Abstract
The activation of adenosine monophosphate-activated protein kinase (AMPK) in skeletal muscle coordinates systemic metabolic responses to exercise1. Autophagy, a lysosomal degradation pathway that maintains cellular homeostasis2, is upregulated during exercise, and a core autophagy protein, beclin 1, is required for skeletal muscle AMPK activation3. Herein, we describe a role for the innate immune sensing molecule, toll-like-receptor 9 (Tlr9)4, and its interaction with beclin 1 in exercise-induced muscle AMPK activation. Mice lacking Tlr9 are deficient in exercise-induced skeletal muscle AMPK activation and GLUT4 plasma membrane localization but not autophagy. Tlr9 binds beclin 1, and this interaction is increased by energy stress (glucose starvation and endurance exercise) and decreased by a BCL2 mutation3, 5 that blocks disruption of BCL2/beclin 1 binding. Tlr9 regulates the assembly of the endolysosomal beclin 1/UVRAG-containing phosphatidylinositol 3-kinase complex (PI3KC3-C2) in skeletal muscle during exercise, and knockout of beclin 1 or UVRAG inhibits glucose starvation-induced cellular AMPK activation. Moreover, Tlr9 functions in a muscle-autonomous fashion in ex vivo contraction-induced AMPK activation, glucose uptake, and beclin 1/UVRAG assembly. These findings reveal a heretofore undescribed role of a toll-like receptor in muscle AMPK activation and glucose metabolism during exercise, as well as unexpected cross-talk between this innate immune sensor and autophagy proteins.
TLR9 senses unmethylated CpG-DNA from bacteria and mitochondrial DNA (mtDNA) to initiate type I interferon and pro-inflammatory cytokine production in immune cells4,6. Exogenously administered TLR9 ligands activate AMPK in cardiomyocytes and neurons and protect against hypoxic injury7. Yet, aside from studies in the heart8, 9, it is unknown whether TLR9 plays a physiological role in non-immune cells in vivo.
We performed a proteomic screen in HeLa cells to identify proteins that interact with a functionally important region of beclin 1, amino acids 267–284 (data not shown). This region of beclin 1, when linked to the cell penetrating leader sequence Tat, is sufficient to induce autophagy and exert beneficial effects in animal models of human disease2. Its flexibility is crucial for conformational changes in beclin 1-containing class III phosphatidylinositol 3-kinase (PI3KC3) complexes that promote their membrane association and lipid kinase activity10. One interactor identified with high-confidence was TLR7, an endosomal TLR that recognizes ssRNA. We confirmed that exogenously expressed TLR7, as well as the related protein TLR9, co-immunoprecipitate with beclin 1 in HeLa cells (Extended Data Fig. 1a, b).
We explored whether there is a regulated interaction between beclin 1 and TLR9 during energy stress, including glucose starvation in cultured cells and endurance exercise in mice. We focused on TLR9 as mtDNA is sensed by endosomal TLR9 rather than TLR76 and exercise induces mitophagy (autophagic degradation of mitochondria)11, which may be an intracellular route for delivery of mtDNA to the lumen of TLR9 containing endolysosomes12. We confirmed that endogenous beclin 1 co-immunoprecipitates with C’ terminal HA-tagged TLR9 in U2OS cells (Fig. 1a). Mutational analyses showed that deletion of amino acids 267–284 (the bait in our proteomic screen) from Flag epitope-tagged beclin 1 or deletion of the cytoplasmic Toll/Interleukin-1 receptor (TIR) domain of TLR9-HA blocks TLR9-HA/beclin 1 binding (Extended Data Fig. 1c, d). The interaction between beclin 1 and the TIR domain of TLR9 is direct (Extended Data Fig. 1e). The TLR9-HA/beclin 1 interaction is upregulated by glucose starvation (a low energy stress condition) but not by other stressors such as amino acid starvation or mitochondrial damaging agents (Fig. 1a, Extended Data 1f, g).
Figure 1. TLR9 interacts with beclin 1 during glucose starvation and exercise.
(a) Co-immunoprecipitation of transfected TLR9-HA with endogenous beclin 1 in U2OS cells cultured in normal or glucose starvation media (1 h). Results representative of three independent experiments. (b, c) Representative western blots (b) and quantitation (c) of beclin 1 co-immunoprecipitated with Tlr9-HA in vastus lateralis (VL) muscles from Tlr9-HA knock-in (KI) mice at indicated duration of exercise. See Extended Data Fig. 2b for p-AMPKα (Thr172)/total AMPKα quantitation. Data are mean ± s.e.m. Unpaired two-tailed t-test with Hommel method. Values at 0 min considered as 1, results expressed as relative arbitrary units (A.U.) and are combined data from three independent experiments. Similar results observed in each experiment. (d) Quantitation of mitochondrial DNA (mtDNA) co-immunoprecipitated with Tlr9-HA from gastrocnemius muscles (Methods). For each mtDNA primer set, value of WT resting considered as 1. Data are mean ± s.e.m. Two-tailed Mann-Whitney test. For c and d, data points, individual mice (sample size indicated in parentheses). WCL, whole cell lysate. For uncropped gels, see Supplementary Fig. 1.
Next, we determined whether beclin 1 interacts with Tlr9 in skeletal muscle during exercise. We confirmed that Tlr9 mRNA is expressed in mouse skeletal muscle at levels similar to those in other non-immune organs (Extended Data Fig. 1h). Additionally, Tlr9 mRNA expression is undetectable in embryonic myoblasts, but markedly increases after differentiation into myotubes (Extended Data Fig. 1i). Given the well-established difficulty generating antibodies that reliably detect endogenous Tlr9, we used CRISPR technology to construct knock-in mice with an HA epitope tag inserted at the C’ terminus of the Tlr9 locus (Extended Data Fig. 1j, k). These mice express Tlr9-HA in all tissues examined (Extended Data Fig. 1l, m). The predominant form is the active ~80 kD cleavage product that consists of a carboxyl-terminal fragment containing a portion of the Tlr9 ectodomain, the transmembrane domain, and the cytoplasmic domain13. This form is generated by endolysosomal proteolysis and mediates ligand recognition and subsequent signaling cascades4, 14.
Endogenous beclin 1 in skeletal muscle co-immunoprecipitated with Tlr9-HA in a time-dependent manner after initiation of treadmill exercise. The interaction was first detected at 10 min, plateaued at 20 min, and was undetectable by 50 min (Fig. 1b, c; Extended Data Fig. 2a). The interaction of beclin 1/Tlr9 at 10 min corresponded to the first time point when increased skeletal muscle AMPK phosphorylation was detected (Fig. 1b, Extended Data Fig. 2b). In spleen (the tissue with highest Tlr9 expression), exercise did not increase beclin 1/Tlr9 interaction or AMPK phosphorylation (Extended Data Fig. 2c–f). Moreover, at 20 min after exercise but not at rest, mtDNA (but not genomic DNA) co-immunoprecipitated with muscle Tlr9-HA (Fig. 1d, Extended Data Fig. 3a). Up to 90 min after exercise, no increase in circulating mtDNA was detectable (Extended Data Fig. 3b), suggesting that mtDNA may be delivered to endolysosomal Tlr9 during exercise via an intracellular route. An exogenous Tlr9 ligand, ODN2395, failed to increase AMPK phosphorylation in mouse muscle explants (Extended Data Fig. 3c–e). Thus, in skeletal muscle during acute exercise, beclin 1 and Tlr9 interact, and simultaneously, an endogenous ligand, mtDNA, associates with the innate immune sensor Tlr9. We cannot exclude the possibility that additional CpG DNA ligands associate with muscle Tlr9 during exercise.
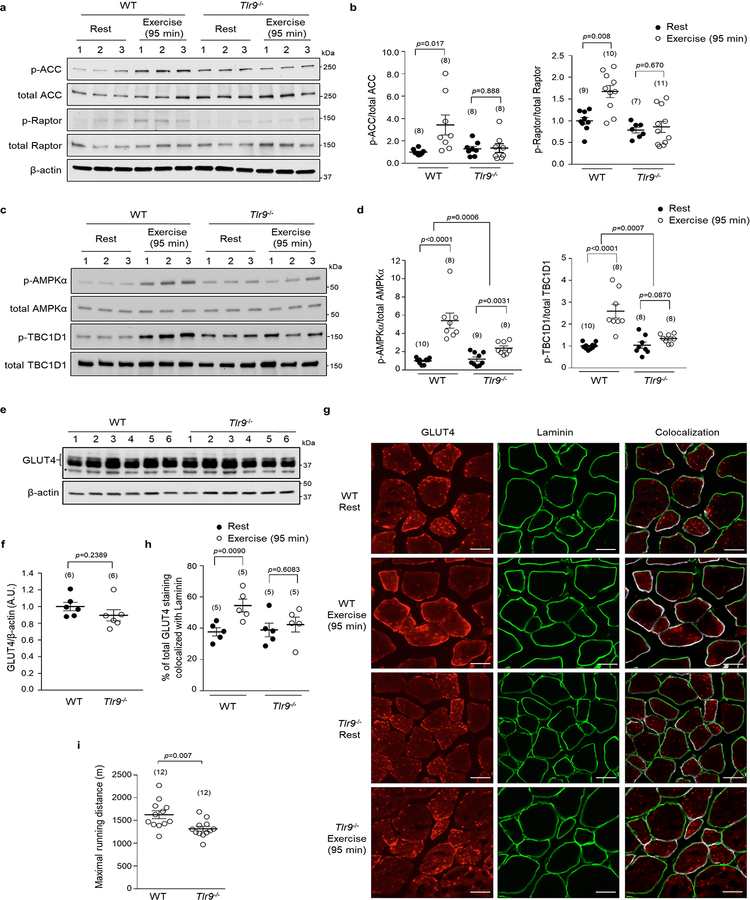
In view of previous links between beclin 1 and exercise-induced muscle AMPK activation3, these findings suggested that Tlr9 might exert unexpected metabolic functions in muscle during exercise. Therefore, we studied the skeletal muscles of Tlr9−/− mice15 during treadmill exercise. Fiber type, mitochondrial respiratory capacity, and capillary density were similar in muscles of wild-type and Tlr9−/− mice (Extended Data Fig. 4a–g). Tlr9−/− and wild-type mice also had similar cardiac function (Extended Data Fig. 4h). However, compared to littermate controls, Tlr9−/− mice were deficient in exercise-induced muscle AMPK activation, as determined by quantitation of phosphorylation of AMPK and its downstream targets, TBC1D1, acetyl-CoA carboxylase (ACC), and Raptor (Fig. 2a, b, Extended Data Fig. 5a–d). Consistent with defective AMPK and TBC1D1 phosphorylation, Tlr9−/− mice failed to exhibit exercise-induced plasma membrane localization of the GLUT4 glucose transporter (Fig. 2c, d; Extended Data Fig. 5e–h), which is essential for exercise-stimulated muscle glucose uptake16. Furthermore, Tlr9−/− mice did not display decreased plasma glucose levels during exercise (Fig. 2e) and they exhibited decreased exercise endurance (Extended Data Fig. 5i).
Figure 2. Tlr9 is required for exercise-induced muscle AMPK activation.
(a, b) Representative western blots (a) and quantitation (b) of p-AMPK(Thr172) and p-TBC1D1(Ser237) in VL muscles. (c, d) Representative images of GLUT4 staining in VL muscles (c) and quantitation (d) of GLUT4/Laminin colocalization in extensor digitorum longus (EDL) muscles (see Extended Data Fig. 5g for EDL representative images). At least 100 muscle fibers per mouse. Arrow, Glut4 localization at plasma membrane. Scale bars, 20 μm. (e) Blood glucose levels. (f, g) Representative western blots (f) and quantitation (g) of AMPK phosphorylation in VL muscles from indicated recipient mice transplanted with indicated donor bone marrow. (h, i) Representative western blots (h) and quantitation (i) of AMPK phosphorylation in EDL muscles. (j) Glucose uptake in EDL muscles. Western blots and images from one representative experiment and quantitation data combined from three independent experiments. Similar results observed for each experiment. In b, d, e, g, i and j, data points, individual mice (b, d, e, g) or muscles (i, j), sample size indicated in parentheses. Data are mean ± s.e.m. In b, d, e, i and j, unpaired two-tailed t-test to compare different conditions per genotype. Two-way ANOVA for magnitude of changes between different conditions in mice of different genotypes. In g, unpaired two-tailed t-test for differences between donor genotypes for each recipient genotype and two-way ANOVA for differences between recipient genotypes. For uncropped gels, see Supplementary Fig. 1.
Muscles of Tlr9−/− and wild-type mice were similar with respect to known regulators of AMPK activation in response to exercise, including increased AMP/ATP and ADP/ATP ratios, decreased glycogen levels, and levels of total LKB1 and LKB1 phosphorylated at serine 42817 (Extended Data Fig. 6a–h). Moreover, downstream AMPK-regulated glucose transport pathways were intact in Tlr9−/− mice, as administration of a direct AMPK activator, PF-73918, to Tlr9−/− and wild-type mice resulted in similar levels of TBC1D1 phosphorylation in skeletal muscle and decline in blood glucose (Extended Data Fig. 6i–k). Thus, Tlr9 is required for skeletal muscle AMPK activation and its effects on glucose metabolism during exercise through a newly defined mechanism.
To evaluate whether Tlr9 in hematopoietic cells contributes to exercise-induced skeletal muscle AMPK activation, we non-competitively transplanted irradiated wild-type or Tlr9−/− recipient mice with bone marrow cells from either wild-type or Tlr9−/− donor mice. Recipient mice exhibited high levels of donor cell chimerism in the peripheral blood (Extended Data Fig. 7a, b). A greater increase in exercise-induced skeletal muscle phosphorylation of AMPK and its substrate, TBC1D1, was observed in wild-type compared to Tlr9−/− recipient mice (Fig. 2f, g; Extended Data Fig. 7c, d). Notably, donor genotype had no effect on AMPK or TBC1D1 phosphorylation in either recipient genotype, indicating that Tlr9 in hematopoietic cells is not required for Tlr9-dependent exercise-induced AMPK activation in skeletal muscle.
To further assess whether Tlr9 functions in a tissue-autonomous manner in skeletal muscle AMPK activation, we performed ex vivo electrical stimulation to induce muscle contraction. Similar to observations during treadmill exercise, muscles from wild-type as compared to Tlr9−/− mice displayed a greater increase in ex vivo contraction-induced AMPK and TBC1D1 phosphorylation (Fig. 2h, i; Extended Data 7e, f). Importantly, there was a corresponding decrease in ex vivo contraction-stimulated glucose uptake in muscles of Tlr9−/− mice (Fig. 2j). Therefore, Tlr9 functions in a muscle cell autonomous manner to regulate AMPK activation and glucose metabolism. This may contribute to alterations in muscle AMPK activation, serum glucose levels, and exercise endurance in Tlr9−/− mice, although other physiological factors may also influence these phenotypes in vivo.
The defect in exercise-induced skeletal muscle AMPK activation, plasma membrane GLUT4 localization, and exercise endurance in Tlr9−/− mice closely resembled phenotypes previously reported in Becn1+/− mice and mice with non-phosphorylatable knock-in mutations in BCL2 (Thr69Ala, Ser70Ala, and Ser87A; termed BCL2 AAA) that prevent exercise-induced disruption of BCL2/beclin 1 binding3. However, unlike Becn1+/− mice or BCL2 AAA mice, Tlr9−/− mice did not exhibit defects in exercise-induced skeletal muscle autophagic flux (Extended Data Fig. 8a, b). Thus, impaired exercise-induced autophagy may not be responsible for defects in AMPK activation in Becn1+/− or BCL2 AAA mice; instead, the functions of beclin 1 in autophagy and AMPK activation may be independent.
We hypothesized that the defect in AMPK activation in BCL2 AAA mice3 may involve decreased interaction of beclin 1 with Tlr9. We crossed BCL2 AAA mice with Tlr9-HA knock-in mice. The exercise-induced increase in beclin 1/Tlr9-HA interaction observed in wild-type mice was not observed in BCL2 AAA mice (Extended Data Fig. 8c, d). Similarly, in cultured cells that stably express BCL2 AAA19, glucose starvation failed to increase beclin 1/TLR9-HA binding (Extended Data Fig. 8e–g). Thus, constitutive binding of beclin 1 by BCL2 blocks not only stress-induced autophagy3, 5, but also glucose starvation- and exercise-induced interaction of beclin 1 with Tlr9.
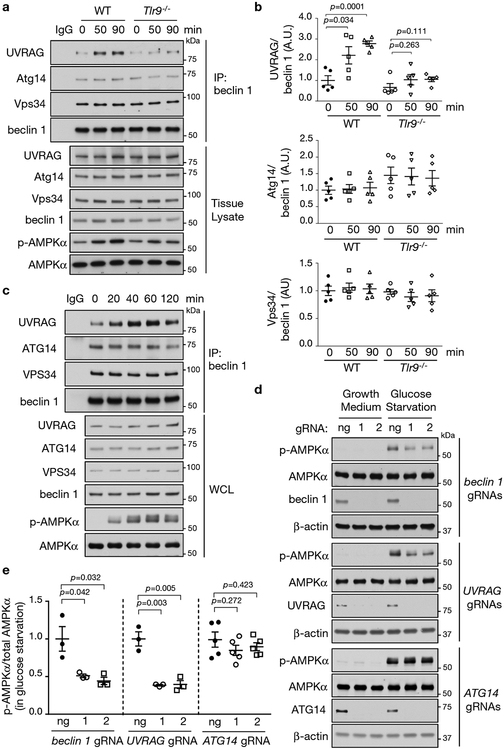
To explore the mechanism by which Tlr9/beclin 1 interaction regulates AMPK activation, we evaluated the effect of Tlr9 knockout on beclin 1 association with other members of PI3KC3 complexes in skeletal muscle during exercise. The two main complexes are PI3KC3-C1, which contains beclin 1, Vps34, Vps15 and Atg14 and functions in autophagic vesicle nucleation, and PI3KC3-C2, which contains beclin 1, Vps34, Vps15 and UVRAG and functions in endolysosomal and autophagolysosomal maturation20. During exercise, increased UVRAG, but not increased Atg14, bound to beclin 1, suggesting enhanced assembly of PI3KC3-C2 (Fig. 3a, b, Extended Data Fig. 9a, b). This increased UVRAG/beclin 1 interaction was blocked in Tlr9−/− mice (Fig. 3a, b). Moreover, increased UVRAG/beclin 1 binding occurred during ex vivo electrical stimulation-induced contraction in muscles from wild-type but not Tlr9−/− mice (Extended Data Fig. 9c, d). Additionally glucose starvation in U2OS cells led to a steady increase in UVRAG/beclin 1, but not ATG14/beclin 1, interaction (Fig. 3c). CRISPR-mediated knockout of beclin 1 and UVRAG, but not ATG14, reduced glucose starvation-induced AMPK phosphorylation (Fig. 3d, e) but not Ser428 phosphorylation or total levels of LKB1 (Extended Data Fig. 10). Thus, the PI3KC3-C2 complex may regulate AMPK activity.
Figure 3. TLR9 is required for beclin 1/UVRAG interaction during exercise.
(a) Co-immunoprecipitation of UVRAG, Atg14 or Vps34 with beclin 1 in tibialis anterior muscles (TA) at indicated duration of exercise. Tissue lysate from WT 0 min sample used for control IgG precipitation. (b) Quantitation of co-immunoprecipitation of UVRAG, Atg14 and Vps34 with beclin 1 in three independent experiments using design shown in (a). Data points, individual mice (n=5). Data are mean ± s.e.m. Unpaired two-tailed t-test comparing 50 min or 90 min exercise versus 0 min condition for each genotype. (c) Co-immunoprecipitation of UVRAG, ATG14 or VPS34 with beclin 1 in U2OS cells at indicated time points after glucose starvation. Similar results observed in three independent experiments. (d, e) Representative blots (d) and quantitation (e) of effects of indicated gene knockout on AMPK activation in U2OS cells cultured in normal or glucose starvation media (1 h). Two independent gRNAs per gene target. Quantitation data from 3 (beclin 1 gRNAs, UVRAG gRNAs) or 5 (ATG14 gRNAs) independent experiments. Data are mean ± s.e.m. Unpaired two-tailed t-test. ng, no gRNA. For uncropped gels, see Supplementary Fig. 1.
Taken together, our data are consistent with a model in which cross-talk between Tlr9 and beclin 1/UVRAG-containing complexes leads to AMPK activation during energy stress. Our findings integrate three heretofore disparate aspects of cell biology – innate immune sensing, protein complexes containing autophagy proteins, and AMPK activation – in skeletal muscle responses to exercise. A previous study in immune cells indicated that autophagy proteins involved in LC3-associated phagocytosis regulate TLR9 responses to DNA immune complexes21. The link between an innate immune receptor capable of recognizing mtDNA and the activation of AMPK may serve as a fundamental mechanism to maintain muscle homeostasis during exercise. The autophagy machinery is an ancient and highly conserved set of proteins that mediates responses to nutritional stress even in unicellular organisms. Teleologically, these proteins may also have evolved to respond to stresses unique to metazoan organisms such as exercise — a process that requires increased metabolic resources, especially in skeletal muscle. The cross-talk between an innate immune sensor that responds to DNA and between autophagy proteins during exercise may represent a more general paradigm in which innate immune sensors of danger and cellular quality control mechanisms intersect to maintain normal mammalian physiology.
METHODS
Mouse strains.
All mice were housed in a pathogen-free facility under 12 h light dark cycles with ad libitum access to food and water. All mouse experimental procedures were approved by the Institutional Animal Care and Use Committee at UT Southwestern Medical Center and performed in accordance with institutional guidelines. Eight- to twelve-week-old male mice were used except for bone marrow transplantation experiments (see section below). Tlr9−/− mice15, BCL2 AAA mice3 and GFP-LC3 transgenic mice22 have been described previously and all strains were rederived in the UTSW Transgenic Mouse facility on a C57BL/6J background and further backcrossed to C57BL/6J for at least 10 generations.
Tlr9-HA knock-in (KI) mice were generated by CRISPR genome editing technology at the Transgenic Core Facility at UT Southwestern Medical Center. Guide RNAs (targeting the Tlr9 genome near the region encoding the stop codon) and the donor oligo were designed, synthesized and validated by Sigma-Aldrich, Inc. The gRNA (antisense) targeted genome sequence is 5’-GGGACCTACAGCAGAATAG-3’. The sequence of the donor oligo is 5’-TTCTGGGCCCAGCTGAGTACAGCCCTGACTAGGGACAACCG CCACTTCTATAACCAGAACTTCTGCAGGGGACCTACAGCAGAATACCCATACGATGTTCCAGATTACGCTTAGCTCAGAGCAACAGCTGGAAACAGCTGCATCTTCATGTCTGGTTCCCGAGTTGCTCTGCCTGCCTTG-3’. After homologous recombination, the antisense PAM sequence (CCG) recognition by Cas9 is diminished with a change of C to A (CAG) and this change also generates a PstI restriction site (CTGCAG) for further genotyping purposes. Genotyping primers were designed to generate a 473 bp PCR product for the WT allele (500 bp PCR product for the KI allele) surrounding the targeting region. The PAM sequence is located 224 bp from the 5’ end for both the WT and KI allele and 246 bp and 273 bp from the 3’ end for the WT and KI allele, respectively. The PCR products were subjected to PstI digestion. The KI allele PCR product is cut into two fragments (226 bp and 278 bp) while the WT allele PCR product is not cleaved. The sequences of genotyping primers are: Forward, 5’-CTGCTGGCTCAGCAGCG-3’; Reverse, 5’-GAACTCCAGTCCTGTTCTGTG-3’.
To generate Tlr9-HA KI mice, C57BL/6J (Jackson Laboratories) mouse zygotes were injected with gRNA, Cas9 mRNA and donor oligo into the pronucleus and cytoplasm and then transferred to surrogate mothers for gestation. Genomic DNA isolated from tail biopsies of F0 founders was used as a template for PCR genotyping. PCR products from each mouse were ligated into TA cloning vectors (Invitrogen) followed by bacterial transformation. Four colonies were selected for each mouse and plasmids were purified and sequenced. An F0 mouse with at least one plasmid carrying the correct KI sequence was considered as a positive founder. Since F0 founders are mosaic, positive founders were bred with wild-type C57BL/6J mice (Jackson laboratory) to isolate the KI allele. Mice from the F1 generation that carried the KI allele were considered to have stable germline transmission and used for further breeding to generate homozygous KI mice for experiments.
Plasmids.
pBICEP vectors encoding Flag tagged-beclin 1 full-length protein or deletion mutants were generated as described23. cDNA encoding full length or amino acids 1–868 of TLR9 were cloned into pCR3.1 and an HA tag sequence was added to the C’ terminus of the TLR9 cDNA. Codon-optimized TLR7 cDNA24 (gift of G. Barton, UC Berkeley) was cloned into pCR3.1 with an HA tag sequence added to the C’ terminus. A lentivirus vector pXPR_023 expressing Cas9 and gRNA (gift of J. Doench, The Broad Institute of Harvard and MIT) was used for CRISPR-mediated gene knockout. The gRNAs targeting individual genes were designed using Broad Institute Genetic Perturbation Platform (GPP) and the cloning of gRNAs into the pXPR_023 vector was performed according to the online instructions at the GPP web portal (https://portals.broadinstitute.org/gpp/public/). Guide RNAs targeted sequences used in the study are: human BECN1, #1, 5’-GAACCTCAGCCGAAGACTGA-3’(sense); #2, 5’-GAAGGTTGCATTAAAGACGT-3’(antisense); human UVRAG, #1, 5’-AGGAATCCCTAAATGAGCTG-3’(sense); #2, 5’-GAGTCTGTTTAATTGCACAC-3’(antisense); and human ATG14, #1, 5’- TTGTTAGGGAGGCTAATCCA-3’(antisense); #2, 5’-AGGAAGTAAAGACGGG TGTG-3’(sense).
Cell culture.
HeLa, U2OS, and Phoenix cell lines were obtained from ATCC and cultured in DMEM (Gibco) containing 10% FBS (Gibco), 2 mM L-glutamine (Gibco) and non-essential amino acids (Gibco). Cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Glucose starvation was performed by replacing the normal growth medium with DMEM (without glucose) (Gibco) containing 10% dialyzed FBS (Gibco) for the indicated time period. Amino acid starvation was performed by replacing the normal growth medium with Earle’s Balanced Salt Solution (Gibco). For treatment with mitochondrial damaging agents and PF-739, cells cultured in normal growth medium were incubated with oligomycin (2.5 μM) + antimycin A (250 nM), carbonyl cyanide 3-chlorophenylhdrazone (CCCP) (5 μM) or PF-739 (5 μM) for 1 h. Lentiviruses carrying Cas9 cDNA and gRNA for infection of U2OS cells to knock out targeted genes were packaged in Phoenix cells (ATCC) by co-transfection of pXPR_023 vector with packaging plasmids. Media containing virus particles were collected 48 h after transfection and used to infect U2OS cells for 3 h in the presence of 8 μg/ml polybrene. Seventy-two hours after infection, puromycin antibiotic selection was added to the media of infected cells and maintained for 4 days before cells were used for experiments.
Lysate preparation and immunoprecipitation assays.
Cells were lysed in lysis buffer containing 50 mM Hepes pH 7.4, 50 mM KCl, 50 mM NaF, 5 mM Na4P2O7, 5 mM β-glycerophosphate, 1 mM EDTA, 1 mM EGTA, 1 mM Na3VO4, 1 mM dithiothreitol supplemented with protease inhibitor cocktail (Roche) and 1% (w/v) Triton X-100. Lysates were precleared with Protein A agarose (Sigma) for 1 h followed by immunoprecipitation of indicated protein. For immunoprecipitation of Flag-beclin 1 or Myc-BCL2, anti-Flag-M2 agarose (Sigma) or anti-Myc agarose (Santa Cruz) was added to the lysate for 3 h. For immunoprecipitation of overexpressed TLR7-HA, TLR9-HA or endogenous beclin 1, anti-HA high affinity antibody (#11867423001, Roche) or anti-beclin 1 antibody (#sc48341, Santa Cruz) was added to the precleared lysate for 3 h before Protein G agarose (Santa Cruz, for anti-HA high affinity antibody) or Protein A agarose (Sigma, for anti-beclin 1 antibody) was added for another 1.5 h. Immunoprecipitates were subjected to SDS-PAGE and western blot analysis.
Muscle tissues were homogenized in ice-cold lysis buffer (1 ml lysis buffer/100 mg tissue) without Triton X-100 using an Ultra-Turrax T10 Disperser (IKA) before Triton X-100 was added to the homogenates to reach a concentration of 1% (w/v). For analyses of Tlr9 expression levels in tissues of Tlr9-HA KI mice, mice were perfused with PBS prior to sacrifice and tissue harvesting. Muscle homogenates were incubated at 4°C for 1 h followed by centrifugation at 20 000 × g for 15 min. The supernatants were recovered as tissue lysates and precleared with Protein A agarose (Sigma) for 1 h followed by immunoprecipitation of Tlr9-HA, beclin 1 or UVRAG. Anti-HA antibody (#3724, Cell signaling) or anti-beclin 1 antibody (#sc48341, Santa Cruz) or anti-UVRAG antibody (#11315, Cell signaling) was added to the lysates for 3 h before Protein A agarose was added for another 1.5 h. Immunoprecipitates were denatured with Laemmli buffer at 37°C for 30 min before SDS-PAGE and western blot analysis.
Protein Purifications.
The maltose-binding domain (MBP)-tagged TLR9 TIR domain (amino acids 868–1032) plasmid (gift of J. Hurley, UC Berkeley) was used to re-clone the MBP-TLR9 TIR domain into the pCAG vector, removing the twin-Strep and Flag tags. One liter of HEK 293S cells (3.0 × 106 cells/ml) was transfected with the MBP-TIR plasmid using polyethylenimine (PEIpro) in the presence of 10 mM sodium butyrate (Sigma-Aldrich). After shaking for 48 h at 37°C and 8% CO2, cells were harvested and lysed by adding 1% Triton X-100, followed by centrifugation at 50,000 x g for 30 min. The clear supernatant was collected, amylose resins (NEB) were added and incubated for 1 h at 4°C. The resins were washed with 15 column volumes of the purification buffer (20 mM Tris pH 8.0, 150 mM NaCl, 1 mM DTT). The protein was eluted with purification buffer containing 10 mM maltose (Sigma-Aldrich) and subjected to size exclusion chromatography. The eluate was pooled, concentrated, flash-frozen by liquid nitrogen and stored at −80°C. The bacterial expression plasmid containing full-length human BECN 1 (gift from Dr. Matthew J. Ranaghan, the Broad Institute of Harvard and MIT) was used to express and purify Beclin 1 protein as described25.
In vitro binding assays.
Five hundred nanograms of MBP protein and MBP-TIR protein were incubated with 250 ng of beclin 1 protein in binding buffer (0.1% Triton, 1mM DTT and 0.1%BSA in PBS) for 2 h at 4°C before adding 50 μl of amylose resin (Bio-Rad) for another 2 h at 4°C. After washing with binding buffer three times and with PBS two times, 100 μl of PBS containing 10 mM maltose was added to the resin for elution at RT for 15 min. The eluates were subjected to western blot analysis.
Western blot analysis.
All primary antibodies used for western blots were purchased from commercial sources. Antibodies from Santa Cruz include: anti-beclin 1 (sc11427), anti-myc HRP (sc40 HRP) and anti-β-actin HRP (sc47778 HRP). Antibodies from Cell Signaling Technology include: anti-UVRAG (#11315), anti-Vps34 (#4263), anti-Atg14 (#96752, to detect mouse Atg14), anti-p-AMPKα(Thr172) (#2535), anti-AMPKα (#2532), anti-p-Raptor(Ser792) (#2083), anti-Raptor (#2280), anti-TBC1D1 (#66433) and anti-HA (#3724, to detect Tlr9-HA in tissue lysates of Tlr9-HA KI mice). Other primary antibodies include: anti-p-ACC(Ser79) (#07–303, Millipore), anti-ACC (#04–322, Millipore), anti-p-TBC1D1(Ser237) (#07–2268, Millipore), anti-ATG14 (M184–3, MBL International, to detect human ATG14), anti-HA high affinity-HRP (#12013819001, Roche, to detect overexpressed TLR9-HA in cells and Tlr9-HA after immunoprecipitation from muscle lysates of the Tlr9-HA KI mice), anti-Flag M2-HRP (A8592, Sigma), and anti-GLUT4 (GT41-A, Alpha Diagnonstic International). To detect beclin 1 in Tlr9-HA immunoprecipitates from muscle lysates by western blot analysis, 10% MINI-PROTEAN TGX Precast gels (Bio-Rad) were used. For detection of all other proteins, 4–20% Precast gels were used.
Exercise studies.
Mouse treadmill exercise experiments were performed as previously described3 with minor modifications. Eight to twelve week-old mice were used for all experiments. Briefly, mice were acclimated to the 10° uphill treadmill for two days. On day one, mice ran for 5 min at a speed of 8 m/min followed by 2 min at a speed of 10 m/min. On day two, mice ran for 5 min at a speed of 10 m/min followed by 2 min at a speed of 12 m/min. On day three, mice were allowed to rest. On day four, mice were subjected to a single bout of running starting at a speed of 10 m/min for 40 min. Afterwards, the speed was increased at a rate of 1 m/min every 10 min for a total of 30 min and then at a rate of 1 m/min every 5 min until reaching the designated time point (for tissue harvesting) or mouse exhaustion (for measurement of maximal running distance). Tissues were snap frozen using liquid nitrogen at desired time points after exercise. For blood glucose measurements after exercise, food was removed from the mouse cages for 4 h before mice were subjected to treadmill exercise on day four. Blood was obtained from the tail vein and blood glucose levels were measured using commercial glucose assay reagents (Sigma G6918, P7119 and F5803). For experiments involving the harvesting of muscle samples for immunoprecipitation, food was removed from the cages of the experimental mice at 6:00 PM on day three, and then mice were refed ad libitum on day four for 3 h before treadmill running experiments were performed.
Bone marrow transplantation.
Bone marrow transplantation experiments were performed as previously described26. Donor bone marrow cells were obtained from 8-week-old WT or Tlr9−/− male mice and non-competitively transplanted into irradiated 9- to 12-week-old WT or Tlr9−/− male recipients. The recipient mice were irradiated using an XRAD 320 X-ray irradiator (Precision X-Ray Inc.) with two doses of 540 rad (1,080 rad in total) delivered at least 3 h apart. Two million donor bone marrow cells were injected into the tail vein of each recipient mouse. Eight weeks after transplantation, high levels of donor cell chimerism in the blood of recipient mice was confirmed using real-time PCR. Specifically, ~80 μl of blood was obtained from the tail vein of recipient mice and DNA was extracted using a QIAamp DNA mini kit (Qiagen). Twenty nanograms of DNA was used for real-time PCR using a SYBR green PCR kit (Qiagen) to determine the relative copy number of WT genome or Tlr9−/− genome using the following primers: for WT genome, Forward, 5’-GAAGGTTCTGGGCTCAATGGTCATGTG-3’; Reverse, 5’-GCAATGGAAAGGACTGTCCACTTTGTG-3’; for Tlr9−/− genome, Forward 5’-ATCGCCTTCTATCGCCTTCTTGACGAG-3’; Reverse, 5’-GCAATGGAAAGGAC TGTCCACTTTGTG-3’. The relative copy numbers of Tlr7 genome was used as an internal control for normalization. The following primers were used for Tlr7 genome: Forward, 5’-AGGGTATGCCGCCAAATCTAAAG-3’; Reverse, 5’-ACCTTTGTGTG CTCCTGGAC-3’. Nine weeks after transplantation, recipient mice were used for exercise studies.
Ex vivo muscle studies.
Ex vivo muscle incubation was performed as previously described27, 28 with certain modifications. Specifically, the feeding status of mice was synchronized by removing food overnight from their cages the day before experiments and replacing food for 4 h on the morning of the day when experiments were performed. Mice were anaesthetized by isoflurane inhalation and EDL muscles were dissected with the tendons at both ends ligated with 5–0 silk suture loops (F.S.T) and suspended between two electrodes on a contraction apparatus at their resting length at 30°C in modified Krebs–Ringer–Henseleit (KRH) buffer (120 mM NaCl, 4.74 mM KCl, 1.25 mM MgCl2, 1.2 mM KH2PO4, 25 mM NaHCO3, 1.25 mM CaCl2, 5.5 mM glucose, 1 mM sodium pyruvate, 5 mM Hepes pH 7.4 and 0.1% bovine serum albumin) constantly gassed with 95% O2/5% CO2. To study the effects of Tlr9 ligand on AMPK activation, muscles were pre-incubated for 45 min before treatment with 1 μM control ODN or ODN 2395 or 1 μM PF-739 (as positive control) for 1 h followed by snap freezing in liquid nitrogen. To study muscle contraction-induced AMPK activation, muscles were pre-incubated for 45 min before they were either left resting or subjected to electrical stimulation (1 Hz, 30 V square wave pulses of 10 ms duration) for 20 min. Pulses were generated by a stimulus isolation amplifier (model 4AD, Getting Instruments) with the control of a waveform generator (model GH-CJDS66, Koolertron). Muscles were removed from the contraction apparatus and snap frozen using liquid nitrogen. For measurement of glucose uptake during contraction, muscles were pre-incubated for 45 min before being subjected to electrical stimulation for 5 min. Muscles were then transferred into fresh KRH medium containing 0.25 μCi/ml [3H] 2-deoxy-glucose (2DG) (PerkinElmer) and 0.1 μCi/ml [14C] mannitol (PerkinElmer) and subjected to electrical stimulation for another 15 min. For each electrically stimulated EDL muscle, the other EDL muscle from the same mouse was left resting for 15 min in the KRH medium containing [3H] 2-deoxy-glucose and [14C] mannitol and used as a resting control. After treatments, muscles were harvested, washed in ice-cold KRH medium, dabbed dry on filter paper and snap frozen in liquid nitrogen. For 2DG uptake analysis, each muscle was homogenized in 600 μl ice-cold lysis buffer using an Ultra-Turrax as described above. Two hundred and fifty microliters of lysate was mixed with 5 ml scintillation liquid (3a70B, RPI) to measure 2DG uptake by scintillation counting using [14C] mannitol for estimation of extracellular space.
Autophagy analyses.
To assess autophagic flux in skeletal muscle in resting and exercise conditions, WT;GFP-LC3 mice and Tlr9−/−;GFP-LC3 mice were acclimated to the treadmill as described above. During the morning of day four, mice were treated with either PBS or chloroquine (50 mg/kg) via intraperitoneal injection. For resting groups, after 3 h of PBS or chloroquine treatment, mice were anaesthetized by inhalational isoflurane and perfused with 30 ml 4% paraformaldehyde (PFA) in PBS at a rate of 4.5 ml/min. Vastus lateralis muscles were collected and processed for frozen sectioning as described22. For exercise groups, after 90 min of PBS or chloroquine treatment, mice were subjected to treadmill running for 90 min before perfusion and muscle collection. Frozen muscle section slides were air-dried at room temperature for 15 min before mounting and imaging by fluorescence microscopy using a Zeiss Axioplan2 microscope. The total number of GFP–LC3 puncta per 2,500 μm2 was counted using image J. More than 15 randomly chosen fields (by an observer blinded to experimental genotype) were used per mouse and an average value was determined.
Immunofluorescence analyses.
For GLUT4 staining, WT and Tlr9−/− mice were subjected to exercise for 95 min before perfusion and harvesting muscles for frozen section preparation as described above. Frozen muscle section slides were heated at 50°C for 15 min, rehydrated in PBS for 30 min, blocked with 1% BSA in PBS for 1 h and probed with anti-GLUT4 antibody (Rabbit, GT41-A, Alpha Diagnostic International) and anti-Laminin-2 antibody (Rat, L0663, Sigma) overnight at 4°C. Alexa Fluor 594 anti-rabbit secondary antibody and Alexa Fluor 488 anti-rat secondary antibody (Invitrogen) were then added for 1 h at room temperature before the slides were mounted and imaged using a Zeiss Axioplan2 microscope. Colocalization of GLUT4 and Laminin were calculated using ImageJ. For muscle fiber type staining and capillary density determination, TA muscles were snap frozen in liquid nitrogen-cooled 2-methylbutane before being placed into OCT compound (Fisher Healthcare) and cryo-sectioned at a thickness of 10 μm. For muscle fiber type staining, sections were blocked with 10% goat serum (Gibco) in PBS for 1 h at room temperature, and then probed with anti-Myosin heavy chain type I (Mouse, BA-D5, Developmental Studies Hybridoma Bank), anti-Myosin heavy chain type IIa (Mouse, SC-71, Developmental Studies Hybridoma Bank), anti-Myosin heavy chain type IIb (Mouse, BF-F3, Developmental Studies Hybridoma Bank) and anti-Laminin (Rabbit, L9393, Sigma) antibodies overnight at 4°C. Alexa Fluor 594 goat anti-mouse IgG1 (Invitrogen), Alexa Fluor 647 goat anti-mouse IgG2b (Invitrogen), Alexa Fluor 488 goat anti-rabbit IgG (H+L) (Invitrogen) and Dylight 405 goat anti-mouse IgM (Jackson ImmunoResearch) secondary antibodies were then added for 1 h at room temperature before the slides were mounted and imaged using a Zeiss LSM780 inverted microscope. For capillary density determination, muscle sections were air dried at room temperature before being put into cold acetone (−20°C) for 15 min. Slides were then washed three times with PBS and blocked with 1% BSA in PBS for 1 h before being probed with biotinylated Lectin I (5 μg/ml, B-1105, Vector Laboratories) and anti-Laminin-2 antibody (Rat, L0663, Sigma) overnight at 4°C. Alexa Fluor 488-conjugated streptavidin and Alexa Fluor 594 anti-rat secondary antibody (Invitrogen) were then added for 1 h at room temperature before the slides were mounted and imaged using a Zeiss Axioplan2 microscope.
Muscle morphology and COX enzymatic activity.
Fresh-frozen muscle sections were prepared as described above for muscle fiber type staining. H&E staining was performed following the protocol from the TREAT-NMD website (http://www.treat-nmd.eu/downloads/file/sops/cmd/MDC1A_M.1.2.004.pdf), and imaged using an Olympus IX83 microscope. COX enzymatic activity was performed as described previously29, and visualized using an Olympus IX83 microscope. COX stain intensity was calculated using ImageJ (NIH).
Mitochondrial DNA and genomic DNA binding to Tlr9-HA.
Anti-HA high affinity antibody (Roche; 1μg/sample), Protein G Dynabeads (Invitrogen; 50 μl/sample) and herring testes DNA (Sigma; 50 μg/sample) were added together and incubated at 4°C overnight. The HA antibody-bound beads were then washed once with lysis buffer before use in immunoprecipitation. Approximately 150 mg of gastrocnemius muscles from WT or Tlr9−/− mice were homogenized as described above using a lysis buffer containing 50 mM Hepes pH 7.4, 50 mM KCl, 50 mM NaF, 5 mM Na4P2O7, 5 mM β-glycerolphosphate, 1 mM EDTA, 1 mM EGTA, 1 mM Na3VO4, 1 mM dithiothreitol, 5% (v/v) glycerol supplemented with protease inhibitor cocktail (Roche) and 1% (w/v) NP40. Lysates were precleared with Protein A agarose for 1 h and incubated with the prepared HA antibody-bound beads at 4°C overnight. The beads were then washed three times with lysis buffer. Approximately one seventh of the beads were saved for western blot analysis to assess the amount of immunoprecipitated Tlr9-HA; the remainder of the beads were resuspended in 80 μl nuclease-free water containing 200 μg/ml Proteinase K (Roche) and incubated at 55°C for 30 min followed by 95°C for 15 min to inactivate Proteinase K. After centrifugation at 12 000 × g for 5 min, the supernatants were collected and 5 μl of each sample was used for real-time PCR with each set of mitochondrial DNA primers using SYBR green PCR kit (Qiagen). The six sets of mouse mitochondrial DNA primers targeting different regions of the mitochondrial genome and two sets of genomic DNA primers have been described previously30. Mitochondrial DNA primer sets are: set1 (targeting nd1), Forward, 5’-CAAACACTTATTACAACCCAAGAACA-3’; Reverse, 5’-TCATATTATGGCTATGGGTCAGG-3’; set2 (targeting nd5), Forward, 5’-AGCATTCGGAAGCATCTTTG-3’; Reverse, 5’-TTGTGAGGACTGGAATGCTG-3’; set3 (targeting nd5), Forward, 5’-CCACGCATTCTTCAAAGCTA-3’; Reverse, 5’-TCGGATGTCTTGTTCGTCTG-3’; set4 (targeting nd6), Forward, 5’-TGGTTTGGGAGATTGGTTG-3’; Reverse, 5’-CACAACTATATATTGCCGCTACCC-3’; set5 (targeting rnr1), Forward, 5’-CCTCTTAGGGTTGGTAAATTTCG-3’; Reverse, 5’-CGAAGATAATTAGTTTGGGTTAATCG-3’; set6 (targeting rnr2), Forward, 5’-AAACAGCTTTTAACCATTGTAGGC-3’; Reverse, 5’-TTGAGCTTGAACGCTTT CTTTA-3’. Genomic DNA primer sets are: set1 (targeting POLG), Forward, 5’-ATGAATGGGCCTACCTTGA-3’; Reverse, 5’-TGGGGTCCTGTTTCTACAGC-3’; set2 (targeting POLG), Forward, 5’-TCCTGGAACAGTTGTGCTTTC-3’; Reverse, 5’-CCATCTACTCAGGACGGAGTTC-3’.
Mitochondrial DNA levels in plasma.
Mouse blood was collected by cardiac puncture into EDTA-coated microtainers (BD). Plasma was obtained after two rounds of centrifugation (3000 × g, 10 min at 4°C). Two hundred and fifty microliters of plasma from each mouse was used to extract circulating DNA using QIAamp circulating nucleic acid kit (Qiagen). One twentieth of the total yield from each plasma sample was used for real-time PCR to assess levels of mitochondrial DNA using six sets of mouse mitochondrial DNA primers as described above.
Real-time PCR.
To assess mRNA levels of Tlr9 in different tissues, real-time PCR analysis was performed on normalized mouse multiple tissue cDNA panels (Takara) using SYBR green PCR kit (Qiagen). To compare mRNA levels of Tlr9 in myoblast and myotubes, mouse primary myoblasts were isolated and differentiated into myotubes as previously described31. RNA was isolated from myoblasts and myotubes using RNeasy mini kit (Qiagen) and reverse-transcribed into cDNA using iScript cDNA synthesis kit (Bio-Rad) before use in real-time PCR analyses. The QuantiTect primer assay for mouse Tlr9 (Qiagen) was used. The following primers were used for β-actin: Forward, 5’-CTGGCTCCTAGCACCATGAAGAT-3’; Reverse, 5’-GGTGGACAGTGAGGCCAGG AT-3’.
AMP, ADP and ATP measurements.
Frozen EDL muscles were homogenized in 500 μl of ice-cold 0.1 M HClO4/40% (v/v) CH3OH using an Ultra-Turrax tissue disintegrator as described above. After centrifugation (20,000 g, 10 min, 4°C), supernatants were neutralized with 1.1 M (NH4)2HPO4, dried and resuspended in HPLC-grade water. Nucleotides were then separated and quantified by HPLC/UV detection as described previously32.
Measurement of muscle glycogen content.
Mouse TA muscles were homogenized with 600 μl of ddH2O using an Ultra-Turrax tissue disintegrator as described above before immediately placing them on a 95°C heat block for 15 min to inactivate enzymes in the samples. After centrifugation (18,000 g, 10 min at 4°C), 5 μl of supernatant was used for the measurement of glycogen content using a Glycogen Assay kit (Abcam).
Mouse PF-739 treatment.
The direct AMPK activator PF-73918 (AOBIOUS) was dissolved in DMSO at a concentration of 200 mg/ml. Before performing mouse experiments, a solution containing 10% 200 mg/ml PF-739 in DMSO/30% PEG 400/60% H2O was prepared for subcutaneous injection into mice (50 μl/10g body weight). A solution containing 10% DMSO/30% PEG 400/60% H2O was injected into control mice. Ninety min after injection, mice were anaesthetized by isoflurane inhalation and blood and tissues were collected for blood glucose measurement and western blot analyses.
Statistical analyses.
Student t-tests and Mann-Whitney tests were performed using Prism software (GraphPad). One-way ANOVA and two-way ANOVA were performed using R Project for Statistical Computing software. Hommel method was used for post-host adjustment for multiple comparisons. The statistical methods used for each analysis are specified in the figure legends.
Extended Data
Extended Data Figure 1. TLR7 and TLR9 interaction with beclin 1 and generation of Tlr9-HA knock-in (KI) mice.
(a, b) Co-immunoprecipitation of Flag-beclin 1 with TLR7-HA (a) or TLR9-HA (b) in transfected HeLa cells. (c) Co-immunoprecipitation of TLR9-HA with Flag-beclin 1 full-length (FL) or deletion mutant proteins in transfected U2OS cells. Flag-Beclin 1 Δaa244–266 is a control deletion mutant. (d) Co-immunoprecipitation of TLR9-HA FL protein or truncation mutant lacking the Toll/Interleukin-1 receptor (TIR) domain with Flag-beclin 1 in transfected U2OS cells. (e) MBP-pull-down of recombinant beclin 1 with an MBP-TLR9 TIR domain fusion protein. (f) Co-immunoprecipitation of indicated TLR9-HA constructs with indicated Flag-beclin 1 constructs in transfected U2OS cells cultured in normal media or subjected to 1 h glucose (-Glu) or amino acid (-AA) starvation. (g) Co-immunoprecipitation of TLR9-HA with Flag-Beclin 1 in transfected U2OS cells cultured in normal media, subjected to glucose starvation (-Glu, 1 h) or treatment for 1 h with mitochondrial damaging agents, oligomycin (2.5 μM) + antimycin A (250 nM) (OA) or carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (5 μM), or direct AMPK activator, PF-739 (5 μM). For a-g, results representative of three independent experiments. (h) Tlr9 mRNA levels in different tissues. Value in lung is considered as 1. (i) Tlr9 mRNA levels in myoblasts and myotubes (i.e. before and after myocyte differentiation, respectively) normalized to β-actin levels. Value in myotubes is considered as 1. In h and i, data are mean ± s.e.m. of triplicate samples. (j) Schematic diagram of the Tlr9 locus and CRISPR-based gene knock-in strategy (see Methods section). (k) Representative genotyping of WT (+/+), heterozygous KI (+/KI) and homozygous KI (KI/KI) mice. (l) Western blots of Tlr9-HA expression in indicated tissues from mice (loading amount for spleen and lung = 20 μg and for heart and muscle = 50 μg). Full-length (FL) (~130 kd) and cleaved (~80 kd) Tlr9 detected in spleen. In muscles and heart, only predominant cleaved form of Tlr9 detected due to lower Tlr9 expression levels. Asterix denotes non-specific band. (m) Western blot indicating that FL and cleaved Tlr9 are detected in muscle lysates after enriching by immunoprecipitation with anti-HA antibody. For k-m, similar results observed in three independent experiments. For uncropped gels, see Supplementary Fig. 1.
Extended Data Fig. 2. Tlr9/beclin 1 interaction and AMPK phosphorylation increases in skeletal muscle but not in spleen during exercise.
(a) Representative western blots of endogenous beclin 1 co-immunoprecipitated with endogenous Tlr9-HA in vastus lateralis (VL) muscles from Tlr9-HA KI mice and WT mice (negative control) at indicated duration of exercise. Asterix, non-specific band also observed in WT mice. Similar results were observed in three independent experiments. (b) Quantitation of p-AMPKα (Thr172)/total AMPKα (representative blots are shown in Fig. 1b) in VL muscles from Tlr9-HA KI mice at indicated duration of exercise. Results are combined data from three independent experiments with similar results in each experiment. (c, d) Representative blots (c) and quantitation (d) of p-AMPKα (Thr172)/total AMPKα in the spleen of Tlr9-HA KI mice at indicated duration of exercise. Results are combined data from two independent experiments with similar results in each experiment. (e, f) Representative western blots (e) and quantitation (f) of beclin 1 co-immunoprecipitated with Tlr9-HA in spleens from Tlr9-HA KI mice at indicated duration of exercise. Results are combined data from two independent experiments with similar results in each experiment. In b, d and f, data points, individual mice (sample size indicated in parentheses). Data are mean ± s.e.m. Values at 0 min are considered as 1. In b, d, and f, unpaired two-tailed t-test with Hommel method. For uncropped gels, see Supplementary Fig. 1.
Extended Data Fig. 3. Levels of genomic DNA associated with Tlr9 in skeletal muscle; levels of plasma mitochondria DNA; and effects of exogenous Tlr9 ligand treatment on skeletal muscle AMPK phosphorylation.
(a) Real time PCR quantitation of genomic DNA bound to Tlr9. Shown are the cycle threshold (Ct) for genomic DNA that co-immunoprecipitates with Tlr9-HA from gastrocnemius muscles of WT or Tlr9−/−HA KI mice at rest or after 20 min exercise. Two sets of genomic DNA primers (see Methods section) were used. (b) Quantitation of mitochondrial DNA (mtDNA) in plasma at serial time points after exercise. The amount of mtDNA was quantified by qPCR using 6 sets of mtDNA primers (see Methods section). For each primer set, the value at the 0 min time point is considered as 1. In a and b, data are mean ± s.e.m., data points, individual mice, n=4. (c, d) Representative blots (c) and quantitation (d) of p-AMPK Thr172/total AMPK in extensor digitorum longus (EDL) muscles incubated ex vivo with 1 μM control ODN or ODN2395 (a TLR9 ligand) for 1 h. Data are mean ± s.e.m. Data points, individual muscles, n=3. Unpaired two-tailed t-test. Western blots from one representative experiment and quantitation data combined from three independent experiments. Similar results observed in each experiment. (e) Western blot of AMPK Thr172 phosphorylation in EDL muscles incubated with or without 1 μM PF-739 (a direct AMPK activator) for 1 h. PF-739 is a positive control for experiments in c and d with respect to AMPK activation in EDL muscles ex vivo. Similar results observed in three independent experiments. For uncropped gels, see Supplementary Fig. 1.
Extended Data Fig. 4. Wild-type and Tlr9−/− mice display similar muscle characteristics and cardiac function.
(a, b) Representative H&E staining (a) and fiber type staining (b) of tibialis anterior (TA) muscles. In b, green, plasma membrane marker (Laminin); pink, type I fibers (MHC I positive); red, type IIa fibers (MHC IIa positive); blue, type IIb fibers (MHC IIb positive); black, type IIx fibers. (c) Relative quantitation of fiber type composition in TA muscles shown in b. Data are mean ± s.e.m. At least 1400 muscle fibers per mouse. (d, e) Representative images (d) and quantitation (e) of staining for cytochrome C oxidase (COX) enzymatic activity (a measure of mitochondrial respiratory capacity) in TA muscles. Data are mean ± s.e.m. At least 480 muscle fibers per mouse. (f, g) Representative images (f) and quantitation (g) of capillary density in TA muscles. Data are mean ± s.e.m. At least 240 muscle fibers per mouse. (h) Cardiac function determined by echocardiographic measurement of ejection fraction. For a, b, d and f, images are representative from one of two independent experiments. Similar results observed in each experiment. Data are mean ± s.e.m. For c, g and h, data points, individual mice. N=5 mice per group for c, e and h, and n=4 mice per group for g. Unpaired two-tailed t-test. Scale bars, 50 μm.
Extended Data Fig. 5. Measurement of exercise-induced muscle AMPK activation, exercise-induce muscle GLUT4 plasma membrane localization, and maximal running distance in wild-type and Tlr9−/− mice.
(a, b) Representative western blots (a) and quantitation (b) of phosphorylation of AMPK substrates (ACC Ser79 and Raptor Ser792) in VL muscles. The samples in (a) and (b) are the same as those used in Fig. 2a and 2b of the main text. (c, d) Representative western blots (c) and quantitation (d) of p-AMPK and p-TBC1D1 in EDL muscles. (e, f) Western blots (e) and quantitation (f) of GLUT4 levels in EDL muscles. Asterix denotes a non-specific band. (g) Representative images of GLUT4 (red) and Laminin (green) immunofluorescent staining (used for quantitation in h and Fig. 2d of main text) in EDL muscles. White denotes colocalization between GLUT4 and Laminin determined by ImageJ software. Scale bars, 20 μm. (h) Percentage of total GLUT4 staining colocalized with Laminin calculated by ImageJ software. At least 100 muscle fibers per mouse. (i) Maximal running distance. Data are combined from three independent experiments. Similar results observed in each experiment. For b, d, f, h, and i, data are mean ± s.e.m., data points, individual mice, sample size indicated in parentheses. In a, b, c d, g and h, western blots and images are one representative experiment and quantitation are combined data from three independent experiments. Similar results were observed in each experiment. In b, d and h, two-tailed t-test to compare different conditions for each genotype and two-way ANOVA to compare magnitude of changes between different conditions in mice of different genotypes. In f and i, unpaired two-tailed t-test. For uncropped gels, see Supplementary Fig. 1.
Extended Data Fig. 6. Similar levels of adenine nucleotides, glycogen, total LKB1 and LKB1 Ser428 phosphorylation and similar response to AMPK allosteric activator in WT and Tlr9−/− muscles.
(a-e) Measurements of ATP (a), ADP (b), AMP (c), ADP/ATP ratio (d) and AMP/ATP ratio (e) in EDL muscles. (f) Glycogen content in TA muscles. (g, h) Representative western blots (g) and quantitation (h) of p-LKB1 Ser428 and total LKB1 in VL muscles. Representative blots are from one experiment and quantitation data are combined from three independent experiments. Similar results were observed in each experiment. (i, j) Representative western blots (i) and quantitation (j) of AMPK activation markers (p-AMPK, p-TBC1D1) in TA muscles of mice 90 min after subcutaneous administration of PF-739 (100mg/5ml/kg body weight). (k) Blood glucose levels 90 min after PF-739 treatment. In i, j and k, western blots are from one representative experiment and quantitation results are combined data from two independent experiments. Similar results were observed in each experiment. Data points, individual mice, sample size indicated in parentheses. Data are mean ± s.e.m. Unpaired two-tailed t-test. For uncropped gels, see Supplementary Fig. 1.
Extended Data Fig. 7. Hematopoietic cells are not responsible for the defect in exercise-induced AMPK activation in Tlr9−/− muscles and Tlr9 is required for ex vivo electrical stimulation-induced AMPK activation.
(a, b) Percentage of WT Tlr9 genome (a) or Tlr9−/− genome (b) in blood cells from mice with indicated donor or recipient genotypes at 8 weeks after bone marrow transplantation as determined by real-time q-PCR. Copy number of Tlr7 genome is internal control. Data are mean ± s.e.m. for five randomly selected mice per group. Data points, individual mice. The mean of WT/WT (a) or Tlr9−/−/Tlr9−/− (b) donor/recipient combinations was considered as 100%. (c, d) Representative western blots (c) and quantitation (d) of TBC1D1 Ser237 phosphorylation in VL muscles of indicated recipient mice transplanted with indicated bone marrow cells at rest and after 90 min exercise. (e, f) Representative western blots (e) and quantitation (f) of TBC1D1 Ser237 phosphorylation in EDL muscles of mice +/− 20 min electrical stimulation. In c-f, western blots are from one representative experiment and quantitation results are combined data from three independent experiments. Similar results were observed for each experiment. Data points, individual mice (d) or muscles (f), sample size indicated in parentheses. Data are mean ± s.e.m. In d, unpaired two-tailed t-test to compare differences between donor genotypes; two-way ANOVA for differences between recipient genotypes. In f, unpaired two-tailed t-test to compare +/−electrical stimulation conditions for each genotype. Two-way ANOVA to compare the magnitude of changes between +/− electrical stimulation conditions in muscles of different genotypes. For uncropped gels, see Supplementary Fig. 1.
Extended Data Fig. 8. Tlr9−/− mice have normal exercise-induced autophagy and BCL2 binding to beclin 1 inhibits exercise- and glucose starvation-induced beclin 1/Tlr9 interaction.
(a, b) Representative images (a) and quantitation (b) of GFP-LC3 puncta in VL muscles of WT;GFP-LC3 and Tlr9−/−;GFP-LC3 mice at rest or after exercise +/− chloroquine pretreatment. Scale bars, 20 μm. Arrows, representative puncta counted in b. More than 15 randomly chosen fields were used per mouse and an average value was determined for each mouse. Images are from one representative experiment and quantitative data are combined results of two independent experiments. Similar results observed in each experiment. Data points, individual mice (sample size in parentheses). (c, d) Representative western blots (c) and quantitation (d) of beclin 1 co-immunoprecipitated with Tlr9-HA at serial time points after exercise in VL muscles of Tlr9-HA KI mice crossed to either BCL2 AAA mice or WT BCL2 littermates. Western blots are from one representative experiment and quantitation data are combined results from three independent experiments. Similar results observed in each experiment. Data points, individual mice (sample size in parentheses). (e) Co-immunoprecipitation of transiently expressed Flag-beclin 1 with Myc-BCL2 in HeLa/BCL2 or HeLa/BCL2AAA cells19 grown in normal media or subjected to 2 h glucose starvation. Similar results were observed in three independent experiments. (f, g) Representative western blots (f) and quantitation (g) of Flag-beclin 1 co-immunoprecipitated with Tlr9-HA in HeLa/BCL2 or HeLa/BCL2AAA cells grown in normal media or subjected to 2 h glucose starvation. Data are mean ± s.e.m. from three experiments; sample size indicated in parentheses. In b, unpaired two-tailed t-test to compare different conditions for each genotype. Two-way ANOVA to compare magnitude of changes between different conditions in mice of different genotypes. In d, one-way ANOVA to compare 0 min and 20 min exercise conditions for each genotype. Two-way ANOVA to compare the magnitude of changes between 0 min and 20 min exercise conditions in mice of different genotypes. In g, unpaired two-tailed t-test. For uncropped gels, see Supplementary Fig. 1.
Extended Data Fig. 9. Beclin 1 and UVRAG interaction increases during exercise and electrical stimulation-induced muscle contraction.
(a, b) Representative blots (a) and quantitation (b) of UVRAG co-immunoprecipitated with beclin 1 in TA muscles of mice at indicated duration of exercise. Western blots are from one representative experiment and quantitation data are combined results from five independent experiments. Similar results observed in each experiment (c, d) Representative blots (c) and quantitation (d) of beclin 1 co-immunoprecipitated with UVRAG in EDL muscles of mice +/− 20 min electrical stimulation. Western blots are from one representative experiment and quantitation data are combined results from two independent experiments. Similar results observed in each experiment. In b and d, data points, individual mice (b) or muscle (d), sample size indicated in parentheses. Data are mean ± s.e.m. In b, unpaired two-tailed t-test. In d, two-tailed t-test to compare +/− electrical stimulation conditions for each genotype. Two-way ANOVA to compare the magnitude of changes +/− electrical stimulation conditions in muscles of different genotypes. Asterix, non-specific band observed in IgG control condition. For uncropped gels, see Supplementary Fig. 1.
Extended Data Fig. 10. Beclin 1, UVRAG, or ATG14 knockout does not affect LKB1 protein levels or glucose starvation-induced LKB1 Ser428 phosphorylation.
Effects of beclin 1 (a), UVRAG (b), or ATG14 (c) knockout in U2OS cells on LKB1 protein levels and LKB1 Ser428 phosphorylation 1 h after glucose starvation. Left panels, representative western blots of p-LKB1, total LKB1 and total AMPKβ in cells with indicated gene knockout (two independent gRNAs per target gene). Right panels, quantitation of total LKB1/total AMPKβ (from 4 independent experiments) and p-LKB1/total LKB1 (from 3 independent experiments). Similar results observed in each experiment. Data are mean ± s.e.m. Sample size indicated in parentheses. Unpaired two-tailed t-test. ng, no gRNA. For uncropped gels, see Supplementary Fig. 1.
Supplementary Material
Acknowledgments.
The authors thank R. Bassel-Duby, C. He, R. Medzhitov, and P. Scherer for helpful discussions; C. Chang, J. Doench, G. Barton, Dr. Jørgen Jensen, J. Hurley, N. Mizushima, M. Ranaghan and F. Yarovinsky for providing critical reagents; L. Nguyen for technical assistance; and L. Bennett and H. Smith for assistance with manuscript preparation. This work was supported by NIH grants RO1 CA109618 (B.L.), U19 AI109725 (B.L), and U19 AI142784 (B.L.); Cancer Prevention Research Institute of Texas grant RP120718 (B.L.); and a Fondation Leducq grant 15CBD04 (B.L). M.J. was supported by a “chargé de recherché” post-doctoral grant from the Belgian F.N.R.S.
Footnotes
Competing interests. B.L. is a Scientific Founder of Casma Therapeutics, Inc.
Data availability.
Full scans for all western blots are provided in Supplementary Figure 1. Source data for all graphs in this manuscript have been provided. All other data are available from the corresponding author on reasonable request.
References
- 1.Kjobsted R et al. AMPK in skeletal muscle function and metabolism. FASEB J 32, 1741–1777 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine B & Kroemer G Biological functions of autophagy genes: A disease perspective. Cell 176, 11–42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He C et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 481, 511–515 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandey S, Kawai T & Akira S Microbial sensing by Toll-like receptors and intracellular nucleic acid sensors. Cold Spring Harb Perspect Biol 7, a016246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei Y, Pattingre S, Sinha S, Bassik M & Levine B JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell 30, 678–688 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West AP & Shadel GS Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat Rev Immunol 17, 363–375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shintani Y et al. TLR9 mediates cellular protection by modulating energy metabolism in cardiomyocytes and neurons. Proc Natl Acad Sci U S A 110, 5109–5114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou DC et al. CpG oligodeoxynucleotide preconditioning improves cardiac function after myocardial infarction via modulation of energy metabolism and angiogenesis. J Cell Physiol 233, 4245–4257 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Omiya S et al. Toll-like receptor 9 prevents cardiac rupture after myocardial infarction in mice independently of inflammation. Am J Physiol Heart Circ Physiol 311, H1485–H1497 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rostislavleva K et al. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science 350, aac7365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan Y, Drake JC & Yan Z Exercise-induced mitophagy in skeletal muscle and heart. Exerc Sport Sci Rev 47, 151–156 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Leo MG et al. Autophagosome-lysosome fusion triggers a lysosomal response mediated by TLR9 and controlled by OCRL. Nat Cell Biol 18, 839–850 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ewald SE et al. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature 456, 658–662 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majer O et al. Release from UNC93B1 reinforces the compartmentalized activation of select TLRs. Nature 575, 371–374 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemmi H et al. A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Zisman A et al. Targeted disruption of the glucose transporter 4 selectively in muscle causes insulin resistance and glucose intolerance. Nat Med 6, 924–928 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Xie Z, Dong Y, Scholz R, Neumann D & Zou MH Phosphorylation of LKB1 at serine 428 by protein kinase C-zeta is required for metformin-enhanced activation of the AMP-activated protein kinase in endothelial cells. Circulation 117, 952–962 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cokorinos EC et al. Activation of skeletal muscle AMPK promotes glucose disposal and glucose lowering in non-human primates and mice. Cell Metab 25, 1147–1159 e1110 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Wei Y et al. The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. Elife 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurley JH & Young LN Mechanisms of autophagy initiation. Annu Rev Biochem 86, 225–244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henault J et al. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity 37, 986–997 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizushima N, Yamamoto A, Matsui M, Yoshimori T & Ohsumi Y In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15, 1101–1111 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shoji-Kawata S et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 494, 201–206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee BL et al. UNC93B1 mediates differential trafficking of endosomal TLRs. Elife 2, e00291 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ranaghan MJ et al. The autophagy-related Beclin-1 protein requires the Coiled-coil and BARA domains to form a homodimer with submicromolar affinity. Biochemistry 56, 6639–6651 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agathocleous M et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 549, 476–481 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aslesen R & Jensen J Effects of epinephrine on glucose metabolism in contracting rat skeletal muscles. Am J Physiol 275, E448–456 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Jensen TE et al. PT-1 selectively activates AMPK-gamma1 complexes in mouse skeletal muscle, but activates all three gamma subunit complexes in cultured human cells by inhibiting the respiratory chain. Biochem J 467, 461–472 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell 141, 280–289 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White MJ et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell 159, 1549–1562 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Millay DP et al. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 499, 301–305 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plaideau C et al. Effects of pharmacological AMP deaminase inhibition and Ampd1 deletion on nucleotide levels and AMPK activation in contracting skeletal muscle. Chem Biol 21, 1497–1510 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.