Abstract
Objective
The αIIbβ3 antagonist anti-platelet drug abciximab is the chimeric antigen-binding fragment (Fab) comprising the variable regions of murine mAb 7E3 and the constant domains of human IgG1 and light chain κ. Previous mutagenesis studies suggested that abciximab binds to the β3 C177-C184 specificity-determining loop (SDL) and Trp129 on the adjacent β1-α1 helix. These studies could not, however, assess whether 7E3 or abciximab prevents fibrinogen binding by steric interference, disruption of either the αIIbβ3-binding pocket for fibrinogen or the β3 SDL (which is not part of the binding pocket, but affects fibrinogen binding), or some combination of these effects. To address this gap, we used cryo-electron microscopy (EM) to determine the structure of the αIIbβ3–abciximab complex at 2.8-Å resolution.
Approach and Results
The interacting surface of abciximab is comprised of residues from all three complementarity-determining regions of both the light and heavy chains, with high representation of aromatic residues. Binding is primarily to the β3 SDL and neighboring residues, the β1-α1 helix, and β3 residues Ser211, Val212 and Met335. Unexpectedly, the structure also indicated several interactions with αIIb. As judged by the cryo-EM model, molecular-dynamics simulations, and mutagenesis, the binding of abciximab does not appear to rely on the interaction with the αIIb residues, and does not result in disruption of the fibrinogen-binding pocket; it does, however, compress and reduce the flexibility of the SDL.
Conclusions
We deduce that abciximab prevents ligand binding by steric interference, with a potential contribution via displacement of the SDL and limitation of the flexibility of the SDL residues.
Keywords: integrin receptor, αIIbβ3, antiplatelet agent, Cell Biology/Structural Biology, Platelets, Myocardial Infarction, Percutaneous Coronary Intervention, Thrombosis
INTRODUCTION
The drug abciximab is the antigen-binding fragment (Fab) of a chimeric monoclonal antibody (mAb) comprising the variable regions of murine mAb 7E3 and the constant domains of human IgG1 and light chain κ.1 Its target is the platelet integrin αIIbβ3 receptor, which is highly expressed on platelets and megakaryocytes and required for normal platelet aggregation.2 Patients who lack functional αIIbβ3 receptors on an inherited basis have Glanzmann thrombasthenia, a serious life-long bleeding disorder characterized by mucocutaneous hemorrhage.3
mAb 7E3 and abciximab potently inhibit fibrinogen binding to αIIbβ3, platelet aggregation induced by multiple agonists that depend on ligand binding to αIIbβ3, and platelet-mediated thrombus formation in vivo.4–8 Abciximab was approved by the U.S. Food and Drug Administration (FDA) for human use in 1994 as an adjunct to percutaneous coronary artery intervention for the prevention of cardiac ischemic complications. The manufacturer has estimated that more than 5 million patients worldwide have been treated with abciximab since its approval (Private Communication, Dr. Elliot Barnathan, September 9, 2019). In more than 40 studies conducted on more than 30,000 patients in randomized Phase 3 and post-market studies of patients undergoing percutaneous coronary interventions, including coronary artery stenting, abciximab decreased the odds of both all-cause mortality at 30 days [odds ratio (OR) 0.79, 95% confidence interval (CI) 0.64–0.97)] and death or myocardial infarction at 30 days (OR 0.66, 95% CI 0.60–0.72).9
mAb 7E3 IgG has the unusual property of binding much more rapidly to activated platelets than unactivated platelets.4 The F(ab’)2 and Fab fragments of 7E3 bind more rapidly to unactivated platelets than does the intact IgG and so their rates of binding are less affected by platelet activation. In a reciprocal manner, dimers, trimers, and tetramers of 7E3 bind less rapidly to unactivated platelets than does the intact IgG, and they demonstrate the greatest increase in binding rate with platelet activation.10 Thus, 7E3 binding is consistent with size-dependent access to its epitope. Despite its relatively slow binding to unactivated αIIbβ3, 7E3 IgG eventually reaches the same maximal binding level as other mAbs to αIIbβ3, suggesting that αIIbβ3 continually undergoes conformational changes that variably limit access of 7E3 to its epitope.
mAb 7E3 reacts with non-human primate and dog platelets in addition to human platelets,11 which allowed for testing its anti-platelet and anti-thrombotic properties in a number of animal models.7, 8, 12–14 mAb 7E3 and abciximab also react with the related integrin receptor αVβ3, which shares the same β integrin subunit.15–17
The binding of mAb 7E3 to αIIbβ3 has been studied biochemically and by site-directed mutagenesis and electron microscopy (EM), leading to the localization of its epitope to discontinuous areas of the β3 subunit head domain near the fibrinogen/Arg-Gly-Asp (RGD) -binding pocket made up of the metal ion-dependent adhesion site (MIDAS) and the adjacent to the metal ion-dependent adhesion site (ADMIDAS), as well as the nearby specificity-determining loop (SDL),18–21 but none of the studies to date have had the resolution to identify the specific antibody–integrin interactions at the atomic level or to assess whether abciximab alters the fibrinogen/RGD-binding pocket itself. To address these gaps in our knowledge, we have utilized cryo-EM to obtain a high-resolution three-dimensional structure of the αIIbβ3–abciximab complex and a combination of molecular dynamics (MD) simulations and site-directed mutagenesis to probe the stability of its inter-molecular interactions.
MATERIALS and METHODS
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Cloning, expression, protein purification, and complex formation
Based on previous studies by Zhu et al.,22 human αIIb integrin (residues 1–963) was fused in frame at its C terminus to a PreScission protease cleavage site, an acidic coiled-coil region, and a Strep II tag, and cloned into the pEF1 vector (Invitrogen). Human β3 integrin (residues 1–690) was fused in frame at its C terminus to a PreScission protease cleavage site, a basic coiled-coil region, and an 8X histidine tag, and cloned into the pcDNA3.1 vector (Invitrogen). The cloned αIIb subunit contains an Leu959Cys mutation, whereas the β3 subunit contains a Pro688Cys mutation; together, these mutations allow for the formation of a disulfide bond between the two integrin subunit chains near the C terminus.
Recombinant αIIbβ3 was expressed in mammalian cell culture using the Expi293 expression system (Thermo Fisher Scientific). Expi293 suspension cells were co-transfected with αIIb- and β3-encoding plasmids using TransIT-PRO reagent (Mirus) according to the manufacturer’s protocol. Transfected cells were cultured for 5–6 days in Expi293 expression medium (Thermo Fisher Scientific) at 37°C, 8% CO2, with shaking at 125 rpm.
The expressing cells were harvested by centrifugation and the supernatant was dialyzed with at least 3 changes against dialysis buffer (20 mM HEPES, pH 7.4, 300 mM NaCl, 2 mM CaCl2, 1 mM MgCl2). The αIIbβ3 complexes were then purified using TALON metal-affinity resin (Takara), followed by gel filtration (Superdex™ Increase 200, GE Healthcare Life Sciences) in a buffer containing 20 mM HEPES, pH 7.4, 150 mM NaCl, 2 mM CaCl2, 1 mM MgCl2.
Abciximab was obtained from Centocor/Lilly. Purified αIIbβ3 was mixed with abciximab at a 1:6 molar ratio (0.74 μM/4.55 μM) in a buffer containing 20 mM HEPES, pH 7.4, 150 mM NaCl, 2 mM CaCl2, 1 mM MgCl2, incubated for 2 hours at 4°C, and subjected to gel filtration (Superdex™ Increase 200). The fractions containing αIIbβ3–abciximab complexes were identified by SDS-PAGE followed by Coomassie blue staining and then imaged by negative-stain electron microscopy (EM).
Protein sequencing
The primary amino-acid sequence of abciximab was determined by de novo peptide sequencing23, 24 using tandem mass spectrometry (MS) by Rapid Novor Inc. Eight in-solution and in-gel digestions of abciximab were prepared with 5 different enzymes (pepsin, trypsin, chymotrypsin, Asp N, and Lys C). Prior to digestion, in-solution samples were reduced and alkylated. For in-gel digestions, samples were reduced, electrophoresed in an SDS-polyacrylamide gel, deglycosylated, reduced again, alkylated, and digested. All digestions were analyzed by LC-MS/MS using Thermo Fisher Q Exactive™, Orbitrap Fusion™ mass spectrometers. Peptides were characterized from LC-MS/MS data using de novo peptide sequencing and then assembled into the antibody sequence.24 Both heavy and light chains were covered 100% with at least 5 peptide scans. The discrimination between the Leu and Ile amino acids was based on three criteria: 1) The W-ion Isoleucine Leucine Determination (WILD™) method (Rapid Novor Inc.), 2) enzyme specificity at Leu and Ile in chymotrypsin and pepsin digestions, and 3) statistical residue distribution and homologous sequences by reference to the international ImMunoGeneTics information system (http://www.imgt.org/). This approach allowed for the discrimination between Leu and Ile among all the Leu/Ile amino-acids, except for heavy chain Leu/Ile 98. That residue was arbitrarily assigned as Leu98.
Negative-stain EM
4-μL aliquots of the αIIbβ3–abciximab complex (0.01 mg/mL) were adsorbed for 30 s onto glow-discharged copper grids covered with a carbon-coated plastic film and negatively stained with 0.75% (w/v) uranyl formate solution as described.25 Specimens were imaged with a CM10 electron microscope (Philips) equipped with a tungsten filament and operated at an acceleration voltage of 100 kV. Micrographs were collected with an XR16L-ActiveVu camera (AMT) at a defocus value of −1.5 μm. The nominal magnification was 52,000X, which corresponds to a calibrated magnification of 41,513X and a pixel size of 2.65 nm at the specimen level. From 49 images, 14,593 particles were automatically picked with Gautomatch (http://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/) and windowed into 128X128-pixel images. After image normalization and particle centering, the particle images were classified into 100 groups using K-means classification procedures implemented in SPIDER.26
Cryo-EM sample preparation and data collection
For cryo-EM grid preparation, vitrified grids were prepared with a Vitrobot Mark IV (Thermo Fisher Scientific) set at 100% humidity and 4°C. 4-μL aliquots were applied to glow-discharged Quantifoil 400 mesh 1.2/1.3 gold grids, which were blotted for 3 s with a blot force setting of −2 and then plunged into liquid ethane.
Image stacks were collected with a Titan Krios electron microscope (Thermo Fisher Scientific) in the Cryo-EM Resource Center at the Rockefeller University operated at an acceleration voltage of 300 kV. Data were collected with a K2 Summit camera (Gatan) in super-resolution counting mode using SerialEM.27 The nominal magnification was 28,000X (calibrated pixel size of 1.0 Å on the specimen level) and the defocus range was −1.2 to −2.5 μm. Exposures of 10 s were dose-fractionated into 40 frames (250 ms per frame), with a dose rate of 6 electrons/pixel/s (~1.5 electrons/Å2/frame), resulting in a total dose of 60 electrons/Å2.
Image processing
Image stacks were motion-corrected, dose-weighted, summed, and binned over 2X2 pixels in MotionCor2.28 The contrast transfer function (CTF) parameters were determined with CTFFIND4.29 A total of 2,970,080 particles were auto-picked from 4,236 micrographs using Gautomatch. The particles were extracted into 240X240-pixel images, rescaled into 80×80-pixel images, and subjected to 2D classification into 150 classes in RELION-3,30 which was used for all further processing steps. After discarding the particles in classes that showed poor averages, the remaining 2,408,289 particles were subjected to 3D classification into 6 classes using as reference an initial model generated in RELION-3. Three classes showed clear secondary structure and the corresponding 1,161,396 particles were combined, re-centered, and re-extracted into 288X288-pixel images. After 3D refinement and post-processing, the overall resolution of the map was 3.0 Å. Subsequent CTF refinement and Bayesian polishing further improved the map to a resolution of 2.8 Å. The local-resolution map was calculated in RELION-3.
Model building and refinement
The crystal structure of αIIbβ3 (PDB: 3FCS)22 and a homology model for abciximab generated with PIGS31 were rigid-body fitted into the cryo-EM density map in UCSF Chimera.32 Residues were manually fitted and adjusted in Coot.33 The built model was refined with phenix.real_space_refine,34 followed by minor manual adjustments. To validate the model, a cross-validation procedure was performed. The model was refined against half map 1. Then, FSC curves were calculate between the refined model and half map 1 (work), half map 2 (free), and the combined map.
Molecular dynamics simulations
Two sets of standard MD simulations were performed starting from the cryo-EM structure of the αIIbβ3–abciximab complex or the 2.55-Å resolution X-ray crystal structure of ligand-free αIIbβ3 integrin in its closed conformation (PDB: 3FCS).22 S For the latter, only the protein segments corresponding to those resolved by cryo-EM were simulated. Since the flexible loop corresponding to β3 amino-acid residues 75 to 78 (GDSS) was not resolved in either the cryo-EM or the crystal structure, it was built using a knowledge-based loop prediction approach within the Bioluminate module of the Schrödinger software package.35–37 Non-protein molecules, except all metal ions, were removed from the simulation setups of both the cryo-EM structure and the crystal structure. All water molecules that were present in the crystal structure were included in the MD simulation setup for this structure. To be able to compare between the two sets of simulations, crystallographic water molecules that coordinated the metal ions in the MIDAS, ADMIDAS, and SyMBS were added to the simulation setup of the cryo-EM structure after structural superimposition of all residues within 4 Å from the ions. After adding hydrogens, the crystal structure of αIIbβ3 and the cryo-EM structure of the αIIbβ3–abciximab complex were immersed in dodecahedral water boxes of TIP3P water38 such that there were at least 10 Å of water molecules on each side of the protein systems. The NaCl concentration was set to 150 mM. The protein was modeled using the CHARMM36m force field,39 with a 12-Å cutoff for non-bonded interactions. Long-range electrostatic interactions were calculated using the particle-mesh Ewald method.40 All bonds involving hydrogen atoms were fixed using the LINCS algorithm,41 and hydrogen masses were increased to 4 amu using a mass repartitioning scheme,42 allowing a time step of 4 fs. The crystallographic and cryo-EM systems were energy-minimized and equilibrated at a temperature of 300 K using the Nosé-Hoover thermostat43, 44 and at a pressure of 1 atm using the Parinello-Rahman barostat.45, 46 After equilibration, 4 independent MD simulations of 500 ns each were run for each system. MD setup and simulations were performed with the Gromacs 2018 software package,47 and the simulation trajectories were analyzed using cpptraj from AmberTools.48
Protein–protein fingerprint analysis
To characterize the type of inter-molecular interactions between the αIIbβ3 integrin protein and abciximab, we calculated interaction fingerprints between side-chain atoms other than hydrogen. A distance cutoff of 4.5 Å between non-polar atoms was used to identify non-polar interactions. Aromatic interactions were defined as those in which the distance between atoms within two aromatic rings was less than 4 Å and further classified by orientation depending on the angle formed by the rings. Those with an angle between rings of 0° were defined as purely face-to-face and those with an angle of 90° were defined as purely edge-to-face. Interaction with angles between 30° and 150° were defined as edge-to-face, and the remaining interactions were defined as face-to-face. Interactions were defined as electrostatic if the distance between two charged atoms on different residues was less than 4 Å. Finally, a hydrogen bond, either direct or water-mediated, was defined by a distance below 3.5 Å between donor and acceptor atoms and an angle of less than 135° between the three atoms forming the bond.
Calculations were performed on 4000 frames from the trajectories (two frames per ns of simulation) using an in-house python script. The average probabilities and errors for each inter-molecular interaction were estimated by counting the transitions from formed to broken (c10) and from broken to formed (c01) in the simulation trajectory, as well as the number of frames in which the interaction did not change its state (c00 and c11). The posterior distribution of the probabilities ϑ that any given contact was formed or broken was estimated from the likelihood
and a Dirichlet prior on the probabilities ϑ∼Dir(b), following Noé et al.28 For each interaction, the posterior of the contact fraction π was calculated from the posterior of the transition probabilities ϑ using the detailed-balance condition
Supplemental Figures VII and VIII report the median and 95% credible interval of these values.
In silico mutation analysis
To characterize the contribution of individual interactions between αIIbβ3 and abciximab to the stability of the complex, we estimated the effect of every possible mutation at each integrin residue involved in inter-molecular interaction by calculating the change in free energy of binding, ΔΔG(bind). The ΔΔG(bind) was calculated as:
where I and I′ represent the native and mutated integrin, respectively, A stands for abciximab, I ⋅ A and I′ ⋅ A refer to abciximab complexes with native and mutated integrin, respectively, and I + A and I′ + A denote isolated integrin (native or mutated) and abciximab. Using these definitions, negative values of ΔΔG(bind) indicate that the mutant integrin binds more strongly than the native integrin. Calculations were performed using the Molecular Mechanics-Generalized Born Surface Area (MM-GBSA) method using an implicit (i.e., continuum) solvation model as implemented in the Residue Scanning panel of the Biologics module of the Schrödinger software package.49–52 After each mutation, the conformations of the mutated residues, as well as those of any residue within 3 Å, were predicted using the Prime side-chain prediction method with backbone sampling followed by energy minimization. To account for protein flexibility in the free-energy estimation, 20 structures were obtained by clustering the combined MD simulation trajectories of the cryo-EM structure into 20 clusters and selecting the structure with the lowest average root-mean-square deviation (RMSD) from the other members in each cluster. The value of ΔΔG(bind) was calculated as an average over the individual values predicted for each representative structure weighted by the relative sizes of individual clusters from which the representative structures were chosen. The error was calculated as a standard error of the average over 20 representative frames. The significance of the change in binding free-energy was assessed for individual mutations using one sample t-test. Mutations that had a non-significant change (p>0.05) are indicated in Supplemental Figure IX. Normality of the data was confirmed using Shapiro-Wilk test,
Analysis of the fraction of time during which αIIbβ3 integrin residues are in contact with abciximab residues
The fraction of time in which side-chain atoms other than hydrogen of αIIbβ3 residues were within 4.5 Å of any side-chain atom other than hydrogen of abciximab was calculated with cpptraj, averaging the values over all simulation frames.
Effect of abciximab binding on the dynamics of select αIIbβ3 residues
To assess the impact of abciximab binding on the dynamics of select αIIbβ3 residues and the SyMBS, MIDAS, and ADMIDAS metal ions, we measured the root-mean-square fluctuations (RMSFs) of the αIIbβ3 residues and ions at the interface with abciximab, and compared them to the dynamics of equivalent residues in the crystal structure of αIIbβ3 alone (PDB: 2FCS). RMSFs were calculated for the four separate 500 ns MD simulations and then plotted as the mean ± SD. We focused our attention on residues that have contact fractions ≥0.5, including those in the SDL and residues involved in coordinating the SyMBS, MIDAS, and ADMIDAS metal ions. For comparison with SDL residues, we performed RMSF calculations on an αIIb loop (residues 118–128) and a β3 loop (residues 334–339) near the abciximab-binding interface, with αIIb Thr119 and Glu120 and β3 Met335 and Asp336 actually making contact with abciximab. The data were analyzed by comparing the results obtained for individual residues in both the abciximab-bound and free receptor using a two-tailed, Student’s t-test. Four RMSF values were calculated for individual residues in both the abciximab-bound and free receptor from each simulation replicate. Normality and uniformity of variance were not tested. The data were analyzed by comparing the results using a two-tailed, Student’s t-test. Residues for which p ≤ 0.05 were considered to have had their dynamics significantly affected by abciximab binding.
Site-directed mutagenesis
Mutant β3 Met335Asp and αllb Arg153Ser, Glu157Ala, and Asp159Val cDNA constructs were generated with the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer’s protocol. Wild-type full-length αllb cDNA in pEF/V5-His A and wild-type full-length β3 cDNA in the pcDNA3.1/Myc-His expression vector were used as the templates. Incorporation of mutations was confirmed by Sanger sequencing of plasmid DNA (Genewiz).
Cell culture and transfection
HEK 293 cells (for stable cell lines) and HEK 293T cells were grown in Dulbecco’s modified Eagle medium (DMEM) (GIBCO) supplemented with 10% fetal bovine serum. Cells were grown to 85–90% confluence and then co-transfected with an equal amount of wild-type or mutant full-length αlIb and wild-type or mutant full-length β3 cDNA using the Lipofectamine™ 2000 Transfection Reagent (Invitrogen). The stable cell lines expressing either wild-type or the β3 Met335Asp mutant integrin were generated by G418 selection. 48 hours post-transfection, G418 (0.8 mg/mL) was added to transfected cells and resistant cells grown under selection for 14 days, followed by sorting for αIIbβ3 expression using mAb 10E5. HEK293T cells transiently expressing αIIb mutant integrins were harvested 48 hours after transfection and studied immediately.
Flow cytometry
Wild-type and mutant cells were harvested with 0.05% trypsin-EDTA (Thermo Fisher Scientific), washed with Dulbecco’s phosphate-buffered saline (DPBS; Thermo Fisher Scientific) and resuspended in HEPES-buffered modified Tyrode’s buffer (HBMT; 10 mM HEPES, pH 7.4, 12 mM NaHCO3,138 mM NaCl, 2.7 mM KCl, 0.4 mM NaH2PO4, 0.35% bovine serum albumin, 0.1% glucose) with 2 mM Ca2+ and 1 mM Mg2+ at a density of 0.5X106 cells/50 μL. Alexa488-conjugated mAb 10E5, mAb 7E3, or abciximab at 10 μg/ml was added to a final concentration of 10–20 μg/ml and incubated for 30 minutes at room temperature. The cells were washed once, resuspended with 450 μL of HBMT containing 2 mM Ca2+ and 1 mM Mg2+, and analyzed by flow cytometry (Becton Dickinson FACSCalibur). Background antibody binding was determined by adding 25-fold excess unlabeled antibody. After the background was subtracted, the geometric mean fluorescence values obtained with mAb 7E3 or abciximab were divided by the value obtained with 10E5 to normalize for differences in αIIbβ3 expression. The data points for each of the three separate analyses that were performed for each sample are reported.
Data availability
The cryo-EM map has been deposited at the Electron Microscopy Data Bank under accession code EMD-21044 and the coordinates have been deposited at the Protein Data Bank under accession code 6V4P. MD simulation data (initial coordinates and topologies, as well as simulation trajectories) have been deposited within the Open Science Framework at https://osf.io/jqcxk (DOI: 10.17605/OSF.IO/JQCXK).
Statistical analysis
Statistical analyses are described in the experimental methods that employed statistical analysis.
RESULTS
Cryo-EM structure of the αIIbβ3–abciximab complex
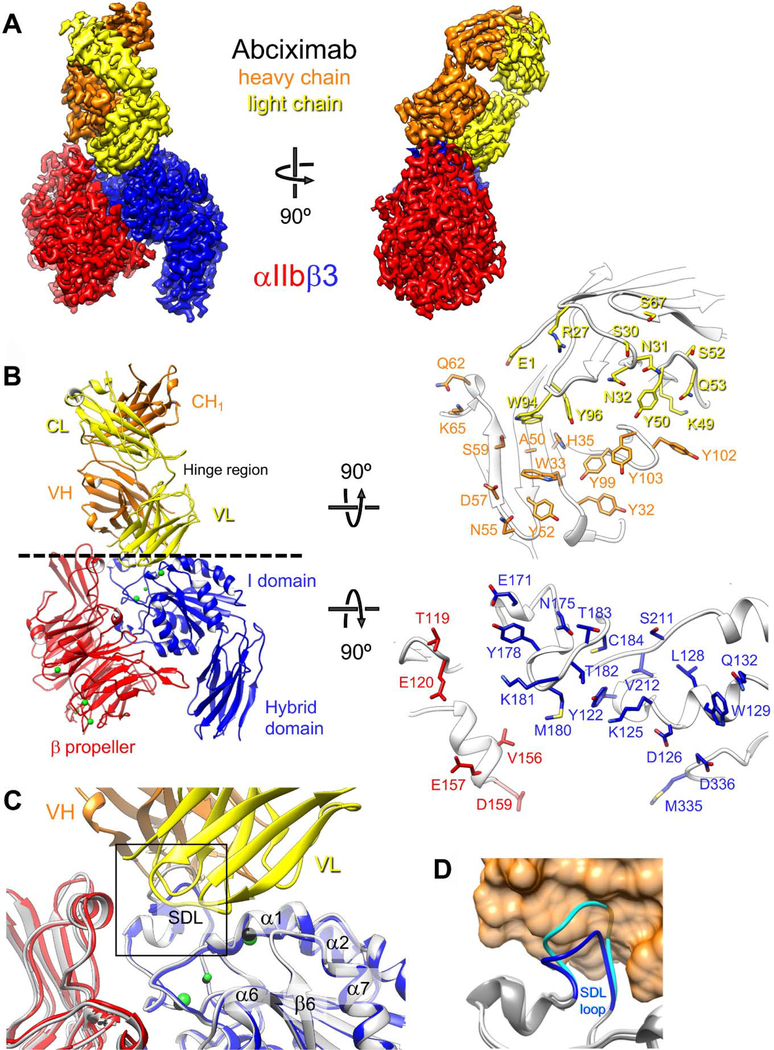
We first prepared a complex of abciximab with human αIIbβ3 ectodomain (αIIb residues 1–963, β3 residues 1–690; stabilized throughout the analysis by a C-terminal coiled coil and disulfide bond) (Supplemental Figure IA) and assessed the quality of the complex by negative-stain EM. Raw images showed a mono-dispersed particle population (Supplemental Figure IB) and 2D-class averages revealed αIIbβ3 in the closed conformation with a Fab fragment extending from the head domain (Supplemental Figure IC). The sample was vitrified and imaged using a K2 Summit direct electron detector on a Titan Krios electron microscope. Image processing (Supplemental Figure II) resulted in a density map of the abciximab-bound αIIbβ3 headpiece at 2.8-Å resolution (Figure 1A and Supplemental Figure III), which allowed us to build an atomic model for αIIb residues 1–451, comprising the β-propeller domain, β3 residues 58–432, comprising the βI and hybrid domains, and abciximab residues 1–221 from the heavy chain (H) and residues 1–214 from the light chain (L) (Figure 1B, left panel). The image processing and model refinement statistics are summarized in Supplemental Table I. A small density occupied the fibrinogen/RGD-binding site but could not be modeled. Alignment of our model of the abciximab-bound αIIbβ3 headpiece with the corresponding residues in the X-ray structure of the unliganded αIIbβ3 ectodomain22 yielded a root-mean-square deviation (RMSD) between equivalent Cα atoms of 0.7 Å. In particular, abciximab binding does not appear to affect the fibrinogen-binding pocket (Figure 1C). The one notable difference between the unliganded and the abciximab-bound αIIbβ3 headpiece is the conformation of the β3 subunit’s SDL, which is compressed upon abciximab binding (Figure 1D), with displacement of Lys181 by 3.4 Å, Met180 by 3.0 Å, and Thr182 by 2.4 Å to prevent steric clashes, mostly of Lys181(β3) with His35(H), and Tyr178(β3) with Trp33(H), Tyr52(H), and Asn55(H).
Figure 1.
Cryo-EM structure of the αIIbβ3–abciximab complex. (a) Density map of the αIIbβ3–abciximab complex at 2.8-Å resolution, segmented and colored according to the individual polypeptide chains. (b) Left panel: Atomic model of the αIIbβ3–abciximab complex colored as in panel (a). The green spheres represent the metal ions in the MIDAS, ADMIDAS and SyMBS. VH: variable domain of the heavy chain; CH1: first constant domain of the heavy chain; VL: variable domain of the light chain; CL: constant domain of the light chain. Hinge region denotes the flexible linkers that connect the variable and constant domains in abciximab. Right panel: Interaction surfaces of abciximab (top) and αIIbβ3 (bottom). Side chains of residues that are within 4.5 Å from a side chain of the interacting protein are shown in stick representation. (c) Fibrinogen/RGD-binding pocket in the cryo-EM structure of the αIIbβ3–abciximab complex (colored as in panel (a)) and in the X-ray crystal structure of the unliganded αIIbβ3 integrin (light grey with metal ions shown as black spheres). The overlay shows that abciximab binding does not induce meaningful differences in the binding pocket. The boxed area is shown in panel d. (d) Conformation of the β3 SDL loop in the cryo-EM structure of the αIIbβ3–abciximab complex (dark grey with SDL in blue) and in the X-ray crystal structure of the unliganded αIIbβ3 integrin (light grey with SDL in cyan). Abciximab is shown as an orange transparent surface to illustrate that the SDL in the conformation seen in the X-ray crystal structure of the unliganded αIIbβ3 integrin (cyan) would clash with abciximab.
The interacting surface of abciximab derived from the model and investigated by MD simulations was comprised of contributions from residues from all three complementarity-determining regions (CDRs) of both the light and heavy chain variable domains, with high representation of aromatic residues (light chain Tyr50, Trpy94, and Tyr96; heavy chain Tyr32, Trp33, Tyr52, Tyr99, Tyr102, and Tyr103) (Figure 1B, right panel). As expected from prior mutagenesis studies, the interacting β3 residues include many residues from the Cys177-Cys184 SDL (Cys177, Tyr178, Met180, Lys181, Thr182, Thr183, and Cys184) and neighboring residues (Glu171 and Asn175), as well as residues on the β1-α1 loop that are adjacent to and/or contribute to the MIDAS and ADMIDAS (Tyr122, Lys125, Asp126, Trp129, and Gln132). Also represented are Ser211 and Val212, and Met335 and Asp336. The Met335 carbonyl contributes to coordinating the ADMIDAS metal ion and this bond is broken when ligand binds to the MIDAS, freeing the α7 helix to initiate its piston-like motion, which is responsible for the dramatic swing-out motion of the hybrid domain away from αIIb that places the receptor in the high-affinity ligand-binding conformation.53 Unexpectedly, two regions of αIIb (Thr119, Glu120 and Val156, Glu157, Asp159) were identified as being within 4.5 Å of an abciximab side-chain atom other than hydrogen. Overall, the interaction between abciximab and αIIbβ3 buried a total of more than 1,200 Å2 of solvent-exposed surface, 218 Å2 with αIIb and 1,040 Å2 with β3.
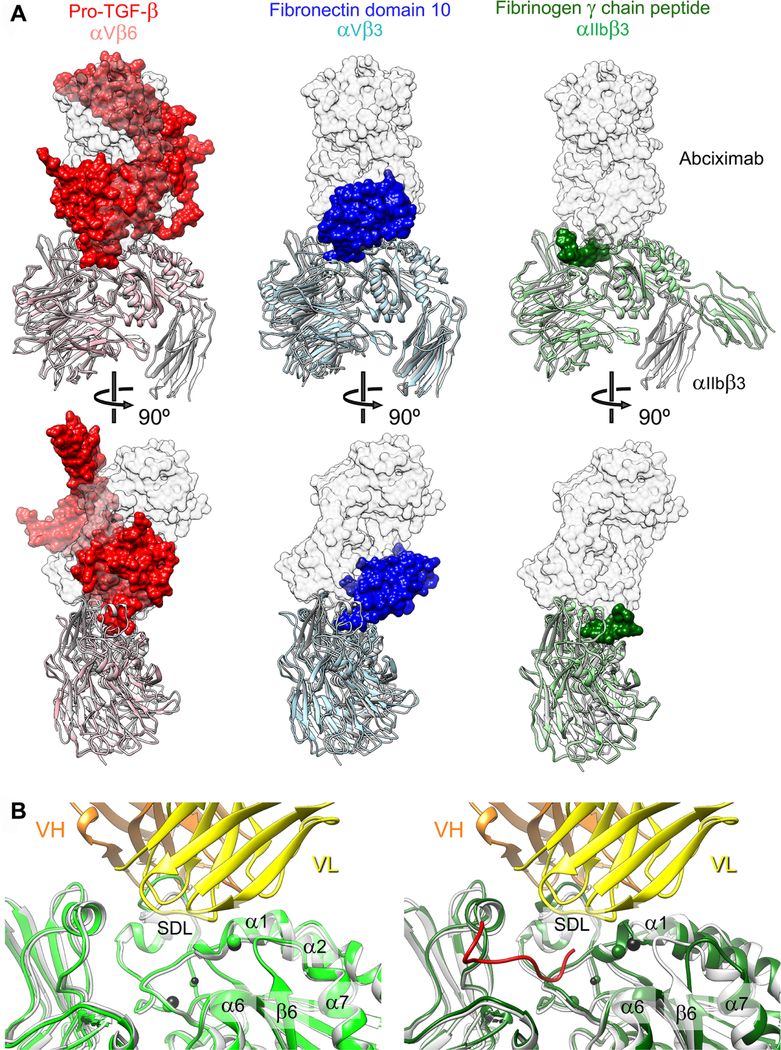
Superimposition of our structure of the αIIbβ3–abciximab complex with structures of other αIIbβ3 and αV integrin–ligand complexes strongly suggests that abciximab bound to the integrin would sterically prevent the binding of physiological ligands to these receptors (Figure 2). The structure of the short fibrinogen γ-chain peptide bound to αIIbβ3 (PDB: 2VDO)54 does not show substantial steric clashes with abciximab (Figure 2), which is consistent with our previous data demonstrating that a small RGD peptide does not prevent the binding of mAb 7E3.55 Nonetheless, by analogy, both mAb and abciximab would likely interfere with the binding of the entire fibrinogen molecule as judged from overlays of our αIIbβ3–abciximab complex structure with structures of αVβ3 in complex with fibronectin domain 10 (PDB: 4MMX)56 and αVβ6 in complex with pro-TGF-β (PDB: 5FFO).19, 57
Figure 2.
Abciximab prevents ligands from access to the integrin ligand-binding pocket. (a) Shown are superimpositions of our structure of the αIIbβ3–abciximab complex (αIIbβ3 in grey ribbon representation and abciximab as transparent grey surface) with other ligand–integrin complexes, which are αVβ6 (salmon) in complex with pro-TGF-β (red) (PDB: 5FFO)38, αVβ3 (light blue) in complex with the tenth domain of fibronectin (dark green) (PDB: 4MMX)37, and αIIbβ3 (light green) in complex with a peptide of the fibrinogen γ-chain (blue) (PDB: 2VDO)35. The overlays show that ligand binding would cause sterical clashes with the bound abciximab. (b) Ligand binding has little effect on the abciximab-binding region of αIIbβ3. Left panel: Superimposition of our αIIbβ3–abciximab complex structure (αIIbβ3 shown in light grey and abciximab in yellow and orange) with the crystal structure of unbound αIIbβ3 (light green) (PDB: 3FCS)22 shows that abciximab does not affect the ligand-binding pocket. Right panel: Superimposition of our αIIbβ3–abciximab complex structure (αIIbβ3 shown in light grey and abciximab in yellow and orange) with the crystal structure of αIIbβ3 (dark green) in complex with a peptide of the fibrinogen γ-chain (red) (PDB: 2VDO)35 shows that peptide binding induces only subtle changes to the abciximab-interacting region of αIIbβ3.
MD simulations
MD simulations were used to investigate the dynamics of the αIIbβ3–abciximab complex, as well as the stability of the inter-molecular interactions seen in the cryo-EM structure. In addition, to evaluate whether bound abciximab affects the integrin’s ability to bind ligand, the αIIbβ3 headpiece was also simulated in the absence of abciximab using as the starting structure the X-ray crystal structure of unliganded αIIbβ3 ectodomain in its closed conformation (PDB: 3FCS).22 To facilitate comparison of the ligand-binding pockets in the simulated cryo-EM and X-ray crystal structures, both simulation setups included the crystallographic water molecules that coordinate the metal ions in the MIDAS, ADMIDAS, and SyMBS. The mass-weighted RMSDs of the structures during the simulations (4X500 ns for each system) from the starting structures showed that the αIIbβ3–abciximab complex as a whole is more flexible than the unliganded integrin (Supplemental Figures IVA and VA). The RMSD values of the individual αIIb and β3 integrin subunits in both sets of simulations (Supplemental Figures IVB, IVC, VB, and VC) and of the variable regions of the abciximab light and heavy chains (Supplemental Figures IVF and IVG) all remained below 2.5 Å. Similarly, the RMSD values of the constant regions of the light and heavy chains of abciximab remained stable around 2.5 Å and 3 Å (Supplemental Figures IVH and IVI). However, the entire light and heavy chains of abciximab showed greater flexibility (Supplemental Figures IVD, and IVE). Therefore, the major contribution to the greater flexibility of the αIIbβ3–abciximab complex compared to the unliganded αIIbβ3 headpiece results from the high flexibility of the hinge regions that connect the constant and variable domains in abciximab.
Notably, the overall structural conformation of the fibrinogen/RGD-binding pocket was similar between the cryo-EM structure of the αIIbβ3–abciximab complex and the X-ray structure of unbound αIIbβ3 as assessed by the all-atom mass-weighted RMSD over time of all residues that have atoms other than hydrogen within 4 Å of the MIDAS, ADMIDAS, or SyMBS metal ions in at least 10% of the MD frames (Supplemental Figure VI).
The nature of the inter-molecular interactions maintained between αIIbβ3 and abciximab residues during simulation was assessed by fingerprint analysis, and the results are shown in Supplemental Figure VII for αIIbβ3 residues and in Supplemental Figure VIII for abciximab residues. The figures also depict the fraction of time a given residue is involved in an intermolecular interaction. The analysis led to the identification of a number of interactions between the β3 integrin subunit and the abciximab heavy or light chains that were maintained during most of the simulated time (contact fraction greater than 0.70; residue labels with dark grey background in Supplemental Figure VII). The involvement of several of these β3 residues (e.g., Lys125, Trp129, and Thr182) in 7E3 binding was previously suggested by mutagenesis studies.19, 58 Fingerprint analysis also indicated that the αIIb subunit participates only weakly in the interaction with abciximab, with only the Asp159(αIIb)–Arg27(L) contact maintained for more than half of the simulation time (contact fraction: 0.64).
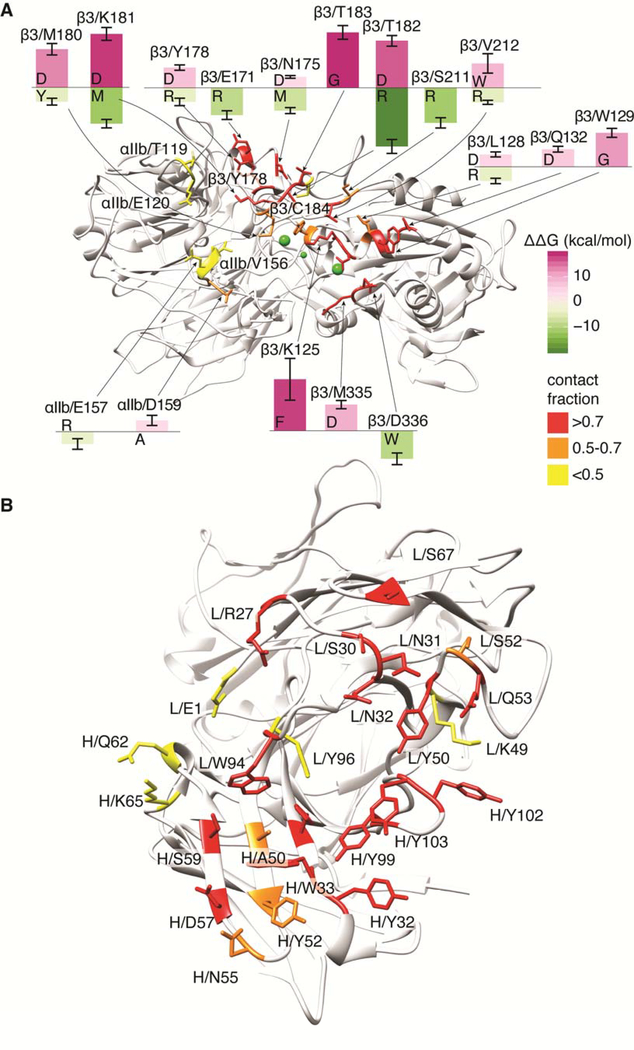
Predictions of mutations that would eventually impact the stability of the αIIbβ3–abciximab complex are shown in Supplemental Figure IX. Specifically, the figure reports mutations with calculated free-energy differences from wild type (ΔΔG(bind)) larger than 3.5 kcal/mol. Figure 3 illustrates the location of αIIbβ3 and abciximab residues involved in intermolecular contacts in at least 10% of the MD simulation frames (see also Supplemental Figures VII and VIII), and visually summarizes the predicted top stabilizing (in green) and destabilizing (in purple) mutations of αIIbβ3 residues (see also Supplemental Figures IX). Mutants predicted to most destabilize the αIIbβ3–abciximab complex (i.e., with the largest positive ΔΔG(bind) values) that had not been previously tested by mutagenesis were considered for experimental testing. Since the interaction of abciximab with the αIIb residues was most unexpected, and the interaction with Met335 in β3 has potential implications for abciximab’s effects on the swing-out of the β3 subunit,53 we focused our attention on these residues.
Figure 3.
Visualization of residues in (a) αIIbβ3 and (b) abciximab involved in inter-molecular contacts in at least 10% of the MD-simulation frames (see also Supplemental Figures VII and VIII). Insets in panel a refer to residues with calculated free-energy differences (ΔΔG(bind)) from wild type that are larger than 3.5 kcal/mol for top stabilizing (in green) and destabilizing (in purple) mutations of αIIbβ3 residues (see also Supplemental Figure IX).
Mutations
To assess the contribution of different αIIb residues to abciximab binding, we focused on Arg153, Glu157, and Asp159. Although Arg153’s interactions did not fulfill our criteria for being at the interface with abciximab, the mutation modeling analysis predicted that an Arg153Ser mutation would create a relatively large positive change in free energy (4.9 ± 2.1 kcal/mol), suggesting that it may affect the binding. Glu157 did meet our criteria for being at the abciximab interface, and although no substitutions of this residue were predicted to produce an increase in free energy ≥3.5 kcal/mol, we selected the Glu157Val mutation because it introduces a non-charged residue that would disrupt the salt bridge of Glu157 with Arg27 in the abciximab light chain. Asp159 had the highest contact fraction among the αIIb residues at the abciximab interface and the Asp159Ala mutation was predicted to produce the largest positive change in free energy (3.7 ± 1.7 kcal/mol).
To adjust for differences in expression levels, we normalized the binding for αIIbβ3 expression as judged by the binding of mAb 10E5. Neither individual αIIb mutant Glu157Val nor Asp159Ala reduced either mAb 7E3 or abciximab binding compared to binding to wild-type αIIbβ3 (Supplemental Figure X). Binding was also unaffected by combining the Arg153Ser and Asp159Ala mutations or by combining the Arg153Ser, Glu157Val, and Asp159Ala mutations. Mutating β3 residue Met335 to aspartic acid, the analogous murine residue and the mutation predicted to most affect the free energy of binding, also did not affect the binding of either mAb 7E3 or abciximab.
Effect of abciximab binding on the dynamics of select αIIbβ3 residues
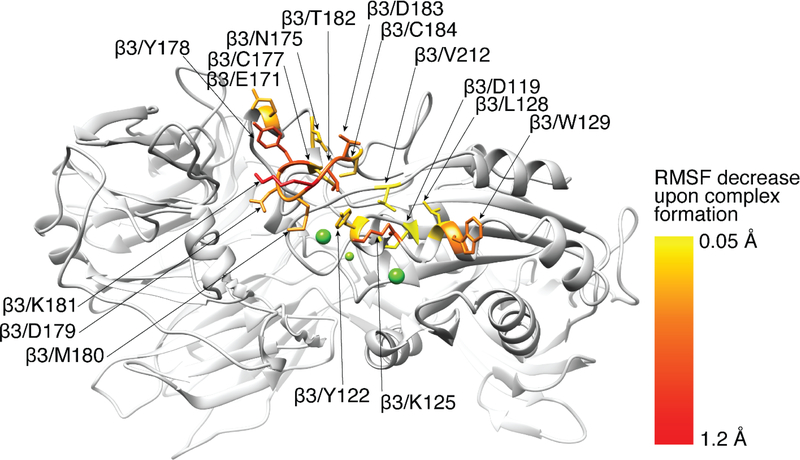
Residues in the αIIb 118–128 and β3 334–339 loops in the absence of abciximab exhibited dynamic motions (measured as RMSFs) that ranged from 0.4 to 2.2 Å (Supplemental Figure XI). Binding of abciximab resulted in an overall trend of reducing the RMSF values during simulation. Figure 4 depicts αIIbβ3 residues whose fluctuations are most affected by complex formation with abciximab. Although binding of abciximab resulted in an overall reduction of the RMSF values of the αIIb 118–128 and β3 334–339 loops, none of the changes were statistically significant. The residues coordinating the metal ions had uniformly lower fluctuations compared to the loops (0.4–0.7 Å) with the exception of Met335 (~1.3 Å) (Supplemental Figure XI). Abciximab binding did not significantly alter the dynamics of any of these residues except for Asp119, whose fluctuation reduction was small but statistically significant (ΔRMSF=0.06 ± 0.03, with p=0.03). The metal ions also had low RMSF values and abciximab binding did not alter them. In sharp contrast, all eight SDL residues, which demonstrated a range of fluctuations similar to those of the αIIb and β3 loops, became significantly less mobile in the presence of abciximab. Among the residues with contact fractions ≥0.5, αIIb Glu157 and Asp159 had relatively large fluctuations (RMSF ~2 Å), both in the absence and presence of abciximab; β3 Lys125 and Trp129 showed the most dramatic reductions in fluctuations with abciximab binding (ΔRMSF = 0.74 ± 0.08 Å with p<0.001 and 0.52 ± 0.14 Å with p<0.001, respectively); residues near the SDL (Glu171,Tyr122, and Asn175) had intermediate reductions in fluctuations (ΔRMSF = 0.38 ± 0.19 with p=0.01, 0.27 ± 0.12 with p=0.01, and 0.25 ± 0.09 with p=0.001, respectively); Leu128 and Val212 had minor reductions in fluctuations (0.16 ± 0.08 with p<0.01 and 0.13 ± 0.07 with p=0.02, respectively); and the fluctuations of Asp126, Gln132, and Ser211 were unaffected.
Figure 4.
Residues of αIIbβ3 whose fluctuations are most affected by complex formation with abciximab are shown in stick representation and color-coded based on the change in RMSF. The RMSF changes are statistically significant in all of the residues depicted in stick representation (see also Supplemental Figure XI).
DISCUSSION
Our high-resolution cryo-EM structure of the αIIbβ3–abciximab complex provides important new insights into how abciximab inhibits ligand binding to αIIbβ3, including unexpected contacts with several αIIb residues, a lack of effect on the fibrinogen/RGD-binding pocket, and effects on the dynamics of the β3 SDL. The results of the current study demonstrate that abciximab binds primarily to a discontinuous epitope on the β3 integrin subunit that includes the Cys177-Cys184 segment in the SDL and the helical region between Tyr122 and Trp129 that contributes to both the MIDAS and ADMIDAS. These data are in full accord with previous studies by others18, 20 and our own studies19 of the binding of mAb 7E3 based on site-directed mutations, as well as our localization of the mAb 7E3 epitope to the αIIbβ3 head domain by negative-stain EM.21
The interactions of abciximab with several αIIb residues was surprising because the above site-directed mutagenesis studies demonstrated that swapping the murine for human Cys177-Cys184 sequence within the SDL essentially eliminates mAb 7E3 binding, and substituting the two murine β3 residues Trp129 and Asn 133 for their human counterparts (Ser and Thr, respectively), results in partial loss of mAb 7E3 binding.18 Furthermore, these same investigators did not find a reduction in mAb 7E3 binding to nine different mouse-human αIIb hybrid receptors, including one (Val156-Trp162) that included three of the residues (Val156, Glu157, and Asp159) that were within 4.5 Å from abciximab residues in the cryo-EM structure. To gain a better understanding of the strength of the interactions between αIIb and abciximab we performed MD simulation studies and found that the majority of the interactions involving the αIIb subunit were lost during most of the simulated time, with only the Asp159(αIIb)–Arg27(L) contact remaining formed for slightly more than half of the time, suggesting that these interactions are relatively weak. To further explore their contribution to abciximab binding, and based on predictions of changes in free energy with different mutations, we made individual αIIb Glu157Val and Asp159Ala substitutions and combined substitutions of αIIb Arg153Ser + Asp159Ala and Arg153Ser + Glu157Val + Asp159Ala, and found that none of these reduced abciximab binding when normalized for αIIbβ3 expression.
The other two αIIb residues identified in the cryo-EM structure as interacting with abciximab (Thr119 and Glu120) had very low contact fractions during the MD simulations and so were not further investigated. Taken together, our results suggest that while abciximab makes some contact with αIIb, these interactions do not make a major contribution to abciximab binding. This conclusion is consistent with previous studies by others and our group demonstrating that 7E3 and abciximab also react with αVβ315, 16, 59 and that 7E3 does not hold the αIIbβ3 receptor together when exposed to EDTA under conditions that dissociate αIIb from β3.11
Our atomic-level structure and MD simulations indicate that while abciximab interacts directly with β3 Asp126, which helps coordinate the ADMIDAS metal ion, it does not alter the fibrinogen/RGD-binding site itself, nor does its binding adjacent to the fibrinogen/RGD-binding site affect the dynamics of the residues that coordinate the three metal ions or the movement of the metal ions themselves. Although we cannot formally exclude the possibility that abciximab might cause allosteric modulation of the fibrinogen/RGD-binding pocket over a longer simulation time scale or different simulation conditions, the lack of an effect on the fibrinogen/RGD binding pocket itself is consistent with our previous finding that a small RGD peptide does not inhibit mAb 7E3 binding to αIIbβ355 and the lack of a major impact of the binding of the small fibrinogen γ-chain peptide on the residues involved in binding abciximab.54 The inferred sterical clashes between abciximab and the larger αVβ3 ligand derived from fibronectin based on the overlap of the crystal structures, as well as the inferred clashes based on the crystal structure of the proTGF-β1-αVβ6 complex, support the hypothesis that abciximab probably primarily inhibits fibrinogen binding by sterical interference.
Our structure also showed that abciximab binds to β3 Tyr122, Lys125, and Thr182, residues that link the MIDAS domain to the SDL.19, 57 Our previous mutagenesis studies suggested that mutating Thr182 to Asn results in a conformational rearrangement of the SDL, with 7E3 only reacting with one of the possible conformations.19 Since the Thr182Asn mutation is predicted to only produce a small increase in free energy (1.9 ± 0.8 kcal/mol), it is likely that the effect of the Thr182Asn mutation on 7E3 binding is caused by a conformational rearrangement of the SDL, although we cannot exclude the possibility of a contribution from the loss of the direct interaction between 7E3 and Thr182.
We previously reported that mutating Lys125 to either Ala or Arg does not alter 7E3 binding whereas the Lys125Gln mutation reduces binding by 50%.19 Of note, in our current studies the Lys125Gln mutation was predicted to increase the change in free energy the most among the mutations (10.2 ± 1.3 kcal/mol vs 7.6 ± 0,8 kcal/mol for the Ala and −1.4 ± 2.4 kcal/mol for the Arg mutations), and its effect may also be due, in part, to its disrupting the link between the α1 helix and the SDL and/or producing a conformational change in the α1 helix (residues 125–127).19
Our data also demonstrate that in addition to binding to the SDL, abciximab distorts the SDL and limits its dynamics. None of the residues in the fibrinogen γ-chain sequence that binds to the fibrinogen/RGD pocket (γ−404–411) interact directly with the SDL,54 but none the less, the SDL may play an important role in defining αIIbβ3 ligand-binding specificity. For example, Takagi et al.60 found that swapping the β3 SDL (Cys177-Cys184) into β1 and then expressing the mutant in combination with αV converted the ligand-binding specificity such that the mutant could bind fibrinogen, von Willebrand factor, vitronectin, and fibronectin, whereas αVβ1 only bound fibronectin. In addition, reverse swapping of the β1 SDL into β3 resulted in a marked reduction in αVβ3 binding to fibrinogen, vitronectin, and von Willebrand factor. The SDLs of other integrin receptors have also been reported to affect ligand binding, including β261 and β4.62 Moreover, the crystal structure of αVβ6 in complex with a pro-TGF-β3 peptide57 demonstrates direct binding of the peptide to the β6 SDL. Additional support for the importance of the connection between the SDL and the fibrinogen/RGD-binding pocket comes from analysis of the two Glanzmann thrombasthenia patients with different missense mutations of β3 Arg214 that did not cause major reductions in expression.63, 64 Arg214 does not interact with any of the γ−404–411 residues, but it does link the SDL to the α2-α3 loop by a salt bridge with Asp179. In both cases, the receptors retained some ability to bind RGD peptides, but not fibrinogen or mAb PAC1, which has an RYD sequence in its variable domain and selectively binds to activated αIIbβ3.65 Thus, collectively, we deduce that abciximab prevents ligand binding by steric interference, with a potential contribution via alteration of the SDL.
Supplementary Material
HIGHLIGHTS.
Abciximab was the first approved αIIbβ3 antagonist and has been administered to millions of patients undergoing percutaneous coronary interventions.
Cryo-electron microscopy of the abciximab-αIIbβ3 complex at 2.8-Å resolution defines abciximab’s epitope and mode of binding.
Abciximab binds adjacent to the Arg-Gly-Asp (RGD) ligand-binding site, does not disrupt the metal ion dependent adhesion site (MIDAS), but does compress and restrict the mobility of the β3 specificity determining loop (SDL).
Abciximab most likely prevents ligand binding by steric interference, with a potential contribution from its effects on the SDL.
ACKNOWLEDGMENTS
We thank Mark Ebrahim and Johana Sotiris at the Evelyn Gruss Lipper Cryo-EM Resource Center of The Rockefeller University for assistance with data collection. Computations were run on resources available through the Scientific Computing Facility at the Icahn School of Medicine at Mount Sinai and the Extreme Science and Engineering Discovery Environment under MCB080077 (to MF), which is supported by National Science Foundation grant number ACI-1053575.
SOURCES OF FUNDING
Supported in part by grant HL19278 from the National Heart, Lung and Blood Institute and grant UL1 TR001866 from the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program, National Institutes of Health; and funds from Stony Brook University.
NONSTANDARD ABBREVIATIONS AND ACRONYMS
- ADMIDAS
adjacent to metal ion-dependent adhesion site
- CDR
complementarity-determining region
- CI
confidence interval
- Cryo-EM
cryo-electron microscopy
- CTF
contrast transfer function
- DMEM
Dulbecco’s modified Eagle medium
- DPBS
Dulbecco’s phosphate-buffered saline
- EM
electron microscopy
- FDA
Food and Drug Administration
- H
heavy chain
- HBMT
HEPES-buffered modified Tyrode’s buffer
- mAb
monoclonal antibody
- L
light chain
- MIDAS
metal ion-dependent adhesion site
- MD
molecular dynamics
- OR
odds ratio
- RGD
Arg-Gly-Asp
- RMSD
root-mean-square deviation
- RMSF
root-mean-square fluctuations
- SDL
specificity-determining loop
- SyMBS
synergistic metal-binding site
Footnotes
DISCLOSURE OF CONFLICTS OF INTEREST
In accord with Federal law and the policies of the Research Foundation of the State University of New York, B.S. Coller has royalty interests in abciximab (Centocor).
References
- 1.Coller BS, Scudder LE, Beer J, Gold HK, Folts JD, Cavagnaro J, Jordan R, Wagner C, Iuliucci J, Knight D, Ghrayeb J, Smith C, Weisman HF and Berger H. Monoclonal antibodies to platelet glycoprotein IIb/IIIa as antithrombotic agents. Annals of the New York Academy of Sciences. 1991;614:193–213. [DOI] [PubMed] [Google Scholar]
- 2.Coller BS and Shattil SJ. The GPIIb/IIIa (integrin αIIbβ3) odyssey: a technology-driven saga of a receptor with twists, turns, and even a bend. Blood. 2008;112:3011–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.George JN, Caen JP and Nurden AT. Glanzmann’s thrombasthenia: The spectrum of clinical disease. Blood. 1990;75:1383–1395. [PubMed] [Google Scholar]
- 4.Coller BS. A new murine monoclonal antibody reports an activation-dependent change in the conformation and/or microenvironment of the platelet glycoprotein IIb/IIIa complex. The Journal of clinical investigation. 1985;76:101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coller BS. GPIIb/IIIa antagonists: Pathophysiologic and therapeutic insights from studies of c7E3 Fab. Thrombosis and haemostasis. 1997;26:285–293. [PubMed] [Google Scholar]
- 6.Taylor FB, Coller BS, Chang ACK, Peer G, Jordan R, Engellener W and Esmon CT. 7E3 F(ab’) 2, a monoclonal antibody to the platelet GPIIb/IIIa receptor, protects against microangiopathic hemolytic anemia and microvascular thrombotic renal failure in baboons treated with C4b binding protein and a sublethal infusion of E scherichia coli Blood. 1997;89:4078–4084. [PubMed] [Google Scholar]
- 7.Coller BS, Folts JD, Scudder LE and Smith SR. Antithrombotic effect of a monoclonal antibody to the platelet glycoprotein IIb/IIIa receptor in an experimental animal model. Blood. 1986;68:783–786. [PubMed] [Google Scholar]
- 8.Coller BS, Folts JD, Smith SR, Scudder LE and Jordan R. Abolition of in vivo platelet thrombus formation in primates with monoclonal antibodies to the platelet GPIIb/IIIa receptor: Correlation with bleeding time, platelet aggregation and blockade of GPIIb/IIIa receptors. Circulation. 1989;80:1766–1774. [DOI] [PubMed] [Google Scholar]
- 9.Bosch X, Marrugat J and Sanchis J. Platelet glycoprotein IIb/IIIa blockers during percutaneous coronary intervention and as the initial medical treatment of non-ST segment elevation acute coronary syndromes. Cochrane Database Syst Rev. 2013;11:CD002130. [DOI] [PubMed] [Google Scholar]
- 10.Coller BS. Activation affects access to the platelet receptor for adhesive glycoproteins. JCell Biol. 1986;103:451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coller BS, Peerschke EI, Seligsohn U, Scudder LE, Nurden AT and Rosa JP. Studies on the binding of an alloimmune and two murine monoclonal antibodies to the platelet glycoprotein IIb-IIIa complex receptor. J Lab Clin Med. 1986;107:384–392. [PubMed] [Google Scholar]
- 12.Coller BS and Scudder LE. Inhibition of dog platelet function by in vivo infusion of F(ab) 2 fragments of a monoclonal antibody. Blood. 1985;66:1456–1459. [PubMed] [Google Scholar]
- 13.Gold HK, Coller BS, Yasuda T, Saito T, Fallon JT, Guerrero JL, Leinbach RC, Ziskind AA and Collen D. Rapid and sustained coronary artery recanalization with combined bolus injection of recombinant tissue-type plasminogen activator and monoclonal anti-platelet GPIIb/IIIa antibody in a dog model. Circulation. 1988;77:670–677. [DOI] [PubMed] [Google Scholar]
- 14.Yasuda T, Gold HK, Fallon JT, Leinbach RC, Guerrero JL, Scudder LE, Kanke M, Shealy D, Ross MJ, Collen D and Coller BS. Monoclonal antibody against the platelet GPIIb/IIIa receptor prevents coronary artery reocclusion following reperfusion with recombinant tissue-type plasminogen activator in dogs. Journal of Clinical Investigation. 1988;81:1284–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charo IF, Bekeart LS and Phillips DR. Platelet glycoprotein IIb-IIIa-like proteins mediate endothelial cell attachment to adhesive proteins and the extracellular matrix. The Journal of biological chemistry. 1987;262:9935–9938. [PubMed] [Google Scholar]
- 16.Tam SH, Sassoli PM, Jordan RE and Nakada MT. Abciximab (ReoPro, chimeric 7E3 Fab) demonstrates equivalent affinity and functional blockade of glycoprotein IIb/IIIa and alpha(v)beta3 integrins. Circulation. 1998;98:1085–1091. [DOI] [PubMed] [Google Scholar]
- 17.Coller BS, Cheresh DA, Asch E and Seligsohn U. Platelet vitronectin receptor expression differentiates Iraqi-Jewish from Arab Patients with Glanzmann thrombasthenia in Israel. Blood. 1991;77:75–83. [PubMed] [Google Scholar]
- 18.Puzon-McLaughlin W, Kamata T and Takada Y. Multiple discontinuous ligand-mimetic antibody binding sites define a ligand binding pocket in integrin alpha(IIb)beta(3). J Biol Chem. 2000;275:7795–7802. [DOI] [PubMed] [Google Scholar]
- 19.Artoni A, Li J, Mitchell B, Ruan J, Takagi J, Springer TA, French DL and Coller BS. Integrin β3 regions controlling binding of murine mAb 7E3: implications for the mechanism of integrin αIIbβ3 activation. Proc Natl Acad Sci U S A. 2004;101:13114–13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takagi J, DeBottis DP, Erickson HP and Springer TA. The role of the specificity-determining loop of the integrin beta subunit I-like domain in autonomous expression, association with the alpha subunit, and ligand binding. Biochemistry. 2002;41:4339–4347. [DOI] [PubMed] [Google Scholar]
- 21.Choi WS, Rice WJ, Stokes DL and Coller BS. Three-dimensional reconstruction of intact human integrin alphaIIbbeta3: new implications for activation-dependent ligand binding. Blood. 2013;122:4165–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu J, Luo BH, Xiao T, Zhang C, Nishida N and Springer TA. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol Cell. 2008;32:849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma B and Johnson R. De novo sequencing and homology searching. Molecular & cellular proteomics : MCP. 2012;11:O111.014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma B Novor: real-time peptide de novo sequencing software. Journal of the American Society for Mass Spectrometry. 2015;26:1885–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohi M, Li Y, Cheng Y and Walz T. Negative staining and image classification - powerful tools in modern electron microscopy. Biological procedures online. 2004;6:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M and Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–9. [DOI] [PubMed] [Google Scholar]
- 27.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. [DOI] [PubMed] [Google Scholar]
- 28.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y and Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature methods. 2017;14:331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohou A and Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E and Scheres SH. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcatili P, Rosi A and Tramontano A. PIGS: automatic prediction of antibody structures. Bioinformatics (Oxford, England). 2008;24:1953–4. [DOI] [PubMed] [Google Scholar]
- 32.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC and Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–12. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Lohkamp B, Scott WG and Cowtan K. Features and development of Coot. Acta crystallographica Section D, Biological crystallography. 2010;66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC and Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66:213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu K, Day T, Warshaviak D, Murrett C, Friesner R and Pearlman D. Antibody structure determination using a combination of homology modeling, energy-based refinement, and loop prediction. Proteins. 2014;82:1646–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salam NK, Adzhigirey M, Sherman W and Pearlman DA. Structure-based approach to the prediction of disulfide bonds in proteins. Protein Eng Des Sel. 2014;27:365–374. [DOI] [PubMed] [Google Scholar]
- 37.Beard H, Cholleti A, Pearlman D, Sherman W and Loving KA. Applying Physics-Based Scoring to Calculate Free Energies of Binding for Single Amino Acid Mutations in Protein-Protein Complexes. Plos One. 2013;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW and Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 39.Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, de Groot BL, Grubmuller H and MacKerell AD Jr. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods. 2017;14:71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darden T, York D and Pedersen L. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 41.Hess B, Bekker H, Berendsen HJC and Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry. 1997;18:1463–1472. [Google Scholar]
- 42.Feenstra KA, Hess B and Berendsen HJC. Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. Journal of Computational Chemistry. 1999;20:786–798. [DOI] [PubMed] [Google Scholar]
- 43.Nose S A Molecular-Dynamics Method for Simulations in the Canonical Ensemble. Mol Phys. 1984;52:255–268. [Google Scholar]
- 44.Hoover WG. Canonical Dynamics - Equilibrium Phase-Space Distributions. Phys Rev A. 1985;31:1695–1697. [DOI] [PubMed] [Google Scholar]
- 45.Parrinello M and Rahman A. Polymorphic Transitions in Single-Crystals - a New Molecular-Dynamics Method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 46.Nose S and Klein ML. Constant Pressure Molecular-Dynamics for Molecular-Systems. Mol Phys. 1983;50:1055–1076. [Google Scholar]
- 47.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B and Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- 48.Roe DR and Cheatham TE. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. Journal of Chemical Theory and Computation. 2013;9:3084–3095. [DOI] [PubMed] [Google Scholar]
- 49.Borrelli KW, Cossins B and Guallar V. Exploring hierarchical refinement techniques for induced fit docking with protein and ligand flexibility. Journal of computational chemistry. 2010;31:1224–1235. [DOI] [PubMed] [Google Scholar]
- 50.Li J, Abel R, Zhu K, Cao Y, Zhao S and Friesner RA. The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins: Structure, Function, and Bioinformatics. 2011;79:2794–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacobson MP, Friesner RA, Xiang ZX and Honig B. On the role of the crystal environment in determining protein side-chain conformations. Journal of Molecular Biology. 2002;320:597–608. [DOI] [PubMed] [Google Scholar]
- 52.Jacobson MP, Pincus DL, Rapp CS, Day TJF, Honig B, Shaw DE and Friesner RA. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. [DOI] [PubMed] [Google Scholar]
- 53.Xiao T, Takagi J, Coller BS, Wang J and Springer TA. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 2004;432:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Springer TA, Zhu J and Xiao T. Structural basis for distinctive recognition of fibrinogen gammaC peptide by the platelet integrin alphaIIbbeta3. The Journal of cell biology. 2008;182:791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beer JH and Coller BS. Immobilized RGD-containing peptides as molecular probes of platelet GPIIb/IIIa receptors. Thrombosis and haemostasis. 1989;62:341. [Google Scholar]
- 56.Van Agthoven JF, Xiong JP, Alonso JL, Rui X, Adair BD, Goodman SL and Arnaout MA. Structural basis for pure antagonism of integrin αVβ3 by a high-affinity form of fibronectin. Nat Struct Mol Biol. 2014;21:383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong X, Hudson NE, Lu C and Springer TA. Structural determinants of integrin beta-subunit specificity for latent TGF-beta. Nat Struct Mol Bio. 2014;21:1091–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takada Y, Kamata T, Puzon-McLaughlin W and Shattil SJ. Multiple discontinuous binding sites for ligand-mimetic antibodies (e.g., PAC-1) define a ligand-binding pocket in integrin alpha IIb beta 3 (CD41/CD61). Blood. 1999;94:372a–372a. [DOI] [PubMed] [Google Scholar]
- 59.Varner JA, Nakada MT, Jordan RE and Coller BS. Inhibition of angiogenesis and tumor growth by murine 7E3, the parent antibody of c7E3 Fab (abciximab, ReoProT). Angiogenesis. 1999;3:53–60. [DOI] [PubMed] [Google Scholar]
- 60.Takagi J, Kamata T, Meredith J, Puzon-McLaughlin W and Takada Y. Changing ligand specificities of alphavbeta1 and alphavbeta3 integrins by swapping a short diverse sequence of the beta subunit. The Journal of biological chemistry. 1997;272:19794–19800. [DOI] [PubMed] [Google Scholar]
- 61.Kamata T, Tieu KK, Tarui T, Puzon-McLaughlin W, Hogg N and Takada Y. The role of the CPNKEKEC sequence in the beta(2) subunit I domain in regulation of integrin alpha(L)beta(2) (LFA-1). Journal of immunology (Baltimore, Md : 1950). 2002;168:2296–301. [DOI] [PubMed] [Google Scholar]
- 62.Tsuruta D, Hopkinson SB, Lane KD, Werner ME, Cryns VL and Jones JC. Crucial role of the specificity-determining loop of the integrin beta4 subunit in the binding of cells to laminin-5 and outside-in signal transduction. The Journal of biological chemistry. 2003;278:38707–14. [DOI] [PubMed] [Google Scholar]
- 63.Bajt ML, Ginsberg MH, Frelinger AL III, Berndt MC and Loftus JC. A spontaneous mutation of integrin αIIbβ3 (platelet glycoprotein IIb-IIIa) helps define a ligand binding site. The Journal of biological chemistry. 1992;267:3789–3794. [PubMed] [Google Scholar]
- 64.Lanza F, Stierle A, Fournier D and et al. A new variant of Glanzmann’s thrombasthenia (Strasbourg I). Platelets with functionally defective glycoprotein IIb-IIIa complexes and a glycoprotein IIIa Arg214Trp mutation. Journal of Clinical Investigation. 1992;89:1995–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taub R, Gould RJ, Garsky VM, Ciccarone TM, Hoxie J, Friedman PA and Shattil SJ. A monoclonal antibody against the platelet fibrinogen receptor contains a sequence that mimics a receptor recognition domain in fibrinogen. The Journal of biological chemistry. 1989;264:259–65. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM map has been deposited at the Electron Microscopy Data Bank under accession code EMD-21044 and the coordinates have been deposited at the Protein Data Bank under accession code 6V4P. MD simulation data (initial coordinates and topologies, as well as simulation trajectories) have been deposited within the Open Science Framework at https://osf.io/jqcxk (DOI: 10.17605/OSF.IO/JQCXK).