Summary
Rheumatoid arthritis (RA) risk has a large genetic component (~60%) that is still not fully understood. This has hampered the design of effective treatments that could promise lifelong remission. RA is a polygenic disease with 106 known genome-wide significant associated loci and thousands of small effect causal variants. Our current understanding of RA risk has suggested cell-type-specific contexts for causal variants, implicating CD4+ effector memory T cells, as well as monocytes, B cells and stromal fibroblasts. While these cellular states and categories are still mechanistically broad, future studies may identify causal cell subpopulations. These efforts are propelled by advances in single cell profiling. Identification of causal cell subpopulations may accelerate therapeutic intervention to achieve lifelong remission.
Keywords: rheumatoid, arthritis, statistical, genetics, polygenic
Introduction
Rheumatoid arthritis (RA) is an autoimmune disease characterized by chronic inflammation of synovial joint tissue. Left untreated, RA leads to destruction of bone and joint tissue which causes varying degrees of immobility. This destruction is caused by an immune response in which chronic persistent inflammation, perhaps triggered by an autoantigen, leads to irreversible joint destruction1. RA is the one of the most common autoimmune diseases, with a global prevalence of up to 0.8%2. Individuals with RA may or may not be reactive for antibodies against citrullinated peptides (ACPA+). For the most part, ACPA status is correlated with rheumatoid factor (RF) status3.
There is currently no cure for RA, and treatments depend on the severity of an individual’s disease and specific treatment response, often requiring life-long administration. For moderate to severe RA, conventional synthetic disease modifying anti-rheumatic drugs (csDMARDs) such as methotrexate are prescribed to suppress the immune system’s inflammatory activity4. When severe RA does not respond to csDMARDs, typically therapeutic strategies target specific components of the immune system, employing engineered proteins referred to as biological DMARDs, which target TNF cytokines (adalimumab), IL1 cytokines (anakinra), IL6 cytokines (tofacitinib), or CD20+ B cells (rituximab). In most instances, treatment is a lifelong commitment. One goal of future treatment of RA is lifelong remission, in which disease-driving cells are specifically targeted and abrogated. In order to achieve this, the field must pin down the causal cell types and their activated, pathogenic states that drive RA. Fortunately, progress in the area of functional genomics and statistical genetics has led to the nomination of several RA-driving cell types and states, the top candidates of which are subpopulations of CD4+ T effector memory cells1,2,5-7. Disease-driving cell states may be implicated when their regulatory elements harbor a disproportionately large amount of RA genetic variation. However, our current understanding of disease-driving cell types and states is limited by our resolution to identify these pathogenic states. With greater resolution of different cell-states, more specific cell-state-targeting therapeutics may one day be developed.
In this review, we focus on the use of statistical genetics and functional genomics in defining the key mechanisms that underpin RA. These studies aimed to identify key cell type players and cellular mechanisms driving the progression of RA. Such strategies constitute three major categories: 1) genetic fine-mapping studies highlighting the action of single variants, which, regardless of pleiotropy, typically act in specific cell-state contexts, potentially revealing candidate drug targets, 2) polygenic studies which may reveal genome-wide mechanisms central to pathogenic cell state regulation, and 3) functional experiments which identify overly abundant cellular populations in individuals with RA defined by particular transcriptomic or proteomic signatures.
Much of our initial understanding of the genetic susceptibility to RA comes from HLA serologic typing studies, linkage studies, and candidate gene studies which implicated the major histocompatibility complex (MHC) locus and a handful of other loci, including PTPN22 and CTLA4. Much of our more recent understanding of the polygenic genetic architecture of RA, and other non-Mendelian diseases or traits, comes from genome-wide association studies (GWAS) and recent advances in statistical genetics. GWAS queries the entire genome agnostically to identify regions harboring common variants that are strongly associated with a disease or trait. Strongly associated loci typically consist of not one but many SNPs in linkage disequilibrium (LD), meaning nearby alleles tend to be inherited together in segments unaffected by recombination. Because a truly causal SNP and a nearby SNP in high LD cannot be easily distinguished, statistical approaches have been developed to prune these loci via conditional analyses, Bayesian inference, or experimental functional validation to reach one or a few putatively causal variants. The genetic risk of RA and other polygenic traits is weakly accounted for by genome-wide significant (p<5×10−8) associations, which led to the controversial issue of “missing heritability”8. Genetic signals that are difficult to detect, such as smaller common variant effects (sample size limits power), larger effect rare variants (population allele frequency limits power), and complex gene-by-environment interactions (accurate measurement and confounding variables limit power) likely account for the remainder of RA genetic susceptibility. This leads us to a second class of statistical methods, which estimate total disease heritability using genome-wide variants and identify possible functional categories of genomic activity, such as cell-type-specific chromatin remodeling or transcription, that are enriched for variants with a true causal effect, although modest9,10. Because this class of methods takes advantage of the continuum of genome-wide associations, without restricting the analysis to confirmed statistically significant loci, there is power to detect putatively causal regulatory mechanisms or pathways, affected by many SNPs of varying effect size. In this review, we will revisit foundational studies of RA that led to current statistical and functional methods that continue to track down increasingly more resolute causal cell states.
Early RA genetic studies
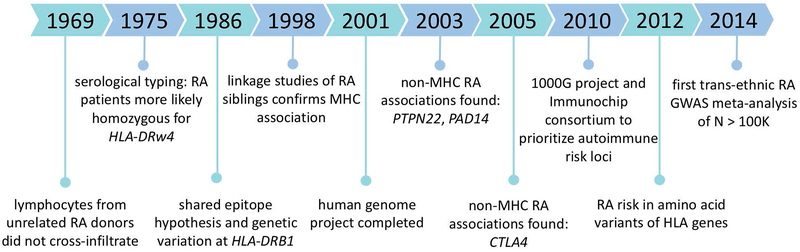
Before considering the current era of RA genetics, in which we may ask questions that might be answered using statistical genetics, such as what genetic associations are shared across a population and which cell types are most enriched for this genetic variation, we must understand the decades of work done by experimental biologists and geneticists that primed us to ask such questions (Figure 1).
Figure 1.
Chronology of milestones in the field of rheumatoid arthritis human genetics leading up to the GWAS boom in 2010.
While rheumatoid arthritis was long postulated to be a genetic disease, in 1950 the Empire Rheumatism Council was the first to support such claims11. The council observed that first-degree relatives of RA-affected individuals are twice as likely to be affected than unrelated controls. Subsequent examinations of patient registries demonstrated that risk for individuals with affected first degree relatives is increased >3 fold12,13. Twin studies were adopted as a classical approach to compare phenotypes between pairs of monozygotic (identical) and dizygotic (fraternal) twins12,14,15. Twin studies provide a unique circumstance where genetic and environmental influences can be studied and quantitatively estimated. Early twin studies estimated that heritability of RA, or proportion of phenotypic variation explained by genetics, was between 40 and 80%16. Heritability and familial aggregation was similar between seropositive and seronegative RA (see below). Reproducible evidence of increased twin concordance and familial clustering spurned geneticists to pursue genetic studies of RA risk and to define the disease mechanisms through which those genetic risk factors act.
As epidemiologists began to understand that RA risk has a heritable component, early experimental work began to identify the genes contributing to RA risk, before twin studies began estimating RA heritability. In 1969, Gonzalo Astorga and Ralph Williams observed that lymphocyte infiltration in peripheral blood was attenuated in mixed samples of lymphocytes from unrelated RA patients, relative to healthy controls17. This was surprising, as in-tissue lymphocyte infiltration was a hallmark of rheumatoid arthritis. Seven years later in 1975, Stastny and colleagues published findings indicating that this in-vitro lymphocyte infiltration is regulated by the HLA (human leukocyte antigen)-D locus and that individuals with RA were more likely to be homozygous for the allele termed HLA-DRw4, subsequently renamed HLA-DR4, than healthy controls18. This genetic commonality between RA patients accounted for the significantly lower reactivity of the lymphocytes. This discovery of a shared genetic factor among individuals with RA, through lymphocytic toxicity typing experiments, opened a new path to study the disease via implicating the HLA genes in the MHC, amidst a research climate in which other genetic findings had been contested.
In 1986, Jack Silver and Sanna Goyert proposed the “shared epitope” hypothesis, in which there are no disease-associated genes, only associated epitopes, or regions of antigens that contact antibodies19. The next year, Peter Gregersen and Robert Winchester along with Silver, formalized this hypothesis20. They observed that while HLA-DR4 was strongly associated with RA individuals, there existed groups of RA patients, specifically of non-European ancestry, who did not share the same HLA-DR4 haplotype, making clear that RA was not regulated simply by one gene. Compared to Mendelian traits, regulated by a single gene and following traditional inheritance patterns, RA was proving to be much more complicated. The HLA-DR4 haplotype harbored 4 tightly linked genes. They observed that a single gene, HLA-DRB1, which had consistent sequence features within different subtypes of this haplotype. This nominated HLA-DRB1 as a candidate RA-driving gene. The shared epitope hypothesis was based on the observation that identical hypervariable sequences at different alleles of HLA-DRB1 encoded epitopes with identical amino acid sequences. Therefore, these sequences had the ability to functionally impact the presentation of antigens due to their prime positioning along the peptide-binding groove. Furthermore, these findings deemed disease associations to be only partially explained by serological typing. As it would turn out in 2005, the shared epitope hypothesis was only relevant to a subset of RA patients, those that were ACPA+21. Here, we note that RA genetic studies that stratified patients by antibody status may use ACPA positivity (seropositive), clinically determined by a blood test for cyclic citrullinated peptide (CCP) antibodies, while other studies may have used rheumatoid factor (RF) to stratify individuals. About 66% of patients are positive for anti-CCP, and it is 90% or more specific for RA22. Since ACPA is rare among healthy individuals, ACPA presence often simplifies RA diagnoses, owing to reported high specificity22. On the other hand, RA patients without these antibodies (ACPA-) showed no association with dosage of the shared epitope allele, leaving RA pathogenesis, in the absence of ACPA, and its connection to the MHC a mystery for almost ten more years. Mechanistically, one possibility is that antigen citrullination leads to an increase in binding affinity between it and the MHC, which in turn elicits an immune response by activating CD4+ T cells. Therefore, HLA-DRB1’s strong RA association led to the nomination of CD4+ T cells as a candidate RA-disease driving cell type. As for the shared epitope hypothesis, decades of research have not yet definitively identified the single putative antigen imagined to drive RA pathogenesis, though many members of the family of citrullinated peptides remain a promising antigen.
As researchers advanced their knowledge of the MHC region, containing what we know today to be a hypervariable region of nine HLA genes all in tight linkage disequilibrium, a complicated nomenclature was adopted23. An allele is named by modifying a gene name by adding two digits, indicating the class of alleles approximately corresponding to its antiquated serological type, such as HLA-DRB1*04, referencing DRw4. An additional two digits correspond to the amino acid sequence of the protein encoded by the allele, such as HLA-DRB1*0401. Current HLA nomenclature with additional digits classify alleles even further, for example, specifying synonymous changes in the HLA sequence.
In the late 1990s and early 2000s, linkage studies of siblings with RA and their families recapitulated the MHC association observed in the past24. In linkage studies, genetic markers were placed at intervals throughout the genome and researchers could track their inheritance. Co-segregation of these chromosomal intervals with the observed familial pattern of RA had the potential to reveal a putatively causal gene locus. While linkage analyses were incredibly useful for Mendelian diseases, such as cystic fibrosis, they revealed no new knowledge beyond the MHC for RA risk. In general, linkage analyses fell short especially for polygenic diseases with few alleles of large effect size, such as both height and RA.
In the ensuing years, research advanced beyond the MHC, due to increasing evidence that the MHC did not fully account for the genetic variation observed in RA patients. In 2003, Botstein and colleagues argued that in order to study the “elusive genes” underpinning polygenic traits, the field must prioritize genome-wide SNP association studies over linkage studies. Linkage studies, although genome-wide, often identified large linkage peaks, implicating many genes which were difficult to prioritize. Moreover, limitations of gene annotations, whose approximate locations were denoted by genetic markers, resulted in bias toward annotated genes25. Genome-wide SNP association studies, on the other hand, provided a framework to test for DNA-level variation in an unbiased fashion, permitting the implication of both genes and their regulatory regions in disease biology. Laying the groundwork for future studies, Botstein and colleagues identified putatively functional SNPs, specifically those lying in coding regions leading to nonsynonymous amino acid substitutions. A year later, Begovich and colleagues followed up on a set of 87 candidate functional SNPs in chromosomal regions identified in RA linkage studies and other candidate genes to perform an association study of these SNPs in nearly 1,000 cases and controls26. This study revealed a significant RA association with the R620W missense SNP in the PTPN22 gene, which was confirmed in a replication study. Other similar studies identified strong non-MHC RA associations within the genes PADI4 and CTLA4, respectively27,28. While the early days of candidate gene studies propelled our understanding of complex traits, such as RA, toward our increasingly thorough understanding of many Mendelian diseases, inconsistencies and lack of reproducibility of these association studies would plague the interpretation of such findings. For example Plenge et al demonstrated that most candidate gene associations were not reproducible28. Confidently identifying statistically significant associations with non-MHC loci with more modest effects required large sample sizes, estimated to be at least thousands by Plenge and colleagues; most candidate gene studies recruited less than 1,000 individuals. Due to this power issue, many findings in the literature were ultimately not easily reproducible.
Large scale RA genetic studies
The late 1990s was a period of bleak prospects for complex traits, such as RA, due to irreproducible linkage analyses and poorly powered candidate gene studies. With the long-awaited Human Genome Project’s completion on the horizon, in 1996, Risch and Merikangas proposed genome-wide comprehensive association analyses29. They proposed that the human genome sequence of many individuals might be used to identify polymorphisms, which could then be tested for association with a disease or trait, using a cohort of cases and controls. The Human Genome Project was completed and progress published in 200130; it revealed for the first time 94% of the base pairs in the human genome and 1.4 million SNPs. With this information, researchers were equipped with new data to develop strategies to investigate genome-wide associations with complex traits.
The International HapMap Project laid the foundation for the approach that many researchers would take to perform genome-wide association studies (GWAS). First, they described an exhaustive approach in which each variant genome-wide would be tested for association with the disease of interest; such a study would require whole genome sequencing on thousands of individuals and be prohibitively expensive. Second, they described the approach suggested by Collins and colleagues in 1997, by which a “very dense map of SNPs” covering diverse regions of the genome, e.g. coding and noncoding, could be used in association testing to identify at least the neighborhood of putatively causal variants31. This neighborhood would be defined by patterns of linkage disequilibrium (LD), indicating that either the associated SNP (index SNP) or any other SNP in strong LD with the index SNP could be causal. Therefore, theoretically only one SNP per LD block, or haplotype, would need to be tested. HapMap followed up on the plan to design such a dense map of SNPs, by selecting haplotypes and SNPs for genotyping across a diverse group of populations. Biotech companies quickly began developing various GWAS chips, some designed for full genome coverage and others designed for denser coverage of known risk loci in particular diseases. One landmark GWAS included the Wellcome Trust Case Control Consortium’s (WTCCC) use of a commercial genotyping chip32. The Affymetrix GeneChip 500K Mapping Array Set covered 500,568 SNPs genome-wide and was well powered to detect SNPs of both larger (80% for odds ratio (OR) of 1.5) and more modest (43% for OR of 1.3) effect size.
This first generation of GWAS revealed many novel genetic underpinnings in RA; here we revisit a few foundational associations. In the landmark WTCCC manuscript, variants in HLA-DRB1 and PTPN22 were confirmed to be associated. In this work, three novel RA associations were identified: 10p15 (IL2RA), 18p11 (PTPN2), 12q24. In 2007, Plenge and colleagues identified a reproducible association at 6q23 (near TNFAIP3 and OLIG3)33. TNFAIP3 is a known inhibitor of NFKB signaling and knockout mice are affected with chronic inflammation. Again in 2007, Plenge and colleagues identified the association of TRAF1 (tumor necrosis factor) and C5 (complement component) with RA34. In 2008, we meta-analyzed existing RA GWAS data and identified an association with CD40 (the encoded protein of which is necessary for antigen presenting cells to become activated) and CCL21 (lymphocyte chemotaxis)35. In 2010, Stahl and colleagues conducted an RA GWAS meta-analysis of more than 41K individuals of European descent36. This study identified 7 novel RA risk variants, near IL6ST, SPRED2, RBPJ, CCR6, IRF5 and PXK genes. In 2014, Okada and colleagues performed the first European and East Asian RA GWAS trans-ethnic meta-analysis with a combined total of cases and controls greater than 100,000 individuals37. This study identified 47 novel risk loci, bringing the total number of associated variants in 2014 to 101, including some population-specific risk loci. This comprehensive study mapped risk variants to likely causal genes. Some of these results seemed to validate current RA drug targets with genetic support. Specifically, several proteins encoded by RA risk genes are targets of currently approved RA drugs, targets shown in parentheses: Etanercept (TNF), Infliximab (TNF), Adalimumab (TNF), Golimumab (TNF-α), Certolizumab pegol (TNF-α), Tocilizumab (IL-6), Abatacept (IL-6), Rituximab (CD20), Sulfasalazine (NFKB), Tacrolimus (Calceineurin), Tofacitinib (JAK/STAT), Iguratimod (NFKB, COX-2), and Prednisolone (glucocorticoid).
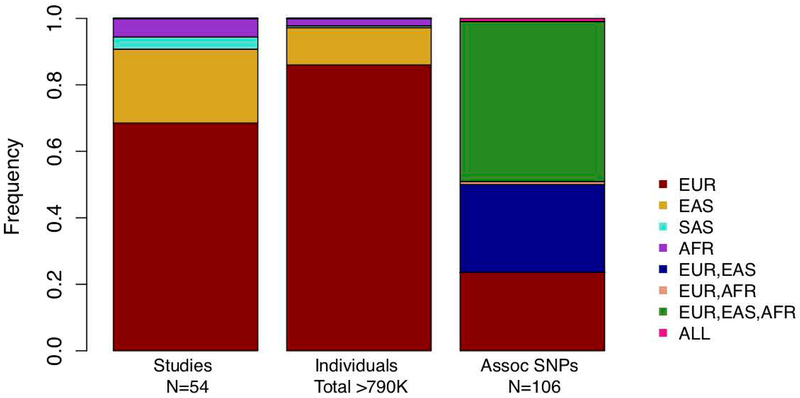
To date, 54 RA GWAS have been performed and published (extracted from the GWAS Catalog in July, 201938) with the most well powered studies accounting for tens of thousands of cases and controls (Figure 2).
Figure 2.
Number of published rheumatoid arthritis GWAS studies and genome-wide significant SNPs reveals bias toward European populations, although there is a greater proportion of SNPs shared between at least two populations than specific to Europeans. EUR (European), EAS (East Asian), SAS (South Asian), AFR (African).
Studies of notably large sample size have identified 106 significant independent associations37,39-42. According to the GWAS Catalog, 37 of RA GWAS have been performed in European populations, 12 in East Asian populations, including Japanese, Korean, and Han Chinese, 3 in populations with African descent including African American, South African, and an African-Arab admixture, and 2 in South Asians. While the prevalence of RA varies among different ancestral populations, the majority of genome-wide significant associations appear to be shared between European, East Asian, and African populations. However, it is difficult to determine population specificity among causal variants due to differences in allele frequency, which can vary across populations. These differences can affect our ability to identify genome-wide associations. Therefore, the different roles of so-called population-specific variants in genetic and environmental RA risk is currently poorly understood. These associations include loci harboring coding variants and noncoding regulatory variants. Included in the strongly associated coding variants, not specific to ACPA+ RA, are HLA-DRB1, HLA-DQA1, PTPN22, IL6R, TYK2, and DNASEIL343. Some genome-wide significant non-coding variants were not specific to ACPA+ RA patients. These included alleles found to lie either in intronic regions or in proximal 5’ or 3’ intergenic regions of immune-mediating genes, for example, IRF8, IRF5, RUNX1, TNFAIP3, STAT4, CCR6, and CTLA4. We include a list of reported genome-wide significant RA loci, represented by lead SNPs, in Table 1 from the following studies37,39-42.
Table 1.
List of 106 rheumatoid arthritis genome-wide significant SNPs. Population indicates in the ancestral diversity of associations according to the GWAS Catalog.
| SNP | CHR | POS | GENES | Population |
|---|---|---|---|---|
| chr1:2523811 | 1 | 2523811 | TNFRSF14-MMEL1 | EUR |
| rs227163 | 1 | 7961206 | TNFRSF9 | EUR,EAS |
| rs761426 | 1 | 17087404 | PADI2 | EUR,EAS,AFR |
| rs2301888 | 1 | 17672730 | PADI4 | EUR,EAS,AFR |
| rs28411352 | 1 | 38278579 | MTF1-INPP5B | EUR,EAS,AFR |
| rs12140275 | 1 | 38633879 | LOC339442 | EUR,EAS,AFR |
| rs2476601 | 1 | 114377568 | PTPN22 | EUR,EAS |
| rs624988 | 1 | 117263790 | CD2 | EUR,EAS,AFR |
| rs2228145 | 1 | 154426970 | IL6R | EUR,EAS,AFR |
| rs2317230 | 1 | 157674997 | FCRL3 | EUR |
| rs4656942 | 1 | 160831048 | LY9-CD244 | EUR |
| rs72717009 | 1 | 161405053 | FCGR2A | EUR |
| chr1:161644258 | 1 | 161644258 | FCGR2B | EUR,EAS,AFR |
| rs2105325 | 1 | 173349725 | LOC100506023 | EUR |
| rs17668708 | 1 | 198640488 | PTPRC | EUR,EAS,AFR |
| rs10175798 | 2 | 30449594 | LBH | EUR,EAS,AFR |
| rs34695944 | 2 | 61124850 | REL | EUR |
| rs13385025 | 2 | 62461120 | B3GNT2 | EUR,EAS,AFR |
| rs1858037 | 2 | 65598300 | SPRED2 | EUR,EAS,AFR |
| rs9653442 | 2 | 100825367 | AFF3 | EUR,EAS |
| rs6732565 | 2 | 111607832 | ACOXL | EUR,EAS,AFR |
| rs11889341 | 2 | 191943742 | STAT4 | EUR,EAS |
| rs6715284 | 2 | 202154397 | CFLAR-CASP8 | EUR,EAS,AFR |
| rs1980422 | 2 | 204610396 | CD28 | EUR,EAS,AFR |
| rs3087243 | 2 | 204738919 | CTLA4 | EUR,EAS,AFR |
| rs4452313 | 3 | 17047032 | PLCL2 | EUR,EAS |
| rs3806624 | 3 | 27764623 | EOMES | EUR,EAS |
| rs73081554 | 3 | 58302935 | DNASE1L3-ABHD6-PXK | EUR,EAS |
| rs9826828 | 3 | 136402060 | IL20RB | EUR,EAS,AFR |
| rs13142500 | 4 | 10727357 | CLNK | EUR,EAS,AFR |
| rs11933540 | 4 | 26120001 | C4orf52 | EUR,EAS |
| rs2664035 | 4 | 48220839 | TEC | EUR |
| rs10028001 | 4 | 79502972 | ANXA3 | EUR |
| rs45475795 | 4 | 123399491 | IL2-IL21 | EUR,EAS,AFR |
| rs7731626 | 5 | 55444683 | ANKRD55 | EUR,EAS,AFR |
| rs2561477 | 5 | 102608924 | C5orf30 | EUR,EAS |
| rs657075 | 5 | 131430118 | IL3-CSF2 | EUR,EAS,AFR |
| rs9378815 | 6 | 426155 | IRF4 | EUR |
| chr6:14103212 | 6 | 14103212 | CD83 | ALL |
| rs9268839 | 6 | 32428772 | HLA-DRB1 | EUR,AFR |
| rs2234067 | 6 | 36355654 | ETV7 | EUR,EAS,AFR |
| rs2233424 | 6 | 44233921 | NFKBIE | EUR,EAS |
| rs9372120 | 6 | 106667535 | ATG5 | EUR,EAS,AFR |
| rs17264332 | 6 | 138005515 | TNFAIP3 | EUR,EAS,AFR |
| rs7752903 | 6 | 138227364 | TNFAIP3 | EUR,EAS |
| rs9373594 | 6 | 149834574 | PPIL4 | EUR,EAS,AFR |
| rs2451258 | 6 | 159506600 | TAGAP | EUR,EAS,AFR |
| rs1571878 | 6 | 167540842 | CCR6 | EUR,EAS |
| rs67250450 | 7 | 28174986 | JAZF1 | EUR,EAS |
| rs73366469 | 7 | 74619286 | GTF2I | EUR |
| rs4272 | 7 | 92236829 | CDK6 | EUR,EAS,AFR |
| rs34130487 | 7 | 100161582 | MIR95-MIR106B | EUR,EAS |
| chr7:128580042 | 7 | 128580042 | IRF5 | EUR,EAS |
| rs2736337 | 8 | 11341880 | BLK | EUR,EAS |
| rs998731 | 8 | 81095395 | TPD52 | EUR,EAS,AFR |
| rs678347 | 8 | 102463602 | GRHL2 | EUR,EAS,AFR |
| rs1516971 | 8 | 129542100 | PVT1 | EUR,EAS,AFR |
| rs11574914 | 9 | 34710338 | CCL19-CCL21 | EUR,EAS,AFR |
| rs10985070 | 9 | 123636121 | TRAF1-C5 | EUR,EAS,AFR |
| rs706778 | 10 | 6098949 | IL2RA | EUR |
| rs947474 | 10 | 6390450 | PRKCQ | EUR,EAS |
| rs3824660 | 10 | 8104722 | GATA3 | EUR,EAS |
| rs12413578 | 10 | 9049253 | 10p14 | EUR,EAS,AFR |
| rs793108 | 10 | 31415106 | ZNF438 | EUR,EAS,AFR |
| rs2671692 | 10 | 50097819 | WDFY4 | EUR,EAS |
| rs71508903 | 10 | 63779871 | ARID5B | EUR |
| rs6479800 | 10 | 64036881 | RTKN2 | EUR |
| rs726288 | 10 | 81706973 | SFTPD | EUR,EAS |
| rs331463 | 11 | 36501787 | TRAF6-RAG1/2 | EUR |
| rs508970 | 11 | 60906450 | CD5 | EUR,EAS,AFR |
| rs968567 | 11 | 61595564 | FADS1-FADS2-FADS3 | EUR |
| rs11605042 | 11 | 72411664 | ARAP1 | EUR,EAS,AFR |
| rs4409785 | 11 | 95311422 | CEP57 | EUR,EAS,AFR |
| chr11:107967350 | 11 | 107967350 | ATM | EUR,EAS,AFR |
| rs10790268 | 11 | 118729391 | CXCR5 | EUR |
| rs73013527 | 11 | 128496952 | ETS1 | EUR,EAS |
| rs773125 | 12 | 56394954 | CDK2 | EUR,EAS,AFR |
| rs1633360 | 12 | 58108052 | CDK4 | EUR,EAS |
| rs10774624 | 12 | 111833788 | SH2B3-PTPN11 | EUR,EAS,AFR |
| rs9603616 | 13 | 40368069 | COG6 | EUR |
| rs3783782 | 14 | 61940675 | PRKCH | EUR,EAS,AFR |
| rs1950897 | 14 | 68760141 | RAD51B | EUR,EAS,AFR |
| rs2582532 | 14 | 105392837 | PLD4-AHNAK2 | EUR,EAS |
| rs8032939 | 15 | 38834033 | RASGRP1 | EUR,EAS,AFR |
| rs8026898 | 15 | 69991417 | LOC145837 | EUR,EAS,AFR |
| rs4780401 | 16 | 11839326 | TXNDC11 | EUR,EAS,AFR |
| rs13330176 | 16 | 86019087 | IRF8 | EUR |
| rs72634030 | 17 | 5272580 | C1QBP | EUR,EAS,AFR |
| rs1877030 | 17 | 37740161 | MED1 | EUR,EAS |
| chr17:38031857 | 17 | 38031857 | IKZF3-CSF3 | EUR,EAS,AFR |
| rs8083786 | 18 | 12881361 | PTPN2 | EUR |
| rs2469434 | 18 | 67544046 | CD226 | EUR,EAS,AFR |
| rs34536443 | 19 | 10463118 | TYK2 | EUR,EAS |
| chr19:10771941 | 19 | 10771941 | ILF3 | EUR |
| rs4239702 | 20 | 44749251 | CD40 | EUR,EAS,AFR |
| rs73194058 | 21 | 34764288 | IFNGR2 | EUR,EAS,AFR |
| chr21:35928240 | 21 | 35928240 | RCAN1 | EUR |
| rs8133843 | 21 | 36738242 | RUNX1-LOC100506403 | EUR,EAS |
| rs1893592 | 21 | 43855067 | UBASH3A | EUR |
| rs2236668 | 21 | 45650009 | ICOSLG-AIRE | EUR,EAS,AFR |
| rs11089637 | 22 | 21624807 | MIR301B | EUR |
| rs11089637 | 22 | 21979096 | UBE2L3-YDJC | EUR,EAS |
| rs3218251 | 22 | 37545505 | IL2RB | EUR |
| rs909685 | 22 | 39747671 | SYNGR1 | EUR,EAS,AFR |
| chrX:78464616 | X | 78464616 | P2RY10 | EUR |
| rs5987194 | X | 153301467 | IRAK1 | EUR,EAS |
Linking RA-associated variants to changes in gene expression
As about 90% of GWAS associations were observed to reside in noncoding sequences, determining the role of these variants turned out to be extremely important, but also challenging. This is in part due to less incomplete annotation of non-coding regions of the genome. Knowledge of the regions of the genome that are essential to regulating genes, particularly in a cell-type specific manner, continues to be developed. To quantitatively address these concerns, a suite of methods were developed to assess whether GWAS disease-associated variants act to influence gene regulation. Between 2010 and 2012, several eQTL studies enhanced our understanding of the function of putatively causal RA variants.
In 2010, Kochi and colleagues performed a high-powered Japanese RA GWAS with 2,303 affected individuals, the majority (~80%) of which were ACPA+, and 3,380 controls44. The authors identified a genome-wide significant variant in the CCR6 gene and further identified that the genotype of a different variant in tight LD with the first variant showed significant correlation with expression of CCR6 in EBV-infected lymphoblastoid cell lines. Furthermore, they observed that IL-17 levels in RA individuals were concordant with the genotype of the eQTL variant, nominating CD4+ Th17 cells, which secrete IL-17, in the pathogenesis of RA in humans.
In 2012, Fairfax and colleagues sought to resolve the functional role of HLA alleles in the pathogenesis of RA and other autoimmune disease45. The authors identified a monocyte-specific trans eQTL association between HLA alleles and the AOAH gene, a lipase that degrades lipopolysaccharide and plays a role in the anti-inflammatory response.
In 2014, we performed a CD4+ memory T cell eQTL study in healthy individuals, in order to investigate mechanisms of RA risk variants and their effects on gene expression. Of 30 RA risk loci considered, we demonstrated that 5 co-localized with eQTLs in either resting or activated T cells. These alleles implicated the BLK, C5orf30, GSDMB, IRF5, PLEK genes.
For several years, such studies involving eQTL fine mapping remained attractive to decipher genetic risk mechanisms. In 2017, Chun and colleagues devised a comprehensive strategy to test if a GWAS association has the same genetic driver as an eQTL, while carefully modeling the LD between the associated variant and the eQTL variant46. Their key improvement over previous studies was statistical deconvolution of associated colocalization signals into causal signals. Their observations attributed a discouragingly low proportion of genome-wide associations to modulations in gene expression. T cell, B cell, and monocyte eQTL effects were associated with only a quarter of autoimmune disease risk loci and only 12% specifically for RA. These findings might be attributable to insufficient eQTL cohort size, missing cellular profiling of other candidate pathogenic cells, or pathogenic mechanisms beyond the context of gene expression. On the other hand, our understanding of eQTL effects of RA risk variants mediated by non-coding RNA remains unclear. Previous reports suggested some contribution of long non-coding RNA and short non-coding RNA, e.g. microRNA, mediating eQTL effects of RA risk variants40,47. Therefore, further construction of eQTL resources regarding non-coding RNA is warranted.
Identifying causal variants among genome-wide associations with fine mapping
One critical challenge for GWAS is the identification of causal variants among associated variants. Loci harboring causal variants also contain significantly associated variants which are statistical artifacts of the causal association due to strong LD. Here, we will discuss genetic fine mapping studies that have elucidated the biological mechanisms underpinning RA: Eyre et al which preceded a GWAS meta analysis with dense fine mapping to improve imputation accuracy and power43, Farh et al which implements a Bayesian statistical framework48, and Westra et al which uses genetic fine mapping and further interrogates putatively causal SNPs with experimental tests of functionality49.
By 2010, more than one thousand loci had been identified via association studies linking our genetic code to complex traits. Despite cataloging diverse human genomes, the field of genetics was largely constrained to association testing with common variants (minor allele frequency (MAF) > 5%). Identification of the link between phenotype and genetic variation attributable to rare (MAF < ~0.1%) and low frequency variants (~0.1% < MAF < 5%) required many more genomes and systematic study. The 1000 Genomes Project Consortium argued that low frequency variants outnumber common variants and so they focused their efforts on characterization of at least 95% of all variants with MAF > 1%50 across five populations. Around this time, the Immunochip Consortium was developed to provide a genotyping chip for 200,000 selected SNPs that might be better powered to find low frequency and common autoimmune risk variants. Since either sample sizes would need to be increased to detect rare variant associations or genotyping chips would need to prioritize a smaller number of putatively causal SNPs, Immunochip intersected hundreds of thousands of SNPs catalogued in the 1000 Genomes project with disease-associated loci determined from previously published GWAS for RA, ankylosing spondylitis, Crohn’s disease, ulcerative colitis, celiac disease, multiple sclerosis, type 1 diabetes, systemic lupus erythematosus, and others. This reselection of SNPs would allow for deep replication and fine-mapping of associated loci in a relatively inexpensive manner compared to genome-wide genotyping of SNPs. Eyre and colleagues performed a GWAS meta-analysis with nearly 50,000 individuals, in which they used the Immunochip array to prioritize 186 autoimmune disease risk loci, equating to 129,464 SNPs43. In a standard GWAS framework, genotypes are imputed; however, in this case denser genotyping at select regions resulted in better imputation. This work identified 14 novel RA loci, and redefined the lead SNP signals in 19 associated loci and in the process prioritized likely individual candidate genes, identified independent secondary associations at 6 loci, and lastly, at 4 loci identified low frequency variant associations which was previously not possible with conventional GWAS. Intriguingly, the authors calculated that imputation without fine-mapping would have implicated different causal regions one-third of the time.
The Farh et al study developed a novel statistical approach, named PICS (probabilistic identification of causal SNPs) to fine-map RA-associated variants, in addition to 20 other autoimmune diseases. PICS is based on the principle that the more an index SNP is in high LD with other SNPs, the more inflated the index SNP’s chi-squared association statistic is in a GWAS, however this relationship is not perfectly linear due to variance. Therefore, the authors implemented a permutation strategy in which the association signal at the putatively causal variant is fixed, meaning the genotype of individuals at that SNP was kept as is, and the case-control status was permuted, meaning SNPs were made neutral while retaining their linked structure. This permutation strategy was implemented 1,000 times to simulate association statistics for these SNPs. Using this framework, they asked what the probability is of the index association driving the observed association pattern throughout the locus. After performing genetic fine mapping on 39 GWAS datasets, each representing a different phenotype, the authors identified nearly 5,000 candidate causal SNPs among 636 associated loci. They further fine-mapped reported GWAS catalog associations; PICS estimated that only 5% of reported lead variants are causal. By this strategy, putatively causal SNPs were strongly enriched for deleterious protein coding variants (>3x).
To better understand causal variation in RA and type 1 diabetes (T1D), our group aimed to fine map causal variants across 76 loci49. We first imputed variant genotypes with the 1000 Genomes European cohort which greatly increased the number of interrogated variants as well as the power to detect putatively causal variants; this two-step approach combining imputation with fine-mapping had not previously been done. The authors then used approximate Bayesian fine-mapping to assign posterior probabilities to variants in associated loci, assuming one causal variant per locus but using conditional analyses to interrogate loci with evidence for more than one causal variant. For each variant, its posterior probability is calculated as the ratio of its approximate Bayesian factor (ABF) to the sum of the ABFs of all variants in the locus. The ABF is defined as the likelihood ratio of the probability of observing an effect size according to a normal distribution (mean 0, variance equal to the variance of all effect sizes) to the probability of observing the same effect size according to a normal distribution with wider variance, e.g. adjusted for a prior of observing an odds ratio of 1.5. Of the RA-associated putatively causal variants, we rediscovered missense variants in PTPN22 and TYK2 and found a novel missense variant rs35677470 in DNASE1L3 (expressed in stromal fibroblasts)51 and noncoding variants in CD28-CTLA4 (rs55686954, rs117701653) and TNFAIP3 (rs35926684). The noncoding variants rs117701653 and rs35926684, near CD28-CTLA4 and TNFAIP3, respectively, exhibited allele-specific protein binding activity (electrophoretic mobility shift assay (EMSA)) and CD4+ T cell specific differential enhancer activity (Luciferase reporter assay).
Colocalization of causal variants with functional cell-type-specific regulatory elements
In the following years, researchers would seek to define which types of genomic features colocalize with causal variants. In a locus of multiple candidate causal variants, information about SNP function (e.g. nonsynonymous exonic, splice variant, regulatory allele) greatly influences how confidently a causal variant can be determined. Since the function of noncoding variants is poorly understood, using functional information in the non-coding genome to prioritize alleles is challenging. For example, causal variants might be located in enhancer elements of a set of genes belonging to one pathway. In this case, this result might implicate a specific molecular mechanism responsible for conferring risk.
A. Colocalization with genes expressed in specific cell types
In 2011, our group hypothesized that genetic risk factors of RA, and other non-systemic autoimmune diseases, manifest in a small set of tissues or cell types. Following this logic, disease-associated genetic variation might impact gene expression specifically in these tissues or cell types. Strategies that linked causal variation with specifically expressed genes within tissues had already proven useful in cancer studies52. In terms of autoimmune diseases, such as RA, nearly all immune cells had been implicated as candidate disease-driving cell types. An approach was needed to unbiasedly and quantitatively assess the enrichment of cell-type-specific gene expression in GWAS risk loci. At that time, large compendiums of human gene expression data in immune cell populations were not readily available. Rather, there was extensive profiling of these cellular populations in mice; at that time, the Immunological Genome (Immgen) Consortium had profiled 223 immune cell populations via microarray. Due to many similarities in immune cell populations and cytokine expression between mice and humans53, we felt that it was reasonable to use this database of gene expression to make inferences about human RA. We developed a strategy to assess cell-type-specific gene expression enrichment in GWAS risk loci6. First, a tissue-specificity score was computed for each gene. Then, using genome-wide significant RA-associated variants to define independently associated loci genome-wide, the most tissue-specific gene per locus was selected. Lastly, significance of tissue-specificity was assessed with comparison to a permutation distribution, created from randomly selecting sets of SNPs matched for confounding genomic features. Applying the strategy to RA, we observed that CD4+ T effector memory cells revealed the strongest independent signal for enrichment in RA risk loci.
B. Colocalization with epigenetic marks
Building on this newly created foundation where cell-type-specific genes harbor disease variants, a study from our group in 201354 advanced this concept to include other data types implicating gene regulation. We hypothesized that strongly associated disease variants are less likely to fall in loci of diverse cell type regulatory activity. Expression QTL studies to date had favored identification of ubiquitously acting variants, rather than cell-type-specific variants. We proposed identifying enrichment of risk variants in cell-type-specifically assayed chromatin marks, which could also help stratify eQTLs by cell type specificity. To this end, they developed a method to score (1) the cell type specificity of a given variant according to its proximity to a cell-type-specific chromatin mark and (2) the cell type specificity of a chromatin mark. In this study, we observed that the 31 genome-wide significant RA-associated variants, known at this time, co-localized specifically with CD4+ regulatory T H3K4me3 chromatin marks, strongly implicating CD4+ Tregs as a key driver of RA. H3K4me3 generally marks active promoters and enhancers; and, in this study was shown to be a highly cell-type-specific mark.
In 2012, Maurano and colleagues investigated co-localization of GWAS variants with DNase hypersensitivity sites (DHSs)55. Their strategy was to correlate DHSs with gene promoter activity; they associated GWAS variants lying in such DHSs with activity of the corresponding gene. For RA, they identified 5 associations with known GWAS variants. The strongest DHS-gene association implicated rs1600249 in the regulation of the CTSB gene, a cysteine proteinase which plays a role in articular erosion.
We now return to the Farh et al study which devised a genetic fine mapping strategy and then used epigenetic colocalization to interpret their results. Using their approach to fine map variants in a collection of 39 GWASes and 21 autoimmune disorders, the authors discovered putatively causal variants mostly alter genomic sequences in unexpected ways. They classified these alterations via epigenetic fine mapping, to which end they created a map of cis-regulatory elements of various epigenetic marks, such as cell-type-specific histone modifications, enhancers, genes, and sequence motifs. First, they found that 60% of putatively causal variants resided in enhancer elements, the majority of which were CD4+ T cell enhancers identified after exposing cells to a stimulus, i.e. Ionomycin, PMA, CD3, and CD28. Here, sequence alteration resulted in expression of noncoding RNA and histone acetylation, which is associated with transcriptional activation. Second, the authors found that rather than disrupting TF binding motif sites, fine-mapped noncoding variants were more likely to alter other nearby sequences without recognizable motifs. While the authors observed an enrichment for RA putatively causal variants in ChIP-seq binding regions of IRF4, they did not observe direct alteration of the sequence motif. They estimate that only 10-20% of noncoding GWAS variants alter a TF motif and cause changes in binding affinity. Other than the IRF4 association, this study did not reveal any novel mechanisms of genetic variation outside the MHC in RA.
These studies demonstrated that noncoding regulatory annotations, from experimental assays that profile protein occupancy or chromatin accessibility, have the potential to provide insight into the mechanisms of putatively causal variants. During this time, the generation of regulatory annotations was rapidly increasing. While these annotations tend to co-localize with one another, it is not always clear which annotation is the most informative for distinguishing causal from non-causal variants. For example, transcription factor (TF) ChIP-seq peaks tend to co-localize with gene promoters. Therefore, enrichment of disease-associated variants in TF ChIP-seq peaks might be the result of unaccounted co-localization with promoters; we must ask if the TF ChIP-seq enrichment is still significant when we condition away the promoter association. This ability to distinguish causal variation using regulatory annotations is necessary to glean accurate insight from functional fine-mapping studies.
To address this problem, Genomic Annotation Shifter (GoShifter) was developed as a statistical approach to quantify how well enriched functional annotations can prioritize putatively causal variants within an associated locus7. We note that only variants exceeding genome-wide significance are considered in this study. This task requires that two types of genomic confounding be accounted for: 1) LD and 2) association with a functional annotation, which only due to co-localization with a truly causal annotation, appears enriched for causal variants. GoShifter accounts for both sources of confounding by assessing enrichment of SNP-annotation overlap compared to a null distribution derived from locally shifting annotations, thereby preserving LD structure and removing the effects of co-localization. The study employing GoShifter resulted in several compelling insights to RA pathogenesis. First, the authors surprisingly found that H3K4me3 ChIP-seq peaks in CD4+ memory T cells were not enriched for a selected set of 88 RA-associated SNPs, known at that time. At first, this result contradicted the previously discussed study from Hu et al. Rather, the authors observed that restriction of the ChIP-seq peak to the summit region, where there is the most read pile up and strongest evidence for regulatory activity, resulted in a significantly enriched annotation. This advanced our knowledge from previous studies by formally claiming that H3K4me3 marks in CD4+ memory T cells could help distinguish causal from non-causal RA variants in RA-associated loci.
Finding the remaining heritability in highly polygenic RA
So far, methods were developed that considered genome-wide significant variants and attempted to functionally characterize their mechanisms of action. However, for polygenic traits, such as RA, these strategies were missing causal variants with smaller effect size, not detected by GWAS. As a consequence, these methods only captured the tip of genetic risk iceberg. The genetic variance, or heritability, in confirmed significant SNP associations from GWAS tend to explain a discouragingly low proportion of phenotypic variation in polygenic traits and diseases. “Heritability” is a term indicating the proportion of phenotypic variation that is explained by genetic variation. Phenotypic variation may be explained by genetic and environmental components, as well as the interaction between the two, formulated by the famous expression: P = G + E + GxE. Due to complications and confounding, current polygenic trait studies tend to focus entirely on explaining the genetic component, which can be almost fully accounted for by genotyping. Conventional heritability models assume independence between SNPs, e.g. no epistasis, and therefore, an additive effect genome-wide; just how much this model differs from the truth likely depends on disease complexity. In the case of monogenic or Mendelian traits, the heritability is fully accounted for typically by a mutation affecting the disease-driving gene. In polygenic traits, this initial incomplete measure of genetic heritability was coined “missing heritability” and might arise for several reasons, including (1) many common variants with small effect sizes that reasonably sized GWAS loci were not well powered to find, (2) heritability quantified from familial studies may overestimate heritability in an unrelated population, (3) low frequency variants carrying modest effect sizes tend to be missing from genotyping chips, which are biased in favor of variants with at least 5% MAF56, (4) misdiagnoses and poor or inconsistent phenotyping, (5) SNPs genotyped for a GWAS neglect rare variants and other forms of genetic variation like copy number variation. Family studies may have overestimated heritability of polygenic traits if relatedness among the individuals in the study violates the assumptions of the additive genetic model. For example, if there is dominance, epistasis, or interactions between SNPs or genes, the model is violated. Moreover, the variance of environmental factors in family studies is less than in the general population, potentially resulting in underestimated environmental contribution and therefore overestimated genetic contribution. Regarding the second point, we may simply be missing the additive effects from variants that aren’t genotyped in a GWAS. Among the missing additive effects are rare (MAF < 0.1%) and low (0.1% < MAF < 5%) frequency variants, some of which might have disproportionately large effect sizes; without enormous sample sizes we cannot confidently measure them.
In 2009, Purcell along with the International Schizophrenia Consortium performed a GWAS involving nearly 7,000 individuals but did not find a single variant that reached genome-wide significance57. This observation suggested that the genetic basis of schizophrenia is likely composed of thousands of common variants of small effect size, one of the reasons for the “missing heritability” phenomenon. In this study, the authors estimated that at least one third of the liability of schizophrenia is due to common polygenic variation. As a result, this study became a landmark manuscript establishing a framework to study polygenic traits where GWAS did not reveal any significant associations.
Three years later in 2012, Stahl and colleagues implemented a Bayesian approach to unraveling the polygenic component of rheumatoid arthritis58 and three other diseases. In this study, the authors used polygenic risk scores, which include all variants genome-wide irrespective of genome-wide significance, to estimate the common polygenic component of RA. They found that 2.5 million common variants account for 20% of RA liability scale heritability, independent of heritability attributed to genome-wide significant associations (estimated to be 25%1).
As the field of statistical genetics began to revisit GWAS data looking for polygenic signals, a natural question emerged regarding whether certain key annotations explained more heritability than others. While previous work from our group54 had shown that genome-wide significant SNPs organized in cell-type-specific ways, it was not unreasonable to think that the variants accounting for this “missing heritability” also organize in such a manner, acting through large regulatory networks to affect gene expression or a different biological intermediate before manifesting in a phenotype. In the following years, statistical methods would be developed to model genome-wide, polygenic SNP, no longer restricting to genome-wide significant associations, permitting the, at least partial, recovery of “missing heritability” and assigning that heritability to functional categories. The goal of these approaches was similar to strategies where investigators asked which functional annotations colocalized with genome-wide significant causal variation. However, they leveraged the power of whole genome signals, rather than a small number of individually confirmed loci.
In 2011, Yang and colleagues constructed a now widely used toolkit called GCTA (genome-wide complex trait analysis)10. GCTA, in contrast to conventional GWAS which estimates the association between each SNP genome-wide and a phenotype, estimates the association between groups of SNPs and a phenotype. They demonstrated that heritability could be partitioned into different segments of the genome. This tool became a gold standard for liability scale heritability estimation. Then in 2012, Lee and colleagues found that heritability attributable to different chromosomes was proportional to the length of the chromosomes59. They used this same approach to understand the heritability of schizophrenia by partitioning the genome into those segments that were near genes specifically expressed in the brain, versus the rest of the genome. Using GCTA to estimate and partition genetic heritability, they found 2,725 genes specifically expressed in the central nervous system were disproportionately enriched for heritability. The authors found that approximately 25% of schizophrenia liability is attributable to common causal SNPs. Subsequently Gusev and colleagues began to use similar strategies to assess the degree to which specific functional annotations are enriched for heritability60. They argued that SNPs within regulatory regions explain disproportionate heritability across 11 polygenic diseases.
As these methods emerged, it became natural for geneticists to ask which SNP annotations explain disproportionate amounts of the heritability across the entire spectrum of genome-wide association statistics, e.g. not restricting to significant loci. Annotations enabled investigators to divide SNPs by which types of genes they were closest to, the extent to which the SNPs were in highly conserved regions, their allele frequencies, and whether or not SNPs were in cell-type specific regulatory elements. For diseases like RA, it became important to determine which cell-types are states harbored the SNPs that were explaining the greatest heritability.
Most recently, advances in statistical genetics have enabled investigators to assess which subsets of SNPs, defined by specific functional annotations, explain disproportionately higher or lower heritability using summary statistics. These methods are both efficient and versatile, and can be easily applied to a wide range of data sets, even when genotype data is not easy to share. Finucane and colleagues developed a method called stratified LD score regression (S-LDSC) to directly quantify the proportion of genetic heritability from a GWAS captured by regulatory categories of SNPs, using only summary statistics9. In their study, the authors confirm and nominate disease-driving cell types by identifying enrichments of cell-type-specific epigenetic marks.
S-LDSC performs a regression of the chi-squared GWAS statistic on LD score. The slope of this regression is the estimated genetic heritability according to the specific GWAS, which can be stratified by functional category. Then, per-SNP heritability may be computed by dividing the category-specific heritability by the number of SNPs in the category. If per-SNP heritability within the category is greater than the per-SNP heritability across all SNPs, which can be assessed with a simple z-score test, the category is enriched for explaining heritability. The other nuance of S-LDSC is that unlike GCTA, an earlier method developed to partition SNP heritability, S-LDSC does not require raw genotyping data, which can be difficult to obtain due to privacy issues and has higher computational cost61. Rather, S-LDSC only requires GWAS summary statistics, or per-SNP effect sizes, computed from regressing phenotype on genotype. The first S-LDSC study reinforced known biology regarding RA, such as strong heritability enrichment in immune and hematopoietic regulatory annotations, the strongest signal of which came from H3K4me1 in CD4+ Th17 cells. While S-LDSC did not nominate any new cell types in the disease etiology of RA, this method has the potential to quantify and differentiate the effects of candidate disease-driving cell types.
Three years later, a second study62 utilized S-LDSC to integrate gene expression with GWAS data, with similar goals as the study done by Hu et al in 2011. In this 2018 study, the authors assigned a tissue specificity score to each gene and subsequently defined tissue-specific gene sets comprised of the top 10% of specifically expressed genes. In order to nominate disease-driving cell types, the authors tested if SNPs within 100 kb of cell-type-specifically expressed genes captured more polygenic trait heritability than other SNPs. Consistent with previous findings, this study observed that sets of genes that are more specifically expressed in B cells, T cells, and myeloid cells were significantly enriched for RA heritability. However, we note that each of these categories are broad. As more functional annotation data is generated and more cell types and cell-states are identified, likely through single cell analyses, S-LDSC will be an invaluable tool to quantify cell-type-specific polygenic effects on RA susceptibility. Since its publication, S-LDSC has become a widely-used method to partition SNP heritability and better understand the underpinnings of polygenic traits. Work from our group has used S-LDSC in the context of understanding CD4+ T cell-state-specific regulation, which we will discuss next.
Transcriptional regulatory elements comprehensively capture RA heritability
While regulatory dynamics in disease may be understood by analyzing histone modifications and specifically expressed genes, transcription factor (TF) binding also plays a large role63. TFs bind in cell type specific contexts in enhancer and promoter regions of genes. Therefore, by analyzing their occupancy and modeling their genome-wide binding targets, we may better understand the mechanisms of noncoding regulatory variants. As most TFs recognize sequence-specific motifs in DNA, an alternative allele which interrupts a motif might have a direct impact on lowering TF binding affinity. As a result of this weakened affinity and weakened transcriptional activation, the downstream gene may not be as highly expressed. Modest effects of risk alleles on TF binding affinity may cumulatively account for a non-negligible proportion of polygenic disease heritability. Moreover, identifying what TFs or families of TFs are implicated in disease risk provide testable mechanistic hypotheses. In 2018, Reshef and colleagues adapted the S-LDSC framework to compute directional enrichments, as an allele may increase or decrease TF binding affinity. The authors note that signed enrichments are more strongly based in causality compared to unsigned enrichments which may be the result of co-localization. Although this study did not find any particularly strong mechanism of TF binding affinity underpinning RA, the concept that TFs orchestrate the regulation of a broad set of genes as opposed to only a few target genes exemplifies their polygenic potential. In light of this, in our own study, we hypothesized that TF binding genome-wide could be used as a basis for learning patterns of regulatory activity which might capture polygenic genetic variation.
Here, we will refer to the different lineages of effector CD4+ T cells as “cell-states”, created after TCR activation and characterized by specific cytokine and gene expression profiles. The most well studied CD4+ T cell-states include Th1, Th2, Th17, and Tregs, while other less often studied states include Th9, Th11, and Th22. For each CD4+ T cell-state, one or several key TFs are known to be essential to promote cell-state differentiation. Specifically, T-BET or STAT4 drives differentiation to Th1s, GATA3 to Th2s, STAT3 or ROR-γ-t to Th17s, and FOXP3 or STAT5 to Tregs. As in Reshef et al, identifying genetic variation within regulatory elements bound by TFs can elucidate the genetic underpinnings of polygenic traits. However, regulatory activity that is important to each of these cell-states is not limited to action of just one or two TFs. Since cell-type-specific TF ChIP-seq is limited and knowledge of important regulators is also limited, we aimed to design a strategy that could infer genome-wide cell-state regulatory activity using a single TF ChIP-seq experiment. If we could know which regulatory elements are active in each CD4+ T cell-state, we might uncover cell-state-specific contexts for RA-associated variants. For instance, a variant residing in a Treg-specific regulatory element would suggest that functional follow up on this variant be performed in Treg cells, as this variant is likely to act in a cell-state-specific context.
In light of this, our group designed a strategy called IMPACT that uses lineage-specifying TF binding as an anchor for cell-type regulatory activity and algorithmically extrapolates patterns of this activity genome-wide64. In order to demonstrate the utility of IMPACT, we used the compelling association of CD4+ T cells with RA and aimed to derive CD4+ T cell-state-specific regulatory annotations based on TF ChIP-seq. IMPACT uses the binding sites of a single key transcription factor (TF) as a gold standard for regions of cell-state regulatory function and then learns a quantitative epigenomic signature across thousands of experimental epigenetic and sequence annotations. IMPACT uses this learned epigenomic signature to annotate the entire genome for cell-state regulatory activity. As Finucane and colleagues had previously shown, gene regulatory mechanisms capture polygenic genetic variation. Therefore, in order to assess how well we were predicting disease-driving cell-state regulatory activity, we could measure how well we captured this polygenic RA genetic variation.
Due to different immunological roles assumed by each CD4+ T cell-state, we hypothesized that each state could account for different proportions of RA polygenic heritability. To this end, we aimed to use IMPACT to correlate the RA effect sizes of a set of variants with their predicted IMPACT regulatory activity across a range of cell types and states. Identifying a strong correlation could nominate disease driving cell-states and quantitatively assess the role of different CD4+ T cell states in RA. This also allowed us to validate our strategy; if IMPACT was capturing genetic variation by modeling regulation in a known disease-driving cell type, then we were successfully predicting cell type regulation beyond the scope of the singular TF used to train the model. To this end, we used S-LDSC to partition RA heritability and observed that the top 5% of annotated SNPs in each of these CD4+ T cell annotations (Th1, Th2, Th17, and Treg) explained on average 85.7% of RA heritability, the most comprehensive explanation of RA heritability to date. Intriguingly, each annotation explained a similar proportion of RA heritability and were statistically indistinguishable. From this study we learned that in silico IMPACT annotations outperform TF ChIP-seq or other single experimentally-derived annotations, such as histone modification ChIP-seq and gene expression at capturing polygenic trait heritability. A separate goal of IMPACT was to predict cell-state-specific regulatory contexts for several putatively causal fine-mapped RA variants. In particular, we found that rs35926684 in the TNFAIP3 locus resides in a Th1-specific IMPACT regulatory element, nominating Th1s as the preferred cell type for follow up validation.
Refining RA associations within the MHC
We now return to the MHC, which continues to be the most important locus for RA risk among ACPA+ individuals. Prior decades of work had focused on the parts of the HLA-DRB1 gene encoding the epitope’s alpha helix, which was readily exposed to antibody interaction, on subunit 1 of the HLA-DR molecule. As GWAS were performed with genotyped SNPs, denser typing of the MHC and its HLA alleles was an attractive concept for testing associations within this highly polymorphic region. However, dense genotyping in large cohorts would be expensive, complex, and would not take advantage of the already performed autoimmune disease GWAS. Therefore, Jia and colleagues developed a strategy, SNP2HLA, to impute classical HLA class I and II alleles, taking advantage of long-range LD of the region and genotyping chip markers; Raychaudhuri and colleagues used this strategy in their analysis65. In addition to SNP2HLA, many other HLA imputation methods have emerged, including HLA*IMPUTE and HIBAG.
Using imputation-based approaches, we tested HLA alleles for an RA association, we observed that in a cohort of nearly 20,000 European individuals5. We observed that SNPs encoding the shared epitope had a weaker association signal than the 11th amino acid in subunit 1 of HLA-DR, with an odds ratio of 3.7 and a p-value less than 10−526. Moreover, this association was independent of the shared epitope and was replicated in the South Korean population. Conditional analysis proceeded to identify positions 71 and 74 as statistically significant independent associations. Intriguingly, although in linear space amino acid 11 is far from 71 and 74, in their 3-D conformation, they are spatially proximal, providing potential insight into the importance of their regulatory role within the peptide binding groove. We demonstrated that specific amino acid positions (positions 11/13, 71, and 74 in HLA-DRB1), explained most of the signal for ACPA+ RA5. Position 11 was in tight linkage with position 13. Both are deep within the peptide binding groove. Positions 13, 71, and 74 together implicated the P4 pocket. More recently in 2018, Ting et al confirmed the importance of the P4 pocket by observing a correlation between self-peptide binding affinity and citrullination of the bound ligand at P466. Moreover, we and others have identified other MHC associations among ACPA+ individuals living with RA, independent of HLA-DRB1, including in HLA-DPB167 and HLA-B68,69.
These findings advanced our understanding of RA pathogenesis beyond the implications of the shared epitope hypothesis. They strongly suggest that antigen binding is the key driver of HLA-mediated disease susceptibility. However, it is still uncertain whether disease risk variability due to DBR1 alleles is due to antigen presentation in the periphery or antigen presentation by thymic epithelial cells influencing the T cell repertoire during thymic selection.
Imputing HLA in worldwide populations
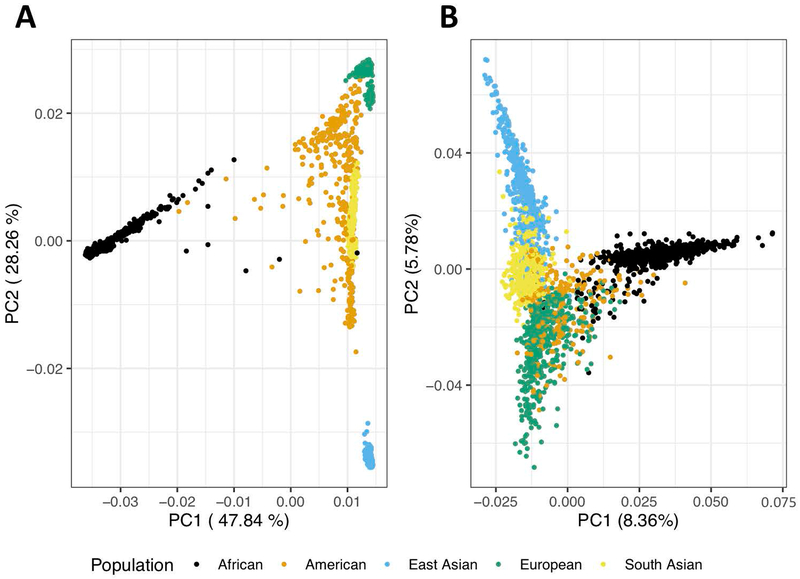
HLA imputation for a given genome, as mentioned above, succeeds only with a representative and well-powered reference panel. The longstanding bias in human genetics experiments within European populations has revealed that HLA allelic patterns are ancestry-specific and as such, imputation in a diverse set of populations with a European reference panel is not a viable approach (Figure 3).50
Figure 3.
A) From 1000 Genomes, first two principal components (PCs) of global genotyping reveals genetic heterogeneity among continental populations. B) From 1000 Genomes, first two PCs of HLA-specific genotyping accounts for a strikingly high proportion of diversity among continental populations.
Many research groups, ours included, have been tackling this problem by designing multi-ethnic reference panels and evaluating the predictive performance of their imputation strategies at HLA class I and II genes. Only then can successes from RA genetic studies begin to indiscriminately benefit the 1% of affected individuals worldwide, rather than just individuals of European descent. In 2017, Karnes and colleagues benchmarked three widely used HLA imputation methods, SNP2HLA65, HIBAG70, and HLA*IMP71, assessing imputation accuracy in a cohort of 2,947 European Americans and 318 African Americans, relative to gold standard Illumina HLA genotyping of these same individuals72. Briefly, SNP2HLA imputes not only HLA alleles but amino acid substitutions using BEAGLE73, HIBAG uses expectation-maximization models, and HLA*IMP uses a graph-based approach which accommodates multiple ancestries. All three methods achieved greater than 93% concordance between gold standard genotyping and imputation in European American individuals. HIBAG, a strategy that uses ensemble bootstrapping and expectation maximization, achieved the greatest concordance (92.9%) between gold standard genotyping and imputation in African Americans, narrowly outperforming SNP2HLA (91.9% concordance) and significantly outperforming HLA*IMP (61.9% concordance), which uses a graph-based approach inspired by a popular phasing algorithm74. However, when accounting for the number of alleles called, SNP2HLA (210 alleles) is a more attractive method compared to both HIBAG (175 alleles) and HLA*IMP (140 alleles). Moreover, for genotyping platforms with lower genomic coverage, SNP2HLA again achieved the highest imputation accuracy. Realizing the importance of ancestry specificity among HLA alleles, all areas of human genetics should be embracing multi-ethnic approaches, with the goal of providing any individual with the opportunity to benefit from the advances in our field.
Practically speaking, integration of population-specific HLA imputation with trans-ethnic RA GWAS has already identified a dosage risk associated with HLA-DOA, a non-classical HLA gene; this association is independent from HLA-DRB1 variants75,76. Therefore, further design of population-specific HLA imputation reference panels are warranted. In addition, multiethnic studies have confirmed the role of position 11/13, 71 and 74 in driving RA susceptibility. For example, in Asian populations Okada and colleagues reproduced the position 13 association, with additional independent signals found at positions 54 and 74. This study revealed that the same amino acid residues confer shared genetic risk to RA77.
Identifying expanded cellular populations in RA patient synovial tissue
While many genetic strategies have indicated that the broad category of T cells are relevant to disease, it has been important to consider which cell populations and subpopulations are playing an active role in RA. One simple approach is to look for cell populations that are altered in individuals with disease, either examining immune population shifts in the blood, or in the inflamed tissue itself. Single cell technologies such as mass cytometry and single cell RNA-seq have enabled investigators in the last five years to redefine the set of immune and stromal cell types that might be germane to RA.
One such approach called MASC (Mixed effects association of single cells) uses patient sample data rather than genetic association data to identify which cell types are expanded among cases versus the controls, while carefully accounting for inter-individual variation78. In one application, our group examined blood from RA cases and controls and applied mass cytometry. Then using MASC to explicitly model case control status, we identified a novel cellular subset CD27− HLA-DR+ effector memory CD4 T cells in RA with an odds ratio of 1.7 in blood compared to controls. This subset most closely resembles Th1, in that it secretes IFN-γ and granzyme A.
Single cell transcriptomic and epigenomic advances
With high resolution profiling of cell types and cell-states coming from single cell data generation, it is imperative that we continue to interrogate the roles of cell-state-specific processes in disease susceptibility. High powered single cell data initiatives profile thousands of cells and have the potential to discover novel regulatory modules and cell subtypes. One such initiative is the Accelerating Medicines Partnership, which in its first phase profiled more than 5,000 cells in 36 RA patients and 15 osteoarthritis (OA) controls. Our group developed a CCA graph-based clustering strategy to identify single cell populations by leveraging correlated regulatory structure with bulk reference samples51. As a result, 3 CD4+ T cell subtypes were identified with distinct expression profiles, CCR7+ T cells, Tregs, and a mix of T peripheral (Tph) and follicular (Tfh) helper cells. Intriguingly, only the Tph/Tfh cells were significantly expanded in RA patients compared to OA controls. There is potential for developing functional annotations based on single cell populations in order to quantify the contribution of cell-specific regulatory elements to RA susceptibility. Notably, this work also identified two classes of monocytes, PLAUR+ pro-inflammatory and IFN-activated, and one class of sublining fibroblast, CD90+ HLA-DR high, expanded in inflamed RA. These identified cell states, or others demonstrating pathogenic regulatory mechanisms, are prominent candidates for future cell-state-targeting drug development. Single cell studies in which we leverage both cellular genetic and functional heterogeneity hold the potential to refine putatively causal risk mechanisms. Moreover, these studies accomodate an experimental framework, in which cells may be sorted, edited, and profiled for function. This type of research has the special advantage to assist human genetics, a fully observational science.
Future for RA genetics, precision medicine, genetic risk scores, and new therapies
Over the last five decades, the field of RA genetics has greatly evolved. As we try to find explanations for the vast polygenic variation we observe, the repertoire of cell type mechanisms contributing to the complexity of RA continues to grow. Moreover, due to multiple candidate causal cell types and pathogenic states, future treatments aiming at lifelong remission will likely require a separate drug for each targeted cell state.
While the catalogue of causal cell states is still being refined and drug development is a lengthy process, the more immediate future may hold more promise for pre-onset individuals at risk for RA. In clinical practice, RA can be detected by the presence of a few biomarkers, such as rheumatoid factor, autoantibodies, and ACPA. However, preventative measures to assess the risk of onset of RA, before detection of these biomarkers, is falling behind successful predictive clinical models of breast cancer, heart disease, and type 1 diabetes79. It is imperative that genetic findings in the field of RA be used to improve risk models so that at-risk individuals can benefit from early treatment and prevention. Such polygenic risk score (PRS) models are currently being developed, which sum up the aggregate risk alleles in one’s genome, weighted by effect size determined from a GWAS. However, the success of PRS and implementation of clinical models depends heavily on disease prevalence, reference populations on which these models are based on, and the genetic divergence of these populations from the individuals in the clinic. The extreme bias of genetics research done with participants of European-descent is deteriorating the possibilities of using genetic findings to benefit individuals across the globe. As we discussed ancestry-specific HLA typing and RA-associated risk as well as ancestry-specific genome-wide significant loci, European-based models of RA will prevent development of beneficial diagnostic and treatment practices in non-European populations, leading to greater healthcare disparities internationally. Diversifying efforts must be made in the genetics community to include already underserved non-European populations.
Therefore, future efforts for identifying therapeutic targets, running RA clinical trials, and developing clinical models must heavily encourage the participation of genetically diverse individuals. Only then can the inevitably impactful findings from RA genetics be used to improve the standard of living and lifespan of individuals at risk for developing this debilitating disease.
Acknowledgements
This work was supported in part by funding from the National Institutes of Health (UH2AR067677, U19AI111224, and 1R01AR063759 (to SR)) and NHGRI HG002295 T32 (to TA). We thank Dr. Laura Donlin for comments and discussion.
Footnotes
Conflict of interest statement: The authors declare no competing interests.
References
- 1.Viatte S, Plant D & Raychaudhuri S Genetics and epigenetics of rheumatoid arthritis. Nature Reviews Rheumatology 9, 141–153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terao C, Raychaudhuri S & Gregersen PK Recent Advances in Defining the Genetic Basis of Rheumatoid Arthritis. Annu. Rev. Genomics Hum. Genet 17, 273–301 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kastbom A, Strandberg G, Lindroos, et al. Anti-CCP antibody test predicts the disease course during 3 years in early rheumatoid arthritis (the Swedish TIRA project). Ann. Rheum. Dis 63, 1085–1089 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aletaha D & Smolen JS Diagnosis and Management of Rheumatoid Arthritis. JAMA 320, 1360 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Raychaudhuri S et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet 44, 291–296 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu X et al. Integrating Autoimmune Risk Loci with Gene-Expression Data Identifies Specific Pathogenic Immune Cell Subsets. Am. J. Hum. Genet 89, 496–506 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trynka G et al. Disentangling the Effects of Colocalizing Genomic Annotations to Functionally Prioritize Non-coding Variants within Complex-Trait Loci. Am. J. Hum. Genet 97, 139–152 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manolio TA et al. Finding the missing heritability of complex diseases. Nature 461, 747 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, Lee H, Goddard ME, et al. GCTA: A Tool for Genome-wide Complex Trait Analysis. (2011). doi: 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis-Faning E Report on an enquiry into the aetiological factors associated with rheumatoid arthritis. (Brit. Med. Assoc, 1950). [Google Scholar]
- 12.Hemminki K, Li X, Sundquist J & Sundquist K Familial associations of rheumatoid arthritis with autoimmune diseases and related conditions. Arthritis Rheum. 60, 661–668 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Frisell T et al. Familial aggregation of arthritis-related diseases in seropositive and seronegative rheumatoid arthritis: a register-based case-control study in Sweden. Ann. Rheum. Dis 75, 183–189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aho K, Koskenvuo M, Tuominen, et al. Occurrence of rheumatoid arthritis in a nationwide series of twins. J. Rheumatol 13, 899–902 (1986). [PubMed] [Google Scholar]
- 15.Silman AJ et al. Twin concordance rates for rheumatoid arthritis: results from a nationwide study. Br. J. Rheumatol 32, 903–907 (1993). [DOI] [PubMed] [Google Scholar]
- 16.MacGregor AJ et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 43, 30–37 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Astorga GP & Williams RC Altered reactivity in mixed lymphocyte culture of lymphocytes from patients with rheumatoid arthritis. Arthritis & Rheumatism 12, 547–554 (1969). [DOI] [PubMed] [Google Scholar]
- 18.Stastny P Mixed lymphocyte cultures in rheumatoid arthritis. J. Clin. Invest. 57, 1148 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silver J & Goyert SM Epitopes Are the Functional Units of HLA Class II Molecules and Form the Molecular Basis for Disease Susceptibility in HLA Class II Antigens 32–48 (Springer; Berlin Heidelberg, 1986). [Google Scholar]
- 20.Gregersen PK, Silver J & Winchester RJ The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 30, 1205–1213 (1987). [DOI] [PubMed] [Google Scholar]
- 21.Huizinga TWJ et al. Refining the complex rheumatoid arthritis phenotype based on specificity of the HLA-DRB1 shared epitope for antibodies to citrullinated proteins. Arthritis & Rheumatism 52, 3433–3438 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Lee DM & Schur PH Clinical utility of the anti-CCP assay in patients with rheumatic diseases. Ann. Rheum. Dis 62, 870–874 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackie SL et al. A spectrum of susceptibility to rheumatoid arthritis within HLA-DRB1: stratification by autoantibody status in a large UK population. Genes & Immunity 13, 120–128 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Cornélis F et al. New susceptibility locus for rheumatoid arthritis suggested by a genome-wide linkage study. Proceedings of the National Academy of Sciences 95, (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Botstein D & Risch N Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat. Genet 33, 228–237 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Begovich AB et al. A Missense Single-Nucleotide Polymorphism in a Gene Encoding a Protein Tyrosine Phosphatase (PTPN22) Is Associated with Rheumatoid Arthritis. Am. J. Hum. Genet 75, 330–337 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki A et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat. Genet 34, 395–402 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Plenge RM et al. Replication of Putative Candidate-Gene Associations with Rheumatoid Arthritis in >4,000 Samples from North America and Sweden: Association of Susceptibility with PTPN22, CTLA4, and PADI4. Am. J. Hum. Genet 77, 1044–1060 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Risch N & Merikangas K The future of genetic studies of complex human diseases. Science 273, 1516–1517 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Lander E et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Collins FS, Guyer MS & Chakravarti A Variations on a Theme: Cataloging Human DNA Sequence Variation. 278, 1580–1581 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Burton PR et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plenge RM et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat. Genet 39, 1477–1482 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plenge RM et al. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N. Engl. J. Med 357, 1199–1209 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raychaudhuri S et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat. Genet 40, 1216–1223 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stahl EA et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet 42, 508–514 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okada Y et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buniello A et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okada Y, Eyre S, Suzuki A, et al. Genetics of rheumatoid arthritis: 2018 status. Ann. Rheum. Dis 78, 446–453 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Sakaue S et al. Integration of genetics and miRNA–target gene network identified disease biology implicated in tissue specificity. Nucleic Acids Res. 46, 11898–11909 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim K et al. Association-heterogeneity mapping identifies an Asian-specific association of the GTF2I locus with rheumatoid arthritis. Sci. Rep 6, 27563 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okada Y et al. Significant impact of miRNA-target gene networks on genetics of human complex traits. Sci. Rep 6, 22223 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eyre S et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet 44, 1336–1340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kochi Y et al. A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility. Nat. Genet 42, 515–519 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Fairfax BP et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat. Genet 44, 502–510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chun S et al. Limited statistical evidence for shared genetic effects of eQTLs and autoimmune disease-associated loci in three major immune cell types. Nat. Genet 49, 600 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Messemaker TC et al. A novel long non-coding RNA in the rheumatoid arthritis risk locus TRAF1-C5 influences C5 mRNA levels. Genes & Immunity 17, 85–92 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Farh KK-H et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Westra H-J et al. Fine-mapping and functional studies highlight potential causal variants for rheumatoid arthritis and type 1 diabetes. Nat. Genet 50, 1366–1374 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Project Consortium G et al. A map of human genome variation from population-scale sequencing The 1000 Genomes Project Consortium*. Nature 467, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang F et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol 20, 928–942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Golub TR et al. Molecular Classification of Cancer: Class Discovery and Class Prediction by Gene Expression Monitoring. Science 286, 531–537 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Schinnerling K, Rosas C, Soto L, et al. Humanized Mouse Models of Rheumatoid Arthritis for Studies on Immunopathogenesis and Preclinical Testing of Cell-Based Therapies. Front. Immunol 10, 203 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trynka G et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet 45, 124–130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maurano MT et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manolio TA Genomewide Association Studies and Assessment of the Risk of Disease. N. Engl. J. Med 363, 166–176 (2010). [DOI] [PubMed] [Google Scholar]
- 57.International Schizophrenia Consortium et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stahl EA et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat. Genet 44, 483–489 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee SH et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat. Genet 44, 247–250 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gusev A et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet 95, 535–552 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gazal S, Marquez-Luna C, Finucane HK, et al. Reconciling S-LDSC and LDAK functional enrichment estimates. Nat. Genet 51, 1202–1204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finucane HK et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet 50, 621–629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reshef YA et al. Detecting genome-wide directional effects of transcription factor binding on polygenic disease risk. Nat. Genet 50, 1483–1493 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amariuta T et al. IMPACT: Genomic Annotation of Cell-State-Specific Regulatory Elements Inferred from the Epigenome of Bound Transcription Factors. Am. J. Hum. Genet 104, 879–895 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jia X et al. Imputing Amino Acid Polymorphisms in Human Leukocyte Antigens. PLoS One 8, e64683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ting YT et al. The interplay between citrullination and HLA-DRB1 polymorphism in shaping peptide binding hierarchies in rheumatoid arthritis. J. Biol. Chem 293, 3236–3251 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ding B et al. Different patterns of associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in the extended major histocompatibility complex region. Arthritis & Rheumatism 60, 30–38 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jawaheer D et al. Dissecting the Genetic Complexity of the Association between Human Leukocyte Antigens and Rheumatoid Arthritis. Am. J. Hum. Genet 71, 585–594 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee H-S et al. Several Regions in the Major Histocompatibility Complex Confer Risk for Anti-CCP-Antibody Positive Rheumatoid Arthritis, Independent of the DRB1 Locus. Molecular Medicine 14, 293–300 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng X et al. HIBAG—HLA genotype imputation with attribute bagging. Pharmacogenomics J. 14, 192–200 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dilthey AT, Moutsianas L, Leslie S, et al. HLA*IMP--an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics 27, 968–972 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karnes JH et al. Comparison of HLA allelic imputation programs. PLoS One 12, e0172444 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Browning BL & Browning SR A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet 84, 210–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li N & Stephens M Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165, 2213–2233 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Okada Y et al. Contribution of a Non-classical HLA Gene, HLA-DOA, to the Risk of Rheumatoid Arthritis. Am. J. Hum. Genet 99, 366–374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okada Y et al. Construction of a population-specific HLA imputation reference panel and its application to Graves’ disease risk in Japanese. Nat. Genet 47, 798–802 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Okada Y et al. Risk for ACPA-positive rheumatoid arthritis is driven by shared HLA amino acid polymorphisms in Asian and European populations. Hum. Mol. Genet 23, 6916–6926 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fonseka CY et al. Mixed-effects association of single cells identifies an expanded effector CD4+ T cell subset in rheumatoid arthritis. Sci. Transl. Med 10, eaaq0305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martin AR et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet 51, 584–591 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]