Abstract
DNA-templated silver clusters (AgC) are fluorescent probes and biosensors whose electronic spectra can be tuned by their DNA hosts. However, the underlying rules that relate DNA sequence and structure to DNA-AgC fluorescence and photophysics are largely empirical. Here, we employ 193 nm activated electron photodetachment (a-EPD) mass spectrometry as a hybrid MS3 approach to gain structural insight into these nanoscale chromophores. Two DNA-AgC systems are investigated with a 20 nt single-stranded DNA (ssDNA) and a 28 nt hybrid hairpin/single-stranded DNA (hpDNA). Both oligonucleotides template Ag10 clusters, but the two complexes are distinct chromophores: the former has a violet absorption at 400 nm with no observable emission, while the latter has a blue-green absorption at 490 nm with strong green emission at 550 nm. Via identification of both apo and holo (AgC-containing) sequence ions generated upon a-EPD and mapping areas of sequence dropout, specific DNA regions that encapsulate the AgC are assigned and attributed to the coordination with the DNA nucleobases. These a-EPD footprints are distinct for the two complexes. The ssDNA contacts the cluster via four nucleobases (CCTT) in the central region of the strand, whereas the hpDNA coordinates the cluster via 13 nucleobases (TTCCCGCCTTTTG) in the double-stranded region of the hairpin. This difference is consistent with prior X-ray scattering spectra and suggests that the clusters can adapt to different DNA hosts. More importantly, the a-EPD footprints directly identify the nucleobases that are in direct contact with the AgC. As these contacting nucleobases can tune the electronic structures of the Ag core and protect the AgC from collisional quenching in solution, understanding the DNA–silver contacts within these complexes will facilitate future biosensor designs.
Keywords: silver, nanoconjugate, DNA, mass spectrometry, UVPD, a-EPD
Graphical Abstract
The production, applications, and fundamental understanding of clusters have advanced tremendously over the past several decades.1,2 DNA-templated silver clusters (AgC) are one class of fluorescent biological probes and sensors with diverse applications in cellular imaging,3–5 detection of cancer- and other disease-related DNA mutations,6–11 enzyme activity sensing,12 live-cell molecular investigations,13 in vitro dopamine quantification,14 single-molecule spectroscopy,15 and low-abundance detection of microRNA sequences.16 These conjugate chromophores form when oligodeoxyribonucleotides with 10–30 residues encapsulate clusters with ~10 silver atoms, and their high fluorescence quantum yield, economical synthesis, and biocompatibility make them an attractive alternative compared to conventional fluorophores.17–19 These complexes fall under the umbrella of atomically precise noble metal nanoclusters with ~102 atoms, which can also be chromophores.2 Their spectra and photophysics are dictated by discrete electronic energy levels that depend on nanocluster stoichiometry, shape, and doping.3,4 We focus on DNA-AgC fluorophores because their emission is strong and tunable.
These chromophores are functional because the DNA scaffold is programmable in two respects. First, DNA sequence tunes the cluster color. Minor sequence changes, even with single nucleobases, yield diverse chromophores whose absorption spectra span the violet to near-infrared.20–23 Second, DNA structure controls the cluster brightness. A DNA strand can be toggled between single- and double-stranded states to reversibly switch cluster adducts between dark and bright isomers, highlighting the profound role of the DNA scaffold on cluster emission and absorption properties.8,25–27 DNA–silver chromophores are synergistic because the valence electrons in the reduced silver atoms establish the electronic structure of the fluorophore, while the DNA nucleobases strongly coordinate the cluster adduct and thus perturb these electronic states.25,27,28 Consequently, understanding the DNA–silver contacts within these complexes is critical to advancing their applications.
Since the mid-1960s, silver has been known to interact specifically with the nucleobases in DNA.29–31 Molecular silver clusters show a similar propensity, especially with cytosine and guanine nucleobases via heteroatom coordination.32–38 DNA sequence and length control DNA-AgC formation; however, the rules that govern cluster binding, size, shape, and electronic properties are empirical.35,39 Recent works in this area have implemented high-throughput, informatics, and machine learning approaches to better understand the complex interplay between DNA sequence and DNA-AgC formation.21,38 One key factor is that the silver clusters, unlike standard small-molecule DNA ligands, are malleable entities, with the ability to change shape and disperse along multiple nucleobases, as suggested by optical spectra that support rod-like clusters, X-ray spectra that support low silver–silver coordination, and crystals with extensive DNA–silver contacts.35,40–43 Recent X-ray scattering studies determined the structure of a DNA-stabilized near-infrared emitting Ag16 cluster.44 This multidendate coordination can be leveraged to create discriminate biosensors in which the cluster adduct inhibits and thus fine-tunes association with target analytes.2,45 At the same time, silver clusters can also be compact, with extensive silver–silver coordination at the expense of silver–DNA coordination.32 The exact binding location and extent of cluster dispersion in these systems remains largely uninvestigated. To address these structural challenges, we have developed mass spectrometry strategies to identify footprints of silver clusters with DNA hosts.
A number of mass spectrometry methods have been used to characterize metal nanoclusters.46 In particular, electrospray ionization MS (ESI-MS) facilitates the analysis of intact DNA-AgC complexes, as it has previously been shown to transfer even noncovalent DNA–ligand complexes from solution phase into the gas phase for subsequent mass spectrometric analysis, preserving both stoichiometry and structure.47,48 Previous studies have shown characterization of DNA–small molecule ligand complexes,49–52 DNA duplex and quadruplex complexes49,50,52,53 i-motif DNA structures,54 and DNA–protein complexes.52 High-resolution and high-mass accuracy measurements establish not only the cluster stoichiometry55 but also its overall charge.32,56,57 Multistage MS methods fragment oligonucleotides to afford more detailed sequence and structural information and enable the localization of ligand-binding sites. Among existing MS/MS methods, collisional activation is the most commonly utilized fragmentation method for nucleic acid characterization. One of the founding studies by McLuckey and co-workers deciphered the fragmentation patterns of DNA upon collisional activation, resulting primarily in the formation of backbone cleavage w and a-B ions (Scheme 1).58 A pioneering study by McLafferty and co-workers built upon the previous framework by extending analysis of DNA to ultraviolet photodissociation (UVPD).59
Scheme 1.
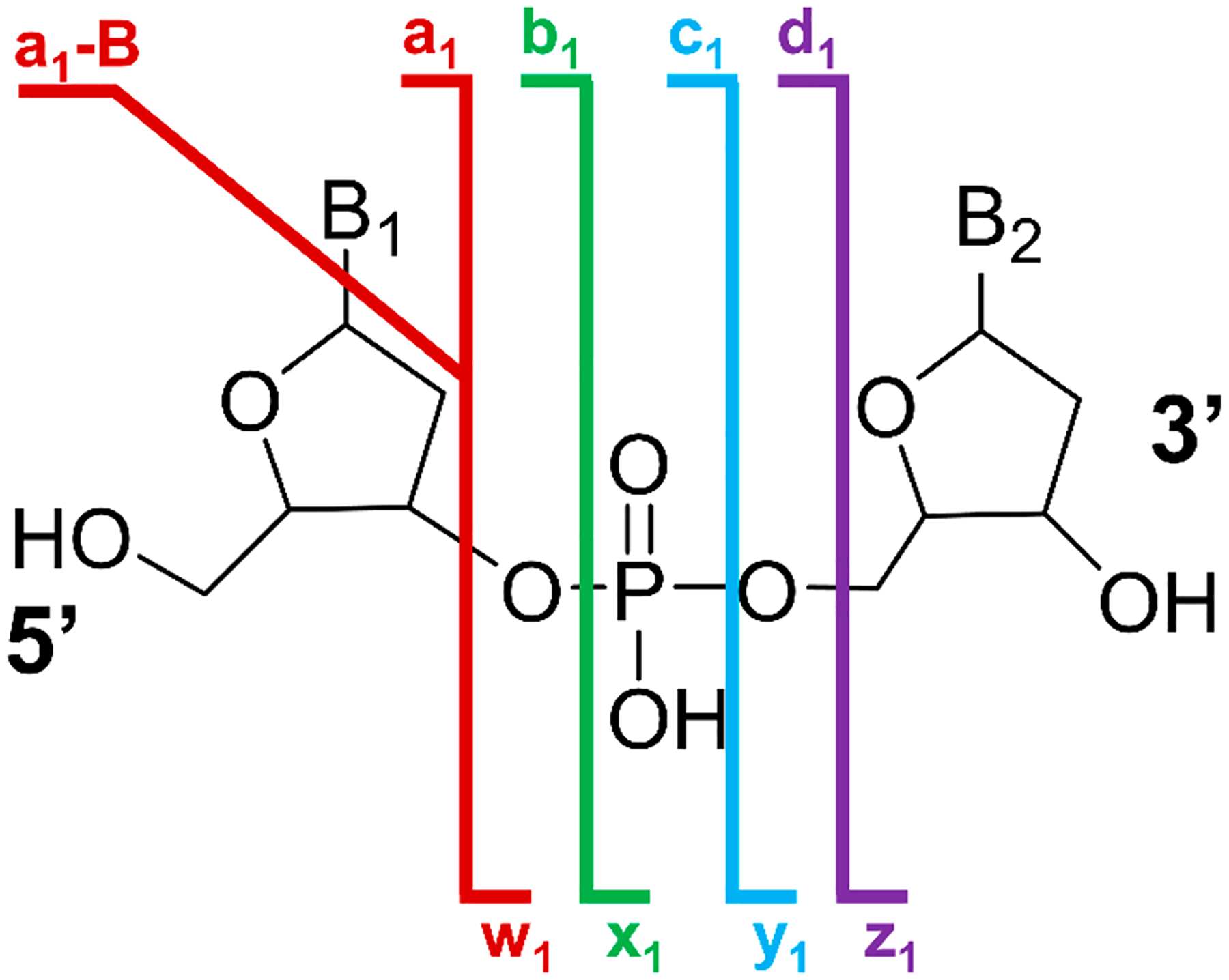
Nomenclature of DNA backbone fragmentation via tandem mass spectrometry methods.
UVPD is an alternative activation method in which high-energy photons (e.g., 6.4 eV per 193 nm photon) are absorbed by and thus dissociate ions. Relative to collisional activation methods, UVPD yields richer and more informative fragmentation patterns that characterize nucleic acids via a diverse array of backbone cleavage ion types including w, x, y, z, a, a-B, b, c, and d ions (Scheme 1).60 In addition to producing an extensive array of backbone cleavage ions, absorption of UV photons by multiply deprotonated DNA ions results in ejection of electrons via a process known as electron photodetachment, or EPD.59 EPD generates an oxidized form of the precursor ion, which can be subsequently isolated and activated for more efficient and informative activation: this hybrid photodissociation/collisional activation MS3 approach is termed activated electron photodetachment, or a-EPD.61 a-EPD of charge-reduced nucleic acids results in abundant production of w, z, a, a-B, and d ions (Scheme 1).
We use a-EPD to investigate two model DNA-AgC complexes: one a single-stranded oligonucleotide (ssDNA) and the other a mixed hairpin/single-stranded DNA (hpDNA). Both constructs are known to assemble and coordinate specific Ag10 clusters, but the two resulting DNA-AgC chromophores have distinct absorption/emission properties.62 Previous studies have shown that the ssDNA-AgC system has a compact shape,25,63 whereas the hpDNA-AgC adopts extended, low-dimensional shapes.41 Here, we employ 193 nm a-EPD as a hybrid MS3 approach to investigate where the Ag clusters bind with their respective DNA hosts based on the patterns of apo ions (DNA fragment ions) and holo ions (DNA fragment ions that contain the entire 10-atom silver cluster). These DNA footprints delineate the cluster binding site and identify the specific nucleobases that are in direct contact with the cluster, providing structural insights into these DNA-AgC systems. Using this technique, we interrogate both the locations and putative sizes of the DNA binding sites of these malleable metal clusters.
RESULTS AND DISCUSSION
In the present study, complexation of a Ag10 cluster with two DNA constructs was evaluated. The resulting conjugates are distinct chromophores, and prior studies suggest that the DNA sequence and structure dictate these electronic spectra. The single-stranded oligonucleotide CCCCAACTCCTTCCCGCCAC preferentially develops a Ag10 adduct with λmax = 400 nm and low emission (Figure S1).1 The hairpin oligonucleotide has the same sequence for the first 18 nucleotides (italicized) but is capped with a six base pair duplex: CCCCAACTCCTTCCCGCCTTTTGGCGGG (stem portion is underlined). The resulting DNA–silver complex is a distinct chromophore with λmax/λex = 490 nm, λem = 550 nm, ϕf = 15%, and τf = 2.4 ns.41 The Ag10 adducts with the ssDNA and hpDNA are specifically favored by controlling the reaction conditions.32,41 Ratios of ≲8:1 (Ag+:DNA) yield only one type of cluster with absorptions at λmax = 400 and 490 nm for the ssDNA and hpDNA scaffolds, respectively (Figure S1A). The Ag+ adducts are reduced under oxygen to eliminate alternate species. The samples have been previously characterized by chromatography, mass spectrometry, and fluorescence spectroscopy.25,41,62 Size exclusion chromatography coupled with UV detection identifies only two types of ssDNA, one without the Ag10 cluster and one with the Ag10 cluster.62 The hpDNA has previously been characterized by fluorescence spectroscopy.41 The emission band does not shift with the excitation wavelength, and the excitation and absorption maxima coincide (Figure S1B).41 These observations support a single type of DNA-bound cluster. With both constructs, the oligonucleotides may fold around their cluster adducts.28,62
The two Ag10 clusters may be isomers because the same cluster with related pairs of single- and double-stranded DNA–silver constructs spectrally interconverts with temperature.25 Our goal is to pinpoint the binding sites and identify the nucleobases that coordinate these two variants of the same cluster. To this end, the negative ionization mode was utilized as DNA ionizes readily in the negative mode owing to the low pKa (<1) of its phosphodiester backbone. This low pKa results in complete deprotonation of DNA in solution; however generally only a fraction of backbone phosphate residues remain deprotonated upon ESI.47 The addition of a volatile salt like ammonium acetate to solution is well known to reduce the abundances of salt adducts in ESI mass spectra of nucleic acids and thus enhances sensitivity.64 More recently, the implementation of submicrometer nanospray emitters was shown to produce a similar effect.65
The MS1 spectra of the ssDNA and the hpDNA in 50 mM ammonium acetate are shown in Figure 1a,c, with the 5–charge state the most abundant for both oligonucleotides. Deconvoluted MS1 spectra are shown in Figures S2 and S3. The ssDNA-AgC and hpDNA-AgC solutions yield similar ESI mass spectra with low charge states and mass shifts of 1069 Da relative to the oligonucleotides alone, highlighting the preferential DNA·Ag10 stoichiometry for both DNA sequences (Figure 1b,d). The 1:10 DNA:Ag stoichiometry matches that observed by elemental analysis for the chromatographically purified complexes2 and is preferential, as no other DNA-AgC species are observed.
Figure 1.
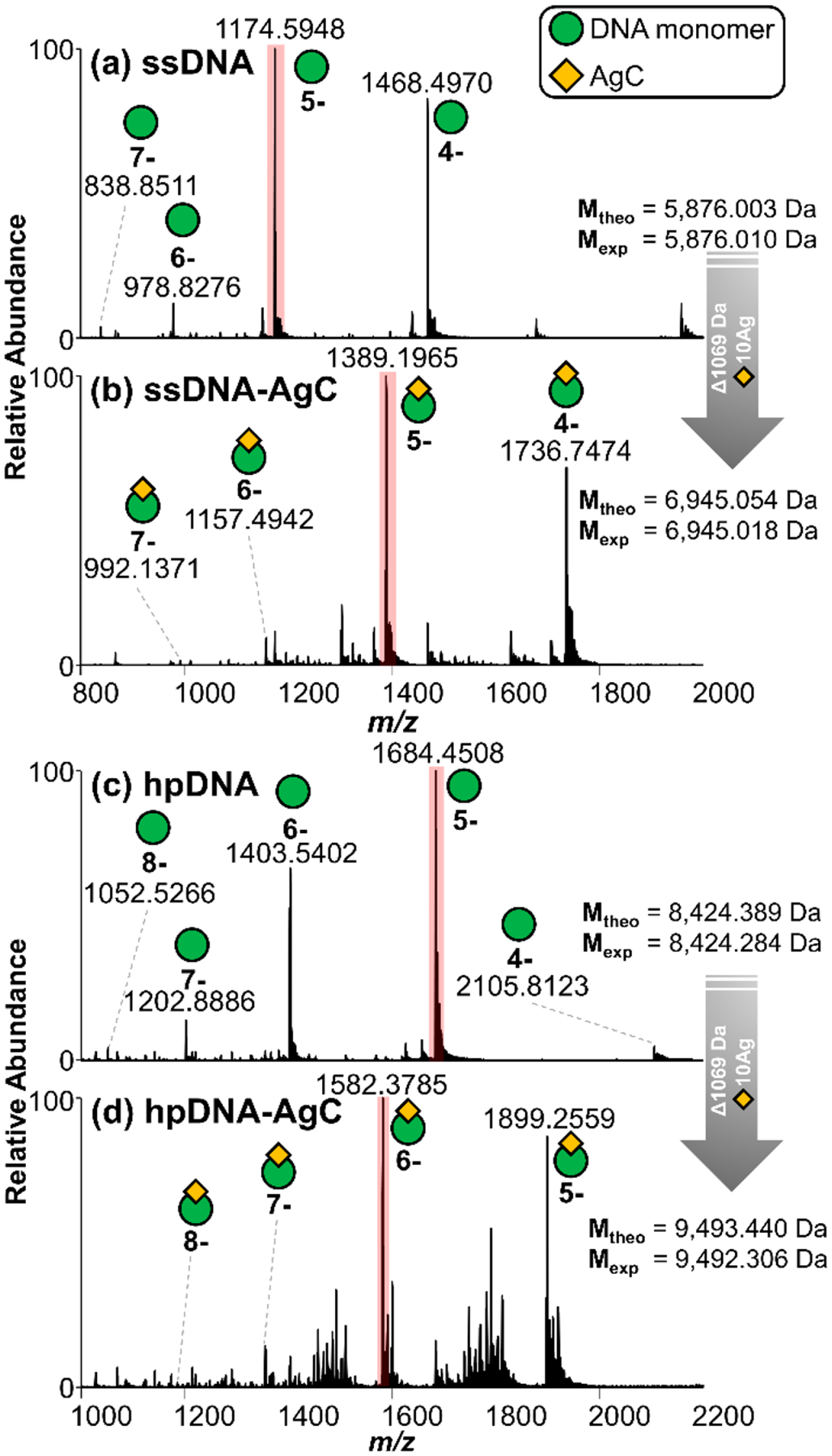
Negative-mode MS1 spectra of ssDNA CCC CAA CTC CTT CCC GCC AC in 50 mM ammonium acetate (a) without and (b) with the AgC and hpDNA CCC CAA CTC CTT CCC GCC TTT TGG CGG G (c) without and (d) with the AgC. The most abundant charge states for each sample are highlighted in pink, and theoretical and experimental values of DNA and DNA-AgC are included, enabling determination of DNA-AgC stoichiometry.
For evaluation of the impact of solvent composition, ESI of a solution containing the ssDNA in 50% methanol and 50 mM ammonium acetate yields the mass spectrum in Figure S4a, displaying a higher and broader range of charge states. Interestingly, the ssDNA-AgC remains intact in the 50% methanol solution, reflecting a strong interaction between the DNA and the AgC (Figure S4b). Figure S4c highlights a few of the impurities in the hpDNA-AgC sample (as observed in Figure 1d), including low levels of bare DNA as well as unidentified degradation products. With respect to ion polarity, the same types of DNA·Ag10 complexes are also observed in the positive mode in which the phosphate backbone is completely protonated and the nucleobases are partially protonated (Figure S5).66 In sum, these results suggest that the stoichiometry and structures of the DNA·Ag10 complexes are faithfully preserved in their transition from solution to the gas phase. For subsequent characterization of the DNA and DNA-AgC complexes by MS/MS, the aqueous solutions containing 50 mM ammonium acetate were analyzed owing to the production of lower charge states (which facilitates deconvolution) and higher MS1 precursor abundances as the signal is split across fewer charge states.
An extensive array of tandem MS methods including higher-energy collisional dissociation (HCD), UVPD, and a-EPD were employed for characterization of the ssDNA·Ag10 and hpDNA·Ag10 complexes. All HCD, UVPD, and a-EPD spectra and deconvoluted mass spectra along with tables of assigned fragment ions (within 20 ppm mass error) are shown in Figures S6–S23. The types and distributions of fragment ions produced by HCD, UVPD, and a-EPD are summarized in Figure 2 for the 4–, 5–, and 6– charge states of ssDNA and ssDNA-AgC (with full tabulation in Table S1).
Figure 2.
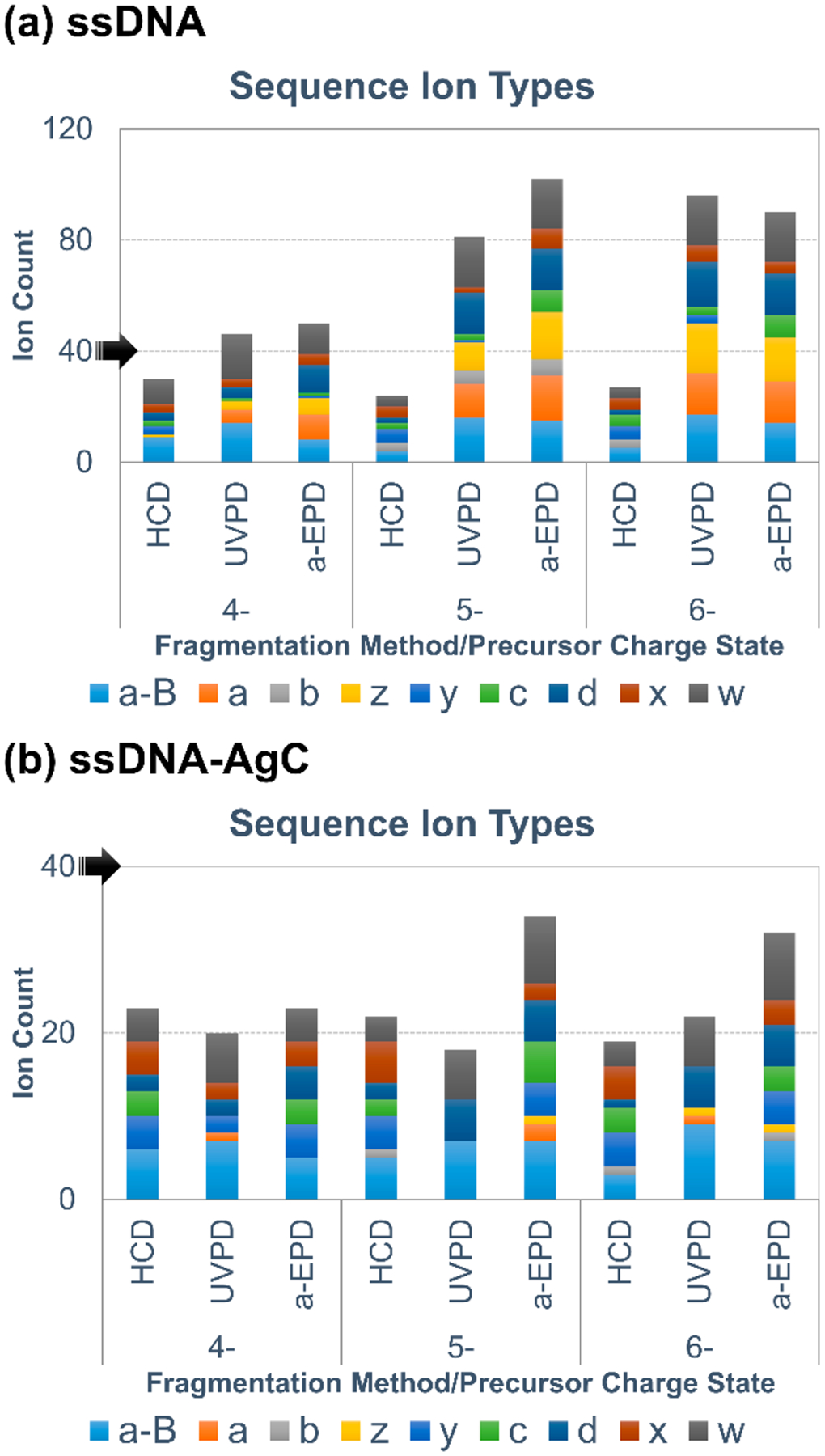
Bar graphs of single-stranded 20-mer oligonucleotide CCC CAA CTC CTT CCC GCC AC backbone sequence ion type distributions and counts for (a) lone DNA and (b) DNA–silver cluster samples. Black arrows are included to highlight scale differences.
UVPD and a-EPD of the ssDNA result in more identified sequence ions and a larger variety of sequence ion types across the 4–, 5–, and 6– charge states compared to HCD (Figure 2a). Additionally, a significant charge-state dependence is observed, as UVPD and a-EPD of the 5– and 6– charge states result in production of more abundant sequence ions compared to the 4– charge state. These observations are consistent with previous studies which have also shown a dependence of sequence coverage on charge state for MS/MS of DNA ions.60
The fragmentation pattern for the ssDNA·Ag10 complex is notably different from that of the ssDNA ions, as all MS/MS methods yield fewer sequence ions, and a-B and w ions dominate the ion-type distributions (Figure 2b; note scale change). Previous studies postulated that metals coordinate and complex with DNA strands to alter their gas-phase fragmentation,67 in alignment with our observations of the difference in ion-type distributions upon silver cluster complexation. Overall, a-EPD results in less congested spectra than does UVPD, while maintaining high sequence coverage for both the DNA and DNA-AgC complexes. The extensive a-B and w ions produced by UVPD, a-EPD, or HCD consistently provide the best sequence coverage of the DNA and DNA-AgC complexes, and the other ions (b, c, d, x, y, and z) generally provide only redundant information or are suppressed for the DNA-AgC complexes (Figure S24). Although often associated with fragmentation of closed-shell ions, the observation of a-B and w ions upon a-EPD (fragmentation of open-shell radical species) aligns with previous studies.61 While a-EPD allows access to higher-energy fragmentation pathways, lower-energy pathways are still accessed with this fragmentation approach and therefore result in the generation of a-B and w ions in addition to an array of other ion types. Taking into account these results, we opted to use a-B and w ions from a-EPD of the 6– charge state for comprehensive analysis of all DNA and DNA-AgC species moving forward.
UV photoactivation of the 6– charge state of the ssDNA or ssDNA·Ag10 complex predominantly results in charge reduction via electron detachment but also produces a wide range of low-abundance sequence ions, including w, x, y, z, a, a-B, b, c, and d ions (Figure 3a,e). Isolation and subsequent collisional activation of the charge-reduced 5– charge state of the DNA or DNA·Ag10 complex yields the a-EPD spectra shown in Figure 3b,f, which are dominated by diagnostic a-B and w ions. The ssDNA·Ag10 complex produces both apo (no AgC) and holo (containing AgC) fragment ions, with the latter retaining the entire 10-atom silver cluster. Fragmentation of the silver cluster was not observed, as DNA fragmentation (with and without retention of the entire AgC) was the major observed fragmentation pathway. This finding is in contrast with previous studies of thiol-protected silver clusters (not DNA-templated), which found that Ag-thiol losses are observed upon collisional dissociation.68 These discrepancies could be explained by both the differences in analyte (thiol-protected AgC vs DNA-AgC) and/or fragmentation method (collisional dissociation vs a-EPD). Assignments of holo ions from deconvoluted spectra are summarized in Table S2.
Figure 3.
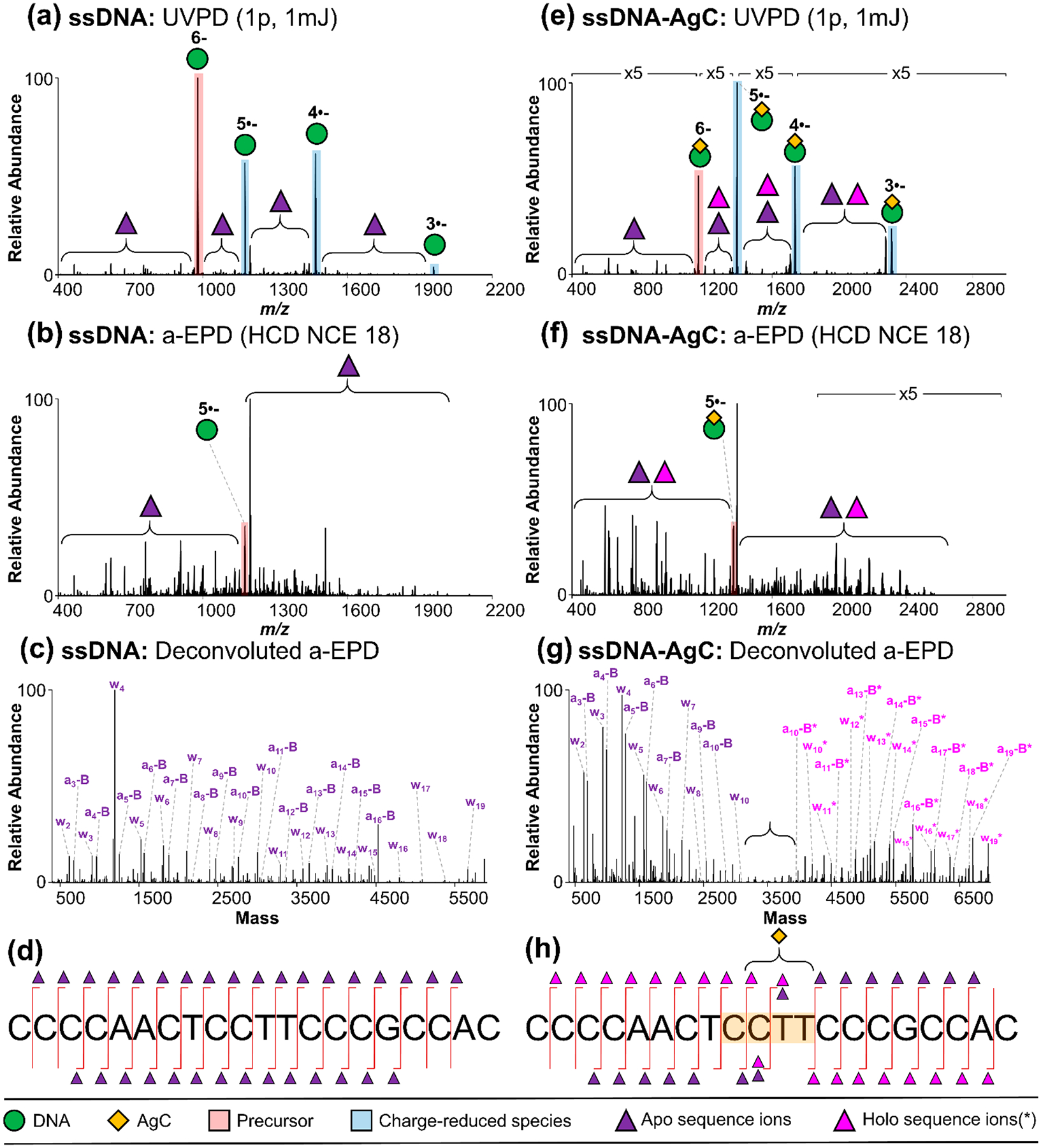
Single-stranded 20-mer oligonucleotide CCC CAA CTC CTT CCC GCC AC without AgC: (a) UVPD spectrum (6– charge state, m/z 978), (b) a-EPD spectrum (5•– charge state of m/z 1174), (c) deconvoluted a-EPD spectrum, and (d) sequence coverage map; with AgC: (e) UVPD spectrum (6– charge state, m/z 1157), (f) a-EPD spectrum (5•– charge state of m/z 1388), (g) deconvoluted a-EPD spectrum, and (h) sequence coverage map. Sequence ion assignments from deconvoluted a-EPD spectra are summarized in Table S2 and Figure S23.
The pattern of apo and holo fragment ions localizes the AgC within its respective DNA scaffold. For example, the deconvoluted a-EPD spectrum of the ssDNA-AgC shows a large dropout in both apo and holo sequence ions in the middle of the DNA strand, with high-abundance apo and holo ions on either side of the dropout region (Figure 3g). This result contrasts the relatively uniform distribution of a-B/w sequence ions for the lone ssDNA (Figure 3c,d). Recent studies have proposed that these ligand-containing holo ions may be produced because the ligand–nucleic acid interactions remain collectively stronger than individual backbone bonds of the nucleic acid upon activation in the gas phase,69 particularly for open-shell radical ions produced upon EPD.70 When mapped onto the DNA sequence, the complementary sets of ssDNA-AgC apo and holo ions unambiguously localize the AgC to the highlighted CCTT region of the DNA strand (Figure 3h). Cytosine strongly coordinates silver clusters, but thymine is a comparably weak ligand, possibly because its N3 is protonated at neutral pH.33,36,71 These thymines may be protected because of the secondary structure of the DNA host. These findings are in agreement with recent studies by Kondo and co-workers, which demonstrated the role of thymine residues in allowing the DNA scaffold sufficient flexibility to fold around a 16-atom silver cluster.44 Thymine is also a flexible junction in folded DNA constructs.72 Size exclusion chromatography studies show that this oligonucleotide folds around its Ag10 cluster,32 and the alternate conformation may hinder the generation of a-B and w sequence ions.
Like its single-stranded analogue, the hpDNA alone similarly fragments into a relatively uniform array of a-B and w sequence ions, as shown in the deconvoluted a-EPD spectrum of the 6– charge state (Figure 4a). However, unlike the ssDNA-AgC, the hpDNA-AgC exhibits a much larger footprint that extends over the 13 nucleobases TTCCCGCCTTTTG (nucleotides 11–23) (Figure 4b). The red lines in Figure 4c depict AgC-coordinated nucleobases. Raw UVPD and a-EPD spectra along with tabulated fragment ion assignments are shown in Figures S25 and S26. One possibility is that the base pairs cross-link silvers.73 These metallobase pairs have been identified and characterized by crystallography, spectroscopy, thermodynamic, and mass spectrometry studies.74–78 Because the Ag10 cluster is partially oxidized with a 6+ charge, as observed from the isotopic distributions (Figures S27 and S28),41 we suggest that the cluster may bind to the duplex portion of the hairpin (if the duplex structure is retained). Another possibility is that the secondary structure of the DNA construct is entirely rearranged upon coordination to the AgC, as supported by recent reports.79,80
Figure 4.
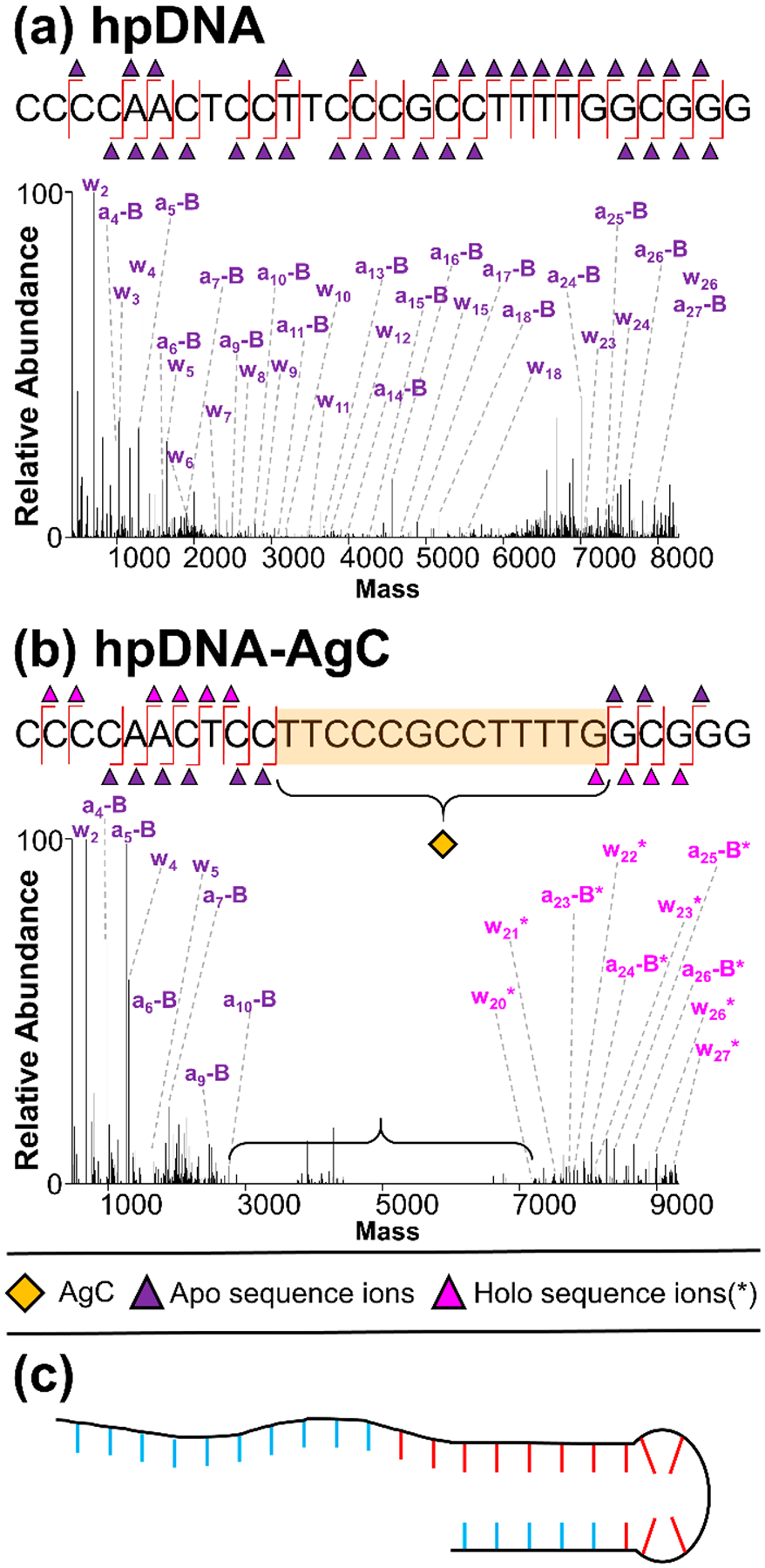
Deconvoluted a-EPD spectra and sequence coverage maps of 6- hpDNA CCCCAACTCCTTCCCGCCTTTTGGCGGG (a) without and (b) with AgC, highlighting localization of the AgC, (c) model of hpDNA-AgC with AgC-coordinating nucleobases in red and noncoordinating nucleobases in blue. Sequence ion assignments from deconvoluted a-EPD spectra are summarized in Figures S25 and S26.
The abundances of apo and holo a-B and w ions from a-EPD data of ssDNA, hpDNA, ssDNA-AgC, and hpDNA-AgC are plotted as a function of DNA sequence (Figure 5). These types of plots offer structural insight about macromolecule–ligand complexes; for example, decreases in fragmentation may reflect changes in secondary structure or the presence of ligand interactions that prevent complementary fragment ions from separating after the cleavage of the backbone, thus impeding their detection.81,82 The binding sites of the silver clusters are determined via the identification of apo and holo ions, not solely via regions of fragmentation suppression. Interestingly, both AgC complexes show a characteristic decrease in sequence coverage at the identified AgC binding locations (Figure 5ac,d) relative to their AgC-free counterparts (Figure 5a,b). The AgC-free DNA ions show more uniform fragmentation across the sequence, although increased fragment ion abundances are observed corresponding to backbone cleavages at many G residues, indicating a potential preferential cleavage of a-EPD at DNA guanine nucleotides. Additionally, a decrease in fragmentation is observed at thymine residues, indicating preferential hampering of this fragmentation pathway at T residues. The putative binding region of the hpDNA-AgC is significantly elongated compared to the ssDNA-AgC, as highlighted by the red boxed region in Figure 5d. Additionally, the abundances of sequence ions in close proximity to the AgC binding location are lower for both the ssDNA-AgC and the hpDNA-AgC (Figure 5c,d), suggesting that the resilient interactions between the AgC and DNA not only suppress production of sequence ions for the portion of the DNA strand in contact with the AgC but also impede fragmentation for regions of the DNA sequence that are in close proximity to the AgC.
Figure 5.
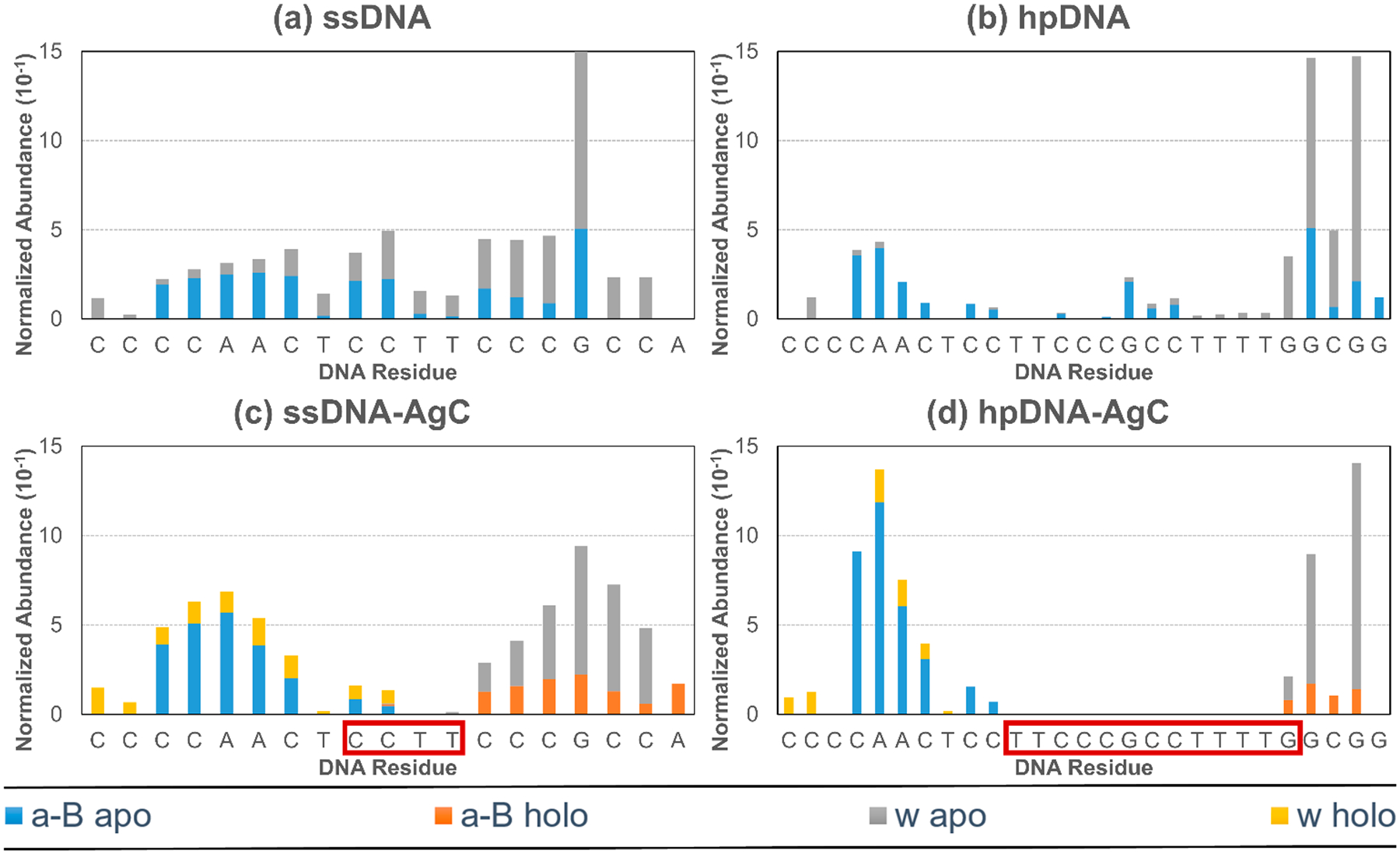
Normalized fragmentation abundances of a-B and w apo and holo ions from a-EPD data of the 6– charge state as a function of DNA sequence for (a) ssDNA, (b) hpDNA, (c) ssDNA-AgC, and (d) hpDNA-AgC. Sum intensities of deconvoluted sequence ion masses were normalized relative to the spectrum TIC prior to deconvolution and are plotted in units of 10−1. Boxed regions of DNA sequence in red indicate AgC binding location.
Importantly, the elongation of the sequence dropout region for the hpDNA-AgC compared to the ssDNA-AgC suggests a significant change in the structure of the complex or mode of interaction of the AgC with the DNA. It is possible that dispersement of the silver cluster along the DNA strand is what causes this shift in binding motif. The size of this sequence dropout area characterized the mode of binding of the AgC to its DNA host, allowing a-EPD to be a “footprint” method for cluster binding.
The distinct DNA footprints for the two Ag10 clusters substantially complement earlier structural studies of the same single-stranded and hairpin DNA complexes.32,41 Extended X-ray absorption fine structure (EXAFS) spectra identified silver–nucleobase and metallic silver–silver interactions within these complexes, but the associated coordination numbers suggest that the Ag10 adducts have different structures. For the single-stranded DNA-Ag10 complex, relatively large silver–silver vs relatively small silver–DNA coordination numbers support a compact, octahedral cluster that is sparsely contacted by its DNA host. For the hairpin DNA-Ag10 complex, these coordination numbers change in opposing directions with fewer silver–silver vs more silver–nucleobase contacts. These opposing changes suggest that the cluster is more dispersed and thus makes more contacts with its hairpin DNA host, as implied from the electronic X-ray spectra of silver cluster–DNA complexes.56 Furthermore, thermodynamic studies show that the same cluster reversibly adopts different isomeric forms depending on the hybridization state of the DNA host and could be spectrally interconverting with temperature.25,28
The MS3 studies presented herein are consistent with both the EXAFS studies,32,41 as the compact cluster with the single-stranded DNA might be expected to have limited contact with its host, whereas the elongated cluster with the hairpin DNA is expected to have more contact with its host. The difference in the ligand contact and coordination not only leads to distinct electronic structures of the AgC but also determines how the chromophore is shielded from water molecules. The power of a-EPD is that the subset of nucleobases within a DNA polymer that are in contact with the silver cluster can be identified. Notably, it is now possible to target these regions. Single nucleobases can profoundly impact the spectra of silver cluster adducts, presumably because the coordination environment can be changed with just a single-nucleotide substitution.23,24 Thus, it may be possible to specifically alter the sequence of a DNA host to elicit specific spectra and photophysical changes.
CONCLUSIONS
In conclusion, a comprehensive and structural MS3 study of two DNA–silver cluster constructs reveals the footprints of silver clusters within their polymeric DNA hosts. a-EPD pinpointed the binding sites and revealed that the area of sequence dropout is substantially elongated for the hpDNA·Ag10 compared to the ssDNA·Ag10. These findings agree with the coordination numbers derived from X-ray spectra, but greatly expand this structural model because specific coordinated nucleobases can be identified. These coordinated nucleobases and their heteroatoms dictate silver cluster spectra, making a-EPD an effective tool for studying the structure of DNA-AgCs and the interaction between silver clusters and their DNA hosts.
METHODS
Materials.
Oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA, USA). Silver nitrate (99.9995%) and sodium borohydride (98+%) used for DNA-AgC synthesis were purchased from Alfa Aesar (Haverhill, MA, USA) and Acros Organics (Waltham, MA, USA), respectively. VivaSpin 15R 2000 MWCO centrifugal concentrators from Sartorius (Goettingen, Germany) were used for centrifugal dialysis.
DNA-AgC Synthesis.
Oligonucleotides CCCCAACTCCTTCCCGCCAC (ssDNA) and CCCCAACTCCTTCCCGCCTTTTGGCGGG (hpDNA) were diluted to 30 μM in water, combined with 8 equivalents of AgNO3, and heated to ~80 °C for 5 min. Solutions were then cooled to room temperature, and the samples were chemically reduced via addition of 4 equivalents of NaBH4 and vortexed for 1 min. Upon reduction, solutions turned yellow due to the formation of Ag clusters. Solutions were then treated with 400 psi O2 for 2–3 h to eliminate alternate clusters.83 Samples were then dialyzed against 100 volumes of solution using centrifugal dialysis to remove impurities. Final DNA and DNA-AgC stock solutions were provided at ~100 μM in water. A list of sequences and masses for all DNA and DNA·Ag10 samples used in this study can be found in Table S3.
Mass Spectrometry.
DNA and DNA-AgC stock solutions were diluted to 10 μM in 50 mM ammonium acetate solution (in water or in 50:50 methanol/water (v/v)). All experiments were performed on a Thermo Fisher Scientific Orbitrap Fusion Lumos Tribrid mass spectrometer modified with a 193 nm excimer laser for 193 nm UVPD.84 All mass spectra were collected with a resolving power of 240 000 at m/z 200, an AGC of 5e5, a spray voltage of 600–800 V, and 1 μscan/scan. MS1 spectra were collected using an average of 25 scans. All MSn spectra were collected using 10 scans of 100 transient averages and isolation widths ranging from 5 to 10 m/z units. HCD was performed using NCE 25, 193 nm UVPD was performed using 1 pulse (5 ns) of 1 mJ, and 193 nm a-EPD was performed via UVPD (1 pulse, 1 mJ) with subsequent HCD collisional activation using NCE 17–18. a-EPD experiments are MS3 experiments in which UVPD is performed at the MS2 stage, and the resulting electron-photodetached precursor is then fragmented with collisional activation (MS3 event). All UVPD experiments were performed in the high-pressure trap of the dual linear ion trap.
UVPD at 213 nm was also explored for a-EPD characterization of DNA due to the recent introduction of a commercially available UVPD option on an Orbitrap platform.85–87 a-EPD experiments at 213 nm were performed on a Thermo Fisher Scientific Orbitrap Fusion Lumos Tribrid mass spectrometer equipped with the 213 nm UVPD option (UVPD occurs in the low-pressure trap of the dual linear ion trap). The performance of 213 nm a-EPD for characterization of DNA-AgC proved similar to 193 nm a-EPD in terms of the types and distributions of fragment ions (Figure S29). a-EPD at 213 nm was performed using a 30 ms photoactivation period (compared to a 2 ms period for 193 nm UVPD) followed by HCD (NCE 18).
Data deconvolution was performed using the Xtract feature in FreeStyle version 1.5 (Thermo Fisher Scientific). Data were deconvoluted using the following parameters: S/N 3, OT analyzer type, nucleotide isotope table; the negative charge box was checked for all negative mode spectra. Data were interpreted manually with the aid of MongoOligo, ChemCalc, and ChemDraw (PerkinElmer). A manual mass tolerance of 20 ppm was used for identification of apo DNA fragment ions.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the following funding sources: NSF (Grant CHE1402753 to J.S.B. and Grant CHE1611451 to J.T.P. and H.C.Y.), the Welch Foundation (Grant F-1155 to J.S.B. and Grant F-1833 to H.C.Y.), and NIH (Grant GM129617 to H.C.Y.). Funding from the UT System for support of the UT System Proteomics Core Facility Network is gratefully acknowledged. We also thank the Furman Advantage program. This work was supported in part by the National Science Foundation EPSCoR Program under NSF Award #OIA-1655740. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect those of the National Science Foundation.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.9b06470.
Additional tables and figures (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Sun Q; Jena P Super Atomic Clusters: Design Rules and Potential for Building Blocks of Materials. Chem. Rev 2018, 118, 5755–5870. [DOI] [PubMed] [Google Scholar]
- (2).Pradeep T; Chakraborty I Atomically Precise Clusters of Noble Metals: Emerging Link between Atoms and Nanoparticles. Chem. Rev 2017, 117, 8208–8271. [DOI] [PubMed] [Google Scholar]
- (3).Wu J; Li N; Yao Y; Tang D; Yang D; Ong’achwa Machuki J; Li J; Yu Y; Gao F DNA-Stabilized Silver Nanoclusters for Label-Free Fluorescence Imaging of Cell Surface Glycans and Fluorescence Guided Photothermal Therapy. Anal. Chem 2018, 90, 14368–14375. [DOI] [PubMed] [Google Scholar]
- (4).Li J; Yu J; Huang Y; Zhao H; Tian L Highly Stable and Multiemissive Silver Nanoclusters Synthesized In Situ in a DNA Hydrogel and Their Application for Hydroxyl Radical Sensing. ACS Appl. Mater. Interfaces 2018, 10, 26075–26083. [DOI] [PubMed] [Google Scholar]
- (5).Lyu D; Li J; Wang X; Guo W; Wang E Cationic-Polyelectrolyte-Modified Fluorescent DNA-Silver Nanoclusters with Enhanced Emission and Higher Stability for Rapid Bioimaging. Anal. Chem 2019, 91, 2050–2057. [DOI] [PubMed] [Google Scholar]
- (6).Choi S; Yu J; Patel SA; Tzeng Y-L; Dickson RM Tailoring Silver Nanodots for Intracellular Staining. Photochem. Photobiol. Sci 2011, 10, 109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Obliosca JM; Liu C; Batson RA; Babin MC; Werner JH; Yeh H-C DNA/RNA Detection Using DNA-Templated Few-Atom Silver Nanoclusters. Biosensors 2013, 3, 185–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Yeh H-C; Sharma J; Han JJ; Martinez JS; Werner JH A DNA–Silver Nanocluster Probe That Fluoresces upon Hybridization. Nano Lett 2010, 10, 3106–3110. [DOI] [PubMed] [Google Scholar]
- (9).Guo W; Yuan J; Dong Q; Wang E Highly Sequence-Dependent Formation of Fluorescent Silver Nanoclusters in Hybridized DNA Duplexes for Single Nucleotide Mutation Identification. J. Am. Chem. Soc 2010, 132, 932–934. [DOI] [PubMed] [Google Scholar]
- (10).Chen Y-A; Obliosca JM; Liu Y-L; Liu C; Gwozdz ML; Yeh H-C NanoCluster Beacons Enable Detection of a Single N6-Methyladenine. J. Am. Chem. Soc 2015, 137, 10476–10479. [DOI] [PubMed] [Google Scholar]
- (11).Yeh H-C; Sharma J; Shih I-M; Vu DM; Martinez JS; Werner JH A Fluorescence Light-Up Ag Nanocluster Probe That Discriminates Single-Nucleotide Variants by Emission Color. J. Am. Chem. Soc 2012, 134, 11550–11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Juul S; Obliosca JM; Liu C; Liu Y-L; Chen Y-A; Imphean DM; Knudsen BR; Ho Y-P; Leong KW; Yeh H-C NanoCluster Beacons as Reporter Probes in Rolling Circle Enhanced Enzyme Activity Detection. Nanoscale 2015, 7, 8332–8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Bossert N; de Bruin D; Götz M; Bouwmeester D; Heinrich D Fluorescence-Tunable Ag-DNA Biosensor with Tailored Cytotoxicity for Live-Cell Applications. Sci. Rep 2016, 6, 37897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Del Bonis-O’Donnell JTD; Thakrar A; Hirschberg JW; Vong D; Queenan BN; Fygenson DK; Pennathur S DNA-Stabilized Silver Nanoclusters as Specific, Ratiometric Fluorescent Dopamine Sensors. ACS Chem. Neurosci 2018, 9, 849–857. [DOI] [PubMed] [Google Scholar]
- (15).Petty JT; Zheng J; Hud NV; Dickson RM DNA-Templated Ag Nanocluster Formation. J. Am. Chem. Soc 2004, 126, 5207–5212. [DOI] [PubMed] [Google Scholar]
- (16).Shah P; Rørvig-Lund A; Chaabane SB; Thulstrup PW; Kjaergaard HG; Fron E; Hofkens J; Yang SW; Vosch T Design Aspects of Bright Red Emissive Silver Nanoclusters/DNA Probes for MicroRNA Detection. ACS Nano 2012, 6, 8803–8814. [DOI] [PubMed] [Google Scholar]
- (17).Gwinn E; Schultz D; Copp SM; Swasey S DNA-Protected Silver Clusters for Nanophotonics. Nanomaterials 2015, 5, 180–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Obliosca JM; Liu C; Yeh H-C Fluorescent Silver Nanoclusters as DNA Probes. Nanoscale 2013, 5, 8443. [DOI] [PubMed] [Google Scholar]
- (19).Petty JT; Story SP; Hsiang J-C; Dickson RM DNA-Templated Molecular Silver Fluorophores. J. Phys. Chem. Lett 2013, 4, 1148–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Richards CI; Choi S; Hsiang J-C; Antoku Y; Vosch T; Bongiorno A; Tzeng Y-L; Dickson RM Oligonucleotide-Stabilized Ag Nanocluster Fluorophores. J. Am. Chem. Soc 2008, 130, 5038–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Copp SM; Bogdanov P; Debord M; Singh A; Gwinn E Base Motif Recognition and Design of DNA Templates for Fluorescent Silver Clusters by Machine Learning. Adv. Mater 2014, 26, 5839–5845. [DOI] [PubMed] [Google Scholar]
- (22).Petty JT; Fan C; Story SP; Sengupta B; Sartin M; Hsiang J-C; Perry JW; Dickson RM Optically Enhanced, Near-IR, Silver Cluster Emission Altered by Single Base Changes in the DNA Template. J. Phys. Chem. B 2011, 115, 7996–8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Obliosca JM; Babin MC; Liu C; Liu Y-L; Chen Y-A; Batson RA; Ganguly M; Petty JT; Yeh H-C A Complementary Palette of NanoCluster Beacons. ACS Nano 2014, 8, 10150–10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Yourston LE; Lushnikov AY; Shevchenko OA; Afonin KA; Krasnoslobodtsev AV First Step Towards Larger DNA-Based Assemblies of Fluorescent Silver Nanoclusters: Template Design and Detailed Characterization of Optical Properties. Nanomaterials 2019, 9, 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Petty JT; Sergev OO; Nicholson DA; Goodwin PM; Giri B; McMullan DR A Silver Cluster–DNA Equilibrium. Anal. Chem 2013, 85, 9868–9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Petty JT; Sengupta B; Story SP; Degtyareva NN DNA Sensing by Amplifying the Number of Near-Infrared Emitting, Oligonucleotide-Encapsulated Silver Clusters. Anal. Chem 2011, 83, 5957–5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Yeh H; Sharma J; Han JJ; Martinez JS; Werner JH A Beacon of Light. IEEE Nanotechnology Magazine 2011, 5, 28–33. [Google Scholar]
- (28).Ganguly M; Bradsher C; Goodwin P; Petty JT DNA-Directed Fluorescence Switching of Silver Clusters. J. Phys. Chem. C 2015, 119, 27829–27837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Daune M; Dekker CA; Schachman HK Complexes of Silver Ion with Natural and Synthetic Polynucleotides. Biopolymers 1966, 4, 51–76. [Google Scholar]
- (30).Eichhorn GL; Butzow JJ; Clark P; Tarien E Interaction of Metal Ions with Polynucleotides and Related Compounds. X. Studies on the Reaction of Silver(I) with the Nucleosides and Polynucleotides, and the Effect of Silver(I) on the Zinc(II) Degradation of Polynucleotides. Biopolymers 1967, 5, 283–296. [DOI] [PubMed] [Google Scholar]
- (31).Jensen RH; Davidson N Spectrophotometric, Potentiometric, and Density Gradient Ultracentrifugation Studies of the Binding of Silver Ion by DNA. Biopolymers 1966, 4, 17–32. [Google Scholar]
- (32).Petty JT; Sergev OO; Ganguly M; Rankine IJ; Chevrier DM; Zhang P A Segregated, Partially Oxidized, and Compact Ag10 Cluster within an Encapsulating DNA Host. J. Am. Chem. Soc 2016, 138, 3469–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Ritchie CM; Johnsen KR; Kiser JR; Antoku Y; Dickson RM; Petty JT Ag Nanocluster Formation Using a Cytosine Oligonucleotide Template. J. Phys. Chem. C 2007, 111, 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Petty JT; Ganguly M; Yunus AI; He C; Goodwin PM; Lu Y-H; Dickson RM A DNA-Encapsulated Silver Cluster and the Roles of Its Nucleobase Ligands. J. Phys. Chem. C 2018, 122, 28382–28392. [Google Scholar]
- (35).Huard DJE; Demissie A; Kim D; Lewis D; Dickson RM; Petty JT; Lieberman RL Atomic Structure of a Fluorescent Ag8 Cluster Templated by a Multistranded DNA Scaffold. J. Am. Chem. Soc 2019, 141, 11465–11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Soto-Verdugo V; Metiu H; Gwinn E The Properties of Small Ag Clusters Bound to DNA Bases. J. Chem. Phys 2010, 132, 195102. [DOI] [PubMed] [Google Scholar]
- (37).Ono A; Cao S; Togashi H; Tashiro M; Fujimoto T; Machinami T; Oda S; Miyake Y; Okamoto I; Tanaka Y Specific Interactions between Silver(I) Ions and Cytosine–cytosine Pairs in DNA Duplexes. Chem. Commun 2008, 0, 4825–4827. [DOI] [PubMed] [Google Scholar]
- (38).Schultz D; Brinson RG; Sari N; Fagan JA; Bergonzo C; Lin NJ; Dunkers JP Structural Insights into DNA-Stabilized Silver Clusters. Soft Matter 2019, 15, 4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).New SY; Lee ST; Su XD DNA-Templated Silver Nanoclusters: Structural Correlation and Fluorescence Modulation. Nanoscale 2016, 8, 17729–17746. [DOI] [PubMed] [Google Scholar]
- (40).Copp SM; Gorovits A; Swasey SM; Gudibandi S; Bogdanov P; Gwinn EG Fluorescence Color by Data-Driven Design of Genomic Silver Clusters. ACS Nano 2018, 12, 8240–8247. [DOI] [PubMed] [Google Scholar]
- (41).Petty JT; Ganguly M; Rankine IJ; Chevrier DM; Zhang P A DNA-Encapsulated and Fluorescent Ag106+ Cluster with a Distinct Metal-Like Core. J. Phys. Chem. C 2017, 121, 14936–14945. [Google Scholar]
- (42).Copp SM; Schultz D; Swasey SM; Faris A; Gwinn EG Cluster Plasmonics: Dielectric and Shape Effects on DNA-Stabilized Silver Clusters. Nano Lett. 2016, 16, 3594–3599. [DOI] [PubMed] [Google Scholar]
- (43).Neidig ML; Sharma J; Yeh H-C; Martinez JS; Conradson SD; Shreve AP Ag K-Edge EXAFS Analysis of DNA-Templated Fluorescent Silver Nanoclusters: Insight into the Structural Origins of Emission Tuning by DNA Sequence Variations. J. Am. Chem. Soc 2011, 133, 11837–11839. [DOI] [PubMed] [Google Scholar]
- (44).Cerretani C; Kanazawa H; Vosch T; Kondo J Crystal Structure of a NIR-Emitting DNA-Stabilized Ag16 Nanocluster. Angew. Chem., Int. Ed 2019, 58, 1–6. [DOI] [PubMed] [Google Scholar]
- (45).Petty JT; Ganguly M; Rankine IJ; Baucum EJ; Gillan MJ; Eddy LE; Léon JC; Müller J Repeated and Folded DNA Sequences and Their Modular Ag106+ Cluster. J. Phys. Chem. C 2018, 122, 4670–4680. [Google Scholar]
- (46).Lu Y; Chen W Application of Mass Spectrometry in the Synthesis and Characterization of Metal Nanoclusters. Anal. Chem 2015, 87, 10659–10667. [DOI] [PubMed] [Google Scholar]
- (47).Rosu F; De Pauw E; Gabelica V Electrospray Mass Spectrometry to Study Drug-Nucleic Acids Interactions. Biochimie 2008, 90, 1074–1087. [DOI] [PubMed] [Google Scholar]
- (48).Breuker K; McLafferty FW Stepwise Evolution of Protein Native Structure with Electrospray into the Gas Phase, 10–12 to 102 S. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 18145–18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Mazzitelli CL; Brodbelt JS Probing Ligand Binding to Duplex DNA Using KMnO4 Reactions and Electrospray Ionization Tandem Mass Spectrometry. Anal. Chem 2007, 79, 4636–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Wilson JJ; Brodbelt JS Infrared Multiphoton Dissociation of Duplex DNA/Drug Complexes in a Quadrupole Ion Trap. Anal. Chem 2007, 79, 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Mazzitelli CL; Brodbelt JS; Kern JT; Rodriguez M; Kerwin SM Evaluation of Binding of Perylene Diimide and Benzannulated Perylene Diimide Ligands to Dna by Electrospray Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom 2006, 17, 593–604. [DOI] [PubMed] [Google Scholar]
- (52).Hofstadler SA; Griffey RH Analysis of Noncovalent Complexes of DNA and RNA by Mass Spectrometry. Chem. Rev 2001, 101, 377–390. [DOI] [PubMed] [Google Scholar]
- (53).Swasey SM; Rosu F; Copp SM; Gabelica V; Gwinn EG Parallel Guanine Duplex and Cytosine Duplex DNA with Uninterrupted Spines of AgI-Mediated Base Pairs. J. Phys. Chem. Lett 2018, 9, 6605–6610. [DOI] [PubMed] [Google Scholar]
- (54).Garabedian A; Butcher D; Lippens JL; Miksovska J; Chapagain PP; Fabris D; Ridgeway ME; Park MA; Fernandez-Lima F Structures of the Kinetically Trapped I-Motif DNA Intermediates. Phys. Chem. Chem. Phys 2016, 18, 26691–26702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Fabris D A Role for the MS Analysis of Nucleic Acids in the Post-Genomics Age. J. Am. Soc. Mass Spectrom 2010, 21, 1–13. [DOI] [PubMed] [Google Scholar]
- (56).Schultz D; Gardner K; Oemrawsingh SSR; Markešević N; Olsson K; Debord M; Bouwmeester D; Gwinn E Evidence for Rod-Shaped DNA-Stabilized Silver Nanocluster Emitters. Adv. Mater 2013, 25, 2797–2803. [DOI] [PubMed] [Google Scholar]
- (57).Koszinowski K; Ballweg KA Highly Charged Ag64+ Core in a DNA-Encapsulated Silver Nanocluster. Chem. - Eur. J 2010, 16, 3285–3290. [DOI] [PubMed] [Google Scholar]
- (58).McLuckey SA; Van Berker GJ; Glish GL Tandem Mass Spectrometry of Small, Multiply Charged Oligonucleotides. J. Am. Soc. Mass Spectrom 1992, 3, 60–70. [DOI] [PubMed] [Google Scholar]
- (59).Guan Z; Kelleher NL; O’Connor PB; Aaserud DJ; Little DP; McLafferty FW 193 Nm Photodissociation of Larger Multiply-Charged Biomolecules. Int. J. Mass Spectrom. Ion Processes 1996, 157–158, 357–364. [Google Scholar]
- (60).Smith SI; Brodbelt JS Characterization of Oligodeoxynucleotides and Modifications by 193 Nm Photodissociation and Electron Photodetachment Dissociation. Anal. Chem 2010, 82, 7218–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Petty JT; Sergev OO; Kantor AG; Rankine IJ; Ganguly M; David FD; Wheeler SK; Wheeler JF Ten-Atom Silver Cluster Signaling and Tempering DNA Hybridization. Anal. Chem 2015, 87, 5302–5309. [DOI] [PubMed] [Google Scholar]
- (62).Antoine R; Lemoine J; Dugourd P Electron Photodetachment Dissociation for Structural Characterization of Synthetic and Bio-Polymer Anions. Mass Spectrom. Rev 2014, 33, 501–522. [DOI] [PubMed] [Google Scholar]
- (63).Petty JT; Giri B; Miller IC; Nicholson DA; Sergev OO; Banks TM; Story SP Silver Clusters as Both Chromophoric Reporters and DNA Ligands. Anal. Chem 2013, 85, 2183–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Xia Z; DeGrandchamp JB; Williams ER Native Mass Spectrometry beyond Ammonium Acetate: Effects of Nonvolatile Salts on Protein Stability and Structure. Analyst 2019, 144, 2565–2573. [DOI] [PubMed] [Google Scholar]
- (65).Kenderdine T; Xia Z; Williams ER; Fabris D Submicrometer Nanospray Emitters Provide New Insights into the Mechanism of Cation Adduction to Anionic Oligonucleotides. Anal. Chem 2018, 90, 13541–13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Sannes-Lowery KA; Mack DP; Hu P; Mei H-Y; Loo JA Positive Ion Electrospray Ionization Mass Spectrometry of Oligonucleotides. J. Am. Soc. Mass Spectrom 1997, 8, 90–95. [Google Scholar]
- (67).Keller KM; Brodbelt JS Charge State-Dependent Fragmentation of Oligonucleotide/Metal Complexes. J. Am. Soc. Mass Spectrom 2005, 16, 28–37. [DOI] [PubMed] [Google Scholar]
- (68).Chakraborty P; Baksi A; Khatun E; Nag A; Ghosh A; Pradeep T Dissociation of Gas Phase Ions of Atomically Precise Silver Clusters Reflects Their Solution Phase Stability. J. Phys. Chem. C 2017, 121, 10971–10981. [Google Scholar]
- (69).Vušurović J; Breuker K Relative Strength of Noncovalent Interactions and Covalent Backbone Bonds in Gaseous RNA–Peptide Complexes. Anal. Chem 2019, 91, 1659–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Paul D; Marchand A; Verga D; Teulade-Fichou M-P; Bombard S; Rosu F; Gabelica V Probing Ligand and Cation Binding Sites in G-Quadruplex Nucleic Acids by Mass Spectrometry and Electron Photodetachment Dissociation Sequencing. Analyst 2019, 144, 3518–3524. [DOI] [PubMed] [Google Scholar]
- (71).Sengupta B; Ritchie CM; Buckman JG; Johnsen KR; Goodwin PM; Petty JT Base-Directed Formation of Fluorescent Silver Clusters. J. Phys. Chem. C 2008, 112, 18776–18782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).He C; Goodwin PM; Yunus AI; Dickson RM; Petty JT A Split DNA Scaffold for a Green Fluorescent Silver Cluster. J. Phys. Chem. C 2019, 123, 17588–17597. [Google Scholar]
- (73).Lippert B; Sanz Miguel PJ The Renaissance of Metal–Pyrimidine Nucleobase Coordination Chemistry. Acc. Chem. Res 2016, 49, 1537–1545. [DOI] [PubMed] [Google Scholar]
- (74).Kondo J; Tada Y; Dairaku T; Hattori Y; Saneyoshi H; Ono A; Tanaka Y A Metallo-DNA Nanowire with Uninterrupted One-Dimensional Silver Array. Nat. Chem 2017, 9, 956–960. [DOI] [PubMed] [Google Scholar]
- (75).Müller J Metal-Mediated Base Pairs in Parallel-Stranded DNA. Beilstein J. Org. Chem 2017, 13, 2671–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Tanaka Y; Kondo J; Sychrovský V; Šebera J; Dairaku T; Saneyoshi H; Urata H; Torigoe H; Ono A Structures, Physicochemical Properties, and Applications of T–HgII–T, C–AgI–C, and Other Metallo-Base-Pairs. Chem. Commun 2015, 51, 17343–17360. [DOI] [PubMed] [Google Scholar]
- (77).Loo K; Degtyareva N; Park J; Sengupta B; Reddish M; Rogers CC; Bryant A; Petty JT Ag+-Mediated Assembly of 5′-Guanosine Monophosphate. J. Phys. Chem. B 2010, 114, 4320–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Swasey SM; Leal LE; Lopez-Acevedo O; Pavlovich J; Gwinn EG Silver (I) as DNA Glue: Ag(+)-Mediated Guanine Pairing Revealed by Removing Watson-Crick Constraints. Sci. Rep 2015, 5, 10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Swasey SM; Karimova N; Aikens CM; Schultz DE; Simon AJ; Gwinn EG Chiral Electronic Transitions in Fluorescent Silver Clusters Stabilized by DNA. ACS Nano 2014, 8, 6883–6892. [DOI] [PubMed] [Google Scholar]
- (80).Swasey SM; Gwinn EG Silver-Mediated Base Pairings: Towards Dynamic DNA Nanostructures with Enhanced Chemical and Thermal Stability. New J. Phys 2016, 18, 045008. [Google Scholar]
- (81).Cammarata MB; Brodbelt JS Structural Characterization of Holo- and Apo-Myoglobin in the Gas Phase by Ultraviolet Photodissociation Mass Spectrometry. Chem. Sci 2015, 6, 1324–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Mehaffey MR; Cammarata MB; Brodbelt JS Tracking the Catalytic Cycle of Adenylate Kinase by Ultraviolet Photodissociation Mass Spectrometry. Anal. Chem 2018, 90, 839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Petty JT; Story SP; Juarez S; Votto SS; Herbst AG; Degtyareva NN; Sengupta B Optical Sensing by Transforming Chromophoric Silver Clusters in DNA Nanoreactors. Anal. Chem 2012, 84, 356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Klein DR; Brodbelt JS Structural Characterization of Phosphatidylcholines Using 193 Nm Ultraviolet Photodissociation Mass Spectrometry. Anal. Chem 2017, 89, 1516–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Fornelli L; Srzentić K; Huguet R; Mullen C; Sharma S; Zabrouskov V; Fellers RT; Durbin KR; Compton PD; Kelleher NL Accurate Sequence Analysis of a Monoclonal Antibody by Top-Down and Middle-Down Orbitrap Mass Spectrometry Applying Multiple Ion Activation Techniques. Anal. Chem 2018, 90, 8421–8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Brodie NI; Huguet R; Zhang T; Viner R; Zabrouskov V; Pan J; Petrotchenko EV; Borchers CH Top-Down Hydrogen–Deuterium Exchange Analysis of Protein Structures Using Ultraviolet Photodissociation. Anal. Chem 2018, 90, 3079–3082. [DOI] [PubMed] [Google Scholar]
- (87).Blevins MS; Klein DR; Brodbelt JS Localization of Cyclopropane Modifications in Bacterial Lipids via 213 Nm Ultraviolet Photodissociation Mass Spectrometry. Anal. Chem 2019, 91, 6820–6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


