ligands are reported: all chains and multiple occurrences of the same ligand in the same experimental structures are considered, as well as multiple nucleosidic/nucleotidic substructures within the same compound. Data have been further deconvoluted on the basis of proximity to orthosteric binding sites and active sites versus additional sites on the protein surface, when appropriate. The results of our analysis are fully reported in SI (SI_results.txt) and in the polar plots in the manuscript and in SI; the puckering values of one representative structure for each protein are shown in Table 1 as an example. We have also evaluated the success of ring-constrained analogues already reported, in light of new structural information.
Table 1.
List of proteins analyzed, arranged by gene name. For analysis in the text, the quadrants of the pseudorotational cycle are approximately defined as (P value, ˚): (N), −45° < P ≤ 45°; (S), 135° < P ≤ 225°; (E), 45° < P ≤ 135°; (W), 225° < P ≤ 315°.
| Gene name | Protein name | representative PDB IDa | ligandb | P (˚) | ν (˚) |
|---|---|---|---|---|---|
| Purinergic Receptors | |||||
| ADORA1 | adenosine receptor A1 | 6D9H (h) | ADN | 33° | 35° |
| ADORA2A | adenosine receptor A2A | 2YDO (h) | ADN | 15° | 30° |
| ADORA2B | adenosine receptor A2B | c | |||
| ADORA3 | adenosine receptor A3 | c | |||
| P2RY1 | P2Y purinoceptor 1 | 4XNW (h) | 2ID | 340° | 25° |
| P2RY2 | P2Y purinoceptor 2 | c | |||
| P2RY4 | P2Y purinoceptor 4 | c | |||
| P2RY6 | P2Y purinoceptor 6 | c | |||
| P2RY11 | P2Y purinoceptor 11 | c | |||
| P2RY12 | P2Y purinoceptor 12 | 4PXZ (h) | 6AD | 182° | 32° |
| P2RY13 | P2Y purinoceptor 13 | c | |||
| P2RY14 | P2Y purinoceptor 14 | c | |||
| P2RX1 | P2X purinoceptor 1 | c | |||
| P2RX2 | P2X purinoceptor 2 | c | |||
| P2RX3 | P2X purinoceptor 3 | 5SVK (h) | ATP | 159° | 33° |
| P2RX4 | P2X purinoceptor 4 | 4DW1 (z) | ATP | 160° | 32° |
| P2RX5 | P2X purinoceptor 5 | c | |||
| P2RX6 | P2X purinoceptor 6 | c | |||
| P2RX7 | P2X purinoceptor 7 | 5U2H (p), 5XW6 (c) | ATP, 128 | 94°, 53° | 11°, 45° |
| Phosphatases, Cyclases, Kinases and other Enzymes | |||||
| ACPP | prostatic acid phosphatase (PAP) | ||||
| ADA1 | adenosine deaminase | 3IAR (h) | 3D1 | 40° | 39° |
| ADA2 | adenosine deaminase 2 | c | |||
| ADA | adenosine deaminase | 3KM8 (m) | 9DI | 40° | 39° |
| Adcy1 | adenylate cyclase type 1 | c | |||
| Adcy2 | adenylate cyclase type 2 | 3C16 (r) | ATP | 86° | 40° |
| Adcy3 | adenylate cyclase type 3 | c | |||
| Adcy4 | adenylate cyclase type 4 | c | |||
| Adcy5 | adenylate cyclase type 5 | c | |||
| Adcy6 | adenylate cyclase type 6 | c | |||
| Adcy7 | adenylate cyclase type 7 | c | |||
| Adcy8 | adenylate cyclase type 8 | c | |||
| Adcy9 | adenylate cyclase type 9 | c | |||
| Adcy10 | adenylate cyclase type 10 | 4USW (h) | ATP | 108° | 22° |
| ADK | adenosine kinase | 1BX4 (h), 5KB5 (m) | ADN, ADN | 97°, 106° | 47°, 40° |
| AHCY | adenosylhomocysteinase | 4PFJ (h), 5AXA (m), 1D4F (r) | ADN, ADN, ADN | 90°, 74°, 77° | 39°, 42°, 45° |
| AK1 | adenylate kinase 1 | 2C95 (h) | B4P | 170° | 44° |
| AK2 | adenylate kinase 2 | 2C9Y (h) | B4P | 172° | 36° |
| AK3 | adenylate kinase 3 | c | |||
| AK4 | adenylate kinase 4 | 2BBW (h) | GP5 | 144° | 34° |
| AK5 | adenylate kinase 5 | 2BWJ (h) | AMP | 249° | 31° |
| AK6 | adenylate kinase 6 | 3IIJ (h) | ADP | 170° | 32° |
| AK7 | adenylate kinase 7 | c | |||
| AK8 | adenylate kinase 8 | c | |||
| AK9 | adenylate kinase 9 | c | |||
| ALPI | intestinal-type alkaline phosphatase | c | |||
| ALPL | alkaline phosphatase, tissue-nonspecific isozyme (TNAP) | c | |||
| ALPP | alkaline phosphatase, placental type | c | |||
| ALPPL2 | alkaline phosphatase, germ cell type | c | |||
| AMPD1 | AMP deaminase1 | c | |||
| AMPD2 | AMP deaminase2 | c | |||
| AMPD3 | AMP deaminase3 | c | |||
| CD38 | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1 | 2I67 (h) | APR | 156° | 35° |
| ENPP1 | ectonucleotide pyrophosphatase/phosphodiesterase family member 1 | 4GTW (m) | AMP | 151° | 36° |
| ENPP2 | ectonucleotide pyrophosphatase/phosphodiesterase family member 2 (autotaxin) | c | |||
| ENPP3 | ectonucleotide pyrophosphatase/phosphodiesterase family member 3 | 6C02 (h), 6F2V (r) | APC, AMP | 93°, 36° | 34°, 36° |
| ENPP4 | ectonucleotide pyrophosphatase/phosphodiesterase family member 4 | 4LQY (h) | AMP | 36° | 39° |
| ENTPD1 | ectonucleoside triphosphate diphosphohydrolase 1 | c | |||
| ENTPD2 | ectonucleoside triphosphate diphosphohydrolase 2 | 3CJ7 (r) | AMP | 158° | 40° |
| ENTPD3 | ectonucleoside triphosphate diphosphohydrolase 3 | c | |||
| ENTPD4 | ectonucleoside triphosphate diphosphohydrolase 4 | c | |||
| ENTPD5 | ectonucleoside triphosphate diphosphohydrolase 5 | c | |||
| ENTPD6 | ectonucleoside triphosphate diphosphohydrolase 6 | c | |||
| ENTPD7 | ectonucleoside triphosphate diphosphohydrolase 7 | c | |||
| ENTPD8 | ectonucleoside triphosphate diphosphohydrolase 8 | c | |||
| NME1 | nucleoside diphosphate kinase A | 1UCN (h) | ADP | 20° | 39° |
| NME2 | nucleoside diphosphate kinase B | 3BBF (h) | GDP | 20° | 41° |
| NME3 | nucleoside diphosphate kinase 3 | 1ZS6 (h) | ADP | 28° | 30° |
| NME4 | nucleoside diphosphate kinase, mitochondrial | c | |||
| NME6 | nucleoside diphosphate kinase 6 | c | |||
| NME7 | nucleoside diphosphate kinase 7 | c | |||
| NT5E | 5′-nucleotidase (CD73) | 4H2G (h) | ADN | 30° | 36° |
| PDE1A | Calcium/calmodulin-dependent 3′,5′-cyclic nucleotide phosphodiesterase 1A | c | |||
| PDE1B | Calcium/calmodulin-dependent 3′,5′-cyclic nucleotide phosphodiesterase 1B | c | |||
| PDE1C | Calcium/calmodulin-dependent 3′,5′-cyclic nucleotide phosphodiesterase 1C | c | |||
| PDE2A | cGMP-dependent 3′,5′-cyclic phosphodiesterase | c | |||
| PDE3A | cGMP-inhibited 3′,5′-cyclic phosphodiesterase A | c | |||
| PDE3B | cGMP-inhibited 3′,5′-cyclic phosphodiesterase B | c | |||
| PDE4A | cAMP-specific 3′,5′-cyclic phosphodiesterase 4A | c | |||
| PDE4B | cAMP-specific 3′,5′-cyclic phosphodiesterase 4B | 1ROR (h) | AMP | 49° | 37° |
| PDE4C | cAMP-specific 3′,5′-cyclic phosphodiesterase 4C | c | |||
| PDE4D | cAMP-specific 3′,5′-cyclic phosphodiesterase 4D | 2PW3 (h) | CMP | 24° | 44° |
| PDE7A | High affinity cAMP-specific 3′,5′-cyclic phosphodiesterase 7A | c | |||
| PDE7B | cAMP-specific 3′,5′-cyclic phosphodiesterase 7B | c | |||
| PDE8A | High affinity cAMP-specific and IBMX-insensitive 3′,5′-cyclic phosphodiesterase 8A | c | |||
| PDE8B | High affinity cAMP-specific and IBMX-insensitive 3′,5′-cyclic phosphodiesterase 8B | c | |||
| PDE10A | cAMP and cAMP-inhibited cGMP 3′,5′-cyclic phosphodiesterase 10A | 2OUY (h) | CMP | 27° | 42° |
| PDE11A | Dual 3′,5′-cyclic-AMP and -GMP phosphodiesterase 11A | c | |||
| PNP | purine nucleoside phosphorylase | 1RCT (h) | NOS | 56° | 16° |
| PRKAR1A | cAMP-dependent protein kinase type I-alpha regulatory subunit | 5KJX (h) | CMP | 42° | 47° |
| PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit | c | |||
| PRKAR1B | cAMP-dependent protein kinase type I-beta regulatory subunit | c | |||
| PRKAR2B | cAMP-dependent protein kinase type II-beta regulatory subunit | 1CX4 (r) | CMP | 34° | 45° |
| RAPGEF1 | Rap guanine nucleotide exchange factor1 | c | |||
| RAPGEF2 | Rap guanine nucleotide exchange factor2 | c | |||
| RAPGEF3 | Rap guanine nucleotide exchange factor3 (EPAC1) | c | |||
| RAPGEF4 | Rap guanine nucleotide exchange factor4 (EPAC2) | 4MGK (m) | CMP | 43° | 40° |
| RAPGEF5 | Rap guanine nucleotide exchange factor5 | c | |||
| RAPGEF6 | Rap guanine nucleotide exchange factor6 | c | |||
| Transporters | |||||
| SLC17A9 | solute carrier family 28 member 9 (VNUT) | c | |||
| SLC28A1 | sodium/nucleoside cotransporter (CNT1) | c | |||
| SLC28A2 | sodium/nucleoside cotransporter (CNT2) | c | |||
| SLC28A3 | solute carrier family 28 member (CNT3) | c | |||
| SLC29A1 | equilibrative nucleoside transporter (ENT1) | c | |||
| SLC29A2 | equilibrative nucleoside transporter (ENT2) | c | |||
| SLC29A3 | equilibrative nucleoside transporter (ENT3) | c | |||
Nucleoside/nucleotide-containing structures. The natural ligand (agonist, antagonist, substrate or product) of the protein, if present, was chosen as representative structure. Otherwise the structure at higher resolution was picked. The species is indicated in parenthesis: human (h), mouse (m), rat (r), zebrafish (z), giant panda (p), chicken (c)
If more than one ligand is present in the structure, the one belonging to the first chain is indicated.
Structure is not reported.
All the genes reported in Table 1 have been searched in the PDB web-site. Our reported results concern the proteins (with the relative species) with a 3D structure of their complex with a nucleoside/nucleotide deposited in the PDB; phosphatases (ALPL, ALPP, ALPPL2, ALPI, ACPP), AMP deaminases and transporters are missing.
Purinergic receptors.
In the adenosine receptors (ARs), both the native agonist (adenosine in A1 and A2A ARs) and three different synthetic A2AAR agonists appear to be bound to the orthosteric binding site in a (N) conformation (Figures 2A and B), where they can interact with the receptor through hydrogen bonds (Figure 3A). A G protein-bound GDP, co-crystallized with A2AAR (PDB ID: 5G53), appears in an East (E) conformation. Curiously, among the known P2YR complex structures, a (N) conformation occurs in one P2Y1R structure (cocrystallized with a methanocarba antagonist ligand, MRS2500), and a (S) conformation occurs in two P2Y12R structures (Figures 2C and D).
Figure 2.
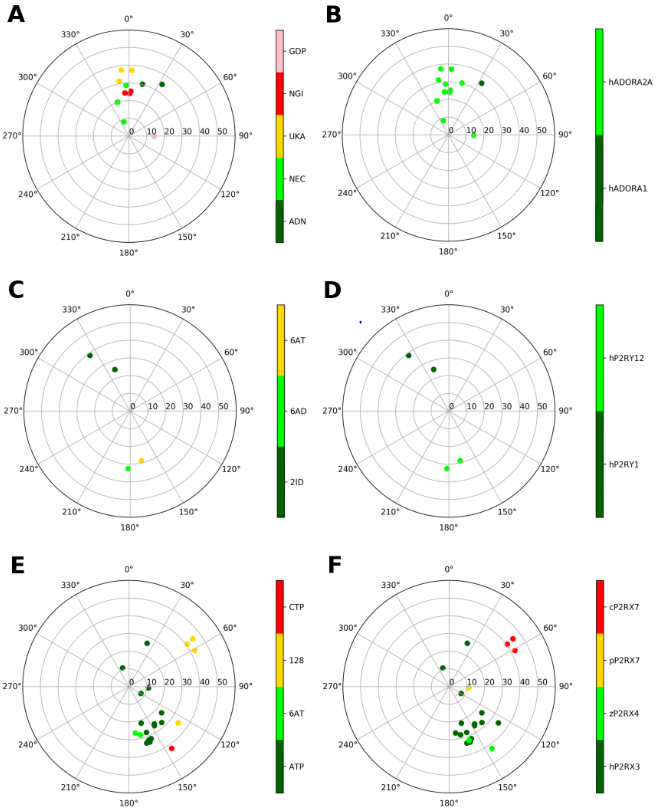
Representation of the ribose conformations of purinergic receptors, shown on the pseudorotational cycle (P on polar axes, νmax on x and y axes). The plots on the left and on the right depict the data on a per-ligand (residue name according to PDB file) and per-protein (species and gene name) basis. A and B. A1 and A2AARs (all human). Nucleoside ligands in (A) are labeled as: ADN (adenosine), NEC (NECA), UKA (UK432097), NGI (CGS21680). GDP is also present in one of the structures (5G53), where it is bound to a Gs protein. C and D. P2Y1 and P2Y12Rs (all human). Nucleotide ligands in (C) are labeled according as: 2ID (MRS2500), 6AD (2MeSADP), 6AT (2MeSATP). E and F. Ribose conformations at P2XRs (all species, as indicated) are shown for human P2RX3, Danio rerio (zebrafish) P2RX4, Ailuropoda melanoleuca (giant panda) P2RX7, Gallus gallus (chicken) P2RX7; with ligands: ATP, 6AT (2MeSATP), 128 (TNP-ATP (spiro-trinitrobenzene-ATP)), CTP.
Figure 3.
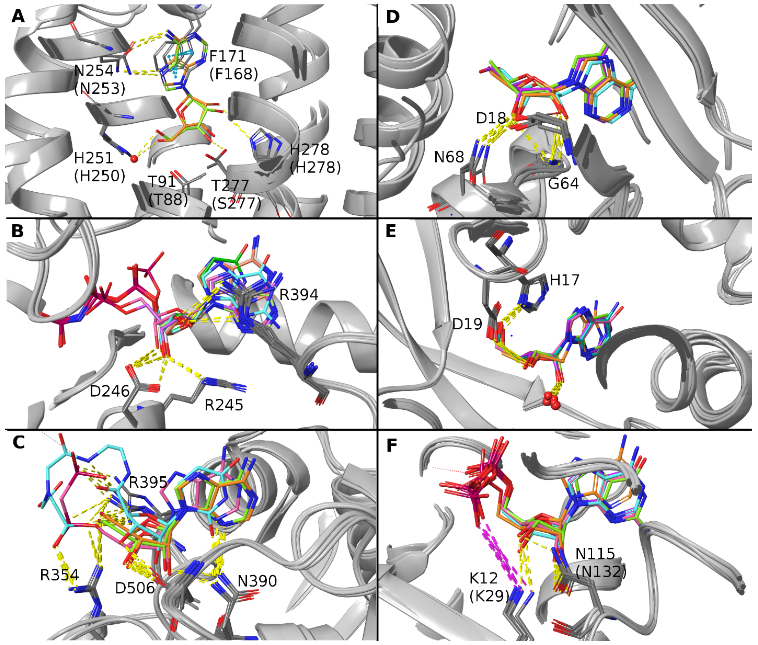
Three dimensional views of selected bound ligands, identified by PDB ID and color in the legend (compound abbreviations are reported in Figure 2, 4 and 5 legends). A. Superposition of ADN-bound human ARs A1 (6D9H, orange) and A2A (2YDO, lime). ADN appears in the (N) conformation. B. Overlay of nucleotides bound to rat E-NTPDase2. The superposition shows the following nucleotide ligands: ANP (4BR5, white), UNP (4BR2, green), AU1 (4BR0, pink), GNP (4BQZ, cyan), ANP (3CJA, orange), AMP (3CJ9, lime), AMP (3CJ7, purple). The compounds appear in the (S) conformation. C. Superposition of the C-terminal domain of human 5′-nucleotidase bound to the ligands: ADN (4H2F, orange; 4H2G, lime), A12 (4H2I, pink), 0YQ (4H1Y, cyan). The compounds appear in the (N) conformation. D. Superposition of adenosine kinase bound to ADN (1BX4, orange; 5KB5, cyan), 5I5 (chain A 2I6A, lime) and HO4 (chain A 4OIL, purple). The compounds appear in the (E) conformation. E. Superposition of hADA1 bound to 3DI (3IAR, orange), mADA bound to 1DA (1ADD, cyan), PUR (1FKW, purple; 1UIP, lime) and 9DI (chain A 3KM8, pink). The compounds are all (N)-(E). The red spheres represent a conserved water molecule interacting with the ribose moiety. F. Superposition of NDPK-A bound to ADP (chain A 1UCN, orange; chain A 2HVD, cyan; chain A 2HVE, purple), NDPK-B bound to GDP (chain A 1NUE, lime; chain A 3BBF, pink), and NDPK-C bound to ADP (chain A 1ZS6, green). The compounds are all (N).
Consistent with the above data, the structure activity relationship (SAR) for modified nucleosides binding to the ARs confirms that an A3AR agonist with (N)-methanocarba modification is >100-fold more potent than the corresponding (S)-methanocarba analogue [23], and this principle has been incorporated into highly selective, hypermodified A3AR agonists [15]. Also, the affinities at A1AR and A2AAR were higher with the (N)-methanocarba pseudoribose ring. Also, nucleosides containing a 2′-methyl group, which favors a (N) conformation, were more potent at ARs than those with a 3′-methyl group [22]. At the P2Y1R, the (N)-methanocarba modification enhances the affinity of both agonists and antagonists, and (N)-methanocarba nucleotide antagonist MRS2500 is present in P2Y1R X-ray structure (with residue name 2ID) [20,24]. At the P2Y12R and related P2Y13R, (N)-methanocarba nucleotides are inactive [40]. The P2Y6R is known to prefer a (S) conformation based on the activity of (S)-methanocarba-UDP [19], but there is no structure yet of this P2Y subtype.
P2XR structures with nucleotides (Figures 2E and F) are determined for three of the seven subtypes, specifically P2RX3 (h), P2RX4 (zebrafish, z), P2RX7 (giant panda, p, and chicken, c). They show principally (S) conformations of the bound nucleoside 5′-triphosphate ligands (e.g. P2RX4 (z), exclusively, with the cases of ATP and CTP), although there are examples of structures in the (E) conformation (TNP-ATP at P2RX7 (c)), in the (C) conformation (e.g. ATP at P2RX7 (p) and two examples at P2X3 (h)) or in the (N) conformation (ATP at P2RX3 (h)). Other P2RX3 structures show a (S) ribose conformation.
In the P2XRs, the ribose portion of the molecule does not make interactions with the receptors, so the frequency of a specific conformation of the ribose ring is not justified by the stabilization of a pattern of interactions, and this may explain why (N), (S), (E) and (C) puckering may occur.
Ectonucleotidases.
Figure 4 shows representations of the ribose conformations associated with enzymes that hydrolyze P2 receptor agonists on the extracellular surface. Among ectonucleoside triphosphate diphosphohydrolases (E-NTPDases), only rat E-NTPDase2 structures with various bound nucleotides are available (Figures 4A and B). The (S) conformation of the ribose is found exclusively, which is compatible with a pattern of hydrogen bonds with conserved amino acids, involving ribose 3′-OH and R245 and D246 and the ribose 2′-OH and R394, as shown in an overlay representation (Figure 3B).
Figure 4.
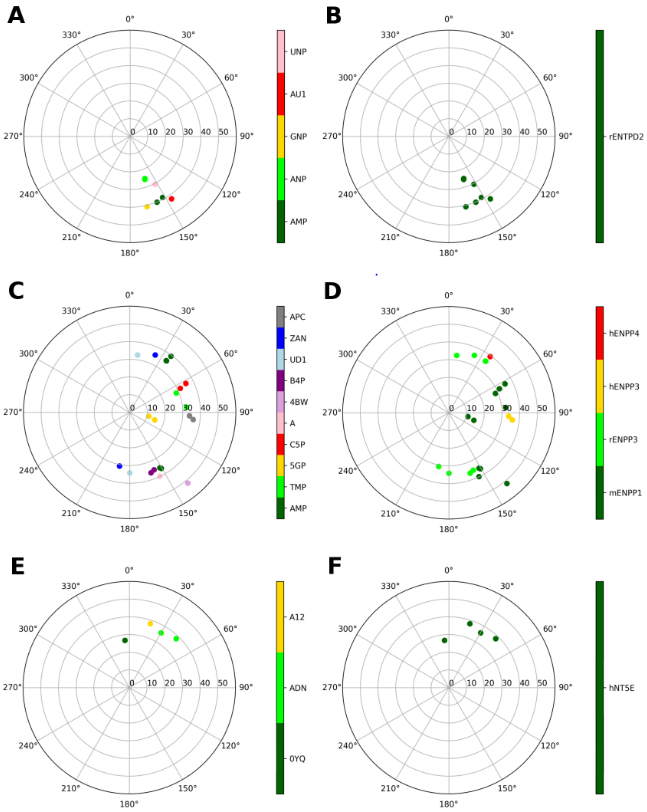
Representations of the ribose conformations associated with enzymes that hydrolyze P2R agonists, shown on the pseudorotational cycle (P on polar axes, νmax on x and y axes). The plots on the left and on the right depict the data on a per-ligand (residue name according to PDB file) and per-protein (species and gene name) basis. A and B. E-NTPDases: Structures of rat ENTPD2 are shown, and the bound nucleotides are: AMP, ANP (phosphonoamino-phosphoryl-oxy-phosphoryl-adenosine), GNP (phosphonoamino-phosphoryl-oxy-phosphoryl-guanosine), AU1 (phosphonoamino-phosphoryl-adenosine), UNP (phosphonoamino-phosphoryl-oxy-phosphoryl-uridine). C and D. Ligands at the nucleotide-binding pocket of E-NPPs: AMP, TMP, 5GP (GMP), C5P (CMP), A (AMP), 4BW (3′3′-cGAMP), B4P (bis(adenosine)-5′-tetraphosphate), UD1 (UDPGlcNAc), ZAN (5′-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]-amino}phosphoryl]adenosine), APC (diphospomethyl phosphonic acid adenosyl ester). ENPP1 data include mouse structures, ENPP3 data include mainly rat structures, with the addition of one human structure (6C02, APC ligand), while ENPP4s include human structures. E and F. 5′-Nucleotidase (all human). Compounds: 0YQ (PSB11552), ADN (adenosine), A12 (phosphomethyl phosphonic acid adenosyl ester).
Among ectonucleotide pyrophosphatase/phosphodiesterases (ENNPs) (Figures 4C and D), various ribose conformations are observed for compounds bound to the nucleotide-binding pocket of the enzyme. There is no conserved pattern of interactions stabilizing a specific conformation of the ribose ring. For example, AMP and GMP bind to the same enzyme to assume a (S) to (E) puckering, and AMP binds to different isozymes to assume a (N) conformation.
In human ecto-5′-nucleotidase (CD73), which hydrolyzes AMP to adenosine, only a (N) conformation is observed (Figure 4E and F). A H-bonding pattern involving D506 with the ribose 2′-OH and 3′-OH, and N390 with the ribose 2′-OH was compatible with the ribose (N) conformation (Figure 3C).
Intracellular enzymes.
Adenosine kinase (mouse and human) structures were analyzed similarly (Figure 5A and B), except that there are two binding sites for nucleosides. Focusing on the substrate-binding site (Figure 5C and D), the compounds assume an (E) conformation, in which the compound is stabilized by a pattern of interactions: O2′ is involved in a hydrogen bond with D18 and G64, and O3′ in a hydrogen bond with D18, G64 and N68, as reported in Figure 3D. The ribose rings of ligands in the ATP/ADP-binding site (Figure 5E and F) adopt a (N) conformation, even if they are not involved in direct interactions with the protein.
Figure 5.
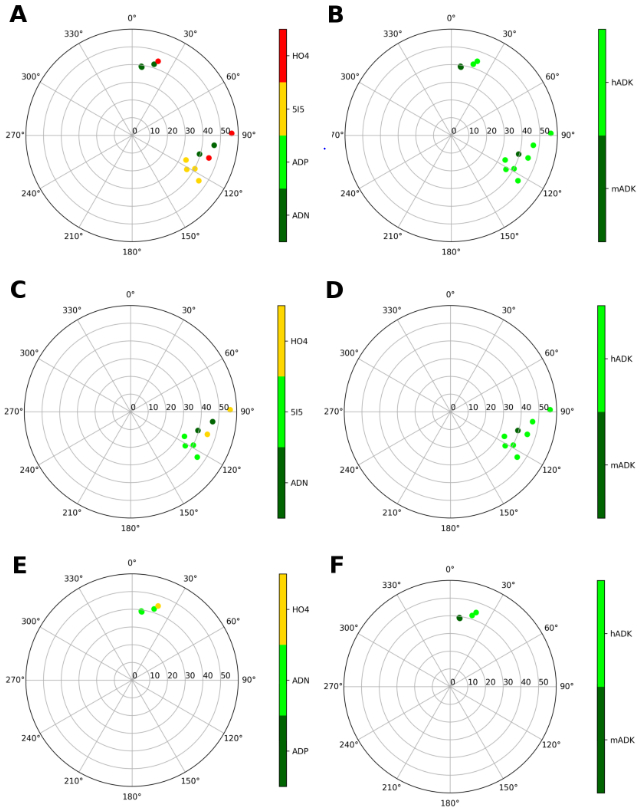
Representation of the ribose conformations of compounds bound to adenosine kinase (human and mouse), shown on the pseudorotational cycle (P on polar axes, νmax on x and y axes). The plots on the left and on the right depict the data on a per-ligand (residue name according to PDB file) and per-protein (species and gene name) basis. A and B. Data relative of all the ligands. C and D. Data of the compounds bound to the substrate-binding site. E and F. Data of the compounds bound to the ATP/ADP-binding site. The compounds are labelled as: ADN (adenosine), ADP, 5I5 (7-(5-deoxy-beta-D-ribofuranosyl)-5-iodo-7H-pyrrolo[2,3-D]pyrimidin-4-amine), HO4 (5-ethynyl-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine)
Conformationally restricted nucleosides have been applied to adenosine kinase [18,29]. Curiously, perhaps consistent with the intermediate (E) conformation associated with the substrate site, Toti et al. found that both (N) and (S)-methanocarba analogues of known nucleoside inhibitors have moderate affinity [18].
Adenosine deaminase (ADA) converts adenosine to inosine, and there is a preference for the (N) substrate sugar ring pucker over the (S) pucker [27,32]. Consistent with this observation, both nucleosidic substrates and inhibitors of ADA that contained conformationally restricted methanocarba rings indicated a strong preference for the (N) conformation [27]. The search for adenosine deaminases gave results for human ADA1 and mouse ADA. All the ligands assume a (N)-(E) conformation (Figure 6A and B), which is compatible with a pattern of hydrogen bonds involving 5′-OH and H17 with D19, 3′-OH with D19 and a water molecule (conserved in all the structures), and 2′-OH with the same conserved water molecule (Figure 3E).
Figure 6.
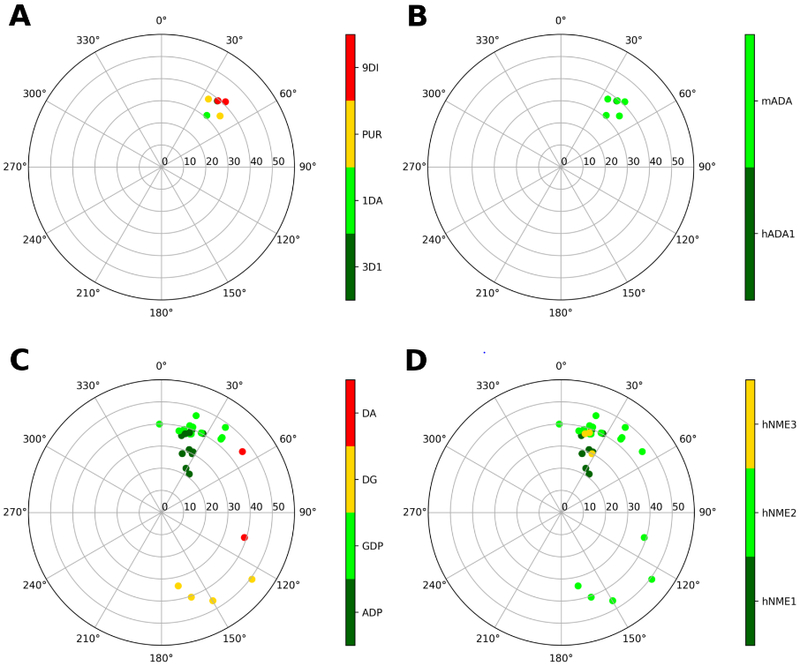
Representation of the ribose conformations of compounds bound to other enzymes, shown on the pseudorotational cycle (P on polar axes, νmax on x and y axes). The plots on the left and on the right depict the data on a per-ligand (residue name according to PDB file) and per-protein (species and gene name) basis. A and B. Data relative to human and and mouse adenosine deaminase. Compounds are labelled as: 3DI (2′-deoxyadenosine), 1DA (1-deaza-adenosine), PUR (purine riboside), 9DI (9-deaza-inosine). C and D. Data relative to human nucleoside-diphosphate kinases, isoforms 1, 2 and 3. Compounds are labelled as: ADP, GDP, DG (2'-deoxyguanosine-5′-monophosphate), DA (2′-deoxyadenosine-5′-monophosphate).
In the case of nucleoside-diphosphate kinases (NDPKs, human), ADP and GDP assume a (N) conformation when they are bound either to the NDPK-A (NME1) or to the NDPK-B (NME2) or to the NDPK-C (NME3) active sites (Figure 6C and D). The ligands’ 2′-OH and 3′-OH groups are stabilized by a H-bonding pattern involving K12 (K29 in NDPK-C) and N115 (N132 in NDPK-C), as shown in Figure 3F. dG and dA show higher variability in the puckering parameters and in the binding mode, assuming a (S) to (E) conformation. However, all these ligands belong to the dinucleotide d(AG) bound NDPK-B, structure 3BBB, where sssssa weak electron density for the dinucleotide was reported, with the dinucleotide d(AG) observed in two of the six chains of the crystal (chains C and E), a single nucleotide dG observed in other two chains (chain B and F) and no nucleotide in the remaining two chains (chains A and D).
Other enzymes (adenylate cyclase, Supplemental Figure S1; adenosylhomocysteinase, Supplemental Figures S2 and S3; adenylate kinases, Supplemental Figure S4; cyclic ADP-ribose hydrolases, Supplemental Figures S5 and S6; phosphodiesterases, Supplemental Figure S7; purine-nucleoside phosphorylase, Supplemental Figures S8 and S9; cAMP-dependent protein kinases, Supplemental Figures S10 and S11; and other GTPases, i.e. Rap guanine nucleotide exchange factors (RAPGEFs), Supplemental Figures S12 and S13) involved in signaling pathways are described in detail in Supporting information. G proteins that couple to GPCRs are not included in this analysis, although a Gs protein is present in one of the A2AAR structures (5G53, Figure 1).
Transporters
Nucleosides are transported across the plasma membrane at concentrative (CNTs) and bidirectionally by equilibrative (ENTs) transporters [28,31]. There is also a nucleotide (ATP) transporter (VNUT) that is associated with neurotransmitter vesicles [30]. Although lacking X-ray structures, (N) and (S)-methanocarba nucleosides have been applied to the nucleoside transporters to demonstrate a strong (N) preference with CNTs or a weak (S) preference with ENTs [33,34].
To summarize the main results, we have categorized the ribose conformations for various proteins, principally human, but also rat, mouse and other mammalian species. We have not analyzed lower species, even when structures are present in the PDB. The majority of the protein structures in the purinergic signaling pathways, so far determined, require a (N) or (S) ribose conformation. This is consistent with a previous survey of ribose ring twists overall in nature, as of 2004 [25]. The associated GPCRs and ligand-gated ion channels displayed either a (N) for adenosine A1/A2ARs and P2Y1R, or (S) for P2Y12R and P2X3,4 (not exclusively at P2X4), with few examples of P2X7Rs in (C) and (E) conformations. Nucleoside transporter (SLC family) structures have not yet been determined. Enzymes that degrade adenosine are either (N-E) for ADA or (E) for ADK (substrate-binding site). AMP deaminase structures bound to nucleosides/nucleotides are unknown. PNP that degrades nucleosides by cleaving the glycosidic bond often contains a flat ribose moiety. Enzymes that hydrolyze phosphates are either (N) for CD73, EPAC2 and several PDEs, or (S) for ENTPD2. Consistently, rat ENTPD2 was shown to hydrolyze the (S)-methanocarba analogue of ATP, but not the (N) isomer [24]. There are examples of hydrolytic enzymes with no preferred conformation, as ENPP1,3 (in contrast to a single ENPP4 structure, with a (N) conformation), CD38 (with a predominance in the South hemisphere) and AHCY ((E) conformation predominant). No nucleoside/nucleotide-bound structures are available for alkaline and acid phosphatases. Enzymes that phosphorylate often prefer an (S) conformation [12], as for adenylate kinases (nucleosidic moieties bound in proximity to ADP of structure 3IIJ), but several (N) examples are seen, such as PRKAR1A/2B regulatory subunits (catalytic subunits not analyzed) and NDPKs. Adenylate cyclases do not display a specific trend of conformations, with a broad distribution between (S) and (E). Methanocarba nucleosides/tides, which pre-establish a protein-preferred conformation, and other locked rings in place of ribose have been applied to only a few of these protein classes [10,12]. Thus, this approach can help guide future synthetic design for small molecule modulators of cell signaling through diverse nucleoside-recognizing protein classes.
CONCLUSIONS
With the determination of many new X-ray and cryo-EM structures that contain bound nucleosides and nucleotides, we present a computational protocol for assembling ribose conformational data from the PDB. We focus on the purinergic signaling pathways, which are of primary research interest to us, and the results suggest the synthesis of novel conformationally constrained ligands in the future. Nevertheless, this script can capture similar information for any set of genes.
We have previously explored modified nucleosides and nucleotides containing ribose ring constraints, such as bicyclo[3.1.0]hexane, to pre-establish a preferred conformation when bound to their target protein. We have demonstrated this approach to the design of ligands showing increased affinity and selectivity within the signaling purinome. Here we have presented a computational method for systematically exploring the preferred ribose conformations in nature, at least for those protein structures so far determined. We have focused on mediators of purinergic signaling, at various extracellular and intracellular levels. Some protein classes prefer exclusively (N) (adenosine and P2Y1 receptors, CD73, phosphodiesterases, adenosine kinase ATP/ADP-binding site, adenosine deaminase, NDPKs, EPAC2, PRKAR1A/2B), others prefer (S) (P2X4 and P2Y12 receptors, E-NTPDase2, adenylate kinase in one of the ADP-binding sites) or (E) (adenosine kinase substrates) conformations, other prefer flat conformations (PNP), while others (P2X3 and P2X7 receptors, adenylate cyclase, adenosylhomocysteinase, CD38, ENPPs) allow various conformations. This analysis will aid in the application of chemically fixed conformations of ribose to the design of ligands for additional protein targets.
Supplementary Material
ACKNOWLEDGMENTS AND FUNDING
We thank the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases (ZIA DK31117) for support.
Footnotes
Declaration of Interest Statement:
None.
REFERENCES
- [1].Burnstock G Purinergic Signalling: Therapeutic Developments. Front. Pharmacol, 2017, 8, 661-x. doi: 10.3389/fphar.2017.00661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fredholm BB; IJzerman AP; Jacobson KA; Linden J; Müller C Nomenclature and classification of adenosine receptors – An update. Pharmacol. Rev., 2011, 63, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Linden J; Koch-Nolte F; Dahl G Purine Release, Metabolism, and Signaling in the Inflammatory Response. Annu. Rev. Immunol, 2019, 37, 325–47. [DOI] [PubMed] [Google Scholar]
- [4].Yegutkin GG Enzymes involved in metabolism of extracellular nucleotides and nucleosides: functional implications and measurement of activities. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 473–497. [DOI] [PubMed] [Google Scholar]
- [5].Musheshe N; Schmidt M; Zaccolo M cAMP: From long-range second messenger to nanodomain signalling. Trends Pharmacol. Sci, 2018, 39, 209–222. [DOI] [PubMed] [Google Scholar]
- [6].Hanover JA; Chen W; Bond MR O-GlcNAc in cancer: An Oncometabolism-fueled vicious cycle. Journal of Bioenergetics and Biomembranes, 2018, 50(3), 155–173. 10.1007/s10863-018-9751-2 [DOI] [PubMed] [Google Scholar]
- [7].Liu H; Adebiyi M; Liu RR; Song A; Manalo J; Wen YE; Wen AQ; Weng T; Ko J; Idowu M; Kellems RE; Eltzschig HK; Blackburn MR; Juneja HS; Xia Y Elevated ecto-5′-nucleotidase: a missing pathogenic factor and new therapeutic target for sickle cell disease. Blood Advances 2018, 2, 1957–1968; doi: 10.1182/bloodadvances.2018015784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Verdin E NAD+ in aging, metabolism, and neurodegeneration. Science, 2015, 350(6265), 1208–1213. 10.1126/science.aac4854 [DOI] [PubMed] [Google Scholar]
- [9]. https://www.rcsb.org.
- [10].Jacobson KA; Tosh DK; Toti KS; Ciancetta A Polypharmacology of conformationally locked methanocarba nucleosides. Drug Disc. Today, 2017, 22, 1782–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Marquez VE et al. Nucleosides with a twist. Can fixed forms of sugar ring pucker influence biological activity in nucleosides and oligonucleotides? J. Med. Chem, 1996, 39, 3739–3747. [DOI] [PubMed] [Google Scholar]
- [12].Marquez VE The properties of locked methanocarba nucleosides in biochemistry, biotechnology and medicine, Chapter 12 In: Modified Nucleosides in Biochemistry Biotechnology and Medicine, Herdewijn P Ed. Wiley-VCH, 2008, pp 307–341. [Google Scholar]
- [13].Tosh DK; Jacobson KA Methanocarba ring as a ribose modification in ligands of G protein-coupled purine and pyrimidine receptors: Synthetic approaches. MedChemComm., 2013, 4, 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tosh DK; Finley A, Paoletta S, Moss SM, Gao ZG, Gizewski E, Auchampach JA, Salvemini D, Jacobson KA In vivo phenotypic screening for treating chronic neuropathic pain: Modification of C2-arylethynyl group of conformationally constrained A3 adenosine receptor agonists. J. Med. Chem, 2014, 57, 9901–9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dunn PM; Kim HS; Jacobson KA; Burnstock G Northern ring conformation of methanocarba-adenosine 5′-triphosphate required for activation of P2X receptors. Drug Devel. Res, 2014, 61, 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Damaraju VL; Mowles D; Smith KM; Yao SY; Young JD; Marquez VE and Cass CE Influence of sugar ring conformation on the transportability of nucleosides by human nucleoside transporters. ChemBioChem 2011, 12, 2774 – 2778. [DOI] [PubMed] [Google Scholar]
- [18].Toti KS; Osborne D; Ciancetta A; Boison D; Jacobson KA South (S)- and North (N)-methanocarba-7-deazaadenosine analogues as inhibitors of human adenosine kinase. J. Med. Chem, 2016, 59, 6860–6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Maruoka H; Barrett MO; Ko H; Tosh DK; Melman A; Burianek LE; Balasubramanian R; Berk B; Costanzi S; Harden TK; Jacobson KA Pyrimidine ribonucleotides with enhanced selectivity as P2Y6 receptor agonists: Novel 4-alkyloxyimino, (S)-methanocarba, and 5′-triphosphate γ-ester modifications. J. Med. Chem, 2010, 53, 4488–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang D; Gao ZG; Zhang K; Kiselev E; Crane S; Wang J; Paoletta S; Yi C; Ma L; Zhang W; et al. Two disparate ligand-binding sites in the human P2Y1 receptor. Nature, 2015, 520, 317–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Altona C; Sundaralingam M J. Am. Chem. Soc, 1972, 94, 8205–8212. [DOI] [PubMed] [Google Scholar]
- [22].Franchetti P; Cappellacci L; Marchetti S; Trincavelli L; Martini C; Mazzoni MR; Lucacchini A; Grifantini M 2′-C-Methyl analogues of selective adenosine receptor agonists: synthesis and binding studies. J. Med. Chem. 1998, 41, 1708–1715. [DOI] [PubMed] [Google Scholar]
- [23].Jacobson KA; Ji X. -d.; Li AH; Melman N; Siddiqui MA; Shin KJ; Marquez VE; Ravi RG Methanocarba analogues of purine nucleosides as potent and selective adenosine receptor agonists. J. Med. Chem, 2000, 43, 2196–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ravi RG; Kim HS; Servos J; Zimmermann H; Lee K; Maddileti S; Boyer JL; Harden TK; Jacobson KA Adenine Nucleotide Analogues Locked in a Northern Methanocarba Conformation: Enhanced Stability and Potency as P2Y1 Receptor Agonists. J. Med. Chem. 2002, 45, 2090–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sun G; Voigt JH; Filippov IV; Marquez VE; Nicklaus MC PROSIT: Pseudo-rotational online service and interactive tool, applied to a conformational survey of nucleosides and nucleotides. J. Chem. Inf. Comput. Sci, 2004, 44, 1752–1762. [DOI] [PubMed] [Google Scholar]
- [26].Azevedo MF; Faucz FR; Bimpaki E; Horvath A; Levy I; de Alexandre RB; Ahmad F; Manganiello V; Stratakis CA Clinical and molecular genetics of the phosphodiesterases (PDEs). Endocr. Rev. 2014, 35, 195–233. doi: 10.1210/er.2013-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ford HJ; Dai F; Mu L; Siddiqui MA; Nicklaus MC; Anderson L; Marquez VE; Barchi JJ Adenosine deaminase prefers a distinct sugar ring conformation for binding and catalysis: kinetic and structural studies. Biochemistry, 2000, 39, 2581–2592. [DOI] [PubMed] [Google Scholar]
- [28].Zhang Y; Zhang Y; Sun K; Meng Z; Chen L The SLC transporter in nutrient and metabolic sensing, regulation, and drug development. J. Mol. Cell Biol, 2018, 11, 1–13. 10.1093/jmcb/mjy052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sjuvarsson E Marquez VE; Eriksson S Selective phosphorylation of South and North-cytidine and adenosine methanocarba-nucleosides by human nucleoside and nucleotide kinases correlates with their growth inhibitory effects on cultured cells. Nucleosides, Nucleotides Nucleic Acids, 2015, 34, 544–564. [DOI] [PubMed] [Google Scholar]
- [30].Sawada K; Echigo N; Juge N; Miyaji T; Otsuka M; Omote H; Yamamoto A; Moriyama Y Identification of a vesicular nucleotide transporter. Proc. Natl. Acad. Sci. USA, 2008, 105, 5683–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Johnson ZL et al. Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters. eLife, 2014, 3, e03604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hernandez S; Ford H Jr.; Marquez VE Is the anomeric effect an important factor in the rate of adenosine deaminase catalyzed hydrolysis of purine nucleosides? A direct comparison of nucleoside analogues constructed on ribose and carbocyclic templates with equivalent heterocyclic bases selected to promote hydration. Bioorg. Med. Chem, 2002, 10, 2723–2730. [DOI] [PubMed] [Google Scholar]
- [33].Lee K; Cass C; Jacobson KA Synthesis using ring closure metathesis and effect on nucleoside transport of a (N)-methanocarba S-(4-nitrobenzyl)thioinosine derivative. Org. Lett, 2001, 3, 597–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Damaraju VL; Mowles D; Smith KM; Yao SYM; Young JD; Marquez VE; Cass CE Influence of sugar ring conformation on the transportability of nucleosides by human nucleoside transporters. ChemBioChem, 2011, 12, 2774 – 2778. [DOI] [PubMed] [Google Scholar]
- [35]. https://www.daylight.com/dayhtml/doc/theory/theory.smarts.html.
- [36].RDKit: Open-source cheminformatics; http://www.rdkit.org
- [37].The Open Babel Package, version 2.3.1. http://openbabel.org.
- [38].O'Boyle NM; Banck M; James CA; Morley C; Vandermeersch T; Hutchison GR Open Babel: An open chemical toolbox. J Cheminform., 2011, 3, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hunter JD Matplotlib: A 2D Graphics Environment. Computing in Science & Engineering, 2007, 9, 90–95 [Google Scholar]
- [40].Chhatriwala M, Ravi RG, Patel RI, Boyer JL, Jacobson KA, Harden TK Induction of novel agonist selectivity for the ADP-activated P2Y1 receptor versus the ADP-activated P2Y12 and P2Y13 receptors by conformational constraint of an ADP analogue. J. Pharm. Exp. Therap, 2004, 311:1038–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


