Abstract
Purpose:
To investigate the potential of contrast enhanced mammography (CEM) and radiomics analysis for the non-invasive differentiation of breast cancer invasiveness, hormone receptor status, and tumor grade.
Procedures:
This retrospective study included 100 patients with 103 breast cancers who underwent pretreatment CEM. Radiomics analysis was performed using MAZDA software. Lesions were manually segmented. Radiomic features were derived from first-order histogram (HIS), co-occurrence matrix (COM), run-length matrix (RLM), absolute gradient, autoregressive model, the discrete Haar wavelet transform (WAV), and lesion geometry. Fisher, probability of error and average correlation (POE+ACC), and mutual information (MI) coefficients informed feature selection. Linear discriminant analysis followed by k-nearest neighbor classification (with leave-one-out cross-validation) were used for pairwise texture-based separation of tumor invasiveness and hormone receptor status using histopathology as the standard of reference.
Results:
Radiomics analysis achieved highest accuracies of: 87.4% for differentiating invasive from noninvasive cancers based on COM+HIS/MI, 78.4% for differentiating HR positive from HR negative cancers based on COM+HIS/Fisher, 97.2% for differentiating human epidermal growth factor receptor 2 (HER2) positive/HR negative from HER2 negative/HR positive cancers based on RLM+WAV/MI, 100% for differentiating triple negative from triple positive breast cancers mainly based on COM+WAV+HIS/POE+ACC, and 82.1% for differentiating triple negative from HR positive cancers mainly based on WAV+HIS/Fisher. Accuracies for differentiating grade 1 vs. grades 2 and 3 cancers were 90% for invasive cancers (based on COM/MI) and 100% for non-invasive cancers (almost entirely based on COM/MI).
Conclusions:
Radiomics analysis with CEM has potential for non-invasive differentiation of tumors with different degrees of invasiveness, hormone receptor status, and tumor grade.
Keywords: Mammography, Breast Cancer, Contrast Media, Biomarkers, Tumors
Introduction
Contrast enhanced mammography (CEM) represents a cutting-edge technique which improves the accuracy of digital mammography by utilization of iodinated contrast given prior to mammography to visualize enhancing neovascularity indicating the site of malignancy. CEM permits to assess morphologic features of breast lesions and, concurrently, to depict the presence of enhancement of a tumor. In the diagnostic setting, CEM has a sensitivity of 96%–100% for the detection of breast cancer similar to dynamic contrast enhanced (DCE) magnetic resonance imaging (MRI) [1–6] and increases the specificity of full-field digital mammography from 42% to 87.7 %. In the screening setting, CEM has improved sensitivity compared with full-field digital mammography [3–4, 7].
Recent advances in medical imaging techniques and the development of high-throughput methods that convert medical images into quantifiable data have heralded radiomics, a new and rapidly evolving field of research. Radiomics analysis extracts and mines imaging features in a non-invasive and cost-effective way—with the central premise that these imaging features in their entirety quantify phenotypic characteristics of the tumor and may accurately reflect the underlying tumor biology and monitor changes over time [8–9]. Whereas invasive tissue sampling can only provide snap-shots of tumor biology and is subject to sampling selection bias, an inherent advantage of radiomics is the ability to non-invasively assess the tumor in its entirety. To date, radiomics research in breast imaging which has been dominated by DCE-MRI [10–14] has successfully extracted morphologic and functional imaging features for the differentiation of molecular breast cancer subtypes, correlation with recurrences scores, and correlation with individual gene signatures, with encouraging results [15–16].
Because CEM relies on the same principal as MRI, i.e., imaging both morphology and neovascularity as a tumor-specific feature, we hypothesized that radiomics analysis of CEM in breast cancer patients could provide similar information. To begin testing this hypothesis, we investigated the potential of radiomics analysis of CEM to differentiate tumors with different breast cancer invasiveness, hormone receptor status, and tumor grade.
Materials and Methods
We conducted a Health Insurance Portability and Accountability Act-compliant, retrospective single-institution study. The institutional review board approved the study and waived the need for informed consent.
Patients
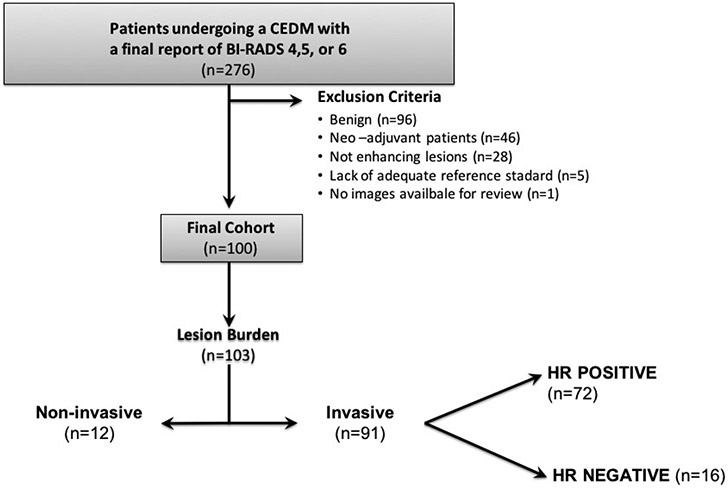
An electronic database of consecutive patients who underwent a CEM examination between January 2010 and March 2018 at our tertiary referral academic center was searched for the presence of a suspicious imaging abnormality (BI-RADS 4/5) or an histopathologically verified breast cancer (BI-RADS 6), yielding a target population of 276 patients. We excluded 176 patients for the following reasons: benign lesion (n = 96), suspicious lesion did not enhance (n=28), prior neo-adjuvant treatment (n=46), lack of histopathology (n = 5), and no images available for review (n = 1). The 28 non-enhancing suspicious lesions were low grade Ductal carcinoma in situ (DCIS) (n = 27) and one was a low-grade invasive cancers with small size (3 mm). Therefore, 100 patients were included in the study (Fig. 1).
Fig. 1.
Flowchart shows study enrollment criteria, final cohort and lesion burden. CEM, contrast enhanced digital mammography; HR+, hormone receptor positive; HR−, hormone receptor negative
Fifty-two patients were previously reported in a different context; the prior publication did not include radiomics analyses of CEM data [5].
CEM imaging technique
CEM was performed using a GE SenoBright mammography unit. CEM technique has been described previously [7]. Briefly the patient receives iodinated contrast material intravenously after which she undergoes mammography with nearly simultaneous low and high energy images. A recombination algorithm is then used to subtract the unenhanced breast tissue using data from the low energy and high energy images, providing subtracted images highlighting areas of contrast enhancement. No severe adverse events occurred due to contrast administration.
Radiomics analysis
All Digital Imaging and Communications in Medicine (DICOM) images were transferred to a database and loaded onto the open source image processing tool OsiriX (OsiriX Foundation). For each patient, lesion laterality, lesion size (maximum diameter in either the CC or MLO view), breast density, and background parenchymal enhancement were recorded. In the case of multifocal disease, the lesion with the maximum diameter was selected. For radiomics analysis, a fellowship-trained radiologist with over four years of experience in breast imaging manually delineated the borders of each lesion to include the entire enhancing lesion and to exclude foreign bodies (e.g., biopsy markers). Contours were delineated on either the cranio-caudal or medio-lateral-oblique view depending on which provided the best visualization of the lesion. Radiomics analysis was performed using a free radiomics platform: MaZda software (Technical University of Lodz), an established methodology as published in prior studies [17–20]. MAZDA allows the computation of almost 300 radiomic features [10]. The list of radiomic features are summarized in Table 1.
Table 1:
Radiomic features from different categories.
| Lesion Geometry (GEO) | First-Order Histogram (HIS) | Absolute Gradient (GRA) | Run-Length Matrix (RLM) | Co-Occurrence Matrix (COM) | Autoregressive Model (ARM) | Wavelet Transform (WAV) |
|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
For further information on the features visit: http://www.eletel.p.lodz.pl/programy/mazda/download/FeaturerList.pdf
Histopathology
Biopsy results were recorded for histology, histological grade and IHC status. Evaluation of IHC status included estrogen receptor, progesterone receptor, HER2 according to standard protocols using an automated Ventana Benchmark XT device (Ventana, Tucson, AZ). Staining results were evaluated according to current ASCO/USCAP guidelines with 1% staining being considered positive [21]. Surgical specimen was considered the standard of reference for histological analysis [22].
Statistical analysis
Three-hundred features were extracted. Fisher, probability of error and average correlation (POE+ACC), as well as mutual information (MI) coefficients were used for feature selection. For details on the features, refer to http://www.eletel.p.lodz.pl/mazda/download/mazda_manual.pdf. Linear discriminant analysis followed by k-nearest neighbor classification (with leave-one-out cross-validation) was used for pairwise texture-based separation of subtypes/hormonal status. Radiomics parameters were correlated with tumor histology (invasive vs non-invasive), hormone receptor status (HR positive vs HR negative, triple negative vs triple positive, triple negative vs HR positive), Her2 positive vs HR negative) and tumor grade (1, 2, or 3). Statistical analyses were performed using SPSS 22.0 (SPSS, IBM). Accuracy is the percentage of cases correctly classified by k-NN, relative to the reference standard and statistical significance was assumed at p-value ≤ 0.05.
Results
Patient population and lesion characteristics
A total of 103 malignant lesions in 100 patients (mean age, 51.5 ± 12 years; age range, 25–79) were detected on CEM and histopathologically verified as breast cancer: 12 (12%) lesions were purely non-invasive cancers (mean size 15.8 ± 10.7 mm, size 3–42 mm) and 91 (88%) lesions were invasive cancers (mean size 22.9 ± 19.4 mm, size 3–97 mm). Of the invasive cancers, 68/91 (75%) were associated with a non-invasive component. Of the invasive cancers, 82/91 (90%) were invasive ductal carcinomas (mean size 22.8 ± 19.5 mm, size range 3–97 mm), 6/91 (7%) were invasive lobular cancers (mean size 27.8 ± 23.3 mm, size range 4–65 mm), and 3/91 (3%) were malignant phyllodes (mean size 18.3 ± 9.1 mm, size range 10–28 mm). Malignant phyllodes were included solely for the analysis of breast cancer invasiveness vs. DCIS. Of the 88 primary breast carcinomas, 72/88 (82%) were HR positive breast cancers, including 68 HR positive/HER2 negative cancers and 4 HR positive/HER2 positive cancers; and 16/88 (18%) were HR negative breast cancers, including 4 HER2 positive cancers and 12 triple negative cancers (Fig. 2 and Fig. 3). Of the 88 primary breast carcinomas, 10 were grade 1 tumors, 51 were grade 2 tumors, and 39 were grade 3 tumors. Among the triple negative tumors, 11/12 (92%) were grade 3 tumors (p = 0.0001).
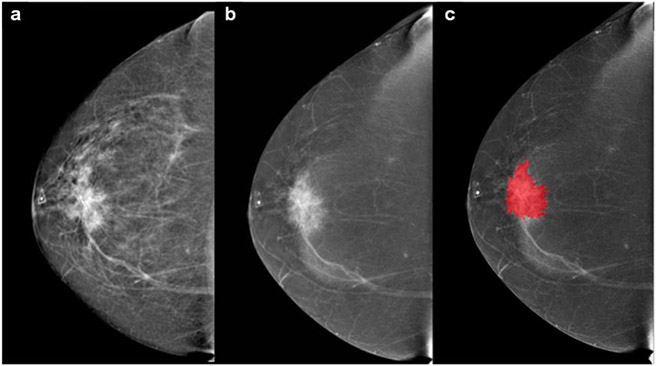
Fig. 2.
55-year-old patient with a palpable mass in the right breast. a Cranio-caudal full-field digital mammography of the right side. Scattered fibroglandular breasts. Corresponding to the palpable finding (triangular marker) at 12:00 in the anterior depth is an irregular shaped, spiculated 4 cm mass that is highly suspicious for malignancy. b Cranio-caudal contrast enhanced mammography. The examination was performed using full breast digital technique after injection of 153 ml of Omnipaque 350. Mild background parenchymal enhancement. The palpable suspicious mass enhances with contrast administration. c The lesions borders were segmented with the MAZDA segmentation tool to run the radiomics analysis. BI-RADS 5: Highly suspicious lesion and biopsy is recommended. The final histology yielded invasive ductal carcinoma (HR positive, HER2 negative, grade 2)
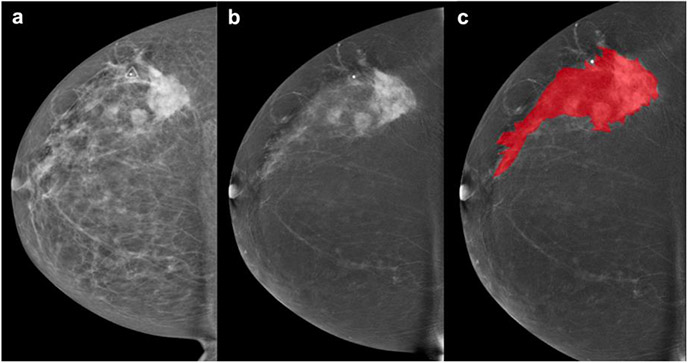
Fig. 3.
44-year-old patient. Palpable left breast abnormality. a Full-field digital mammography and b contrast enhanced mammography of the left breast in the cranio-caudal view. Scattered fibroglandular breasts. Minimal background parenchymal enhancement is present. In the left upper outer quadrant are multiple masses with associated focal asymmetry and suspicious enhancement extending anteriorly towards the nipple spanning approximately 13.3 cm. c Lesion segmentation for the radiomics analysis. BI-RADS 5: Highly suspicious lesions. Final histology: Invasive ductal cancer (HR negative, HER2 negative, grade 3)
Details on lesion characteristics stratified by invasiveness, hormonal status, and tumor grade are summarized in Table 2.
Table 2:
Lesion characteristics stratified by tumor hormonal status and grade
| DIAGNOSIS | HR positive (n=72) |
HR negative (n=16) |
Non- invasive cancers (n=12) |
|||
|---|---|---|---|---|---|---|
| HR positive/ HER2 negative (n=68) |
HR positive/ HER2 positive (n=4) |
HR negative/ HER2 positive (n=4) |
TN (n=12) |
|||
| Invasive cancers (n=88) * |
IDC (n=82) |
62 | 4 | 4 | 12 | - |
| ILC (n=6) |
6 | - | - | - | - | |
| Tumor grade (n=100) * |
G1 (n=10) |
8 | - | - | - | 2 |
| G2 (n=51) |
39 | 1 | 1 | 1 | 9 | |
| G3 (n=39) |
21 | 3 | 3 | 11 | 1 | |
G, grade; HER2, human epidermal growth factor receptor 2; HR, hormone receptor; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; TN, triple negative
Total number of invasive breast cancers excluding malignant phyllodes
Radiomics analysis
The most frequently explored radiomic features in our study are detailed in Table 1.
Radiomics analysis yielded the following accuracies for differentiation between invasive and non-invasive breast cancers: 79.6% (82/103, based on COM+HIS/Fisher), 86.4% (89/103, mainly through COM+WAV/POE+ACC), and 87.4% (90/103, based on COM + HIS/MI).
The highest accuracies for the differentiation of cancers based on tumor invasiveness, association with a non-invasive component, and breast cancer subtypes, respectively, are reported in Table 3.
Table 3:
Diagnostic accuracies for radiomic features based on cancer invasiveness and subtypes
| Types of breast cancers |
Invasive tumors |
Invasive tumors with a non- invasive component |
HR negative b | HR negative/ HER2 positive |
HR negative/ HER2 negative (triple negative) |
|---|---|---|---|---|---|
| Non-invasive tumors | 87.4% (MI) |
- | - | - | - |
| Invasive tumors without a non-invasive component | - | 70.3% (MI) |
- | - | - |
| HR positive a | - | - | 78.4% (Fisher) |
- | 82.1% (Fisher) |
| HR positive/HER2 negative | - | - | - | 97.2% (MI) |
- |
| HR positive/HER2 positive (triple positive) | - | - | - | - | 100% (POE+ACC/MI) |
HR, hormone receptor; HER2, human epidermal growth factor receptor 2; MI, mutual information; POE+ACC, Probability of error and average correlation
[HR positive/HER2 negative or HR positive/HER2 positive]
[HR negative/HER2 positive or HR negative/HER2 negative (triple negative)]
For the differentiation of HR positive from HR negative cancers, we achieved the following diagnostic accuracies: 78.4% (69/88, based on COM+HIS/Fisher), 73.9% (65/88, based on WAV+COM+HIS+RLM/POE+ACC), and 75% (66/88, based on COM +WAV+HIS/MI).
For the differentiation of HER2 positive/HR negative from HER2 negative/HR positive cancers, we achieved the following diagnostic accuracies: 97.2% (70/72, based on RLM+WAV/MI), 95.8% (69/72, mainly based on WAV+RLM/Fisher), 90.3% (65/72, based on RLM+COM+WAV/POE+ACC), although the results are limited by small sample size of first group (n = 4).
For the differentiation of triple negative from triple positive cancers, we achieved the following diagnostic accuracies: 100% (16/16, mainly based on COM+WAV+HIS/POE+ACC), 87.5% (14/16, based on HIS+WAV/Fisher), and 81.3% (13/16, mainly based on COM/MI); results are again limited by small sample size.
Accuracies for the differentiation of triple negative from HR positive cancers were as follows: 82.1% (69/84, WAV+HIS/Fisher), 78.6% (66/84, WAV+HIS+COM+RLM/POE+ACC), 81% (68/84, COM+HIS/MI).
For the differentiation of invasive cancers with and without non-invasive component, we achieved diagnostic accuracies of 70.3% (64/91, based on HIS+COM+WAV/MI), 68.1% (62/91, based on HIS+COM+WAV/Fisher), and 64.8% (59/91, based on HIS+COM+WAV/POE+ACC).
The differentiation of grade 1 invasive tumors from grades 2 and 3 invasive tumors was successful with the following accuracies: 90% (79/88, based on COM/MI), 87.5% (77/88, mainly based on COM/Fisher), and 87.5% (77/88, mainly COM+WAV/POE+ACC). Similarly, the differentiation of grade 1 non-invasive from grades 2 and 3 non-invasive tumors was successful, with the following accuracies: 100% (12/12, mainly based on COM+HIS+WAV/POE+ACC), 100% (12/12, almost entirely based on COM/MI), and 91.7% (11/12, mainly based on WAV+COM+ autoregressive model/Fisher), although these results are limited by small sample size of grade 1 non-invasive tumors (n = 2). Whereas the analysis of grades 1 and 2 non-invasive tumors vs grade 3 non-invasive tumors was not possible due to the limited number of grade 3 non-invasive tumors (n = 1), the differentiation of grades 1 and 2 invasive tumors from grade 3 invasive tumors yielded the following accuracies: 65.9% (58/88, mainly WAV+COM/Fisher), 62.5% (55/88, mainly WAV+RLM+HIS/POE+ACC), 62.5% (55/88, mainly through WAV+HIS+COM/MI) (see Table 4).
Table 4:
Diagnostic accuracies for radiomic features based on cancer grading
| Tumor grade | G1 | G2 | G3 | |
|---|---|---|---|---|
| Invasive tumors | G1 | - | - |
65.9% (MI) |
| G2 | 90% (MI) |
- | ||
| G3 | - | - | ||
| Non-invasive tumors | G1 | - | - | n/a* |
| G2 | 100% (POE+ACC/MI) |
- | ||
| G3 | - | - | ||
G1, grade 1; G2, grade 2; G3, grade 3; MI, mutual information; POE+ACC, Probability of error and average correlation
analysis not possible.
Discussion
Breast cancer is a heterogeneous disease with different outcomes and treatment responses based on tumor subtypes and tumor heterogeneity.
To our knowledge, no study has focused on the potential of radiomics analysis of CEM for the non-invasive differentiation of invasive from non-invasive disease, tumors of different hormone receptor status, or tumors of different grades. Our results indicate that radiomics analysis of CEM using combinations of radiomic features can differentiate between invasive and non-invasive breast cancers with accuracies of 79.6%–87.4%.
There is a strong argument for the development of supplemental means to evaluate the tumor in its entirety which could provide prognostic and predictive information. In this context, there is a unique opportunity to couple breast imaging with radiomics analysis [23–25].
So far, radiomics for the determination of prognostic and predictive factors in breast imaging has been almost exclusively dominated by DCE-MRI, which has yielded encouraging results [16, 23–27]. Bhooshan, et al. [26–27] investigated the ability of breast MRI radiomic features to determine tumor invasiveness, achieving an accuracy of 83%. However, MRI may not be an option for every woman due to its high cost and low availability or patient ineligibility (claustrophobia, pacemakers, metallic implants, and renal insufficiency). CEM is a technology approved by the Food and Drug Administration that utilizes iodinated contrast for imaging neo-angiogenesis similar to MRI; CEM achieves sensitivities similar to DCE-MRI, ranging from 96–100% [28]. It therefore stands to reason that radiomic analysis could also be performed using CEM to non-invasively obtain data that could provide prognostic and predictive indicators in breast cancer patients.
To date, there are limited radiomics data in patients with CEM. Patel, et al. [29] evaluated the use of computer-aided diagnosis with CEM to improve the diagnostic performance of the interpreting breast imagers. In this study, the authors extracted morphologic and textural features from the low-energy and recombined images of 50 lesions. Coupling radiomics analysis with machine learning, they constructed a predictive model using a support vector machine classifier. Based on this classifier, CEM with computer-aided diagnosis achieved an overall diagnostic accuracy of 90% that outperformed the diagnostic accuracy of expert radiologist review. The authors concluded that CEM with computer-aided diagnosis can provide information that is complementary to radiologist interpretation and reduce false-positive findings. Danala, et al. [30] used CEM with computer-aided diagnosis and a multilayer perception machine learning system for breast mass classification, proving that CEM with a computer-aided diagnosis and machine learning tool increased accuracy in mass region segmentation and consequently diagnostic performance.
In breast cancer patients, HR status in conjunction with other factors have relevant predictive and prognostic implications and are routinely used to guide therapy decisions. In this study, we also aimed to differentiate between HR positive and HR negative breast cancer and demonstrated that radiomics with a combined set of features, i.e., COM and HIS, succeeded in differentiating HR positive and HR negative breast cancer subtypes with accuracies ranging of 73.9%–78.4% and even better accuracies of 90.3%–97.2%) for the differentiation of HR positive/HER2 negative and HR negative/HER2 positive. Similar results were achieved for the differentiation of triple negative from triple positive breast cancers with accuracies up to 100%, as well as for the differentiation of triple negative and HR positive breast cancers with accuracies ranging from 78.6%–82.1%. However, it must be noted that that for triple negative vs triple positive and triple negative vs HR positive breast cancers, the sample size of triple negative cancers was small and further studies with larger numbers are warranted to confirm these excellent results.
Although we achieved high accuracies for the differentiation of tumors with different tumor invasiveness and hormone receptor status, the differentiation of tumors with different tumor grades was less successful. For both invasive and non-invasive breast cancer, we were able to differentiate low and high grade invasive breast cancers with a combination of WAV, HIS, and RLM radiomic features. We had higher accuracies when we combined grades 2 and 3 breast cancers for differentiation from grade 1 tumors, with good results for both invasive (87.5%–90%) and non-invasive breast cancers (91.7%–100%). Moderate accuracies of 65.9% were achieved when separating grade 3 invasive tumors from grades 1 and 2 invasive tumors. For non-invasive tumors, such analysis was not possible as there was only one case of a non-invasive grade 3 tumor.
Our early results indicate that with further experience, radiomics with CEM might provide additional imaging biomarkers for the non-invasive characterization of breast cancers. It must be noted that this methodology can practically be applied by most breast imaging centers as the software used is free and readily available.
This preliminary work needs to be validated with multi-institutional studies and must include larger numbers of patients. Fortunately, the adoption of CEM is steadily increasing. This will also enable better determination of which radiomic features and combinations thereof are the most relevant in each setting.
This study has some limitations. Inherent to the CEM technology, we could only evaluate tumor radiomic features from 2D images, which might not be as representative of tumor heterogeneity as a 3D volumetric assessment such as MRI. However, despite this limitation, we achieved good results for the differentiation of tumors with different tumor receptor status as well as invasiveness, indicating that valuable information can also be derived from 2D image analysis. Furthermore, we performed segmentation manually on subtracted images and in only the one view in which the tumor was seen best. These factors may have limited the reproducibility of our results in a field prone to inter-observer variability. Our ongoing studies will be multireader studies we will perform the suggested analysis only limiting the analysis to manual segmentation. Furthermore, we acknowledge the possibility of overfitting of the selected features. The free radiomics platform MAZDA software is able to adopt a feature filtration approach, ranking the features and selecting the most discriminative for the analysis. However, the overfitting of the selected features might still be present as a consequence of the limited dataset and further larger scale studies will be necessary to validate these preliminary results.
Twenty- eight breast cancers were not included in the radiomics analysis of tumor neovascularity as they showed no tumor enhancement. These breast cancers were solely detected on the low-energy mammograms and were primarily small, non-invasive tumors. In this proof of concept study there are relatively small numbers of patients in some subgroups. We therefore did not divide our study population into a training dataset and a validation dataset but used a k-nearest neighbor classifier with leave-one-out cross-validation which does not require two separate datasets and which has been used successfully in numerous other studies [31–33]. All CEM examinations were performed with the same machine which assures standardized image acquisition, yet our results might not be equally applicable across vendor platforms. Larger studies including CEM machines from all vendors are warranted to validate these results.
Conclusions
In conclusion, our early results demonstrate that radiomics analysis with CEM has the potential for non-invasive characterization of potentially heterogeneous tumors with different breast cancer invasiveness, tumor markers, hormone receptor status, and tumor grade. Larger studies are required to validate these data and determine the optimal radiomics criteria to further improve the results presented here.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the support in manuscript writing and editing from Joanne Chin.
Sources of Funding
This study was funded by the National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30 CA008748 and a grant from the Breast Cancer Research Foundation.
Footnotes
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Statement of Human and Animal Rights
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Diekmann F, Bick U (2007) Tomosynthesis and contrast-enhanced digital mammography: recent advances in digital mammography. Eur Radiol 17:3086–3092. [DOI] [PubMed] [Google Scholar]
- 2.Dromain C, Thibault F, Muller S, et al. (2011) Dual-energy contrast-enhanced digital mammography: initial clinical results. Eur Radiol 21:565–574. [DOI] [PubMed] [Google Scholar]
- 3.Lobbes MBI, Lalji U, Houwers J, et al. (2014) Contrast-enhanced spectral mammography in patients referred from the breast cancer screening programme. Eur Radiol 24:1668–1676. [DOI] [PubMed] [Google Scholar]
- 4.Lalji UC, Houben IPL, Prevos R, et al. (2016) Contrast-enhanced spectral mammography in recalls from the Dutch breast cancer screening program: validation of results in a large multireader, multicase study. Eur Radiol 26:4371–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jochelson MS, Dershaw DD, Sung JS, et al. (2013) Bilateral contrast-enhanced dual-energy digital mammography: feasibility and comparison with conventional digital mammography and MR imaging in women with known breast carcinoma. Radiology 266:743–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fallenberg EM, Dromain C, Diekmann F, et al. (2014) Contrast-enhanced spectral mammography versus MRI: Initial results in the detection of breast cancer and assessment of tumour size. Eur Radiol 24:256–264. [DOI] [PubMed] [Google Scholar]
- 7.Jochelson MS, Pinker K, Dershaw DD, et al. (2017) Comparison of screening CEDM and MRI for women at increased risk for breast cancer: A pilot study. Eur J Radiol 97:37–43. [DOI] [PubMed] [Google Scholar]
- 8.Valdora F, Houssami N, Rossi F, Calabrese M, Tagliafico AS (2018) Rapid review: radiomics and breast cancer. Breast Cancer Res Treat 169:217–229. [DOI] [PubMed] [Google Scholar]
- 9.Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: Images Are More than Pictures, They Are Data. Radiology 278:563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinker K, Shitano F, Sala E, et al. (2018) Background, current role, and potential applications of radiogenomics. J Magn Reson Imaging JMRI 47:604–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimm LJ (2016) Breast MRI radiogenomics: Current status and research implications. J Magn Reson Imaging JMRI 43:1269–1278. [DOI] [PubMed] [Google Scholar]
- 12.Dong Y, Feng Q, Yang W, et al. (2018) Preoperative prediction of sentinel lymph node metastasis in breast cancer based on radiomics of T2-weighted fat-suppression and diffusion-weighted MRI. Eur Radiol 28:582–591. [DOI] [PubMed] [Google Scholar]
- 13.Saha A, Harowicz MR, Mazurowski MA (2018) Breast cancer MRI radiomics: An overview of algorithmic features and impact of inter-reader variability in annotating tumors. Med Phys 45:3076–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saha A, Harowicz MR, Wang W, Mazurowski MA (2018) A study of association of Oncotype DX recurrence score with DCE-MRI characteristics using multivariate machine learning models. J Cancer Res Clin Oncol 144:799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sutton EJ, Oh JH, Dashevsky BZ, et al. (2015) Breast cancer subtype intertumor heterogeneity: MRI-based features predict results of a genomic assay. J Magn Reson Imaging JMRI 42:1398–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Zhu Y, Burnside ES, et al. (2016) MR Imaging Radiomics Signatures for Predicting the Risk of Breast Cancer Recurrence as Given by Research Versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Radiology 281:382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fetit AE, Novak J, Rodriguez D, et al. (2018) Radiomics in paediatric neuro-oncology: A multicentre study on MRI texture analysis. NMR Biomed 31:1. [DOI] [PubMed] [Google Scholar]
- 18.Larue RTHM, Defraene G, De Ruysscher D, et al. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br J Radiol 90:20160665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Fried DV, Fave XJ, Hunter LA, Yang J, Court LE (2015) IBEX: An open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys 42:1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szczypiński PM, Strzelecki M, Materka A, Klepaczko A (2009) MaZda--a software package for image texture analysis. Comput Methods Programs Biomed 94:66–76. [DOI] [PubMed] [Google Scholar]
- 21.Hammond MEH, Hayes DF, Wolff AC, et al. (2010) American society of clinical oncology/college of american pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Oncol Pract 6:195–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guiu S, Michiels S, André F, et al. (2012) Molecular subclasses of breast cancer: how do we define them? The IMPAKT 2012 Working Group Statement. Ann Oncol Off J Eur Soc Med Oncol 23:2997–3006. [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi K, Abe H, Newstead GM, et al. (2015) Intratumoral heterogeneity of the distribution of kinetic parameters in breast cancer: comparison based on the molecular subtypes of invasive breast cancer. Breast Cancer Tokyo Jpn 22:496–502. [DOI] [PubMed] [Google Scholar]
- 24.Blaschke E, Abe H (2015) MRI phenotype of breast cancer: Kinetic assessment for molecular subtypes. J Magn Reson Imaging JMRI 42:920–924. [DOI] [PubMed] [Google Scholar]
- 25.Mazurowski MA, Zhang J, Grimm LJ, Yoon SC, Silber JI (2014) Radiogenomic analysis of breast cancer: luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology 273:365–372. [DOI] [PubMed] [Google Scholar]
- 26.Bhooshan N, Giger ML, Jansen SA, Li H, Lan L, Newstead GM (2010) Cancerous breast lesions on dynamic contrast-enhanced MR images: computerized characterization for image-based prognostic markers. Radiology 254:680–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhooshan N, Giger M, Edwards D, et al. (2011) Computerized three-class classification of MRI-based prognostic markers for breast cancer. Phys Med Biol 56:5995–6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jochelson M (2014) Contrast-Enhanced Digital Mammography. Radiol Clin North Am 52:609–616. [DOI] [PubMed] [Google Scholar]
- 29.Patel BK, Ranjbar S, Wu T, et al. (2018) Computer-aided diagnosis of contrast-enhanced spectral mammography: A feasibility study. Eur J Radiol 98:207–213. [DOI] [PubMed] [Google Scholar]
- 30.Danala G, Patel B, Aghaei F, et al. (2018) Classification of Breast Masses Using a Computer-Aided Diagnosis Scheme of Contrast Enhanced Digital Mammograms. Ann Biomed Eng 46:1419–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fruehwald-Pallamar J, Czerny C, Holzer-Fruehwald L, et al. (2013) Texture-based and diffusion-weighted discrimination of parotid gland lesions on MR images at 3.0 Tesla. NMR Biomed 26:1372–1379 [DOI] [PubMed] [Google Scholar]
- 32.Mayerhoefer ME, Schima W, Trattnig S, Pinker K, Berger-Kulemann V, Ba-Ssalamah A (2010) Texture-based classification of focal liver lesions on MRI at 3.0 Tesla: a feasibility study in cysts and hemangiomas. J Magn Reson Imaging JMRI 32:352–359. [DOI] [PubMed] [Google Scholar]
- 33.Li Z, Mao Y, Li H, Yu G, Wan H, Li B (2016) Differentiating brain metastases from different pathological types of lung cancers using texture analysis of T1 postcontrast MR. Magn Reson Med 76:1410–1419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.